Abstract
Most tumor suppressor genes are commonly inactivated in the development of colorectal cancer (CRC). The activation of tumor suppressor genes may be beneficial to suppress the development and metastasis of CRC. This study analyzed genes expression and methylation levels in different stages of CRC. Genes with downregulated mRNA expression and upregulated methylation level in advanced CRC were screened as the potential tumor suppressor genes. After comparing the methylation level of screened genes, we found that MBD1 gene had downregulated mRNA expression and upregulated methylation levels in advanced CRC and continuously upregulated methylation level in the progression of CRC. Enrichment analysis revealed that genes expression in accordance with the elevated expression of MBD1 mainly located on chromosomes 17p13 and 17p12 and 8 tumor suppressor genes located on chromosome 17p13. Further enrichment analysis of transcription factor binding site identified that SP1 binding site had higher enrichment and could bind with MBD1. In conclusion, MBD1 may be a tumor suppressor gene in advanced CRC and affect the development and metastasis of CRC by regulating 8 tumor suppressor genes through binding with SP1.
1. Introduction
The occurrence and development of malignant tumors are mainly caused by the activation of oncogene and inactivation of tumor suppressor genes. In colorectal cancer (CRC), tumor suppressor genes are significantly inhibited with the progression of cancer. Screening genes with the function of inhibiting tumor growth in advanced CRC is helpful to find the way of suppressing the progression and metastasis of tumor. A gene is a locus (or region) of DNA which is made up of nucleotides and produces biological function through gene expression and protein coding. In the progression of cancer, the expression of oncogenes is enhanced, while the expression of tumor suppressor gene is inhibited. Methylation modification is essential for gene expression. DNA methylation can shut down the activity of certain genes, while demethylation promotes the activity of genes. Hence, the change of methylation levels is a critical index in the process of screening tumor suppressor genes. In advanced CRC, tumor cell can metastasize to lymph nodes and even distant organ. In this stage, the activity of tumor suppressor gene is significantly inhibited. Therefore, the activation of tumor suppressor genes may be the potential way for the treatment of CRC. This study analyzed genes expression and methylation levels in the different stage of CRC to find out genes with decreased expressions and increased methylation levels in advanced CRC. By comparing methylation levels in different stage, gene with continuously upregulation methylation levels in the progression of CRC was screened. After screening, MBD1 may be a tumor suppressor gene in advanced CRC. In order to figure out the potential mechanism of MBD1 in the process of tumor inhibition, this study further screened genes related to MBD1 high expression in CRC gene expression profile.
2. Methods and Materials
2.1. Data Source
DNA methylation data of CRC was from Level 3 Cancer Genome Altas (TCGA) [1], with Illumina Human Methylation 27 as the data chip platform, including 37 cases of normal CRC data and 166 cases of CRC data. The 166 cases of CRC data included 32 cases of stage I data, 64 of stage II data, 45 of stage III data, 24 of stage IV data, and 1 case with unclear stage. This study mainly analyzed the methylation data of the 165 CRC cases.
CRC gene expression profile data were taken from TCGA, with Agilent G4502A as the data chip platform, including 174 cases of expression profile data (19 cases of normal CRC and 155 cases of CRC) (Table 1).
Table 1.
The distribution of clinical characteristics of methylation and expression profile in CRC patients.
