ABSTRACT
Citrus canker, caused by Xanthomonas citri pv. citri, is a serious disease of citrus plants worldwide. Earlier phylogenetic studies using housekeeping genes revealed that X. citri pv. citri is related to many other pathovars, which can be collectively referred as Xanthomonas citri pathovars (XCPs). From the present study, we report the genome sequences of 18 XCPs and compared them with four XCPs available in the public domain. In a tree based on phylogenomic marker genes, all the XCPs form a monophyletic cluster, suggesting their origin from a common ancestor. Phylogenomic analysis using the type strain further established that all the XCPs belong to one species. Clonal analysis of the core genome revealed the presence of two major lineages within this monophyletic cluster consisting of some clonal variants. Incidentally, the majority of these XCPs were first noticed in India, corroborating their clonal relationship and their common origin. Comparative analysis revealed an open pan-genome and the role of interstrain genomic flux of these XCPs since their diversification from a common ancestor. Even though there are wide variations in type III gene effectomes, we identified three core effectors which can be valuable in resistance-breeding programs. Overall, genomic examination of ecological relatives allowed us to dissect the tremendous genomic potential of X. citri species to rapidly evolve into specialized strains infecting diverse crop plants.
IMPORTANCE Host specialization is one of the characteristic features of highly evolved pathogens such as the Xanthomonas group of phytopathogenic bacteria. Since the hosts involve staple crops and economically important fruits such as citrus, detailed understanding of the diversity and evolution of such strains infecting diverse plants is important for quarantine purposes. In the present study, we carried out genomic investigation of members of a phylogenetically and ecologically defined group of Xanthomonas strains pathogenic to diverse plants, including citrus. This group includes the oldest Xanthomonas pathovars and also recently emerged pathovars in a particular country where they are endemic. Our high-throughput genomic study has provided novel insights into the evolution of a unique lineage consisting of serious pathogens and their ecological relatives, suggesting the nature, scope, and pattern of rapid and recent diversification. Further, from the level of species to that of clonal variants, the study revealed interesting genomic patterns in diversification of a Xanthomonas lineage and perhaps will inspire careful study of the host range of the included pathovars.
KEYWORDS: ecology, evolution, hypervariation, India, pathovar, Xanthomonas
INTRODUCTION
Xanthomonas spp. represent an economically important and complex group of phytopathogenic bacteria, infecting 125 monocots and 268 dicots (1–3). One of the bases of classification of the members of the genus Xanthomonas is their characteristic host specificity (2, 4, 5). However, there are still controversies regarding classification of the various host/tissue-specific pathogenic variants, also known as pathovars. This can be attributed to high conservation of the 16S rRNA gene sequence, the use of single genes or only a few genes for analysis, and variations in DNA-DNA hybridization results due to differences in the methods used for reassociation studies (spectrophotometric or S1 nuclease technique) (5–8). Hence, there is a need to pursue systematic understanding of the relationships and differences among the members of a phylogenetically defined set of pathovars at the whole-genome level.
With the advent of next-generation sequencing (NGS) technologies, genomics is revolutionizing the field of microbial research (9). Unlike data obtained by previous phylogenetic techniques using single or multiple loci, genome sequence data can resolve the phylogeny beyond the species or clonal complex, i.e., to the strain and clone levels (10). Genome-derived criteria such as average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) (11, 12) are emerging as modern standards for establishing species status. Genealogical study of a group of strains by considering mutation and recombination will also allow us to infer their true phylogeny.
X. citri pv. citri is a serious pathogen of citrus plants and has been assigned quarantine organism status (13–16). It has been known for over a century, and the disease caused by the pathogen was first noticed in India (17). Owing to its importance in quarantine studies and with the advent of molecular biology, various PCR- and quantitative PCR (qPCR)-based diagnostic methods were developed to distinguish X. citri pv. citri from other strains (18–22). These techniques targeted some plasmid-borne genes, general or pathogenicity regulatory factors, or rRNA gene sequences. Further, these qPCR assays were recently transferred to the droplet digital PCR (ddPCR) format for quantitative detection of pathogens, resulting in higher degrees of accuracy and sensitivity (23). Interestingly, according to the results of a single-locus (gyrB) phylogenetic study that included all xanthomonad species, X. citri is closely related to 28 pathovars infecting diverse plants, which are denoted here as X. citri pathovars (XCPs) (24) (Table 1).
TABLE 1.
Metadata of various Xanthomonas strains used in the present study
| Strain no. | Pathovar | Isolation Yr | Location | Host | Host taxonomy | Host region(s) | Reference |
|---|---|---|---|---|---|---|---|
| 1 | X. campestris pv. vitiswoodrowii LMG 954 | 1961 | India | Cissus woodrowii | Vitaceae, Vitales, Rosids, Eudicots | India | 25 |
| 2 | X. axonopodis pv. bauhiniae LMG 548 | 1961 | India | Bauhinia racemosa | Fabaceae, Fabales, Rosids, Eudicots | Southeast Asia | 26 |
| 3 | X. axonopodis pv. martyniicola LMG 9049 | 1958 | India | Martynia diandra | Martyniaceae, Lamiales, Asterids, Eudicots | Mexico | 27 |
| 4 | X. campestris pv. vitiscarnosae LMG 939 | 1962 | India | Cayratia trifolia | Vitaceae, Vitales, Rosids, Eudicots | Australia and Asia | 27 |
| 5 | X. campestris pv. viticola LMG 965 | 1972 | India | Vitis vinifera | Vitaceae, Vitales, Rosids, Eudicots | Mediterranean region | 28 |
| 6 | X. campestris pv. vitistrifoliae LMG 940 | 1961 | India | Cayratia trifolia | Vitaceae, Vitales, Rosids, Eudicots | Australia and Asia | 29 |
| 7 | X. axonopodis pv. khayae LMG 753 | 1957 | Sudan | Khaya senegalensis | Meliaceae, Sapindales, Rosids, Eudicots | Africa | 30 |
| 8 | P. cissicola LMG 21719 | 1974 | Japan | Cayratia japonica | Vitaceae, Vitales, Rosids, Eudicots | Australia and Asia | 31 |
| 9 | X. axonopodis pv. melhusii LMG 9050 | 1961 | India | Tectona grandis | Lamiaceae, Lamiales, Asterids, Eudicots | Southeast Asia | 32 |
| 10 | X. campestris pv. bilvae NCPPB 3213 | 1982 | India | Aegle marmelos | Rutaceae, Sapindales, Rosids, Eudicots | India and Bangladesh | 33 |
| 11 | X. campestris pv. azadirachtae LMG 543 | 1971 | India | Azadirachta indica | Meliaceae, Sapindales, Rosids, Eudicots | India and subcontinent | 34 |
| 12 | X. axonopodis pv. punicae LMG 859 | 1959 | India | Punica granatum | Lythraceae, Myrtales, Rosids, Eudicots | Iran | 35 |
| 13 | X. campestris pv. durantae LMG 696 | 1956 | India | Duranta repens | Verbenaceae, Lamiales, Asterids, Eudicots | United States | 36 |
| 14 | X. citri pv. citri LMG 9322 | 1915 | North America | Citrus aurantifolia | Rutaceae, Sapindales, Rosids, Eudicots | India | 37 |
| 15 | X. axonopodis pv. cajani LMG 558 | 1950 | India | Cajanus cajan | Fabaceae, Fabales, Rosids, Eudicots | India | 38 |
| 16 | X. axonopodis pv. clitoriae LMG 9045 | 1974 | India | Clitoria sp. | Fabaceae, Fabales, Rosids, Eudicots | Southeast Asia and Madagascar | 39 |
| 17 | X. campestris pv. mangiferaeindicae LMG 941 | 1948 | India | Mangifera indica | Anacardiaceae, Sapindales, Rosids, Eudicots | India | 40 |
| 18 | X. campestris pv. centellae LMG 9044 | 1979 | India | Centella asiatica | Apiaceae, Apiales, Asterids, Eudicots | Wetlands in Asia | 41 |
| 19 | X. citri pv. glycines LMG 712 | 1956 | Sudan | Glycine max | Fabaceae, Fabales, Rosids, Eudicots | East Asia | 42 |
| 20 | X. citri pv. malvacearum LMG 761 | 1958 | Sudan | Gossypium sp. | Malvaceae, Malvales, Rosids, Eudicots | Tropical and subtropical regions | 43 |
| 21 | X. campestris pv. thespesiae LMG 9057 | 1978 | India | Thespesia populnea | Malvaceae, Malvales, Rosids, Eudicots | India | 44 |
| 22 | X. campestris pv. leeana LMG 9048 | 1967 | India | Leea edgeworthii | Vitaceae, Vitales, Rosids, Eudicots | Asia | 45 |
From the present study, we report the genomes of 18 reference pathovar strains of XCPs and compare them with those of 4 publicly available XCPs (46–49). However, these XCPs have historically been classified into three different species of Xanthomonas and one of these strains has been assigned to another genus, Pseudomonas (Table 1). The majority of these pathovars infect rosids and a few asterids, including commercially important crops. Interestingly, 17 of 22 pathovars were first reported from India and some, e.g., X. axonopodis pv. punicae LMG 859, causing a devastating disease in pomegranate, are still endemic in India (35, 50). India is the center of diversification for many plants due to the presence of favorable climatic conditions and a large amount of agricultural land. In the last century, numerous Xanthomonas pathogens were reported from India (17, 24). To date, there have been limited numbers of comparative genomic studies of closely related strains of Xanthomonas that are phylogenetically defined rather than phenotypically defined. Owing to its importance, there is a need to gain insights into the emergence, diversification, and evolution of various ecological and evolutionary relatives of X. citri. Furthermore, such a genomic resource and knowledge of X. citri will be valuable in management of the pathogen.
