Abstract
Impacts of introgressive hybridisation may range from genomic erosion and species collapse to rapid adaptation and speciation but opportunities to study these dynamics are rare. We investigated the extent, causes and consequences of a hybrid zone between Anopheles coluzzii and Anopheles gambiae in Guinea-Bissau, where high hybridisation rates appear to be stable at least since the 1990s. Anopheles gambiae was genetically partitioned into inland and coastal subpopulations, separated by a central region dominated by A. coluzzii. Surprisingly, whole genome sequencing revealed that the coastal region harbours a hybrid form characterised by an A. gambiae-like sex chromosome and massive introgression of A. coluzzii autosomal alleles. Local selection on chromosomal inversions may play a role in this process, suggesting potential for spatiotemporal stability of the coastal hybrid form and providing resilience against introgression of medically-important loci and traits, found to be more prevalent in inland A. gambiae.
Interspecific hybridisation is an important, yet probably underestimated, force in the evolution of animal biodiversity with possible impacts ranging from erosion of species divergence and ultimate species collapse1 to rapid adaptation or even speciation2,3. The latter ‘hybrid speciation’ is predicted to arise via accumulation of genetic variation through introgression, resulting in admixed populations that are ecologically divergent from the parental lineages3. More generally, whilst rare hybridization can undoubtedly lead to adaptive introgression4,5, the adaptive potential of very frequent interspecific hybridization is far less clear. Systems in which uncharacteristically extreme hybridisation rates are found offer the opportunity to study such evolutionary processes. The Anopheles coluzzii-Anopheles gambiae malaria vector species-pair in western Africa provides an ideal opportunity to study speciation under extreme hybridization6,7.
Members of the Anopheles gambiae complex include the most important malaria vectors in Sub-Saharan Africa. Early evidence of genetic discontinuities in West Africa within the nominal species of the complex, A. gambiae, came from cytogenetic studies, which led to the description of five chromosomal forms, the FOREST and SAVANNA forms widespread in west, central and east Africa, the MOPTI form in northern west African savannah areas, and the BAMAKO and BISSAU forms, which are found only in localized areas of Mali and the far-west coast, respectively. Each is characterized by different arrangements of paracentric inversions on chromosome-2 and differing larval ecologies8,9, and studies have associated specific inversion polymorphisms with multiple environmental adaptions8,9,10. However, following the discovery of fixed differences in ribosomal DNA on the pericentromeric region of chromosome-X, which overlapped only partially with the chromosomal forms, focus shifted from chromosomal to ‘molecular forms’, termed M and S11. These forms have been recently named Anopheles coluzzii and A. gambiae12, respectively and the species-pair has become a model system for speciation with gene flow13 due to their closely related genomes14 characterized by three large highly divergent ‘genomic islands’ near the centromeres of each chromosome15,16. Subsequent analyses have shown that divergence is more widespread around the genome than first thought5,17 and the involvement of two of the three islands of divergence in early-stage speciation became controversial5,18,19,20. However, a recent laboratory study has demonstrated that the chromosome-X island of divergence is associated with assortative mating between A. coluzzii and A. gambiae21 confirming this genomic region as a primary candidate location for genes involved in reproductive isolation. These species have also shown differences in at least two genes which contribute to medically important phenotypes. First, mutations associated with knockdown resistance (kdr) to DDT and pyrethroid insecticides have been initially found at much higher frequency in A. gambiae than in A. coluzzii even when the species are sympatric22. There is evidence that recent adaptive introgression has subsequently levelled kdr frequencies between species5,20,23, although this imbalance appears to persist in the far western region24. Second, the allelic variant r1 of the immune-related tep1 gene, which has been linked to resistance to infection by Plasmodium falciparum malaria parasites, was found to be exclusive to A. coluzzii25.
Reproductive isolation between A. coluzzii and A. gambiae occurs at the adult stage by incompletely-understood pre-mating mechanisms26, while no intrinsic postzygotic isolating mechanisms are observed between the two species27. At the larval stage, ecological niche partitioning and selection against hybrids are likely to play important roles in species segregation28. Anopheles gambiae typically breeds in rain-dependent ephemeral pools, while A. coluzzii is more successful in breeding sites created by irrigation (e.g. rice-fields) or other human activities (e.g. water reservoirs), or in urban polluted environments29. There is also evidence that A. coluzzii larvae display greater salinity tolerance, which may permit better exploitation of marginal habitats30. Despite broad sympatry between A. coluzzii and A. gambiae across West and Central Africa, hybrid rates, as detected by X-linked species-species diagnostic markers11,31, are usually below 1%32. However, assortative mating may periodically be disrupted and temporally-unstable bursts of elevated hybridisation have been reported19. The ‘typical’ situation of low hybridisation breaks down most clearly in a hybrid zone between A. coluzzii and A. gambiae on the western edge of the species’ distribution6,33. Although, as yet undetected, stable high hybridisation may occur elsewhere, the far-west region has become an important case study area, with hybridisation rates >20% being recorded persistently for almost 20 years in Guinea Bissau6,7,34. Hybridization appears to involve asymmetric introgression from A. coluzzii to A. gambiae7 and to have eroded the major genomic islands of divergence on chromosomes-2 and -3 but to a far lesser extent on chromosome-X19,35,36,37.
The temporal stability and spatial extent of the hybrid zone are of both academic interest and medical concern. Whilst continued introgression might ultimately lead to erosion of divergence or even species collapse1 in the hybrid zone, available evidence from the analysis of temporal samples in the area is equivocal. Gordicho et al.34 pointed to an apparent decline of A. coluzzii in coastal Guinea Bissau, from a frequency of 23–44% between 1993 and 1996 to 8.0% and 4.5% in 2007 and 2010, respectively. However, a frequency close to 25% was recorded for A. coluzzii in a sample collected in the same locality in 20097, indicating that stochastic temporal instability cannot be ruled out. Moreover, the available temporal data is based on a relatively limited number of collections and without accounting for eventual seasonal variations.
Another possibility is that introgression may be creating a pool of aberrantly high diversity facilitating adaptation to new or marginal niches2,3. Intriguingly, the hybrid zone overlaps with one of the best examples of intraspecific structuring based on inversion polymorphisms. The BISSAU chromosomal form (characterized by a low frequency of 2Rb and 2La arrangements, and a high frequency of 2Rd) and the SAVANNA chromosomal form (characterized by high frequency of 2Rb and 2La, sometimes with increasing complexity of karyotypes due to the presence of other inversions such as 2Rj and 2Rd) were shown to intergrade from coastal areas to inland, suggesting ecologically-driven genetic divergence due to adaptation to coastal habitats (e.g. brackish water habitat and/or competition with the euryhaline sympatric Anopheles melas sibling species)8,38,39,40.
Other major uncertainties remain about the extreme hybrid zone and its significance. Though documented in neighbouring countries33,41, the spatial extent of the hybrid zone is still unclear, as are the links to ecology and the impact on traits of medical importance. Although a significantly higher Plasmodium infection rate was detected in A. gambiae when compared with A. coluzzii and hybrids in coastal Guinea Bissau42, the potential impact of the hybrid zone on Plasmodium transmission and other traits of medical importance, such as insecticide resistance and host preferences, has received limited attention.
In this study, we examined the following hypotheses: (1) The hybrid zone is confined to the coastal region and differentiated from inland regions at chromosomal inversion polymorphisms, which may link to local adaptation; (2) Asymmetric introgression from A. coluzzii to A. gambiae is promoting intraspecific divergence between coastal and inland populations of A. gambiae, potentially leading to radiation of a distinct hybrid form; (3) The establishment of a hybrid form builds the potential for aggregation of medically important mutations and phenotypes.
