ABSTRACT
Marine algae produce a variety of glycans, which fulfill diverse biological functions and fuel the carbon and energy demands of heterotrophic microbes. A common approach to analysis of marine organic matter uses acid to hydrolyze the glycans into measurable monosaccharides. The monosaccharides may be derived from different glycans that are built with the same monosaccharides, however, and this approach does not distinguish between glycans in natural samples. Here we use enzymes to digest selectively and thereby quantify laminarin in particulate organic matter. Environmental metaproteome data revealed carbohydrate-active enzymes from marine flavobacteria as tools for selective hydrolysis of the algal β-glucan laminarin. The enzymes digested laminarin into glucose and oligosaccharides, which we measured with standard methods to establish the amounts of laminarin in the samples. We cloned, expressed, purified, and characterized three new glycoside hydrolases (GHs) of Formosa bacteria: two are endo-β-1,3-glucanases, of the GH16 and GH17 families, and the other is a GH30 exo-β-1,6-glucanase. Formosa sp. nov strain Hel1_33_131 GH30 (FbGH30) removed the β-1,6-glucose side chains, and Formosa agariphila GH17A (FaGH17A) and FaGH16A hydrolyzed the β-1,3-glucose backbone of laminarin. Specificity profiling with a library of glucan oligosaccharides and polysaccharides revealed that FaGH17A and FbGH30 were highly specific enzymes, while FaGH16A also hydrolyzed mixed-linked glucans with β-1,4-glucose. Therefore, we chose the more specific FaGH17A and FbGH30 to quantify laminarin in two cultured diatoms, namely, Thalassiosira weissflogii and Thalassiosira pseudonana, and in seawater samples from the North Sea and the Arctic Ocean. Combined, these results demonstrate the potential of enzymes for faster, stereospecific, and sequence-specific analysis of select glycans in marine organic matter.
IMPORTANCE Marine algae synthesize substantial amounts of the glucose polymer laminarin for energy and carbon storage. Its concentrations, rates of production by autotrophic organisms, and rates of digestion by heterotrophic organisms remain unknown. Here we present a method based on enzymes that hydrolyze laminarin and enable its quantification even in crude substrate mixtures, without purification. Compared to the commonly used acid hydrolysis, the enzymatic method presented here is faster and stereospecific and selectively cleaves laminarin in mixtures of glycans, releasing only glucose and oligosaccharides, which can be easily quantified with reducing sugar assays.
KEYWORDS: marine microbes, algal bloom, β-glucan, carbon cycle, diatoms, glycobiology, glycoside hydrolase, laminarin, laminarinase, organic matter
INTRODUCTION
Marine algae are thought to contribute ∼50% of the global primary production, converting more carbon dioxide into biomass through photosynthesis than tropical forests (1). Major outputs of carbon fixation are glycans, which can amount to 80% of the algal biomass (2) and have a variety of functions in energy and carbon storage, as cell wall building material, in cell communication, as adhesives, and as organic templates for biomineralization processes (3–5). Among the most abundant glycans produced by algae are β-glucans, including laminarin, which is a linear polysaccharide of ∼20 to 30 β-1,3-linked glucose residues with β-1,6-linked side chains consisting of a single glucose molecule. Laminarin is produced by many algal species that are characterized by high productivity and rapid turnover (3, 6–12). Consequently, laminarin may play a major role in the marine carbon cycle. Diatoms alone are thought to produce ∼40% of the 45 to 50 gigatons (1 gigaton = 109 tons) of organic carbon annually in the sea (2, 13, 14). Alderkamp et al. estimated that the global annual production of laminarin, also known as chrysolaminarin, by these microalgae amounts to 5 to 15 gigatons (15). To place this into perspective, the current annual increase in atmospheric CO2 is 4 gigatons (16). Hence, new methods are required to better monitor the turnover of this abundant glycan in the sea and to improve our understanding of the impact of laminarin on the marine carbon cycle.
Structural and quantitative analysis of glycans in marine samples such as marine snow (sinking aggregated detritus), fecal pellets, algal cell walls, particulate organic matter (POM), and dissolved organic matter remains complicated, because in seawater glycans occur as complex mixtures in which individual components may occur at low concentrations (17). Glycans are built from different sugar monomers, which can be linked in various ways and configurations into linear or branched macromolecules, frequently hosting substitutions with diverse chemical groups (18). A common method for compositional glycan analysis involves acid hydrolysis to identify and to quantify the monomers with reducing sugar assays, chromatographic assays, or spectroscopy (19). However, information concerning linkage types, sequences, and stereochemistry remains difficult to obtain with this commonly used method. This problem especially applies to marine samples, in which the abundance of polysaccharides is often too low to permit chromatographic purification for more sophisticated nuclear magnetic resonance (NMR) or mass spectrometry analysis, which would allow structural elucidation of the types of glycans in the mixture (17). Therefore, new stereospecific and linkage-specific approaches are required to identify polysaccharides in natural samples, preferably with little or no need for additional purification. Enzymes evolved to overcome the structural diversity of glycans present in nature; in plant and mammalian systems, enzymes are used to dissect and to analyze the structures of complex cell wall and mammalian glycans in situ (20, 21). Moreover, enzymes are less prone to side reactions, which are common with acid hydrolysis, leading to conversions of monosaccharides (22). Enzymes may also be powerful tools for the analysis of ecologically relevant marine glycans, which often have different chemical structures than plant glycans and thus require different sets of enzymes.
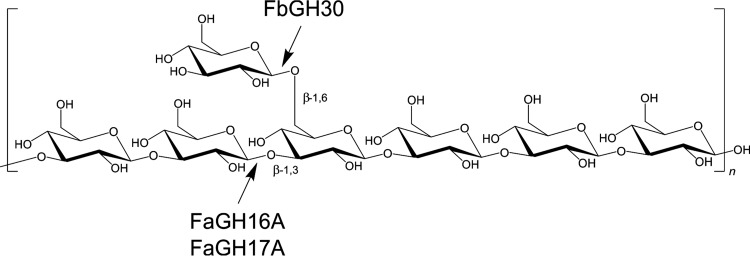
Bacteria depolymerize algal polysaccharides with glycoside hydrolases (GHs) and polysaccharide lyases (PLs), which are classified in the Carbohydrate-Active Enzymes (CAZy) database into over 130 GH families and over 20 PL families (23). Depolymerization of polysaccharides, including laminarin, involves systems of enzymes with different specificities and modes of action. Laminarinases are classified into endo-β-1,3-glucanases (laminarinases) (EC 3.2.1.6 and EC 3.2.1.39), which cleave randomly within the chain, hydrolyzing the β-1,3 backbone into glucose and oligosaccharides, and exo-β-1,3-glucanases (EC 3.2.1.58), which cleave at the nonreducing ends, hydrolyzing laminarin oligosaccharides into glucose (Fig. 1). Endo-acting laminarinases belong to different glycoside hydrolase families, mainly GH16, GH17, GH55, GH64, and GH81, and exo-acting laminarinases belong to the GH3 family. Exo- and endo-acting enzymes specific for the β-1,3 linkage have been described (24); however, the enzymes responsible for cleaving the β-1,6-linked side chains in laminarin remain unknown, to the best of our knowledge. The side chains can limit the activity of endo-acting enzymes, owing to steric hindrance, and thereby limit the complete hydrolysis of laminarin (25). Therefore, an enzyme system for complete hydrolysis of laminarin into measurable glucose would require a mixture of endo- and exo-acting enzymes with different specificities, accounting for the two types of linkages present in laminarin.
FIG 1.
Laminarin structure and enzyme activities. The linear storage glucan consists of a β-1,3-d-glucose polysaccharide with β-1,6-linked monomer side chains. The characterization of three enzymes from marine Bacteroidetes, belonging to the GH16, GH17, and GH30 families, presented here showed that the GH30 enzyme hydrolyzed the β-1,6-linked side chain and the GH17 and GH16 enzymes hydrolyzed the β-1,3-d-linked main chain of laminarin.
Metagenome and proteome data on marine blooms of algae and bacteria represent a source of ecologically relevant enzymes and support the notion that laminarin represents a crucial carbon and energy source for prokaryotic heterotrophs (26). During spring microalgal blooms in the North Sea (2009), diatoms supported the growth of heterotrophic Bacteroidetes, which consumed glycans and other molecules produced by the algae. Protein expression data showed that putative laminarinases were among the proteins with the highest levels of expression, which suggested that laminarin represented a major food resource for marine Bacteroidetes. The GH16 and GH30 families were among the most highly expressed proteins, which suggests that these enzymes may mediate the turnover of laminarin from microalgae and therefore may represent suitable tools for marine glycan analysis.
