Abstract
N2-Alkyl-2′-deoxyguanosine triphosphate (N2-alkyl-dGTP) derivatives with methyl, butyl, benzyl, 4-ethynylbenzyl substituents were prepared and tested for substrates for human DNA polymerases. N2-Benzyl-dGTP was equal to dGTP as a substrate for DNA polymerase κ, but was a poor substrate for pol β, δ, η, ι, or ν. In vivo reactivity was evaluated by incubation of N2-4-ethynylbenzyl-dG with wild-type and pol κ deficient mouse embryonic fibroblasts. CuAAC click reaction with 5(6)-FAM-azide demonstrate that only pol κ containing cells were able to incorporate N2-4-ethynylbenzyl-dG into the nucleus. This is the first instance of a Y-family-polymerase-specific dNTP and this method can be used to probe the activity of pol κ in vivo.
Keywords: DNA polymerase, DNA modification, fluorescent labelling, nucleotides, bioconjugation
Graphical Abstract
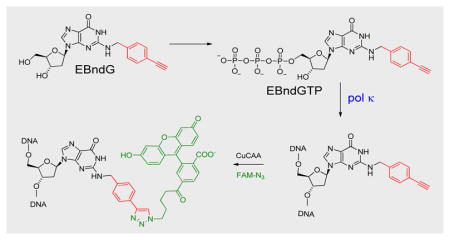
Synthetic nucleotide analogs are widely used tools in chemical biology, diagnostics, and therapeutics. Modifications on the Watson-Crick hydrogen-bonding face have been employed to probe the value of Watson-Crick hydrogen bonds in DNA replication,[1] and create extra-biological base pairs.[2] Minor groove modifications are used to elucidate critical protein-DNA interactions,[3] while major groove modifications have proved to be useful in exploring polymerase enzymology[4] and cellular reactivity.[5] Inhibitors of viral reverse transcriptases are in clinical use for HIV, hepatitis B, and hepatitus C therapy.[6] Human DNA polymerase inhibitors are also in use for cancer chemotherapy. Gemcitabine, a cytidine analog that is incorporated into the DNA but then inhibits DNA synthesis, is used to treat pancreatic cancer, non-small cell lung cancer, breast cancer, and bladder cancer.[6c] Gemcitabine is effective because it affects rapidly growing tumor cells more than normal tissue. More recently, specialized polymerases that are over expressed in tumors have been the target of inhibition studies.[7] Identification of nucleotide analogs as selective substrates or inhibitors for specific polymerases is challenging because all polymerases utilize the four cononocial dNTPs, and correct base pairing is mostly dependent on the polymerase recognizing Watson-Crick geometry. Recently, engineered polymerases were utilized to create a modifed nucleotide that can recognize a carcinogen-modified DNA template,[8] and expanded size dNTPs (dxNTPs) have been shown to have some selectivity for human DNA polymerase θ.[9] In this report, we utilize the known reactivity of DNA polymerase κ to rationally design N2-benzyl-dGTPs that are highly specific substrates for pol κ. These triphosphates are the first reports of nucleotide triphosphates that are highly selective substrates for a human Y-family polymerase. These compounds can be utilized to probe for the reactivity of pol κ in vivo, and potentially be modified to be selective inhibitors of pol κ.
