Abstract
Phagocytosis is a critical process to maintain tissue homeostasis. In the retina, photoreceptor cells renew their photo-excitability by shedding photoreceptor outer segments (POSs) in a diurnal rhythm. Shed POSs are phagocytosed by retinal pigment epithelial (RPE) cells to prevent debris accumulation, retinal degeneration and blindness. Phagocytosis ligands are the key to understanding how RPE recognizes shed POSs. Here we characterized mesoderm development candidate 2 (Mesd or Mesdc2), an endoplasmic reticulum (ER) chaperon for low-density lipoprotein receptor-related proteins (LRPs), to extrinsically promote RPE phagocytosis. The results showed that Mesd stimulated phagocytosis of fluorescence-labeled POS vesicles by D407 RPE cells. Ingested POSs were partially degraded within 3 h in some RPE cells to dispense undegradable fluorophore throughout the cytoplasm. Internalized POSs were colocalized with phagosome biomarker Rab7, suggesting that Mesd-mediated engulfment is involved in a phagocytosis pathway. Mesd also facilitated phagocytosis of POSs by primary RPE cells. Mesd bound to unknown phagocytic receptor(s) on RPE cells. Mesd was detected in the cytoplasm, but not nuclei, of different retinal layers and is predominantly expressed in the ER-free cellular compartment of POSs. Mesd was not secreted into medium from healthy cells but passively released from apoptotic cells with increased membrane permeability. Released Mesd selectively bound to the surface of POS vesicles and apoptotic cells, but not healthy cells. These results suggest that Mesd may be released from and bind to shed POSs to facilitate their phagocytic clearance.
Keywords: Mesd, Mesdc2, phagocytosis, retinal pigment epithelial cells, RPE
Introduction
Phagocytosis or engulfment of apoptotic cells and cellular debris is an important biological process to maintain tissue homeostasis and innate immune balance (Erwig and Henson, 2007; Poon et al., 2014). The event is initiated by phagocyte recognition of extracellular targets via multiple ligand-receptor interactions. Phagocytic receptors can recognize targets directly via abnormal “eat-me” signals, such as phosphatidylserine and calreticulin, on target surface or indirectly through bridging molecules (Gardai et al., 2005; Hochreiter-Hufford and Ravichandran, 2013). Phagocytosis ligands are composed of both eat-me signals and bridging molecules and are the key to elucidating the mechanisms by which phagocytosis targets are recognized and selected for clearance (Li, 2012). Identification of these signaling molecules will help understand the mechanisms of diseases related to phagocytosis dysfunction (Gal et al., 2000; Jonsson et al., 2013; Neumann and Takahashi, 2007) and may eventually lead to therapeutic intervention by modulating engulfment in disease conditions (Brown and Neher, 2012; Fricker et al., 2012; Poon et al., 2014).
Retinal pigment epithelial (RPE) cells are one of the most active phagocytes in our body to maintain retinal homeostasis (Li, 2012). Photoreceptor outer segments (POSs) are capable of converting light to electric impulses but are susceptible to photo-oxidative damage. For renewal, photoreceptor cells shed the oldest POSs with the most damage from their tips in a diurnal rhythms (Kevany and Palczewski, 2010; Nandrot et al., 2004). Shed POS vesicles are phagocytosed by adjacent RPE for nutrient recycle and regeneration of POSs at their bases near the connecting cilia (Strauss, 2005). The delicate balance between POS shedding and regeneration is pivotal to maintain the precise length of POSs and their close interaction with RPE (Strauss, 2005). This interaction is crucial to proper function of photoreceptors, such as the visual cycle (Strauss, 2005). Defect in RPE phagocytosis, such as Royal College of Surgeon (RCS) rats with a mutation in phagocytic receptor MerTK, may lead to accumulation of unphagocytosed debris, retinal degeneration and blindness (Dowling and Sidman, 1962; Gal et al., 2000).
Phagocytosis ligands are traditionally identified on a case-by-case basis with technical challenges. We recently developed an innovative technology of open reading frame phage display (OPD) for the unbiased identification of phagocytosis ligands in the absence of receptor information (Caberoy and Li, 2012; Caberoy et al., 2010a; Li, 2012). We further combined OPD with next generation DNA sequencing (NGS) as the only technology of “ligandomics” to globally identify cell-wide phagocytosis ligands (Ding et al., 2015; Guo et al., 2015a). One of the identified proteins for microglial phagocytosis is mesoderm development candidate 2 (Mesd or Mesdc2) (unpublished data, Ding et al.).
Mesd is an endoplasmic reticulum (ER) chaperone to facilitate the folding of the low-density lipoprotein receptor-related protein (LRP) family (Hsieh et al., 2003). Mutations in Mesd are associated with developmental defects (Hsieh et al., 2003). LRPs play diverse roles in cell signaling and endocytosis (Li et al., 2000; Schneider and Nimpf, 2003). Mesd was reported to bind to LRP5/6 on cell surface, competitively block the binding of Wnt ligands to LRP5/6 and inhibit Wnt/β-catenin signaling (Li et al., 2005; Lin et al., 2013; Lu et al., 2010). However, it is unclear how Mesd strictly expressed in the ER may regulate LRP5/6 on cell surface (Li et al., 2005).