RESULTS
Whole-genome sequencing and assembly of XCPs.
The genome sequencing of 18 XCPs, including the X. citri pv. citri type strain LMG 9322T, was performed. The raw reads were assembled de novo into genomes with a minimum contig size of 500 bp and coverage ranging from 89× to 168×. The XCPs have N50 values of ∼42 kbp to 203 kbp. The genome size for all the strains (approximately 5 Mb) was typical for the species, indicating that no reductive evolution has taken place in these reference pathovar strains. The statistics and general features of the assembled data are summarized in Table 2.
TABLE 2.
General genomic features of Xanthomonas strains sequenced in the present study
| Strain no. | Pathovar | Genome size (Mb) | No. of contigs | Coverage (×) | N50 (bp) | No. of genes | No. of tRNAs | NCBI accession no. |
|---|---|---|---|---|---|---|---|---|
| 1 | X. campestris pv. vitiswoodrowii LMG 954 | 5.0 | 102 | 163 | 190,811 | 4,234 | 53 | LOKG00000000 |
| 2 | X. axonopodis pv. bauhiniae LMG 548 | 5.2 | 192 | 113 | 64,306 | 4,463 | 52 | LOKR00000000 |
| 3 | X. axonopodis pv. martyniicola LMG 9049 | 5.0 | 76 | 164 | 203,290 | 4,185 | 52 | LOJX00000000 |
| 4 | X. campestris pv. vitiscarnosae LMG 939 | 5.0 | 105 | 113 | 139,613 | 4,257 | 53 | LOKI00000000 |
| 5 | X. campestris pv. vitistrifoliae LMG 940 | 5.0 | 184 | 115 | 58,206 | 4,325 | 53 | LOKH00000000 |
| 6 | X. axonopodis pv. khayae LMG 753 | 4.9 | 354 | 104 | 42,454 | 4,326 | 52 | LOKN00000000 |
| 7 | P. cissicola LMG 21719 | 5.2 | 313 | 130 | 46,664 | 4,522 | 51 | LOJT00000000 |
| 8 | X. axonopodis pv. melhusii LMG 9050 | 5.1 | 101 | 168 | 112,987 | 4,337 | 52 | LOJW00000000 |
| 9 | X. campestris pv. azadirachtae LMG 543 | 5.2 | 236 | 107 | 55,793 | 4,498 | 52 | LOKS00000000 |
| 10 | X. campestris pv. durantae LMG 696 | 5.4 | 187 | 138 | 95,928 | 4,672 | 52 | LOKP00000000 |
| 11 | X. citri pv. citri LMG 9322 | 5.2 | 206 | 142 | 79,989 | 4,713 | 52 | MDJT00000000 |
| 12 | X. axonopodis pv. cajani LMG 558 | 5.4 | 312 | 100 | 65,061 | 4,713 | 52 | LOKQ00000000 |
| 13 | X. axonopodis pv. clitoriae LMG 9045 | 5.1 | 91 | 114 | 126,410 | 4,266 | 53 | LOKA00000000 |
| 14 | X. campestris pv. centellae LMG 9044 | 5.2 | 315 | 158 | 53,813 | 4,473 | 49 | LOJR00000000 |
| 15 | X. citri pv. glycines LMG 712 | 5.2 | 142 | 89 | 96,827 | 4,497 | 49 | LOKO00000000 |
| 16 | X. citri pv. malvacearum LMG 761 | 5.1 | 223 | 155 | 60,648 | 4,392 | 52 | LOKM00000000 |
| 17 | X. campestris pv. thespesiae LMG 9057 | 5.0 | 93 | 144 | 108,468 | 4,154 | 50 | LOJU00000000 |
| 18 | X. campestris pv. leeana LMG 9048 | 5.0 | 92 | 153 | 145,937 | 4,170 | 52 | LOJY00000000 |
Phylogenetic analysis using marker genes: monophylogenetic origin of XCPs.
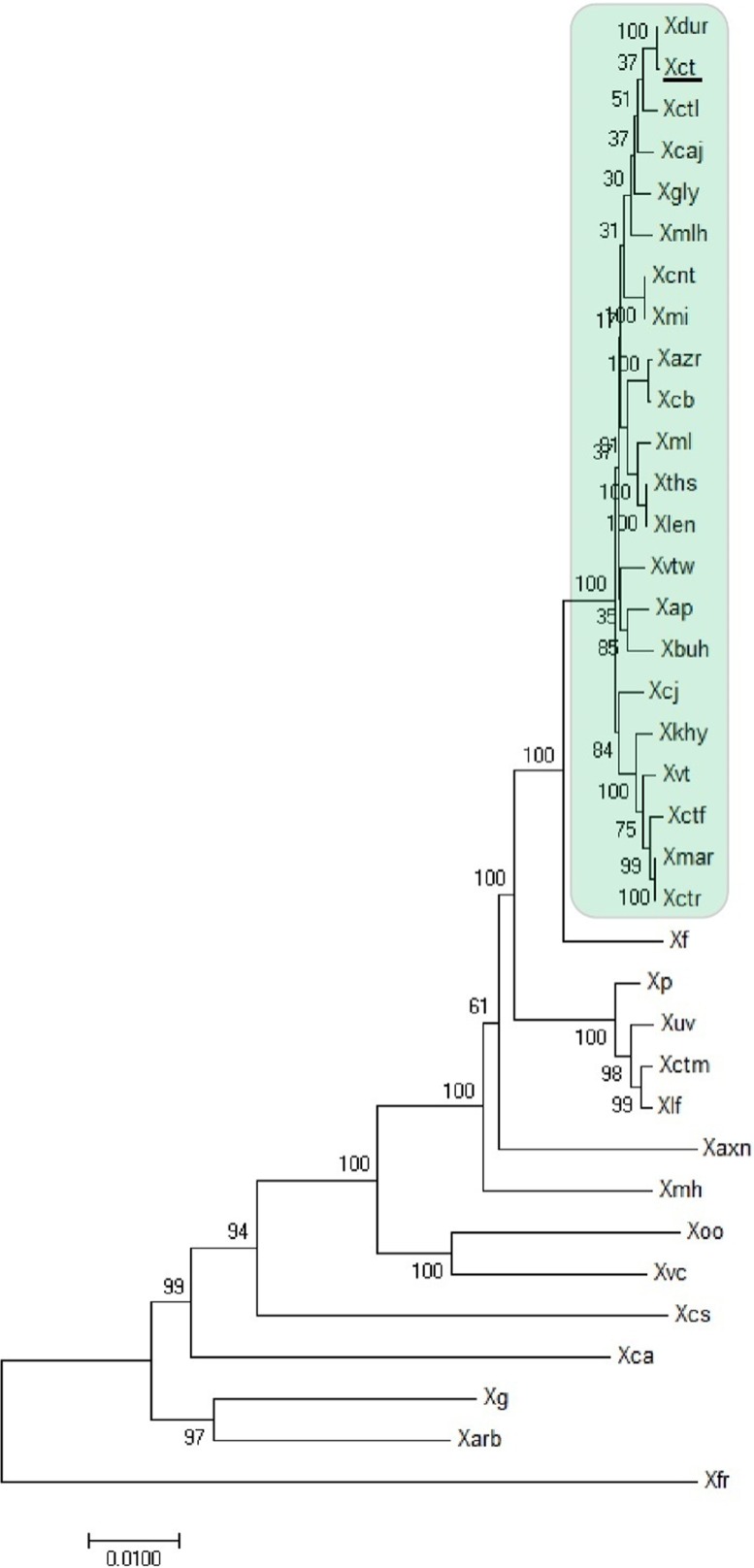
Phylogenetic analysis was performed for 22 XCPs using 28 reference genes (51) and 14 publically available type/representative strains known to be phylogenetically related to X. citri pv. citri (Fig. 1) (52). Interestingly, 22 strains (including 18 strains sequenced in this study and 4 from the database) formed a monophyletic group with X. citri pv. citri LMG 9322 (type strain) and not with the type strain X. axonopodis pv. axonopodis LMG 3585 or the type strain X. campestris pv. campestris ATCC 33913. X. fuscans NCPPB 381 represents the outgroup closest to the XCPs.
FIG 1.