To investigate these hypotheses, we conducted: (i) a countrywide southwest-northeast transect study using species-specific molecular markers and microsatellites spanning coastal to far-inland Guinea-Bissau, in order to quantify the extent of the hybrid zone; (ii) a focal investigation of chromosomal inversion polymorphisms, in order to determine karyotype differences between coast and inland; (iii) a focal analysis by whole genome sequencing and ancestry informative markers of individuals classified as A. gambiae, to determine intraspecific genome-wide divergence between coastal and inland populations of this species and including a comparison with a geographically distant A. gambiae sample from a low hybridisation region; (iv) an investigation of markers and traits important for malaria transmission and control in A. gambiae from coast and inland.
Results
Species distribution and admixture along the transect follow ecological zonation
A total of 687 specimens collected in eight localities of Guinea Bissau were analysed using 19 microsatellite loci (Supplementary Table 1). Polymorphism at chromosome-3 loci was higher than chromosome-X loci, whether measured as allelic richness (Ar) or expected heterozygosity (He) (Mann-Whitney Tests: Ar: z = 4.24, P < 0.001; He: z = 4.35, P < 0.001) (Supplementary Table 2). None of the loci showed a significant heterozygote deficit across collection sites, which could indicate null alleles, and there was no consistent signal across loci, which could indicate within-sample substructure.
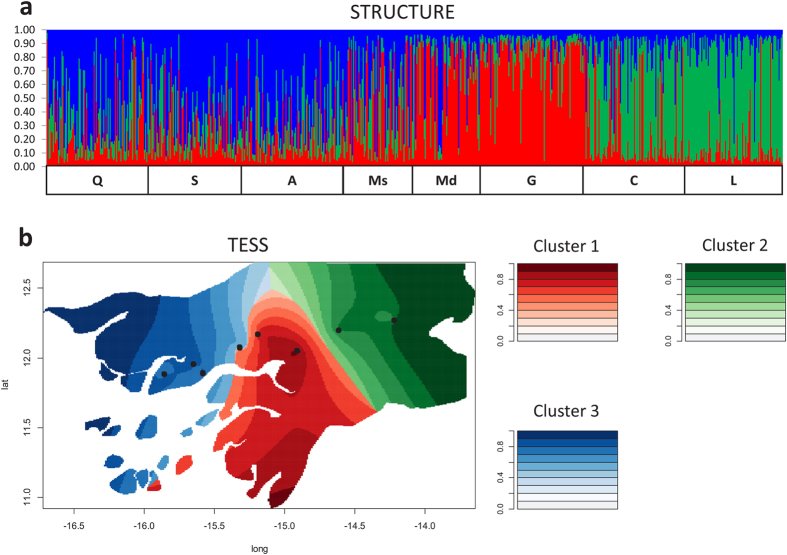
Both spatial (TESS) and non-spatial (STRUCTURE) clustering analyses partitioned the overall sample into three distinct genetic clusters (Fig. 1, Supplementary Fig. 1). These clusters differed in their species composition (Table 1) and geographic distribution (Fig. 2). Cluster 1 (hereafter referred to as cluster “A. coluzzii”) was dominated by A. coluzzii and was located in the central region of the country. In contrast, cluster 2 (referred to as “A. gambiae-inland”) was dominated by A. gambiae and prevailed in the two eastern inland localities (Comuda and Leibala). Cluster 3 (referred to as “A. gambiae-coast”) was comprised almost entirely of A. gambiae and IGS/SINE-admixed individuals, and was found mostly in the three western coastal localities (Quinhamel, Antula and Safim). The degree of cross-cluster genetic admixture also varied geographically (Fig. 2). The proportion of admixed individuals (i.e. those with qi < 0.50 for all clusters determined by STRUCTURE) was higher in coastal localities (14.3–20.4%) than in the central part of the country (1.5–11.3%) and in the eastern inland localities (4.3–6.6%).
Figure 1. Microsatellite-based Bayesian clustering (STRUCTURE) and spatially explicit analysis (TESS).
(a) Graph of individual assignment probabilities of belonging to each of the three clusters detected by STRUCTURE. Red: cluster 1 (A. coluzzii), green: cluster 2 (A. gambiae-inland), blue: cluster 3 (A. gambiae-coast). The lower squares delimit mosquito samples according to collection site, from coast to inland (Q: Quinhamel, S: Safim, A: Antula, Ms: Mansoa, Md: Mandingará, G: Ga-Mbana, C: Comuda, L: Leibala). (b) Map of Guinea Bissau showing assignment probability densities for the optimal number of clusters obtained in TESS. The map was created by the R-script of software POPS version 1.262 (http://membres-timc.imag.fr/Olivier.Francois/pops.html). Black dots represent the localities sampled.
Table 1. Association between Bayesian genetic clusters (STRUCTURE and TESS) and molecular identification of species by IGS and SINE.
| Structure |
Tess |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | Admixed | Cluster 1 | Cluster 2 | Cluster 3 | Admixed | ||
| IGS/SINE | A. coluzzii | 0.882 | 0.022 | 0.022 | 0.104 | 0.914 | 0.006 | 0.026 | 0.111 |
| Admixed | 0.062 | 0.062 | 0.398 | 0.463 | 0.062 | 0.000 | 0.436 | 0.111 | |
| A. gambiae | 0.057 | 0.916 | 0.580 | 0.433 | 0.024 | 0.994 | 0.538 | 0.778 | |
| N | 211 | 178 | 231 | 67 | 210 | 163 | 305 | 9 | |
Values represent the relative proportions of each species (determined by IGS/SINE markers) within each STRUCTURE or TESS cluster. Highest proportions are highlighted in bold. N: total number of specimens assigned to each cluster based on a probability threshold (Tq) of 0.50.
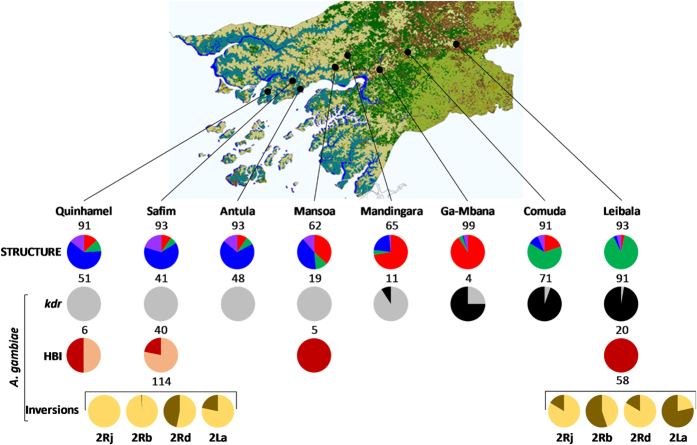
Figure 2. Map of localities surveyed showing microsatellite-based genetic cluster composition (STRUCTURE), and kdr frequency, human blood index and chromosome inversion frequency for Anopheles gambiae.
Map: GlobCover 2009 land cover map of Guinea Bissau. Copyright notice: © ESA 2010 and UCLouvain. Available at: http://www.fao.org/geonetwork/srv/en/metadata.show?id=37189&currTab=simple (Date of access: 27/11/2015). STRUCTURE: proportion of individuals assigned to each genetic cluster (assignment threshold Tq = 0.5). Red: cluster 1 (A. coluzzii), green: cluster 2 (A. gambiae-inland), blue: cluster 3 (A. gambiae-coast), purple: admixed. kdr: allele frequency of the knockdown resistance-associated 1014F allele (in black); HBI: human blood index (in red); inversions: frequency of the inverted arrangement in dark yellow at each of four inversions scored. Sample sizes are above each pie chart. Data on kdr, HBI and inversions are shown for A. gambiae only, identified by IGS/SINE at each site.
