ABSTRACT
The exopolysaccharide alginate, produced by the opportunistic human pathogen Pseudomonas aeruginosa, confers a survival advantage to the bacterium by contributing to the formation of characteristic biofilms during infection. Membrane-anchored proteins Alg8 (catalytic subunit) and Alg44 (copolymerase) constitute the alginate polymerase that is being activated by the second messenger molecule bis-(3′, 5′)-cyclic dimeric GMP (c-di-GMP), but the mechanism of activation remains elusive. To shed light on the c-di-GMP-mediated activation of alginate polymerization in vivo, an in silico structural model of Alg8 fused to the c-di-GMP binding PilZ domain informed by the structure of cellulose synthase, BcsA, was developed. This structural model was probed by site-specific mutagenesis and different cellular levels of c-di-GMP. Results suggested that c-di-GMP-mediated activation of alginate polymerization involves amino acids residing at two loops, including H323 (loop A) and T457 and E460 (loop B), surrounding the catalytic site in the predicted model. The activities of the respective Alg8 variants suggested that c-di-GMP-mediated control of substrate access to the catalytic site of Alg8 is dissimilar to the known activation mechanism of BcsA. Alg8 variants responded differently to various c-di-GMP levels, while MucR imparted c-di-GMP for activation of alginate polymerase. Furthermore, we showed that Alg44 copolymerase constituted a stable dimer, with its periplasmic domains required for protein localization and alginate polymerization and modification. Superfolder green fluorescent protein (GFP) fusions of Alg8 and Alg44 showed a nonuniform, punctate, and patchy arrangement of both proteins surrounding the cell. Overall, this study provides insights into the c-di-GMP-mediated activation of alginate polymerization while assigning functional roles to Alg8 and Alg44, including their subcellular localization and distribution.
IMPORTANCE The exopolysaccharide alginate is an important biofilm component of the opportunistic human pathogen P. aeruginosa and the principal cause of the mucoid phenotype that is the hallmark of chronic infections of cystic fibrosis patients. The production of alginate is mediated by interacting membrane proteins Alg8 and Alg44, while their activity is posttranslationally regulated by the second messenger c-di-GMP, a well-known regulator of the synthesis of a range of other exopolysaccharides in bacteria. This study provides new insights into the unknown activation mechanism of alginate polymerization by c-di-GMP. Experimental evidence that the activation of alginate polymerization requires the engagement of specific amino acid residues residing at the catalytic domain of Alg8 glycosyltransferase was obtained, and these residues are proposed to exert an allosteric effect on the PilZAlg44 domain upon c-di-GMP binding. This mechanism is dissimilar to the proposed mechanism of the autoinhibition of cellulose polymerization imposed by salt bridge formation between amino acid residues and released upon c-di-GMP binding, leading to activation of polymerization. On the other hand, conserved amino acid residues in the periplasmic domain of Alg44 were found to be involved in alginate polymerization as well as modification events, i.e., acetylation and epimerization. Due to the critical role of c-di-GMP in the regulation of many biological processes, particularly the motility-sessility switch and also the emergence of persisting mucoid phenotypes, these results aid to reach a better understanding of biofilm-associated regulatory networks and c-di-GMP signaling and might assist the development of inhibitory drugs.
KEYWORDS: Pseudomonas aeruginosa, alginate, polymerases
INTRODUCTION
Alginate is a secreted anionic exopolysaccharide composed of various proportions of 1,4-linked β-d-mannuronic acid (M) and its C-5 epimer α-l-guluronic acid (G), which is naturally produced by some bacteria and brown seaweeds (1). Understanding alginate biosynthesis has been significant for the scientific community due to its role in bacterial pathogenesis as well as the possibility to produce tailor-made alginates exhibiting material properties suitable for various medical and industrial applications. In the context of pathogenesis, Pseudomonas aeruginosa is the leading cause of chronic bronchopulmonary infection in cystic fibrosis patients and many nosocomial infections, associated with an extraordinary capability to form multicellular aggregates embedded in extracellular polymeric substances (EPS) known as biofilms. In bacteria, the secretion of EPS leads to the formation of a developed biofilm protecting embedded bacteria from environmental stresses as well as the immune system and antibiotic treatments (2). Alginate production by P. aeruginosa creates a survival advantage as it mediates the formation of persistent biofilms during chronic infections (1, 3–5). It reduces the diffusion of antibiotics, scavenges free radicals released by triggered mechanisms in the immune system, and interferes with cell-mediated killing of P. aeruginosa (6–8). For many years, alginate biosynthesis has been studied in P. aeruginosa as a model organism. Briefly, bacterial alginate is synthesized by an envelope-spanning multiprotein complex consisting of proteins Alg8 and Alg44 (polymerization), AlgG (epimerization), AlgX (acetylation), and AlgK and AlgE (secretion), with the possible involvement of AlgI-AlgJ-AlgF (acetylation) and AlgL (alginate degradation) (9–11). These proteins are responsible for processive polymerization of alginate, translocation of nascent polymer (poly-M) across the periplasm, modification events in the periplasm (i.e., epimerization of M to G residues and acetylation), and secretion of modified alginate (poly-MG) through the outer membrane (9–11).
However, alginate biosynthesis is under the control of the second messenger bis-(3′, 5′)-cyclic dimeric GMP (c-di-GMP). c-di-GMP signaling is very complex and almost ubiquitous in bacteria, controlling various biological processes during motility-sessility switch and biofilm formation (12). Adjustment of cellular levels of c-di-GMP are critical for binding to specific receptor/effector proteins, mainly via the PilZ domain in order to reach a desired physiological output such as exopolysaccharide production (13, 14). In this regard, the molecular mechanism of posttranslational activation of alginate polymerization by c-di-GMP is still unknown. The interacting cytoplasmic membrane-anchored proteins Alg8 and Alg44 constitute the catalytic unit of the alginate polymerase, which is activated by binding of a dimeric form of c-di-GMP to the Alg44 PilZ domain in the cytoplasm (11, 15).
In this study, we investigated the molecular mechanism of the c-di-GMP-mediated activation of alginate polymerization by applying an in silico model of Alg8 and PilZAlg44 domain to mimic the structure of BcsA protein (cellulose synthase). BcsA was chosen because it has the c-di-GMP-sensing PilZ domain mediating the activation of the polymerization of cellulose exopolysaccharide in Rhodobacter sphaeroides, which is controlled by a proposed autoinhibition mechanism (16). This mechanism is referred to the steric hindrance by salt bridge formation in proximity to the catalytic site of BcsA, which is formed between the first arginine of the PilZ domain's R580XXXR584 motif and E371 preceding the RW motif (a signature of the glycosyltransferase family 2). Consequently, the BcsA gating loop adopts a resting state and blocks the catalytic site. This steric hindrance was proposed to be eliminated upon c-di-GMP binding to R580, opening the gate for substrate to enter into the catalytic site (16, 17). To this end, various site-specific mutageneses were performed, informed by predicted models. The impacts of respective Alg8 variants on alginate production were tested in PDO300 with either single or double deletions of alg8 and alg44, respectively, at high and low cellular levels of c-di-GMP mediated by the activity of c-di-GMP metabolizing enzymes MucR and RocR. Furthermore, the role of Alg44 proposed as alginate copolymerase was assessed through site-specific mutagenesis with respect to in vivo alginate polymerase activity, alginate modification (i.e., acetylation and epimerization), and protein localization. Subcellular localization of Alg8 and Alg44 was visualized by generating translational fusions with a superfolding derivative of green fluorescent protein (sfGFP) and confocal laser scanning microscopy.
RESULTS
How is alginate polymerization regulated by a c-di-GMP-dependent mechanism?
The second messenger c-di-GMP is required for alginate production via interaction with the PilZ domain of membrane-anchored Alg44, which itself interacts with Alg8 glycosyltransferase (alginate polymerase) (9, 11). Previously, it was shown that alteration of intracellular levels of c-di-GMP is critical for engaging c-di-GMP receptor/effector proteins to trigger specific outputs (13, 18). In the regulation of alginate biosynthesis, RocR (a phosphodiesterase degrading c-di-GMP) and MucR (a diguanylate cyclase synthesizing c-di-GMP) are important in c-di-GMP turnover, and they negatively and positively regulate alginate biosynthesis, respectively (19). However, the activation mechanism of alginate polymerization via Alg8 glycosyltransferase as a target in response to different c-di-GMP levels remains unknown.
c-di-GMP-mediated activation of cellulose synthesis via the BcsA protein in R. sphaeroides provides currently the best-understood mechanistic model (16). Furthermore, bioinformatic analysis showed that both Alg8 (PA3541) and BcsA (accession number 5EIY_A; Rhodobacter sphaeroides 2.4.1) belong to the glycosyltransferase family 2 (GT-2), and they share the same conserved signature motifs and residues (13.7% identity and 23.6% similarity), which were experimentally shown to be critical for production of alginate and cellulose, respectively (11, 17).
This experiment aimed at unraveling the molecular mechanism of c-di-GMP-mediated activation of alginate polymerization in comparison to the c-di-GMP-dependent activation of cellulose synthesis while also considering the importance of different levels of c-di-GMP. Informed by the BcsA structure, an in silico structural model of Alg8 (amino acids [aa] 1 to 494) C-terminally fused to the PilZAlg44 domain (aa 1 to 122 of Alg44) was developed by using the Phyre2 Protein Fold Recognition Server (20). The Phyre2 server relies on the key principle that protein structure is more conserved than protein sequence in evolution (21). Hence, this prediction server enables reliable prediction of the three-dimensional structure of a given protein by using a library of well-known protein structures and advanced remote homology detection (21).