Here we report that Mesd can be released from apoptotic cells and that extracellular Mesd binds to shed POSs and stimulates their phagocytosis by RPE cells. The detection of endogenous Mesd with shed POS vesicles suggests that released Mesd may extrinsically facilitate the clearance of shed POSs by RPE cells.
Materials and Methods
Cell culture
D407 human RPE and HEK293 cell lines were previously described (Caberoy et al., 2009) and cultured in Dulbeccos's modified essential medium (DMEM) supplemented with 10% FBS and 2 mM L-glutamine (Mediatech, Manassas, VA).
Plasmids
The coding sequence of Mesd was amplified by reverse transcription-PCR (RT-PCR) from mouse brain, as described (Caberoy et al., 2010b). Briefly, mice (C57BL/6, 6-8 weeks old) were euthanized by CO2 inhalation, followed by cervical dislocation. The brain was isolated. Total RNA was extracted using TRIzol reagent (Life Technologies, Grand Island, NY). Mesd coding sequence with a C-terminal FLAG was amplified by RT-PCR, digested and cloned in to pEGFP-N1 plasmid (Clonetch, Mountain View, CA) at BglII and NotI sites to replace green fluorescent protein (GFP). Additionally, Mesd coding sequence was amplified by PCR, digested with BglII and XhoI, and cloned into pMal-c4E plasmid (New England Biolab, Ipswich, MA) in frame at BamHI and SalI sites. The resulting Mesd-FLAG and maltose-binding protein (MBP)-Mesd plasmids were verified by sequencing. GFP-FLAG plasmid was previously described (Caberoy et al., 2010a).
All animal procedures in this study were approved by the Institutional Animal Care and use Committee at the University of Miami and complied with the Guide for the Care and Use of Laboratory Animals published by the United States National Institutes of Health (NIH).
Recombinant Mesd
MBP-Mesd fusion protein and MBP control were expressed in BL21(DE3) bacteria and purified using amylose columns, as described (Guo et al., 2015a). Purified MBP-Mesd and MBP were dialyzed against phosphate-buffered saline (PBS), analyzed for purity by SDS-PAGE and verified by Western blot using affinity-purified anti-Mesd antibody (Thermo Fisher Scientific, Waltham, MA).
Primary RPE cells
Primary RPE cells were prepared, as previously described (Guo et al., 2015a). Briefly, C57BL/6 mice at postnatal day 10 were euthanized by CO2 inhalation. Eyes were enucleated. After the removal of the cornea, lens and retina, eyecups were digested with trypsin/EDTA. RPE cells were collected by pipetting and centrifugation. RPE spheres were cultured in Minimum Essential Medium Eagle (MEM) Alpha Modification (Sigma, St. Louis, MO). The medium was supplemented with 10% FBS, 2 mM L-Glutamine, 1X nonessential amino acids, 1X penicillin/streptomycin, bFGF (10 ng/ml), EGF (1 ng/ml), 1X N1 supplement and THT (taurine, 210 ng/ml; hydrocortisone 1.2 μg/ml; triiodo-thyronine, 60 ng/ml) (Sigma) (Salero et al., 2012). RPE spheres were dissociated with trypsin, washed and cultured as monolayer in the same medium without bFGF and EGF for 3 days before phagocytosis assay.
POS vesicle preparation
POS vesicles were prepared from fresh bovine retina and labeled with pHrodo (Caberoy et al., 2012; Guo et al., 2015b). Briefly, fresh bovine eyes within 24 h postmortem were purchased from Pel-Freez Biologicals (Rogers, AR) and received on ice. Retinas were isolated and gently shaken at 4°C for 15 min in PBS containing 2.5% sucrose to detach POSs. After removal of retinas, detached POS vesicles were collected and washed twice by centrifugation at 38,700 × g for 30 min. Purified POS vesicles (500 μg protein) were incubated with pHrodo succinimidyl ester (20 ng/ml, Life Technologies) for 30 min at room temperature with end-over-end rotation. The reaction was stopped by incubation with 1% BSA in PBS for 15 min. Labeled vesicles were washed twice with PBS by centrifugation at 16,000 × g for 30 min.
RPE Phagocytosis assay
D407 RPE cells or RPE primary cells were seeded on coverslips precoated with poly-L-lysine (Sigma) in 12-well plates and cultured overnight. pHrodo-labeled POSs (50 μg/ml) were added to RPE cells for phagocytosis assay in the presence of MBP-Mesd or MBP control at indicated concentrations for 3 h at 37°C. After washing, cells were fixed with 4% buffered paraformaldehyde for 10 min, mounted with DAPI and analyzed by confocal microscopy. Z-stack images of pHrodo signal were obtained using a Leica SP5 confocal microscope with emission wavelength at 551-644 nm (Caberoy et al., 2010a; Caberoy et al., 2010b). Nuclei were stained by DAPI (0.5 μg/ml) and detected with emission wavelength at 415-472 nm. Intracellular pHrodo signals of confocal images were quantified by ImageJ software (NIH) and normalized against cell number (i.e., DAPI spot number) per viewing field. Alternatively, cells were collected by trypsin digestion, washed and analyzed by flow cytometry.