Chromosomal form composition contrasts between coastal and inland samples
A total of 260 half-gravid females collected in coastal Safim (N = 202) and inland Leibala (N = 58) were karyotyped successfully (Fig. 2, Table 2). Anopheles gambiae from Safim were characterized by five karyotypes on chromosome 2R, based on the 2Rb and 2Rd polymorphisms, while the sample from Leibala comprised of 10 karyotypes based on 2Rb, 2Rd inversions and exclusive presence of 2Rj inversion. The high frequencies of 2Rb and 2La inverted arrangements and the high number of karyotypes observed in A. gambiae from Leibala (Fig. 2, Table 2) suggest predominance of the SAVANNA chromosomal form, common across West Africa8,9. Conversely, the higher frequency of the 2 Rd inverted arrangement (χ2 = 42.06, d.f. = 2, P < 0.001; Fig. 1) coupled with the lower frequency of 2La (χ2 = 92.12, d.f. = 2, P < 0.001) and the virtual lack of 2Rb (χ2 = 126.28, d.f. = 2, P < 0.001) inverted arrangements observed in Safim is consistent with predominance of the BISSAU chromosomal form8,9, occurring in both A. coluzzii and A. gambiae, which exhibited similar 2Rd and 2La frequencies (Fisher’s exact-test, P = 0.23 and P = 0.71, respectively).
Table 2. Karyotype frequencies (%) in Safim (coast) and Leibala (inland), Guinea Bissau.
| Chromosome-2R karyotypes | Safim |
Leibala | ||
|---|---|---|---|---|
| A. coluzzii | Admixed | A. gambiae | A. gambiae | |
| +/+ | 25.0 | 23.7 | 6.9 | |
| +/d | 66.7 | 56.6 | 51.8 | 8.6 |
| d/d | 16.7 | 17.1 | 22.8 | |
| +/b | 29.3 | |||
| b/b | 17.2 | |||
| bd/+, b/d | 16.7 | 0.9 | 8.6 | |
| jb/+, j/b | 10.3 | |||
| jbd/+, j/bd, jb/d | 6.9 | |||
| jb/b | 3.4 | |||
| jbd/b, jb/bd | 5.2 | |||
| jb/d, jd/b, j/bd | 3.4 | |||
| bd/d | 0.9 | |||
| d/u | 1.3 | |||
| Total | 12 | 76 | 114 | 58 |
| Chromosome-2L karyotypes | ||||
| +/+ | 66.7 | 61.8 | 61.4 | 5.2 |
| +/a | 33.3 | 32.9 | 36.8 | 32.8 |
| a/a | 5.3 | 1.8 | 62.1 | |
| Total | 12 | 76 | 114 | 58 |
Whole genome sequencing reveals extensive autosomal introgression in coastal A. gambiae
Anopheles gambiae sequences were obtained from 12 individuals from Antula, five from Safim and four from Leibala (Guinea Bissau), and from four additional specimens collected approximately 1,500 km away in Accra, Ghana, a region representative of the low interspecific hybridisation observed in most of the species range (Supplementary Table 3).
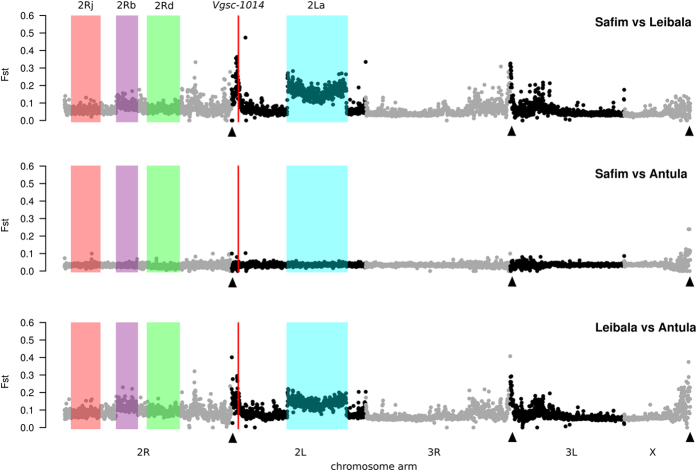
Anopheles gambiae coastal samples (Safim vs. Antula) showed very low genomic divergence, as expected for a within-species comparison (Fig. 3). However, divergence was elevated in the comparisons between each of these coastal sites and A. gambiae from the Leibala inland site. Differentiation (FST) was especially pronounced at pericentromeric regions on each chromosome, which are known to segregate between A. coluzzii and A. gambiae (e.g. Neafsey et al.17), with 22% of FST values within the top 5% of genome-wide FST values located therein (χ2 = 383, P = 1 × 10−85). Similarly, the 2La inversion region was exceptionally divergent, with 68% of FST values in the top 5% (χ2 = 1029, P = 1 × 10−226). However, the other genomic regions corresponding to the karyotypic differences observed among the sampling locations (Fig. 2, Table 2) were not significantly divergent in sequence comparisons (Fig. 3), with no windows in any of the 2R inversion regions located within the top 5% of FST values.
Figure 3. Manhattan plots of mean pairwise FST between Anopheles gambiae samples of Guinea Bissau.
Plots show the three pairwise comparisons of genome-wide differentiation between A. gambiae samples from Safim (coastal), Antula (coastal) and Leibala (inland). No A. coluzzii individuals were included in the analysis. Arrows indicate the location of the centromeres. The red vertical line indicates position 1014 of the voltage-gated sodium channel gene which corresponds to the kdr locus. The four inversion systems scored are also shown by the shaded areas. A 50 kb stepping window was used to generate the mean FST.
To analyse evidence of introgression from A. coluzzii within the samples of A. gambiae sequenced, we used Ancestry Informative Markers (AIMs) retrieved from a genome-wide A. coluzzii vs. A. gambiae comparison carried out in a region of infrequent hybridization17. This yielded 236 Single Nucleotide Polymorphisms (SNPs) on chromosome-X and 93 SNPs on the autosomes in the A. gambiae Guinea Bissau samples (Supplementary Fig. 2; see also Supplementary Dataset 1). The proportionate AIM-estimated ancestries for these markers are shown in Fig. 4. Coastal A. gambiae from Antula and Safim showed much higher numbers of (putatively introgressed) autosomal A. coluzzii AIMs than the inland sample from Leibala (χ2 = 333.1, d.f. = 4, P < 0.001). Whilst the proportion of A. coluzzii AIMs was also higher in coastal sites for chromosome-X (χ2 = 88.6, d.f. = 4, P < 0.001), this proportion was much lower than on the autosomes (χ2 = 124.8, d.f. = 4, P < 0.001). Indeed, there was a stretch (>3 Mb) of ‘pure’ A. gambiae homozygous AIMs located in the pericentromeric region of chromosome-X (Fig. 4), that was found in all individuals irrespective of sampling location.
Figure 4. Percentage ancestry based on ancestry informative markers (AIMs) in Anopheles gambiae individuals from Guinea Bissau.