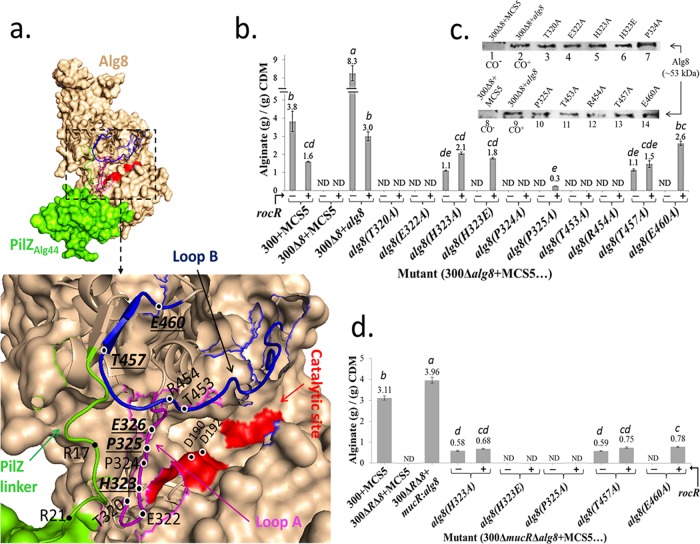
The overall Phyre2 output was a structural model homologous to BcsA (confidence, 100%; coverage, 84%) with the C-terminal part aligned with the crystal structure of the PilZ domain from Alg44 (15) (Fig. 1a; see also Fig. S1 in the supplemental material). This model showed the full incorporation of the PilZ domain into the last C-terminal transmembrane helix domain of Alg8 (TM4) through a linker and a two-stranded β-sheet formation with one strand located at the C-terminal end of a long loop of Alg8 running across the catalytic pocket (Fig. 1a; Fig. S1). This loop, designated here loop B and comprising residues I441 to G462, was homologous to the gating loop (residues R499 to I517) in the BcsA structure, which also contained BcsA's T511 residue conferring the autoinhibition mechanism found in cellulose synthesis. This loop is also critical for relocating the precursor of cellulose into the catalytic site at open-rest transition (16). Another predicted loop of Alg8-PilZAlg44, designated here loop A and comprising residues T320 to E326, was homologous to BcsA's loop harboring E371 of the glycosyltransferase domain, which further stabilizes the autoinhibition mechanism by salt bridge formation (16). Based on this model, we hypothesized that the amino acid residues located within predicted loops A and B surrounding the Alg8 catalytic site potentially affect the response of Alg8 to c-di-GMP binding at its different cellular levels, resulting in the alteration of the alginate production. To test this hypothesis, substitutions of alanine for cytoplasmic amino acid residues surrounding the catalytic site of Alg8 were conducted. These included T320, E322, H323, P324, and P325 from loop A and T453, R454, T457, and E460 from loop B (Fig. 1a; Fig. S1). In addition, the mutation H323E (for the effect of opposite charge substitution) and combinational mutations E322A/H323E, P324A/P325A, T453A/T457A, and R454A/T457A were introduced and analyzed. Then, all the above-mentioned site-specific alg8 mutants were expressed in trans, i.e., using respective pBBR1MCS-5 derivatives, in PDO300Δalg8 to assess alginate production in the absence or presence of overproduced c-di-GMP-degrading RocR, providing permissive (high level of c-di-GMP) and nonpermissive (low level of c-di-GMP) conditions, respectively.
FIG 1.
Highly conserved amino acids of Alg8 are involved in c-di-GMP-dependent regulation of alginate polymerization. (a) The in silico fusion of Alg8-PilZAlg44 was modeled using the Phyre2 server. Residues selected for site-specific mutagenesis were shown on loop A (magenta), loop B (blue), and the PilZ domain (green). Mutations of bolded and underlined residues were responsive to the absence and presence of RocR overproduction (i.e., reduced levels of c-di-GMP), while for other shown residues alginate production was abolished independent of RocR. (b) Alginate quantification of PDO300Δalg8 transformants harboring various plasmids containing respective site-specific mutants of alg8 with (+) and without (−) the rocR gene. (c) Immunoblot analysis of envelope fractions developed using an anti-Alg8 antibody showed that none of mutations affected the Alg8 localization to the envelope fraction (lanes 3 to 7 and lanes 10 to 14). Lanes 1 to 8 and 2 to 9 represent negative and positive controls, respectively. For estimating relative protein amounts, the protein band intensity was analyzed by the ImageJ software. The protein band based on genomic expression derived from 300Δ8+alg8 (lanes 2 and 9) was set as 1.0 in density, and relative densities of other bands were calculated as follows: lane 3 (1.14), lane 4 (0.937), lane 5 (1.06), lane 6 (1.03), lane 7 (1.48), lane 10 (0.9), lane 11 (0.848), lane 12 (0.817), lane 13 (0.985), and lane 14 (0.987). (d) Highly conserved amino acids of Alg8 whose replacement with alanine decoupled alginate polymerization from c-di-GMP-dependent and MucR-dependent regulation. Alginate quantification was performed for PDO300ΔmucRΔalg8 transformants with plasmids harboring respective site-specific mutants of alg8 with (+) and without (−) the rocR gene. The data in histograms in panels b and d represent the means ± SD for four independent repetitions, and treatments with different lowercase italic letters above the bars are significantly different (post hoc Tukey's HSD test, P < 0.05). CDM, cell dry mass; 300, PDO300; ND, not detectable; MCS5, pBBR1MCS-5.
Alginate quantification data (Fig. 1b; see also Fig. S2 in the supplemental material) showed the following: (i) alanine substitutions of Alg8 amino acid residues T320, E322, and P324 (on loop A) and T453 and R454 (on loop B) abolished alginate polymerization independent of the c-di-GMP level, i.e., the presence or absence of overproduced RocR; (ii) alanine substitutions of H323 and P325 (on loop A) and T457 and E460 (on loop B) resulted in either abolished or significantly reduced alginate production in the absence of RocR (i.e., permissive levels of c-di-GMP), but they were positively responsive to the presence of RocR (i.e., lower nonpermissive levels of c-di-GMP) as shown by increased alginate production. Remarkably, the point mutation E460A on loop B abolished alginate production while it restored alginate production upon the presence of RocR overproduction to a level similar to that obtained for the nonmutated control strain PDO300Δalg8 (pBBR1MCS-5:alg8:rocR) (Fig. 1b; Fig. S2).
Interestingly, the replacement of H323 with negatively charged glutamic acid, PDO300Δalg8(pBBR1MCS-5:alg8) (H323E), abolished alginate production in the absence of RocR overproduction, while alginate production was restored to 60% of the nonmutated strain PDO300Δalg8(pBBR1MCS-5:alg8:rocR) and to 86% of the H323A mutant in the presence of RocR overproduction (Fig. 1b; Fig. S2).
We also applied the c-di-GMP nonbinding variant of Alg44 (i.e., R17A and R21A) in combination with the responsive group of Alg8 variants (i.e., H323A, H323E, P325A, T457A, and E460A) (Table 1) to assess the possibility of alginate production independent of c-di-GMP in the PDO300Δalg8Δalg44 mutant (Table 1). None of these Alg8 variants could restore alginate production (data not shown), indicating that binding of c-di-GMP to the PilZ domain of Alg44 is required for activation of alginate production, presumably via conformational change of the PilZAlg44 domain as observed by Whitney and coworkers (15).
TABLE 1.
Strains, plasmids, and primers used in this study
| Strains, plasmids, and primers | Descriptiona | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PDO300 | mucA22 isogenic mutant derived from PAO1, Alg+ | 53 |
| PDO300Δalg8 | Isogenic alg8 deletion mutant derived from PDO300, Alg− | 26 |
| PDO300Δalg44 | Isogenic alg44 deletion mutant derived from PDO300, Alg− | 23 |
| PDO300Δalg8Δalg44 | Isogenic alg8 and alg44 deletion mutant derived from PDO300, Alg− | 11 |
| PDO300ΔmucR | Isogenic mucR deletion mutant derived from PDO300 | 19 |
| PDO300ΔmucRΔalg8 | Isogenic mucR and alg8 deletion mutant derived from PDO300, Alg− | This study |
| E. coli | ||
| Top10 | Cloning strain; F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| XL1 Blue | Cloning strain; recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 Δ(lac-proAB) [F′ proAB lacIq lacZΔM15 Tn10(Tetr)] | 54 |
| S17-1 | Donor strain in transconjugation; thi-1 proA hsdR17 (rK− mK+) recA1; tra gene of plasmid RP4 integrated in chromosome | 55 |
| SM10 | Donor strain for pFLP2 plasmid; thi-1 thr-1 leuB6 supE44 tonA21 lacY1 recA::RP4-2-Tc::Mu Kmr | 55 |
| BL21(DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| C41(DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Lucigen |
| C43(DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Lucigen |
| Origami (DE3) | F− ompT hsdSB (rB− mB−) gal dcm lacY1 ahpC (DE3) gor522::Tn10 trxB (Kanr Tetr) | Novagen |
| ClearColi | F− ompT hsdSB (rB− mB−) gal dcm lon λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) msbA148 ΔgutQ ΔkdsD ΔlpxL ΔlpxM ΔpagP ΔlpxP ΔeptA | Lucigen |
| Rosetta | F− ompT hsdSB (rB− mB−) gal dcm pRARE (Camr) | Novagen |
| Plasmids | ||
| pETDuet-1 | Ampr; coexpression of two target genes; carries the pBR322-derived ColE1 replicon lacI gene PT7 | Novagen |
| pETDuet-1:alg44-12His | NcoI-BamHI fragment comprising alg44-12× histidine tag fusion and the codon-optimized E. coli | This study |
| pBBR1MCS-5 | Gmr; broad-host-range vector; Plac | 46 |
| pBBR1MCS-5:alg8 | HindIII-PstI fragment comprising alg8 inserted into vector pBBR1MCS-5 | 26 |
| pBBR1MCS-5:alg44 | HindIII-BamHI fragment comprising alg44 inserted into vector pBBR1MCS-5 | 23 |
| pBBR1MCS-5:alg44-6his | Translational alg44-hexahistidine tag fusion inserted into vector pBBR1MCS-5 | 23 |
| pBBR1MCS-5:mucR | KpnI-ClaI fragment comprising mucR inserted into vector pBBR1MCS-5 | 19 |
| pBBR1MCS-5:mucR:alg8 | KpnI-ClaI fragment comprising mucR and HindIII-BamHI fragment comprising alg8 inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:pilN | HindIII-EcoRI fragment comprising pilN inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(Δstop):gfp | HindIII-EcoRI fragment comprising alg8 without stop codon and PstI-XbaI fragment comprising gfp inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(Δstop):sfgfp | HindIII-EcoRI fragment comprising alg8 without stop codon and PstI-XbaI fragment comprising sfgfp inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(Δstop):gfp | HindIII-EcoRI fragment comprising alg44 without stop codon and PstI-XbaI fragment comprising gfp inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(Δstop):sfgfp | HindIII-EcoRI fragment comprising alg44 without stop codon and PstI-XbaI fragment comprising sfgfp inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:pilN(Δstop):sfgfp | HindIII-EcoRI fragment comprising pilN without stop codon and PstI-XbaI fragment comprising sfgfp inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T320A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T320A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T320A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T320A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(H323A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis H323A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(H323A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis H323A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(P324A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis P324A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(P324A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis P324A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(P325A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis P325A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(P325A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis P325A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(P325A):alg44(R17A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis P325A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R17A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(P325A):alg44(R17A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis P325A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R17A and XbaI-SacI fragment encoding RocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(P325A):alg44(R21A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis P325A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R21A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(P325A):alg44(R21A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis P325A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R21A and XbaI-SacI fragment encoding RocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T453A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T453A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T453A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T453A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(R454A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis R454A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(R454A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis R454A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T457A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T457A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T457A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T457A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T457A):alg44(R17A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T457A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R17A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T457A):alg44(R17A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T457A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R17A inserted into vector pBBR1MCS-5 and XbaI-SacI fragment encoding RocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T457A):alg44(R21A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T457A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R21A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T457A):alg44(R21A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T457A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R21A inserted into vector pBBR1MCS-5 and XbaI-SacI fragment encoding RocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E460A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E460A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E460A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E460A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E460A):alg44(R17A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E460A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R17A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E460A):alg44(R17A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E460A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R17A inserted into vector pBBR1MCS-5 and XbaI-SacI fragment encoding RocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E460A):alg44(R21A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E460A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R21A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E460A):alg44(R21A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E460A and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R21A inserted into vector pBBR1MCS-5 and XbaI-SacI fragment encoding RocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(H323E) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis H323E inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(H323E):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis H323E and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(H323E):alg44(R17A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis H323E and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R17A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(H323E):alg44(R17A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis H323E and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R17A inserted into vector pBBR1MCS-5 and XbaI-SacI fragment encoding RocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(H323E):alg44(R21A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis H323E and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R21A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(H323E):alg44(R21A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis H323E and ClaI-HindIII fragment comprising alg44 encoding site-directed mutagenesis R21A inserted into vector pBBR1MCS-5 and XbaI-SacI fragment encoding RocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E322A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E322A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E322A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E322A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E322A/H323E) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E322A and H323E inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(E322A/H323E):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis E322A and H323E and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T453A/T457A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T453A and T457A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(T453A/T457A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis T453A and T457A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(R454A/T457A) | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis R454A and T457A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg8(R454A/T457A):rocR | HindIII-PstI fragment comprising alg8 encoding site-directed mutagenesis R454A and T457A and XbaI-SacI fragment comprising rocR (PA3947) inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(Q258A)-6his | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis Q258A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(M259A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis M259A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(K260A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis K260A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(T264A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis T264A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(S265A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis S265A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(P266A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis P266A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(C267A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis C267A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(D268A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis D268A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(C269A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis C269A inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:alg44(C267A/C269A)-6His | HindIII-BamHI fragment comprising alg44 encoding translational Alg44-hexahistidine tag fusion with site-directed mutagenesis C267A and C269A inserted into vector pBBR1MCS-5 | This study |
| Mini-CTX-lacZ | Chromosomal integration vector at the CTX phage att site on the P. aeruginosa chromosome Tcr | 56 |
| Mini-CTX:Palg44-6his | PstI-HindIII fragment encoding alginate operon promoter (PalgD) and HindIII-BamHI fragment encoding C-terminally hexahistidine-tagged Alg44 inserted into vector Mini-CTX-lacZ | 11 |
| Primers | ||
| Alg8Fw(T453A) | aagcaagaagatctacgtgttcttccgcctcgaccggcagtcctggGCGcgccagccgaccaagctggagcgcggcctggcca | This study |
| Alg8Fw(T457A) | aagcaagaagatctacgtgttcttccgcctcgaccggcagtcctggacccgccagccgGCGaagctggagcgcggcctggcca | This study |
| Alg8Fw(T453A-T457A) | aagcaagaagatctacgtgttcttccgcctcgaccggcagtcctggGCGcgccagccgGCGaagctggagcgcggcctggcca | This study |
| Alg8Fw(R454A) | aagcaagaagatctacgtgttcttccgcctcgaccggcagtcctggaccGCgcagccgaccaagctggagcgcggcctggcca | This study |
| Alg8Fw(R454A-T457A) | aagcaagaagatctacgtgttcttccgcctcgaccggcagtcctggaccGCgcagccgGCGaagctggagcgcggcctggcca | This study |
| Alg8Fw(E460A) | aagcaagaagatctacgtgttcttccgcctcgaccggcagtcctggacccgccagccgaccaagctgGCGcgcggcctggccagcttccagcgctggtt | This study |
| Alg8Rev-Alg8End | aagcaagactgcagaaatcatacgatggtcagcagcacggcgacgaagatgctggcgg | This study |
| AlgFw-AatII | aagcaagagacgtcgagaacgaccacctggagcactggcgcctgggtcgct | This study |
| Alg8Rv(P324A) | aagcaagagaagctcttctccggCGCgtgctcgaccgtgttgatcgccgcgtcgggcacgtagaaggt | This study |
| Alg8Rv(P325A) | aagcaagagaagctcttctcCGCcgggtgctcgaccgtgttgatcgccgcgtcgggcacgtagaaggt | This study |
| Alg8Rv(P324A-P325A) | aagcaagagaagctcttctcCGCCGCgtgctcgaccgtgttgatcgccgcgtcgggcacgtagaaggt | This study |
| Alg8Rv(H323E) | aagcaagagaagctcttctccggcggCTCctcgaccgtgttgatcgccgcgtcgggcacgtagaaggt | This study |
| Alg8Rv(E322H+H323E) | aagcaagagaagctcttctccggcggCTCGTGgaccgtgttgatcgccgcgtcgggcacgtagaaggt | This study |
| Alg8Rv(T320A) | aagcaagagaagctcttctccggcgggtgctcgacCGCgttgatcgccgcgtcgggcacgtagaaggt | This study |
| Alg8Rv(T320A+E322A) | aagcaagagaagctcttctccggcgggtgCGCgacCGCgttgatcgccgcgtcgggcacgtagaaggt | This study |
Lowercase letters in primer sequences are unchanged sequences; uppercase letters are changed sequences for the generation of site-specific mutagenesis.
To assess whether the resulting different levels of alginate production obtained from various Alg8 variants were due to impaired function of Alg8 rather than impaired protein localization or protein production level, the strains producing the respective Alg8 variants were subjected to protein analysis. The envelope fractions of the cells were isolated and assessed via immunoblotting using anti-Alg8 antibodies. The results showed that corresponding protein bands were detected for all Alg8 variants exhibiting an intensity similar to that seen with the positive control, i.e., PDO300Δalg8(pBBR1MCS-5:alg8) (Fig. 1c). This result indicated that the replacement of any of the amino acid residues tested did not affect the overall structure of Alg8, suggesting an important role in polymerization rather than Alg8 localization.
Overall, these results indicated that predicted loops A and B of Alg8 and amino acid residues H323, P325, T457, and E460 surrounding the catalytic site might be involved in the c-di-GMP-mediated activation of alginate polymerization.
Furthermore, the replacement of the catalytic domain (residues 71 to 381) loops A and B of P. aeruginosa Alg8 with the algal (Ectocarpus siliculosus) homologous sequences did not restore alginate production either with or without RocR overproduction (see Fig. S1c in the supplemental material, underlined sequences). In addition, hybrid genes encoding various fusion proteins of Alg8-Alg44-PilZAlg44 could not restore alginate production in the PDO300Δalg8Δalg44 mutant (see Fig. S3 in the supplemental material). However, it cannot be excluded that the lack of functionality is due to instability or impaired localization of the fusion proteins.
Decoupling alginate polymerization from MucR-dependent regulation.
The GGDEF-EAL motif present in MucR was proposed to specifically regulate alginate synthesis by imparting a localized c-di-GMP pool for the PilZ domainAlg44-mediated activation of alginate polymerization in response to environmental stimuli (19, 22). We hypothesized that the c-di-GMP-responsive group of Alg8 variants (i.e., H323A, H323E, P325A, T457A, and E460A) might also respond to the absence of MucR and the associated elimination of the proposed c-di-GMP pool. Therefore, the impact of these variants on alginate production was analyzed in nonmucoid mutant PDO300ΔmucRΔalg8 with and without RocR overproduction (Fig. 1d).
The variants H323E and P325A with RocR overproduction could not restore alginate production in PDO300ΔmucRΔalg8 to a detectable level, while they did in PDO300Δalg8 as described above (i.e., when MucR was present) (Fig. 1b and d; see also Fig. S2 and S4 in the supplemental material). However, the variants H323A, T457A, and E460A were able to restore alginate production in the absence of MucR in PDO300ΔmucRΔalg8, but interestingly, the respective alginate production levels were not significantly different from each other. On the other hand, compared with their activity in PDO300Δalg8 (i.e., in the presence of MucR), they were significantly reduced, up to 3.4-fold (Fig. 1b and d; Fig. S2 and S4).
These results confirmed that MucR specifically regulates alginate polymerization and that alanine substitution of H323, T457, and E460 of Alg8 decoupled activation of alginate polymerization from c-di-GMP. However, the analysis of other existing c-di-GMP-synthesizing and -degrading enzymes in P. aeruginosa and their impact on c-di-GMP levels could provide further insight into conditions required for c-di-GMP-mediated activation of alginate production.
A schematic representation of mutual and combinational effects of Alg8 variants MucR and RocR on alginate polymerization is provided in Fig. S3. This model shows the various Alg8 variants' interplay with the various levels of c-di-GMP in view of activation of alginate polymerization. Remarkably, high c-di-GMP levels caused inhibition in H323E and E460A variants (see Fig. S5, row a, in the supplemental material), while RocR-mediated reduction of c-di-GMP levels favored the activation of alginate polymerization (Fig. S5, row b). Interestingly, H323A, T457A, and E460A variants retained functionality independent from c-di-GMP depletion due to the absence of MucR and the presence of RocR overproduction (Fig. S5, row d).
The periplasmic domain of Alg44 copolymerase is important for alginate polymerization and modification events.