In Bacteroidetes, the carbohydrate-active enzymes involved in the depolymerization of glycans are often clustered in genetic islands termed polysaccharide utilization loci (PUL) (27, 28). We identified a putative PUL for laminarin catabolism in Formosa agariphila, and a homolog of this PUL was upregulated by laminarin in Gramella forsettii (29). These PUL contain orthologues of the GH16 and GH30 enzymes that were highly expressed during the North Sea algal bloom in 2009; in addition, they contain GH17 enzymes. Therefore, we focused on these conserved PUL of marine Formosa spp. and characterized putative GH16, GH17, and GH30 enzymes, showing that they are active laminarinases. In subsequent experiments, we used the enzymes to quantify laminarin in diatom species grown in the laboratory. We also used the enzymes to measure laminarin in environmental samples by digesting particulate organic matter collected on filters from North Sea and Arctic Ocean waters. Our results demonstrate that carbohydrate-active enzymes have strong potential for the specific quantification of select types of glycans in marine organic matter.
RESULTS
Heterologous expression and initial characterization of FaGH16A, FaGH17A, and FbGH30 enzymes.
We cloned and expressed three enzymes, i.e., GH16 and GH17 enzymes from a genetic island of Formosa agariphila KMM 3901T (29) and a GH30 enzyme from Formosa sp. nov strain Hel1_33_131 (30); both species are involved in laminarin catabolism. Two endo-acting laminarinases from Zobellia galactanivorans, with ∼44% and 43% sequence identity with GH16A of F. agariphila (FaGH16A), have recently been identified and characterized. Although these findings suggest that FaGH16A may be a suitable tool for the analysis of laminarin (25, 31), the enzymes also hydrolyzed related glucans with β-1,4 linkages, showing that they are of limited specificity. We chose to include a GH16 enzyme in our analysis to illustrate the activity of a promiscuous laminarinase, as a control, and to highlight the advantages of specific enzymes. Enzymes with higher specificity than the GH16 enzyme may be needed for highly selective laminarin analysis. We hypothesized that such alternative enzymes might be from the GH17 and GH30 families. To our knowledge, GH17 enzymes have been described only from terrestrial microbes and plants. They are endo-type enzymes, which are highly specific for nondecorated stretches of β-1,3-glucans (32). This suggests that orthologous genes from marine bacteria may have a similar function. The GH30 family contains enzymes with known β-1,6-glucanase activity (33, 34); hence, we reasoned that this enzyme may hydrolyze the β-1,6-linked side chains of laminarin in an exo-type manner and may be required, together with an endo-type enzyme, for efficient depolymerization of laminarin.
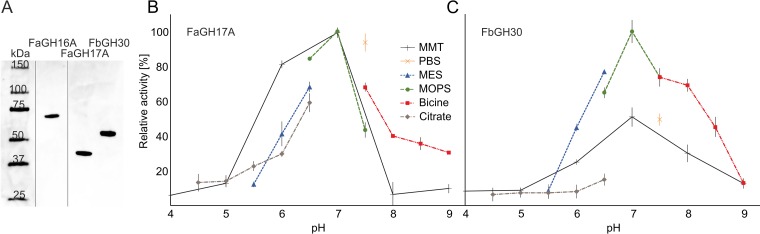
After immobilized metal affinity chromatography and size exclusion chromatography, the typical yields of purified enzyme were ∼20 mg liter−1 for FaGH16A and ∼50 mg liter−1 for FaGH17A. Because the GH30 enzyme from F. agariphila was targeted to inclusion bodies without detectable amounts of soluble protein, we chose to clone and to produce an orthologous enzyme from Formosa sp. nov. strain Hel1_33_131 (i.e., FbGH30), which was isolated from Helgoland in the North Sea and shares 70% identity with FaGH30. FbGH30 was produced as a soluble protein in Escherichia coli, and overexpression yielded ∼30 mg liter−1 of purified protein (Fig. 2A). Size exclusion chromatography and SDS-PAGE data revealed molecular masses of 59 kDa (FaGH16A), 42 kDa (FaGH17A), and 55 kDa (FbGH30), which are close to the theoretical values of 58.5 kDa (FaGH16A), 44.8 kDa (FaGH17A), and 54.7 kDa (FbGH30), suggesting that all three enzymes occur as monomers in solution. This notion was supported by dynamic light scattering (DLS) measurements, confirming that the two enzymes used for analytical assays remained monodisperse and stable for at least 4 months at 4°C. Over a period of 120 days, FaGH17A and FbGH30 maintained hydrodynamic radii of 3.63 ± 0.23 nm (mean ± standard deviation [SD]) and 3.78 ± 0.12 nm, respectively.
FIG 2.
Recombinant glycoside hydrolases from marine Bacteroidetes showing greatest activity in MOPS buffer at neutral pH. (A) SDS-PAGE analysis of purified enzymes. One gel is shown. Unimportant lanes were intentionally omitted, as indicated by vertical lines. Approximately 0.5 μg of each protein was loaded on the gel. All proteins were run on the same gel, and lanes were spliced for clarity. (B and C) Enzymatic rates were measured with 0.1% (wt/vol) laminarin, which was hydrolyzed by 100 nM (∼5.0 μg ml−1) purified FaGH17A (B) or FbGH30 (C) at 37°C for 30 min in 50 mM buffer. The greatest activity rate was observed in MOPS buffer at pH 7.0 and was set as the 100% reference value. MES, morpholineethanesulfonic acid; MMT, malic acid-MES-Tris base.
Temperature stability and pH optima of FaGH17A and FbGH30.
Melting curve measurements with dynamic light scattering revealed the mesophilic character of FaGH16A, FaGH17A, and FbGH30. The melting curves showed that FaGH17A started to aggregate at ∼40°C, FbGH30 at ∼42°C, and FaGH16A at ∼55°C, as measured with three different protein concentrations (see Fig. S1 in the supplemental material). Because FaGH17A and FbGH30 showed higher specificity for laminarin (see below), we concentrated our analysis of stability and activity in different buffer systems on these two enzymes and tested the activity of FaGH17A and FbGH30 with different organic and inorganic buffers and pH values (covering a pH range of 4 to 9), using laminarin as the substrate. Bell-shaped activity profiles were obtained for both enzymes, with the highest enzymatic activities at a pH optimum of 7 (Fig. 2B and C). Both enzymes had the highest activities in 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer. Therefore, a reaction temperature of 37°C and a buffer system of MOPS or phosphate-buffered saline (PBS), with a pH of 7, were chosen for subsequent experiments.
Activity measurements of FaGH16A, FaGH17A, and FbGH30 with glucan polysaccharides and oligosaccharides.
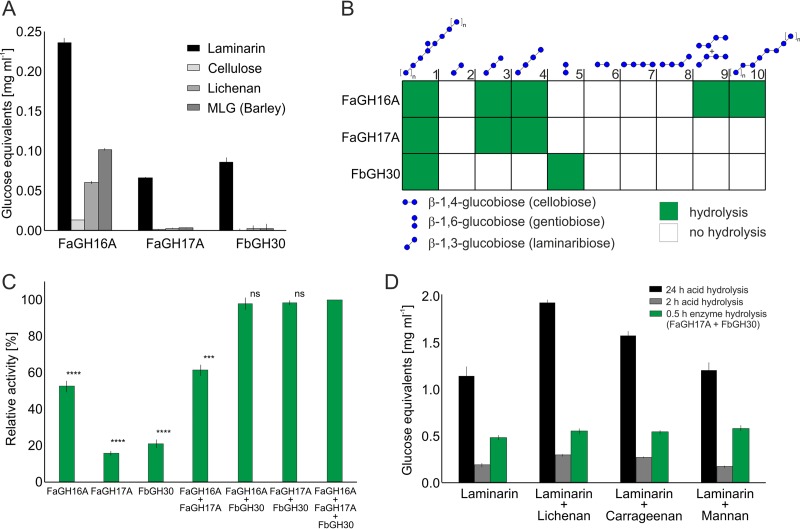
Activity measurements with defined substrates revealed that FaGH17A, FbGH30, and FaGH16A were active with laminarin, while having different specificities and modes of action. To test their modes of action, we used fluorophore-assisted carbohydrate polyacrylamide gel electrophoresis (FACE) (35). We digested laminarin and analyzed samples taken during the time course of the reaction. FaGH16A and FaGH17A hydrolyzed laminarin into oligosaccharides of different sizes, creating a ladder-type profile (Fig. S2A), which is typical for endo-acting glycoside hydrolases (25). Time course analysis of the products released by FbGH30 revealed the accumulation of glucose during all phases of the enzyme assay (Fig. S2B), a pattern that is typical for an exo-acting enzyme (25, 31).