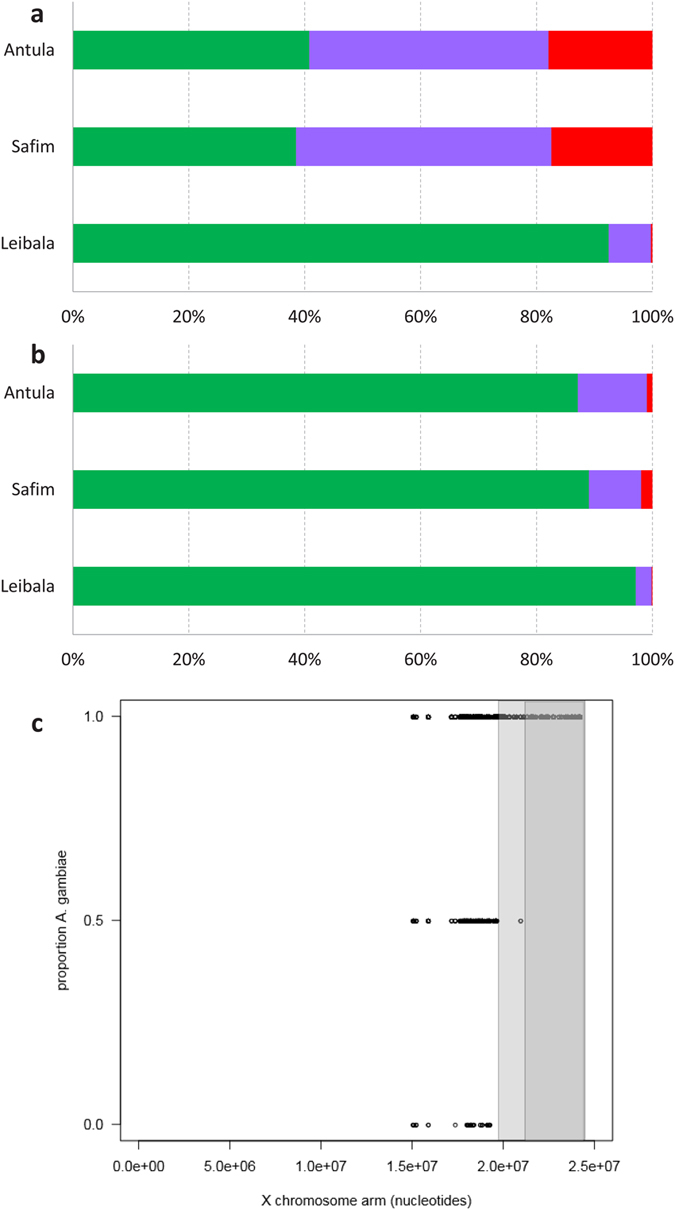
Stacked bars show percentage of ancestry found in whole genome sequenced individuals sampled in two coastal (Safim N = 5, Antula N = 12) and one inland locality (Leibala, N = 4) estimated using AIMs identified from genome-wide comparisons between samples from a more typical region of infrequent hybridization17. Percentage ancestry is based on 93 autosomal (a) and 236 chromosome-X (b) AIMs, each scored as being homozygous for A. gambiae (green) or A. coluzzii (red) or heterozygous/admixed (purple). (c) Position and genotype of chromosome-X AIMs. Each AIM was genotyped as 0 = A. coluzzii homozygote, 0.5 = A. coluzzii/gambiae heterozygote or 1 = A. gambiae homozygote. Shaded areas correspond to the >3 Mb region (dark grey) homozygous for for A. gambiae AIMs, which increases to >4 Mb (light grey) if a single heterozygous AIM is included.
Genome sequence differentiation between Guinea Bissau and the low hybridisation region of Ghana is shown in the PCA plot of Fig. 5, which was based on the analysis of chromosome-3L SNPs, though 3R SNPs gave a near identical profile (Supplementary Fig. 3). Principal component (PC) 1 explained by far the greatest variance and substitution of alternative secondary principal components did not alter patterns (Supplementary Fig. 3). The PCA clearly separated all coastal Guinea Bissau specimens from a group including the inland Guinea Bissau sample of Leibala and Ghanaian specimens (Fig. 5).
Figure 5. Principal Components Analysis based on chromosome arm 3L variants.
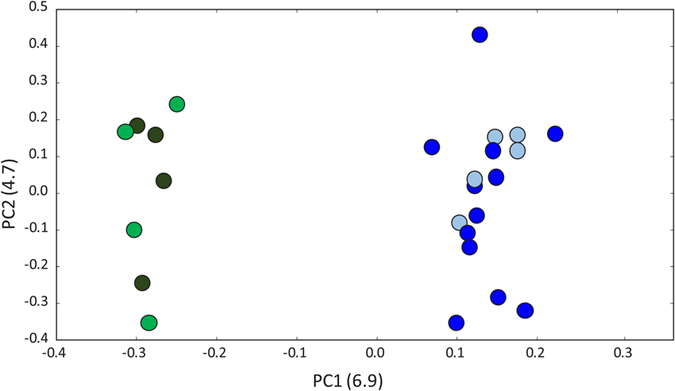
Dots represent the relationship between PC1 and PC2 of whole genome sequenced individuals collected in Antula (blue), Safim (light blue), Leibala (green) and Accra-Ghana (dark green).
Medically-relevant loci and traits differ between inland and coastal A. gambiae
Two major insecticide resistance loci were genotyped in all specimens that had been analysed using microsatellites. The Vgsc-1014F (kdr) allele (associated with DDT and pyrethroid resistance) was detected at very low frequency (<5%) in the two central localities of Ga-Mbana and Mandingara but was common in the inland localities, exceeding 95% in Leibala (Fig. 2). This kdr allele was entirely absent from all three coastal samples. Among the clusters identified by STRUCTURE, the 1014F allele frequency was 77% in A. gambiae-inland but was below 5% in A. gambiae-coast and A. coluzzii clusters (Table 3). The ace-1 119S allele (associated with carbamate and organophosphate resistance) was detected only in inland sites, albeit at low frequencies (Comuda: 2.1%; Leibala: 3.3%) and always as heterozygotes, but was absent elsewhere (Table 3).
Table 3. Distribution of insecticide resistance-associated alleles and human blood index among Bayesian genetic clusters (STRUCTURE).
| Cluster 1 (A. coluzzii) | Cluster 2 (A. gambiae-inland) | Cluster 3 (A. gambiae-coast) | Admixed | |
|---|---|---|---|---|
| kdr f(1014F) | 0.019 | 0.767 | 0.045 | 0.179 |
| [0.009–0.039] | [0.714–0.809] | [0.029–0.070] | [0.120–0.257] | |
| (211) | (176) | (231) | (67) | |
| ace-1 f(119S) | 0.002 | 0.014 | 0.000 | 0.022 |
| [0.000–0.015] | [0.005–0.034] | [0.000–0.010] | [0.006–0.069] | |
| (211) | (178) | (231) | (67) | |
| HBI | 0.385 | 0.824 | 0.308 | 0.391 |
| [0.151–0.677] | [0.648–0.926] | [0.202–0.435] | [0.205–0.612] | |
| (13) | (34) | (65) | (23) |
Values represent proportions and 95% confidence intervals in square brackets of the resistance-associated alleles in the case of kdr and ace-1 and blood meals taken from humans, in the case of the human blood index (HBI), respectively. Sample sizes are in parenthesis.
Blood-fed females were collected in Safim (N = 93) and Antula (N = 8) in the coastal region; Mansoa (at the border between coastal and central regions; N = 12) and Leibala (inland region; N = 30) (Fig. 2). Among STRUCTURE-based genetic clusters, the Human Blood Index (proportionate feeding on humans, HBI) was more than two-fold greater for A. gambiae-inland than for A. gambiae-coast and A. coluzzii (χ2 = 24.29, d.f. = 2, P < 0.001; Table 3), which exhibited a relatively high frequency of bovine blood meals (30%) (Supplementary Table 4).
