ABSTRACT
Cyanobacteria are emerging as alternative crop species for the production of fuels, chemicals, and biomass. Yet, the success of these microbes depends on the development of cost-effective technologies that permit scaled cultivation and cell harvesting. Here, we investigate the feasibility of engineering cell morphology to improve biomass recovery and decrease energetic costs associated with lysing cyanobacterial cells. Specifically, we modify the levels of Min system proteins in Synechococcus elongatus PCC 7942. The Min system has established functions in controlling cell division by regulating the assembly of FtsZ, a tubulin-like protein required for defining the bacterial division plane. We show that altering the expression of two FtsZ-regulatory proteins, MinC and Cdv3, enables control over cell morphology by disrupting FtsZ localization and cell division without preventing continued cell growth. By varying the expression of these proteins, we can tune the lengths of cyanobacterial cells across a broad dynamic range, anywhere from an ∼20% increased length (relative to the wild type) to near-millimeter lengths. Highly elongated cells exhibit increased rates of sedimentation under low centrifugal forces or by gravity-assisted settling. Furthermore, hyperelongated cells are also more susceptible to lysis through the application of mild physical stress. Collectively, these results demonstrate a novel approach toward decreasing harvesting and processing costs associated with mass cyanobacterial cultivation by altering morphology at the cellular level.
IMPORTANCE We show that the cell length of a model cyanobacterial species can be programmed by rationally manipulating the expression of protein factors that suppress cell division. In some instances, we can increase the size of these cells to near-millimeter lengths with this approach. The resulting elongated cells have favorable properties with regard to cell harvesting and lysis. Furthermore, cells treated in this manner continue to grow rapidly at time scales similar to those of uninduced controls. To our knowledge, this is the first reported example of engineering the cell morphology of cyanobacteria or algae to make them more compatible with downstream processing steps that present economic barriers to their use as alternative crop species. Therefore, our results are a promising proof-of-principle for the use of morphology engineering to increase the cost-effectiveness of the mass cultivation of cyanobacteria for various sustainability initiatives.
KEYWORDS: cyanobacteria, cell morphology, biomass recovery, biotechnology, cell division, Min system
INTRODUCTION
While cyanobacteria and algae offer many potential benefits relative to traditional land plants, the commercialization of such photosynthetic crop species has been limited due to the technical problems of scaled cultivation. Cyanobacteria exhibit rapid division times, high photosynthetic efficiencies, and the capacity to be cultivated in nonpotable water supplies on nonarable lands, and they are readily genetically manipulated, all of which are features that make them of considerable interest as alternative crop species (1–3). Yet, these advantages are overshadowed by several economic considerations that have prevented the widespread adoption of cyanobacteria and algae as alternative crop species (4–6). In contrast to the technology for plants that has been under development for millennia, cyanobacterial mass cultivation is relatively underdeveloped. Therefore, improvements in infrastructure, strains, and equipment that decrease costs are essential for economic viability, particularly for the production of commodity goods such as biofuels. While there have been considerable efforts in the academic sphere to engineer cyanobacteria and algae for improved photosynthesis or increased production of target metabolites, there has been less emphasis on modifying them to decrease cultivation costs (7).
Three of the highest costs associated with cyanobacterial cultivation are related to pumping/mixing the liquid culture broth, to the recovery and dewatering of cell biomass, and to the treatment or lysis of cells to release nonsecreted metabolites (4–7). Water processing and biomass dewatering have substantial costs limiting the economic feasibility of scaled cyanobacterial cultivation in part because incident solar light is inherently a diffuse energy source. Therefore, cultures are dilute relative to heterotrophic bacteria grown in media supplemented with organic carbon sources. Furthermore, cyanobacterial cells are small and have densities near that of water, and so mechanical processes, including centrifugation, filtration, or chemical flocculation are frequently required to remove cells from suspension. The capital costs of industrial centrifuges are high, as are the energetic operational costs, which further contribute to increases in the net life cycle carbon footprint (4). Filtration requires periodic replacement of microfilters and also has considerable operational costs, especially for many cyanobacterial species with very small cell sizes (7). Chemical flocculation involves the addition of ions or polymers that counteract the repulsive charge at cell surfaces, causing cells to aggregate into larger bodies that are more readily removed. However, the compounds typically remain associated with the biomass, which can be problematic for downstream applications (e.g., feeds or aquaculture), and the process can be inefficient, requiring additional dewatering steps (5).
Cyanobacteria with improved recovery characteristics might decrease these costs and improve their outlook for biotechnology applications. The ideal cyanobacteria would remain small and buoyant during growth to minimize mixing costs, yet be readily precipitated from solution during harvest; this dichotomy complicates simple bioprospecting for unusually large or dense cyanobacterial species. Alternatively, genetic approaches could be used to gain control over the physical shape of bacteria, facilitating their recovery and processing at the time of harvest (Fig. 1A) (8–10). Additional genetic alterations that decrease the costs associated with biomass processing and lysis without compromising the viability of the cells would further improve scalability (7, 11). One approach that could mitigate harvest and lysis costs is user-regulated control over bacterial cytoskeletal components that alter cell morphology and/or integrity (8).
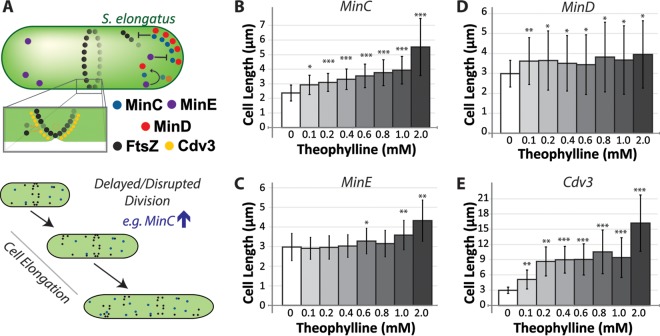
FIG 1.