Maximum likelihood tree of different Xanthomonas species and pathovars infecting diverse hosts constructed using 28 reference genes with 500 bootstrap replications. The scale bar shows the number of nucleotide substitution per site. The “type strain,” X. citri pv. citri LMG 9322T (Xct), is underlined. The strains in the green box consist of Xanthomonas pv. strains forming a clade with X. citri pv. citri LMG 9322 (denoted as XCPs). Abbreviations: Xvtw, X. campestris pv. vitiswoodrowii LMG 954; Xbuh, X. axonopodis pv. bauhiniae LMG 548; Xmar, X. axonopodis pv. martyniicola LMG 9049; Xctr, X. campestris pv. vitiscarnosae LMG 939; Xvt, X. campestris pv. viticola LMG 965; Xctf, X. campestris pv. vitistrifoliae LMG 940; Xkhy, X. axonopodis pv. khayae LMG 753; Xcj, P. cissicola LMG 21719; Xmlh, X. axonopodis pv. melhusii LMG 9050; Xcb, X. campestris pv. bilvae NCPPB 3213; Xazr, X. campestris pv. azadirachtae LMG 543; Xap, X. axonopodis pv. punicae LMG 859; Xdur, X. campestris pv. durantae LMG 696; Xct, X. citri pv. citri LMG 9322; Xcaj, X. axonopodis pv. cajani LMG 558; Xctl, X. axonopodis pv. clitoriae LMG 9045; Xmi, X. campestris pv. mangiferaeindicae LMG 941; Xcnt, X. campestris pv. centellae LMG 9044; Xgly, X. citri pv. glycines LMG 712; Xml, X. citri pv. malvacearum LMG 761; Xths, X. campestris pv. thespesiae LMG 9057; Xlen, X. campestris pv. leeana LMG 9048; Xf, X. fuscans pv. fuscans NCPPB 381; Xp, X. perforans 91-118; Xuv, X. euvesicatoria LMG27970; Xctm, X. axonopodis pv. citrumelo F1; Xlf, X. alfalfae subsp. alfalfae LMG 495; Xaxn, X. axonopodis DSM 3585; Xmh, X. axonopodis pv. manihotis LMG 784; Xoo, X. oryzae ATCC 35933; Xvc, X. vasicola NCPPB 2417; Xcs, X. cassavae CF BP 4642; Xca, X. campestris pv. campestris ATCC 33913; Xg, X. gardneri ATCC 19865; Xarb, X. arboricola pv. juglandis strain CF BP 2528; Xfr, X. fragariae LMG 25863. Except for the 18 genomes sequenced in-house (Table 2), all of the genomes were from databases.
Phylogenomic analysis—establishing the species status of XCPs as X. citri.
To confirm the species status of XCPs, we examined the ANI and dDDH values with respect to the type strain X. citri pv. citri LMG 9322, as shown in Table 3. The ANI and dDDH values for XCPs were above 98% and 86%, respectively, well above the species cutoff values (95% and 70%, respectively) (11, 12). It may be reliably inferred from the data that all of these strains belong to X. citri, irrespective of their previous classification.
TABLE 3.
Average nucleotide identity and digital DNA-DNA hybridization values of pathovars, taking X. citri pv. citri LMG 9322T as the referencea
| Strain | ANI (%) | dDDH (%) |
|---|---|---|
| X. campestris pv. vitiswoodrowii LMG 954 | 98.57 | 88.2 |
| X. axonopodis pv. bauhiniae LMG 548 | 98.6 | 89.1 |
| X. axonopodis pv. martyniicola LMG 9049 | 98.62 | 88.6 |
| X. campestris pv. vitiscarnosae LMG 939 | 98.61 | 88.7 |
| X. campestris pv. viticola LMG 965 | 98.58 | 88.2 |
| X. campestris pv. vitistrifoliae LMG 940 | 98.56 | 88.6 |
| X. axonopodis pv. khayae LMG 753 | 98.27 | 86.3 |
| P. cissicola LMG 21719 | 98.29 | 86.9 |
| X. axonopodis pv. melhusii LMG 9050 | 98.68 | 89.6 |
| X. campestris pv. bilvae NCPPB 3213 | 98.45 | 89.9 |
| X. campestris pv. azadirachtae LMG 543 | 98.6 | 89.1 |
| X. axonopodis pv. punicae LMG 859 | 98.67 | 89 |
| X. campestris pv. durantae LMG 696 | 99.41 | 98.4 |
| X. citri pv. citri LMG 9322T | 100 | 100 |
| X. axonopodis pv. cajani LMG 558 | 98.92 | 92.2 |
| X. axonopodis pv. clitoriae LMG 9045 | 98.97 | 92.2 |
| X. campestris pv. mangiferaeindicae LMG 941 | 98.72 | 90.5 |
| X. campestris pv. centellae LMG 9044 | 98.57 | 90.8 |
| X. citri pv. glycines LMG 712 | 98.68 | 90 |
| X. citri pv. malvacearum LMG 761 | 98.29 | 86.9 |
| X. campestris pv. thespesiae LMG 9057 | 98.65 | 88.7 |
| X. campestris pv. leeana LMG 9048 | 98.62 | 88.7 |
| X. fuscans pv. fuscans NCPPB 381T | 96.09 | 67.4 |
| X. campestris pv. campestris str. ATCC 33913T | 84.63 | 29.6 |
| X. axonopodis DSM 3585T | 92.76 | 49.6 |
ANI, average nucleotide identity; dDDH, digital DNA-DNA hybridization. ANI and dDDH values for the XCPs are highlighted in bold.
Genealogical analysis—role of recombination in pathovar diversification.
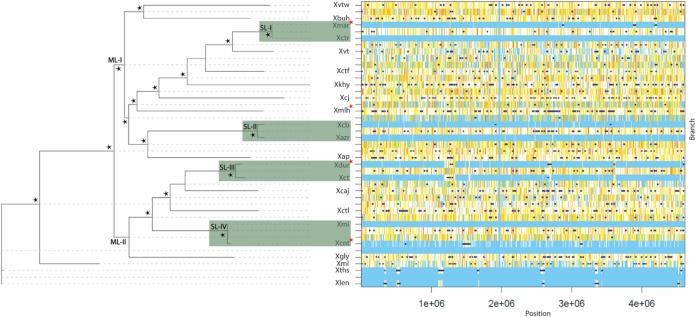
Since all 22 XCPs belong to one species, recombination may be playing a key role in the functional diversification of the strains. Hence, there is a need to construct a tree by considering regions that are not affected by recombination, so we subjected the core genome to ClonalFrameML analysis (see Materials and Methods) (Fig. 2). For the given data, the ratio of recombination and mutation rates (R/θ) is 0.0349, the mean length of imports (δ) is 116.77, and the average distance of the imports, or the divergence between donor and recipient (ν), is 0.55. Thus, the ratio representing the relative effects of recombination (r) and mutation (m) is 2.24 (r/m = R/θ × δ × ν). It is evident from the values that, though the occurrence of recombination is very low compared to that of mutation, the impact of recombination is twice that of mutation.
FIG 2.
XCP genealogy and graphical representation of recombinational events as inferred by ClonalFrameML tree. The major lineages obtained are designated ML-I and ML-II. The sublineages (SL-I, SL-II, SL-III, and SL-IV) are highlighted in green. Strains isolated from asterids are marked with a red star, and the remaining strains were isolated from rosids. Here, the variations detected by comparing each clade with its most recent common ancestor are depicted in the graph. Substitutions are represented by vertical lines and recombination events by dark blue horizontal bars. Light blue vertical lines represent the absence of substitutions, and white lines refer to nonhomoplasic substitutions. All other colors represent homoplastic substitutions, with increases in homoplasy associated with increases in the degree of redness (from white to red). The branches with ≥99 bootstrap values (obtained by the use of the initial PhyML tree) are marked by black stars.
The whole-genome-based tree obtained after correction for recombination factors for the XCPs showed that 19 of 22 reference pathovars were forming two major lineages (ML), ML-I and ML-II. ML-I consists of 12 XCPs (X. campestris pv. vitiswoodrowii LMG 954, X. axonopodis pv. bauhiniae LMG 548, X. axonopodis pv. martyniicola LMG 9049, X. campestris pv. vitiscarnosae LMG 939, X. campestris pv. viticola LMG 965, X. campestris pv. vitistrifoliae LMG 940, X. axonopodis pv. khayae LMG 753, Pseudomonas cissicola LMG 21719, X. axonopodis pv. melhusii LMG 9050, X. campestris pv. bilvae NCPPB 3213, X. campestris pv. azadirachtae LMG 543, and X. axonopodis pv. punicae LMG 859) and ML-II consists of 7 XCPs (X. campestris pv. durantae LMG 696, X. citri pv. citri LMG 9322, X. axonopodis pv. cajani LMG 558, X. axonopodis pv. clitoriae LMG 9045, X. campestris pv. mangiferaeindicae LMG 941, X. campestris pv. centellae LMG 9044, and X. citri pv. glycines LMG 712). The remaining three reference pathovars, X. citri pv. malvacearum LMG 761, X. campestris pv. thespesiae LMG 9057, and X. campestris pv. leeana LMG 9048, were not included in ML-I and ML-II. Further, there are some pathovars displaying very low levels of variability within these lineages. We have marked them as sublineages (SL). There are two sublineages in each of the major lineage; ML-I consists of SL-I (X. axonopodis pv. martyniicola LMG 9049 and X. campestris pv. vitiscarnosae LMG 939) and SL-II (X. campestris pv. bilvae NCPPB 3213 and X. campestris pv. azadirachtae LMG 543), and ML-II consists of SL-III (X. citri pv. citri LMG 9322 and X. campestris pv. durantae LMG 696) and SL-IV (X. campestris pv. mangiferaeindicae LMG 941 and X. campestris pv. centellae LMG 9044). Interestingly, of 22 XCPs, 18 pathovars infect rosids and only 4 pathovars can infect asterids; their distribution in the two lineages is indicated by red stars in Fig. 2. A graphical representation of recombination events in XCPs is shown in Fig. 2. It is clear from the light blue vertical lines in the graphical output that very few mutations occurred among the sublineages. However, there were some recombination events in the sublineages (shown by dark blue horizontal bars); hence, they may be referred to as clonal variants.