To map the specificity of the enzymes, we used different β-glucans from terrestrial plants and marine algae and measured the reaction products with reducing sugar assays. We tested activity with β-1,4-linked carboxy-methyl cellulose, β-1,3/β-1,4-mixed-linked lichenan, β-1,3/β-1,4-mixed-linked glucan (MLG) from barley, and laminarin. Hydrolysis was quantified with the p-hydroxybenzoic acid hydrazide (PAHBAH) assay (36), which detects the reducing end, i.e., the aldehyde group. Figure 3A shows that FaGH16A released the greatest amount of reducing sugar equivalents (0.23 ± 0.006 mg ml−1 [mean ± SD]) with laminarin as the substrate, 3.6-fold more than FaGH17A (0.064 ± 0.001 mg ml−1) and 2.8-fold more than FbGH30 (0.083 ± 0.006 mg ml−1) (P < 0.0001). FaGH16A also hydrolyzed lichenan (0.058 ± 0.004 mg ml−1) and mixed-linked glucan from barley (0.099 ± 0.006 mg ml−1), which contain β-1,3 and β-1,4 linkages. FaGH17A and FbGH30 did not release significant amounts of reducing sugar equivalents when these glucans were used as the substrates (P < 0.0001). These results pointed to FaGH17A and FbGH30 being more specific than FaGH16A, which appeared to have greater activity with laminarin, however.
FIG 3.
Glycoside hydrolases showing different levels of specificity for laminarin and related glucans. (A) Activity tests with glucan polysaccharides and oligosaccharides containing β-1,3, β-1,4, and β-1,6 linkages, based on the PAHBAH reducing sugar assay. Shown are mean and standard deviation (SD) values from three technical replicates. (B) Enzyme specificity tested with defined oligosaccharide substrates, using thin-layer chromatography. The results are presented as a heatmap (see Fig. S3 in the supplemental material). The substrates at 0.1% (wt/vol) were hydrolyzed for 30 min at 37°C by 100 nM (∼5 μg ml−1) purified enzyme in PBS buffer at pH 7.5. (C) Mixtures of FbGH30 and FaGH17A or FbGH30 and FaGH16A, showing greater activity than the individual enzymes. The highest activity level with all three enzymes was set to 100%, and all other samples were compared to that value. Laminarin at 0.1% (wt/vol) was hydrolyzed for 30 min at 37°C by 100 nM (∼5.0 μg ml−1) of each purified enzyme in PBS buffer at pH 7.5, and hydrolysis was measured with the PAHBAH assay. Shown are mean and SD values from three technical replicates. ****, P < 0.0001; ***, P < 0.001, independent two-sample Student's t test. ns, not significant. (D) Comparison of hydrolysis yields of enzymatic, partial acid, and total acid hydrolysis of different polysaccharides. Lichenan, carrageenan, and mannan at 0.1% (wt/vol) were added to 0.1% (wt/vol) laminarin and were hydrolyzed for 30 min at 37°C with 100 nM purified enzyme (∼5 μg ml−1 of FaGH16A, FaGH17A, or FbGH30) in 50 mM MOPS buffer. Boiling for 5 min at 100°C stopped the reaction. Partial acid hydrolysis was conducted for 2 h at 20°C with 50 mM H2SO4. Total acid hydrolysis was carried out for 24 h at 100°C with 1 M HCl. The reaction mixtures were analyzed with the PAHBAH assay. All experiments were carried out in triplicate. Shown are mean and SD values from three technical replicates.
To validate these results, we used different glucans (oligosaccharides and polysaccharides) as substrates and analyzed the reaction products by thin-layer chromatography (TLC) (Fig. S3). The substrates included laminarin oligosaccharides (biose, triose, and tetraose of the β-1,3 series), cellooligosaccharides (biose and tetraose of the β-1,4 series), mixed-linked cellotetraose with internal β-1,3 linkages at different positions, gentiobiose (a β-1,6-linked biose), and β-1,3/β-1,4-mixed-linked glucan oligosaccharides (Fig. 3B). Of the three enzymes, FaGH16A hydrolyzed most substrates, digesting laminarin and β-1,3 oligosaccharides into glucose and laminaribiose. It also hydrolyzed MLG, which was not hydrolyzed by the other enzymes. FaGH16A hydrolyzed mixed-linked cellotetraoligosaccharides with an internal β-1,4 linkage, an activity that was shown previously for the GH16 laminarinases from Zobellia galactanivorans (25, 31). FaGH17A was exclusively active with laminarin polysaccharides and oligosaccharides, producing glucose and laminaribiose as the major products, which confirmed the results described above. These experiments confirmed that the highly selective activity of terrestrial enzymes in the GH17 family for β-1,3-glucans (32, 37, 38) could be extended to this marine GH17 enzyme. The β-1,3-trisaccharide was the smallest substrate hydrolyzed into glucose and laminaribiose by FaGH17A. FbGH30 hydrolyzed laminarin and produced glucose as the major product, but it did not show activity with the other substrates containing β-1,3 or β-1,4 linkages. Instead, FbGH30 was the only enzyme that hydrolyzed gentiobiose, a β-1,6-linked disaccharide. In aggregate, these experiments indicated that FaGH16A is a promiscuous enzyme that cleaves laminarin and mixed-linked glucans, FaGH17A is a specific endo-β-1,3-glucanase, and FbGH30 is a specific exo-β-1,6-glucanase.
Increased hydrolysis of laminarin with mixtures of enzymes.
The preferences of FaGH16A, FaGH17A, and FbGH30 for different types of linkages suggested that combinations of these enzymes might improve laminarin hydrolysis and consequently the sensitivity of the analytical assays. We tested this by measuring the activity of enzyme mixtures with laminarin as the substrate in the PAHBAH assay, which is sensitive and feasible and has a limit of detection of 2 nM (39). As noted previously, FaGH16A had the highest activity, compared to FaGH17A and FbGH30. In contrast, combinations of FaGH17A with FaGH16A increased reducing sugar concentrations (Fig. 3C). This effect increased when FbGH30 was added to either enzyme. Together, FaGH16A and FbGH30 increased the yield by a factor of 1.9 ± 0.04 (P < 0.0001) and FaGH17A and FbGH30 increased the yield by a factor of 6.2 ± 0.43, compared to FaGH17A alone. Because the mixture of FaGH17A and FbGH30 achieved the same total yield as FaGH16A and FbGH30 but showed greater specificity, we chose this enzyme mixture for the enzymatic analysis of laminarin in microalgae.
Next, we investigated how the enzyme mixture performed in comparison with acid when other polysaccharides were present. To evaluate the ability of the enzymes to hydrolyze laminarin in the presence of other polysaccharides, we incubated different glycan mixtures with enzymes or acid and compared the product yields. The mixture of FaGH17A and FbGH30 was incubated with laminarin alone or with added reference polysaccharides, i.e., lichenan, carrageenan, and mannan. The laminarin concentration was kept constant; hence, the product concentrations obtained with the enzymes after 30 min of hydrolysis remained the same in all tested mixtures. This result indicated that only laminarin and none of the other substrates was cleaved by the enzyme mixture (Fig. 3D). Interestingly, the moderate acid hydrolysis protocol that is commonly used for laminarin quantification in microalgae (14) delivered lower values than the enzyme method even after 2 h of hydrolysis, which showed that enzymes react faster. Moreover, the acid-derived values varied when other polysaccharides were present, owing to the nonspecific hydrolysis of mannan, lichenan, and carrageenan, which showed that acid is less specific. Acid hydrolysis for 24 h completely hydrolyzed the laminarin into glucose; the yield of reducing ends with enzymes was ∼50% of the total acid hydrolysis, indicating that the enzyme mixture cleaved about 50% of the glycosidic linkages in laminarin (Fig. S4). However, this longer acid hydrolysis also strongly hydrolyzed the other polysaccharides, leading to highly inflated sugar-reducing signals, compared to the enzyme method. This experiment highlights the superior specificity of enzymes, which hydrolyzed laminarin more quickly and more accurately than the commonly used acid hydrolysis approach.
Enzymatic quantification of laminarin in diatom batch cultures and environmental samples.