Discussion
We have genetically characterised the species-pair A. coluzzii and A. gambiae in the zone of extreme hybridization at the far-west of their distribution, in order to investigate the extent of the hybrid zone, possible determinants and consequences of massive introgression for population divergence and malaria transmission and control.
Hypothesis 1: The hybrid zone is limited to the coastal region and differentiated from inland populations by chromosomal inversions
Results from species diagnostic markers (IGS and SINE) and microsatellites supported the hypothesis that in Guinea Bissau the hybrid zone is confined to the coastal region. Three clusters were detected from the microsatellite data, irrespective of analysis method, comprising of a coastal region dominated by individuals typing as A. gambiae or admixed, a central region dominated by A. coluzzii and an inland region of A. gambiae, suggesting that the core of the hybrid zone is confined to a stretch of no more than 110 km from the coast. Genetic partitioning of coastal and inland A. gambiae populations separated by a region dominated by A. coluzzii has also been reported along the Gambia river, a few hundred kilometres north of Guinea Bissau40.
Based on prior observations from Guinea Bissau39, Senegal and The Gambia34,38 we also proposed that chromosomal inversion polymorphisms, frequently implicated in environmental adaptation8,9,10, would differ between inland and coastal regions. This proved to be the case and we suggest that this is likely to reflect a difference in the frequency of two chromosomal forms. Anopheles gambiae at the coast are characterized by a high frequency of the 2Rd inversion and low frequency of 2Rb and 2La, compatible with that described for the BISSAU chromosomal form8,9 and this inversion pattern was shared with the sympatric A. coluzzii population. Inland A. gambiae is characterized by higher complexity along chromosome-2R, including the exclusive presence of 2Rj inversion and by high frequency of 2Rb and 2La inversions. This pattern also characterises inland populations from The Gambia38,40 and is typical of contiguous populations surrounding the Fouta-Djalon massif, which incompletely intergrade with neighbouring populations of the SAVANNA form9,32,40. The concurrence of the BISSAU form and the hybridization zone may not be coincidental but rather linked to the adaptive potential of variation within the 2Rd inversion for exploitation of coastal habitats8,38,40. The alternate frequency of the 2La inversion between the more humid south-western coast (Safim) and the drier north-eastern inland (Leibala) (Supplementary Fig. 4) is in line with previous findings in the region39,40, and agrees with the well-known latitudinal variation of the 2La inverted karyotype in association with aridity and the savannah-forest transition in West Africa8,10.
Inland vs. coastal differentiation was also clearly evident in genome sequence data for the 2La inversion, which shows large areas of fixed or near fixed differences between orientations43. However, no such correspondence of karyotypic and genome sequence variation was evident for the 2Rd inversion region, or others on chromosome arm 2R, suggesting far more sharing of polymorphism between orientations as a result of more recent origin and/or a more diffuse nature of the putatively adaptive variants.
Hypothesis 2: a distinct hybrid form is being created in the coastal zone
Whilst inland A. gambiae is genomically similar to A. gambiae from areas of low hybridisation (here represented by the Ghanaian population5,36), the genomes of coastal A. gambiae display signs of extensive introgression from A. coluzzii, as shown by the AIM analysis. A similar contrast was also noted in a comparison between samples from distinct inland and coastal localities in Guinea-Bissau, using a 15 SNP panel19, indicating that this pattern is not sample site dependent. The single and notable exception to the pattern is the chromosome-X pericentromeric island of interspecific divergence which retains an apparently unbroken A. gambiae haplotype >3 Mb long. While the pattern of autosomal introgression from A. coluzzii into A. gambiae agrees with the asymmetric introgression pattern demonstrated in the coastal region7, AIM analysis showed that introgression can only partially penetrate chromosome-X. Although sequence data comes from a small number of A. gambiae specimens selected in advance for consistency between IGS and SINE, this observation coupled with proximity to the centromere agree with a lower recombination and/or stronger selection against recombinants at the X-pericentromeric island as compared to the rest of the genome. This is concordant with results from a study genotyping hemizygous males from the same geographic region37, which detected limited recombination in the X-pericentromeric island.
Given the geographical zonation of species detected, opportunities for introgressive backcrossing of the coastal admixed population appear to be diminishing. Therefore, rather than a putative decline of A. coluzzii or progressive erosion of divergence by introgression in the coastal region, what we appear to be observing is creation of a novel hybrid form by strong asymmetric introgression, characterised by an admixed genome and an A. gambiae like X-pericentromeric island. Merging of the A. coluzzii genome by introgression into A. gambiae but with an apparently highly conserved stretch of A. gambiae genome on the heterogametic sex chromosome – which has recently been implicated in partial reproductive isolation between the species21 – represents an extraordinary example of genomic flexibility, which may be characteristic of the species complex14. Further temporal monitoring is required to examine the stability of divergence of this putative hybrid form and the extent to which fluctuations, e.g. related to seasonality, may affect its occurrence.
The finding of a hybrid form typing as A. gambiae by species-specific markers located in the X-pericentromeric island raises the question of whether this form might also be found ‘hidden’ elsewhere throughout the sympatric range of the species-pair. This may be true but probably to a limited extent because: (i) the stable elevated hybrid rates reported in the “far-west” African region have never been reported outside this region; (ii) although available genome-wide data remains geographically sparse, in low hybridization regions additional species diagnostic markers on chromosome-3L are often found in linkage disequilibrium with those on chromosome-X19; (iii) coastal areas, which represent a relatively small fraction of the range of the species-pair, may have peculiar characteristics favouring the evolution of a new form better adapted to elevated breeding site salinity to which A. coluzzii, and especially A. gambiae, have limited tolerance, by comparison to the brackish water-breeding west African species A. melas29. Nevertheless, given that recent introgression is detectable even in low hybridization countries19, genomic examination of coast-to-inland transects from elsewhere would certainly be of interest.
The timing and mechanisms underlying the breakage in reproductive isolation between A. coluzzii and A. gambiae require further investigation. However, it is interesting to note that environmental changes associated with the implementation of rice cultivation occurred in Guinea Bissau within a relatively recent time scale. Mangrove swamp rice cultivation was established in the 15th century by farmers of the Balanta ethnic group in the central part of the Mansoa river valley and subsequently expanded westwards to the coast along the major rivers of the country44. The establishment of rice fields in coastal areas could have provided the means for A. coluzzii – known to be better adapted to agricultural larval habitats26 and slightly more salinity tolerant than A. gambiae30 – to expand its range into the salt-water mangrove swamp coastal areas of Guinea Bissau. A recent expansion (i.e. 500 year ago) could have promoted contact between A. coluzzii and an already-present coastal A. gambiae population, providing the opportunity for the establishment of a secondary contact zone. This would present concomitant potential for introgression to create novelty for adaptation to the coastal region. A similar pattern of A. coluzzii predominance in rice-cultivated central regions and increased hybridisation westwards towards the coast was also found in The Gambia40. In one of the brackish water-breeding specialists, the east African species A. merus, salinity tolerance maps to multiple QTL including the large 2Rop inversion region45, which encompasses the 2Rd inversion in A. coluzzii and A. gambiae. Though speculative at present, involvement of the 2Rd inversion in salinity tolerance is consistent with its observed distribution and may be a key adaptation in more coastally adapted A. coluzzii30. The extent to which anthropogenic activity may have influenced the establishment of a secondary contact zone and introgression is unclear, but introgression between closely related taxa associated with human-mediated environmental change has been described for a number of other organisms46, and can affect not only the genetic integrity of species but also their capacity to adapt to altered environments3.