| Characteristics | Colorectal cancer | |
|---|---|---|
| Methylation | Expression | |
| Sum | 166 | 155 |
| Gender | ||
| Male | 83 | 79 |
| Female | 83 | 76 |
| Age at initial pathologic diagnosis | ||
| <50 | 12 | 10 |
| ≥50 | 154 | 145 |
| Anatomic neoplasm subdivision | ||
| Ascending colon | 32 | 28 |
| Cecum | 33 | 29 |
| Descending colon | 6 | 6 |
| Hepatic flexure | 8 | 9 |
| Sigmoid colon | 69 | 66 |
| Splenic flexure | 2 | 2 |
| Transverse colon | 15 | 14 |
| TNM | ||
| T1 + T2 + T3 + T4 | 166 | 155 |
| N1 + N2 | 67 | 60 |
| M1 | 24 | 23 |
| AJCC pathologic tumor_stage | ||
| Stage I | 32 | 29 |
| Stage II | 64 | 63 |
| Stage III | 45 | 39 |
| Stage IV | 24 | 23 |
| Histologic diagnosis | ||
| Colon adenocarcinoma | 140 | 131 |
| Colon mucinous adenocarcinoma | 24 | 22 |
| Family history colorectal cancer | ||
| Yes | 26 | 23 |
| No | 140 | 132 |
| History of another malignancy | ||
| Yes | 9 | 9 |
| No | 157 | 146 |
| History colon polyps | ||
| Yes | 85 | 84 |
| No | 81 | 71 |
| Tumor status | ||
| Tumor free | 10 | 10 |
| With tumor | 155 | 144 |
| Vascular invasion indicator | ||
| Yes | 37 | 34 |
| No | 106 | 107 |
| Lymphovascular invasion indicator | ||
| Yes | 87 | 78 |
| No | 70 | 72 |
2.2. Gene Screening with Differential Methylation and Expression
According to AJCC stage, early CRC data (stage I and stage II) and metastatic CRC data (stage III and stage IV) were compared with 37 cases of normal CRC data to screen genes with differential methylation level. In the analysis of gene expression, based on the 155 cases of CRC expression profile data, early CRC data (stage I data and stage II) and metastatic CRC data (stage III and stage IV) were compared with 19 cases of normal CRC data to screen genes with differential expression. Unpaired t-test was used for gene screening. Bonferroni FWER [2] was performed for adjusting p value to control false discovery rate (FDR). p value less than 0.05 was considered significant difference.
CRC methylation data were divided into four groups (I, II, III, and IV stage) in order to screen genes with continuous increase or decrease of methylation level at the four stages. Multiple comparisons were performed for differential gene expression screening followed by paired comparison (stage II versus stage I; stage III versus stage II; stage IV versus stage III) by using One-way ANOVA. The folder change of differential gene was 1 time. Bonferroni FWER was performed to adjust p value. p value less than 0.05 was considered significant difference. The intersection was taken from the three paired differential genes. Based on the progression of CRC, genes with continuous increase or decrease of methylation level at the four stages were selected.
Bonferroni FWER was a step-wise process, in an individual correction way for each p value and multiplying each gene's p value by its number on gene lists. It shows significance if the corrected p value was less than the error ratio. The corrected p value equals the multiplication of p value by gene number and is less than 0.05. Therefore, if 1000 genes were tested each time, single p value accepted was 0.00005 at most, thus making correction strict. We did so by using Bonferroni FWER to correct p value to ensure reliability of the screened out difference genes, especially when colorectal cancer tissues had less difference.
2.3. Regulation Mechanism of MBD1 in CRC
Expression profile data GSE39582 (including 566 cases of CRC tissues) taken from Gene Expression Omnibus (GEO) [3] were used to analyze the potential mechanism of MBD1 in CRC. To clarify the effect of high and low expression of MBD1 on CRC expression profile, 566 cases of expression profile data were divided into high and low groups according to the expression of MBD1 in CRC: expression of MBD1 was ordered from high to low and divided into two groups (MBD1 high expression and MBD1 low expression) with 283 cases in each group; GSEA [4, 5] was used for enrichment analysis between the two group based on chromosome location (gene set, version V5.1).
3. Results
3.1. Screening Results of Tumor Suppressor Gene
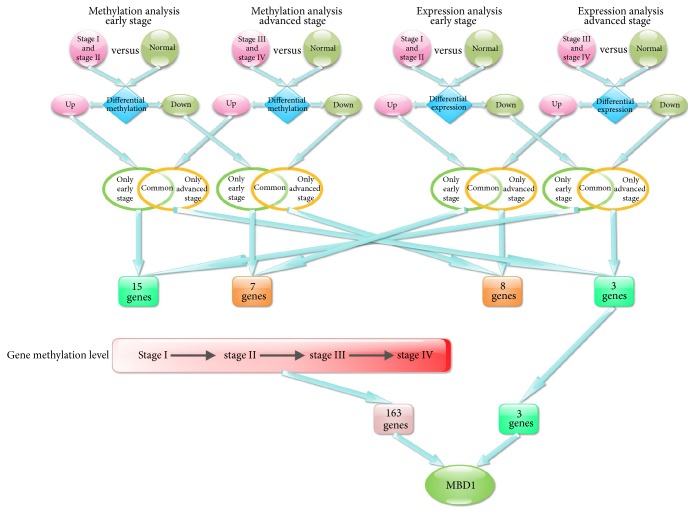
To screen genes with the function of tumor inhibition in advanced CRC, only genes with downregulated mRNA expression and upregulated methylation level in metastatic CRC were selected. Based on the data from TCGA database, early CRC data (stage I data and stage II) and metastatic CRC data (stage III and stage IV) were compared with normal CRC data and finally 1470 genes with upregulated methylation level, 2677 genes with downregulated methylation level, 1984 genes with upregulated expression, and 3255 genes with downregulated expression were obtained in early CRC. There were 1164 genes with upregulated methylation level, 2569 genes with downregulated methylation level, 1882 genes with upregulated expression, and 3143 genes with downregulated expression obtained in advanced CRC.
