ABSTRACT
Diverse bacteria inhabit amphibian skin; some of those bacteria inhibit growth of the fungal pathogen Batrachochytrium dendrobatidis. Yet there has been no systematic survey of anti-B. dendrobatidis bacteria across localities, species, and elevations. This is important given geographic and taxonomic variations in amphibian susceptibility to B. dendrobatidis. Our collection sites were at locations within the Appalachian Mountains where previous sampling had indicated low B. dendrobatidis prevalence. We determined the numbers and identities of anti-B. dendrobatidis bacteria on 61 Plethodon salamanders (37 P. cinereus, 15 P. glutinosus, 9 P. cylindraceus) via culturing methods and 16S rRNA gene sequencing. We sampled co-occurring species at three localities and sampled P. cinereus along an elevational gradient (700 to 1,000 meters above sea level [masl]) at one locality. We identified 50 anti-B. dendrobatidis bacterial operational taxonomic units (OTUs) and found that the degree of B. dendrobatidis inhibition was not correlated with relatedness. Five anti-B. dendrobatidis bacterial strains occurred on multiple amphibian species at multiple localities, but none were shared among all species and localities. The prevalence of anti-B. dendrobatidis bacteria was higher at Shenandoah National Park (NP), VA, with 96% (25/26) of salamanders hosting at least one anti-B. dendrobatidis bacterial species compared to 50% (7/14) at Catoctin Mountain Park (MP), MD, and 38% (8/21) at Mt. Rogers National Recreation Area (NRA), VA. At the individual level, salamanders at Shenandoah NP had more anti-B. dendrobatidis bacteria per individual (μ = 3.3) than those at Catoctin MP (μ = 0.8) and at Mt. Rogers NRA (μ = 0.4). All salamanders tested negative for B. dendrobatidis. Anti-B. dendrobatidis bacterial species are diverse in central Appalachian Plethodon salamanders, and their distribution varied geographically. The antifungal bacterial species that we identified may play a protective role for these salamanders.
IMPORTANCE Amphibians harbor skin bacteria that can kill an amphibian fungal pathogen, Batrachochytrium dendrobatidis. Some amphibians die from B. dendrobatidis infection, whereas others do not. The bacteria that can kill B. dendrobatidis, called anti-B. dendrobatidis bacteria, are thought to influence the B. dendrobatidis infection outcome for the amphibian. Yet how anti-B. dendrobatidis bacterial species vary among amphibian species and populations is unknown. We determined the distribution of anti-B. dendrobatidis bacterial species among three salamander species (n = 61) sampled at three localities. We identified 50 unique anti-B. dendrobatidis bacterial species and found that all of the tested salamanders were negative for B. dendrobatidis. Five anti-B. dendrobatidis bacterial species were commonly detected, suggesting a stable, functional association with these salamanders. The number of anti-B. dendrobatidis bacteria per individual varied among localities but not among co-occurring salamander species, demonstrating that environment is more influential than host factors in structuring the anti-B. dendrobatidis bacterial community. These anti-B. dendrobatidis bacteria may serve a protective function for their salamander hosts.
KEYWORDS: Batrachochytrium dendrobatidis, pathogen inhibition, salamanders, skin bacteria, symbionts
INTRODUCTION
The skin microbiome of vertebrates serves as a barrier against pathogens (1–3) and can mediate disease risk (4, 5). Lower disease risk in vertebrates has been associated with different characteristics of the microbiome, such as high bacterial species richness (6–8), specific microbial community assemblages (2, 7, 9, 10), and the presence of microbes that produce metabolites that inhibit growth of pathogens (1, 11, 12). For amphibians, inter- and intraspecies-specific variations in the skin microbiome (13–15) may contribute to variation in responses to infection by the deadly fungal pathogen Batrachochytrium dendrobatidis (16, 17). Yet we know little about how microbial diversity differs among amphibian host species and environments and how this relates to B. dendrobatidis-associated disease risk. Characterizing these patterns is a step toward understanding evolutionary and ecological processes structuring functionally important microbial skin assemblages, which may aid in development of conservation strategies.
B. dendrobatidis has caused mass die-offs and extirpations of susceptible amphibians on a global scale (18–21), and yet no effective treatments exist for amphibians in the wild. An active line of research has been to identify bacteria that inhibit B. dendrobatidis growth, here referred to as anti-B. dendrobatidis bacteria, and to use their geographic distribution to predict fungal disease outcome in the field (11, 22, 23) or to use them in trials of efficiency of bioaugmentation to mitigate B. dendrobatidis-associated disease symptoms (24–27). To date, roughly 255 anti-B. dendrobatidis bacterial operational taxonomic units (OTUs) that have been identified from the skin of 37 amphibian species have been deposited in a reference database (28). Nine of these bacterial species have been used in bioaugmentation trials (2, 24, 25, 29, 30).
Bioaugmentation trials using anti-B. dendrobatidis bacteria have had mixed success. For instance, three studies found no effect of an anti-B. dendrobatidis bacterial species, Janthinobacterium lividum, in reducing B. dendrobatidis-associated disease symptoms (29, 31, 32), even though J. lividum had previously been shown to be effective against B. dendrobatidis (25–27). This suggests that a bacterial species may not mitigate the impact of B. dendrobatidis on host health for all host species or in all environments.
Host and environmental factors may influence anti-B. dendrobatidis bacterial composition or whether specific bacterial species are inhibitory. For instance, Bresciano et al. (33) found several anti-B. dendrobatidis bacterial species at high-elevation sites but did not detect these anti-B. dendrobatidis bacteria at low elevations, suggesting that environmental factors impact the geographic distribution of anti-B. dendrobatidis bacteria. If an amphibian population does not naturally harbor a bacterial species, that bacterial species may not colonize or persist on the host if used in bioaugmentation trials (34). Further, temperature can impact whether specific bacterial taxa produce inhibitory metabolites, and this may also depend on the species of host from which the bacteria were isolated (35, 36).
We quantified differences in the number and identity of anti-B. dendrobatidis bacteria on three Plethodon species at three localities and along an elevational gradient in the central Appalachian Mountains. We chose Plethodon salamanders because previous sampling had indicated a low prevalence of B. dendrobatidis even though B. dendrobatidis often occurs on other amphibians in the region (37). This contrasts with widespread population declines caused by B. dendrobatidis in closely related plethodontid salamanders in Central America (38). The skin microbiome of Plethodon salamanders has been shown to limit B. dendrobatidis infection (24, 27, 37, 39), and we expected anti-B. dendrobatidis bacteria to be prevalent. We also expected that different salamander species and populations would harbor different anti-B. dendrobatidis bacterial species given known species-level (14, 40) and population-level (13, 15, 33) differences in amphibian skin bacterial communities. However, we expected a few anti-B. dendrobatidis bacterial species, such as J. lividum, to be widely distributed across species and localities given previous reports of these taxa being present on Plethodon salamanders (41, 42). Our overall objectives were to (i) describe the geographic and taxonomic distribution of anti-B. dendrobatidis bacteria, (ii) determine how host and site factors influence the number of anti-B. dendrobatidis bacteria detected, and (iii) determine if the B. dendrobatidis inhibition score was related to bacterial phylogenetic relatedness. The inhibitory properties of anti-B. dendrobatidis bacteria may inform research on other emerging fungal pathogens such as B. salamandrivorans, which also poses a significant threat to global amphibian biodiversity (43, 44).
RESULTS
We did not detect B. dendrobatidis on any of the salamanders.
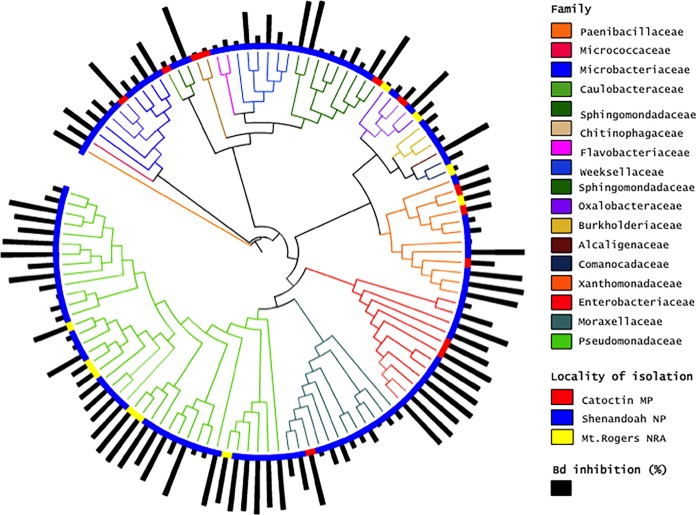
We collected and tested 341 bacterial isolates from 61 Plethodon salamanders at three localities (Catoctin MP, n = 103; Shenandoah NP, n = 187; Mt. Rogers NRA, n = 51), with an average of 6 (range, 0 to 17) bacterial morphotypes per individual (Table 1). We identified 119 of those bacterial isolates, representing a total of 50 anti-B. dendrobatidis bacterial OTUs across all salamanders (Table 1), as anti-B. dendrobatidis (Fig. 1). The 50 anti-B. dendrobatidis bacterial isolates belonged to four bacterial phyla (Actinobacteria [n = 5], Proteobacteria [n = 33], Bacteroidetes [n = 11], and Firmicutes [n = 1]), with B. dendrobatidis inhibition levels ranging from 1% to 100% (Fig. 1). We found that the more closely related anti-B. dendrobatidis bacterial species did not share similar B. dendrobatidis inhibition scores (Fig. 1) (P = 0.865 [Mantel]; Mantel statistic r = −0.023).