Control over Min system expression enables modification of cyanobacterial cell size. (A) Cartoon schematic of the function of the Min system of S. elongatus, which regulates the position and assembly of the FtsZ ring required for cell division. MinD (red) binds to the pole, recruiting MinC (blue) to repress assembly of FtsZ (black). MinE (purple) dynamically displaces MinCD, resulting in a pole-to-pole oscillation. Cdv3 (yellow) localizes to the developing divisome where it acts through unknown mechanisms to spatially regulate FtsZ assembly and/or cell constriction. (B to E) Average cell lengths for S. elongatus strains induced to upregulate MinC (B), MinE (C), MinD (D), or Cdv3 (E) from a theophylline-inducible riboswitch integrated at neutral site 2 (NS2) 24 h after addition of inducer. Each of the indicated Min proteins was encoded with an mTurquoise2 tag (see Materials and Methods). Error bars represent standard deviations from ≥3 independent experiments with ≥200 measured cells for each experiment. P values are indicated from two-tailed unequal variances t tests relative to the uninduced controls; *, P < 0.01; **, P < 10−5; ***, P < 10−8.
FtsZ is a bacterial cytoskeletal polymer that is critical for the establishment of the divisome and the regulation of cell division; in turn, the Min system regulates FtsZ assembly and positioning (12, 13). Synechococcus elongatus has functional homologs of the minC, minD, and minE genes that compose the classic Min system in Escherichia coli, and these genes are widely conserved in cyanobacteria (14–16). MinC acts in conjunction with MinD to inhibit FtsZ assembly, while MinE is a negative regulator of MinCD. The emergent dynamics between these proteins act to position the FtsZ ring (Z ring) at the midzone of elongating bacteria (Fig. 1A) (12, 13, 16). Cyanobacteria also express Cdv3, a protein that possesses key features of another Min protein, DivIVA, found in Bacillus subtilis, which lacks MinE. DivIVA is a key factor regulating Z-ring placement and constriction in B. subtilis, and Cdv3 is also important for cyanobacterial divisome placement and functioning (14, 17), although the exact mechanisms underlying Cdv3's activity remain to be elucidated. Altering the activity of any of the Min system factors can disrupt Z-ring formation and/or positioning, thereby generating changes in cell size (13, 15, 16, 18). Therefore, careful tuning of the activity of these proteins may provide a way to alter cell morphology in a predictive manner that could be useful for cyanobacterial cultivation and harvesting.
In this work, we modulated the expression levels of each of the four Min system genes found within the model cyanobacterium Synechococcus elongatus PCC 7942 and examine the resulting impact on cell morphology. Our experimental design places an emphasis on examining the potential of this genetic approach for engineering cyanobacterial cell morphology in ways that might improve cyanobacterial biomass recovery and processing. Specifically, we analyzed the potential for FtsZ-regulatory genes to be controlled in a manner that would alter cell division, resulting in programmable changes in cell size. Further, our approach examined the plasticity of cell morphology that is tolerated in rod-shaped cyanobacteria without significant losses in cell viability or photosynthetic productivity. Finally, through the analysis of cell sedimentation rates and susceptibility to lysis, we observed the effects of changing the cell length of S. elongatus on properties that are relevant to the recovery and processing of cyanobacterial cell biomass. As our results provide direct evidence that S. elongatus can be engineered to inducibly elongate by nearly three orders of magnitude and that hyperelongated cyanobacteria exhibit favorable sedimentation and lysis properties, we conclude with a discussion of how such genetic tools may be of use for scaled cyanobacterial cultivation.
RESULTS
Expression of Min components from a tunable riboswitch alters cyanobacterial length.
To explore the effects of altered Min activity on cell shape, we generated cyanobacterial strains with an additional integrated copy of minC, minD, minE, or cdv3 under the control of a riboswitch. This inducible system is tightly off in the absence of the riboresponse regulator, theophylline, and exhibits a tunable induction pattern that is largely linear in response to theophylline (see Fig. S1 in the supplemental material) (19). Therefore, this expression system provides a robust platform for investigating the range of morphological engineering that can be achieved through altering the expression of key Min system genes.
To readily assess the expression and localization of the introduced genes, we tagged each introduced Min gene with mTurquoise2 (mTurq) (20). MinC, MinD, and MinE were N-terminally tagged, while Cdv3 was C-terminally tagged. We have previously demonstrated that tagged MinC or Cdv3 can fully replace the endogenous copy without causing phenotypes associated with ΔminC or Δcdv3 backgrounds (16). Furthermore, both mTurq-MinD and mTurq-MinC retained the capacity to oscillate within the cell. This suggests that the reporter constructs mTurq-MinC, mTurq-MinD, and Cdv3-mTurq retain many, if not all, of their functional properties. Due to the difficulty of generating fully penetrant ΔminE lines of S. elongatus (16), we do not have conclusive evidence indicating that mTurq-MinE can replace the endogenous (untagged) copy.
We first examined the dose-responsive impact of increasing protein levels of each tagged Min component on cell morphology. We exposed separate cultures to increasing concentrations of theophylline, from 0 μM to 2 mM, and measured the dimensions of cells after 24 h of induction. As expected, we observed that mTurq fluorescence was directly related to the amount of theophylline added, and no mTurq signal was observed in uninduced cultures (Fig. S1).
Each of the Min constructs produced changes in cell length that were responsive to the amount of theophylline added to the culture. The expression of mTurq-MinC correlated with increases in the average cell length (from 20% to ∼100% relative to uninduced controls), and these size increases were proportional to the amount of inducer added (Fig. 1B). This increase in size is anticipated, as MinC disrupts FtsZ polymerization, thereby delaying the establishment of a divisome and onset of cell septation (16, 21, 22). Upregulation of mTurq-MinE also increased cell size (Fig. 1C), consistent with our previous results (16). The mTurq-MinE-induced elongation was relatively mild (∼40%) and was observed only at the highest induction levels (2 mM) (Fig. 1C). By contrast, upregulation of mTurq-MinD increased the variability of cell lengths in the population while only minimally increasing average cell size (Fig. 1D). Altered activity of MinD has been previously associated with increased cell size variability due to the mispositioning of the divisome, which results in subsequent asymmetries in cell division (18, 23). The most dramatic increases in cell length were associated with the upregulation of Cdv3-mTurq. Twenty-four hours after inducing Cdv3 fusions, the average cell size increased by 100% to 400% (Fig. 1E). Again, the degree of cell elongation correlated with the level of induction, but the relationship was not linear as it was in cells with elevated MinC levels (Fig. 1B).