Hypothesis 3: Establishment of a hybrid form may promote aggregation of medically important mutations and phenotypes
Our hypothesis that hybridisation might influence traits related to malaria transmission and control through aggregation of medically important mutations and phenotypes was not met for the traits investigated here (i.e. insecticide resistance associated mutations and host preference). Previously we found an elevated frequency of an introgressed A. coluzzii specific tep1r1 Plasmodium-resistant allele in coastal A. gambiae47. Whilst evidence for an increased zoophilic tendency detected in A. gambiae coupled with the introgression of the tep1r1 parasite-resistant allele might predict a lower contribution of this species to malaria transmission in the coastal area, P. falciparum infection rates were actually higher in A. gambiae than in sympatric A. coluzzii collected in coastal Guinea Bissau in 200942. Rather than hybridization, it is the partitioning of A. gambiae into two genetically distinct subpopulations that currently appears to have public health implications. This genetic partitioning into coastal and inland populations probably involves restrictions to gene flow arising from physical isolation by the central region (represented by Mansoa, Mandigara and Ga-Mbana) where A. coluzzii predominates. There was evidence for marked differences between coastal and inland A. gambiae subpopulations in both host preference and in the frequency of the kdr insecticide resistance allele.
The contrasting kdr frequencies between coast and inland subpopulations could also have implications for vector control. Different insecticide selective pressures could hypothetically explain this observation but there is no evidence of higher insecticide pressure in inland Guinea Bissau. Insecticide treated nets are the only anti-vector measure used by the malaria control programme of Guinea Bissau48 and these would exert a highest pressure in coastal areas, where most population of the country lives. There have also been reports associating kdr resistance with insecticide usage in cotton crops49. However, cotton is not among the most important produces of Guinea Bissau50. A more plausible explanation is that inland A. gambiae is part of the larger western African A. gambiae population known to display high kdr frequency22, which agrees with the grouping of this population with individuals from Ghana in the whole genome sequence PCA.
Conclusion
Genomic introgression, coupled with restrictions to gene flow due to habitat segregation in the central region of the country, are promoting divergence between two genetically distinct A. gambiae populations in Guinea Bissau. One is a coastal, A. coluzzii-introgressed population, characterized by lower inversion polymorphism (excepting the 2Rd inversion), greater zoophilic tendency and absence of kdr mutations; the other is an inland, more typical, anthropophilic A. gambiae population potentially more adapted to aridity and displaying high kdr frequency. In other hybrid zones, asymmetrical introgression is often driven by mate choice (see While et al.51 and references therein). At present in the Guinea Bissau hybrid zone the drivers of this asymmetry are not clear, and further investigation is now warranted. Epidemiological studies are required to determine how this zonation is impacting malaria transmission. Monitoring of the stability of the genetic partitioning is also crucial, both from the perspective of providing a window into the timescale of what may represent contemporary adaptive introgression, perhaps on a trajectory towards hybrid speciation, and for the potential that disruption of the current partitioning could lead to a salinity tolerant, insecticide resistant hybrid form with a higher capacity to transmit malaria in the densely-populated coastal region of Guinea Bissau and neighbouring west African countries.
Methods
Mosquito sampling
Mosquito collections took place between 10th and 30th October 2010 in eight localities along a 180 km southwest-northeast transect covering three major regions of Guinea Bissau (Fig. 2, Supplementary Fig. 4): (i) a south-western coastal region, characterised mainly by mixed flooded forests and croplands and with a mean aridity index >1.0 (localities of Quinhamel, Safim and Antula); (ii) a central region, where large patches of evergreen forest are present (Mansoa, Mandingara and Ga-Mbana) and mean aridity index between 0.8 and 1.0; (iii) a north-eastern inland region where shrubland and open deciduous forest appear to prevail and with a mean aridity index between 0.6 and 0.8 (Comuda and Leibala).
Host-seeking mosquitoes were collected indoors (19:00–07:00) by CDC miniature light traps. These mosquitoes were stored individually in tubes filled with silica gel and cotton for DNA analyses. In addition, indoor resting collections were performed. The midgut contents of blood fed females caught resting were squashed onto filter paper (Whatman® Grade 1) for blood meal analysis. The ovaries of half-gravid females were preserved in Carnoy’s fixative (absolute ethanol: glacial acetic acid, 3:1) and stored at −20 °C for polytene chromosome preparations. The remainder of the carcasses of half-gravid and blood fed females was also kept dry in silica gel until DNA extraction.
Molecular identification
DNA extraction from individual females was performed using a phenol: chloroform protocol52. Molecular identification of species was performed by PCR-RFLP targeting species-specific polymorphisms at the intergenic spacer (IGS) of the ribosomal DNA53 and by a PCR assay targeting the SINE 200X6.1 retrotransposon insertion31. Specimens were identified as A. coluzzii or A. gambiae if they had coincident species-specific patterns for both markers. Specimens exhibiting either a consistent A. coluzzii/A. gambiae heterozygous pattern for both IGS and SINE or a discordant result between markers were classified as admixed.
Microsatellite genotyping
Nineteen microsatellites, nine mapping on chromosome-X and ten on chromosome-3, were genotyped (Supplementary Table 1). Each locus was PCR amplified with fluorescently labelled primers52 and fragment analysis was performed on an automated sequencer at the Science Hill DNA Analysis Facility, Yale University. To control for variation between capillary runs, the PCR products of two A. coluzzii (SUAKOKO strain) specimens from a laboratory colony were used in all runs. One additional positive control (DNA template from a colony mosquito) and one negative control (no template) were also included to assess PCR quality. Allele sizes were scored using GENEMARKER® (SoftGenetics, PA, USA).
Genetic diversity was assessed by estimates of allele richness (AR), expected heterozygosity (He) and inbreeding coefficient (FIS) available in FSTAT v. 2.9.3.254. Departures from Hardy-Weinberg proportions were tested by exact tests using ARLEQUIN v.3.555. Presence of null alleles was tested using MICRO-CHECKER with the complete range option56. Whenever multiple tests were performed the nominal significance level (α = 0.05) was adjusted by a sequential Bonferroni procedure.
The Bayesian clustering analysis method implemented in STRUCTURE 2.3.357 was used to infer the number of genetic clusters (K) without prior information of sampling locations. A model with correlated allele frequencies within populations (λ = 1) with the option of admixture (α was allowed to vary) was used. For each value of K (K = 1 to 10) 10 independent runs were performed with a burn-in period of 100,000 iterations followed by 200,000 iterations. Two ad hoc approaches implemented in Structure Harvester v.0.6.9458 were used to determine K: (i) an estimation of ln[Pr(X|K)]57; and ii) the ΔK statistic59. Upon selecting K, data across runs were optimally aligned with CLUMPP60 using the Greedy algorithm. Assignment of individual mosquitoes in to genetic clusters was performed based on a probability threshold (Tq) of 0.50.
Spatially-explicit genetic clustering analysis was conducted using TESS v.2.361. Individual coordinates for each specimen were randomly generated within a 10 km radius circle around the geographic coordinate of each locality. The two admixture models (CAR and BYM) were used in the analysis. Ten independent runs were carried out with a burn-in period of 100,000 iterations and 100,000 data-collection iterations for each Kmax (K = 2 to K = 9). The Deviance Information Criterion (DIC) was used to select and to infer the number of clusters. The best performing admixture model and optimal Kmax were selected from plots of the DIC and Kmax as the lowest value at which the DIC curve reached a plateau. Individual membership probabilities of the ten runs for the optimal Kmax were averaged using the greedy algorithm in CLUMPP. The R-script available in POPS62 was used to display spatial interpolations of the Q matrices obtained with TESS based on kriging methods.