TABLE 1.
Summary of salamanders sampleda
| Locality | No. of sites | Elevation(s) (m) | Species | No. of salamanders sampled | No. of salamanders with anti-B. dendrobatidis bacteria | Avg no. of isolates per salamander (range) | Avg no. of anti-B. dendrobatidis isolates per salamander (range) |
|---|---|---|---|---|---|---|---|
| Catoctin | 1 | 404 | P. cinereus | 7 | 4 | 8.9 (5–14) | 1 (0–2) |
| 1 | 404 | P. glutinosus | 7 | 3 | 5.7 (2–9) | 0.6 (0–2) | |
| Subtotal | 14 | 7 | 7.3 | 0.8 | |||
| Shenandoah | 1 | 702 | P. cinereus | 4 | 3 | 5.5 (1–10) | 2.3 (0–3) |
| 2 | 797 ± 6 | P. cinereus | 6 | 6 | 9 (5–14) | 3.7 (1–7) | |
| 2 | 881 ± 18 | P. cinereus | 4 | 4 | 6.3 (4–11) | 3.8 (1–7) | |
| 1 | 974 | P. cinereus | 5 | 5 | 9.4 (6–16) | 4.4 (3–6) | |
| 4 | 697–974 | P. cylindraceus | 7 | 7 | 5.6 (2–8) | 2.4 (1–4) | |
| Subtotal | 26 | 25 | 7.2 | 3.3 | |||
| Mt. Rogers | 2 | 997, 1,053 | P. cinereus | 11 | 5 | 2.7 (1–6) | 0.6 (0–2) |
| 1 | 997 | P. glutinosus | 8 | 2 | 2 (0–3) | 0.25 (0–1) | |
| 1 | 1,053 | P. cylindraceus | 2 | 1 | 2 (0–4) | 0.5 (0–1) | |
| Subtotal | 21 | 8 | 2.4 | 0.4 | |||
| Total | 61 | 40 | 5.6 | 1.7 |
Data represent numbers of salamanders with at least one anti-B. dendrobatidis bacterial species and total numbers of bacterial isolates and anti-B. dendrobatidis bacterial isolates detected per salamander. The salamanders at Shenandoah NP had a higher (96%) prevalence of anti-B. dendrobatidis bacteria and a higher number (mean = 3.3) of anti-B. dendrobatidis bacterial species per individual than the salamanders at Catoctin Mountain Park, MD (50%; mean = 0.8) or Mt. Rogers National Recreation Area, VA (38%; mean = 0.4), regardless of species. At Shenandoah NP, P. cinereus had similar proportions of anti-B. dendrobatidis bacterial isolates along an elevational gradient.
FIG 1.
Phylogenetic distribution of B. dendrobatidis inhibition by anti-B. dendrobatidis bacteria across 17 families of bacteria. Branches represent individual isolates that were cultured from Plethodon salamanders sampled at three localities. Each bar represents the relative percent B. dendrobatidis inhibition (1% to 100%) by each isolate. We found that B. dendrobatidis inhibition values were not related to the phylogenetic relationship of the anti-B. dendrobatidis bacteria (P > 0.05 [Mantel test]), suggesting functional trait redundancy. Bd, B. dendrobatidis.
Most (40/61, 66%) salamanders harbored at least one anti-B. dendrobatidis bacterial species, with a higher proportion at Shenandoah NP (25/26, 96%) than at Catoctin MP (7/14, 50%) and Mt. Rogers NRA (8/21, 38%) (χ2 contingency table test: χ2 = 12.9, df = 2, P < 0.0001) (Table 1).
Most anti-B. dendrobatidis bacteria were rare; 35 anti-B. dendrobatidis bacterial species were found on only one salamander individual. Ten anti-B. dendrobatidis bacterial species were detected either on one salamander species or at one locality. A few anti-B. dendrobatidis bacterial species were widespread; specifically, five anti-B. dendrobatidis bacterial species (Acinetobacter rhizosphaerae, Luteibacter rhizovicinus, two Pseudomonas spp., and one Stenotrophomonas sp.) were found in multiple localities and on multiple salamander species (Table 2). These five bacterial species have also been commonly detected on other amphibian species globally (Woodhams et al. 2015 [28]) (Table 2). An OTU corresponding to Luteibacter rhizovicinus was the only anti-B. dendrobatidis bacterial OTU found at all three sampled localities, and a Pseudomonas sp. OTU was the only anti-B. dendrobatidis bacterial OTU found on all three salamander species. No anti-B. dendrobatidis bacteria were found at all localities and on all salamander species.
TABLE 2.
Summary of the five common anti-B. dendrobatidis bacterial species that we detected on at least two salamander species and in at least two localitiesa
| Anti-B. dendrobatidis bacterial OTU | Mean % inhibition (range) | Localities | No. of sites | Salamander species | No. of salamanders | No. of other amphibian species in Woodhams et al. database (2015) |
|---|---|---|---|---|---|---|
| Acinetobacter rhizosphaerae (denovo45) | 28 (1–99) | Catoctin, Shenandoah | 6 | P. cylindraceus, P. cinereus | 9 | 4 |
| Luteibacter rhizovicinus (denovo18) | 35 (11–58) | Catoctin, Shenandoah, Mt. Rogers | 4 | P. cylindraceus, P. cinereus | 4 | 2 |
| Pseudomonas sp. (denovo13) | 58 (4–100) | Shenandoah, Mt. Rogers | 7 | P. cylindraceus, P. cinereus, P. glutinosus | 14 | 25 |
| Pseudomonas sp. (denovo20) | 63 (1–100) | Shenandoah, Mt. Rogers | 7 | P. cylindraceus, P. cinereus | 11 | 26 |
| Stenotrophomonas sp. (denovo22) | 89 (45–100) | Catoctin, Shenandoah | 4 | P. cylindraceus, P. cinereus | 4 | 1 |
We indicate the number of amphibian species on which the same anti-B. dendrobatidis bacterial OTUs have been detected in previous studies using the database of Woodhams et al. (28) (total, 37 species).
We identified 13 novel anti-B. dendrobatidis bacterial species that did not match any bacteria previously identified on amphibian skin (28). Twelve of those bacterial species occurred on only a single individual, while one was detected on two heterospecific individuals at one site.
Locality was the only significant predictor of the proportion of anti-B. dendrobatidis bacterial species per individual (generalized linear mixed-effects model [GLMM]: LR Chisq = 12.0107, df = 2, P = 0.0025). The average proportion of anti-B. dendrobatidis bacterial species per individual was higher at Shenandoah NP, with 46% (3.3/7.2) of bacterial species isolated being anti-B. dendrobatidis bacteria, while 11% (0.8/7.3) at Catoctin MP and 17% (0.4/2.4) at Mt. Rogers NRA were anti-B. dendrobatidis bacteria (Table 1). Among the host (species, sex, body condition, cover object) and site (leaf litter depth, soil pH, substrate temperature) characteristics that we examined, none were significant predictors of the proportion of anti-B. dendrobatidis bacteria per individual (GLMM: P > 0.05). On P. cinereus in Shenandoah NP, the proportions of anti-B. dendrobatidis bacteria per individual were similar among all elevations (GLM: t-value = 0.566, P = 0.58) (Table 1).
DISCUSSION
Geographic and taxonomic distribution of anti-B. dendrobatidis bacteria.