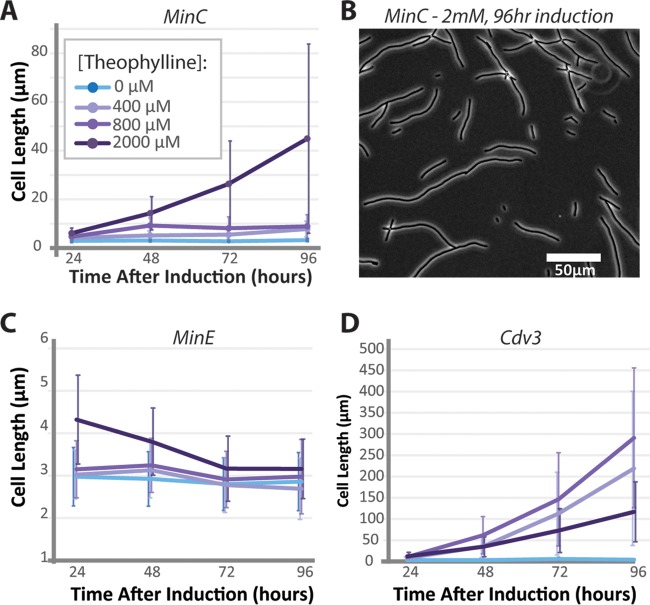
At short time periods, increased levels of MinC, MinE, or Cdv3 resulted in cell elongation; therefore, these proteins are candidates to be explored for engineering the morphology of the cyanobacterial population toward larger cells. By contrast, as an increase in MinD only changed the variability in cell length in the culture, we did not consider it a good candidate for engineering cells toward a specific morphology. To more completely assay the capacity of MinC, MinE, and Cdv3 to alter cell morphology over longer time periods, we followed mTurq-MinC, mTurq-MinE, and Cdv3-mTurq lines for multiple days after induction. MinC cells induced at the highest theophylline concentrations elongated by about 20-fold, reaching average cell lengths of 45 μm after 96 h (Fig. 2A). These cells were variable in size (Fig. 2B), whereas lower induction of mTurq-MinC resulted in cell populations that reached a steady-state cell length at 48 h that was 2- to 3-fold longer than in uninduced cultures. Therefore, modest induction in the levels of MinC might represent an approach for making relatively subtle increases in cell size, while greatly increased MinC protein levels might result in larger changes in gross cell size within a subpopulation of cells. Long-term upregulation of MinE did not stably increase cell lengths (Fig. 2C); at later time points, all cells returned to a baseline length of ∼3 μm. This suggests that the modest increases in cell length at 24 h after 2 mM theophylline induction of mTurq-MinE (Fig. 1C) might be due to an artifact of overexpression or toxicity of the inducer. Supporting this interpretation, we observed a small but significant increase in the doubling time of wild-type cells incubated with 2 mM theophylline that was absent at lower concentrations of the inducer (see Fig. S2). This is similar to, albeit slightly lower than, previous reports of toxic effects of theophylline on S. elongatus higher than (but not at) 2 mM (19). We therefore limited our later analysis to induction conditions containing ≤1 mM theophylline.
FIG 2.
Long-term cell morphology changes following induction of Min proteins. S. elongatus cultures were exposed to increasing concentrations of theophylline for 4 days and imaged to analyze cell lengths. (A) Average lengths for NS2::riboswitch::mTurq-MinC strains. (B) A representative field of cells 96 h after induction with 2,000 μM theophylline. Average cell lengths for NS2::riboswitch::mTurq-MinE (C) and NS2::riboswitch::Cdv3-mTurq (D) strains induced with the indicated concentrations of theophylline. Error bars are standard deviations from 3 independent experiments.
Division-arrested S. elongatus cells exhibit dramatic cell elongation.
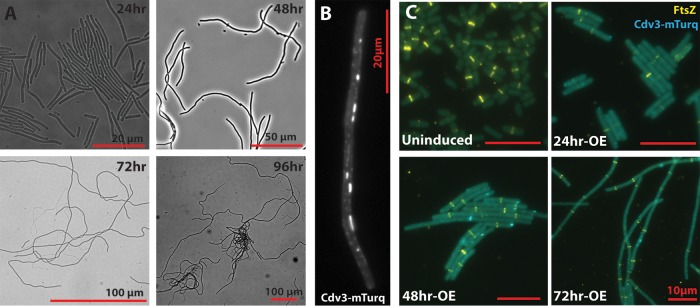
Cells overexpressing Cdv3 elongated at an accelerating rate over time, with most cells reaching average lengths >100 μm after 3 to 4 days of induction (Fig. 2D and 3A). S. elongatus seems exquisitely sensitive to overexpression of Cdv3-mTurq, and increases in cell length of at least 30-fold were observed regardless of the concentration of theophylline at these later time points. These relatively large increases in cell size over short time periods suggest that division in induced cells may be largely arrested, but that growth continues. Consistent with this, the average cell length in Cdv3-mTurq induced lines followed an exponential trajectory (Fig. 2D and 3A), where the average cell length doubled every 12.7 ± 1.5 h within the first 48 h after induction.
FIG 3.
Cells with upregulated Cdv3-mTurq activity exhibited radical elongation and disrupted FtsZ organization. (A) NS2::riboswitch::Cdv3-mTurq strains were induced with 400 μM theophylline and representative bright-field images from each day following induction are presented. (B) Representative localization pattern of Cdv3-mTurq in live elongated S. elongatus cells treated as in panel A for 48 h. Cdv3-mTurq concentrates into puncta and filaments along the length of the cell. (C) Immunolocalization of FtsZ (yellow) in representative cells induced as in panel A for the indicated number of hours. Formation of Z rings is delayed in Cdv3-mTurq-expressing (blue) lines (24hr-OE), while multiple mispositioned Z rings are evident in highly elongated cells (48 to 72 h postinduction) without clear indications of constriction. Bars = 10 μm.