Alg44 is essential for alginate polymerization through its interaction with Alg8 and binding to c-di-GMP (11). Alg44 has been proposed as an alginate copolymerase that consists of three distinct domains including the c-di-GMP-binding cytoplasmic PilZ domain, a transmembrane domain, and a periplasmic domain (Fig. 2a) (23, 24). The periplasmic domain has been proposed to contribute to the periplasmic scaffold bridging the inner and outer membranes, but its functional role in alginate biosynthesis remains unexplored.
FIG 2.
Role of conserved amino acid residues of Alg44 proposed to be localized to the periplasmic domain in alginate biosynthesis and purification of the Alg44 dimer produced by recombinant P. aeruginosa. (a) Uniprot analysis of the periplasmic domain of Alg44 shows the alignment of highly conserved regions among different alginate-producing species P. aeruginosa (PA), Azotobacter vinelandii (AZ), Pseudomonas fluorescens (PF), Pseudomonas putida (PP), and Pseudomonas syringae (PS). Dashed lines show the positions of these highly conserved regions in the Alg44 model predicted by the Phyre2 server. Alginate quantification showed that P266A, C267A, and/or C269A completely abolished alginate production and that for other residues it was significantly reduced. The data represent the means ± SD for four repetitions, and treatments with different lowercase italic letters above the bars are significantly different (post hoc Tukey's HSD test, P < 0.05). (b) Production of free uronic acids in liquid culture mediated by variants of Alg44 (flowthrough samples were obtained using filters with a 10-kDa cutoff), indicating that alginate polymerization was impaired by site-specific mutagenesis of highly conserved periplasmic amino acid residues of Alg44. The data represent the means ± the SD for four repetitions, and asterisks indicate pairs of significantly different values (post hoc Tukey's HSD test: *, P < 0.05; ***, P < 0.001). (c) Immunoblot analysis of envelope fractions developed using anti-His tag antibodies showed that mutations M259A, P266A, C267A, D268A, and C269A completely disrupted Alg44 localization to the envelope fraction (lane 2 and lanes 6 to 10). The intensity of other protein bands was consistent with the amount of alginate produced by complemented mutants (lanes 1, 3 to 5, and 12). For estimating relative protein amounts, the protein band intensity was analyzed by the ImageJ software. The band of genomic expression of alg44-6His (lane 11) was set as 1.0 in density, and the relative densities of other bands were calculated as follows: lane 1 (0.76), lane 3 (0.76), lane 4 (0.29), lane 5 (0.13), and lane 12 (2.1). Lanes 13 and 14 represent negative controls. (d) Composition of alginates impacted by various Alg44 variants (see also Tables S1 and S2 in the supplemental material). Ac.%, percentage of acetylation; FG, molar fraction of guluronate (G) residue. (e) Purification of the Alg44 dimer. SDS-PAGE gel (stained with Coomassie brilliant blue) (lanes 1 to 3) and an immunoblot (lanes 4 to 6) of peak I (Fig. S7) showed a very stable dimer plus the monomer bands of Alg44. TM, transmembrane domain; CDM, cell dry mass; 300, PDO300; MCS5, pBBR1MCS-5; ND, not detectable.
Bioinformatic analysis using the Universal Protein Resource (UniProt) server showed highly conserved clustered residues 258QMKGTLTSPCDC269 in the periplasmic domain of Alg44 of alginate-producing Pseudomonas and Azotobacter species (Fig. 2a). Therefore, these amino acid residues were subjected to site-specific mutagenesis to assess their impact on alginate polymerization, Alg44 production, and localization in PDO300Δalg44 mutant (Table 1).
Site-specific mutagenesis of these residues with alanine either abolished or significantly reduced alginate production compared to its levels in the wild-type strain and in PDO300Δalg44(pBBR1MCS-5:alg44-6His) (Fig. 2a; see also Fig. S6 in the supplemental material). Except for variant D268A, which produced small amounts of alginate, alanine substitution in each residue in the PCDC (residues 266 to 269) cluster abolished alginate production. Additionally, alanine substitution of M259 resulted in a significant decrease in alginate production (Fig. 2a; Fig. S6).
We hypothesized that if polymerization occurs but the alginate is misguided into the periplasm, it must be secreted as uronic acid as a consequence of periplasmic alginate lyase AlgL-mediated degradation (25). To test this hypothesis, an alginate oligomer/uronic acid detection assay was used. Statistical analyses did not show a significant difference between the total uronic acid amount (i.e., high- and low-molecular-mass alginate) and the low-molecular-mass fraction (filtrate, 10-kDa cutoff) for all variants, except for S265A (P < 0.05), which suggested that the reduction in alginate production was due to impaired polymerization (Fig. 2b). For P266A, C267A, and C269A, the total uronic acid amount and the low-molecular-mass fraction were equal, indicating that alginate polymerization was completely abolished (Fig. 2b). The ratio of high- to low-molecular-mass alginate produced by the wild-type strain and PDO300Δalg44(pBBR1MCS-5:alg44) was significantly (P < 0.001) different from PDO300Δalg44 producing Alg44 variants (Fig. 2b). Furthermore, the impact of amino acid replacements on protein localization was analyzed. Immunoblotting using anti-His tag antibodies showed that no Alg44 variant harboring alanine substitutions of P266, C267, and C269, separately, was detected (Fig. 2c). However, Alg44 variants M259A and D268A, which mediated very low alginate yields on solid culture, were not detectable by immunoblotting (Fig. 2a and c). These results indicated that these amino acids contribute to the integrity of the proposed periplasmic scaffold, which is necessary for accomplishment of polymerization.
Furthermore, the possibility that the two cysteine residues (residues 267 and 269) mediate disulfide bond formation for localization of protein or inter- and intramolecular interactions was investigated. Immunoblotting analysis of dithiothreitol (DTT)-treated and untreated membrane fractions of PDO300Δalg44 complemented with alg44-6His either in cis or in trans expression showed a higher-molecular-mass band (∼70 kDa) when Alg44 was encoded in cis and in the absence of DTT, while it was missing when treated with DTT, i.e., under reducing conditions (see Fig. S7 in the supplemental material). This difference equals to ∼25 kDa, and the identity of this interacting partner could not be confirmed because protein quantities were too low. This band was not detected when Alg44 was encoded by the respective plasmids in trans (Fig. S7). The lack of Alg44 in samples corresponding to alanine substitution of cysteine residues reconfirmed their role in the localization of Alg44 (Fig. S7).
Alg44 regulates modification of alginate through its periplasmic domain.
Previously, our in vivo assessment had shown that alginate polymerization is linked with modification events, including acetylation and epimerization (11). Alg44 overproduction was shown to have an impact on alginate composition by affecting modification events while it structurally interacts with Alg8 (alginate polymerase) and the periplasmic subunits (i.e., AlgX and AlgK) to constitute the proposed periplasmic scaffold of the multiprotein complex (9, 11). We hypothesized that impaired polymerization of alginates observed in previous experiments must impact the modification events too. In effect, compositional analysis of alginates produced by Alg44 variants grown on solid media showed that the replacement of respective residues with alanine (i.e., Q258A, M259A, K260A, T264A, S265A, and D268A) decreased the acetylation degree to 34% to 52%, whereas in wild-type Alg44, the acetylation rate was 64% (Fig. 2d; see also Tables S1 and S2 in the supplemental material). Among them, M259A and D268A, which also showed impaired localization, revealed the highest impact on acetylation degree with 38% and 34%, respectively (Fig. 2d; Tables S1 and S2).
Additional copy numbers of Alg44 contributed to a lower molar fraction of G residues (FG = 0.14), which was consistent with previous results (11). Interestingly, S265A, whose impact on polymerization was less than by the other variants, did not change the molar fraction of G residues, while other amino acid residue substitutions increased the molar fraction of G residues to 0.2 to 0.23 (Fig. 2d; Tables S1 and S2). M259A showed the highest epimerization degree (FG = 0.3) (Fig. 2d; Tables S1 and S2). These data demonstrated that the periplasmic domain of Alg44 has a regulatory impact on alginate modification events. Additional copy numbers of Alg44 boosted acetylation and lowered epimerization (Tables S1 and S2), while alanine substitution of periplasmic residues impaired this regulatory effect.
Alg44 forms a homodimer in P. aeruginosa.
In order to analyze the quaternary structure of Alg44, both heterologous and homologous productions of the full-length alg44-12His gene were assessed. Briefly, due to weak production and significant truncation of Alg44 in Escherichia coli strains, heterologous production was found to be not suitable (see Fig. S8 in the supplemental material). However, homologous overproduction of Alg44 in P. aeruginosa was continued using the mutant PDO300Δalg44 complemented with the integration of alg44-6His into the genome. Importantly, initial protein analysis using immunoblotting showed that the presence of 5 mM EDTA could significantly reduce proteolytic truncation of Alg44 (see Fig. S9 in the supplemental material). This suggested that metalloproteases might have mediated the degradation. Membrane fractions of disrupted cells were subjected to affinity chromatography purification in the presence of a cocktail of protease inhibitor, 5 mM EDTA, and DTT. Interestingly, two distinct bands corresponding to the molecular masses of the monomer and dimer of Alg44 were detected using immunoblotting (Fig. S9).
To further purify Alg44, size exclusion chromatography was employed. The chromatogram showed two major peaks (see Fig. S10 in the supplemental material). Interestingly, when protein samples were treated with 0.2% alginate solution and DTT, impurities were dissociated from the two major peaks, in contrast to what was seen in untreated samples (Fig. S10a and b). Protein analysis showed that the first major peak (I) with coverage of retention volume (Rv) of 11 to 13 ml corresponded to an apparent molecular mass of ∼160 to 370 kDa (Fig. S10a and b), indicating the presence of either high-oligomeric states of Alg44 or detergent. However, under denaturing conditions using SDS-PAGE analysis and subsequent immunoblotting, a protein band corresponding to an apparent molecular mass of ∼85 kDa was detected in peak I fractions, which suggested the presence of a stable Alg44 dimer (Fig. 2e). No peak belonging to Alg44 monomer was detectable. On the other hand, when Triton X-100 was replaced with n-dodecyl β-d-maltoside (DDM), a distinct peak at an Rv of 14.5 ml was detected, which was calculated to exhibit an apparent molecular mass of 83.7 kDa, corresponding to the Alg44 dimer (Fig. S10c). This peak was further analyzed by immunoblotting, which suggested the presence of the stable Alg44 dimer with an apparent molecular mass of ∼85 kDa (see Fig. S11 in the supplemental material). Interestingly, replacement of Triton X-100 with DDM detergent caused loss of the stability of the dimer along with protein truncation (Fig. S11). Protein sequencing of the corresponding purified protein band confirmed the identity of Alg44 (coverage, 67.61%; number of unique peptides, 24) along with the presence of the proposed signal peptide sequence (Fig. S10d). Besides achieving full-length Alg44 purification, these experiments indicated that this protein forms at least a very stable dimer. These results suggested that Alg44 dimerization is not dependent on disulfide bond formation because treatment with DDT did not disrupt the dimer. This result was consistent with previous findings suggesting Alg44 dimerization based on in vivo chemical cross-linking (11) as well as that the purified PilZAlg44 domain existed in dimer form after crystallization (15).