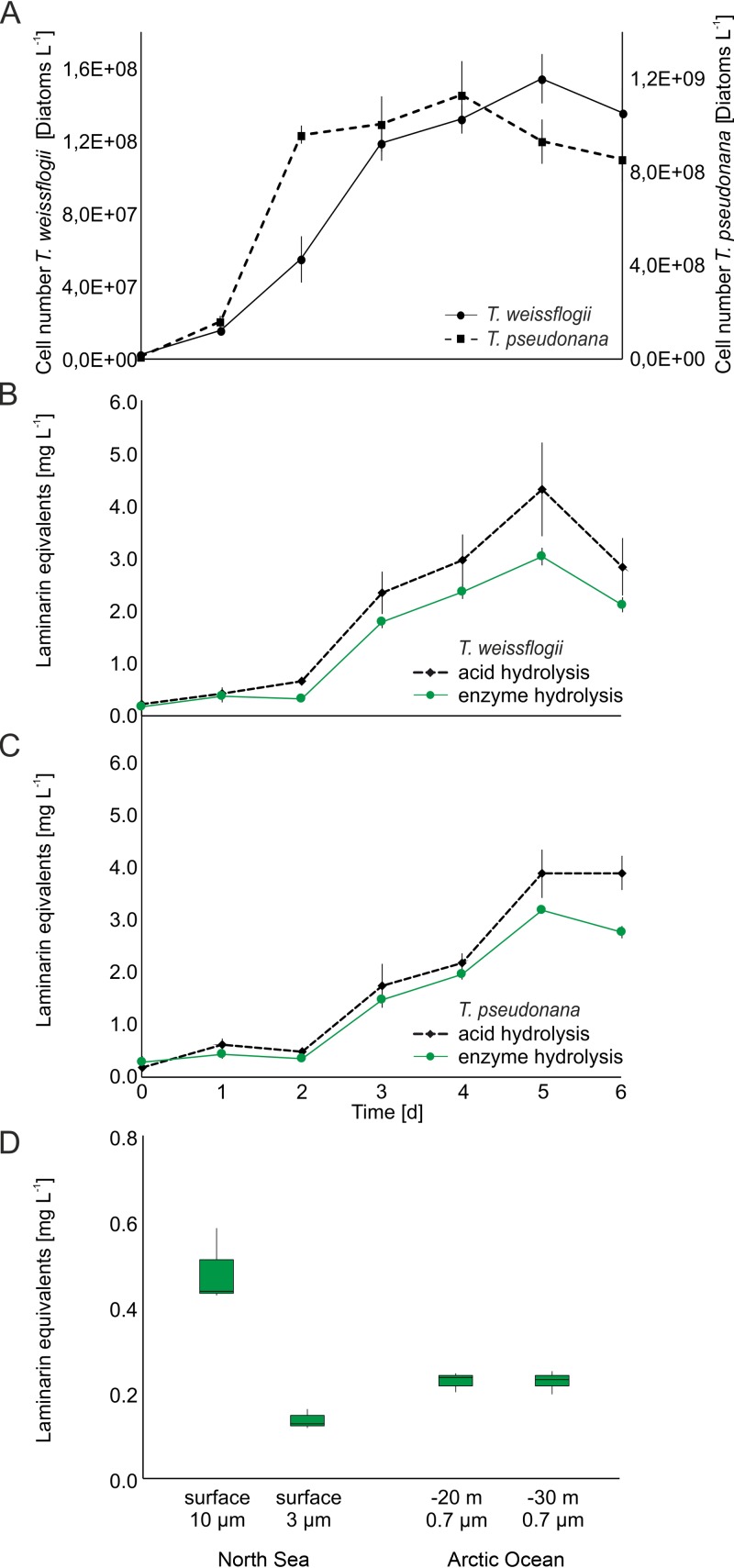
We quantified laminarin in microalgae with the enzyme mixture (FaGH17A and FbGH30) and with the conventional acid-based method (14). The two species Thalassiosira weissflogii and Thalassiosira pseudonana displayed typical growth behavior (Fig. 4A). Repeated extraction of the retentate with MOPS buffer did not increase the total yield, which confirmed that one step is generally sufficient to extract laminarin (40). To account for the fact that the enzymes hydrolyzed only about 50% of the glycosidic linkages, we prepared a calibration curve with laminarin that had been digested under the same conditions as the natural samples, which revealed a linear relationship between laminarin concentrations and reducing sugar signals. This calibration curve enabled the quantification of laminarin despite its incomplete hydrolysis (Fig. S5). The correlation coefficients for both the acid hydrolysis and the enzymatic hydrolysis were higher than 0.99.
FIG 4.
Laminarin quantification in Thalassiosira weissflogii and T. pseudonana laboratory cultures and in environmental samples. (A) Numbers of diatoms cultured in modified ESAW/HESNW medium at 15°C, with a 12-h/12-h light/dark cycle, for 6 days. (B) Quantification of laminarin extracted from T. weissflogii by acid (dashed black line) or enzyme (solid green line) hydrolysis. (C) Quantification of laminarin extracted from T. pseudonana by acid (dashed black line) or enzyme (solid green line) hydrolysis. (D) Laminarin contents of particulate organic matter, which was concentrated by filtering seawater from the North Sea near Helgoland (spring 2009) and from the Arctic Ocean near Svalbard (summer 2015) and enzymatically hydrolyzed. Water-soluble extracts were hydrolyzed for 30 min at 37°C with 100 nM (∼5.0 μg ml−1) FaGH17A and FbGH30, with 1 mg ml−1 BSA, in 50 mM MOPS buffer at pH 7. Alternatively, the extracted polysaccharides were hydrolyzed twice for 2 h at 20°C with 50 mM H2SO4, with shaking at 1,500 rpm. The products were quantified with the PAHBAH reducing sugar assay. The calibration curve was prepared with laminarin hydrolyzed with enzyme or acid, and the reducing sugar signals were measured as described above (see Fig. S5).
The conventional acid hydrolysis and the new enzymatic approach gave comparable glucan quantities over the time course of the growth experiment. The acid method gave higher values than the enzyme method, i.e., 8 to 23 pg and 0.3 to 3.9 pg of glucan per cell for T. weissflogii and T. pseudonana, respectively, compared to 6 to 20 pg and 0.4 to 3.5 pg of glucan per cell, respectively, via enzymatic digestion. While the cell densities and laminarin concentrations correlated well for T. weissflogii during the growth experiment (Fig. 4A and B), this was not the case for T. pseudonana (Fig. 4A and C). At the beginning of the growth period and during the exponential growth phase, cell numbers increased independently of the laminarin concentrations, which was noted previously for this diatom (2). Table 1 compares previously established glucan quantities in diatoms derived by acid hydrolysis with values obtained in our experiments, showing that they are in a similar range as the enzyme-based measurements.
TABLE 1.
Glucan amounts of various diatom species, related to dry weight and growth phase
| Species | Sampling growth phase | Mediuma | Temperature (°C) | Light cycle (light h/dark h) | Irradiance (μmol photons m−2 s−1) | Glucan wt (pg cell−1) | Hydrolysis method | Reference |
|---|---|---|---|---|---|---|---|---|
| T. weissflogii | Mean over 60 days | F/2 | 20 | 24/0 | 300 | 45.2 | Acid | Li (61) |
| T. pseudonana | Late exponential | F/2 | 20 | 12/12 | 75 | 2.4 | Acid | Brown (58) |
| Skeletonema marinoi | Late exponential | F/2 | 20 | 12/12 | 75 | 2.5 | Acid | Brown (58) |
| Chaetoceros debilis | Late stationary | F/2 | 18 | 24 | 200 | 15.8 | Acid | Størseth et al. (59) |
| Chaetoceros affinis | Exponential | F/10 | 13 | 14/10 | 40.0 | Acid | Myklestad and Haug (60) | |
| S. marinoi | Stationary | F/10 | 13 | 14/10 | 200 | 375.0 | Acid | Granum et al. (13) |
| T. weissflogii | Late exponential | NEPCC | 15 | 12/12 | 140 | 16.0 | Acid | This study |
| T. pseudonana | Late exponential | NEPCC | 15 | 12/12 | 140 | 1.3 | Acid | This study |
| T. weissflogii | Late exponential | NEPCC | 15 | 12/12 | 140 | 15.0 | Enzymatic | This study |
| T. pseudonana | Late exponential | NEPCC | 15 | 12/12 | 140 | 1.5 | Enzymatic | This study |
We also analyzed the laminarin content in particulate organic matter (POM) obtained from two marine sampling sites (Fig. 4D). POM was obtained by filtering seawater during a diatom-dominated algal bloom in the North Sea near the island of Helgoland and in the Fram Strait in the Arctic Ocean. The retentate from the North Sea corresponds to a time point shortly after the spring bloom of diatoms peaked in a longer time series (26). The proteome data set for bacterioplankton from the same day showed that orthologues of the laminarinases that we used here for the assay were highly expressed in situ. Accordingly, we were able to determine laminarin concentrations in the POM fraction by using the enzyme assay. The 10-μm POM fraction of the Helgoland samples collected from 1 liter of seawater contained over 0.48 ± 0.09 mg and the 3-μm fraction contained over 0.13 ± 0.02 mg of laminarin (Fig. 4D). Acid hydrolysis of these two samples yielded a slightly greater signal (data not shown). This discrepancy is similar to what we observed with the cultivated diatoms. The enzyme mixture hydrolyzed laminarin in samples from the Arctic Ocean, providing nearly the same signals for the two water depths; about 0.22 ± 0.03 mg of laminarin was contained within the POM collected from 1 liter of seawater. Altogether, these data show that these enzymes can detect and quantify laminarin in POM without additional purification.