We identified a large number of anti-B. dendrobatidis bacterial taxa on the three Plethodon species, but few were widespread. Other studies have also found high diversity and little overlap in anti-B. dendrobatidis bacteria among individual amphibians (11, 14, 22). High taxonomic turnover across the landscape with functional trait redundancy has been reported in other microbial communities (45, 46). Turnover in the regional pool of microorganisms is likely due to different environmental conditions driving environmental filtering of microbial species but not microbial traits (45, 47, 48). This indicates that selection for certain bacteria may be driven by the function of the bacteria and not species identity. For instance, Loudon et al. (49) found that bacteria with anti-B. dendrobatidis properties are more likely to colonize a host than those that lack anti-B. dendrobatidis properties, suggesting that certain bacteria may be more prevalent on amphibian skin given their functional properties regardless of species identity.
Five anti-B. dendrobatidis bacterial species were widely distributed across multiple localities and multiple salamander species, suggesting a symbiotic association between these anti-B. dendrobatidis bacteria and Plethodon salamanders. Specifically, Pseudomonas spp. were the most widespread in both our study and a previously published study (28). Several ecological and physiological factors could explain this relationship, including (i) their ability to utilize available glucose (50) or to produce siderophores (51), both of which would reduce resource availability for competitors, as well as (ii) selection by the host for pseudomonads given their capacity to produce numerous antimicrobial compounds (52, 53). If generalist anti-B. dendrobatidis bacteria, such as pseudomonads, are important as an amphibian defense mechanism against B. dendrobatidis, then the abundance of these bacterial species may be a key factor in protection (54, 55). In some cases, high cell density is needed to produce inhibitory metabolites because the metabolites are produced in high quantities only when bacteria form a biofilm and undergo quorum sensing (56, 57). B. dendrobatidis likely reduces the growth of some anti-B. dendrobatidis bacteria (58), and augmenting these bacterial populations on susceptible amphibians during B. dendrobatidis epidemics may prove helpful.
We expected J. lividum to be common because it was previously detected, using molecular approaches, on most individuals in two of three P. cinereus populations in Virginia (27, 42, 59). However, using culture methods, we found anti-B. dendrobatidis bacterial OTUs in the Janthinobacterium genus on only three P. cinereus salamanders at two of our three sampled localities (Catoctin MP and Shenandoah NP). Janthinobacterium spp. may be widely distributed but not always common across the range of P. cinereus. Alternatively, Janthinobacterium or other anti-B. dendrobatidis bacterial species may commonly occur but may be in low abundance (59) and less likely to be cultured.
Influence of host and site factors on anti-B. dendrobatidis bacterial distribution.
We found no support for the notion of host species factors influencing the number of anti-B. dendrobatidis bacteria. Other studies have shown that levels of skin bacterial richness differ among amphibian species (2, 15, 40). However, the processes that structure the antifungal bacterial community may differ from those structuring the total bacterial community, given different selection pressures (60, 61). For instance, bacteria can diversify more in the presence of a parasite (62), and even a single gene locus change can modify community structure to the same extent as the loss of an apex predator (63). Alternatively, the host biology of Plethodon salamanders may explain the differences (64). We studied terrestrial, direct-developing amphibian species within a single genus that are ecologically similar throughout their lifetime (65), in contrast to previous studies that compared species from different families of amphibians that have both aquatic and terrestrial life stages (15, 40, 66).
Location was a key predictor of the prevalence and number of anti-B. dendrobatidis bacteria, indicating environmental differentiation. However, none of the site characteristics that we examined (substrate temperature, soil pH, and leaf litter depth) explained this pattern. Across the three localities, salamanders at Shenandoah NP had a higher number of anti-B. dendrobatidis bacteria than salamanders at Catoctin MP or Mt. Rogers NRA. Other environmental properties such as long-term trends in temperature may have influenced salamander bacterial diversity (67), as both Catoctin MP and Mt. Rogers NRA have experienced higher rates of warming than Shenandoah NP (68).
Spatial scale may explain variations in the number of anti-B. dendrobatidis bacteria among localities but not variations along the elevational gradient. Environmental differences (e.g., temperature, moisture) of localities 150 km apart (e.g., Catoctin MP and Shenandoah NP) may be more likely to drive diversity patterns than the small-scale variations in environmental conditions from 700 meters above sea level (masl) to 1,000 masl within Shenandoah NP. For instance, Sunagawa et al. (45) found temperature, rather than other environmental factors (e.g., salinity, nutrients) or geography, to be the most important factor driving ocean microbiome richness, but only when large temperature differences were examined. In other systems, differences in geochemistry, such as pH (45, 69), total nitrogen (70), or dissolved organic matter (71), can influence bacterial composition more than climate or elevation. We suggest future research on habitat chemistry to identify the factors driving environmental differentiation.
We did not detect B. dendrobatidis in this study, but B. dendrobatidis has been previously detected at all three of the localities that we sampled (37, 72; K. R. Lips, unpublished data). B. dendrobatidis is commonly detected in amphibian communities throughout the eastern United States, with prevalence estimates of 10% to 40% (reviewed in reference 37). This suggests that these salamanders may be exposed to B. dendrobatidis and that anti-B. dendrobatidis bacteria or other factors (e.g., immunogenetic properties) (73, 74) may be limiting B. dendrobatidis infection on these salamanders.
We found evidence for potential herd immunity to B. dendrobatidis (11, 75) at Shenandoah NP, with 96% of salamanders harboring at least one anti-B. dendrobatidis bacterial species. The concept of herd immunity suggests that an infectious disease will be less likely to affect a population if the proportion of protected individuals is above a threshold value, generally between 80% and 95% (75). Woodhams et al. (11) suggested that this value is approximately 80% in the amphibian-B. dendrobatidis system, as a population of Rana muscosa with 86% of individuals harboring anti-B. dendrobatidis bacteria persisted with B. dendrobatidis, whereas a population with 62% died off following B. dendrobatidis invasion. We found that 50% and 38% of salamanders at Catoctin MP and Mt. Rogers NRA, respectively, had anti-B. dendrobatidis bacteria; more anti-B. dendrobatidis bacteria could have been present, but we were unable to detect them due to the limitations of culturing (76).
Association between B. dendrobatidis inhibition and anti-B. dendrobatidis bacterial phylogeny.
B. dendrobatidis inhibition strength was not related to bacterial phylogeny, indicating that phylogenetic information is not predictive of B. dendrobatidis inhibition strength. Nonetheless, certain genera contain a large proportion of anti-B. dendrobatidis bacterial species, chiefly Pseudomonas and Stenotrophomonas, and detection of bacterial sequences in those genera likely indicates that many of those species have anti-B. dendrobatidis traits (94). Two possible, nonexclusive mechanisms may explain the lack of phylogenetic conservatism of B. dendrobatidis inhibition: (i) genes involved in inhibition are widely distributed among bacterial phylogenies, and (ii) horizontal gene transfer of antifungal genes may occur among unrelated bacteria. First, functions involving few genes are often widely distributed among bacterial phylogenies (77), and relatively few genes can be involved in the biosynthetic pathways that produce secondary metabolites. For instance, the sequences that produce the anti-B. dendrobatidis metabolites violacein (78) and 2,4-diacetylphloroglucinol (79) consist of five and eight genes, respectively (80, 81). Second, horizontal gene transfer of genes important in B. dendrobatidis inhibition may be occurring among unrelated bacteria, as homologous recombination can occur among bacterial lineages with levels of DNA divergence as high as 25% (82). For instance, the transfer of genes encoding antifungal compounds between different bacterial species was previously observed (83, 84). While we cannot determine which mechanism(s) operated, we found that anti-B. dendrobatidis bacterial inhibition strength was phylogenetically dispersed among the members of at least four bacterial phyla, demonstrating the wide taxonomic breadth of this trait.
Conservation implications.
There are no proven strategies to prevent infection, mortality, or population declines of amphibians caused by B. dendrobatidis. The use of anti-B. dendrobatidis bacterial augmentation has been championed as a conservation strategy (34, 85), but results have been inconsistent, in part because environmental conditions may control bacterial communities on amphibian skin. We found that individual salamanders had a mostly unique anti-B. dendrobatidis bacterial profile, with a few shared bacterial species, suggesting that enhancing the functional traits of the existing anti-B. dendrobatidis bacterial community (86) and using only widely distributed anti-B. dendrobatidis bacteria may be beneficial bioaugmentation strategies (34). One method to enhance the functional traits of a community is to use prebiotics, which selectively promote growth or activity in the community. For instance, polysaccharide β-glucan has been commercially used as a prebiotic to improve resistance of fish to infection by pathogens (87). Additionally, using ubiquitous anti-B. dendrobatidis bacterial species in bioaugmentation trials, such as those identified here, may increase the likelihood that the bacteria will persist for longer time periods on target amphibians (34). In two bioaugmentation studies, introduction of anti-B. dendrobatidis bacteria onto amphibian skin did not affect the skin microbial community (2, 88), indicating that adding probiotics is likely not detrimental to the host in terms of their resident microbial community. Yet augmented anti-B. dendrobatidis bacterial populations often do not persist on amphibian hosts for longer than a few months (25, 27), and reinoculation over time may be needed (89).