The intersection analysis was performed based on the differential expression genes and differential methylation genes and divided into four groups: (1) intersection was obtained from genes with upregulated methylation level in early stage and in advanced stage; 355 specific genes with upregulated methylation in early stage; and 49 specific genes with upregulated methylation in advanced stage and 1115 common genes were collected; (2) intersection was obtained from genes with downregulated methylation level in early stage and in advanced stage; 522 specific genes with downregulated methylation in early stage; and 414 specific genes with downregulated methylation in advanced stage and 2155 common genes were collected; (3) intersection was obtained from genes with upregulated gene expression in early stage and in advanced stage; 315 specific genes with upregulated expression in early stage, and 213 specific genes with upregulated expression in advanced stage and 1669 common genes were collected; (4) intersection was obtained from genes with downregulated gene expression in early stage and in advanced stage; 428 specific genes with downregulated expression in early stage, and 316 specific genes with downregulated expression in advanced stage and 2827 common genes were collected.
Intersection was performed in genes with upregulated methylation and downregulated gene expression in early stage, and 15 genes were obtained. Intersection was performed in genes with downregulated methylation and upregulated expression in early stage, and 7 genes were obtained. Intersection was performed in genes with downregulated methylation and upregulated expression in advanced stage, and 8 genes were obtained. Intersection was performed in genes with upregulated methylation and downregulated expression in advanced stage, and 3 genes were obtained.
Upregulation of gene expression can promote the performance of gene function, while upregulation of methylation can inhibit gene function. Hence, only the 15 genes with upregulated methylation and downregulated expression in early stage might be the tumor suppressor genes. The 7 genes with downregulated methylation and upregulated expression in early stage might be the cancer-promoting genes. The 8 genes with downregulated methylation and upregulated expression in advanced stage might be the cancer-promoting genes. The 3 genes with upregulated methylation and downregulated expression in advanced stage might be the tumor suppressor genes. From the above results, we found that tumor suppressor gene was more while cancer-promoting gene was less in early stage. In advanced stage, cancer-promoting gene was more while tumor suppressor gene was less.
From the above analysis, we only found 3 genes with upregulated methylation and downregulated gene expression in advanced CRC. In order to screen genes with sustained change of methylation in the four stages of CRC, we compared genes with differential methylation in three paired groups: stage II versus stage I, stage III versus stage II and stage IV versus stage III and obtained the intersection. A total of 163 genes with sustained increasing methylation and 222 genes with sustained decreasing methylation were screened in I, II, III, and IV stages of CRC. Intersection was obtained among the 163 genes with increased methylation in CRC and 3 genes with upregulated methylation and downregulated gene expression in advanced CRC, and finally MBD1 was screened (Figure 1).
Figure 1.
Flow chart of screening differentially methylated genes and differentially expressed genes.
3.2. Survival Analysis of MBD1 in CRC
MBD1 had upregulated methylation level and downregulated gene expression in advanced CRC and had sustained increasing methylation in the four stages of CRC. Hence, MBD1 might be a tumor suppressor gene in advanced CRC. For further confirmation, expression profile data GSE17536 taken from GEO database was used for survival analysis of MBD1. GSE17536 data includes 177 CRC tissues with determined survival time, with Affymetrix Human Genome U133 Plus 2.0 Array as the chip platform [6]. We extracted 177 expression values of MBD1, ordered and divided into MBD1 high-expression group and MBD1 low-expression group. Each group included 88 cases of data, with the median data (1 case) being deleted. Kaplan-Meier [7] was used to analyze the survival rate of the two groups with the survival curve drawn (Figure 2). The results showed that, with the change of MBD1 expression, the difference of survival rate was significant (p = 0.0227). Patients with high-expression MBD1 had higher survival rate than patients with low-expression MBD1.