Chromosomal analysis
Polytene chromosome preparations were obtained from the ovaries of half-gravid females and banding patterns were scored under a phase-contrast microscope (400X magnitude). Paracentric inversion karyotypes were named as follows: standard (non-inverted, i.e. 2R+j, +b, +d and 2L+a) and inverted arrangements (2Rj, b, d and 2La) were considered as alternative alleles of a bi-allelic locus (where locus = chromosomal region containing an inversion polymorphism and allele = inversion orientation arrangement). BISSAU and SAVANNA chromosomal forms were defined according to the criteria of Bryan et al.38, Coluzzi et al.8 and Petrarca et al.39.
Whole genome sequencing
Mosquitoes identified as A. gambiae by IGS/SINE from three sites in Guinea Bissau were sequenced using Illumina® technology at the Wellcome Trust Sanger Institute (Hinxton, UK). Individuals from Safim and Leibala were sequenced specifically for this study. Genomic DNA library preparation (input DNA amount >50 ng), cluster generation and sequencing were undertaken according to the manufacturer’s protocol for paired-end 100 bp sequence reads. In addition, a sample from Antula was retrieved from the genomes available at the Anopheles gambiae 1000 genomes consortium63. Additional A. gambiae and also A. coluzzii specimens were sequenced but did not yield sufficient quality sequence for inclusion after conservative filtering (see below). Finally, A. gambiae genomes from Accra, Ghana, were taken from Clarkson et al.5.
To ensure that all samples were comparable and that there was high confidence in the variants scored, raw SNPs called using the Unified Genotyper algorithm from the GATK package64 were conservatively hard filtered using a custom Python script. Unlike simple ‘hard filtering’, the script allowed the DP annotation distribution to be considered for each individual and variants were rejected if they possessed the following annotation parameters: GQ < 40, DP < 14, DP < median DP/2 or DP > median DP x 2, MQ < 40, QD < 5, HRun > 3. The final two filtering rules were taken from the Human 1000 Genomes Project65. The additional rules were trialled against an unpublished data set of A. gambiae laboratory crosses and were found to reduce Mendelian errors compared to just the human filtering rules.
To further increase the quality of these datasets, positions with missing genotypes in any sample were removed using VCFtools package (v0.1.12a)66. In analyses where only Guinea Bissau sequences were compared, all filtered SNP calls were merged and positions missing data in any individual were removed using VCFtools to create a Guinea Bissau-missingless VCF file. For analyses which included samples from Ghana, the four additional individuals’ VCF files were merged with the Guinea Bissau-missingless VCF (using VCFtools) and missing sites were removed again to produce an all-sample-missingless VCF file. This was carried out to preserve the high resolution of variants in the Guinea Bissau-only analyses owing to the higher sequencing depth of those samples. Guinea Bissau samples had a mean coverage of 32.3x compared to 23.8x for Ghanaian samples (Supplementary Table S3).
Pairwise population FST estimates were calculated using weir-fst-pop available in VCFtools. FST means, standard errors and 95% confidence intervals were then calculated using custom Perl and R scripts and Manhattan plots were created with the R package qqman67.
As A. coluzzii individual whole genome sequence data were not available, an ancestry informative marker (AIM) approach was used5,19,20. Informative markers were obtained from a 400k SNP chip used to characterise divergence between Malian A. gambiae and A. coluzzii17. SNPs with allele frequency differences >0.9 between species were selected as being ancestry informative. Percentage ancestry was assessed by scoring all Guinea Bissau individuals at each marker as being of homozygous A. gambiae, homozygous A. coluzzii or heterozygous (admixed) ancestry. Autosomes and chromosome-X were examined separately because chromosome-X has been shown to be less susceptible to introgression14 and because it has a higher number of AIMs than all the autosomes combined5,15.
To produce a maximally informative dataset for principal component analysis (PCA), singletons were removed and linkage disequilibrium (LD) pruning was conducted using 500 SNP windows, sliding 100 SNP at a time, with an LD (r2) threshold of 0.1. A low r2 threshold was used due to the high genetic diversity and low linkage disequilibrium found in these mosquito species17. Calculations were done using the all-sample-missingless VCF and the software PLINK (v1.9)68. PCA was then performed on the 3L and 3R chromosome arms using the smartpca function in the EIGENSOFT (6.0.1) package69. Chromosome-3 was chosen to better reflect relation between samples with respect to gene flow, because this chromosome is less affected by confounding factors such as large polymorphic inversions (i.e. such as those found on chromosome arms 2L and 2R) and regions related to speciation (i.e. such as the centromeric region on chromosome-X)14.
Insecticide resistance genes
Position 1014 at the voltage-gated sodium channel gene (VGSC1014), in which two mutations (L1014F70 and L1014S71) are associated with knockdown resistance (kdr), was genotyped by PIRA-PCR72. The G119S mutation in the acetylcholinesterase-1 (ace-1) gene associated with resistance to carbamates and organophosphates was genotyped by PCR-RFLP73.
Blood meal identification
A two-site ELISA74 was used to identify the origin of blood meal in engorged females. Blood meals were tested for the presence of chicken, cow, dog, goat/sheep, horse/donkey, human, pig, and rabbit immunoglobulin G (IgG). Four positive controls (blood from the tested host) and 14 negative controls (two blood samples from the other seven hosts) were used in every 96-well microplate. Absorbance was read at 492 nm wave length and cut-off values were calculated for each plate as the mean plus three times the standard deviation of the negative controls.
Additional Information
How to cite this article: Vicente, J. L. et al. Massive introgression drives species radiation at the range limit of Anopheles gambiae. Sci. Rep. 7, 46451; doi: 10.1038/srep46451 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
To the people of Guinea Bissau for their hospitality during mosquito collections. We are grateful to Cesar Capinha (GHTM/IHMT) for preparing the aridity index map presented in the supplementary materials. We thank the Anopheles gambiae 1000 Genomes consortium for providing some of the whole genome sequences here analysed. This study received financial support from FP7/EC funded INFRAVEC project (contract agreement no. 228421), FCT Portugal/FEDER (through Program COMPETE) co-funds (PTDC/BIA-EVF/120407/2010), MIUR-FIRB “Futuro in Ricerca 2010” grant to BC (Grant No. RBFR106NTE) and University of Rome SAPIENZA “AWARDS 2013” project. TA was funded by a Sir Henry Wellcome Postdoctoral fellowship (WT100066MA).
Footnotes
The authors declare no competing financial interests.
Author Contributions Research design: A.d.T., A.R., D.W. and J.P.; mosquito collections: B.C., C.A.S., J.D., J.L.V., J.P. and M.P.; molecular identification, genotyping and ELISA: B.C., C.A.S., E.M. and J.L.V.; whole genome sequencing: A.M., D.M., D.P.K., E.D., J.S. and M.J.D.; karyotyping: B.C., M.P. and V.P., data analysis: B.G., C.S.C., D.W., G.B., J.L.V., J.P. and T.A.; Manuscript writing: A.d.T., B.C., C.S.C., D.W., J.P. and M.J.D.