Inhibitory metabolites produced by anti-B. dendrobatidis bacteria likely have general antimicrobial properties (52) and may provide a general defensive benefit against pathogens to the host (9, 13, 23, 42). Therefore, our results may also be useful if the deadly, closely related fungal pathogen Batrachochytrium salamandrivorans (43) reaches this salamander biodiversity hot spot (44).
MATERIALS AND METHODS
Field sampling.
We sampled three species of terrestrial woodland salamanders, Plethodon cinereus, P. glutinosus, and P. cylindraceus, at three localities within a 497-km stretch in the central Appalachians in spring 2012 (Table 1). We chose the three localities, Catoctin Mountain Park (MP), MD, Shenandoah National Park (NP), VA, and Mt. Rogers National Recreation Area (NRA), VA, because they were within the range of localities where we had previously tested these species for B. dendrobatidis and found <1% B. dendrobatidis prevalence (37) and because they were within the distribution of P. cinereus and either P. glutinosus or its sister species P. cylindraceus (65). At Catoctin MP, we sampled P. cinereus (n = 7) and P. glutinosus (n = 7) salamanders at one site. At Shenandoah NP, we sampled 19 P. cinereus salamanders at six sites along an elevational gradient and sampled 7 P. cylindraceus salamanders at four of those sites (Table 1). At Mt. Rogers NRA, we sampled two P. cylindraceus and six P. cinereus salamanders at one site and eight P. glutinosus and five P. cinereus salamanders at a second site (Table 1). We had permits from state and federal agencies for handling and swabbing live amphibians (DNR permit no. 50269 [Maryland], VDGIF permit no. 042151 [Virginia], and NPS permit no. SHEN-2011-SCI-0014 [Shenandoah National Park]), and we received approval for the research from the University of Maryland Institutional Animal Care and Use Committee (R-11-11).
We collected two skin swabs from each salamander, one to culture bacteria and one to test for the presence of B. dendrobatidis. We used a new pair of nitrile powder-free gloves to handle each salamander and rinsed each salamander twice for 30 s with sterile water to remove transient microbes (42). We placed each individual salamander into a new plastic bag and swabbed it 20 times on the left side (dorsal/ventral sides and front/back limbs [five strokes each]) with a MW-113 swab (Medical Wire, United Kingdom). We stored the swab in a 1.5-ml tube on ice until we returned to the laboratory, where samples were stored in a −80°C freezer until analysis for B. dendrobatidis quantification. Then, we swabbed the right side of each salamander 20 times and immediately streaked the swab onto a R2-A nutrient agar plate in a zigzag fashion and wrapped the plate in Parafilm in the field. We stored plates at ambient temperature (12°C to 23°C) throughout incubation.
We recorded GPS coordinates, leaf litter depth, and soil pH (Kelway soil tester) at each site. For each individual, we (i) measured the substrate temperature at capture (Fluke infrared thermometer), (ii) recorded the cover object, (iii) identified the species and sex, and (iv) measured mass and snout-to-vent length (SVL) to quantify their body condition. From previous research on their effects on microbial diversity, we expected that a number of variables, such as the sources of the environmental microbes (e.g., leaf litter depth and cover object [59, 90]), pH (70), temperature (45), species (40), sex (91), and body condition (92), might influence the distribution of anti-B. dendrobatidis bacteria.
Microbiology procedures.
We isolated morphologically distinct bacterial colonies into pure cultures based on color, form, elevation, margin, substance, and opacity. We preserved isolates in 20% glycerol in a −80°C freezer until challenge assays were conducted.
We conducted bacterium-B. dendrobatidis challenge assays with each bacterial isolate (n = 341) and B. dendrobatidis isolate JEL 404 (Maine, USA) using a modified version of a protocol by Bell et al. (93). To begin, we grew cryopreserved bacteria on 1% tryptone plates for 3 days, passaged the bacteria in 3 ml of 1% tryptone broth, and grew the bacteria on a shaker at 100 rpm for an additional 3 days. Then, we made bacterial isolate/B. dendrobatidis cocultures, B. dendrobatidis monocultures, and negative controls in 3-ml culture tubes. For the bacterial isolate/B. dendrobatidis cocultures, we added 100 μl of a bacterial isolate and 100 μl of B. dendrobatidis to 1 ml of 1% tryptone broth. We grew bacteria and B. dendrobatidis together to mimic natural conditions, as bacteria and B. dendrobatidis likely interact on amphibian skin. Nonetheless, Becker et al. (94) demonstrated that testing B. dendrobatidis/bacterial isolate cocultures versus bacterial monocultures against B. dendrobatidis had no effect on the magnitude of B. dendrobatidis inhibition. For B. dendrobatidis monocultures, we added 100 μl of B. dendrobatidis to 1.1 ml of 1% tryptone broth. For a negative control, we added 1.2 ml of 1% tryptone broth to the 3-ml tubes. We grew these cultures on a shaker at 100 rpm for 3 days. To obtain microbially produced metabolites, termed cell-free supernatant (CFS [93]), we centrifuged the bacterial isolate/B. dendrobatidis cocultures, B. dendrobatidis monocultures, and negative controls at 10,000 rpm for 5 min. Then, we used 18-gauge hypodermic needles attached to 3-ml syringes (B. dendrobatidis Vacutainer no. 309657) to remove the supernatant and filtered the supernatant through 0.22-μm-pore-size filters in 13-mm-diameter-syringe-filter holders (Millipore GSWP01300/SX0001300). We harvested B. dendrobatidis zoospores by flooding 1-week-old B. dendrobatidis plates grown at room temperature (RT) with 1% tryptone and filtering the broth through a sterilized coffee filter in a glass funnel.
To set up the assays, we added 50 μl of approximately 1 × 106 zoospores/ml of JEL 404 (counted with a hemocytometer; approximately 50,000 zoospores in each well) to a 96-well plate in all wells except those designated for use as negative controls. Into the bacterium-B. dendrobatidis sample wells, we added 50 μl of the CFS from each bacterium-B. dendrobatidis sample in each of four wells. We used four controls (two negative controls and two positive controls [PC]) in each 96-well assay using four wells for each control. The positive controls were as follows: 50 μl of B. dendrobatidis zoospores plus 50 μl of B. dendrobatidis CFS (PC) and 50 μl of B. dendrobatidis zoospores plus 50 μl of water (nutrient-depleted positive control [NDPC]). The negative controls were as follows: 50 μl of B. dendrobatidis zoospores heat-killed (HK) at 60°C for 60 min plus 50 μl of B. dendrobatidis CFS (HK B. dendrobatidis) and 50 μl of 1% tryptone broth plus 50 μl of B. dendrobatidis CFS. We measured the level of absorbance (optical density at 492 nm [OD492]) in each well using a microplate reader on days 0, 1, 4, 7, 8, and 10 of the experiment.
B. dendrobatidis inhibition score calculations.
We used the day of maximum growth (day 7 or 8) of the PC for each assay as the day to quantify B. dendrobatidis inhibition. To calculate B. dendrobatidis inhibition scores, we divided the average OD reading from the bacterium-B. dendrobatidis sample (n = 4) by the average OD reading from the NDPC for that assay (n = 4), after correcting for the average OD of the heat-killed B. dendrobatidis (HK, n = 4), and subtracted that value from 1. We subtracted the OD of the HK to remove the value corresponding to the baseline absorbance of B. dendrobatidis zoospores. The equation we used was as follows: inhibition score = 1 – [(sampleOD – HKOD)/(NDPCOD – HKOD)]. We interpreted values greater than zero as being representative of anti-B. dendrobatidis bacteria, indicating that the bacterium-B. dendrobatidis sample wells had less growth than the NDPC wells as measured by OD. Prior to the OD readings, we visually inspected each well using an inverted microscope and coded the visual observations of the sample wells as no, weak, moderate, or strong inhibition in comparison to the NDPC wells.
To identify anti-B. dendrobatidis bacteria, we used a conservative approach: (i) both the OD readings and visual observations had to support inhibition, and (ii) we used the NDPC rather than the PC to quantify B. dendrobatidis inhibition strength. We used the NDPC rather than the PC because the experimental wells in challenge assays contained 50 μl of CFS in which bacteria and B. dendrobatidis had previously been cultured, while the PC wells contained 50 μl of previously unused medium. The growth of B. dendrobatidis in experimental wells could have been affected by depletion of the nutrients used by the bacteria and B. dendrobatidis. Therefore, comparing the experimental wells to the NDPC wells accounts for the issue of nutrient depletion and is a more conservative approach in identifying anti-B. dendrobatidis bacteria.