The Alg8-Alg44 complex appears in a nonuniform, punctate, and patchy arrangement in the envelope surrounding the P. aeruginosa cell.
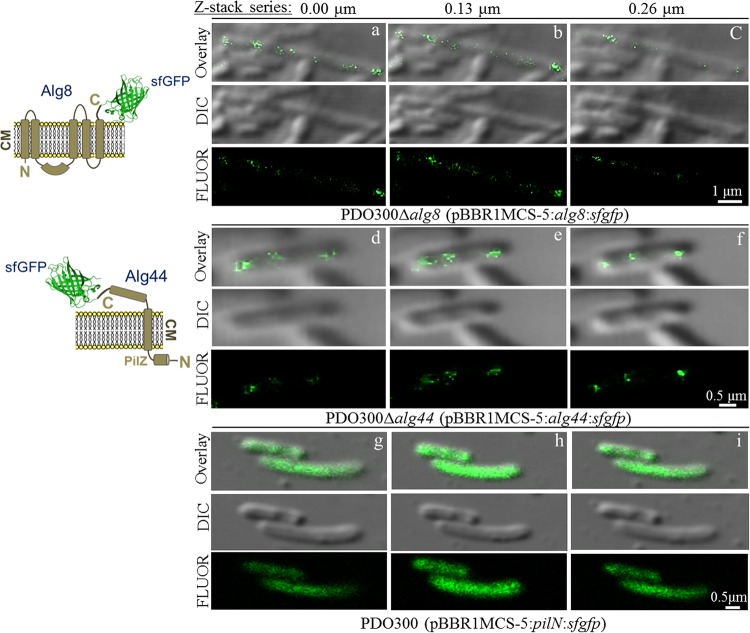
In previous studies, it was shown that interacting proteins Alg8 and Alg44 localize to the cytoplasmic membrane of P. aeruginosa, while the C termini of both proteins were proposed to be exposed to the periplasm (24, 26, 27). How the proposed alginate biosynthesis multiprotein complex localizes and distributes in the cells remains elusive. Here, membrane-anchored Alg8 and Alg44 forming alginate polymerase were selected to visualize the cellular localization and distribution. Due to the presence of the N-terminal signal peptides and the localization of the C-terminal part of these proteins in the oxidizing environment of the periplasm, a superfolding derivative of GFP (sfGFP) (28–30) was translationally fused to the respective C termini of Alg8 and Alg44. Genes encoding these fusion proteins were transferred into respective negative mutants, resulting in recombinant strains PDO300Δalg8(pBBR1MCS-5:alg8:sfgfp) and PDO300Δalg44(pBBR1MCS-5:alg44:sfgfp). As regular GFP is not fluorescent in the oxidizing environment of the periplasm, strains PDO300Δalg8(pBBR1MCS-5:alg8:gfp) and PDO300Δalg44(pBBR1MCS-5:alg44:gfp) were used as negative controls to further confirm the periplasmic localization of the C termini of both proteins (data not shown). Furthermore, PDO300(pBBR1MCS-5:pilN:sfgfp) was used as an additional control to rule out technical artifacts. PilN protein is part of a multiprotein complex responsible for the biogenesis of type IV pili in P. aeruginosa. PilN is anchored in the cytoplasmic membrane with its C-terminal domain exposed to the periplasm (31). All strains were grown under the same growth conditions on PIA medium to rule out the effects of cultivation conditions on the level of gene expression, and experiments were repeated three times.
Initially, functional protein localization of sfGFP fusion proteins was assessed by confirming restoration of alginate production in the respective isogenic knockout mutants. The mucoid phenotype and alginate quantification showed that PDO300Δalg8(pBBR1MCS-5:alg8:sfgfp) and PDO300Δalg44(pBBR1MCS-5:alg44:sfgfp) were complemented and produced alginate (5.16 ± 0.1 and 1.1 ± 0.02 g/g of CDM, respectively).
The sfGFP fusion protein- and alginate-producing P. aeruginosa cells were observed by confocal laser scanning microscopy (CLSM). By employing IMARIS image analysis software (Bitplane), respective images of bacteria included the cells that appeared with a coplanar orientation based on fluorescent distribution and were analyzed by including various numbers of optical sections (z-stack series), which were separated by 0.13 μm. As expected, PDO300Δalg8(pBBR1MCS-5:alg8:gfp) and PDO300Δalg44(pBBR1MCS-5:alg44:gfp) were not fluorescent, while both Alg8-sfGFP and Alg44-sfGFP led to fluorescent cells with punctate fluorescent foci that were distributed surrounding the cells in nonuniform and patchy patterns (Fig. 3, columns a to c and columns d to f). Analysis of z-stack series showed that the intensity and arrangement of distributed fluorescent foci were different between optical sections (Fig. 3, columns a to c and columns d to f).
FIG 3.
Alg8 and Alg44 proteins appear localized and distributed in a nonuniform, punctate, and patchy arrangement in the cell envelope of P. aeruginosa. Schematic membrane models of the C-terminal fusions of Alg8 and Alg44 with sfGFP protein are presented on the left. In addition to the C-terminal fusion of PilN with sfGFP protein, all respective images of bacteria that appeared with coplanar orientation based on fluorescent distribution were visualized using CLSM and analyzed using IMARIS image analysis software (Bitplane), each at three independent times. Analysis of various numbers of optical sections (z-stack series), which were separated by 0.13 μm, showed fluorescent foci of PDO300Δalg8(pBBR1MCS-5:alg8:sfgfp) (columns a to c) and PDO300Δalg44(pBBR1MCS-5:alg44:sfgfp) (columns d to f) are arrayed in particular puncta and patchy patterns around the cells, while fluorescent foci in PDO300 (pBBR1MCS-5:pilN:sfgfp) (columns g to i) were uniformly distributed and not in any particular pattern, which ruled out possible technical artifacts. CM, cytoplasmic membrane; FLUOR, fluorescent; DIC, differential interference contrast.
These results were compared with a translational fusion of PilN (PA5043) with sfGFP [PDO300(pBBR1MCS-5:pilN:sfgfp)] in order to rule out artifacts that might be mediated by sfGFP or the microscopic technique. No punctate or particular arrangement was observed in optical sections for PilN-sfGFP distribution by z-stack series analysis, and fluorescent foci were uniformly distributed (Fig. 3, columns g to i). This result further supported that Alg8 and Alg44 are distributed in specific and nonuniform arrangements around the cell. However, to visualize this distribution at higher resolution, alternative techniques such as stimulated emission depletion (STED) microscopy and immunoelectron microscopy might be employed for further analysis.
DISCUSSION
c-di-GMP signaling in bacteria is one the most complex signaling systems that regulate many physiological activities of bacteria leading to adaptation and fitness in varying environments. Molecular mechanisms of effector proteins for c-di-GMP signaling are still poorly understood in bacteria. Until now, the molecular mechanisms that have been profoundly analyzed for a specific output are limited to the membrane-anchored BcsA-BcsB complex (cellulose synthase) and the cytosolic FleQ protein regulating the expression of flagellar and exopolysaccharide biosynthesis genes, both in response to c-di-GMP (16, 32).
Indeed, the response threshold of c-di-GMP receptor/effector proteins is dependent on their binding affinity to c-di-GMP, i.e., at different cellular levels of c-di-GMP; in particular, c-di-GMP receptor/effector proteins such as Alg44 will be engaged to mediate a well-orchestrated physiological response (18). The c-di-GMP levels are tightly regulated by c-di-GMP-synthesizing and -degrading proteins (i.e., PDE/DCG proteins) in response to environmental stimuli (13). For example, it was recently shown that the membrane-anchored MucR is a nitrate-sensing protein with PDE/DCG activity involved in regulation of alginate polymerization (19). Here, the underlying molecular mechanism of c-di-GMP-mediated activation of alginate polymerization was studied. We developed a structural model of Alg8 in silico fused to the c-di-GMP binding PilZ domain of Alg44 in resemblance to the cellulose synthase, BcsA (Fig. 1; Fig. S1). This structural model and recent findings unraveling the molecular mechanism of c-di-GMP-mediated activation of cellulose synthesis informed the identification of amino acid residues involved in activation of alginate polymerization. Amino acid residues of Alg8 predicted to interact with the c-di-GMP receptor/effector PilZAlg44 domain were subjected to site-specific mutagenesis.
Critical amino acid residues of Alg8 whose replacement with alanine impacted the c-di-GMP level required for activation of alginate polymerization were identified, while H323A, T457A, and E460A resulted in decoupling of alginate synthesis from both general (RocR controlled) and localized (MucR mediated) levels of c-di-GMP (Fig. 1, S1, and S5).
These residues were distributed in loops A and B of Alg8, which were modeled by the Phyre2 server according to BcsA structure and are homologous to those loops of BcsA involved in activation of cellulose polymerization by c-di-GMP (Fig. 1 and S1). Here, we provided experimental evidence that H323 and P325 from loop A and T457 and E460 from loop B are involved in the c-di-GMP-dependent mechanism that regulates alginate polymerization (Fig. 1). Since not all of these critical Alg8 residues align with conserved amino acid residues of BcsA, we propose that the molecular mechanism of alginate polymerization activation is different from the autoinhibition mechanism as described for activation of cellulose polymerization.