The Cdv3-mTurq fusion localized in small puncta and large filaments along the length of the cell (Fig. 3B; see also Fig. S1). This localization pattern is unlike that of endogenous Cdv3, which we have previously shown to be concentrated at the cell midzone (16). We examined the FtsZ staining pattern in Cdv3-overexpressing cells by immunolocalization to determine how Z-ring formation was influenced by Cdv3-mTurq expression (Fig. 3C). Twenty-four hours after induction, many elongated cells lacked a clearly defined Z ring, and those that were visible exhibited faint FtsZ staining. By comparison, in uninduced controls, nearly all cells >2.5 μm had a single, brightly staining Z ring localized at the cell midzone (Fig. 3C). Cells that were induced to express Cdv3-mTurq for 48 to 72 h commonly exhibited one or more brightly staining Z rings along the length of the cell, but there was no obvious sign of visible cell constriction at the resolution of the light microscope. Collectively, these results suggest that Cdv3-mTurq induction delays the onset of Z-ring formation and also causes functional changes within the divisome that prevent septation. Because of the division arrest that leads to extreme cell elongation, the manipulation of Cdv3 represents an attractive target for testing the effectiveness of cell morphology engineering as it applies to cyanobacterial harvest and lysis.
Hyperelongated cells are more prone to sediment.
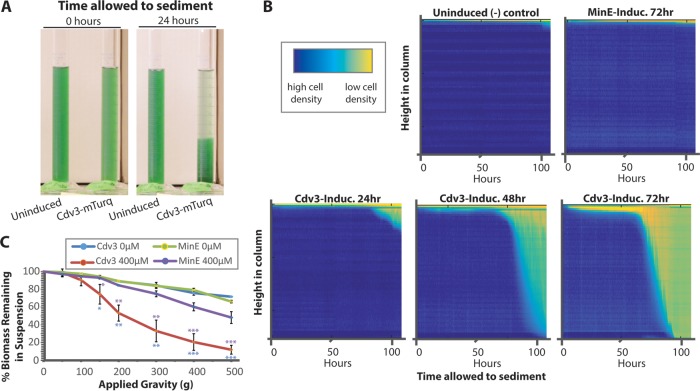
To examine the capacity for cell elongation to improve biomass recovery, we monitored sedimentation rates of cyanobacterial cells after Cdv3-mTurq expression. We first examined cell sedimentation under normal gravity by recording the positions of suspended cells in a stationary water column over time. Time-lapse imaging of cell suspensions revealed that hyperelongated Cdv3-expressing cells spontaneously settled to the bottom of the water column under normal gravity (Fig. 4A; see also Movie S1 and Fig. S3). To best represent this temporal data of cell sedimentation, we converted time-lapse data into a heatmap of cell positions along the length of the water column over time (Fig. 4B). A direct correlation between the average cell length at the time cells were seeded into cylinders and the rapidity of sedimentation was observed (Fig. 4B and 2D). Uninduced controls or cells induced to accumulate mTurq-MinE did not exhibit sedimentation by gravity (Fig. 4A and B), indicating the increased sedimentation rates were correlated with cell elongation rather than theophylline addition. Similarly, we examined the clearance of cell biomass from suspension under mild centrifugal forces and found that elongated cells pelleted more rapidly (Fig. 4C).
FIG 4.
Enhanced sedimentation in hyperelongated cells. (A) Cdv3-mTurq lines of S. elongatus that were induced with 400 μM theophylline for 72 h settle under natural gravity more rapidly than uninduced control samples when unperturbed over time. (B) Heatmaps of cyanobacterial density along the water column over time when induced. Cdv3-mTurq strains induced with 400 μM for the indicated period of time (bottom) and uninduced strains and MinE-overexpressing negative controls (top). Additional example experiments are displayed in Fig. S3 and Movie S1 in the supplemental material. (C) Quantification of biomass that remains in suspension following mild centrifugation of S. elongatus for strains induced to overexpress mTurq-MinE (purple) or Cdv3-mTurq (red) or uninduced controls (green, blue) for 72 h. P values are from an unequal variances t test against the uninduced Cdv3-mTurq control (blue) or against the corresponding 400 μM-induced mTurq-MinE control (purple). *, P ≤ 0.05; **, P ≤ 0.01, ***, P ≤ 0.001.
Hyperelongated cells are more readily lysed by mechanical force.
Beyond enhanced cell sedimentation rates in cyanobacteria with elevated Cdv3 levels, the increase in cell lengths might be expected to have a direct impact on the vulnerability of these cells to disruptive agents and torsional/shear forces that are sometimes employed to lyse cells for bioproduct recovery. Cell elongation increases the cell surface-to-volume ratio and, more importantly, can be expected to exacerbate the torsional force experienced by a cell under sheer stress due to (i) increased torque from additional length and/or (ii) structural defects in the cell wall caused by abnormal cell extension.
We examined cell lysis of normal and elongated cells by tracking populations of cyanobacterial cells by flow cytometry before and after passage through a cell disrupter. After the induction of Cdv3-mTurq, the forward scatter and chlorophyll α autofluorescence of cells continued to increase at 24 and 48 h after theophylline addition (Fig. 5A), in agreement with the increasing size of these cells (Fig. 1E, 2D, and 3). It should be noted that a population of cells (∼30%) displayed normal scatter and autofluorescence, consistent with the subpopulation of smaller cells that we observed by microscopy beginning 48 h after Cdv3-mTurq induction (Fig. 3A). Cyanobacterial populations were subjected to relatively mild pressures in a cell disrupter and examined for cell lysis by flow cytometry (Fig. 5B). We observed a significant decrease in the proportion of elongated cells after passage through the cell disrupter at even the lowest pressure that could be programmed (20,000 to 30,000 lb/in2 is the manufacturer's suggestion for bacterial lysis) and a nearly complete lysis of the elongated population at 8,000 lb/in2 (Fig. 5B to D). By contrast, uninduced or MinE-overexpressing cells were lysed at rates similar to those of the uninduced controls at the lowest pressure, and likewise were incompletely disrupted at 4,000 or 8,000 lb/in2 (Fig. 5C).
FIG 5.
Hyperelongated cells are preferentially vulnerable to mild shear forces. (A) NS2::riboswitch::Cdv3-mTurq strains were analyzed by flow cytometry 0, 24, or 48 h after induction with 800 μM theophylline. Increased forward scatter and chlorophyll-associated red autofluorescence were observed, and these changes were correlated with the increased cell size of Cdv3-overexpressing cells. Cell counts are gated into wild-type length (blue box) and elongated (red box). (B) Uninduced (top) and Cdv3-overexpressing (bottom) cyanobacterial cultures were exposed to increasing pressure and lytic forces on a cell disruptor, and the population was monitored by flow cytometry. The proportions of elongated cells (red) relative to WT lengths (blue) are represented in pie charts for each condition. (C) Quantification of the proportions of intact cells remaining after cell disruption with increasing pressures. P values from two-tailed unequal variances t tests against the equivalent uninduced control are reported above the relevant bars wherever P < 0.05. (D) Quantification as in panel C, but where only the proportions of elongated cells are tracked.