We attempted to minimize the variation of bacterial population size and growth stage by allowing each culture to grow for 3 days. At approximately 3 days, most bacterial cultures reached the later phases of growth, when inhibitory metabolites are produced (95). This is a commonly accepted practice and is currently the best method for testing a large number of bacterial isolates (35, 60, 93, 94).
Molecular procedures.
We sequenced the 16S rRNA gene of all anti-B. dendrobatidis bacterial isolates using Sanger sequencing. We selected a single colony of each isolate from agar plates using a sterilized toothpick, added the colony to 25 μl of sterile deionized water, and boiled the mixture for 10 min at 95°C to extract bacterial DNA. We used PCR to amplify a 1,037-bp fragment of the 16S rRNA gene using the 8F/1045R primer set. Each 25-μl PCR assay mixture consisted of 1.25 U of AmpliTaq Gold DNA polymerase (ThermoFisher) in proprietary buffer, 2.5 μM MgCl2, 200 nM concentrations of deoxynucleoside triphosphates (dNTPs), a 600 nM concentration of each primer, and 2 μl DNA template. PCR conditions were 95°C for 8 min, followed by 30 cycles of 95°C for 45 s, 52°C for 30 s, and 72°C for 30 s and a final extension (72°C for 3 min). We followed the sequencing methods outlined by Muletz et al. (37) to sequence the cleaned PCR products. Individual sequences were assembled and edited in Sequencher 5.1 to obtain a consensus sequence per anti-B. dendrobatidis bacterial isolate.
We tested all sampled salamanders for B. dendrobatidis using quantitative PCR (qPCR). We extracted DNA from swabs using a MoBio PowerSoil DNA extraction kit following the manufacturer's protocol. We used primers developed by Boyle et al. (96) and used iTaq supermix with Rox (Bio-Rad) following their qPCR protocol. We ran all DNA samples in duplicate and used standards of 100, 10, 1, and 0.1 zoospore genomic equivalents (ZGEs) developed from a Puerto Rican B. dendrobatidis isolate, JEL 427. If one of the duplicates returned a positive signal, it was run a third time. Samples were considered positive if they amplified twice before 0.1 ZGEs.
Sequence analysis.
We identified anti-B. dendrobatidis bacteria to the lowest taxonomic level with MacQIIME 1.9.1 (97) using the pick_de_novo_otus.py command with default parameters. This command clustered sequences into operational taxonomic units (OTUs) using UCLUST based on 97% pairwise identity and assigned taxonomy using UCLUST (Edgar 2010) with the Greengenes reference database (98) (release 13_8). We aligned the bacterial isolate sequences using the Ribosomal Database Project (http://rdp.cme.msu.edu) alignment tools following Dunitz et al. (99), including an archaeal outgroup in the alignment. We built a phylogenetic tree using Fasttree (100) implemented in MacQIIME and used the package “ape” (101) in the R environment (102) to root the tree and trim the outgroup.
Comparison of identified anti-B. dendrobatidis bacteria to published database entries.
We compared our anti-B. dendrobatidis bacterial 16S rRNA sequences to those in a published database (28) to identify common and novel anti-B. dendrobatidis bacteria. As described above, we identified bacterial OTUs using the pick_de_novo_otus.py command with default parameters in MacQIIME 1.9.1. We then split the OTU table using the split_otu_table.py command to ensure that it would contain only anti-B. dendrobatidis bacteria and filtered out zeros (not anti-B. dendrobatidis bacteria) using the filter_otus_from_otu_table.py command. We summed the number of anti-B. dendrobatidis bacterial OTUs in the Woodhams database, which contains a total of 255 OTUs. Next, we used a custom blast analysis in Geneious 8.1 (103) to query our anti-B. dendrobatidis bacterial isolate sequences against those 255 anti-B. dendrobatidis bacterial OTUs. We used a megablast program, having Geneious return results as query-centered alignment data only and returning only the top hit. We considered anti-B. dendrobatidis bacterial sequences in our data set to be novel if all sequences in that OTU had <97% pairwise identity to the OTUs in the Woodhams data set. For common anti-B. dendrobatidis bacteria in our data set, we counted the number of amphibians in the Woodhams data set that had OTUs that matched at >97% pairwise identity. Note that while these OTUs matched antifungal database entries, such a result does not necessarily indicate that these bacteria exhibit antifungal activity but does indicate that they are strong candidates for exhibiting antifungal activity.
Statistical analysis.
All statistical analyses were performed in R 3.1.3 (102).
To determine if the numbers of salamanders with at least one anti-B. dendrobatidis bacterial OTU differed among the three localities, we used a χ2 contingency table test. To determine if the number of anti-B. dendrobatidis bacteria per salamander was related to host or environmental characteristics, we used a generalized linear mixed-effects model (GLMM) with a binomial distribution in the package “lme4” (104). We included species and locality as the main explanatory variables, along with additional covariates of host (sex, body condition, cover object) and site (leaf litter depth, soil pH, substrate temperature). As the response variable, we used the proportion of anti-B. dendrobatidis bacteria per salamander (i.e., the total number of anti-B. dendrobatidis bacteria divided by the total number of bacteria isolated) to account for variations in the number of isolates cultured per individual. We also ran this analysis using the raw counts of anti-B. dendrobatidis bacterial OTUs as the response variable with a Poisson distribution, and our results were the same. We included site as a random effect in the model to account for pseudoreplicating sites at localities. To quantify salamander body condition, we used the residuals of the linear regression of body mass on SVL for P. cinereus and for the P. glutinous complex (P. cylindraceus and P. glutinosus) separately, as P. cinereus is smaller in size than the P. glutinosus complex. To determine which variables in the model significantly predicted the proportion of anti-B. dendrobatidis bacteria per salamander, we used the Anova function in the “car” package (105) with the type III margin sum of squares reported. Then, we used Tukey's honestly significant difference (HSD) test for post hoc comparisons to determine significant differences between categorical levels of fixed effects using the lsmeans function in the package “lsmeans” (106). We assessed model goodness of fit by visually inspecting residuals.
We used a generalized linear model to determine if the number of anti-B. dendrobatidis bacteria per salamander was related to elevation for P. cinereus sampled along an elevational gradient in Shenandoah NP. As described above, we used the proportion of anti-B. dendrobatidis bacteria per salamander as the response variable. We assessed goodness of fit for the model by visually inspecting residuals and used a quasibinomial distribution to account for overdispersion.
We tested if B. dendrobatidis inhibition scores were phylogenetically conserved across anti-B. dendrobatidis bacteria using a Mantel test. We generated a distance matrix of B. dendrobatidis inhibition scores among anti-B. dendrobatidis bacterial isolates using Euclidean distances (94) and a corresponding distance matrix of percent sequence identity from the anti-B. dendrobatidis bacterial isolate sequence alignment. We used the “vegan” package (107) to conduct the Mantel test with 10,000 permutations. We visualized inhibition scores of anti-B. dendrobatidis bacteria on the bacterial phylogeny using iTol (108) (http://itol.embl.de/).
Accession number(s).
We deposited the 16S rRNA sequences for the 119 anti-B. dendrobatidis bacterial isolates in GenBank (accession numbers KU738912 to KU739030). All other data from this study, including inhibition data and sequence alignments, have been deposited on figshare (https://figshare.com/s/f96dafa91c0b15291818).
ACKNOWLEDGMENTS
We thank Nick Caruso, Cameron Houser, and Joe Hoyt for field assistance. We thank Sam Barnett, Megan Miraglia, and Nancy McInerney for laboratory assistance. We thank Sara Bell, Andrew Loudon, and Matt Becker for guidance in developing a bacterial B. dendrobatidis challenge assay protocol.
This project was funded by a National Park Service (NPS) George M. Wright Climate Change Fellowship and an Environmental Protection Agency (EPA) STAR Fellowship (no. F13B20412) awarded to C.R.M.-W. The views expressed in this publication are solely those of the authors, and NPS or EPA does not endorse any products or commercial services mentioned in this publication.
This is contribution number 575 of the Amphibian Research and Monitoring Initiative (ARMI) of the U.S. Geological Survey.