RT-PCR
Retinas isolated from adult C56BL/6 mice in the above experiment were used to extract total RNA using TRIzol reagent, as described (Caberoy et al., 2010b). RT-PCR was performed using the following primers. Mesd primers: 5′-TCAGATCTATGGCGGACACTCCGGGCGAGG-3′ and 5′-TTCTCGAGCTAAAGGTCTTCTCTTCTGCTCCC-3′ (underlined sequences for Mesd, GenBank Accession # NM_023403); GAPDH primers: 5′-TGGTGAAGCAGGCATCTGAG-3′ and 5′-GTGCAGCGAACTTTATTGATGG-3′ (Accession # NM_008084).
Western blot
After POS detachment in the above experiment, bovine retinas were collected, homogenized in RIPA buffer (Thermo Fisher Scientific, Waltham, MA), and analyzed by Western blot using anti-Mesd antibody or anti-β-actin antibody (Cell Signaling, Danvers, MA) (Guo et al., 2015a).
Immunocytochemistry
D407 RPE cells with engulfed pHrodo-labeled POSs in the above phagocytosis assay were fixed with 10% buffered formalin, permeabilized with PBS containing 1% Triton X-100, and incubated with anti-Rab7 antibody (Abcam, Cambridge, MA) (Guo et al., 2015a). After washing, bound antibody was detected using Alexa Fluor 488-labeled secondary antibody. Nuclei were visualized by DAPI. The fluorescence signals of pHrodo, Alexa Fluor 488 and DAPI were detected by confocal microscopy.
Immunohistochemistry
Immunohistochemistry was performed, as previously described (Guo et al., 2015a). Briefly, C57BL/6 mice (6-8 weeks old) were anesthetized by intraperitoneal injection of ketamine (90 μg/g) and xylazine (8 μg/g). Anesthetized mice were intracardially perfused with 4% paraformaldehyde. Eyes were enucleated from euthanized mice and fixed with the same solution overnight at 4°C. After removal of the cornea and lens, eyecups were incubated with sucrose gradient solutions (10% and 20% for 3 h each; 30% for overnight) at 4°C. After 3 rounds of freeze-thaw, eyecups were embedded in OCT (optimal cutting temperature compound). Frozen tissue sections in 7-μm thickness were incubated with rabbit anti-Mesd and mouse anti-rhodopsin antibodies (Millipore, Billerica, MA), followed by Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa 594-labeled goat anti-mouse IgG antibodies. The nuclei were visualized with DAPI. The fluorescence signals were analyzed using a fluorescence microscope.
Extracellular trafficking
HEK293 cells in 6-well plates were transfected with Mesd-FLAG plasmid using jetPRIME reagents (Polyplus Transfection, Illkirch, France). After 48 h, cells were cultured in 293 SFM II medium (Life Technologies) and treated with or without etoposide (200 μM) for 16 h. The conditioned media of apoptotic and healthy cells with released Mesd-FLAG were collected and centrifuged at 200 × g for 10 min at room temperature. Supernatants were filtered through 0.2-μm syringe filters (Pall Life Sciences, Port Washing, NY, low-protein binding), concentrated with Amicon centrifugal filter units (10 kDa, Millipore), and analyzed by Western blot using anti-FLAG M2 monoclonal antibody (Sigma) (Ding et al., 2015).
Binding to POS and D407 cells
Mesd-FLAG or GFP-FLAG plasmid was transfected into HEK293 cells, as described above. Cells were harvested at 48 h post transfection, washed and resuspended in PBS. After 3 cycles of freeze-thaw, cell lysates were prepared by centrifugation at 21,100 × g for 20 min at 4°C and filtration through 0.2-μm syringe filters. D407 cells were collected by mild digestion with trypsin/EDTA. D407 cells or POS vesicles were incubated with the cell lysates at 4°C for 1 h. After washing, cells or vesicles were incubated with anti-FLAG mAb, labeled with Alexa Fluor 594-conjugated secondary antibody, washed and analyzed by flow cytometry.
Binding to apoptotic or healthy cells
Purified MBP-Mesd and MBP were labeled with FITC, as described with minor modifications (Caberoy et al., 2010b). Briefly, proteins were washed in Amicon centrifugal filter unit, resuspended in 0.1 M sodium bicarbonate buffer (pH 8.3), and incubated with FITC-5-EX succinimidyl ester (500 μg/ml, Life Technologies). The labeling reaction was stopped by adding 1/10 volume of 1 M Tris buffer (pH8.0) and incubating for 15 min at room temperature. Labeled proteins were purified using Bio-Spin columns with Bio-Gel P-30 (Bio-Rad, Hercules, CA). For binding analysis, HEK392 cells were treated with or without etoposide for 16 h as above and harvested. FITC-labeled MBP-Mesd or MBP was incubated with apoptotic or healthy cells for 1 h at 4°C, washed, fixed with 4% buffered paraformaldehyde and analyzed by confocal microscopy. Apoptotic cells were labeled with propidium iodide, and nuclei were visualized by DAPI.
Statistical analysis
Data were expressed as means ± s.e.m. and analyzed by Student’s t-test. Data were considered significant when P < 0.05.