The mammalian cell utilizes sixteen DNA polymerases to replicate DNA, the four high-fidelity enzymes that duplicate the bulk of genomic and mitochondrial DNA, together with specialized DNA polymerases that perform roles in the DNA damage response. Translesion DNA synthesis (TLS) polymerases are a subset of the specialized polymerases that are involved in the bypass of DNA damage.[10] TLS polymerases include the Y-family DNA polymerases, pol η, pol ι, pol κ, and REV1, the B-family pol ζ, [10b] and perhaps other pols such as λ, ν, θ, and PrimPol.[11] These polymerases have unique DNA binding sites that enable the polymerases to bypass a variety of DNA damage. However, polymerases that participate in lesion bypass also perform other functions. For example, while DNA polymerase κ (pol κ) is the most active polymerase in the accurate bypass of bulky N2-dG adducts,[12] pol κ also bypasses the structurally divergent 8-oxo-dG,[13] participates in nucleotide excision repair,[14] replicates non-B-DNA sequences,[15] and it’s polymerase activity is involved in the initiation of the ATR checkpoint signal.[16] In addition, abnormal expression of pol κ correlates with increased mutations in tumors.[17] Further complicating the matter, many of these function are not unique to pol κ, as both pol ι and η[18] can bypass N2-dG adducts, pol δ is the major NER polymerase, and pol η can also replicate non-B-DNA sequences.[19]
The roles of individual polymerases in the cell are difficult to resolve, in part, because all polymerases utilize undamaged DNA and the four dNTPs as substrates. To help elucidate the many roles of pol κ, we have designed a triphosphate that is a highly specific substrate for pol κ. Here we describe the synthesis of N2-benzyl-dGTP (N2-Bn-dGTP), and show that it is a substrate for purified pol κ but a very poor substrate for pol β, δ, η, ι, and ν. We also show that N2-p-ethynylbenzyl-dG (EBndG), when given to cells, is incorporated into the DNA only in the presence of pol κ.
The rationale for the design of a pol κ-specific dNTP substrate is illustrated in Figure 1. Pol κ bypasses hydrophobic N2-alkyl-dG adducts ranging from methyl to (benzo[a]pyren-6-yl)methyl,[20] as well as the carcinogenic N2-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo [a] pyren-10-yl-dG (N2-BP-dG) (Figure 1a) with kinetic parameters approaching those of undamaged DNA.[21] While high fidelity polymerases utilize interactions with the minor groove of the DNA to enhance fidelity, pol κ has an opening on the minor groove side of the DNA that can accommodate bulky alkyl groups attached to the N2-position of dG in the template base.[22] We hypothesized that this binding pocket can also accommodate the N2-alkyl groups when bound to the incoming dGTP (Figure 1b). We investigated the specificity of the dNTPs by examining the in vitro reactivity with an array of human polymerases that includes pols κ, β, δ, η, ι, and ν. These enzymes include members of the A-, B-, X- and Y-polymerase families.
Figure 1.
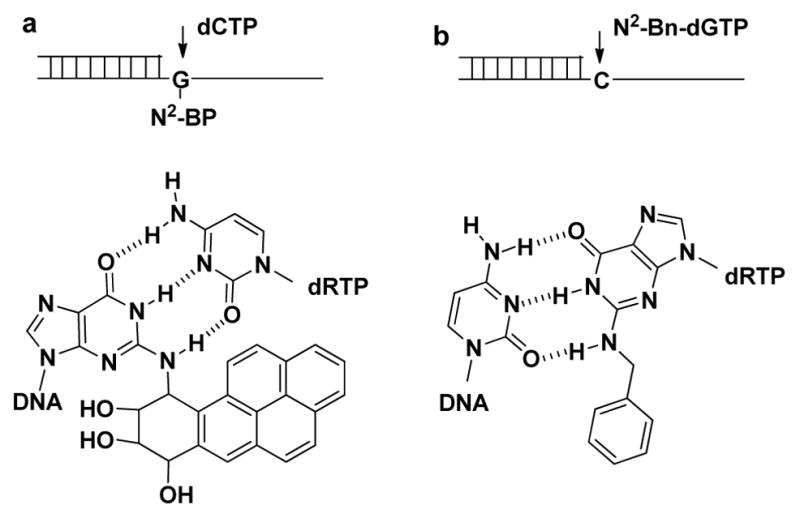
Rationale for design of pol κ specific dNTP substrate. a, Incorporation of dCTP opposite N2-BP-dG. b, Incorporation of N2-Bn-dGTP opposite dC.
The in vitro activities of purified human polymerases with N2-alkyl-dGTPs were analyzed by primer extension assays, as illustrated in Figure 2. The panels on the left show that pol κ, η and ι readily inserted dGTP opposite dC. The panels on the right demonstrate that only pol κ rapidly incorporated N2-Bn-dGTP. Even at a higher N2-Bn-dGTP concentration and longer time periods, pol η and ι were much less efficient at utilizing N2-Bn-dGTP. Figure S1 (Supporting Information) shows representative gel images of all the polymerases examined. The in vitro reactivity of six purified polymerases were examined as described in the Supporting Information. The kinetic parameters are presented in Table S1 and the reactivities are summarized in Figure 3.