REFERENCES
- 1.Gallo RL, Nakatsuji T. 2011. Microbial symbiosis with the innate immune defense system of the skin. J Invest Dermatol 131:1974–1980. doi: 10.1038/jid.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker MH, Walke JB, Cikanek S, Savage AE, Mattheus N, Santiago CN, Minbiole KPC, Harris RN, Belden LK, Gratwicke B. 2015. Composition of symbiotic bacteria predicts survival in Panamanian golden frogs infected with a lethal fungus. Proc Biol Sci 282:pii: 20142881. doi: 10.1098/rspb.2014.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowrey L, Woodhams DC, Tacchi L, Salinas I. 2015. Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl Environ Microbiol 81:6915–6925. doi: 10.1128/AEM.01826-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenthal M, Goldberg D, Aiello A, Larson E, Foxman B. 2011. Skin microbiota: microbial community structure and its potential association with health and disease. Infect Genet Evol 11:839–848. doi: 10.1016/j.meegid.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flórez LV, Biedermann PH, Engl T, Kaltenpoth M. 2015. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep 32:904–936. doi: 10.1039/C5NP00010F. [DOI] [PubMed] [Google Scholar]
- 6.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 7.Verhulst NO, Qiu YT, Beijleveld H, Maliepaard C, Knights D, Schulz S, Berg-Lyons D, Lauber CL, Verduijn W, Haasnoot GW, Mumm R, Bouwmeester HJ, Claas FHJ, Dicke M, van Loon JJA, Takken W, Knight R, Smallegange RC. 2011. Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS One 6:e28991. doi: 10.1371/journal.pone.0028991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longo AV, Savage AE, Hewson I, Zamudio KR. 15 July 2015. Seasonal and ontogenetic variation of skin microbial communities and relationships to natural disease dynamics in declining amphibians. R Soc Open Sci doi: 10.1098/rsos.140377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Federici E, Rossi R, Fidati L, Paracucchi R, Scargetta S, Montalbani E, Franzetti A, La Porta G, Fagotti A, Simonceli F, Cenci G, Di Rosa I. 2015. Characterization of the skin microbiota in Italian stream frogs (Rana italica) infected and uninfected by a cutaneous parasitic disease. Microbes Environ 30:262–269. doi: 10.1264/jsme2.ME15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schieber AMP, Lee YM, Chang MW, Leblanc M, Collins B, Downes M, Evans RM, Ayres JS. 2015. Disease tolerance mediated by microbiome E-coli involves inflammasome and IGF-1 signaling. Science 350:558–563. doi: 10.1126/science.aac6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodhams DC, Vredenburg VT, Simon M-A, Billheimer D, Shakhtour B, Shyr Y, Briggs CJ, Rollins-Smith LA, Harris RN. 2007. Symbiotic bacteria contribute to innate immune defenses of the threatened mountain yellow-legged frog, Rana muscosa. Biol Conserv 138:390–398. doi: 10.1016/j.biocon.2007.05.004. [DOI] [Google Scholar]
- 12.Hol WHG, Garbeva P, Hordijk C, Hundscheid MPJ, Gunnewiek PJAK, van Agtmaal M, Kuramae EE, de Boer W. 2015. Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology 96:2042–2048. doi: 10.1890/14-2359.1. [DOI] [PubMed] [Google Scholar]
- 13.Lauer A, Simon MA, Banning JL, Lam BA, Harris RN. 2008. Diversity of cutaneous bacteria with antifungal activity isolated from female four-toed salamanders. ISME J 2:145–157. doi: 10.1038/ismej.2007.110. [DOI] [PubMed] [Google Scholar]
- 14.Flechas SV, Sarmiento C, Cárdenas ME, Medina EM, Restrepo S, Amézquita A. 2012. Surviving chytridiomycosis: Differential anti-Batrachochytrium dendrobatidis activity in bacterial isolates from three lowland species of Atelopus. PLoS One 7:e44832. doi: 10.1371/journal.pone.0044832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kueneman JG, Parfrey LW, Woodhams DC, Archer HM, Knight R, McKenzie VJ. 2014. The amphibian skin-associated microbiome across species, space and life history stages. Mol Ecol 23:1238–1250. doi: 10.1111/mec.12510. [DOI] [PubMed] [Google Scholar]
- 16.Crawford AJ, Lips KR, Bermingham E. 2010. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc Natl Acad Sci U S A 107:13777–13782. doi: 10.1073/pnas.0914115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piovia-Scott J, Pope K, Worth SJ, Rosenblum EB, Poorten T, Refsnider J, Rollins-Smith LA, Reinert LK, Wells HL, Rejmanek D, Lawler S, Foley J. 2015. Correlates of virulence in a frog-killing fungal pathogen: evidence from a California amphibian decline. ISME J 9:1570–1578. doi: 10.1038/ismej.2014.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, Slocombe R, Ragan MA, Hyatt AD, McDonald KR, Hines HB, Lips KR, Marantelli G, Parkes H. 1998. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci U S A 95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosch J, Martinez-Solano I, Garcia-Paris M. 2001. Evidence of a chytrid fungus infection involved in the decline of the common midwife toad (Alytes obstetricans) in protected areas of central Spain. Biol Conserv 97:331–337. doi: 10.1016/S0006-3207(00)00132-4. [DOI] [Google Scholar]
- 20.Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, Carey C, Livo L, Pessier AP, Collins JP. 2006. Emerging infectious disease and the loss of biodiversity in a neotropical amphibian community. Proc Natl Acad Sci U S A 103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vredenburg VT, Knapp RA, Tunstall TS, Briggs CJ. 2010. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc Natl Acad Sci U S A 107:9689–9694. doi: 10.1073/pnas.0914111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam BA, Walke JB, Vredenburg VT, Harris RN. 2010. Proportion of individuals with anti-Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana muscosa. Biol Conserv 143:529–531. doi: 10.1016/j.biocon.2009.11.015. [DOI] [Google Scholar]
- 23.Kueneman JG, Woodhams DC, Van Treuren W, Archer HM, Knight R, McKenzie VJ. 13 November 2015. Inhibitory bacteria reduce fungi on early life stages of endangered Colorado boreal toads (Anaxyrus boreas). ISME J doi: 10.1038/ismej.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris RN, Lauer A, Simon MA, Banning JL, Alford RA. 2009. Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis Aquat Organ 83:11–16. doi: 10.3354/dao02004. [DOI] [PubMed] [Google Scholar]
- 25.Harris RN, Brucker RM, Walke JB, Becker MH, Schwantes CR, Flaherty DC, Lam BA, Woodhams DC, Briggs CJ, Vredenburg VT, Minbiole KPC. 2009. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J 3:818–824. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]
- 26.Vredenburg VT, Briggs CJ, Harris RN. 2011. Host pathogen dynamics of amphibian chytridiomycosis: the role of the skin microbiome in health and disease, p 342–355. In Olsen LE, Choffnes DA, Pray L (ed), Fungal diseases: an emerging challenge to human, animal and plant health. The National Academies Press, Washington, DC. [Google Scholar]
- 27.Muletz CR, Myers JM, Domangue RJ, Herrick JB, Harris RN. 2012. Soil bioaugmentation with amphibian cutaneous bacteria protects amphibian hosts from infection by Batrachochytrium dendrobatidis. Biol Conserv 152:119–126. doi: 10.1016/j.biocon.2012.03.022. [DOI] [Google Scholar]
- 28.Woodhams DC, Alford RA, Antwis RE, Archer H, Becker MH, Belden LK, Bell SC, Bletz M, Daskin JH, Davis LR, Flechas SV, Lauer A, Gonzalez A, Harris RN, Holden WM, Hughey MC, Ibáñez R, Knight R, Kueneman J, Rabemananjara F, Reinert LK, Rollins-Smith LA, Roman-Rodriguez F, Shaw SD, Walke JB, McKenzie V. 2015. Antifungal isolates database of amphibian skin-associated bacteria and function against emerging fungal pathogens. Ecology 96:595–595. doi: 10.1890/14-1837.1. [DOI] [Google Scholar]
- 29.Nebergall EE. 2013. Ecology and applications of cutaneous mechanisms of resistance to amphibian chytridiomycosis. Master's thesis. Oregon State University, Corvallis, OR. [Google Scholar]
- 30.Woodhams DC, Geiger CC, Reinert LK, Rollins-Smith LA, Lam B, Harris RN, Briggs CJ, Vredenburg VT, Voyles J. 2012. Treatment of amphibians infected with chytrid fungus: learning from failed trials with itraconazole, antimicrobial peptides, bacteria, and heat therapy. Dis Aquat Organ 98:11–25. doi: 10.3354/dao02429. [DOI] [PubMed] [Google Scholar]
- 31.Bletz M. 2013. Probiotic bioaugmentation of an anti-Bd bacteria, Janthinobacterium lividum, on the amphibian, Notophthalmus viridescens: transmission efficacy and persistence of the probiotic on the host and non-target effects of probiotic addition on ecosystem components. Master's thesis. James Madison University, Harrisonburg, VA. [Google Scholar]