Pan-genome analysis of XCPs.
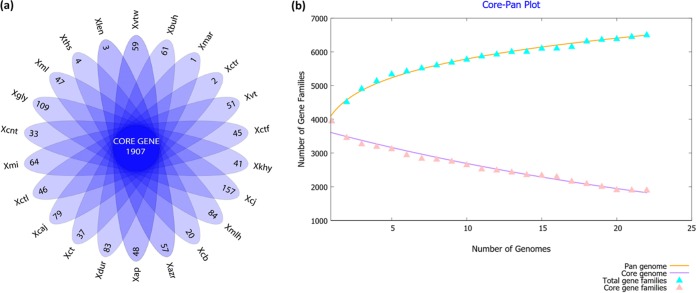
To study the interpathovar differences in detail, we carried out detailed gene-content analysis, particularly considering the variable or accessory part of the genomes, which was not present in all the strains under study. Pan-genome analysis provided the core and variable gene pool among the XCPs as shown in Fig. 3a. It is evident from the core pan-plot that the size of the pan-genome increased unboundedly with the addition of the new genomes (Fig. 3b). Sequential addition of 22 genomes resulted in the addition of approximately 6,500 nonredundant gene families, and the plot has yet to attain a plateau. This analysis showed that 1,907 genes constitute the core gene pool of the XCPs. The total variable gene pool comprised more than half of the whole gene pool, or the pan-genome.
FIG 3.
Pan-genome analysis of XCPs. (a) Floral plot showing the number of unique genes in each XCP in the petals and the number of orthologous core sets in the center. (b) Pan-genome profile analysis showing the pan-genome curve generated by plotting the total number of distinct gene families in the pan-genome and the core genome against the number of genomes considered.
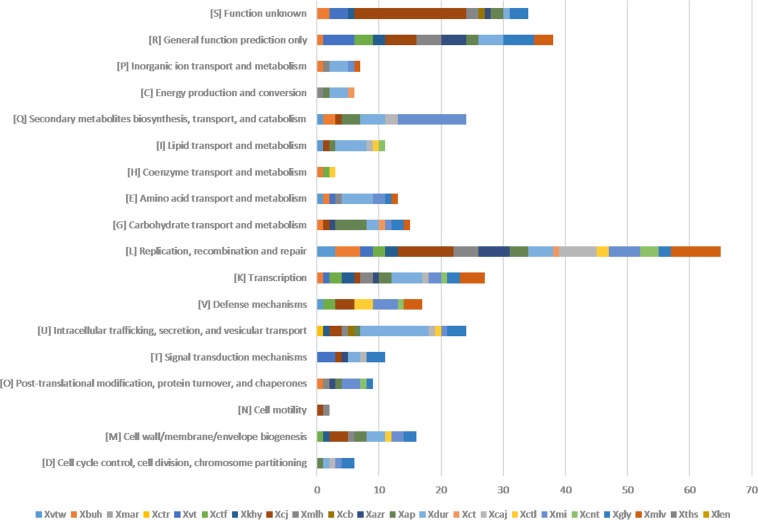
The unique genes identified for each genome are listed along with their GC content in Data Set S1 in the supplemental material. Overall, the proportion of unique genes having atypical GC content (i.e., not within 64.5 ± 2.5%) was approximately 69%. The number of unique genes ranged from as low as below 5 in X. axonopodis pv. martyniicola LMG 9049, X. campestris pv. vitiscarnosae LMG 939, X. campestris pv. leeana LMG 9048, and X. campestris pv. thespesiae LMG 9057 to 157 in strain P. cissicola LMG 21719. Since genes unique to each pathovar may provide us with clues of their ecological diversification, these genes were further annotated and functionally classified using Clusters of Orthologous Groups (COG) classification. Most (∼70%) of the unique genes were not assigned to any COG class, suggesting their role in the functional diversification of strains. For pathovars X. axonopodis pv. martyniicola LMG 9049, X. campestris pv. thespesiae LMG 9057, and X. campestris pv. leeana LMG 9048, no functional classes were obtained. An overview of the unique genes classified into COG functional categories is shown in Fig. 4. Overall, of 25 COG classes, 18 were assigned to the unique genes of XCPs. Interestingly, their distribution among the pathovars is not uniform. Functional classes such as “replication, recombination, and repair” (3Rs), “transcription,” “intracellular trafficking, secretion, and vesicular transport,” “secondary metabolite biosynthesis, transport, and catabolism,” “defense mechanisms,” and “cell wall/membrane/envelope biogenesis” are dominant among 18 classes. Further, genes for primary metabolites such as carbohydrates, lipids, and amino acids were also present at a moderate level.
FIG 4.
Distribution of COG-based functional categories of unique genes of the XCPs represented by bar graphs. Here, the x axis represents the number of genes and the y axis represents the functional categories. Each strain is represented by a colored box.
Understanding genome dynamics in pathovar diversity.
The atypical GC content of unique genes indicates that horizontal gene transfer (HGT) has played a key role in the acquisition of novel genes from distant relatives. Hence, to understand the genomic flux in a better way, it is important to see the clustering of unique genes, as HGT generally leads to the acquisition of genes in the clusters as genomic islands. In order to know the location and dynamics of genomic flux, we concentrated on large (>10-kb) regions harboring unique and variable genes, which we refer to here as large dynamic regions (LDRs).
Such an analysis revealed LDRs in 10 of 22 XCPs. Details of these regions are given in Data Set S2 and in abridged form in Table 4. All the LDRs displayed atypical GC content ranging from 49.5% to 64.2%, suggesting their acquisition through horizontal gene transfer. Apart from the low levels of GC content, the presence of tRNAs, phage, transposons, and integrase genes correlates with their dynamic and mobile nature. Interestingly, most of the LDRs were found in XCPs belonging to major lineages (ML-I and ML-II) whereas no LDRs were found for X. citri pv. malvacearum LMG 761, X. campestris pv. thespesiae LMG 9057, and X. campestris pv. leeana LMG 9048 (XCPs not included in MLs). In fact, except one, all the XCPs in ML-II have LDRs and only 4 of 12 XCPs in ML-I have LDRs. These LDRs have also played a role in the diversification of XCPs in sublineages, i.e., SL-III (i.e., X. campestris pv. durantae LMG 696 and X. citri pv. citri LMG 9322) and SL-IV (X. campestris pv. mangiferaeindicae LMG 941 and X. campestris pv. centellae LMG 9044) of ML-II.
TABLE 4.