Results
Mesd stimulates POS phagocytosis by D407 RPE cells
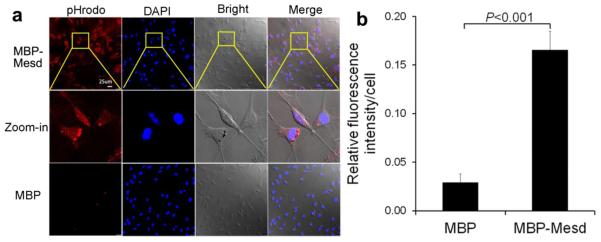
We expressed and purified recombinant Mesd as a fusion protein of MBP, which was verified by SDS-PAGE and Western blot (Fig. 1a). Purified MBP-Mesd promoted phagocytosis of pHrodo-labeled POS vesicles by D407 RPE cells. (Fig. 1b). pHrodo is a pH-sensitive fluorogenic dye that is not fluorescent at neutral pH but is activated in acidic phagosomes to generate red fluorescence (Caberoy et al., 2012). This enabled reliable detection of engulfed POS vesicles. In addition, pHrodo and DAPI signals on the same confocal z-stack images were superimposed with the cognate bright fields to show internalized targets within the boundary of RPE cells. Zoom-in merged images (middle-row images, Fig. 1b) indicated that some ingested pHrodo signals were released from degraded POSs and dispensed throughout the cytoplasm within 3 h. Other pHrodo-POSs remained as undegraded granules. As a control, MBP failed to stimulate RPE phagocytosis (bottom-row images, Fig. 1b). Quantification of engulfed pHrodo signals by confocal microscopy indicated that MBP-Mesd significantly induced RPE phagocytosis of POS vesicles (P<0.001, Fig. 1c). These results were verified by flow cytometry to quantify engulfed pHrodo signals (Fig. 1d).
Fig. 1.
Mesd facilitates RPE phagocytosis. a Purification of MBP-Mesd. MBP-Mesd and MBP control were expressed in bacteria, purified and analyzed by SDS-PAGE to determine purity (left panel). MBP-Mesd was verified by Western blot using anti-Mesd antibody (right panel). b Mesd stimulates phagocytosis of pHrodo-labeled POS vesicles by D407 RPE cells. The same confocal z-stack images of pHrodo and DAPI are superimposed with the cognate bright fields to reveal engulfed POSs inside the boundary of RPE cells. Images in the top and middle rows are for MBP-Mesd (100 nM). MBP (bottom row, 100 nM) was included as a negative control. Arrows indicate undegraded pHrodo-labeled targets inside RPE cells. Arrowheads indicate pHrodo signals released from degraded POSs and dispensed throughout the cytoplasm. Bar = 25 μm. c The relative fluorescence intensity per cell in b was quantified. d Engulfed pHrodo signal in b was quantified by flow cytometry. e RPE phagocytosis was analyzed with increased concentrations of MBP-Mesd or MBP control and quantified. Mean ± s.e.m., n=5, Student’s t-test. d Engulfed pHrodo signal in b was quantified by flow cytometry.
Phagocytosis studies with increased concentrations of MBP-Mesd and MBP indicated that Mesd significantly induced RPE phagocytosis at 50, 100 and 200 nM with maximal activity at 100 nM (Fig. 1e).
Mesd expression in the retina
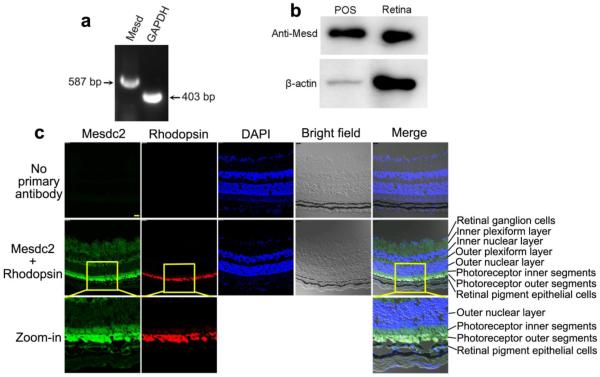
We detected Mesd expression in the retina by RT-PCR (Fig. 2a) and verified its expression in the retinal homogenate and shed POSs by Western blot (Fig. 2b). To detect whether Mesd is presence in POSs, we performed immunohistochemistry. The results showed that Mesd is expressed in retinal ganglion cells, inner and outer plexiform layers, photoreceptor inner segments and RPE (Fig. 2c). Few signals were found in the inner and outer nuclear layer. This distinct expression pattern is consistent with the reported Mesd expression in the ER but not in the nuclei. Surprisingly, the strongest Mesd signals were detected in POSs and colocalized with POS biomarker rhodopsin.
Fig. 2.
Mesd expression in the retina. a RT-PCR to detect Mesd expression in mouse retina. GAPDH was included as a positive control. b Western blot analysis of Mesd expression in the retina and shed POSs. 300 μg protein/lane for retinal homogenate or POS vesicles. c Immunohistochemistry to detect the expression pattern of Mesd in the retina. Mesd is expressed in the retinal ganglion cells, inner and outer plexiform layers, photoreceptor inner segments and RPE. However, the highest level of Mesd expression was detected in the POSs and colocalized with rhodopsin. Few signals were detected in the inner and outer unclear layers.