- 32.Becker MH, Harris RN, Minbiole KPC, Schwantes CR, Rollins-Smith LA, Reinert LK, Brucker RM, Domangue RJ, Gratwicke B. 2011. Towards a better understanding of the use of probiotics for preventing chytridiomycosis in Panamanian golden frogs. Ecohealth 8:501–506. doi: 10.1007/s10393-012-0743-0. [DOI] [PubMed] [Google Scholar]
- 33.Bresciano JC, Salvador CA, Paz-y-Mino C, Parody-Merino AM, Bosch J, Woodhams DC. 2015. Variation in the presence of anti-Batrachochytrium dendrobatidis bacteria of amphibians across life stages and elevations in Ecuador. Ecohealth 12:310–319. doi: 10.1007/s10393-015-1010-y. [DOI] [PubMed] [Google Scholar]
- 34.Bletz MC, Loudon AH, Becker MH, Bell SC, Woodhams DC, Minbiole KPC, Harris RN. 2013. Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol Lett 16:807–820. doi: 10.1111/ele.12099. [DOI] [PubMed] [Google Scholar]
- 35.Daskin JH, Bell SC, Schwarzkopf L, Alford RA. 2014. Cool temperatures reduce antifungal activity of symbiotic bacteria of threatened amphibians - implications for disease management and patterns of decline. PLoS One 9:e100378. doi: 10.1371/journal.pone.0100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodhams DC, Brandt H, Baumgartner S, Kielgast J, Kuepfer E, Tobler U, Davis LR, Schmidt BR, Bel C, Hodel S, Knight R, McKenzie V. 2014. Interacting symbionts and immunity in the amphibian skin mucosome predict disease risk and probiotic effectiveness. PLoS One 9:e96375. doi: 10.1371/journal.pone.0096375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muletz C, Caruso NM, Fleischer RC, McDiarmid RW, Lips KR. 2014. Unexpected rarity of the pathogen Batrachochytrium dendrobatidis in Appalachian Plethodon salamanders: 1957–2011. PLoS One 9:e103728. doi: 10.1371/journal.pone.0103728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng TL, Rovito SM, Wake DB, Vredenburg VT. 2011. Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci U S A 108:9502–9507. doi: 10.1073/pnas.1105538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker MH, Harris RN. 2010. Cutaneous bacteria of the redback salamander prevent morbidity associated with a lethal disease. PLoS One 5:e10957. doi: 10.1371/journal.pone.0010957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenzie VJ, Bowers RM, Fierer N, Knight R, Lauber CL. 2012. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J 6:588–596. doi: 10.1038/ismej.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris RN, James TY, Lauer A, Simon MA, Patel A. 2006. Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of amphibian species. EcoHealth 3:53. doi: 10.1007/s10393-005-0009-1. [DOI] [Google Scholar]
- 42.Lauer A, Simon MA, Banning JL, André E, Duncan K, Harris RN. 2007. Common cutaneous bacteria from the Eastern red-backed salamander can inhibit pathogenic fungi. Copeia 2007:630–640. doi: 10.1643/0045-8511(2007)2007[630:CCBFTE]2.0.CO;2. [DOI] [Google Scholar]
- 43.Martel A, Blooi M, Adriaensen C, Van Rooij P, Beukema W, Fisher MC, Farrer RA, Schmidt BR, Tobler U, Goka K, Lips KR, Muletz C, Zamudio KR, Bosch J, Lötters S, Wombwell E, Garner TWJ, Cunningham AA, Spitzen-van der Sluijs A, Salvidio S, Ducatelle R, Nishikawa K, Nguyen TT, Kolby JE, Van Bocxlaer I, Bossuyt F, Pasmans F. 2014. Recent introduction of a chytrid fungus endangers Western Palearctic salamanders. Science 346:630–631. doi: 10.1126/science.1258268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yap TA, Koo MS, Ambrose RF, Wake DB, Vredenburg VT. 2015. Averting a North American biodiversity crisis. Science 349:481–482. doi: 10.1126/science.aab1052. [DOI] [PubMed] [Google Scholar]
- 45.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, Cornejo-Castillo FM, Costea PI, Cruaud C, d'Ovidio F, Engelen S, Ferrera I, Gasol JM, Guidi L, Hildebrand F, Kokoszka F, Lepoivre C, Lima-Mendez G, Poulain J, Poulos BT, Royo-Llonch M, Sarmento H, Vieira-Silva S, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Bowler C, de Vargas C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Jaillon O, Not F, Ogata H, Pesant S, Speich S, Stemmann L, Sullivan MB, Weissenbach J, Wincker P, Karsenti E, Raes J, Acinas SG, Bork P, et al. 2015. Ocean plankton. Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 46.Vega Thurber R, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale E, Kelly L, Rohwer F. 2009. Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 47.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. 2009. The merging of community ecology and phylogenetic biology. Ecol Lett 12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- 49.Loudon AH, Venkataraman A, VanTreuren W, Woodhams D, Parfrey LW, McKenzie V, Knight R, Schmidt T, Harris R. 2016. Vertebrate hosts as islands: dynamics of selection, immigration, loss, persistence and potential function of bacteria on salamander skin. Front Microbiol 7:333. doi: 10.3389/fmicb.2016.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walke JB, Harris RN, Reinert LK, Rollins-Smith LA, Woodhams DC. 2011. Social immunity in amphibians: evidence for vertical transmission of innate defenses. Biotropica 43:396–400. doi: 10.1111/j.1744-7429.2011.00787.x. [DOI] [Google Scholar]
- 51.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 99:7072–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leisinger T, Margraff R. 1979. Secondary metabolites of the fluorescent pseudomonads. Microbiol Rev 43:422–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ligon JM, Hill DS, Hammer PE, Torkewitz NR, Hofmann D, Kempf HJ, van Pée KH. 2000. Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag Sci 56:688–695. doi:. [DOI] [Google Scholar]
- 54.Wei H-L, Zhang L-Q. 2006. Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek 89:267–280. doi: 10.1007/s10482-005-9028-8. [DOI] [PubMed] [Google Scholar]
- 55.Yasumiba K, Bell S, Alford R. 2016. Cell density effects of frog skin bacteria on their capacity to inhibit growth of the chytrid fungus, Batrachochytrium dendrobatidis. Microb Ecol 71:124–130. doi: 10.1007/s00248-015-0701-9. [DOI] [PubMed] [Google Scholar]
- 56.Pantanella F, Berlutti F, Passariello C, Sarli S, Morea C, Schippa S. 2007. Violacein and biofilm production in Janthinobacterium lividum. J Appl Microbiol 102:992–999. [DOI] [PubMed] [Google Scholar]
- 57.Barnard AML, Bowden SD, Burr T, Coulthurst SJ, Monson RE, Salmond GPC. 2007. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Philos Trans R Soc Lond B Biol Sci 362:1165–1183. doi: 10.1098/rstb.2007.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jani AJ, Briggs CJ. 2014. The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. Proc Natl Acad Sci U S A 111:E5049–E5058. doi: 10.1073/pnas.1412752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V, Harris RN. 2014. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J 8:830–840. doi: 10.1038/ismej.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antwis RE, Preziosi RF, Harrison XA, Garner TW. 2015. Amphibian symbiotic bacteria do not show a universal ability to inhibit growth of the global panzootic lineage of Batrachochytrium dendrobatidis. Appl Environ Microbiol 81:3706–3711. doi: 10.1128/AEM.00010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S. 2013. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knope ML, Forde SE, Fukami T. 2012. Evolutionary history, immigration history, and the extent of diversification in community assembly. Front Microbiol 2:273. doi: 10.3389/fmicb.2011.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McClean D, McNally L, Salzberg LI, Devine KM, Brown SP, Donohue I. 2015. Single gene locus changes perturb complex microbial communities as much as apex predator loss. Nat Commun 6:8235. doi: 10.1038/ncomms9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong ACN, Chaston JM, Douglas AE. 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J 7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petranka JW. 1998. Salamanders of the United States and Canada. Smithsonian Books, Washington, DC. [Google Scholar]
- 66.Walke JB, Becker MH, Loftus SC, House LL, Cormier G, Jensen RV, Belden LK. 2014. Amphibian skin may select for rare environmental microbes. ISME J 8:2207–2217. doi: 10.1038/ismej.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meyer EA, Cramp RL, Hernando Bernal M, Franklin CE. 2012. Changes in cutaneous microbial abundance with sloughing: possible implications for infection and disease in amphibians. Dis Aquat Organ 101:235–242. doi: 10.3354/dao02523. [DOI] [PubMed] [Google Scholar]