Large dynamic regions in XCPsa
| Strain LDR | % GC content | LDR size (kbp) | Function, phenotype, or product(s) (no. of genes) |
|---|---|---|---|
| X. campestris pv. vitiswoodrowii LMG 954 W1 | 57.2 | 23.4 | Transposition (TniA [2]; TniB); virulence regulator; ATP-dependent DNA helicase UvrD/PcrA (2); hypothetical protein (21) |
| X. campestris pv. vitistrifoliae LMG 940 F1 | 54.6 | 12.5 | Phage-related protein (phage-related integrase; phage-related regulatory protein cII); succinoglycan biosynthesis protein; hypothetical protein (9) |
| P. cissicola LMG 21719 J1 | 60.2 | 80 | Restriction-modification system (modification methylase [2]; endonuclease; cytosine-specific DNA methyltransferase); secretion system protein (3); tRNA (tRNA-Gly-GCC [2]); PFGI-1-like cluster (8); integrase regulator R; integrase (4); sensor kinase (diguanylate cyclase/phosphodiesterase with PAS/PAC sensor[s]; sensor kinase); phage protein; mobile element protein (4); transposon-related functions; TraU protein (2); hypothetical protein (53) |
| P. cissicola LMG 21719 J2 | 58.4 | 16.7 | DNA repair protein RadC; YciE; VirB1; DNA helicase-related protein; DNA topoisomerase I; mycofactocin radical SAM maturase; genomic island nu Sa beta2; hypothetical protein (11) |
| X. axonopodis pv. melhusii LMG 9050 H1 | 52.9 | 37 | Cyclic beta-1,2-glucan synthase; 4-carboxymuconolactone decarboxylase; d-alanine-d-alanine ligase (2); dehydrogenases with different specificities; O-succinyl benzoic acid-CoA ligase; MchC, MchD protein; adenosylmethionine-8-amino-7-oxononanoate aminotransferase; 3-oxoacyl-[acyl-carrier-protein] synthase, KASII; colicin V secretion ABC transporter ATP-binding protein; HlyD family secretion protein; transposase and inactivated derivatives; superfamily I DNA and RNA helicases; phage integrase; transposon-related functions (2); mobile element protein (5); hypothetical protein (46) |
| X. axonopodis pv. melhusii LMG 9050 H2 | 57.7 | 28.3 | Kinesin-related protein K4; anaerobic dimethyl sulfoxide reductase chain A; ATP-dependent DNA helicase UvrD/PcrA; type III restriction-modification system methylation subunit; chaperone protein DnaJ; DNA repair protein RadC; mycobacteriophage Barnyard protein gp56; RepB/MobA-like protein; MobA/MobL protein; mobile element protein (2); hypothetical protein (18) |
| X. campestris pv. durantae LMG 696 | 61.0 | 27 | Core genes for TIVSS (virB1, virB3, virB4, virB5, virB6, virB8, virB9, virB10, and virB11); parA; trwC; trwB; kfrA; cell filamentation protein Fic; hypothetical protein (13) |
| X. citri pv. citri LMG 9322 C1 | 38.0 | ~17 | Flagellum biosynthesis proteins (FliM, FlgK); DNA replication and repair protein (4); two-component system histidine kinase DccS; methyl-accepting chemotaxis protein TlpB; disulfide interchange protein Dsb; ABC transporters for amino acid, zinc, and long-chain fatty acid; proteins involved in biosynthesis of fatty acid (enoyl-[acyl-carrier-protein] reductase [FMN]); leucine (3-isopropylmalate dehydratase); cofactor (ubiquinone/menaquinone biosynthesis methyltransferase UbiE); thymidylate (ThyX); electron transfer agents (2); hypothetical protein (12) |
| X. citri pv. citri LMG 9322 C2 | 54.7 | 15.5 | TPR domain protein; sensory box histidine kinase/response regulator; general stress protein; hypothetical protein (8) |
| X. axonopodis pv. cajani LMG 558 A1 | 58.6 | 22 | Restriction-modification system (2); phage integrase; hypothetical protein (16) |
| X. axonopodis pv. cajani LMG 558 A2 | 56.7 | 18.6 | Restriction-modification system (3); AAA_5 ATPase associated with various cellular activities; succinoglycan biosynthesis protein; phage integrase; hypothetical protein (13) |
| X. axonopodis pv. clitoriae LMG 9045 L1 | 49.5 | 15.4 | Type II restriction enzyme, methylase subunit YeeA; transcriptional regulator; bipolar DNA helicase HerA; phage integrase; virulence regulator; plasmid mobilization protein; hypothetical protein (12) |
| X. campestris pv. mangiferaeindicae LMG 941 M1 | 64.2 | 95.9 | NRPS-PKS cluster |
| X. citri pv. glycines LMG 712 G1 | 59.4 | 12.5 | Chitinase; transcriptional regulator lacI family; sensory box/GGDEF family protein; mobile element protein (3); hypothetical protein (8) |
CoA, coenzyme A; LDR, large dynamic region; SAM, S-adenosylmethionine; TIVSS, type IV secretion system.
The presence of a large number of hypothetical genes is a marked feature of LDRs, suggesting unknown functions from distant sources. However, these hypothetical protein-encoding gene clusters are also interspersed with known genes and gene clusters. Prominent among them are regulatory gene(s) in LDRs of six XCPs. A gene encoding succinoglycan biosynthesis protein is present in LDRs of two XCPs, i.e., X. campestris pv. vitistrifoliae LMG 940 and X. axonopodis pv. cajani LMG 558, suggesting a possible role of variant polysaccharides in functional diversification. Similarly, the presence of ABC transporter genes in LDRs of two XCPs (X. axonopodis pv. melhusii LMG 9050 and X. citri pv. citri LMG 9322) also indicates the importance of such genes with respect to their functional diversification. There is also a set of unique genes present in a LDR that might be required for one kind of function. Interestingly, such an arrangement is seen in the case of flagellar biosynthesis genes and also for a chemotaxis gene as seen in X. citri pv. citri LMG 9322.
More conspicuous are the cases of the well-known clusters in X. campestris pv. mangiferaeindicae LMG 941 (SL-IV) and X. campestris pv. durantae LMG 696 (SL-III) that make sublineages in ML-II. First, X. campestris pv. mangiferaeindicae LMG 941 had a unique genomic region which could not be detected even in its closest relative, X. campestris pv. centellae LMG 9044. This region was found to correspond to nonribosomal peptide synthetases and polyketide synthases (NRPS-PKS) as previously reported (49). Second, among the XCPs, a region (D1) was found to be exclusively present in X. campestris pv. durantae LMG 696. Interestingly, most of the open reading frames (ORFs) of this region were predicted to be known core genes for the type IV secretion system (T4SS) and genes encoding ATPases for generating energy for setting the whole assembly of T4SS in the peptidoglycan (virB1, virB3, virB4, virB5, virB6, virB8, virB9, virB10, and virB11). Genes such as parA (resolvase), trwCb (relaxase), trwB, and kfrA were also predicted. In addition to these genes, a putative fic gene was also present in D1. fic was recently reported to represent an important class of effectors that bacterial pathogens can use to interfere with host cell signaling pathways (53), hence giving an advantage to X. campestris pv. durantae LMG 696 as a pathogen.
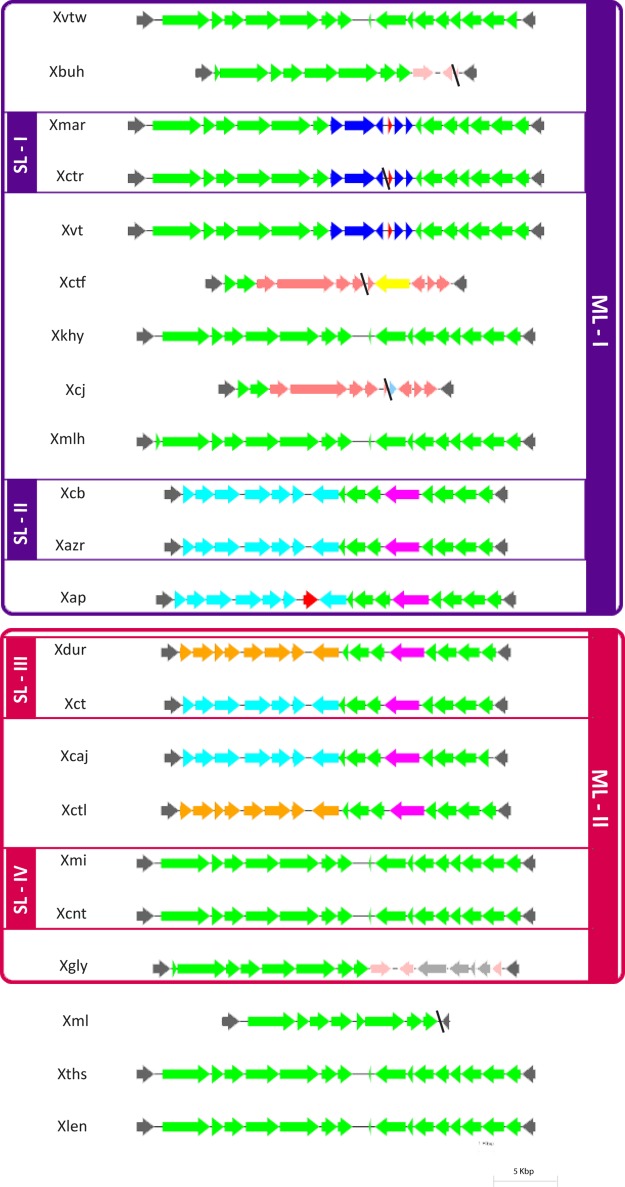
Lipopolysaccharide (LPS) cluster analysis.
The LPS locus is known for its hypervariability and its role in virulence of pathogenic bacteria. In Xanthomonas, the LPS locus is highly variable and is confined between two highly conserved housekeeping genes encoding cystathionine gamma lyase (metB) and an electron transport flavoprotein (etfA) (54). A schematic representation of LPS loci in XCPs is shown in Fig. 5. Atypical GC content of the cassette (ranging from 53% to 59%) was observed. There are two types of LPS cassettes in XCPs: type I (the X. campestris pv. mangiferaeindicae LMG 941 type) and type II (the P. cissicola LMG 21719 type). Most of the XCPs have a type I or chimeric type I cassette which may be considered an ancestral cassette. The chimeric type I cassette harbors some genes of type I, and the remaining genes are novel genes. The type II cassette is present in only two XCPs of ML-I (X. campestris pv. vitistrifoliae LMG 940 and P. cissicola LMG 21719). Interestingly, strains in sublineages have the same cassette, except for the strains of SL-III (X. citri pv. citri LMG 9322 and X. campestris pv. durantae LMG 696).
FIG 5.
Comparison of nucleotide sequences of XCPs encoding LPS cassettes. LPS cassettes of ML-I and ML-II are indicated in purple and red boxes, respectively, and sublineages are also indicated. The color coding of ORFs indicates the levels of homology among different LPS cassettes. All maps are approximately to scale. Red ORFs represent transposable elements. Contig breaks are indicated by black diagonal lines.