Mesd induces POS phagocytosis by primary RPE cells
To further verify the finding, we isolated primary RPE cells from mouse retinas at neonatal day 10. Mesd activity to facilitate phagocytosis of POS vesicles by primary RPE was analyzed. The results showed that MBP-Mesd significantly stimulated phagocytosis by primary RPE cells (P<0.001, Fig. 3a,b). MBP had minimal activity to induce the phagocytosis.
Fig. 3.
Mesd promotes phagocytosis of POS by primary RPE cells. a RPE phagocytosis was performed with primary RPE cells in the presence of MBP-Mesd or MBP (100 nM), as described in Fig. 1b. b Quantification of engulfed pHrodo signals in a, as described in Fig. 1c. Bar = 25 μm. Mean ± s.e.m., n = 5, t-test.
Mesd-mediated engulfment through phagosomes
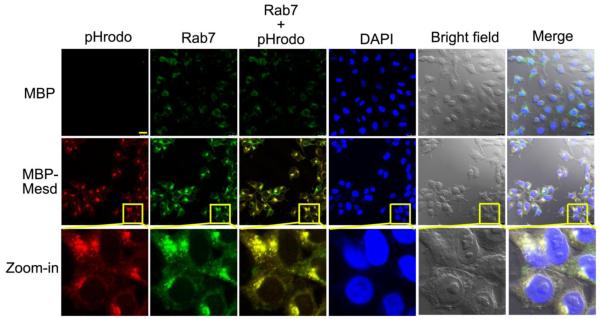
To investigate whether Mesd-mediated internalization of POSs by RPE is through phagocytosis pathway, we analyzed the colocalization of ingested pHrodo signals with phagosome marker Rab7 in D407 cells. The data indicated that Mesd-mediated engulfment of POSs induced aggregation of Rab7, which was colocalized with ingested pHrodo signals (Fig. 4). This colocalization suggests that Mesd promoted POS internalization through RPE phagocytosis pathway.
Fig. 4.
Mesd facilitates POS internalization through RPE phagosome pathway. D407 RPE cells with internlized pHrodo-POSs in Fig. 1b were fixed, permeabilized and analyzed for intracellular phagosome marker Rab7 by immunocytochemistry using anti-Rab7 antibody and Alexa Fluor 488 secondary antibody. Mesd-mediated engulfment induced aggregation of Rab7. Aggregated Rab7 is colocalized with internalized pHrodo. Bar = 25 μm.
Mesd selectively binds to POSs and apoptotic cells
Phagocytosis ligands should selectively recognize extracellular targets, such as apoptotic cells and shed POSs, but not healthy cells. This selective recognition is critical to avoid engulfment of healthy cells (Brown and Neher, 2014). To investigate whether Mesd meets this criterion as a phagocytosis ligand, we quantified the binding of Mesd-FLAG and GFP-FLAG to POS vesicles by flow cytometry and demonstrated that Mesd-FLAG bound to POSs (Fig. 5a). As a control, GFP-FLAG had background POS-binding activity. Furthermore, flow cytometric analysis indicated that FITC-labeled MBP-Mesd preferentially bound to apoptotic but not healthy HEK293 cells (Fig. 5b). This finding was independently verified by confocal analysis (Fig. 5c). Confocal z-stack images showed that FITC-Mesd bound to the surface of apoptotic cells like rings, colocalizing with the plasma membrane (Zoom-in, middle row, Fig. 5c). As a control, FITC-MBP failed to bind to either apoptotic or healthy HEK293 cells. These data suggest that Mesd selectively recognizes phagocytosis targets but not healthy cells.
Fig. 5.
Mesd binds to POS vesicles and apoptotic cells. a Mesd-FLAG or GFP-FLAG cell lysate without detergent was incubated with POS vesicles. After washing, bound FLAG-tagged proteins were detected by flow cytometry using Alexa Fluor 594-labeled anti-FLAG antibody. b,c Mesd binds to apoptotic but not healthy cells. Apoptotic and healthy HEK293 cells were incubated with FITC-labeled MBP-Mesd or MBP, washed and analyzed by flow cytometry (b) or confocal microscopy (c). Apoptotic cells were labeled with propidium iodide. Confocal z-stack images showed that FITC-Mesd bound to the surface of apoptotic cells like rings in c (Zoom-in, middle row). Bar = 25 μm.
Mesd extracellular trafficking
All phagocytosis ligands should have extracellular access to phagocytosis targets and their receptors on phagocyte surface (Li, 2012). Mesd is a well-known ER chaperon that is not found in culture medium (Li et al., 2005). How can Mesd an ER chaperone extrinsically regulate RPE pahgocytosis? Many cytoplasmic proteins, such as annexin I, Lyar and ABCF1, can be released from apoptotic cells and function as phagocytosis ligands (Arur et al., 2003; Guo et al., 2015a; Guo et al., 2015b). We investigated possible release of Mesd from apoptotic cells. The results showed that Mesd-FLAG was not detected in the medium of healthy HEK293 cells (Fig. 6a). However, it was present in the conditioned medium of apoptotic cells, suggesting that Mesd can be released from apoptotic cells.
Fig. 6.