Figure 2.
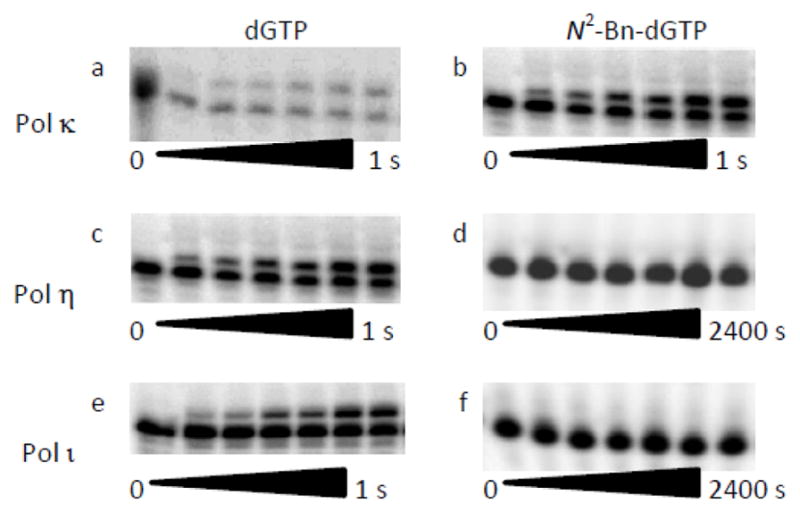
Comparison of the reactivity of pol κ, η and ι with N2-Bn-dGTP. In the primer extension assay, DNA (10 nM) and the polymerase (100 nM) were reacted with 5 μM dGTP (a, c, e) or N2-Bn-dGTP (b, d, f) at a concentration of 5 μM (b), or 25 μM (d, f) for the indicated amount of time. The substrates and products were separated by PAGE. The lower bands on the polyacrylamide gels are the 15-mer starting material, while the upper bands are the 16-mer products.
Figure 3.
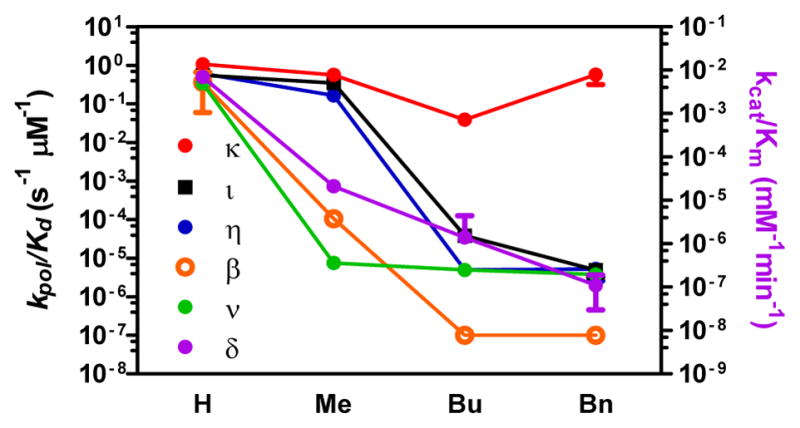
Reactivity of dGTP (H), N2-methyl-dGTP (Me), N2-butyl-dGTP (Bu) and N2-Bn-dGTP (Bn) with DNA polymerase κ. ι, η, β, ν, and δ/PCNA.