DISCUSSION
Highly specific, stable, and active enzymes that do not hydrolyze other polysaccharides enable the selective hydrolysis and quantification of laminarin even in crude algal samples. The enzymes are stereospecific and function in high concentrations of salt, which limits the sensitivity of alternative methods commonly used to analyze marine organic matter, such as proton NMR or mass spectrometry (41). Three enzymes, from the GH16, GH17, and GH30 families, were characterized, revealing various specificities and different potentials as tools for the analysis of marine glucans. Compared to FaGH16A, with broad activity, FaGH17A showed narrow specificity for β-1,3-glucan, which may be due to the enzyme architecture. Active sites of GH17 enzymes are known to be longer than those found in GH16 enzymes (42, 43) and to require longer stretches of nondecorated β-1,3-glucan chain for productive binding, a relationship that was observed previously among agarases of the marine bacterium Zobellia galactanivorans (44). The longer active sites of GH17 enzymes increase the selectivity for nondecorated β-1,3-glucans and decrease the activity of GH17 enzymes with β-1,6-branched laminarin and β-1,3/1,4-mixed-linked glucans; the shorter active site of GH16 enables hydrolysis of mixed-linked glucans and native laminarin, as indicated by greater activity with those substrates. Labourel et al. (25) proposed an alternative hypothesis, i.e., GH16 laminarinases may be able to cleave β-1,6-decorated laminarin due to the presence of small pockets lining the active sites, which can host the side chain glucose residues. These pockets and shorter active sites may explain the greater activity with decorated laminarin, compared to GH17 enzymes.
Combining FbGH30 with FaGH17A was key for efficient hydrolysis of laminarin and significantly improved the final yield. It should be noted that the improved hydrolysis with FbGH30 is a consequence of its ability to remove the β-1,6-glucose side chains. After this debranching, FaGH17A can cleave the nondecorated backbone; together, the two enzymes achieve increased laminarin hydrolysis yields. The TLC data showed that FbGH30 was active with gentiobiose, which has only one β-1,6 linkage, while it was inactive with oligosaccharides with β-1,3 and β-1,4 linkages, indicating that FbGH30 is a highly stereospecific enzyme. The only β-1,6 linkage in laminarin is the side chain, which indicates that FbGH30 removes the side chains of the polysaccharide and in this way opens new recognition sites for the GH17 enzyme. The comparative analysis of these enzymes and their different specificities emphasizes that a sensitive choice of carbohydrate-active enzymes is paramount for the quantitative analysis of marine glycans.
The enzyme mixture of FaGH17A and FbGH30 is more specific and faster and can quantify laminarin despite not hydrolyzing it completely. Thin-layer chromatography experiments revealed that the GH17 enzyme did not cleave laminaribiose (Fig. 3B), indicating that the enzymes did not completely hydrolyze laminarin; comparison with the product yield obtained with acid hydrolysis revealed that the enzymes hydrolyzed about 50% of the glycosidic linkages. This result implies that additional enzymes, such as β-glucosidases, are required to complete the hydrolysis of the remaining laminaribiose and larger oligosaccharides. However, we hydrolyzed different laminarin concentrations with the enzymes, producing a linear calibration curve to account for incomplete hydrolysis. This calibration curve established the amounts of laminarin in diatom extracts and environmental samples. Moreover, our results showed that the acid hydrolysis method based on the method described by Granum and Myklestad (14) produced lower values than the enzyme method even after 2 h of acid hydrolysis, showing that enzyme hydrolysis can be faster. Increasing the reaction time, acid concentration, or temperature led to nonspecific hydrolysis of cooccurring polysaccharides. This nonselective hydrolysis of other polysaccharides or other compounds may explain the slightly higher values measured with the acid method. Although we cannot exclude the possibility that the enzymes may actually underestimate the laminarin content, they evolved to be specific and therefore it is clear that they provide a more conservative measure of laminarin, especially when polysaccharides and other labile compounds are present in the sample.
Applying the enzyme method to environmental samples demonstrated the successful quantification of laminarin. The North Sea retentate included material from the same time and location in the algal bloom at which the orthologues of the laminarinases used here for the assay were highly expressed in the metaproteome data set. We measured concentrations between 0.13 ± 0.02 and 0.48 ± 0.09 mg liter−1 in the particulate organic matter fraction, revealing that laminarin was indeed the elicitor of enzyme expression in the North Sea. To put this into perspective, van Oijen et al. measured total carbohydrate concentrations in Antarctic Ocean water samples by using acid hydrolysis (45). Compared to chlorophyll concentrations that are usually measured during a spring bloom in the North Sea (9.3 μg liter−1), this water body contained a significantly lower chlorophyll concentration (0.7 μg liter−1), and smaller amounts of biomass yielded lower carbohydrate concentrations of up to 15 μg liter−1. In contrast, Pakulski and Benner showed similar or higher concentrations of total carbohydrates in the surface water of the North Atlantic Ocean, the equatorial Pacific Ocean, the Gulf of Mexico, and the Antarctic Ocean (46).
Moran and colleagues recently stated that, in light of an anthropogenically altered carbon cycle (47), a more thorough understanding of organic matter, with its fluxes and intertwined relationships between molecules and metabolizing organisms, requires advancing technologies in different fields of environmental sciences. One important question addresses which molecules represent the largest conduits of carbon flux through the labile marine dissolved organic matter pool. Glycans belong to the most abundant molecules in surface waters, where they are produced by microalgae, but their concentrations seem to decrease rapidly with time and depth; this has been inferred by measuring the glucose concentrations of acid-hydrolyzed samples (48–50). Those studies, together with radiocarbon dating of glycan-rich dissolved organic matter (51), support the notions that heterotrophs rapidly consume glycans and they belong to the most reactive macromolecules. It is possible that laminarin belongs to these abundant but quickly metabolized compounds, which fuel catabolic activity in ocean surface waters. This enzymatic method might be a suitable approach to address these questions by unambiguously measuring concentrations of β-glucans, such as laminarin, in dissolved and particulate organic matter.
MATERIALS AND METHODS
Cloning.
The putative laminarinase genes (FAGH16A locus tag, BN863_18740; FaGH17A locus tag, BN863_18720) were amplified from the genomic DNA of Formosa agariphila (KMM 3901T) (GenBank accession number HG315671.1), which was obtained from the Leibniz Institute DMSZ. Primers were designed to amplify the coding regions corresponding to the catalytic module and a polycystic kidney disease (PKD) domain (52) in the case of FaGH16A (FaGH16Aforward, CTGGTGCCGCGCGGCAGCCATATGGCTAGCGAAGACGATGTGTCCATAG; FaGH17Aforward, CTGGTGCCGCGCGGCAGCCATATGGCTAGCTTAGGATGTAACAATAAAACTCAGATTGCG; FaGH16Areverse, ATCTCAGTGGTGGTGGTGGTGGTGCTCGAGTTATTGGTAAACTCTTAC; FaGH17Areverse, ATCTCAGTGGTGGTGGTGGTGGTGCTCGAGTTAATTTTTAATTGGTGGCACTGCAACG). The PCR was performed using standard PCR amplification protocols. The primers were designed to include ∼30-bp overhangs complementary with the pET28a(+) vector, for Gibson assembly (53), which results in an N-terminal hexahistidine tag in the recombinant protein. A restriction site was included to allow for alternative restriction enzyme-based cloning. The NEB Gibson cloning kit was used, according to the manufacturer's instructions. Colonies were screened for positive clones by using T7 primers and colony PCR. Because the GH30 enzyme from F. agariphila was targeted to inclusion bodies without detectable amounts of soluble protein, we chose to clone and to produce an orthologous enzyme (FbGH30) from Formosa sp. nov. strain Hel1_33_131, which was isolated from Helgoland in the North Sea (30) and shares 70% identity with FaGH30. The FbGH30 gene (locus tag, FORMB_24730) was synthesized and cloned into a pET28a(+) vector by the company GenScript. The vectors with insert were purified from 2-ml Luria-Bertani (LB) broth precultures using the Qiagen Miniprep kit. For protein expression, the vectors were used to transform E. coli BL21(DE3). The cells were stored at −80°C until further use. For common use, we deposited the expression vectors for FaGH17A (Addgene plasmid 86462) and FbGH30 (Addgene plasmid 86463) in the Addgene plasmid repository (http://www.addgene.org).
Overexpression and purification.