DISCUSSION
Our results indicate that cell length in the model cyanobacterium S. elongatus can be manipulated by disrupting cell division, thereby separating the process of division from growth. By controlling protein levels of the Min system proteins MinC and Cdv3, we show that cell length can be tunably increased by 20 to 500% and that cells can elongate by over two orders of magnitude through division arrest. These proteins represent promising genetic targets to control cell morphology in rod-shaped cyanobacteria. Furthermore, we show that elongated cells more readily sediment from solution and are more vulnerable to mechanical lysis. These properties might be useful for decreasing the operating costs associated with cyanobacterial biomass recovery and the processing for scaled applications.
Placing MinC and Cdv3 under the control of tunable riboswitches enable us to adjust the activities of these proteins across a dynamic range in vivo (Fig. 1 and Fig. S1 in the supplemental material). The mechanism by which MinC inhibits FtsZ polymerization has been well studied in other bacteria and confirmed to play an analogous role in cyanobacteria (15, 16, 18). We observed that the induction of mTurq-MinC led to a titratable increase in cell length that was maintained over time (Fig. 1B and 2A). More extreme elongated cell phenotypes were observed following the induction of Cdv3, a protein that is emerging as an important regulator of cyanobacterial division. The addition of minimal amounts of theophylline to induce Cdv3-mTurq translation produces a phenotype largely consistent with cell division arrest, suggesting that S. elongatus' division is relatively sensitive to Cdv3 protein levels. Furthermore, previous reports indicate that knockout of cdv3 causes highly elongated cell phenotypes or death (14, 16, 17), suggesting that Cdv3 must be maintained in a relatively narrow window of activity to promote normal division. However, the mechanism of division arrest in cells with elevated Cdv3 levels cells remains unclear, in part because the mechanism of Cdv3's function is not well understood.
There is some conservation of the sequence of Cdv3, and it is predicted to share structural features with DivIVA, a well-characterized division protein in B. subtilis; although, Cdv3 exhibits sufficient differences to preclude its characterization as a functional homolog (14, 16, 17, 24). Previous studies in Synechocystis sp. strain PCC 6803 have found that Cdv3 interacts with Ftn2 (also called ZipN), which directly binds to FtsZ and is localized to the midzone (17). Consistent with this, we have previously demonstrated that endogenous Cdv3 localizes to the midzone in S. elongatus, colocalizing to FtsZ rings, and is required for the recruitment of a midzone-localized pool of MinC (16). As Ftn2 and MinC are important regulators of cyanobacterial division (15, 18, 25, 26), we speculate that elevating the levels of Cdv3 may lead to sequestration of these or other divisome regulators from their normal sites of action, resulting in delayed Z-ring formation and impaired remodeling of the divisome to allow for septation. While we have shown in another recent report that a C-terminal mNeonGreen fusion of Cdv3 (Cdv3-mNG) (27) can fully replace the endogenous gene and also causes cell elongation when overexpressed, it is possible that the C-terminal fusion of mTurquoise to Cdv3 may act as a dominant-negative inhibitor of endogenous Cdv3 function (16). Regardless, Cdv3 and MinC may be only 2 representative genes that can be genetically manipulated in this fashion to disrupt divisome assembly and to promote cell elongation; other FtsZ-regulatory genes (28) may also be promising targets for this purpose. Furthermore, the addition of chemical inhibitors of FtsZ might be expected to have similar results (29), although they may not be practical for scaled applications.
Elongation in our division-arrested cells was exponential, and proceeded at timescales similar to the doubling time of actively dividing cells. Within the first 48 h, the time for the average Cdv3-mTurq-expressing cell to double in length was 12.7 ± 1.5 h; this is similar to the doubling time of actively dividing cells (Fig. S2). At 48 h after Cdv3-mTurq induction, a bimodal distribution of cell lengths emerges (Fig. 2D and 5A), with a substantial proportion of smaller cells. This indicates that there is incomplete arrest of division in Cdv3-overexpressing strains, which leads to the regeneration of some shorter cells within the population. Dry cell biomass production in Cdv3-overexpressing strains does not decrease relative to uninduced controls (see Fig. S4), directly suggesting that there is no loss of photosynthetic productivity in hyperelongated cells. Instead, it appears that some induced cells undergo “leaky” division, while other cells appear to exponentially elongate without any division events after Cdv3-mTurq induction. We routinely observed 250 to 600 μm cells (∼10% of cells measured) at 72 h and cells approaching 1 mm in length 96 h after Cdv3-mTurq induction. Collectively, these results suggest that hyperelongation of cells does not negatively impact photosynthetic productivity, although identification of an engineering strategy with a more penetrant cell division arrest might be preferable for some applications. It is possible that upregulation of more than one cell division inhibitory factor would lead to a more complete arrest.
The concept of cell morphology engineering for bioindustrial purposes is relatively recent (8) and has potential to improve the economic feasibility of scaled processes by conferring cultivated organisms with processing-compatible properties at the cellular level. In a recent example, Halomonas TD01 strains with increased MinCD activity were observed to sediment more rapidly (9, 10). Genetic control over sedimentation rates is arguably of great interest for the feasibility of scaled cyanobacterial and algal cultures, because cell recovery and dewatering expenses are a major proportion of total cultivation costs (4–7). The manipulation of the intrinsic geometric features of cells could be used to customize biomass for a given set of processing equipment, rather than vice versa. This engineering could take the form of either constitutive expression of morphology-altering genes to permanently alter cell shape or inducible systems that could be activated at a desired time (e.g., just prior to harvest). The latter strategy may be particularly important for cyanobacterial cultivation, as sedimentation rates should be minimized during growth to decrease the energetic costs for liquid handling that are required to sufficiently mix cells during growth. Theophylline is a relatively inexpensive chemical and has been proposed as a feasible inducer for scaled applications (19, 30). However, the inexpensive chemicals could even present substantial costs if needed at high concentrations, and we observe mild toxicity effects of theophylline at ≥2 mM. Thus, strains should be engineered to minimize the amount of exogenous inducer required. An attractive alternative approach might link genetic circuits to signal transduction pathways (e.g., quorum sensing) to enable autoinduction of division-repressing genes under specific conditions, thereby reducing input costs. Finally, the natural cyanobacterial growth rate is expected to influence the length of time required for elongation, and so the optimization of culture conditions to promote shorter division times or the use of rapidly growing strains might make this approach more effective. For example, the related cyanobacterial strain Synechococcus elongatus UTEX 2973 has recently been reported as a promising bioindustrial strain and exhibits a maximal autotrophic doubling time of 1.9 h (31). At this rate, cells could feasibly reach lengths >200 μm in 12 h (the natural light period in a typical day), which is a length at which we observe significantly enhanced cell sedimentation and susceptibility to mechanical lysis (Fig. 4 and 5). Rapidly growing strains such as these might also mitigate potential harvesting problems (e.g., increased spurious cell lysis) because the time between induction of cell elongation and potential harvest would be minimized.