Pol κ reacted rapidly with the three dGTP analogs (Figure S2, S3). The methyl and benzyl substitutions did not impact the kpol, while the Kd rose by a factor of 2. N2-butyl-dGTP was a slightly poorer substrate than dGTP, with a 20-fold decrease in kpol. The Y-family pols η and ι, while not affected by the methyl substitutions, experienced a 10,000-fold reduction in kpol with the butyl substitution (Figures S4–S7). Pol ι reacted poorly with N2-Bn-dGTP, while the reactivity of pol η further decreased due to a decrease in kpol and an increase in Kd. In spite of the ability of pol η to bypass N2-(2-naphthyl)methyl-dG in the template, the polymerase in unable to utilize N2-Bu-dGP or N2-Bn-dGTP as substrates. Of the Y-family polymerases, only pol κ reacts with N2-Bn-dGTP at rates similar to unmodified dNTPs.
Next, we examined the reactivity with representatives of the A-, B- and X-polymerase families. Humans have three A-family polymerases, the high fidelity pol γ, responsible for mitrochondirial replication, and the low fidelity pol θ and ν. Pol θ has a role in alternative NHEJ while pol ν is implicated in the repair of intrastrand crosslinks and translesion DNA synthesis. Despite the fact that pol ν has low fidelity, it incorporated the N2-alkyl-dGTPs very poorly (Figure S10, S11). The kpol/Kd for N2-Bn-dGTP is decreased > 105–fold when compared with dGTP. The reactivity of N2-Bn-dGTP with pol γ and θ still needs to be determined. Humans have four B-family polymerases α, δ and ε are involved in high fidelity DNA replication, while pol ζ is involved in TLS. Pol δ is a B-family polymerase implicated in lagging strand replication. We examined the reactivity of pol δ/PCNA in steady-state kinetics (Figure S12). The reaction is sensitive to small modifications as demonstrated by the 300-fold decreased in kcat/Km for N2-Me-dGTP relative to dGTP (Table S1). Increasing the size of the substitution further decreased reactivity, with a 105-fold decrease in kcat/Km for pol δ with N2-Bn-dGTP. Similarities among B-family polymerases suggest that both pol α and ε will be equally poor at utilizing N2-Bn-dGTP as a substrate. It was previously shown that N2-butylphenyl-dGTP was not a substrate for pol α, γ and β, but was an inhibitor of pol α.[23] Humans have three X-family polymerases, β, λ, and μ. Pol β is the primary polymerase involved in base excision repair, while pol λ and μ are involved in NHEJ. The preferred substrate for pol β is a duplex DNA with a single-nucleotide gap. Pol β was very inefficient at the incorporation of nucleotides opposite the N2-BP-dG.[24] Unsurprisingly, we found that pol β did not accommodate steric bulk at the N2-position of the dNTP (Figures S8, S9). Methyl substitution decreased kpol/Kd values over 1000-fold. Further increases in size of the alkyl group lead to a decrease in kpol/Kd of greater than 6-orders of magnitude.
The fidelity of the pol κ with N2-Bn-dGTP is slightly better than for dGTP. We investigated the fidelity of the incorporation of N2-Bn-dGTP opposite dA, dC, dG, and dT as the template base with Michaelis-Menten kinetics. As summarized in Table S2, the preference for dC is higher with N2-Bn-dGTP than for dGTP. The increased selectivity for dC as template is due to the decreased rate of mispair formation. The finc for dGTP was 0.07 for each nucleotide, while finc averaged 0.015 for N2-Bn-dGTP.
The in vitro reactions have shown that N2-Bn-dGTP reacts with pol κ, but not pol β, δ, η, ι, or ν. To determine if this reaction occurs in vivo, we synthesized N2-(4-ethynylbenzyl)-dG (EBdG) as described in the Supplementary Information. Attachment of the ethynyl moiety to the benzyl group enables the use of copper(I)-catalyzed azide-alkyne cycloaddition chemistry to attach a fluorescent group to the nucleoside and thus determine the amount and location of EBdG in the cell.[5] The labelling of the nuclei is shown in Figure 4a–c, in which mouse embryonic fibroblasts (MEF) were incubated with 10 μM EBdG for 24h. The cells were fixed and the ethynyl groups were reacted with azido-FAM, and the nuclei identified with DAPI-staining of the DNA. The complete overlap DAPI (Figure 4a) and FAM (Figure 4b) signals indicates that the cells incorporated EBdG into the nucleus.
Figure 4.