Proteins were expressed in E. coli BL21(DE3) cells by using 1 liter of autoinduction medium (54), which was inoculated with 1 ml of an overnight culture at 37°C in LB medium supplemented with 50 μg ml−1 kanamycin. Bacteria were grown for 3 days at 20°C in ZYP autoinduction medium with 100 μg ml−1 kanamycin, with shaking at 150 rpm. Cells were harvested by centrifugation at 4,500 × g for 25 min at 4°C, and the pellet was stored at −20°C. After thawing, the cells were suspended in 15 ml IMAC buffer A (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 40 mM imidazole). Resuspended cells were disrupted in a high-pressure homogenizer (Constant Systems) at 1.5 kbar. After the addition of 10 mg DNase (Sigma) and mixing for 10 min at 20°C, the lysate was centrifuged at 16,000 × g for 45 min at 4°C. The supernatant was loaded onto a 5-ml HiTrap IMAC HP column (GE Healthcare), which was charged with 1 column volume (CV) of 500 mM NiSO4 and equilibrated with 5 CVs of IMAC buffer A. After sample injection, the column was washed with IMAC buffer A (15 CVs), and the protein was eluted with 5 CVs of a linear gradient from IMAC buffer A to IMAC buffer B (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 500 mM imidazole), at a flow rate of 5 ml min−1, at 20°C. The 1-ml fractions were analyzed by SDS-PAGE. Fractions corresponding to a band at the expected size were pooled and concentrated in an ultrafiltration stirred cell (Amicon, Millipore), on a polyethersulfone membrane with a 10-kDa cutoff value (Millipore). An aliquot of 1 to 2 ml was loaded onto a 120-ml HiPrep 16/60 Sephacryl S-200 HR column that had been equilibrated previously with 3 CVs of SEC buffer (20 mM Tris-HCl [pH 7.5]) at 20°C. The protein was eluted with 1 CV of SEC buffer, and the fractions were analyzed by SDS-PAGE. Fractions with a single band at the expected size and at an elution time corresponding to monomeric or dimeric protein were combined and concentrated. The concentration and hydrodynamic radius were determined by using a BioSpectrometer (Eppendorf) and dynamic light scattering (DLS).
Dynamic light scattering.
A DynaPro plate reader II (Wyatt Technology), in combination with 384-well plates (Aurora Microplates), was used for measurements of hydrodynamic radii via dynamic light scattering. The 0.2-μm-filtered protein samples were measured in triplicate, after removal of bubbles by centrifugation at 2,000 × g for 2 min in the plate. Samples were measured in SEC buffer (20 mM Tris-HCl [pH 7.5]). Ten measurements with 5-s acquisition time were taken from every well at 25°C. For analysis and calculation of diffusion coefficients and hydrodynamic radii via the Stokes-Einstein equation, Dynamics software (v.7.1.9.3; Wyatt Technologies) was used. For determination of melting curves, we added one drop of paraffin oil to each well, to prevent evaporation. The temperature protocol increased the temperature stepwise from 25 to 80°C, at a rate of 0.25°C min−1.
Comparative hydrolysis of different polysaccharides with FaGH16A, FaGH17A, and FbGH30 enzymes.
To evaluate the substrate specificity of the enzymes, laminarin from Eisenia bicyclis (Carbosynth), carboxy-methyl cellulose, lichenan, and mixed-linked glucan from barley (MLG) (all at 0.1% [wt/vol], from Megazyme) were hydrolyzed for 30 min at 37°C with 100 nM purified enzyme (∼5 μg ml−1 FaGH16A, FaGH17A, or FbGH30) in 1× phosphate-buffered saline (PBS) buffer. The reaction was stopped by boiling the samples for 5 min at 100°C. The reaction mixtures were used for reducing sugar assays and thin-layer chromatography.
In order to compare the hydrolysis yields of enzymatic hydrolysis and partial and total acid hydrolysis, different polysaccharides, including laminarin from E. bicyclis (Carbosynth), lichenan (Megazyme), carrageenan (Sigma), and mannan (Megazyme) (all at 0.1% [wt/vol]), were hydrolyzed for 30 min at 37°C with 100 nM purified enzyme (∼5 μg ml−1 of FaGH16A, FaGH17A, or FbGH30) in 50 mM MOPS buffer. The reaction was stopped by boiling the samples for 5 min at 100°C. Partial acid hydrolysis was conducted in the same manner as for the algal samples, i.e., hydrolysis twice for 2 h at 20°C with 50 mM H2SO4, with shaking at 1,500 rpm (14), followed by centrifugation for 15 min at 12,000 rpm and collection of the supernatant. Total acid hydrolysis was carried out for 24 h at 100°C with 1 M HCl, with shaking at 1,500 rpm. The reaction mixtures were measured with the reducing sugar assay described below. All experiments were carried out in triplicate.
Quantification of product production with PAHBAH reducing sugar assay.
One milliliter of a freshly prepared 9:1 mixture of reagent A (0.3 M 4-hydroxybenzhydrazide, 0.6 M HCl) and reagent B (48 mM trisodium citrate, 10 mM CaCl2, 0.5 M NaOH) was added to 0.1 ml of sample. The mixture was heated for 5 min at 100°C. Absorbance was determined at 410 nm using a BioSpectrometer (Eppendorf). In order to calculate the amount of released reducing ends as glucose reducing end equivalents, a calibration curve with glucose (0, 0.03125, 0.0625, 0.125, 0.25, 0.5, 1, and 2 mg ml−1) was determined for every experiment. Calibration with laminarin was conducted by hydrolysis and measurement of laminarin from E. bicyclis (Carbosynth) at different concentrations (0, 0.03125, 0.0625, 0.125, 0.25, 0.5, 1, and 2 mg ml−1). For the samples that were used to establish the calibration curve, enzyme- or acid-based hydrolysis was carried out in the same manner as for the samples from the wild.
Product profiling with thin-layer chromatography.
In order to produce defined band-like spots, we spotted three spots of 0.4 μl of the hydrolysis products of the following substrates, each at a concentration of 1 mg ml−1, directly next to each other on a silica gel 60 TLC plate (Merck-Millipore). We used laminarin from E. bicyclis (Carbosynth), laminaribiose, laminaritriose, laminaritetraose, laminaripentaose, laminarihexaose, cellobiose, cellotetraose, mixed-linked cellotetraose 1 (cellotriosyl-β-1,3-d-glucose), mixed-linked cellotetraose 2 (cellobiosyl-β-1,3-d-cellobiose plus glucosyl-β-1,3-d-cellotriose), mixed-linked glycan from barley (all from Megazyme), and gentiobiose (AppliChem). Ethyl acetate/acetic acid/methanol/formic acid/water (8:4:1:1:1) was used as the solvent, as reported by Jeng et al. (43). The plate was developed for 10 min at 100°C with 10% sulfuric acid in ethanol.
Product profiling with fluorophore-assisted carbohydrate electrophoresis.
Laminarin from E. bicyclis (0.1% [wt/vol]; Carbosynth) was hydrolyzed for 30 min at 37°C with 100 nM purified enzyme (∼5 μg ml−1 of FaGH16A, FaGH17A, or FbGH30) in 1× PBS buffer. Aliquots of 200 μl were taken at 0 s, 5 min, 20 min, 40 min, and 60 h. The reaction was stopped by boiling the sample for 5 min at 100°C, and the samples were dried in a vacuum concentrator plus (Eppendorf). Derivatization and electrophoresis were performed as described previously (35). Two microliters of 0.15 M 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) and 5 μl of 1 M NaBH3CN were added to the dried oligosaccharides. The reaction mixture was incubated overnight at 37°C and again dried under vacuum. The oligosaccharides were resuspended in 20 μl of 25% glycerol, and 5-μl aliquots were loaded onto a standard 36% tris-glycine-acrylamide gel without SDS or a 4 to 20% Tris-HCl Ready Gel (Bio-Rad). Electrophoresis was performed at 200 V at 4°C using running buffer without SDS (192 mM glycine, 25 mM Tris [pH 8.5]).
Algal growth, extraction, and hydrolysis.
The diatoms Thalassiosira weissflogii and T. pseudonana were cultivated in triplicate batch cultures with modified ESAW/HESNW culture medium (55). They were kept in T75 suspension cell culture flasks at a constant temperature of 15°C, with a 12-h/12-h light/dark cycle, without stirring. A total volume of 250 ml was inoculated with 7 ml of a 25-ml culture that had been grown for 7 days. The growth medium was inoculated with 600,000 cells of T. weissflogii and 2,000,000 cells of T. pseudonana. Illumination was applied with five Sylvania F36W/GRO tubes, resulting in an irradiance of ∼140 μmol photons m−2 s−1. Fifteen milliliters of the cultures was harvested every day. Cultivation was continued until the stationary phase was reached. Diatom abundance was measured by cell counting using a Sedgewick-Rafter counting chamber. The algae were harvested by filtration at 200 mbar on a GF/F glass microfiber filter and were stored at −20°C until extraction. Glycans were extracted from one-half of the filter with 50 mM MOPS buffer at 60°C for 60 min, with shaking at 1,500 rpm (adapted from the method described by Zha et al. [56]), followed by centrifugation for 15 min at 12,000 rpm and collection of the supernatant. A second extraction was performed to confirm the extraction efficiency. The samples were hydrolyzed for 30 min at 37°C with 100 nM purified enzyme (∼5 μg ml−1 of FaGH17A or FbGH30), after the addition of 1 mg ml−1 bovine serum albumin (BSA), in 50 mM MOPS buffer. The reaction was stopped by boiling the samples for 5 min at 100°C. The glycans on the other one-half of the filters were extracted and hydrolyzed twice for 2 h at 20°C with 50 mM H2SO4, with shaking at 1,500 rpm (14), followed by centrifugation for 15 min at 12,000 rpm and collection of the supernatant. All experiments were carried out in triplicate. Both enzymatically hydrolyzed and acid-hydrolyzed samples were used for reducing sugar assays.