Figure 2.
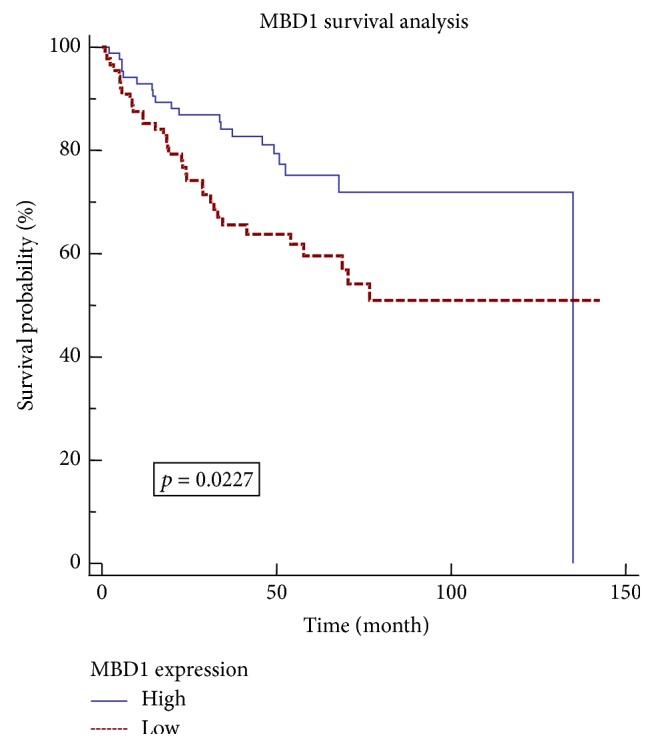
Survival analysis diagram of MBD1.
3.3. Regulation Mechanism of MBD1 in CRC
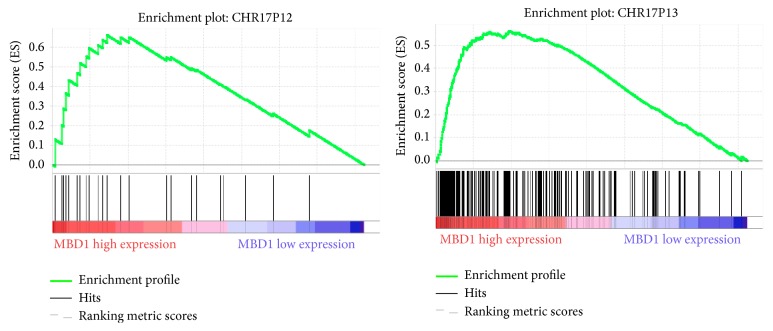
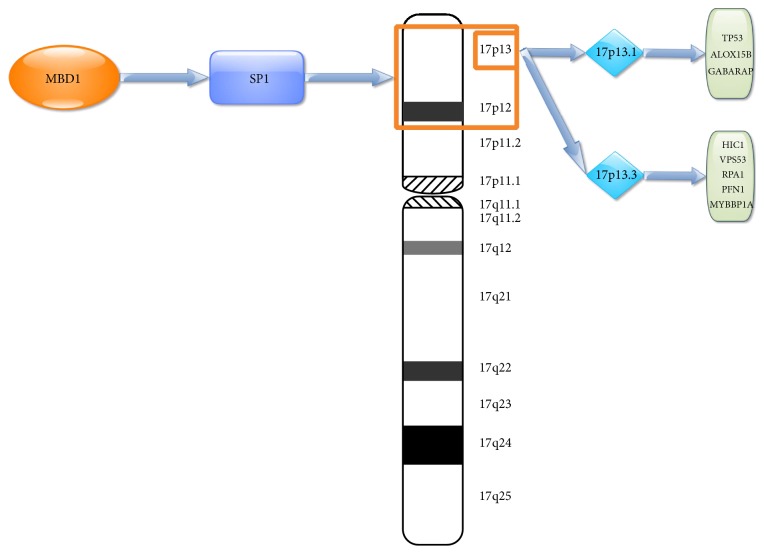
In order to further confirm the molecular mechanism of MBD1 in CRC, expression profile data from 566 CRC cases at different stages were divided in the MBD1 high-expression group and MBD1 low-expression group and GSEA enrichment analysis was performed based on chromosome location. The results revealed that two gene sets with high enrichment in the MBD1 high-expression group were located in chromosomes 17P13 (p = 0.002) and 17P12 (p = 0.002) (Figure 3). The expressions of the two gene set were upregulated in the MBD1 high-expression group, suggesting that 17P13 and 17P12 might be the chromosome zone where MBD1 worked. A total of 89 genes contributed to the enrichment of 17P13 gene set and 12 genes contributed to the enrichment of 17P12 gene set. A total of 1217 known tumor suppressor genes was searched from the tumor suppressor gene database (TSGene 2.0) [8, 9]. Intersection was performed among the 1217 genes and previously mentioned 89 genes, and 8 tumor suppressor genes (HIC1, VPS53, RPA1, ALOX15B, TP53, PFN1, MYBBP1A, and GABARAP) were obtained. No intersection was found among the 1217 genes and previously mentioned 12 genes. Because of all the 8 genes located 17P13 zone, a total of 89 genes in the region of 17P13 were performed through CYTOBAND database of DAVID [10, 11] and 17P13.3 and 17P13.1 had higher enrichment. 17P13.3 enrichment included 22 genes, including 5 tumor suppressor genes (HIC1, VPS53, RPA1, PFN1, and MYBBP1A). 17P13.1 enrichment included 27 genes, including 3 tumor suppressor genes (ALOX15B, TP53, and GABARAP). In order to clarify the potential mechanism of the 8 tumor suppressor genes, enrichment analysis of transcription factor binding sites was performed by using TFM-Explorer [12] in 22 genes in 17P13.3 zone and 27 genes in 17P13.1 zone. A total of 29 proteins interacted with MBD1 after being verified by BioGRID [13] database search. Intersection was obtained based on the 29 genes and the enrichment of transcription factors, SP1, was screened. The binding site of SP1 had higher enrichment in the upstream regulation region of 17P13.3 and 17P13.1 and the upstream regulation region of the 8 tumor suppressor genes all found SP1 binding site. Moreover, SP1 could bind with MBD1. Hence, MBD1 may be a tumor suppressor gene in advanced CRC and affect the development and metastasis of CRC by regulating 8 tumor suppressor genes through binding with SP1 (Figure 4).
Figure 3.
Enrichment figure of gene chromosomes 17 p12 and 17 p13 region that related to MBD1 expression.