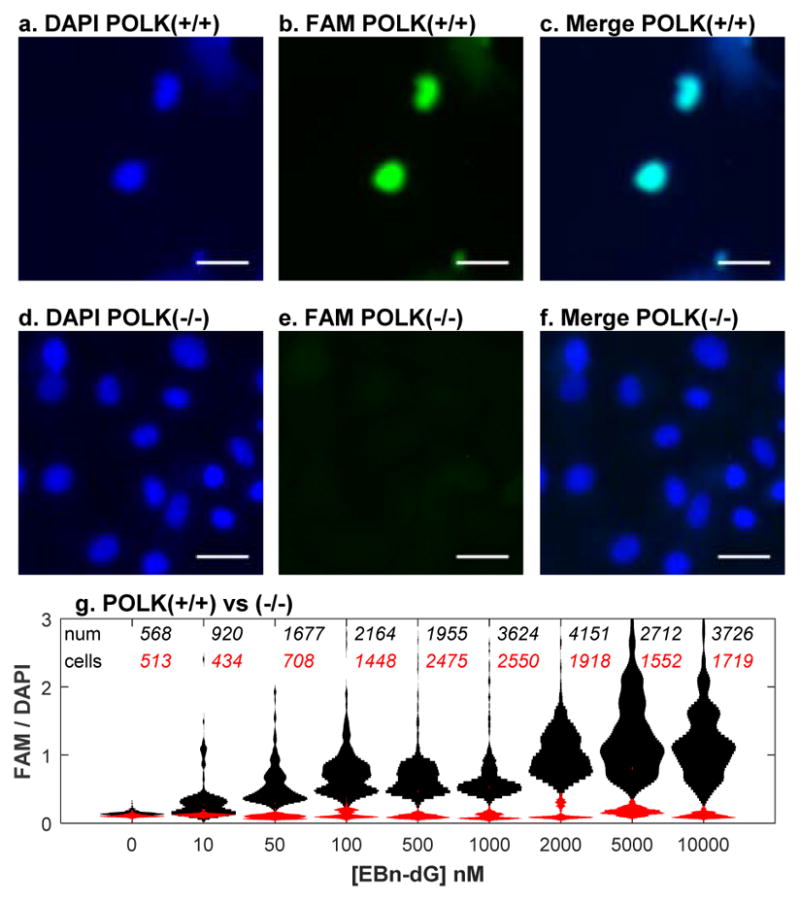
Detection of EBdG in nucleus of pol κ containing cells. Wild-type (a–c) and POLK(−/−) (d–f) cells were treated with 10 μM EBdG for 24 h. DNA is visualized with DAPI-staining (a,d) and EBdG visualized by Click reaction with FAM-N3 (b,e). The white bar is 40 μm. g, Levels of FAM-EBdG conjugate in nuclei of MEF POLK(+/+) (black) and POLK(−/−) (red) cells. The incubation with EBdG was performed three times. The total number of cells in violin plots are described for wildtype (black) and POLK(−/−)(red) cells.
To determine if pol κ is the polymerase that incorporates EBdG into DNA in mammalian cells we incubated POLK deficient MEF cells with the nucleoside and then analyzed the incorporation by visualization after the Click reaction. Figure 4d–f shows that POLK deficient MEF cells were unable to incorporate EBdG into the nucleus. The relative intensities of FAM signal to the DAPI signal was measured for each nucleus. The first lane of Figure 4g shows the background level of FAM fluorescence in the nucleus. At 10 nM EBdG, approximately half of the wild-type cells incorporated a small amount of the nucleoside. Above this concentration, all cells incorporated EBdG. In cells without pol κ, EBdG is not incorporated into the DNA. These results indicate that EBdG is able to diffuse into the nucleus, be converted into the triphosphate, and be incorporated into the DNA. In addition, the incorporation into the DNA is dependent on the presence of pol κ.