The use of morphology engineering might provide an alternative method for improving cyanobacterial biomass recovery that is complementary to other recently reported attempts to engineer cells for improved sedimentation. For example, cyclic di-GMP has been shown to play a role in biofilm formation and cell buoyancy (32), and spurious activation of pathways that lead to high cytosolic di-GMP also lead to increased sedimentation in the cyanobacterium Fremyella diplosiphon (33). Similarly, genetic knockout of factors that regulate biofilm formation or extracellular lipopolysaccharide composition leads to increased aggregation and sedimentation in S. elongatus (34, 35). It is possible that combinatorial control over regulatory pathways for both cell morphology and biofilm development might provide additive effects on the rate of cell sedimentation relative to relying on one approach alone.
Cell morphology engineering may be useful for applications beyond harvest and lysis. For example, work in other bacteria has shown that cells with altered morphology accumulate a higher proportion of their total biomass as a target compound or within inclusion bodies (9, 10, 36). Additionally, morphology engineering may have useful applications for increasing the resistance of cyanobacteria to grazing species that frequently contaminate open-air ponds and which have preferences for bacteria with specific dimensions due to physical feeding constraints (e.g., size of mouth/digestive tract) (37–39). Finally, a methodology for gentle cell lysis of cyanobacteria might enable the recovery of sensitive complexes or structures that can be destroyed under harsh conditions used routinely for cyanobacterial lysis (40).
MATERIALS AND METHODS
Cyanobacterial culture conditions.
S. elongatus (41) was obtained from the American Type Culture Collection (ATCC 33912). Cultures of S. elongatus were routinely grown in 125 ml baffled flasks (Corning) containing 50 ml BG-11 medium (Sigma) buffered with 1 g/liter HEPES, pH 8.3. Media was supplemented with 12.5 μg/ml kanamycin for routine subculturing of riboswitch lines, and antibiotics were omitted for all other experiments. Flasks were cultured in a Multitron II (Infors HT, Switzerland) incubation system with 80 μmol m−2 s−1 white light (Sylvania 15W Gro-Lux fluorescent bulbs) at 32°C, 2% CO2, and a 130-rpm shake speed. Prior to beginning any experiment, cultures were initially back-diluted to an optical density at 750 nm (OD750) of 0.2 daily for at least 2 days so as to maintain logarithmic growth and standardize conditions across flasks. On initiation of an experiment, cells were first inoculated into fresh media at an OD750 of 0.2 (i.e., at time 0). For extended time-course experiments, cultures were back-diluted every 24 h to an OD750 of 0.2. Cells were inoculated with the indicated concentrations of theophylline from a 10 mM stock dissolved in BG-11 medium.
Construct cloning and cyanobacterial transformation.
Riboswitch::mTurq-MinC, riboswitch::mTurq-MinD, riboswitch::mTurq-MinE, and riboswitch::Cdv3-mTurq constructs were generated by backbone PCR amplification of plasmid pEYN12 (19) without luciferase. PCR was performed on minC, minD, minE, and cdv3 from wild-type (WT) S. elongatus cells, and mTurquoise2 was synthesized as a gBlock (IDT DNA; see Table S1 in the supplemental material for the primer list). Isothermal assembly (42) was performed to assemble all 4 constructs. Plasmids were cloned using E. coli DH5α chemically competent cells (Invitrogen). All cyanobacterial transformations were performed as previously described (43), resulting in the integration of the genetic constructs into the endogenous chromosomes by homologous recombination at neutral site 2 (NS2), where the constructs are permanently maintained. To select for transformants, cells were plated on BG-11 agar with 12.5 μg ml−1 kanamycin and were grown on solid plates under 80 μmol m−2 s−1 light at 32°C and 2% CO2 as described above. Single colonies were picked and placed into 96-well plates containing 300 μl of BG-11 medium with 12.5 μg ml−1 kanamycin. Clonal subpopulations were verified for complete gene replacement via PCR.
Microscopy and cell length measurements.
All images were collected using a Zeiss Axio Observer A1 microscope (100×, 1.46 numerical aperture [NA] objective) with an Axiocam ICc5 camera. To fix cells for immunofluorescence or for the long-term storage of samples, 2 ml of cells were fixed with 2.5% glutaraldehyde/2.5% formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) (Electron Microscopy Sciences) at room temperature for 30 min, and then washed with phosphate-buffered saline (PBS). Cell length and average fluorescence measurements were acquired using manual tools in Zeiss Zen software. To limit bias, all measurements were limited to a defined area of a field and all cells within the designated area were measured. Three independent experiments were conducted, and at least 10 random fields for each same-day experiment were captured with an average of ∼25 cells/field. All recorded cell length averages include ≥500 measured cells per strain per condition.
Immunofluorescence staining.
MinCE and Cdv3 overexpression strains were inoculated in 50 ml BG-11 medium containing the indicated levels of theophylline. Two milliliters of cells were fixed with 2.5% glutaraldehyde/2.5% formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) (Electron Microscopy Sciences) at room temperature for 30 min, and were then washed with PBS plus 0.01% Tween 20. After treatment with 0.05% Triton X-100 and 0.01% Tween 20 in PBS for 15 min, the cells were further permeabilized for 30 min at 37°C with 20 μg ml−1 lysozyme dissolved in Tris-HCl (pH 7.5, 10 mM EDTA), were washed, and then were blocked with 5% bovine serum albumin (Sigma-Aldrich) in PBS (blocking buffer) for 60 min. After blocking, the cells were incubated overnight at 4°C with anti-Anabaena FtsZ antibodies (Agrisera Antibodies) diluted 1:250 in blocking buffer and then washed with PBS. Secondary staining was conducted with 1:1,000 goat anti-rabbit IgG Dylight 488 (Life Technologies) in blocking buffer.