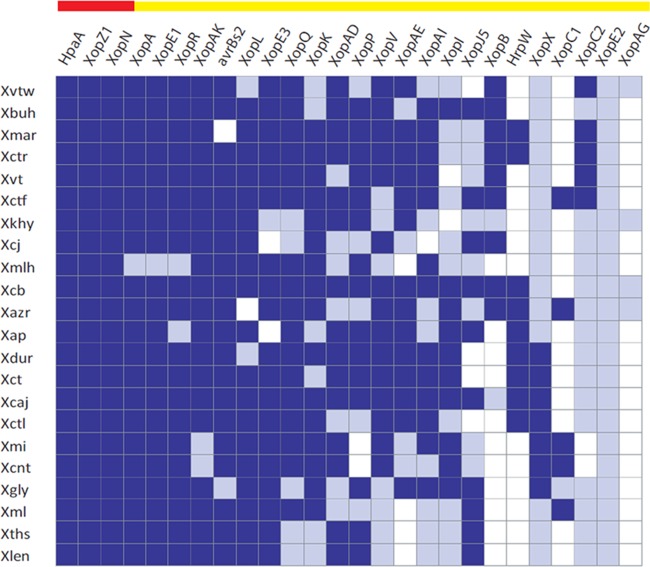
Effectome—the role of variation in effector genes with respect to diversification of XCPs.
Type III effectors (T3Es), being the major pathogenicity determinants (55), play a key role in the determination of host range at the species and even subspecies levels (56, 57). We were particularly interested in the effectome variations reflecting the differences in host range. Due to the repetitive nature of TAL (transcription activator-like) effectors, it is difficult to study them in draft genomes. Hence, we analyzed only the non-TAL effectors of XCPs. Analysis of the various loci associated with T3Es (Fig. 6) suggests that of 26, only 3 T3Es (hpaA, xopZ1, and xopN) compose the core effectome, as they are conserved throughout various host-specific pathovars of this group. The remaining genes showed variability in their conservation; 7 genes (xopA, xopE1, xopR, xopAK, avrBs2, xopL, and xopE3) are present in most of the pathovars, with a few (2 to 3) exceptions, while the other 16 T3Es (xopQ, xopK, xopAD, xopP, xopV, xopAE, xopAI, xopI, xopJ5, xopB, hrpW, xopX, xopC1, xopC2, xopE2, and xopAG) are disrupted or absent in XCPs.
FIG 6.
Type III effectome among XCPs. The presence of a type III effector gene is indicated by blue coloring and its absence by white coloring, and effectors with incomplete sequences, a contig break, or a frameshift mutation resulting in a truncated gene product are indicated by gray coloring. A core set of T3Es is represented by red and a variable set by the yellow horizontal bar on the top.
DISCUSSION
Genomic basis of diversity in XCPs.
The taxonomic and phylogenetic status of the genus Xanthomonas has been subjected to considerable debate, with various genus-wide and subspecific reclassifications proposed (5, 6, 58, 59). This can be attributed to the limited DNA markers used in traditional approaches of classification (5–8). However, genome sequencing has enabled us to address such problematic issues in a serious pathogen such as X. citri and its relatives. Only 3 of 22 XCPs were classified as pathovars of the X. citri species (X. citri pv. glycines LMG 712 and X. citri pv. malvacearum LMG 761, including the type strain, X. citri pv. citri LMG 9322), and the rest of the strains were misclassified at the species level. For instance, 11 of 19 strains (X. campestris pv. vitiswoodrowii LMG 954, X. campestris pv. vitiscarnosae LMG 939, X. campestris pv. viticola LMG 965, X. campestris pv. vitistrifoliae LMG 940, X. campestris pv. bilvae NCPPB 3213, X. campestris pv. azadirachtae LMG 543, X. campestris pv. durantae LMG 696, X. campestris pv. mangiferaeindicae LMG 941, X. campestris pv. centellae LMG 9044, X. campestris pv. thespesiae LMG 9057, and X. campestris pv. leeana LMG 9048) were classified as pathovars of X. campestris and 7 of 19 (X. axonopodis pv. bauhiniae LMG 548, X. axonopodis pv. martyniicola LMG 9049, X. axonopodis pv. khayae LMG 753, X. axonopodis pv. melhusii LMG 9050, X. axonopodis pv. punicae LMG 859, X. axonopodis pv. cajani LMG 558, and X. axonopodis pv. clitoriae LMG 9045) as pathovars of X. axonopodis. Interestingly, P. cissicola LMG 21719 was classified as another genus altogether. In contrast, modern genome-based criteria such as ANI and dDDH had established the species status of all XCPs as X. citri. High genomic relatedness suggests the origin of XCPs from a common ancestor in the recent past. Hence, functional diversification of strains of numerous pathovars has happened at the group or species level. This genomic inference suggests that Xanthomonas also has the genetic capability to diversify even below the species level, which probably is a prerequisite for being a successful and specialized phytopathogenic bacterium.
Genomic patterns of pathovar diversification of XCPs.
Owing to the exceptional functional diversification of XCPs, they represent an interesting model for the study of the evolution of Xanthomonas at the species and strain levels. This is an important case for studying genome-level recombination and mutation rates among the XCPs across their genomes to understand their clonal nature. Interestingly, such an analysis reveals that most (19/22) XCPs are evolving in two major lineages (ML-I and ML-II), suggesting success and selection at the subspecies level. Further, strains in sublineages may represent clonal variants isolated from diverse hosts.
One of the striking compositional differences of the two major lineages of XCPs is that ML-II consists of well-known pathovars that infect major commercial plants such as citrus fruit and mango. Incidentally, they are the oldest pathovars under study, having been reported before the 1950s, while, on the other hand, ML-I largely consists of pathovars of plants that are not highly cultivated and of two recently reported pathovars from India that infect commercial fruit plants, i.e., grape and pomegranate. Interestingly, the pathovar that infects pomegranate is an endemic problem in India. Furthermore, large-scale cultivation of grape and pomegranate started in southern states of India only after the 1970s. In addition, the presence of strains infecting rosids and asterids in both the major lineages suggests that this does not represent a lineage-specific potential.
Role of interstrain genome dynamics in emergence and evolution of XCPs.
The advent of NGS has not only aided in establishing relationships up to the strain level but also revealed differences among strains on the whole-genome scale. Bacteria display interstrain variations that result in diversification of the strains into different lineages and sublineages. This may be attributed to continuous horizontal gene transfer and gene loss. Since phylogenomic and clonal analysis established the monospecies and monophyletic nature of XCPs, genomic flux at the interstrain level may have been crucial in the success of XCPs. Indeed, the open nature of pan-genomes suggests that a huge pool of variable genes has played a major role in emergence and evolution of XCPs. In this context, unique genes that are exclusively found in a particular pathovar or are missing in its next closest relative may be of relevance. Further, large variations in a number of unique genes in each of the pathovars suggest a possible role of gene acquisition and loss in the evolution of these XCPs. Nearly 70% of unique genes with atypical GC content also have “unknown functions,” suggesting both their acquisition from distant organisms through HGT and their potential novel role.
Acquisition of genes by HGT is found to be a major force in genomic diversity of XCPs. The ability to acquire and maintain foreign DNA is reflected in the second major class of unique genes, which are related to “replication, recombination, and repair” (3R). This is also in accordance with our genealogical studies, which showed that the impact of recombination is twice that of mutation. Further, defense mechanisms and secondary metabolites are among the prominent classes, aiding in the ability to counteract defense responses of host, toxicity, fitness, and virulence regulation of the pathogen. For instance, the presence of a unique NRPS-PKS cluster in X. campestris pv. mangiferaeindicae LMG 941 is possibly involved in the production of a novel metabolite (49).
One of the determining features seems to operate at the level of host-pathogen interaction, such as that of identifying critical signals from host environment (such as reactive oxygen species, nutrients, etc.) and elicitors derived from the cell membrane of pathogen (such as LPS molecules) or secreting effectors in the environment (or host) for alteration of the physiology of the host cell (60). Accordingly, among the major classes of unique genes in XCPs are those belonging to the groups of genes encoding proteins involved in cell wall/membrane biogenesis and signal transduction mechanisms. The presence of highly evolved major lineages and sublineages, variable LPS loci, and condensed core type III effectomes of XCPs may be attributed to the diversity of host plants. Host-pathovar interactions are controlled genetically in a gene-for-gene manner by TAL effectors. However, due to the draft nature of genomes, TAL genes were not assembled; therefore, we focused our study on non-TAL effectors. In addition to these, a novel type IV secretion system might have played an important role in the diversification of X. campestris pv. durantae LMG 696, a clonal variant of X. citri pv. citri LMG 9322. Unique genes related to transcription form a third major class after unknown and 3R functions. Hence, it is not just the acquisition of novel genes from distant organisms that is required for the success of pathogens but also the regulation of these and their functions related to interaction, pathogenicity, and fitness.
Moreover, acquisition of or the presence of unique or dynamic genes in a large genomic region may have a role in the regulation and movement of LDRs. This may be important for the success of a pathogen or group of pathogens, as in case of XCPs. However, because of the draft nature of genomes, it is not possible to know definitely whether a LDR is on a plasmid. Hence, there is a need to obtain complete genome sequence of these XCPs. Also, further genetic characterization and transcriptomic studies are required to know the role of LDRs in virulence of XCPs.
Concluding remarks.