- 68.Caruso NM, Sears MW, Adams DC, Lips KR. 2014. Widespread rapid reductions in body size of adult salamanders in response to climate change. Glob Change Biol 20:1751–1759. doi: 10.1111/gcb.12550. [DOI] [PubMed] [Google Scholar]
- 69.Fierer N, McCain CM, Meir P, Zimmermann M, Rapp JM, Silman MR, Knight R. 2011. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology 92:797–804. doi: 10.1890/10-1170.1. [DOI] [PubMed] [Google Scholar]
- 70.Shen C, Xiong J, Zhang H, Feng Y, Lin X, Li X, Liang W, Chu H. 2013. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211. doi: 10.1016/j.soilbio.2012.07.013. [DOI] [Google Scholar]
- 71.Wang J, Soininen J, Zhang Y, Wang B, Yang X, Shen J. 2011. Contrasting patterns in elevational diversity between microorganisms and macroorganisms. J Biogeogr 38:595–603. doi: 10.1111/j.1365-2699.2010.02423.x. [DOI] [Google Scholar]
- 72.Hossack BR, Adams MJ, Campbell Grant EH, Pearl CA, Bettaso JB, Barichivich WJ, Lowe WH, True K, Ware JL, Corn PS. 2010. Low prevalence of chytrid fungus (Batrachochytrium dendrobatidis) in amphibians of US headwater streams. J Herpetol 44:253–260. doi: 10.1670/09-058.1. [DOI] [Google Scholar]
- 73.Ellison AR, Savage AE, DiRenzo GV, Langhammer P, Lips KR, Zamudio KR. 2014. Fighting a losing battle: vigorous immune response countered by pathogen suppression of host defenses in the chytridiomycosis-susceptible frog Atelopus zeteki. G3 (Bethesda) 4:1275–1289. doi: 10.1534/g3.114.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Savage AE, Zamudio KR. 2011. MHC genotypes associate with resistance to a frog-killing fungus. Proc Natl Acad Sci U S A 108:16705–16710. doi: 10.1073/pnas.1106893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson RM, May RM. 1990. Immunization and herd immunity. Lancet 335:641–645. doi: 10.1016/0140-6736(90)90420-A. [DOI] [PubMed] [Google Scholar]
- 76.Walke JB, Becker MH, Hughey MC, Swartwout MC, Jensen RV, Belden LK. 2015. Most of the dominant members of amphibian skin bacterial communities can be readily cultured. Appl Environ Microbiol 81:6589–6600. doi: 10.1128/AEM.01486-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martiny AC, Treseder K, Pusch G. 2013. Phylogenetic conservatism of functional traits in microorganisms. ISME J 7:830–838. doi: 10.1038/ismej.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brucker RM, Harris RN, Schwantes CR, Gallaher TN, Flaherty DC, Lam BA, Minbiole KPC. 2008. Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J Chem Ecol 34:1422–1429. doi: 10.1007/s10886-008-9555-7. [DOI] [PubMed] [Google Scholar]
- 79.Brucker RM, Baylor CM, Walters RL, Lauer A, Harris RN, Minbiole KPC. 2008. The identification of 2,4-diacetylphloroglucinol as an antifungal metabolite produced by cutaneous bacteria of the salamander Plethodon cinereus. J Chem Ecol 34:39–43. doi: 10.1007/s10886-007-9352-8. [DOI] [PubMed] [Google Scholar]
- 80.Sánchez C, Braña AF, Méndez C, Salas JA. 2006. Reevaluation of the violacein biosynthetic pathway and its relationship to indolocarbazole biosynthesis. Chembiochem 7:1231–1240. doi: 10.1002/cbic.200600029. [DOI] [PubMed] [Google Scholar]
- 81.Moynihan JA, Morrissey JP, Coppoolse ER, Stiekema WJ, O'Gara F, Boyd EF. 2009. Evolutionary history of the phl gene cluster in the plant-associated bacterium Pseudomonas fluorescens. Appl Environ Microbiol 75:2122–2131. doi: 10.1128/AEM.02052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cohan FM. 2001. Bacterial species and speciation. Syst Biol 50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- 83.Kinashi H, Shimaji M, Sakai A. 1987. Giant linear plasmids in Streptomyces which code for antibiotic biosysthesis genes. Nature 328:454–456. doi: 10.1038/328454a0. [DOI] [PubMed] [Google Scholar]
- 84.Ravel J, Wellington EMH, Hill RT. 2000. Interspecific transfer of Streptomyces giant linear plasmids in sterile amended soil microcosms. Appl Environ Microbiol 66:529–534. doi: 10.1128/AEM.66.2.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woodhams DC, Bosch J, Briggs CJ, Cashins S, Davis LR, Lauer A, Muths E, Puschendorf R, Schmidt BR, Sheafor B, Voyles J. 2011. Mitigating amphibian disease: strategies to maintain wild populations and control chytridiomycosis. Front Zool 8:8. doi: 10.1186/1742-9994-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mueller UG, Sachs JL. 2015. Engineering microbiomes to improve plant and animal health. Trends Microbiol 23:606–617. doi: 10.1016/j.tim.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 87.Song SK, Beck BR, Kim D, Park J, Kim J, Kim HD, Ringo E. 2014. Prebiotics as immunostimulants in aquaculture: a review. Fish Shellfish Immunol 40:40–48. doi: 10.1016/j.fsi.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 88.Küng D, Bigler L, Davis LR, Gratwicke B, Griffith E, Woodhams DC. 2014. Stability of microbiota facilitated by host immune regulation: informing probiotic strategies to manage amphibian disease. PLoS One 9:e87101. doi: 10.1371/journal.pone.0087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ackleh AS, Carter J, Chellamuthu VK, Ma B. 2016. A model for the interaction of frog population dynamics with Batrachochytrium dendrobatidis, Janthinobacterium lividum and temperature and its implication for chytridiomycosis management. Ecol Modell 320:158–169. doi: 10.1016/j.ecolmodel.2015.09.015. [DOI] [Google Scholar]
- 90.Fitzpatrick BM, Allison AL. 2014. Similarity and differentiation between bacteria associated with skin of salamanders (Plethodon jordani) and free-living assemblages. FEMS Microbiol Ecol 88:482–494. doi: 10.1111/1574-6941.12314. [DOI] [PubMed] [Google Scholar]
- 91.Krynak KL, Burke DJ, Benard MF. 2016. Landscape and water characteristics correlate with immune defense traits across Blanchard's cricket frog (Acris blanchardi) populations. Biol Conserv 193:153–167. doi: 10.1016/j.biocon.2015.11.019. [DOI] [Google Scholar]
- 92.Ruiz-Rodriguez M, Soler JJ, Lucas FS, Heeb P, Jose Palacios M, Martín-Gálvez D, De Neve L, Pérez-Contreras T, Martínez JG, Soler M. 2009. Bacterial diversity at the cloaca relates to an immune response in magpie Pica pica and to body condition of great spotted cuckoo Clamator glandarius nestlings. J Avian Biol 40:42–48. doi: 10.1111/j.1600-048X.2008.04471.x. [DOI] [Google Scholar]
- 93.Bell SC, Alford RA, Garland S, Padilla G, Thomas AD. 2013. Screening bacterial metabolites for inhibitory effects against Batrachochytrium dendrobatidis using a spectrophotometric assay. Dis Aquat Organ 103:77–85. doi: 10.3354/dao02560. [DOI] [PubMed] [Google Scholar]
- 94.Becker MH, Walke JB, Murrill L, Woodhams DC, Reinert LK, Rollins-Smith LA, Burzynski EA, Umile TP, Minbiole KPC, Belden LK. 2015. Phylogenetic distribution of symbiotic bacteria from Panamanian amphibians that inhibit growth of the lethal fungal pathogen Batrachochytrium dendrobatidis. Mol Ecol 24:1628–1641. doi: 10.1111/mec.13135. [DOI] [PubMed] [Google Scholar]
- 95.Bérdy J. 2005. Bioactive microbial metabolites - a personal view. J Antibiot 58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 96.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. 2004. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Organ 60:141–148. doi: 10.3354/dao060141. [DOI] [PubMed] [Google Scholar]
- 97.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dunitz MI, Lang JM, Jospin G, Darling AE, Eisen JA, Coil DA. 2015. Swabs to genomes: a comprehensive workflow. Peerj 3:e960. doi: 10.7717/peerj.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 102.R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 103.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. 67:1–48. J Stat Softw doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 105.Fox J, Weisberg S. 2011. An R companion to applied regression, 2nd ed Sage, Thousand Oaks, CA. [Google Scholar]
- 106.Lenth R. 2015. lsmeans: least-squares means. R package version 2.20-23. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 107.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Wagner H. 2015. vegan: community ecology package. R package version 2.3-1. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 108.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]