In the Phyre2 structural model (Fig. 1 and S1), T457 and E460 flank two-β-sheet strands, which are formed by loop B (homologous to the gating loop of BcsA) and the N-terminal end of PilZAlg44 as previously discussed by Whitney and coworkers (15). This segment was not structurally elucidated, and it remained disordered in solution, whereas it became ordered upon binding to c-di-GMP (15). These results suggested that this region of Alg8, which comprises T457 and E460, adds another interface with the N-terminal end of PilZAlg44. The region comprising H323 was previously proposed to interface with the PilZ domain (11) and was found to be critical for regulation of alginate polymerization upon binding to c-di-GMP (Fig. 1). Alanine replacement of P325, E460, and H323E abolished alginate production in the presence of c-di-GMP levels, strongly inducing alginate production in the control strains PDO300(pBBR1MCS-5) and PDO300Δalg8(pBBR1MCS-5:alg8), while E460A and H323E restored alginate production to a level close to that of the wild type when c-di-GMP levels were decreased by overproduction of the c-di-GMP-degrading enzyme RocR (Fig. 1). Alignment of loops A and B with homologous counterparts of Alg8 from various bacteria showed that these loops are highly conserved, while E460 is less conserved (see Fig. S12 in the supplemental material). Interestingly, H322, which is highly conserved among alginate-producing Gammaproteobacteria, is replaced with a negatively charged glutamic acid or aspartic acid in several representatives from Alphaproteobacteria (belonging to the Rhizobiaceae family) and Epsilonproteobacteria (Fig. S12c). No bacteria from the latter group have been reported to produce alginate, while bacteria belonging to Rhizobiaceae are well-known cellulose producers.
Although the cytosolic PilZ domain of Alg44, which binds c-di-GMP, has been well characterized, the full-length Alg44 was not purified and the role of its periplasmic domain was not studied. Alanine substitutions in highly conserved periplasmic residues caused a significant reduction or abolishment of alginate production (Fig. 2a) by affecting the polymerization process or Alg44 localization to the membrane (Fig. 2b and c). These residues comprise a region homologous to biotinyl-lipoyl-domains of biotin/lipoyl attachment proteins with carboxylase/decarboxylase and acyltransferase activity (33). Furthermore, the cluster of PCDC (residues 266 to 269) was identified for being critical for Alg44 localization (Fig. 2c). The role of the CXC motif on the localization of other proteins has been reported, such as the outer membrane lipoprotein CsgG exposed to the periplasm, copper chaperones in the mitochondrial intermembrane space, and its functional importance for copper trafficking by CopC protein across the bacterial periplasm (34–36). In Alg44, both cysteine residues were required for protein localization. The roles of these residues in intra- and intermolecular interactions are still not clear. However, here experimental evidence that disulfide bond formation might be involved in protein-protein interaction was obtained (Fig. S7). This was observed only when Alg44 was produced from a single gene integrated into the genome, i.e., at physiological copy number, suggesting that the stoichiometry of Alg44 and interacting proteins is important for this function (Fig. S7).
Consistent with previous results that showed Alg44 boosts acetylation of alginate (11), replacement of periplasmic amino acid residues impaired the boosting effect on acetylation while causing a higher level of epimerization (Fig. 2d; see also Tables S1 and S2 in the supplemental material). These data suggested that Alg44 plays a role in alginate modifications through its periplasmic domain, possibly by linking polymerization with modification events.
For further biochemical characterization, full-length Alg44 was produced in P. aeruginosa containing the alg44-6His gene integrated into the genome. Alg44 was purified as a stable dimer (Fig. 2e and S9 to S11). These results suggested that Alg44 forms at least a stable dimer, as had been suggested on the basis of in vivo cross-linking experiments (11). In addition, the dimer of only the cytoplasmic PilZAlg44 domain has been demonstrated (15).
Bioinformatic prediction using the PRED-TAT server (37) showed that protein subunits constituting alginate biosynthesis/modification/secretion multiprotein complex comprise Sec-dependent signal peptides for secretion and localization. In this secretion pathway, unfolded proteins are secreted across the membrane through a protein-conducting channel (37, 38). Both Alg8 and Alg44 are membrane anchored, and they interact with each other, constituting the alginate-polymerizing component of the multiprotein complex. In order to assess the cell surface localization and distribution of the alginate synthesis complex, we employed translational fusion of Alg8 or Alg44 to the sfGFP. In contrast to GFP, the sfGFP properly folds in the oxidative environment such as the periplasm, particularly when the Sec-dependent pathway mediates secretion and localization (28, 29). Alg8 or Alg44 fused to the N terminus of either GFP or sfGFP, respectively, was functional, as it restored alginate production in the respective isogenic knockout mutants. CLSM analysis revealed that sfGFP fusions were fluorescent but GFP fusions were not. This confirmed the localization of both C termini to the periplasm. In addition, CLSM analysis indicated that both Alg8 and Alg44 sfGFP fusion proteins were localized as nonuniform and patchy distributions of fluorescent foci surrounding the P. aeruginosa cell (Fig. 3). As both proteins had been shown to interact with each other and since Alg44 interacts with AlgX and AlgK (periplasmic subunits of the multiprotein complex) as well as with the outer membrane pore AlgE, visualization of the distribution of these proteins presumably indicates the cell surface arrangement of the entire alginate producing multiprotein complex (9–11). Using an immunogold labeling approach, a recent study showed that AlgE was not randomly distributed over the entire outer membrane but colocalized with the proposed multiprotein complex (39). Previously, it was shown that the Psl polysaccharide, which is produced by P. aeruginosa at the initial stages of biofilm development, is helically attached to the cell surface (40). It was suggested that this helical distribution of Psl may promote the formation and stabilization of a polysaccharide matrix and in turn enhances cell-cell interactions that are critical for biofilm development (40). However, our result did not show a typical helical distribution for alginate polymerase, while its possibility cannot be ruled out due to limitations of the imaging techniques used in this study. Nevertheless, the observed nonuniform and patchy distribution might provide some insights into a possible link of the distribution of alginate biosynthesis multiprotein complex with the proposed asymmetrical and spatial distribution of c-di-GMP pools essential for activation of specific proteins such as alginate polymerase (19, 41). An asymmetrical, but nonstochastic, distribution of c-di-GMP in P. aeruginosa was previously observed upon cell division mediated by the activity of c-di-GMP-metabolizing enzymes for regulating the activity of c-di-GMP-binding proteins (41, 42). At least 40 proteins directly synthesize and/or degrade c-di-GMP in P. aeruginosa, which controls the cellular level of this molecule in response to perceived stimuli. However, due to spatial distributions of this molecule for specific outputs, cognate receptor/effector proteins variably respond to c-di-GMP cellular level mainly via the PilZ domain (43). Therefore, the nonuniform and patchy distribution of alginate polymerase (Fig. 3) might be a nonstochastic event and might link to the spatial distribution of c-di-GMP metabolizing enzymes for controlling c-di-GMP turnover when alginate overproduction is required as a specific output. To test this hypothesis, we suggest to apply a similar approach to visualize the distribution of the c-di-GMP providers for alginate polymerase, such as MucR.
Overall, this study provided experimental evidence toward elucidating the molecular mechanism of c-di-GMP-mediated activation of alginate polymerization and the role of Alg44 in alginate polymerization and modifications as well as the cell surface distribution of the respective alginate synthesis multiprotein complexes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
P. aeruginosa and Escherichia coli strains (Table 1) were cultivated in Luria broth Lennox medium, consisting of enzymatic digest of casein (10 g/liter), yeast extract (5 g/liter), and sodium chloride (5 g/liter) (pH 7.3 ± 0.2) (Acumedia; Neogen Corporation), supplemented by appropriate antibiotics. Depending on the selection marker on the plasmid, 300 μg/ml of gentamicin (AppliChem) (for P. aeruginosa) or 10 μg/ml of gentamicin or 100 μg/ml of ampicillin (AppliChem) (for E. coli) was used. Cultivated cultures were grown at 37°C overnight unless otherwise stated. Alginate production and phenotypic assessments were performed using Difco Pseudomonas isolation agar (PIA) medium supplemented by 300 μg/ml of gentamicin. E. coli strains, including C41(DE3), C43(DE3), BL21(DE3), Origami (DE3), ClearColi BL21(DE3), and Rosetta strains applied for heterologous production of protein, were purchased from Lucigen or Novagen (Table 1). Large-scale cell culture preparations were performed in Luria broth medium. All chemicals were purchased from Sigma-Aldrich unless otherwise mentioned. All applied restriction enzymes used for cloning were manufactured by New England BioLabs GmbH. Enzymes for PCR and gene sequencing were purchased from Invitrogen. Alginate powder (from brown algae; Sigma-A2158) was purchased from Sigma-Aldrich.
Construction of isogenic mutants with knockout of alg8 and/or alg44 and mucR and/or alg8 genes.
The P. aeruginosa PDO300 was used to generate isogenic single-and double-gene knockout mutants in alg8 and/or alg44 and mucR and/or alg8 genes as described previously (19, 23). Briefly, this was performed through two events of homologous recombination using suicide plasmid pEX100T. This plasmid contained knockout genes that were disrupted by the aacC1 gene (encoding gentamicin acetyltransferase) flanked by two FLP recombination target (FRT) sites. Removing the FTR-aacC1-FRT cassette was performed by transfer the flippase recombinase-encoding vector pFLP2 (44) into presumable knockout mutants resulting in P. aeruginosa PDO300Δalg8, PDO300Δalg44, PDO300Δalg8Δalg44, PDO300ΔmucR, and PDO300ΔmucRΔalg8 mutants, which were confirmed using antibiotic sensitivity screening and PCR with alg8 up/down, alg44 up/down, and mucR up/down primers (23, 26, 45).
In trans and in cis complementation of knockout mutant in alg8 and/or alg44 and mucR and/or alg8 genes and E. coli strain transformations.
The genes encoding Alg8/Alg44 or Alg44-6His and MucR/Alg8 were transferred into generated mutants using pBBR1MCS-5 (46) plasmid. For incorporation into the genome, mini-CTX-lacZ plasmid was used (47) as described previously in detail (9, 11, 23). The vector pETDuet-1 (Novagen) was applied to construct pETDuet-1:alg44-12His for heterologous production of Alg44-12His (Table 1). Relevant encoding genes (i.e., alg8, alg8-12His, alg44, and alg44-12His) for heterologous production were optimized based on E. coli codon usage and synthesized by GenScript.
Site-specific mutations of alg8 and alg44 genes.