Figure 4.
Regulating schema diagram of 8 tumor suppressor genes that are regulated by MBD1 through binding with SP1.
4. Discussion
MBD1, named methyl-CpG binding protein 1, is a transcriptional repressor and belongs to one member of methyl-CpG binding domain (MBD) family. MBD1 can negatively regulate the transcriptional activity of RNA polymerase II promoter and be involved in chromosome silencing dependent by methylation. Hence, MBD1 can regulate both gene expression and methylation level. Previous study reported that the upregulation of MBD1 enhanced the epithelial mesenchymal transition and invasion of pancreatic cancer cells [14], which suggested that MBD1 contributed to the tumor growth in pancreatic cancer. Another study reported that deletion of 18q21 chromosome where MBD1 was located occurred frequently in CRC [15, 16], which suggested that genes in 18q21 chromosome zone might inhibit tumor growth in CRC. The deletion of 18q21 chromosome could result in the abnormal activation of cancer genes. MBD1 might be a tumor suppressor gene for its location. Previous studies demonstrated that the polymorphism of MBD1 was related to the risk of lung cancer [17, 18], which provided evidence that MBD1 might inhibit tumor growth in lung cancer. Hence, MBD1 expression may have different biological effect in different cancer. In this study, MBD1 had upregulated methylation and downregulated gene expression in advanced CRC and also had sustained upregulated methylation in the four stages of CRC, which suggested that MBD1 was a tumor suppressor gene for advanced CRC. In the study of expression profile of CRC, we found that high expression of MBD1 was closely related to genes in chromosomes 17P12 and 17P13 and the two chromosome zones included many known tumor suppressor genes, such as HIC1, VPS53, RPA1, ALOX15B, TP53, PFN1, MYBBP1A, and GABARAP. Many studies have been reported that TP53 was a tumor suppressor gene. Further study found that the upstream regulation region of the 8 tumor suppressor genes all found SP1 binding site. Moreover, SP1 could bind with MBD1. Hence, MBD1 may be a tumor suppressor gene in advanced CRC and affect the development and metastasis of CRC by regulating 8 tumor suppressor genes through binding with SP1. This study determined that MBD1 may be a tumor suppressor gene in advanced CRC from the aspect of gene expression and methylation level and speculated that MBD1 could regulate chromosomes 17P12 and 17P13 zones to further regulate the expressions of tumor suppressor genes. Therefore, the activation of MBD1 in advanced CRC may inhibit and even reverse the development and metastasis of CRC.