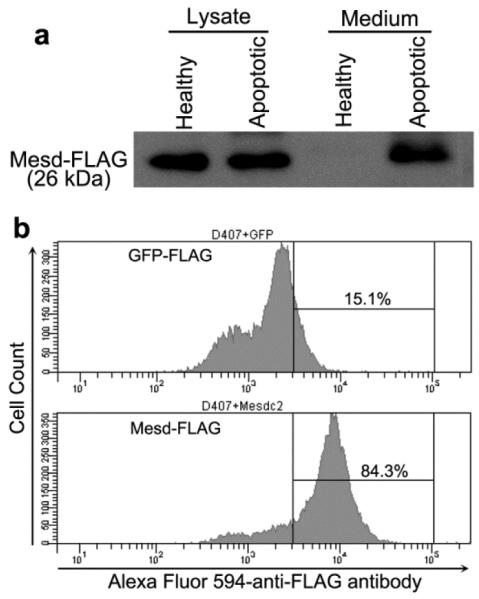
Extracellular trafficking of Mesd and its binding to RPE phagocytes. a Mesd is released into culture medium from apoptotic cells but not healthy cells. Mesd-FLAG was transfected into HEK293 cells. After 48 h, cells were treated with or without etoposide to induce apoptosis. Conditioned media were concentrated and analyzed by Western blot using anti-FLAG antibody. b Mesd binds to RPE phagocytes. Mesd-FLAG or GFP-FLAG lysates was prepared as in Fig. 6a and incubated with D407 RPE cells. After washing, bound FLAG-tagged proteins were detected by flow cytometry using Alexa Fluor 594-labeled anti-FLAG antibody.
Mesd binds to RPE cells
Phagocytosis ligands should bind to their receptor(s) on phagocytes. To investigate whether Mesd binds to RPE cells, we analyzed the binding of Mesd-FLAG and GPF-FLAG to D407 RPE cells by flow cytometry. The results showed that Mesd but not GFP bound to RPE cells (Fig. 6b), suggesting that Mesd interacts with its unknown phagocytic receptor(s) on RPE surface.
Discussion
Mesd is a well-characterized molecular chaperon to facilitate the folding and cell surface expression of LRPs. For example, Mesd facilitates surface expression of LRP1, 2, 4, 5 and 6 (Hoshi et al., 2013; Hsieh et al., 2003; Lighthouse et al., 2011). Deletion of Mesd leads to aberrant expression of LRP5/6 and disruption of mouse embryonic polarity (Hsieh et al., 2003). In conjunction with Frizzled, LRP5/6 are coreceptors for canonical Wnt signaling. Besides its function as a molecular chaperon, Mesd is also capable of binding to mature LRP5/6 on cell surface (Li et al., 2005), thereby inhibiting Wnt/β-catenin signaling (Hsieh et al., 2003; Lin et al., 2013; Lu et al., 2010).
LRPs are well known for their endocytic activity and signal transduction (Li et al., 2001). For example, LRP1 is a multifunctional scavenger receptor (Herz and Strickland, 2001). LRP1 interacts with at least 30 different ligands and is involved in lipoprotein metabolism and cellular entry of viruses and a bacterial toxin.
Despite the classical signal peptide at its N-terminus, Mesd is strictly expressed in the endoplasmic reticulum. Our finding of its absence in the medium of healthy cells (Fig. 6a) is consistent with the previous report that Mesd is not a secretory protein (Li et al., 2005). Although Mesd was reported to bind to and inhibit LRP5/6 on cell surface (Li et al., 2005; Lin et al., 2013; Lu et al., 2010), the lack of the extracellular expression raises a question of how Mesd may access to LRP5/6 on cell surface and inhibit their function.
This study characterized Mesd to extrinsically regulate RPE phagocytosis. All phagocytosis ligands must have extracellular access to their receptors on phagocyte surface and phagocytosis targets (Li, 2012). A similar question is how can Mesd in the ER extrinsically regulate phagocytosis? One of the interesting observations in this study is the release of Mesd from apoptotic cells (Fig. 6a). This finding also implicates that released Mesd may regulate the function of LRP5/6 on cell surface.
About half of the engulfed pHrodo signals were engulfed and undegraded in the cytoplasm of D407 RPE cells in Fig. 1b (zoom-in images). However, some engulfed vesicles were degraded within 3 hours. It appears that pHrodo was not degradable and was released from phagosomes to form diffused red fluorescent signal in local regions of cytoplasm. It also appears that pHrodo was not released from cytoplasm (Fig. 1b, zoom-in, #3), suggesting that cytoplasmic and phagosomal membranes have distinct permeability, at least to pHrodo.