Sampling and processing of environmental samples.
We used environmental samples derived from two different sites. In 2009, samples were collected during a North Sea spring algal bloom that was dominated by diatoms (57). Details about the sampling were described by Teeling et al. (26). In short, samples were taken at the long-term ecological research site near the North Sea island of Helgoland (Kabeltonne station, at 54°11′03″N, 7°54′00″E) on 31 March 2009. During the campaign, biomass from the sea surface was sampled by using sequential filtration through 142-mm-diameter, 10-μm- and 3-μm pore-size, TCTP polycarbonate filters (Millipore), with peristaltic pumping. The filters were frozen at −80°C. The filters were cut into pieces of equal size, and triplicates were treated in the same way as the laboratory cultures. Additionally, we analyzed samples obtained during the expedition PS93 cruise (summer 2015) to the Fram long-term ecological research site near Svalbard in the Arctic Ocean. Via a rosette water sampler on board the research vessel Polarstern, samples were taken at station EG-IV (78°50′07″N, 2°47′95″W) on 30 July 2015. Two liters of sampled water was filtered onto a GF/F glass microfiber filter and stored at −20°C until extraction. The samples were extracted for 60 min at 60°C with 5 ml of 50 mM MOPS buffer, with shaking at 1,500 rpm, followed by centrifugation for 15 min at 12,000 rpm and collection of the supernatant. The samples were hydrolyzed for 30 min at 37°C with 100 nM purified enzyme (∼5 μg ml−1 FaGH17A or FbGH30), with 1 mg ml−1 BSA, and the reaction was stopped by boiling the samples for 5 min at 100°C. The samples were measured using the PAHBAH reducing sugar assay.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Deutsche Forschungsgemeinschaft (grant HE 7217/1-1 to J.-H.H.) and by the Max Planck Society. This work was partially supported by a grant from the U.S. Department of Energy (grant DE-SC0008743 to M.F.P.).
We thank Rudolf Amann for reading and improving the manuscript and providing us with samples from the North Sea. We thank Hanno Teeling for supplying us with the FbGH30 sequence and Carol Arnosti for providing critical insight. Furthermore, we thank Morten Iversen for providing samples from the Arctic Ocean and the crew of the RV Polarstern for helping with sample acquisition.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03389-16.
REFERENCES
- 1.Field C, Behrenfeld M, Randerson J, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Myklestad S. 1974. Production of carbohydrates by marine planktonic diatoms. I. Comparison of nine different species in culture. J Exp Mar Biol Ecol 15:261–274. [Google Scholar]
- 3.Painter TJ. 1983. Algal polysaccharides, p 195–285. In Aspinall GO. (ed), The polysaccharides. Academic Press, New York, NY. [Google Scholar]
- 4.Senni K, Pereira J, Gueniche F, Delbarre-Ladrat C, Sinquin C, Ratiskol J, Godeau G, Fischer AM, Helley D, Colliec-Jouault S. 2011. Marine polysaccharides: a source of bioactive molecules for cell therapy and tissue engineering. Mar Drugs 9:1664–1681. doi: 10.3390/md9091664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gal A, Wirth R, Kopka J, Fratzl P, Faivre D, Scheffel A. 2016. Macromolecular recognition directs calcium ions to coccolith mineralization sites. Science 353:590–593. doi: 10.1126/science.aaf7889. [DOI] [PubMed] [Google Scholar]
- 6.Percival E, Ross A. 1951. The constitution of laminarin. Part II. The soluble laminarin of Laminaria digitata. J Chem Soc 720–726. [Google Scholar]
- 7.Chizhov AO, Dell A, Morris HR, Reason AJ, Haslam SM, McDowell RA, Chizhov OS, Usov AI. 1998. Structural analysis of laminarans by MALDI and FAB mass spectrometry. Carbohydr Res 310:203–210. doi: 10.1016/S0008-6215(98)00177-3. [DOI] [Google Scholar]
- 8.Read SM, Currie G, Bacic A. 1996. Analysis of the structural heterogeneity of laminarin by electrospray-ionisation-mass spectrometry. Carbohydr Res 281:187–201. doi: 10.1016/0008-6215(95)00350-9. [DOI] [PubMed] [Google Scholar]
- 9.Menshova R, Ermakova S, Anastyuk S, Isakov V, Dubrovskaya Y, Kusaykin M, Um B, Zvyagintseva T. 2014. Structure, enzymatic transformation and anticancer activity of branched high molecular weight laminaran from brown alga Eisenia bicyclis. Carbohydr Polym 99:101–109. doi: 10.1016/j.carbpol.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 10.Shin H, Oh S, Kim S, Won Kim H, Son J. 2009. Conformational characteristics of β-glucan in laminarin probed by terahertz spectroscopy. Appl Phys Lett 94:111911. doi: 10.1063/1.3100778. [DOI] [Google Scholar]
- 11.Usui T, Toriyama T, Mizuno T. 1979. Structural investigation of laminaran of Eisenia bicyclis. Agric Biol Chem 43:603–611. doi: 10.1271/bbb1961.43.603. [DOI] [Google Scholar]
- 12.Maeda M, Nishizawa K. 1968. Fine structure of laminaran of Eisenia bicyclis. J Biochem 63:199–206. doi: 10.1093/oxfordjournals.jbchem.a128762. [DOI] [PubMed] [Google Scholar]
- 13.Granum E, Kirkvold S, Myklestad S. 2002. Cellular and extracellular production of carbohydrates and amino acids by the marine diatom Skeletonema costatum: diel variations and effects of N depletion. Mar Ecol Prog Ser 242:83–94. doi: 10.3354/meps242083. [DOI] [Google Scholar]
- 14.Granum E, Myklestad S. 2002. A simple combined method for determination of β-1,3-glucan and cell wall polysaccharides in diatoms. Hydrobiologia 477:155–161. doi: 10.1023/A:1021077407766. [DOI] [Google Scholar]
- 15.Alderkamp A, van Rijssel M, Bolhuis H. 2007. Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol Ecol 59:108–117. doi: 10.1111/j.1574-6941.2006.00219.x. [DOI] [PubMed] [Google Scholar]
- 16.Ciais P, Sabine C, Bala G, Bopp L, Brovkin V, Canadell J, Chhabra A, DeFries R, Galloway J, Heimann M, Jones C, Le Quéré C, Myneni RB, Piao S, Thornton P. 2013. Carbon and other biogeochemical cycles, p 465–570. In Intergovernmental Panel on Climate Change (ed), Climate change 2013: the physical science basis. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 17.Hedges J, Baldock J, Gélinas Y, Lee C, Peterson M, Wakeham S. 2001. Evidence for non-selective preservation of organic matter in sinking marine particles. Nature 409:801–804. doi: 10.1038/35057247. [DOI] [PubMed] [Google Scholar]
- 18.Laine R. 1994. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 × 1012 structures for a reducing hexasaccharide: the Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology 4:759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- 19.Engel A, Händel N. 2011. A novel protocol for determining the concentration and composition of sugars in particulate and in high molecular weight dissolved organic matter (HMW-DOM) in seawater. Mar Chem 127:180–191. doi: 10.1016/j.marchem.2011.09.004. [DOI] [Google Scholar]
- 20.Gilbert H. 2010. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol 153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward OP, Moo-young M, Venkat K. 1989. Enzymatic degradation of cell wall and related plant polysaccharides. Crit Rev Biotechnol 8:237–274. doi: 10.3109/07388558909148194. [DOI] [PubMed] [Google Scholar]
- 22.Mäki-Arvela P, Salmi T, Holmbom B, Willför S, Murzin DY. 2011. Synthesis of sugars by hydrolysis of hemicelluloses: a review. Chem Rev 111:5638–5666. doi: 10.1021/cr2000042. [DOI] [PubMed] [Google Scholar]
- 23.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The Carbohydrate-Active Enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hehemann J-H, Boraston AB, Czjzek M. 2014. A sweet new wave: structures and mechanisms of enzymes that digest polysaccharides from marine algae. Curr Opin Struct Biol 28:77–86. doi: 10.1016/j.sbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Labourel A, Jam M, Legentil L, Sylla B, Hehemann JH, Ferrières V, Czjzek M, Michel G. 2015. Structural and biochemical characterization of the laminarinase ZgLamCGH16 from Zobellia galactanivorans suggests preferred recognition of branched laminarin. Acta Crystallogr D Biol Crystallogr 71:173–184. doi: 10.1107/S139900471402450X. [DOI] [PubMed] [Google Scholar]