Acknowledgments
This work was supported by the National Key Basic Research Program of China (973 Program, 2015CB554002) and Project of the National Natural Science Foundation of China supported by NSFC-Guangdong Joint Fund (U1201226).
Conflicts of Interest
The authors have declared that no conflicts of interest exist.
References
- 1.Deng M., Brägelmann J., Schultze J. L., Perner S. Web-TCGA: an online platform for integrated analysis of molecular cancer data sets. BMC Bioinformatics. 2016;17(1, article no. 72) doi: 10.1186/s12859-016-0917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bland J. M., Altman D. G. Multiple significance tests: the Bonferroni method. British Medical Journal. 1995;310(6973, article 170) doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett T., Suzek T. O., Troup D. B., et al. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Research. 2005;33(1):D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J. P. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 5.Damian D., Gorfine M., Mootha V. K., et al. Statistical concerns about the GSEA procedure. Nature Genetics. 2004;36(7):p. 663. doi: 10.1038/ng0704-663a. [DOI] [PubMed] [Google Scholar]
- 6.Harbig J., Sprinkle R., Enkemann S. A. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic acids research. 2005;33(3, article e31) doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stel V. S., Dekker F. W., Tripepi G., Zoccali C., Jager K. J. Survival analysis I: the Kaplan-Meier method. Nephron - Clinical Practice. 2011;119(1):c83–c88. doi: 10.1159/000324758. [DOI] [PubMed] [Google Scholar]
- 8.Zhao M., Kim P., Mitra R., Zhao J., Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for Tumor Suppressor Genes. Nucleic Acids Research. 2016;44(1):D1023–D1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao M., Sun J., Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Research. 2013;41(1):D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang D. W., Sherman B. T., Tan Q., et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Research. 2007;35(2):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis G., Jr., Sherman B. T., Hosack D. A., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(5):p. P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 12.Tonon L., Touzet H., Varré J.-S. TFM-explorer: mining cis-regulatory regions in genomes. Nucleic Acids Research. 2010;38(2):W286–W292. doi: 10.1093/nar/gkq473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatr-Aryamontri A., Breitkreutz B.-J., Oughtred R., et al. The BioGRID interaction database: 2015 update. Nucleic Acids Research. 2015;43(1):D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J., Zhu W., Xu W., et al. Up-regulation of MBD1 promotes pancreatic cancer cell epithelial-mesenchymal transition and invasion by epigenetic down-regulation of E-cadherin. Current Molecular Medicine. 2013;13(3):387–400. [PubMed] [Google Scholar]
- 15.Derks S., Bosch L. J. W., Niessen H. E. C., et al. Promoter CpG island hypermethylation- and H3K9me3 and H3K27me3-mediated epigenetic silencing targets the deleted in colon cancer (DCC) gene in colorectal carcinogenesis without affecting neighboring genes on chromosomal region 18q21. Carcinogenesis. 2009;30(6):1041–1048. doi: 10.1093/carcin/bgp073. [DOI] [PubMed] [Google Scholar]
- 16.Bader S., Walker M., McQueen H. A., et al. MBD1, MBD2 and CGBP genes at chromosome 18q21 are infrequently mutated in human colon and lung cancers. Oncogene. 2003;22(22):3506–3510. doi: 10.1038/sj.onc.1206574. [DOI] [PubMed] [Google Scholar]
- 17.Liu H., Jin G., Wang H., et al. Methyl-CpG binding domain 1 gene polymorphisms and lung cancer risk in a Chinese population. Biomarkers. 2008;13(6):607–617. doi: 10.1080/13547500802168031. [DOI] [PubMed] [Google Scholar]
- 18.Jang J.-S., Lee S. J., Choi J. E., et al. Methyl-CpG binding domain 1 gene polymorphisms and risk of primary lung cancer. Cancer Epidemiology, Biomarkers & Prevention. 2005;14(11):2474–2480. doi: 10.1158/1055-9965.epi-05-0423. [DOI] [PubMed] [Google Scholar]