Xanthomonas is a complex bacterium and is classified into numerous pathovars based on the host it infects. In the present report, we provide evolutionary insights into the nature and pattern of diversification in a large group of Xanthomonas pathovars which are related to X. citri but infect diverse plants. The genomic knowledge and resources will be valuable in surveillance of X. citri and its relatives.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A collection of 22 genomes of strains within the X. citri pv. citri lineage were used in this study, comprising 18 strains sequenced in-house and 4 genomes from the NCBI database. The required strains were procured from the Belgian Co-ordinated Collection of Microorganisms (BCCM). Except in the case of the type strain of X. citri pv. citri LMG 9322, only genomic DNA was procured from BCCM. All isolates were grown on the media and under the conditions recommended by the culture collection center.
Genome sequencing, assembly, and annotation.
Genomic DNA was extracted by using a ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research). DNA quality checking was done using a NanoDrop 1000 instrument (Thermo Fisher Scientific) and agarose gel electrophoresis. Quantitation of DNA was performed using a Qubit 2.0 fluorometer (Life Technologies). Illumina paired-end sequencing libraries (read length, 2 × 250) of genomic DNA were prepared using Nextera XT sample preparation kits (Illumina, Inc., San Diego, CA, USA) with dual indexing adapters. Illumina sequencing libraries were sequenced in-house using an Illumina Miseq platform (Illumina, Inc., San Diego, CA, USA) and company-supplied paired-end sequencing kits. Adapter trimming was done automatically by MiSeq control software (MCS), and additional adapter contamination identified by the NCBI server was removed by manual trimming. Raw reads were assembled de novo using CLC Genomics Workbench v7.5 (CLC bio, Aarhus, Denmark) with default settings. Annotation was done using the NCBI PGAP pipeline.
Phylogenomic analysis.
To determine the relatedness of XCPs to one another, a phylogeny was determined using 28 universal housekeeping genes (51). These housekeeping genes from Xanthomonas genomes were retrieved, concatenated, and aligned. A maximum likelihood tree was constructed using the general time-reversible (GTR) model and the gamma distribution with invariant sites (G + I) method with 500 bootstrap replications using MEGA7 (61).
Whole-genome comparison.
The intergenomic distances between the XCPs and the type strain (X. citri pv. citri LMG 9322) were determined using BLAST-based average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH). Taking X. citri pv. citri LMG 9322 as a reference, pairwise ANI was calculated using JSpecies1.2.1 (11) and dDDH (12) was calculated using Web tool GGDC 2.0 (http://ggdc.dsmz.de/distcalc2.php).
ClonalFrameML.
Recombination events play an important role in generating novel diversity during the course of evolution of a bacterium. ClonalFrameML analysis (62) generates a phylogenetic tree applying a correction to the branch length accounting for recombination. For obtaining a tree reflecting recombination events, genomes of 22 strains were aligned using MAUVE 2.3.1 (63). A maximum likelihood tree was obtained from a core genome by using PhyML 3.1 (64). ClonalFrameML analysis was carried out with the MAUVE alignment and the PhyML tree (as the starting tree) as input files, performing 100 simulations (emsim = 100) to estimate uncertainty in the results.
Comparative analysis.
Pan-genome analysis was carried out to estimate the number of shared genes (core genome) and the number of unique genes (accessory or variable genome), using the Bacterial Pan Genome Analysis (BPGA) tool (65). USEARCH, which is an integral component of BPGA, was used for clustering analysis. A minimum identity of 50% was used as the cutoff value, and the input sequence was defined as the member sequence and the seed sequence (the seed sequence defines the cluster). The number of combinations of genomes considered for the analysis was chosen to be 20, which signifies the maximum number of clusters possible for a USEARCH run. The flower pot diagram used to represent the core and unique genes was drawn using a python script of Matplotlib (66). Unique genes not having GC content within 64.5% ± 2.5% were considered to have atypical content. All the unique genes were functionally assigned to a specific Clusters of Orthologous Groups (COG) family by searching against the COGs database using the online tool WebMGA (67).
Further, large (>10-kb) dynamic regions were detected by using BRIG-0.95 (68). Dynamic genomic regions were reannotated using the RAST pipeline (69) and reinspected for homology by blastp and assessment of function.
LPS cluster analysis.
The full-length LPS cassette was retrieved from the genomic data using two highly conserved metB and etfA genes. Prokka 1.11 was used to annotate the gene clusters (70). Easyfig 2.2.2 was used to generate the schematic figure representing LPS clusters (71).
Type III effectome.
The effectome of XCPs was determined by sequence-based similarity searches using T3Es from www.xanthomonas.org as the query. Frameshift mutations and truncations in the T3E genes were identified by analyzing the translated protein length and position of a stop codon in the resulting protein sequence using tblastn searches.
Accession number(s).
The genomic sequences determined in this study have been submitted to NCBI GenBank and are available under the accession numbers given in Table 2.
Supplementary Material
ACKNOWLEDGMENT
K.B. is supported by a fellowship from University Grant Commission (UGC); S.M. and S.K. are supported by fellowships from Council of Scientific and Industrial Research (CSIR). We acknowledge the funding from CSIR-IMTECH and CSIR projects Plant-Microbe and Soil Interaction (PMSI) BSC0117 and Expansion and Modernization of the Microbial Type Culture Collection and Gene Bank (MTCC) BSC0402. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02993-16.
REFERENCES
- 1.Leyns F, De Cleene M, Swings J-G, De Ley J. 1984. The host range of the genus Xanthomonas. Botanical Rev 50:308–356. doi: 10.1007/BF02862635. [DOI] [Google Scholar]
- 2.Hayward A. 1993. The hosts of Xanthomonas, p 1–119. In Xanthomonas. Springer, New York, NY. [Google Scholar]
- 3.Chan JW, Goodwin PH. 1999. The molecular genetics of virulence of Xanthomonas campestris. Biotechnol Adv 17:489–508. doi: 10.1016/S0734-9750(99)00025-7. [DOI] [PubMed] [Google Scholar]
- 4.Dye D, Bradbury J, Goto M, Hayward A, Lelliott R, Schroth M. 1980. International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev Plant Pathol 59:153–168. [Google Scholar]
- 5.Schaad NW, Postnikova E, Lacy GH, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK. 2005. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv. malvacearum (ex Smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and and Jones, 1935) Dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst Appl Microbiol 28:494–518. doi: 10.1016/j.syapm.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Vauterin L, Hoste B, Kersters K, Swings J. 1995. Reclassification of Xanthomonas. Int J Syst Evol Microbiol 45:472–489. [DOI] [PubMed] [Google Scholar]
- 7.Hauben L, Vauterin L, Swings J, Moore E. 1997. Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int J Syst Evol Microbiol 47:328–335. doi: 10.1099/00207713-47-2-328. [DOI] [PubMed] [Google Scholar]
- 8.Murata N, Starr MP. 1973. A concept of the genus Xanthomonas and its species in the light of segmental homology of deoxyribonucleic acids. J Phytopathol 77:285–323. doi: 10.1111/j.1439-0434.1973.tb04137.x. [DOI] [Google Scholar]
- 9.Loman NJ, Pallen MJ. 2015. Twenty years of bacterial genome sequencing. Nat Rev Microbiol 13:787–794. doi: 10.1038/nrmicro3565. [DOI] [PubMed] [Google Scholar]
- 10.Maiden MC, van Rensburg MJJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auch AF, von Jan M, Klenk H-P, Göker M. 2010. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottwald TR, Graham JH, Schubert TS. 2002. Citrus canker: the pathogen and its impact. Plant Health Prog 10:32. doi: 10.1094/PHP-2002-0812-01-RV. [DOI] [Google Scholar]
- 14.Brunings AM, Gabriel DW. 2003. Xanthomonas citri: breaking the surface. Mol Plant Pathol 4:141–157. doi: 10.1046/j.1364-3703.2003.00163.x. [DOI] [PubMed] [Google Scholar]
- 15.Graham JH, Gottwald TR, Cubero J, Achor DS. 2004. Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol Plant Pathol 5:1–15. doi: 10.1046/j.1364-3703.2004.00197.x. [DOI] [PubMed] [Google Scholar]
- 16.Gottwald TR, Irey M. 2007. Post-hurricane analysis of citrus canker II: predictive model estimation of disease spread and area potentially impacted by various eradication protocols following catastrophic weather events. Plant Health Prog 10. doi: 10.1094/PHP-2007-0405-01-RS. [DOI] [Google Scholar]