- 26.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glöckner FO, Schweder T, Amann R. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 27.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tancula E, Feldhaus MJ, Bedzyk LA, Salyers AA. 1992. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J Bacteriol 174:5609–5616. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann AJ, Hahnke RL, Huang S, Werner J, Xing P, Barbeyron T, Huettel B, Stüber K, Reinhardt R, Harder J, Glöckner FO, Amann RI, Teeling H. 2013. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl Environ Microbiol 79:6813–6822. doi: 10.1128/AEM.01937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahnke RL, Bennke CM, Fuchs BM, Mann AJ, Rhiel E, Teeling H, Amann R, Harder J. 2015. Dilution cultivation of marine heterotrophic bacteria abundant after a spring phytoplankton bloom in the North Sea. Environ Microbiol 17:3515–3526. doi: 10.1111/1462-2920.12479. [DOI] [PubMed] [Google Scholar]
- 31.Labourel A, Jam M, Jeudy A, Hehemann J-H, Czjzek M, Michel G. 2014. The β-glucanase ZgLamA from Zobellia galactanivorans evolved a bent active site adapted for efficient degradation of algal laminarin. J Biol Chem 289:2027–2042. doi: 10.1074/jbc.M113.538843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varghese JN, Garrett TP, Colman PM, Chen L, Høj PB, Fincher GB. 1994. Three-dimensional structures of two plant β-glucan endohydrolases with distinct substrate specificities. Proc Natl Acad Sci U S A 91:2785–2789. doi: 10.1073/pnas.91.7.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reese E, Parrish F, Mandels M. 1962. β-d-1,6-Glucanases in fungi. Can J Microbiol 8:327–334. doi: 10.1139/m62-045. [DOI] [PubMed] [Google Scholar]
- 34.Oyama S, Yamagata Y, Abe K, Nakajima T. 2002. Cloning and expression of an endo-1,6-β-D-glucanase gene (neg1) from Neurospora crassa. Biosci Biotechnol Biochem 66:1378–1381. doi: 10.1271/bbb.66.1378. [DOI] [PubMed] [Google Scholar]
- 35.Jackson P. 1990. The use of polyacrylamide-gel electrophoresis for the high-resolution separation of reducing saccharides labelled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid. Biochem J 270:705–713. doi: 10.1042/bj2700705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lever M. 1972. A new reaction for colorimetric determination of carbohydrates. Anal Biochem 47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- 37.Woodward JR, Fincher GB. 1982. Substrate specificities and kinetic properties of two (1→3),(1→4)-β-d-glucan endo-hydrolases from germinating barley (Hordeum vulgare). Carbohydr Res 106:111–122. doi: 10.1016/S0008-6215(00)80737-5. [DOI] [Google Scholar]
- 38.Chen L, Sadek M, Stone BA, Brownlee RT, Fincher GB, Høj PB. 1995. Stereochemical course of glucan hydrolysis by barley (1→3)- and (1→3,1→4)-β-glucanases. Biochim Biophys Acta 1253:112–116. doi: 10.1016/0167-4838(95)00157-P. [DOI] [PubMed] [Google Scholar]
- 39.Moretti R, Thorson J. 2008. A comparison of sugar indicators enables a universal high-throughput sugar-1-phosphate nucleotidyltransferase assay. Anal Biochem 377:251–258. doi: 10.1016/j.ab.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiovitti A, Molino P, Crawford SA, Teng R, Spurck T, Wetherbee R. 2004. The glucans extracted with warm water from diatoms are mainly derived from intracellular chrysolaminaran and not extracellular polysaccharides. Eur J Phycol 39:117–128. doi: 10.1080/0967026042000201885. [DOI] [Google Scholar]
- 41.Kelly AE, Ou HD, Withers R, Dötsch V. 2002. Low-conductivity buffers for high-sensitivity NMR measurements. J Am Chem Soc 124:12013–12019. doi: 10.1021/ja026121b. [DOI] [PubMed] [Google Scholar]
- 42.Qin Z, Yan Q, Lei J, Yang S, Jiang Z, Wu S. 2015. The first crystal structure of a glycoside hydrolase family 17β-1,3-glucanosyltransferase displays a unique catalytic cleft. Acta Crystallogr D Biol Crystallogr 71:1714–1724. doi: 10.1107/S1399004715011037. [DOI] [PubMed] [Google Scholar]
- 43.Jeng W-Y, Wang N-C, Lin C-T, Shyur L-F, Wang AHJ. 2011. Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for -1,3- and -1,4-glucan selection. J Biol Chem 286:45030–45040. doi: 10.1074/jbc.M111.271213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hehemann J-H, Correc G, Thomas F, Bernard T, Barbeyron T, Jam M, Helbert W, Michel G, Czjzek M. 2012. Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans. J Biol Chem 287:30571–30584. doi: 10.1074/jbc.M112.377184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Oijen T, van Leeuwe MA, Gieskes WWC. 2003. Variation of particulate carbohydrate pools over time and depth in a diatom-dominated plankton community at the Antarctic Polar Front. Polar Biol 26:195–201. [Google Scholar]
- 46.Pakulski JD, Benner R. 1994. Abundance and distribution of carbohydrates in the ocean. Limnol Oceanogr 39:930–940. doi: 10.4319/lo.1994.39.4.0930. [DOI] [Google Scholar]
- 47.Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A, Crump BC, Dorrestein PC, Dyhrman ST, Hess NJ, Howe B, Longnecker K, Medeiros PM, Niggemann J, Obernosterer I, Repeta DJ, Waldbauer JR. 2016. Deciphering ocean carbon in a changing world. Proc Natl Acad Sci U S A 113:3143–3151. doi: 10.1073/pnas.1514645113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borch NH, Kirchman DL. 1997. Concentration and composition of dissolved combined neutral sugars (polysaccharides) in seawater determined by HPLC-PAD. Mar Chem 57:85–95. doi: 10.1016/S0304-4203(97)00002-9. [DOI] [Google Scholar]
- 49.McCarthy M, Hedges J, Benner R. 1996. Major biochemical composition of dissolved high molecular weight organic matter in seawater. Mar Chem 55:281–297. doi: 10.1016/S0304-4203(96)00041-2. [DOI] [Google Scholar]
- 50.Keith S, Arnosti C. 2001. Extracellular enzyme activity in a river-bay-shelf transect: variations in polysaccharide hydrolysis rates with substrate and size class. Aquat Microb Ecol 24:243–253. doi: 10.3354/ame024243. [DOI] [Google Scholar]
- 51.Repeta DJ, Aluwihare LI. 2006. Radiocarbon analysis of neutral sugars in high-molecular-weight dissolved organic carbon: implications for organic carbon cycling. Limnol Oceanogr 51:1045–1053. doi: 10.4319/lo.2006.51.2.1045. [DOI] [Google Scholar]
- 52.Bycroft M. 1999. The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease. EMBO J 18:297–305. doi: 10.1093/emboj/18.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibson D, Young L, Chuang R, Venter J, Hutchison C, Smith H. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 54.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Harrison PJ, Waters RE, Taylor FJR. 1980. A broad spectrum artificial sea water medium for coastal and open ocean phytoplankton. J Phycol 16:28–35. doi: 10.1111/j.0022-3646.1980.00028.x. [DOI] [Google Scholar]
- 56.Zha X, Xiao J, Zhang H, Wang J, Pan L, Yang X, Luo J. 2012. Polysaccharides in Laminaria japonica (LP): extraction, physicochemical properties and their hypolipidemic activities in diet-induced mouse model of atherosclerosis. Food Chem 134:244–252. doi: 10.1016/j.foodchem.2012.02.129. [DOI] [Google Scholar]
- 57.Wiltshire KH, Dürselen C-D. 2004. Revision and quality analyses of the Helgoland Reede long-term phytoplankton data archive. Helgol Mar Res 58:252–268. doi: 10.1007/s10152-004-0192-4. [DOI] [Google Scholar]
- 58.Brown MR. 1991. The amino-acid and sugar composition of 16 species of microalgae used in mariculture. J Exp Mar Biol Ecol 145:79–99. doi: 10.1016/0022-0981(91)90007-J. [DOI] [Google Scholar]
- 59.Størseth TR, Kirkvold S, Skjermo J, Reitan KI. 2006. A branched β-d-(1→3,1→6)-glucan from the marine diatom Chaetoceros debilis (Bacillariophyceae) characterized by NMR. Carbohydr Res 341:2108–2114. doi: 10.1016/j.carres.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 60.Myklestad S, Haug A. 1972. Production of carbohydrates by the marine diatom Chaetoceros affinis var. willei (Gran) Hustedt. I. Effect of the concentration of nutrients in the culture medium. J Exp Mar Biol Ecol 9:125–136. doi: 10.1016/0022-0981(72)90041-X. [DOI] [Google Scholar]
- 61.Li WKW. 1979. Cellular composition and physiological characteristics of the diatom Thalassiosira weissflogii adapted to cadmium stress. Mar Biol 55:171–180. [Google Scholar]
- 62.Guillard RR, Ryther JH. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can J Microbiol 8:229–239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.