Site-specific mutagenesis of Alg8 was performed by amplifying the fragments of alg8 by PCR using genomic DNA of POA1 strain and the primers carrying point mutations (Table 1). Amplified fragments were ligated into pBBR1MCS-5:alg8 at AatII/XmnI or BglII/PstI sites resulting in pBBR1MCS-5:alg8(T320A), pBBR1MCS-5:alg8(H323A), pBBR1MCS-5:alg8(P324A), pBBR1MCS-5:alg8(P325A), pBBR1MCS-5:alg8(T453A), pBBR1MCS-5:alg8(R454A), pBBR1MCS-5:alg8(T457A), pBBR1MCS-5:alg8(E460A), pBBR1MCS-5:alg8(H323E), pBBR1MCS-5:alg8(E322A), pBBR1MCS-5:alg8(E322A/H323E), pBBR1MCS-5:alg8(T453A/T457A), and pBBR1MCS-5:alg8(R454A/T457A). Resultant constructs were confirmed by sequencing at Massey Genome Service, Massey University. Also, pBBR1MCS-5:alg8(P325A):alg44(R17A), pBBR1MCS-5:alg8(P325A):alg44(R21A), pBBR1MCS-5:alg8(T457A):alg44(R17A), pBBR1MCS-5:alg8(T457A):alg44(R21A), pBBR1MCS-5:alg8(E460A):alg44(R17A), pBBR1MCS-5:alg8(E460A):alg44(R21A), pBBR1MCS-5:alg8(H323E):alg44(R17A), and pBBR1MCS-5:alg8(H323E):alg44(R21A) were generated. Combinations of all constructs with rocR (PA3947) were also provided (Table 1).
For site-specific mutagenesis of Alg44, each residue of the clusters QMK (residues 258 to 260) and TSPCDC (residues 264 to 269) was mutated to alanine using DNA synthesis (GenScript) and ligated into its corresponding region on alg44 using SapI and BamHI sites. The resultant plasmids were pBBR1MCS-5:alg44(Q258A)-6His, pBBR1MCS-5:alg44(M259A)-6His, pBBR1MCS-5:alg44(K260A)-6His, pBBR1MCS-5:alg44(T264A)-6His, pBBR1MCS-5:alg44(S265A)-6His, pBBR1MCS-5:alg44(P266A)-6His, pBBR1MCS-5:alg44(C267A)-6His, pBBR1MCS-5:alg44(D268A)-6His, pBBR1MCS-5:alg44(C269A)-6His, and pBBR1MCS-5:alg44(C267A/C269A)-6His.
Protein localization assessment in the envelope fraction of the cells.
Transformants of P. aeruginosa were grown in LB supplemented with 300 μg/ml gentamicin overnight at 37°C as inoculum. Each PIA plate supplemented with antibiotic was inoculated with 200 μl of washed inoculum and incubated for 36 h at 37°C. Grown bacteria on plates were scraped off and washed twice with buffer W (10 mM HEPES, 150 mM NaCl [pH 7.4]). Pelleted cells were lysed in buffer L (10 mM HEPES, 150 mM NaCl, 10% glycerol, 10 mM EDTA, 0.5 mg/ml lysozyme, Roche EDTA-free complete protease inhibitor [pH 7.4]) for 20 min and were subjected to sonication for disruption. Unbroken cells were separated by centrifugation at 8,000 × g for 30 min at 4°C. Supernatants were subjected to ultracentrifugation at 100,000 × g for 90 min at 4°C to isolate the envelope fraction. Pellets were solubilized in SDS containing loading buffer supplemented with 0.2% Triton X-100 and incubated for 1 h at 4°C. Before protein analysis, protein concentration of samples was assessed via densitometry assessment using various concentrations of bovine serum albumin.
Alginate purification and quantification and free-uronic acid assay.
Two milliliters of bacterial overnight culture grown in LB medium supplemented with the appropriate antibiotic was sedimented, and cells were washed twice with saline solution. Cells were suspended in 1 ml of saline solution, and 200 μl of cell suspension (optical density at 600 nm [OD600], 3.0) was plated onto a thick layer of PIA medium (40 ml in each plate) containing 300 μg/ml of gentamicin and then incubated at 37°C for 72 h, an incubation period that had previously been described and found to be optimal for cell growth and alginate quantification, avoiding nutrient deprivation and desiccation stress (11). Grown cells were scraped off from agar plates and suspended in saline solution until a homogenous suspension was formed. Then, suspensions were pelleted and supernatants containing alginate were precipitated with equal volumes of ice-cold isopropanol. The alginate precipitants were freeze-dried and then redissolved in 50 mM Tris-HCl (pH 7.4)–10 mM MgCl2 to a final concentration of 0.5% (wt/vol), followed by incubation with 15 μg/ml DNase I and 15 μg/ml RNase I at 37°C for 6 h. Then, pronase E was added to a final concentration of 20 μg/ml, and the mixture was incubated at 37°C for a further 18 h. Alginate solutions were dialyzed (molecular mass cutoff, 12 to 14 kDa; ZelluTrans/Roth mini dialyzer; Carl Roth GmbH & Co.) against 5 liters of ultrapure H2O for 48 h. Finally, alginates were precipitated with equal volumes of ice-cold isopropanol and freeze-dried for uronic acid assay and biochemical analysis, which was previously described in detail (9, 11, 48).
Assessment of free uronic acids was performed using 2 ml of overnight liquid culture. Cells were pelleted, and supernatants were filtered through a Vivaspin-500 (GE Healthcare) filter device with a molecular mass cutoff of 10 kDa. The uronic acids in the flowthrough, which consist of free uronic acids and short alginate degradation products, and total sample were measured according to a previously described procedure (49).
Analysis of composition of the alginates.
Compositional analysis of alginate samples was done by using 1H-nuclear magnetic resonance (1H-NMR) for two different sets of original and deacetylated samples. Preparation of samples was done according to a previously described method (11, 50, 51). Partially hydrolyzed samples (10 mg) were dissolved in 2 ml of D2O (99.96%) with shaking for 2 h and lyophilized. This deuteriation step was repeated twice. The deuterium-exchanged alginates (6 mg) were dissolved in 0.7 ml of D2O. 1H-NMR spectra were recorded at 85°C using 500 MHz Bruker Avance spectrometers. Spectra were interpreted as previously described by Grasdalen et al. (11, 52).
Protein production, cell disruption, and preparation of membrane fractions.
The strains of E. coli with pETDuet-1:alg44-12His were grown overnight. The main cultures of LB medium supplemented with 70 μg/ml of ampicillin were inoculated with 1% (vol/vol) of overnight cultures and incubated at 37°C until the OD600 reached 0.5 to 0.6. Then, cultures were induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM and subsequently grown at 25°C. In cis-complemented PDO300Δalg44 with alg44-6His was grown at 37°C overnight for mass production of cells.
Cells were harvested and washed twice with sodium phosphate buffer. After disruption using sonication or a microfluidizer, unbroken cells and cellular debris were removed using centrifugation at 8,000 × g for 45 min at 4°C. Supernatants were subjected to ultracentrifugation at 100,000 × g for 90 min at 4°C to isolate the envelope fraction. Pelleted membrane fractions were dissolved at 4°C for 2 h with buffer A (800 mM NaCl, 50 mM NaH2PO4, 10% glycerol, 5 mM EDTA, 1.5% Triton X-100, 5 mM imidazole, and Roche EDTA-free complete protease inhibitor [pH 7.7]). The insoluble part was pelleted again at 100,000 × g for 30 min at 4°C. Supernatants were subjected to protein purification and analysis.
Protein purification and analysis.
The His-spin protein miniprep kit (Zymo Research) was employed for preliminary purification assessment. Roche cOmplete His tag purification resin (EDTA compatible) was employed for large-scale purification of His-tagged protein. The solubilized membrane fraction was mixed with 5 ml of preequilibrated resin with buffer A and incubated at 4°C for 6 h with gentle shaking. The mixture was packed into columns for gravity flow and washed with 10 column volumes (cv) of buffer A. Elution was performed with 5 cv of buffer B (similar to buffer A with 500 mM imidazole, pH 7.7). A 250-μl volume of eluent was loaded onto a Superdex 200 Increase 10/300 GL column. Two column volumes of buffer C (300 mM NaCl, 50 mM NaH2PO4, 10% glycerol, 5 mM EDTA, 0.02% Triton X-100 [pH 7.8]) was passed through the column at 0.6 ml min−1, and the absorbance at 280 nm was monitored.
A standard curve of retention volume (Rv) versus apparent molecular mass was applied by measuring the elution volume for low- and high-molecular-mass protein standards (Sigma). The plot was obtained for log MW versus Rv, where MW is the known apparent molecular mass of the standard, including the following: (i) ferritin (∼440 kDa), (ii) alcohol dehydrogenase (∼150 kDa), (iii) bovine serum albumin (∼66 kDa), and (iv) carbonic anhydrase (∼29 kDa). The curve was approximately linear at R2 = 0.9891 over the range of standards from MW of 29 to 440 kDa. This curve is presented in the supplemental material.
Fractions were collected in 0.5-ml steps and subsequently assessed by SDS-PAGE (8% acrylamide gels) either with staining with Coomassie brilliant blue solution or immunoblotting. Immunoblotting was performed using an iBlot dry-blotting system (Invitrogen) as described previously (9). For the detection of His-tagged protein, a HisProbe-HRP kit (Thermoscientific) was used according to the manufacturer's instruction. For detection of Alg8, anti-Alg8 (2:5,000) in 2% bovine serum albumin-fraction V (Gibco/Invitrogen) was used. Immunoblotting was followed by resolving with SuperSignal West Pico chemiluminescent substrate (Thermoscientific) and developing on X-ray film (Kodak, Rochester, NY). Protein sequencing was performed by The Centre for Protein Research (CPR) of Otago University.
Imaging of fluorescent proteins and analysis.
The encoding sequencse of GFP and sfGFP were synthesized (GenScript) and ligated at the C-terminal end of alg8, alg44, and pilN genes (without the stop codon) using XbaI and SacI sites, resulting in pBBR1MCS-5:alg8:gfp, pBBR1MCS-5:alg8:sfgfp, pBBR1MCS-5:alg44:gfp, pBBR1MCS-5:alg44:sfgfp, pBBR1MCS-5:pilN:gfp, and pBBR1MCS-5:pilN:sfgfp. Constructs were electroporated into PDO300, PDO300Δalg8, and PDO300Δalg44. Complemented and mucoid colonies were selected for alginate quantification. Microscopic analysis was performed using the cells growing on solid media within 24-h biofilm (mucoid colony), followed by analysis of various numbers of optical sections (z-stack series), separated by 0.13 μm, using confocal laser scanning microscopy (Leica SP5 DM6000B). This experiment was repeated three times to analyze the cells. Images were analyzed using IMARIS image analysis software (Bitplane).