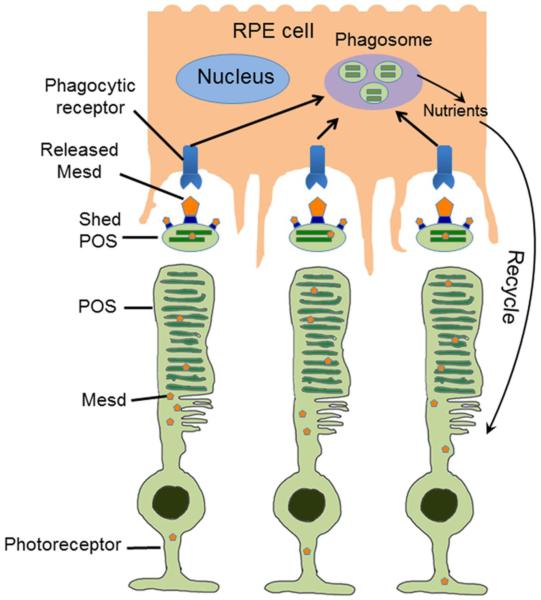
The detection of endogenous Mesd with shed POSs is of importance to its biological relevance to regulate RPE phagocytosis. Macrophages and microglia are mobile phagocytes and can be chemotactically attracted toward apoptotic cells via “find-me” signals (Hochreiter-Hufford and Ravichandran, 2013). In contrast, RPE is stationary phagocytes to engulf POS vesicles, which are shed right into microvilli pockets on the surface of RPE cells in a diurnal rhythm (Fig. 7) (Kevany and Palczewski, 2010; Strauss, 2005). Conceivably, phagocytosis regulators exclusively expressed in retinal ganglion cells or inner plexiform layer may not rapidly travel to the interface of RPE through multiple retinal layers to facilitate the clearance of shed POSs in time. Thus, biologically-relevant regulators for RPE phagocytosis should be expressed in photoreceptors, RPE or other cells (e.g., Muller cells) with direct access to the subretinal space. The detection of endogenous Mesd in shed POSs provides the evidence for the protein to involve in RPE phagocytosis. This is confirmed by immunohistochemistry. Mesd is predominantly expressed in POSs, which is an ER-free cellular compartment (Pearring et al., 2013). The mechanisms to transport Mesd to POSs is yet to be delineated. Thus, Mesd could be released from shed POSs with increased membrane permeability and then binds to the surface of shed POSs in an autocrine manner to facilitate the phagocytic clearance.
Fig. 7.
Mesd is an extrinsic regulator of RPE phagocytosis. Mesd released from apoptotic cells or shed POSs with increased membrane permeability binds to the surface of POS vesicles to facilitate their phagocytic clearance by RPE for the maintenance of retinal homeostasis.
In summary, this study expands the role of Mesd, a well-known molecular chaperone in ER, for extrinsic regulation of RPE phagocytosis. Mesd can be released from apoptotic cells or shed POSs. Released Mesd can facilitate phagocytic clearance of shed POSs by RPE to maintain retinal homeostasis (Fig. 7).
Acknowledgements
We thank Gabriel Gaidosh for the confocal service; Weiwen Wang, Chen Shen and Yanli Ji for technical help. This work was supported by NIH R01GM094449 (W.L.), R21HD075372 (W.L.), BrightFocus Foundation M2012026 (W.L.), Special Scholar Award from Research to Prevent Blindness (RPB) (W.L.), American Heart Association 14PRE18310014 (M.E.L) and 16PRE27250308 (M.E.L), P30-EY014801 and an institutional grant from Research to Prevent Blindness. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- ER
Endoplasmic reticulum
- LRP
Low-density lipoprotein receptor-related protein
- Mesd
Mesoderm development candidate 2
- Mesdc2
Mesoderm development candidate 2
- POS
Photoreceptor outer segment
- RPE
retinal pigment epithelial cell
Footnotes
Competing Interests: The authors declare that they have no competing interests.
References
- Arur S, Uche UE, Rezaul K, Fong M, Scranton V, Cowan AE, Mohler W, Han DK. Annexin I is an endogenous ligand that mediates apoptotic cell engulfment. Dev Cell. 2003;4:587–598. doi: 10.1016/s1534-5807(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Brown GC, Neher JJ. Eaten alive! Cell death by primary phagocytosis: 'phagoptosis'. Trends in biochemical sciences. 2012;37:325–332. doi: 10.1016/j.tibs.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Brown GC, Neher JJ. Microglial phagocytosis of live neurons. Nat Rev Neurosci. 2014;15:209–216. doi: 10.1038/nrn3710. [DOI] [PubMed] [Google Scholar]
- Caberoy NB, Alvarado G, Li W. Tubby regulates microglial phagocytosis through MerTK. J Neuroimmunol. 2012;252:40–48. doi: 10.1016/j.jneuroim.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Li W. Unraveling the molecular mystery of retinal pigment epithelium phagocytosis. Advances in experimental medicine and biology. 2012;723:693–699. doi: 10.1007/978-1-4614-0631-0_88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Maiguel D, Kim Y, Li W. Identification of tubby and tubby-like protein 1 as eat-me signals by phage display. Exp Cell Res. 2010a;316:245–257. doi: 10.1016/j.yexcr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Zhou Y, Li W. Can phage display be used as a tool to functionally identify endogenous eat-me signals in phagocytosis? J Biomol Screen. 2009;14:653–661. doi: 10.1177/1087057109335679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Zhou Y, Li W. Tubby and tubby-like protein 1 are new MerTK ligands for phagocytosis. EMBO J. 2010b;29:3898–3910. doi: 10.1038/emboj.2010.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Caberoy NB, Guo F, LeBlanc ME, Zhang C, Wang W, Wang F, Chen R, Li W. Reticulocalbin-1 facilitates microglial phagocytosis. PLoS One. 2015;10:e0126993. doi: 10.1371/journal.pone.0126993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling JE, Sidman RL. Inherited retinal dystrophy in the rat. J Cell Biol. 1962;14:73–109. doi: 10.1083/jcb.14.