In conclusion, we have found that N2-alkyl-dGTPs are substrates for DNA pol κ. The kinetic parameters for N2-Bn-dGTP are only slightly slower than dGTP. The fidelity of the pol κ catalyzed incorporation is slightly better for N2-Bn-dGTP than it is for dGTP. We also found that N2-Bn-dGTP reacted slowly with representatives from the A-family (ν), B-family (δ), or an X-family (β); the kpol/Kd or kcat/Km values are <105 that of pol κ. We also found that the ethynyl nucleoside, N2-EBn-dG is incorporated into the DNA by cells by pol κ. These reagents will be very useful at elucidating the cellular activities of pol κ, and can be the basis for the selective inhibition of pol κ.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences (ES 021762). We thank Cyrus Vasiri for the gift of wild-type and POLK(−/−) MEF cells.
Footnotes
Supporting information for this article is given via a link at the end of the document
Contributor Information
A. S. Prakasha Gowda, Department of Biochemistry and Molecular Biology, Pennsylvania State University, 500 University Dr., Hershey, PA 17033
Marietta Lee, Department of Biochemistry and Molecular Biology, New York Medical College, Valhalla, NY 10595.
Thomas E. Spratt, Department of Biochemistry and Molecular Biology, Pennsylvania State University, 500 University Dr., Hershey, PA 17033
References
- 1.a) Kool ET, Sintim HO. Chem Commun (Camb) 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]; b) Kim TW, Delaney JC, Essigmann JM, Kool ET. Proc Natl Acad Sci USA. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kool ET. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 2.a) Sefah K, Yang Z, Bradley KM, Hoshika S, Jiménez E, Zhang L, Zhu G, Shanker S, Yu F, Turek D, Tan W, Benner SA. Proceedings of the National Academy of Sciences. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lavergne T, Degardin M, Malyshev DA, Quach HT, Dhami K, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2013;135:5408–5419. doi: 10.1021/ja312148q. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Benner SA. Acc Chem Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]; d) Li L, Degardin M, Lavergne T, Malyshev DA, Dhami K, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2014;136:826–829. doi: 10.1021/ja408814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Meyer AS, Blandino M, Spratt TE. J Biol Chem. 2004;279:33043–33046. doi: 10.1074/jbc.C400232200. [DOI] [PubMed] [Google Scholar]; b) Morales JC, Kool ET. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]; c) Xia S, Christian TD, Wang J, Konigsberg WH. Biochemistry. 2012;51:4343–4353. doi: 10.1021/bi300416z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Hottin A, Marx A. Acc Chem Res. 2016;49:418–427. doi: 10.1021/acs.accounts.5b00544. [DOI] [PubMed] [Google Scholar]; b) Hocek M. J Org Chem. 2014;79:9914–9921. doi: 10.1021/jo5020799. [DOI] [PubMed] [Google Scholar]
- 5.a) Sirbu BM, Couch FB, Cortez D. Nat Protocols. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sirbu BM, McDonald WH, Dungrawala H, Badu-Nkansah A, Kavanaugh GM, Chen Y, Tabb DL, Cortez D. J Biol Chem. 2013;288:31458–31467. doi: 10.1074/jbc.M113.511337. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bergen K, Steck AL, Strütt S, Baccaro A, Welte W, Diederichs K, Marx A. J Am Chem Soc. 2012;134:11840–11843. doi: 10.1021/ja3017889. [DOI] [PubMed] [Google Scholar]
- 6.a) Clark DN, Hu J. Antiviral Res. 2015;123:132–137. doi: 10.1016/j.antiviral.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Margolis AM, Heverling H, Pham PA, Stolbach A. J Med Toxicol. 2014;10:26–39. doi: 10.1007/s13181-013-0325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jordheim LP, Durantel D, Zoulim F, Dumontet C. Nature Reviews Drug Discovery. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]; d) Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, Symonds WT, Hindes RG, Berrey MM. N Engl J Med. 2013;368:34–44. doi: 10.1056/NEJMoa1208953. [DOI] [PubMed] [Google Scholar]; e) Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HM, Niu C, Otto MJ, Furman PA. J Med Chem. 2010;53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- 7.Korzhnev DM, Hadden MK. J Med Chem. 2016;59:9321–9336. doi: 10.1021/acs.jmedchem.6b00596. [DOI] [PubMed] [Google Scholar]