Statistical analysis.
Alginate quantification and uronic acid assay were conducted using four independent repetitions, and data are presented as means ± standard deviations (SD). The Shapiro-Wilk test was used to evaluate the normality of distribution of the data and indicated that the data were normally distributed (P > 0.05). The results were statistically analyzed by analysis of variance (ANOVA) followed by post hoc Tukey's honestly significant difference (HSD) test for pairwise comparisons using XLSTAT statistical add-in software for Microsoft Excel 14.0. The lowercase italic letters on the histograms provide a graphical representation for post hoc pairwise comparisons (Tukey's HSD). A P value less than or equal to 0.05 was considered significant in all evaluations.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Bio-NMR facility of Massey University, Patrick Edwards, the Manawatu Microscopy and Imaging Centre (MMIC) of Massey University, and especially Matthew Savoian for assistance in using microscopic techniques.
This work was supported by grants from the Massey University Research Fund (New Zealand) to B.H.A.R. M.F.M. and S.G. are funded by a Massey University doctoral scholarship.
B.H.A.R. and M.F.M. conceived the project. M.F.M. and S.G. performed experiments. M.F.M. and B.H.A.R. wrote the manuscript. All authors reviewed the manuscript.
We declare that there are no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03499-16.
REFERENCES
- 1.Rehm BHA, Valla S. 1997. Bacterial alginates: biosynthesis and applications. Appl Microbiol Biotechnol 48:281–288. doi: 10.1007/s002530051051. [DOI] [PubMed] [Google Scholar]
- 2.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghafoor A, Hay ID, Rehm BHA. 2011. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol 77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood SR, Firoved AM, Ornatowski W, Mai T, Deretic V, Timmins GS. 2007. Nitrosative stress inhibits production of the virulence factor alginate in mucoid Pseudomonas aeruginosa. Free Radic Res 41:208–215. doi: 10.1080/10715760601052610. [DOI] [PubMed] [Google Scholar]
- 5.May TB, Shinabarger D, Maharaj R, Kato J, Chu L, DeVault JD, Roychoudhury S, Zielinski NA, Berry A, Rothmel RK. 1991. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev 4:191–206. doi: 10.1128/CMR.4.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slack MPE, Nichols WW. 1981. The penetration of antibiotics through sodium alginate and through the exopolysaccharide of a mucoid strain of Pseudomonas aeruginosa. Lancet ii:502–503. [DOI] [PubMed] [Google Scholar]
- 7.Simpson JA, Smith SE, Dean RT. 1989. Scavenging by alginate of free radicals released by macrophages. Free Radic Biol Med 6:347–353. doi: 10.1016/0891-5849(89)90078-6. [DOI] [PubMed] [Google Scholar]
- 8.Bayer AS, Speert DP, Park S, Tu J, Witt M, Nast CC, Norman DC. 1991. Functional role of mucoid exopolysaccharide (alginate) in antibiotic-induced and polymorphonuclear leukocyte-mediated killing of Pseudomonas aeruginosa. Infect Immun 59:302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehman ZU, Wang Y, Moradali MF, Hay ID, Rehm BH. 2013. Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl Environ Microbiol 79:3264–3272. doi: 10.1128/AEM.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hay ID, Schmidt O, Filitcheva J, Rehm BH. 2012. Identification of a periplasmic AlgK-AlgX-MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl Microbiol Biotechnol 93:215–227. doi: 10.1007/s00253-011-3430-0. [DOI] [PubMed] [Google Scholar]
- 11.Moradali MF, Donati I, Sims IM, Ghods S, Rehm BH. 2015. Alginate polymerization and modification are linked in Pseudomonas aeruginosa. mBio 6:3e00453-15. doi: 10.1128/mBio.00453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Römling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 13.Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. 2012. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol 86:1424–1440. doi: 10.1111/mmi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay ID, Wang Y, Moradali MF, Rehman ZU, Rehm BH. 2014. Genetics and regulation of bacterial alginate production. Environ Microbiol 16:2997–3011. doi: 10.1111/1462-2920.12389. [DOI] [PubMed] [Google Scholar]
- 15.Whitney JC, Whitfield GB, Marmont LS, Yip P, Neculai AM, Lobsanov YD, Robinson H, Ohman DE, Howell PL. 2015. Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem 290:12451–12462. doi: 10.1074/jbc.M115.645051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan JL, McNamara JT, Zimmer J. 2014. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496. doi: 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan JLW, Strumillo J, Zimmer J. 2013. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou SH, Galperin MY. 2016. Diversity of c-di-GMP-binding proteins and mechanisms. J Bacteriol 198:32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hay ID, Remminghorst U, Rehm BH. 2009. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol 75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 21.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Hay ID, Rehman ZU, Rehm BH. 2015. Membrane-anchored MucR mediates nitrate-dependent regulation of alginate production in Pseudomonas aeruginosa. Appl Microbiol Biotechnol 99:7253–7265. doi: 10.1007/s00253-015-6591-4. [DOI] [PubMed] [Google Scholar]
- 23.Remminghorst U, Rehm BHA. 2006. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett 580:3883–3888. doi: 10.1016/j.febslet.2006.05.077. [DOI] [PubMed] [Google Scholar]
- 24.Oglesby LL, Jain S, Ohman DE. 2008. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154:1605–1615. doi: 10.1099/mic.0.2007/015305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain S, Ohman DE. 2005. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect Immun 73:6429–6436. doi: 10.1128/IAI.73.10.6429-6436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remminghorst U, Hay ID, Rehm BHA. 2009. Molecular characterization of Alg8, a putative glycosyltransferase, involved in alginate polymerisation. J Biotechnol 140:176–183. doi: 10.1016/j.jbiotec.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Remminghorst U, Rehm BHA. 2006. In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl Environ Microbiol 72:298–305. doi: 10.1128/AEM.72.1.298-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aronson DE, Costantini LM, Snapp EL. 2011. Superfolder GFP is fluorescent in oxidizing environments when targeted via the sec translocon. Traffic 12:543–548. doi: 10.1111/j.1600-0854.2011.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dinh T, Bernhardt TG. 2011. Using superfolder green fluorescent protein for periplasmic protein localization studies. J Bacteriol 193:4984–4987. doi: 10.1128/JB.00315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 31.Ayers M, Sampaleanu LM, Tammam S, Koo J, Harvey H, Howell PL, Burrows LL. 2009. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J Mol Biol 394:128–142. doi: 10.1016/j.jmb.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 32.Matsuyama BY, Krasteva PV, Baraquet C, Harwood CS, Sondermann H, Navarro MV. 2016. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E209–E218. doi: 10.1073/pnas.1523148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jitrapakdee S, Wallace JC. 2003. The biotin enzyme family: conserved structural motifs and domain rearrangements. Curr Protein Pept Sci 4:217–229. doi: 10.2174/1389203033487199. [DOI] [PubMed] [Google Scholar]
- 34.Taylor JD, Matthews SJ. 2015. New insight into the molecular control of bacterial functional amyloids. Front Cell Infect Microbiol 5:33. doi: 10.3389/fcimb.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palumaa P. 2013. Copper chaperones. The concept of conformational control in the metabolism of copper. FEBS Lett 587:1902–1910. doi: 10.1016/j.febslet.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 36.Puig S, Rees EM, Thiele DJ. 2002. The ABCDs of periplasmic copper trafficking. Structure 10:1292–1295. doi: 10.1016/S0969-2126(02)00863-8. [DOI] [PubMed] [Google Scholar]
- 37.Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. 2010. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 38.du Plessis DJ, Nouwen N, Driessen AJ. 2011. The Sec translocase. Biochim Biophys Acta 1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Maleki S, Almaas E, Zotchev S, Valla S, Ertesvag H. 2015. Alginate biosynthesis factories in Pseudomonas fluorescens: localization and correlation with alginate production level. Appl Environ Microbiol 82:1227–1236. doi: 10.1128/AEM.03114-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulasekara BR, Kamischke C, Kulasekara HD, Christen M, Wiggins PA, Miller SI. 2013. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. Elife 2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan RP, Fouhy Y, Lucey JF, Dow JM. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J Bacteriol 188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 45.Schlegel HG, Kaltwasser H, Gottschalk G. 1961. A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch Mikrobiol 38:209–222. doi: 10.1007/BF00422356. [DOI] [PubMed] [Google Scholar]
- 46.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer HP, Chuanchuen R. 2001. Small broad-host-range lacZ operon fusion vector with low background activity. Biotechniques 31:1258–1262. [DOI] [PubMed] [Google Scholar]
- 48.Hay ID, Gatland K, Campisano A, Jordens JZ, Rehm BH. 2009. Impact of alginate overproduction on attachment and biofilm architecture of a supermucoid Pseudomonas aeruginosa strain. Appl Environ Microbiol 75:6022–6025. doi: 10.1128/AEM.01078-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hay ID, Rehman ZU, Rehm BH. 2010. Membrane topology of outer membrane protein AlgE, which is required for alginate production in Pseudomonas aeruginosa. Appl Environ Microbiol 76:1806–1812. doi: 10.1128/AEM.02945-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donati I, Paoletti S. 2009. Material properties of alginates, p 1–53. In Rehm BHA. (ed), Alginates: biology and applications. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 51.Skjak-Braek G, Grasdalen H, Larsen B. 1986. Monomer sequence and acetylation pattern in some bacterial alginates. Carbohydr Res 154:239–250. doi: 10.1016/S0008-6215(00)90036-3. [DOI] [PubMed] [Google Scholar]
- 52.Grasdalen H, Larsen B, Smidsrod O. 1979. Pmr study of the composition and sequence of uronate residues in alginates. Carbohydr Res 68:23–31. doi: 10.1016/S0008-6215(00)84051-3. [DOI] [Google Scholar]
- 53.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Hoiby N, Kharazmi A. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145(Part 6):1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 54.Bullock WO, Fernandez JM, Short JM. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. Biotechniques 5:376–379. [Google Scholar]
- 55.Simon RPU, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 56.Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–950, 952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.