Cell disruption and lysis.
Sheer stress tests were conducted using a One Shot system (Constant Systems Limited). Cells were inoculated as stated above at 250-ml volumes. Next, 50-ml samples were passed through the disrupter that was set to the indicated level of pressure (0 lb/in2, 4,000 lb/in2, or 8,000 lb/in2) and passed through the sapphire aperture for two passes. For “0 lb/in2” treatments, the instrument was set to the lowest possible pressure (0 lb/in2), although monitoring of the internal pressure indicated that cells experienced ∼1,000 to 1,500 lb/in2 maximally as the piston compressed the sample (i.e., true “zero” pressure is not possible on this system).
Flow cytometry to measure lysis of hyperelongated versus wild-type length cells.
Flow cytometry was performed using a BD Accuri C6 flow cytometer (653119; BD Biosciences). Gating was set so that 95% of the control (uninduced, undisrupted) population was included and potential contaminants that fell outside the gate were excluded. Cell counts were collected for 30 s using a 20-mW 488-nm solid-state blue laser with particle events monitored using 533/30 nm (fluorescein isothiocyanate [FITC]/green fluorescent protein [GFP]) and FL3 >670 nm optical filters for chlorophyll-α fluorescence. All cell disruption experiments were conducted with an n value of 3, where independent experiments were executed on different days. For each flow cytometry measurement, at least 180,000 cell events were recorded.
Sedimentation assays.
Samples were back-diluted into BG-11 medium containing the indicated concentrations of theophylline every 24 h for up to 72 h in 50-ml volumes. At each time point, 25 ml of culture was transferred to a 25-ml graduated cylinder and imaged every 15 min to generate a time-lapse video that illustrated gravity sedimentation. Time-lapse images were captured by a Canon EOS 500D digital camera with a 1/10 s at F9 exposure. The cell densities were estimated by calculating the absorbances over the central axis of each graduated cylinder using the red channel of the images, normalized to the maximal absorbance when the cylinders were set up. For centrifuge-assisted sedimentation assays, the OD750 of each culture was measured, and then 10 ml of each cultivar was centrifuged for 2 min on a Sorvall Legend RT benchtop centrifuge (round bucket rotor, 75006445) at the indicated speed. The OD750 of the supernatants were measured again to estimate the proportions of cells remaining in suspension.
Biomass accumulation and growth rate analysis.
To evaluate dry cell biomass after inducing cell elongation, cultures of NS2::riboswitch::Cdv3-mTQ S. elongatus in exponential phase were back-diluted to an OD750 of 0.2. The resulting cultures were split into identical biological replicates containing 60 ml each, where one culture was induced with a final concentration of 400 μM theophylline and a BG-11 blank of an equivalent volume was added to the paired negative control. Cultures were incubated in 250-ml baffled flasks under identical conditions as above, and 30 ml of each culture was withdrawn at time points 36 h and 60 h after induction. Collected cells were pelleted at 6,000 rpm in a SS-34 Sorvall rotor, and pellets were completely dried in a vacuum desiccator before weighing the dry cell mass with an analytical balance (Sartorius A120S).
For the analysis of cell growth rate as a function of theophylline concentrations, wild-type S. elongatus cells in exponential phase were back-diluted to an OD750 of 0.2, and the indicated levels of theophylline were added (0 to 2,000 μM). The optical densities of cultures were measured 24 h after theophylline addition, and the approximate doubling time over this time period was calculated as [24 h × log(2)]/[log(final OD750) − log(0.2)].
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation award number 1517241 with additional support for infrastructure and equipment from the Department of Energy (DE-FG02-91ER20021).
A.J., J.C., J.S.M., J.H., and D.C.D. designed and executed the experiments and analyzed the experimental data. A.J., J.S.M., K.W.O., and D.C.D. conceived the project and prepared the manuscript.
J.S.M., K.W.O., and D.C.D. are listed as coinventors on the provisional patent application entitled “Genetic control of cell size” (serial no. 62/377,964), which describes the manipulation of FtsZ-regulatory proteins for control over cyanobacterial cell morphology and harvest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00053-17.
REFERENCES
- 1.Ducat DC, Way JC, Silver PA. 2011. Engineering cyanobacteria to generate high-value products. Trends Biotechnol 29:95–103. doi: 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC. 2008. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol 19:235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Pittman JK, Dean AP, Osundeko O. 2011. The potential of sustainable algal biofuel production using wastewater resources. Bioresour Technol 102:17–25. doi: 10.1016/j.biortech.2010.06.035. [DOI] [PubMed] [Google Scholar]
- 4.Chisti Y. 2013. Constraints to commercialization of algal fuels. J Biotechnol 167:201–214. doi: 10.1016/j.jbiotec.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 5.Uduman N, Qi Y, Danquah MK, Forde GM, Hoadley A. 2010. Dewatering of microalgal cultures: a major bottleneck to algae-based fuels. J Renew Sustain Energy 2:012701. doi: 10.1063/1.3294480. [DOI] [Google Scholar]
- 6.Singh J, Gu S. 2010. Commercialization potential of microalgae for biofuels production. Renew Sustain Energy Rev 14:2596–2610. doi: 10.1016/j.rser.2010.06.014. [DOI] [Google Scholar]
- 7.Zeng X, Guo X, Su G, Danquah MK, Chen XD, Lin L, Lu Y. 2016. Harvesting of microalgal biomass, p 77–89. In Bux F, Chisti Y (ed), Algae biotechnology. Springer, Berlin, Germany. [Google Scholar]