- 8.a) Wyss LA, Nilforoushan A, Eichenseher F, Suter U, Blatter N, Marx A, Sturla SJ. J Am Chem Soc. 2015;137:30–33. doi: 10.1021/ja5100542. [DOI] [PubMed] [Google Scholar]; b) Wyss LA, Nilforoushan A, Williams DM, Marx A, Sturla SJ. Nucleic Acids Res. 2016;44:6564–6573. doi: 10.1093/nar/gkw589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Kent T, Rusanov TD, Hoang TM, Velema WA, Krueger AT, Copeland WC, Kool ET, Pomerantz RT. Nucleic Acids Res. 2016;44:9381–9392. doi: 10.1093/nar/gkw721. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Winnacker M, Kool ET. Angew Chem Int Ed. 2013;52:12498–12508. doi: 10.1002/anie.201305267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Yang W, Woodgate R. Proc Natl Acad Sci USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Prakash S, Johnson RE, Prakash L. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]; c) Lange SS, Takata K, Wood RD. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Sale JE, Lehmann AR, Woodgate R. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Skosareva LV, Lebedeva NA, Rechkunova NI, Kolbanovskiy A, Geacintov NE, Lavrik OI. DNA Repair (Amst) 2012;11:367–373. doi: 10.1016/j.dnarep.2012.01.002. [DOI] [PubMed] [Google Scholar]; b) Takata K-i, Shimizu T, Iwai S, Wood RD. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]; c) Yousefzadeh MJ, Wood RD. DNA Repair (Amst) 2013;12:1–9. doi: 10.1016/j.dnarep.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, Taylor E, Stevanovic I, Green AJ, Stracker TH, Lindsay HD, Doherty AJ. Mol Cell. 2013;52:566–573. doi: 10.1016/j.molcel.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. J Biol Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 13.Vasquez-Del Carpio R, Silverstein TD, Lone S, Swan MK, Choudhury JR, Johnson RE, Prakash S, Prakash L, Aggarwal AK. PLoS One. 2009;4:e5766. doi: 10.1371/journal.pone.0005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogi T, Lehmann AR. Nature Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 15.a) Baptiste BA, Eckert KA. Environ Mol Mutagen. 2012;53:787–796. doi: 10.1002/em.21721. [DOI] [PubMed] [Google Scholar]; b) Walsh E, Wang X, Lee MY, Eckert KA. J Mol Biol. 2013;425:232–243. doi: 10.1016/j.jmb.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bétous R, Pillaire MJ, Pierini L, van der Laan S, Recolin B, Ohl-Séguy E, Guo C, Niimi N, Grúz P, Nohmi T, Friedberg E, Cazaux C, Maiorano D, Hoffmann JS. The EMBO Journal. 2013;32:2172–2185. doi: 10.1038/emboj.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillaire M-J, Betous R, Conti C, Czaplicki J, Pasero P, Bensimon A, Cazaux C, Hoffmann J-S. Cell Cycle. 2007;6:471–477. doi: 10.4161/cc.6.4.3857. [DOI] [PubMed] [Google Scholar]
- 18.a) Choi JY, Guengerich FP. J Biol Chem. 2006;281:12315–12324. doi: 10.1074/jbc.M600112200. [DOI] [PubMed] [Google Scholar]; b) Choi JY, Guengerich FP. J Mol Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 19.Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DS, Monnat RJ, Jr, Cazaux C, Hoffmann JS. Mol Cell Biol. 2009;29:3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi JY, Angel KC, Guengerich FP. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 21.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 22.Jha V, Bian C, Xing G, Ling H. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a) Khan NN, Wright GE, Dudycz LW, Brown NC. Nucleic Acids Res. 1984;12:3695–3706. doi: 10.1093/nar/12.8.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Khan NN, Wright GE, Dudycz LW, Brown NC. Nucleic Acids Res. 1985;13:6331–6342. doi: 10.1093/nar/13.17.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chary P, Beard WA, Wilson SH, Lloyd RS. Chem Res Toxicol. 2012;25:2744–2754. doi: 10.1021/tx300368f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