- 17.Raychaudhuri S, Verma J, Nariani T, Sen B. 1972. The history of plant pathology in India. Annu Rev Phytopathol 10:21–36. doi: 10.1146/annurev.py.10.090172.000321. [DOI] [Google Scholar]
- 18.Hartung JS, Daniel JF, Pruvost OP. 1993. Detection of Xanthomonas campestris pv. citri by the polymerase chain reaction method. Appl Environ Microbiol 59:1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cubero J, Graham JH, Gottwald TR. 2001. Quantitative PCR method for diagnosis of citrus bacterial canker. Appl Environ Microbiol 67:2849–2852. doi: 10.1128/AEM.67.6.2849-2852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cubero J, Graham JH. 2005. Quantitative real-time polymerase chain reaction for bacterial enumeration and allelic discrimination to differentiate xanthomonas strains on citrus. Phytopathology 95:1333–1340. doi: 10.1094/PHYTO-95-1333. [DOI] [PubMed] [Google Scholar]
- 21.Golmohammadi M, Cubero J, Peñalver J, Quesada JM, López MM, Llop P. 2007. Diagnosis of Xanthomonas axonopodis pv. citri, causal agent of citrus canker, in commercial fruits by isolation and PCR-based methods. J Appl Microbiol 103:2309–2315. doi: 10.1111/j.1365-2672.2007.03484.x. [DOI] [PubMed] [Google Scholar]
- 22.Mavrodieva V, Levy L, Gabriel DW. 2004. Improved sampling methods for real-time polymerase chain reaction diagnosis of citrus canker from field samples. Phytopathology 94:61–68. doi: 10.1094/PHYTO.2004.94.1.61. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Xia Q, Yin Y, Wang Z. 2016. Comparison of droplet digital PCR and quantitative PCR assays for quantitative detection of Xanthomonas citri subsp. citri. PLoS One 11:e0159004. doi: 10.1371/journal.pone.0159004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkinson N, Cowie C, Heeney J, Stead D. 2009. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int J Syst Evol Microbiol 59:264–274. doi: 10.1099/ijs.0.65825-0. [DOI] [PubMed] [Google Scholar]
- 25.Patel MK, Kulkarni YS. 1951. A new bacterial leaf spot on Vitis woodrowii Stapf. Curr Sci 20:132. [PubMed] [Google Scholar]
- 26.Padhya AC, Patel MK, Kotasthane WV. 1965. A new bacterial leaf-spot disease of Bauhinia racemosa Lamk. Curr Sci 34:224–225. [Google Scholar]
- 27.Moniz L, Patel MK. 1958. Three new bacterial diseases of plants from Bombay State. Curr Sci 27:494–495. [Google Scholar]
- 28.Nayadu MV. 1972. Pseudomonas viticola sp. nov., incitant of a new bacterial disease of grape vine. Phytopathol Z 73:183–186. doi: 10.1111/j.1439-0434.1972.tb02539.x. [DOI] [Google Scholar]
- 29.Padhya AC, Patel MK, Kotasthane WV. 1965. A new bacterial leaf-spot on Vitis trifolia. Curr Sci 34:462–463. [Google Scholar]
- 30.Sabet KA. 1959. Studies in the bacterial diseases of Sudan crops II. Bacterial leaf-spot and canker disease of mahogany (Khaya senegalensis (Desr.) A Juss and K grandifoliola C DC). Ann Appl Biol 47:49–56. [Google Scholar]
- 31.Takimoto S. 1939. Bacterial leaf spot of Cissus japonica Willd. Ann Phytopathol Soc Jpn 9:41–43. doi: 10.3186/jjphytopath.9.41. [DOI] [Google Scholar]
- 32.Patel MK, Kulkarni YS, Dhande GW. 1952. Some new bacterial diseases of plants. Curr Sci 21:345–346. [Google Scholar]
- 33.Chakravarti BP, Sarma B, Jain KL, Prasad CKP. 1984. A bacterial leaf spot of bael (Aegle marmelos Correa) in Rajasthan and a revived name of the bacterium [wood apple, India]. Short communication. Curr Sci 53:488. [Google Scholar]
- 34.Desai SG, Gandhi AB, Patel MK, WVK. 1966. A new bacterial leaf-spot and blight of Azadirachta indica A. Juss. Indian Phytopathol 19:322–323. [Google Scholar]
- 35.Hingorani M, Singh N. 1960. Xanthomonas punicae sp. nov. on Punica granatum L. Indian J Agric Sci 29:45–48. [Google Scholar]
- 36.Srinivasan MC, Patel MK. 1957. Two new phytopathogenic bacteria on verbenaceous hosts. Curr Sci 26:90–91. [Google Scholar]
- 37.Hasse CH. 1915. Pseudomonas citri, the cause of citrus canker. J Agric Res 4:97–100. [Google Scholar]
- 38.Kulkarni YS, Patel MK, Abhyankar SG. 1950. A new bacterial leaf-spot and stem canker of pigeon pea. Curr Sci 19:384. [PubMed] [Google Scholar]
- 39.Pandit VM, Kulkarni YS. 1979. Bacterial leaf-spot of Clitoria biflora Dalz. Biovigyanam 5:9–20. [Google Scholar]
- 40.Patel MK, Moniz L, Kulkarni YS. 1948. A new bacterial disease of Mangifera indica L. Curr Sci 17:189. [PubMed] [Google Scholar]
- 41.Basnyat SR, Kulkarni YS. 1979. New bacterial leafspot of Centella asiatica L. Urban Biovigyanam 5:179–180. [Google Scholar]
- 42.Nakano K. 1919. Soybean leaf spot. J Plant Prot (Tokyo) 6:217–221. [Google Scholar]
- 43.Smith EF. 1901. The cultural characters of Pseudomonas hyacinthi, Ps. campestris, Ps. Phaseoli, and Ps. Stewarti—four one-flagellate yellow bacteria parasitic on plants. U.S. Government Printing Office, Washington, DC. [Google Scholar]
- 44.Patil AS, Kulkarni YS. 1981. A new bacterial leaf-spot disease of Thespesia populnea Sol. ex Corr. Curr Sci 50:1040–1041. [Google Scholar]
- 45.Patel AM, Kotasthane WV. 1969. Bacterial blight of Leea edgeworthii incited by Xanthomonas leeanum, nov. sp. Curr Sci 38:519–520. [Google Scholar]
- 46.Gordon JL, Lefeuvre P, Escalon A, Barbe V, Cruveiller S, Gagnevin L, Pruvost O. 2015. Comparative genomics of 43 strains of Xanthomonas citri pv. citri reveals the evolutionary events giving rise to pathotypes with different host ranges. BMC Genomics 16:1098. doi: 10.1186/s12864-015-2310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma V, Midha S, Ranjan M, Pinnaka AK, Patil PB. 2012. Genome sequence of Xanthomonas axonopodis pv. punicae strain LMG 859. J Bacteriol 194:2395–2395. doi: 10.1128/JB.00181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Midha S, Ranjan M, Sharma V, Pinnaka AK, Patil PB. 2012. Genome sequence of Xanthomonas citri pv. mangiferaeindicae strain LMG 941. J Bacteriol 194:3031. doi: 10.1128/JB.00433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Midha S, Patil PB. 2014. Genomic insights into the evolutionary origin of Xanthomonas axonopodis pv. citri and its ecological relatives. Appl Environ Microbiol 80:6266–6279. doi: 10.1128/AEM.01654-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benagi VI, Ravi Kumar MR. 2011. Present status of pomegranate bacterial blight and its management. Acta Hortic 890:475–480. doi: 10.17660/ActaHortic.2011.890.66. [DOI] [Google Scholar]
- 51.Wu M, Eisen JA. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol 9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Constantin EC, Cleenwerck I, Maes M, Baeyen S, Van Malderghem C, De Vos P, Cottyn B. 2015. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol 65:792–806. doi: 10.1111/ppa.12461. [DOI] [Google Scholar]
- 53.Molloy S. 2012. Bacterial pathogenesis: controlling Fic proteins. Nat Rev Microbiol 10:160. doi: 10.1038/nrmicro2757. [DOI] [PubMed] [Google Scholar]
- 54.Patil PB, Sonti RV. 2004. Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice. BMC Microbiol 4:40. doi: 10.1186/1471-2180-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol 60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 56.Rohmer L, Guttman DS, Dangl JL. 2004. Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in the plant pathogen Pseudomonas syringae. Genetics 167:1341–1360. doi: 10.1534/genetics.103.019638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarkar SF, Gordon JS, Martin GB, Guttman DS. 2006. Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174:1041–1056. doi: 10.1534/genetics.106.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ah-You N, Gagnevin L, Chiroleu F, Jouen E, Neto JR, Pruvost O. 2007. Pathological variations within Xanthomonas campestris pv. mangiferaeindicae support its separation into three distinct pathovars that can be distinguished by amplified fragment length polymorphism. Phytopathology 97:1568–1577. doi: 10.1094/PHYTO-97-12-1568. [DOI] [PubMed] [Google Scholar]
- 59.Bui Thi Ngoc L, Vernière C, Jouen E, Ah-You N, Lefeuvre P, Chiroleu F, Gagnevin L, Pruvost O. 2010. Amplified fragment length polymorphism and multilocus sequence analysis-based genotypic relatedness among pathogenic variants of Xanthomonas citri pv. citri and Xanthomonas campestris pv. bilvae. Int J Syst Evol Microbiol 60:515–525. doi: 10.1099/ijs.0.009514-0. [DOI] [PubMed] [Google Scholar]
- 60.Ryan RP, Vorhölter F-J, Potnis N, Jones JB, Van Sluys M-A, Bogdanove AJ, Dow JM. 2011. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol 9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 61.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaudhari NM, Gupta VK, Dutta C. 2016. BPGA—an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373. doi: 10.1038/srep24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 67.Wu S, Zhu Z, Fu L, Niu B, Li W. 2015. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics 12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alikhan N-F, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aziz RK, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 71.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.