- 8.Jiang X-R, Chen G-Q. 2016. Morphology engineering of bacteria for bio-production. Biotechnol Adv 34:435–440. doi: 10.1016/j.biotechadv.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Tan D, Wu Q, Chen J-C, Chen G-Q. 2014. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates. Metabol Eng 26:34–47. doi: 10.1016/j.ymben.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X-R, Wang H, Shen R, Chen G-Q. 2015. Engineering the bacterial shapes for enhanced inclusion bodies accumulation. Metabol Eng 29:227–237. doi: 10.1016/j.ymben.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Curtiss R. 2009. Nickel-inducible lysis system in Synechocystis sp. PCC 6803. Proc Natl Acad Sci U S A 106:21550–21554. doi: 10.1073/pnas.0911953106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margolin W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nature Rev Mol Cell Biol 6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyagishima SY, Wolk CP, Osteryoung KW. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol Microbiol 56:126–143. doi: 10.1111/j.1365-2958.2005.04548.x. [DOI] [PubMed] [Google Scholar]
- 15.Mazouni K, Domain F, Cassier-Chauvat C, Chauvat F. 2004. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol Microbiol 52:1145–1158. doi: 10.1111/j.1365-2958.2004.04042.x. [DOI] [PubMed] [Google Scholar]
- 16.MacCready JS, Schossau J, Osteryoung KW, Ducat DC. 2017. Robust Min-system oscillation in the presence of internal photosynthetic membranes in cyanobacteria. Mol Microbiol 103:483–503. doi: 10.1111/mmi.13571. [DOI] [PubMed] [Google Scholar]
- 17.Marbouty M, Saguez C, Cassier-Chauvat C, Chauvat F. 2009. ZipN, an FtsA-like orchestrator of divisome assembly in the model cyanobacterium Synechocystis PCC6803. Mol Microbiol 74:409–420. doi: 10.1111/j.1365-2958.2009.06873.x. [DOI] [PubMed] [Google Scholar]
- 18.Rowlett VW, Margolin W. 2013. The bacterial Min system. Curr Biol 23:R553–R556. doi: 10.1016/j.cub.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 19.Nakahira Y, Ogawa A, Asano H, Oyama T, Tozawa Y. 2013. Theophylline-dependent riboswitch as a novel genetic tool for strict regulation of protein expression in Cyanobacterium Synechococcus elongatus PCC 7942. Plant Cell Physiol 54:1724–1735. doi: 10.1093/pcp/pct115. [DOI] [PubMed] [Google Scholar]
- 20.Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, van Weeren L, Gadella TW Jr, Royant A. 2012. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun 3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Boer P, Crossley RE, Rothfield LI. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol 174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci U S A 96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Boer PA, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 24.Miyagishima SY. 2011. Mechanism of plastid division: from a bacterium to an organelle. Plant Physiol 155:1533–1544. doi: 10.1104/pp.110.170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelova O, Baulina O, Rasmussen U, Koksharova O. 2013. The pleiotropic effects of ftn2 and ftn6 mutations in cyanobacterium Synechococcus sp. PCC 7942: an ultrastructural study. Protoplasma 250:931–942. doi: 10.1007/s00709-012-0479-2. [DOI] [PubMed] [Google Scholar]
- 26.Koksharova OA, Wolk CP. 2002. A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J Bacteriol 184:5524–5528. doi: 10.1128/JB.184.19.5524-5528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz C, Natale P, Cueto L, Vicente M. 2016. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev 40:57–67. doi: 10.1093/femsre/fuv040. [DOI] [PubMed] [Google Scholar]
- 29.Hurley KA, Santos TM, Nepomuceno GM, Huynh V, Shaw JT, Weibel DB. 2016. Targeting the bacterial division protein FtsZ. J Med Chem 59:6975–6998. doi: 10.1021/acs.jmedchem.5b01098. [DOI] [PubMed] [Google Scholar]
- 30.Seeliger JC, Topp S, Sogi KM, Previti ML, Gallivan JP, Bertozzi CR. 2012. A riboswitch-based inducible gene expression system for mycobacteria. PLoS One 7:e29266. doi: 10.1371/journal.pone.0029266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Liberton M, Cliften PF, Head RD, Jacobs JM, Smith RD, Koppenaal DW, Brand JJ, Pakrasi HB. 2015. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep 5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agostoni M, Waters CM, Montgomery BL. 2016. Regulation of biofilm formation and cellular buoyancy through modulating intracellular cyclic di-GMP levels in engineered cyanobacteria. Biotechnol Bioeng 113:311–319. doi: 10.1002/bit.25712. [DOI] [PubMed] [Google Scholar]
- 34.Parnasa R, Nagar E, Sendersky E, Reich Z, Simkovsky R, Golden S, Schwarz R. 2016. Small secreted proteins enable biofilm development in the cyanobacterium Synechococcus elongatus. Sci Rep 6:32209. doi: 10.1038/srep32209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simkovsky R, Daniels EF, Tang K, Huynh SC, Golden SS, Brahamsha B. 2012. Impairment of O-antigen production confers resistance to grazing in a model amoeba-cyanobacterium predator-prey system. Proc Natl Acad Sci U S A 109:16678–16683. doi: 10.1073/pnas.1214904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Wu H, Jiang X, Chen G-Q. 2014. Engineering Escherichia coli for enhanced production of poly (3-hydroxybutyrate-co-4-hydroxybutyrate) in larger cellular space. Metab Eng 25:183–193. doi: 10.1016/j.ymben.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Day JG. 2013. Grazers: the overlooked threat to the sustained production of future algal biofuels. Biofuels 4:459–461. doi: 10.4155/bfs.13.29. [DOI] [Google Scholar]
- 38.Jezberová J, Komárková J. 2007. Morphological transformation in a freshwater Cyanobium sp. induced by grazers. Environ Microbiol 9:1858–1862. doi: 10.1111/j.1462-2920.2007.01311.x. [DOI] [PubMed] [Google Scholar]
- 39.Hahn MW, Höfle MG. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol Ecol 35:113–121. doi: 10.1111/j.1574-6941.2001.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 40.Frain KM, Gangl D, Jones A, Zedler JA, Robinson C. 2016. Protein translocation and thylakoid biogenesis in cyanobacteria. Biochim Biophys Acta 1857:266–273. doi: 10.1016/j.bbabio.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Shestakov S, Khyen NT. 1970. Evidence for genetic transformation in blue-green alga Anacystis nidulans. Mol Gen Genet 107:372–375. doi: 10.1007/BF00441199. [DOI] [PubMed] [Google Scholar]
- 42.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 43.Clerico EM, Ditty JL, Golden SS. 2007. Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol Biol 362:155–171. doi: 10.1007/978-1-59745-257-1_11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.