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig LP, Henson PM. Immunological consequences of apoptotic cell phagocytosis. Am J Pathol. 2007;171:2–8. doi: 10.2353/ajpath.2007.070135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker M, Neher JJ, Zhao JW, Thery C, Tolkovsky AM, Brown GC. MFG-E8 mediates primary phagocytosis of viable neurons during neuroinflammation. J Neurosci. 2012;32:2657–2666. doi: 10.1523/JNEUROSCI.4837-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG, Apfelstedt-Sylla E, Vollrath D. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet. 2000;26:270–271. doi: 10.1038/81555. [DOI] [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, Henson PM. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123:321–334. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Guo F, Ding Y, Caberoy N, Alvarado G, Wang F, Chen R, Li W. ABCF1 extrinsically regulates retinal pigment epithelial cell phagocytosis. Molecular biology of the cell. 2015a;26:2311–2320. doi: 10.1091/mbc.E14-09-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Ding Y, Caberoy NB, Alvarado G, Liu R, Shen C, Yu J, Zhou Y, Salero E, LeBlanc ME, et al. Lyar Is a New Ligand for Retinal Pigment Epithelial Phagocytosis. Journal of cellular biochemistry. 2015b;116:2177–2187. doi: 10.1002/jcb.25089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochreiter-Hufford A, Ravichandran KS. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harbor perspectives in biology. 2013;5:a008748. doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T, Tezuka T, Yokoyama K, Iemura S, Natsume T, Yamanashi Y. Mesdc2 plays a key role in cell-surface expression of Lrp4 and postsynaptic specialization in myotubes. FEBS Lett. 2013;587:3749–3754. doi: 10.1016/j.febslet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Lee L, Zhang L, Wefer S, Brown K, DeRossi C, Wines ME, Rosenquist T, Holdener BC. Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell. 2003;112:355–367. doi: 10.1016/s0092-8674(03)00045-x. [DOI] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology (Bethesda) 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Eat-me signals: Keys to molecular phagocyte biology and "Appetite" control. J Cell Physiol. 2012;227:1291–1297. doi: 10.1002/jcp.22815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cam J, Bu G. Low-density lipoprotein receptor family: endocytosis and signal transduction. Mol Neurobiol. 2001;23:53–67. doi: 10.1385/MN:23:1:53. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen J, Lu W, McCormick LM, Wang J, Bu G. Mesd binds to mature LDL-receptor-related protein-6 and antagonizes ligand binding. J Cell Sci. 2005;118:5305–5314. doi: 10.1242/jcs.02651. [DOI] [PubMed] [Google Scholar]
- Li Y, Marzolo MP, van Kerkhof P, Strous GJ, Bu G. The YXXL motif, but not the two NPXY motifs, serves as the dominant endocytosis signal for low density lipoprotein receptor-related protein. J Biol Chem. 2000;275:17187–17194. doi: 10.1074/jbc.M000490200. [DOI] [PubMed] [Google Scholar]
- Lighthouse JK, Zhang L, Hsieh JC, Rosenquist T, Holdener BC. MESD is essential for apical localization of megalin/LRP2 in the visceral endoderm. Dev Dyn. 2011;240:577–588. doi: 10.1002/dvdy.22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Lu W, Zhang W, Londono-Joshi AI, Buchsbaum DJ, Bu G, Li Y. The C-terminal region Mesd peptide mimics full-length Mesd and acts as an inhibitor of Wnt/beta-catenin signaling in cancer cells. PLoS One. 2013;8:e58102. doi: 10.1371/journal.pone.0058102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Liu CC, Thottassery JV, Bu G, Li Y. Mesd is a universal inhibitor of Wnt coreceptors LRP5 and LRP6 and blocks Wnt/beta-catenin signaling in cancer cells. Biochemistry. 2010;49:4635–4643. doi: 10.1021/bi1001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, Finnemann SC. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking alphavbeta5 integrin. J Exp Med. 2004;200:1539–1545. doi: 10.1084/jem.20041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Takahashi K. Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J Neuroimmunol. 2007;184:92–99. doi: 10.1016/j.jneuroim.2006.11.032. [DOI] [PubMed] [Google Scholar]
- Pearring JN, Salinas RY, Baker SA, Arshavsky VY. Protein sorting, targeting and trafficking in photoreceptor cells. Prog Retin Eye Res. 2013;36:24–51. doi: 10.1016/j.preteyeres.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. 2014;14:166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salero E, Blenkinsop TA, Corneo B, Harris A, Rabin D, Stern JH, Temple S. Adult human RPE can be activated into a multipotent stem cell that produces mesenchymal derivatives. Cell Stem Cell. 2012;10:88–95. doi: 10.1016/j.stem.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Schneider WJ, Nimpf J. LDL receptor relatives at the crossroad of endocytosis and signaling. Cell Mol Life Sci. 2003;60:892–903. doi: 10.1007/s00018-003-2183-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]