References
- Kleindorfer S. et al. Species collapse via hybridization in Darwin’s tree finches. Am. Nat. 183, 325–341 (2014). [DOI] [PubMed] [Google Scholar]
- Roy D., Lucek K., Walter R. P. & Seehausen O. Hybrid ‘superswarm’ leads to rapid divergence and establishment of populations during a biological invasion. Mol. Ecol. 24, 5394–5411 (2015). [DOI] [PubMed] [Google Scholar]
- Nolte A. W. & Tautz D. Understanding the onset of hybrid speciation. Trends Genet. 26, 54–58 (2010). [DOI] [PubMed] [Google Scholar]
- Pardo-Diaz C. et al. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet. 8, e1002752, doi: 10.1371/journal.pgen.1002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson C. S. et al. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat. Commun. 5, 4248, doi: 10.1038/ncomms5248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E. et al. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J. Med. Entomol. 45, 1057–1063 (2008). [DOI] [PubMed] [Google Scholar]
- Marsden C. D. et al. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol. Ecol. 20, 4983–4994 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M., Petrarca V. & Di Deco M. A. Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae. Boll. Zool. 52, 45–63 (1985). [Google Scholar]
- Coluzzi M., Sabatini A., della Torre A., di Deco M. A. & Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298, 1415–1418 (2002). [DOI] [PubMed] [Google Scholar]
- Ayala D., Ullastres A. & González J. Adaptation through chromosomal inversions in Anopheles. Front. Genet. 5, 129, doi: 10.3389/fgene.2014.00129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Torre A. et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol. Biol. 10, 9–18 (2001). [DOI] [PubMed] [Google Scholar]
- Coetzee M. et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619, 246–724 (2013). [PubMed] [Google Scholar]
- Feder J. L., Egan S. P. & Nosil P. The genomics of speciation-with-gene-flow. Trends Genet. 28, 342–350 (2012). [DOI] [PubMed] [Google Scholar]
- Fontaine M. C. et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347, 1258524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Hahn M. W. & Nuzhdin S. V. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3, e285, doi: 10.1371/journal.pbio.0030285 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J., Cheng C., Simard F., Costantini C. & Besansky N. J. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol. Ecol. 19, 925–939 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey D. E. et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330, 514–517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank T. E. & Hahn M. W. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 23, 3133–3157 (2014). [DOI] [PubMed] [Google Scholar]
- Lee Y. et al. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc. Natl Acad. Sci. USA 110, 19854–19859 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris L. C. et al. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc. Natl. Acad. Sci. USA 112, 815–820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboagye-Antwi F. et al. Experimental swap of Anopheles gambiae’s assortative mating preferences demonstrates key role of X-chromosome divergence island in incipient sympatric speciation. PLoS Genet. 11, e1005141, doi: 10.1371/journal.pgen.1005141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F. et al. Distribution of knock-down resistance mutations in Anopheles gambiae molecular forms in west and west-central Africa. Malar. J. 7, 74, doi: 10.1186/1475-2875-7-74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main B. J. et al. Complex genome evolution in Anopheles coluzzii associated with increased insecticide usage in Mali. Mol Ecol. 24, 5145–5157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opondo K. O. et al. Does insecticide resistance contribute to heterogeneities in malaria transmission in The Gambia? Malar J. 15, 166, doi: 10.1186/s12936-016-1203-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J. et al. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc. Natl Acad. Sci. USA 108, 244–249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diabate A. et al. Evidence for divergent selection between the molecular forms of Anopheles gambiae: role of predation. BMC Evol. Biol. 8, 5, doi: 10.1186/1471-2148-8-5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diabate A., Dabire R. K., Millogo N. & Lehmann T. Evaluating the effect of postmating isolation between molecular forms of Anopheles gambiae (Diptera: Culicidae). J. Med. Entomol. 44, 60–64 (2007). [DOI] [PubMed] [Google Scholar]
- Lehmann T. & Diabaté A. The molecular forms of Anopheles gambiae: A phenotypic perspective. Infect. Genet. Evol. 8, 737–746 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdem C. et al. Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PLoS ONE 7, e39453, doi: 10.1371/journal.pone.0039453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene Fossog B. et al. Habitat segregation and ecological character displacement in cryptic African malaria mosquitoes. Evol. Appl. 8, 326–345 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F. et al. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar. J. 7, 163, doi: 10.1186/1475-2875-7-163 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Torre A., Tu Z. J. & Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem. Mol. Biol. 35, 755–769 (2005). [DOI] [PubMed] [Google Scholar]
- Caputo B. et al. Anopheles gambiae complex along The Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malar. J. 7, 182, doi: 10.1186/1475-2875-7-182 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordicho V. et al. First report of an exophilic Anopheles arabiensis population in Bissau City, Guinea-Bissau: recent introduction or sampling bias? Malar. J. 13, 423, doi: 10.1186/1475-2875-13-423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo B. et al. The “far-west” of Anopheles gambiae molecular forms. PLoS One 6, e16415, doi: 10.1371/journal.pone.0016415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D., Wilding C. S., Steen K., Pinto J. & Donnelly M. J. Gene flow-dependent genomic divergence between Anopheles gambiae M and S Forms. Mol. Biol. Evol. 29, 279–291 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo B. et al. The last bastion? X chromosome genotyping of Anopheles gambiae species pair males from a hybrid zone reveals complex recombination within the major candidate ‘genomic island of speciation’. Mol. Ecol. 25, 5719–5731 (2016). [DOI] [PubMed] [Google Scholar]
- Bryan J. H., di Deco M. A., Petrarca V. & Coluzzi M. Inversion polymorphism and incipient speciation in Anopheles gambiae s.str. in The Gambia, West Africa. Genetica 59, 167–176 (1982). [Google Scholar]
- Petrarca V., Carrara G. C., Di Deco M. A. & Petrangeli G. The Anopheles gambiae complex in Guinea Bissau. Parassitologia 25, 29–39 (1983). [PubMed] [Google Scholar]
- Caputo B. et al. Prominent intraspecific genetic divergence within Anopheles gambiae sibling species triggered by habitat discontinuities across a riverine landscape. Mol. Ecol. 23, 4574–4589 (2014). [DOI] [PubMed] [Google Scholar]
- Nwakanma D. C. et al. Breakdown in the process of incipient speciation in Anopheles gambiae. Genetics 193, 1221–1231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford M. R. et al. Plasmodium falciparum infection rates for some Anopheles spp. from Guinea-Bissau, West Africa [version 2; referees: 2 approved]. F1000Research 3, 243, doi: 10.12688/f1000research.5485.2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J. et al. The population genomics of trans-specific inversion polymorphisms in Anopheles gambiae. Genetics 183, 275–288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle S. Rice sector policy options in Guinea Bissau. Working paper (Charles H Dyson School of Applied Economics and Management, Cornell University, Ithaca, New York (2015). [Google Scholar]
- Smith H. A. et al. Genome-wide QTL mapping of saltwater tolerance in sibling species of Anopheles (malaria vector) mosquitoes. Heredity 115, 471–479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispo E., Moore J. S., Lee-Yaw J. A., Gray S. M. & Haller B. C. Broken barriers: human-induced changes to gene flow and introgression in animals: an examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. Bioessays 33, 508–518 (2011). [DOI] [PubMed] [Google Scholar]
- Mancini E. et al. Adaptive potential of hybridization among malaria vectors: introgression at the immune locus TEP1 between Anopheles coluzzii and A. gambiae in ‘Far-West’ Africa. PLoS One 10, e0127804, journal.pone.0127804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Malaria Report, 2014 (World Health Organization, Geneva, 2014).
- Yadouleton A. et al. Cotton pest management practices and the selection of pyrethroid resistance in Anopheles gambiae population in northern Benin. Parasit. Vectors 4, 60, doi: 10.1186/1756-3305-4-60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. Guinea Bissau country profile: Major food and agriculture productions. http://faostat.fao.org/DesktopDefault.aspx?PageID=339&lang=en&country=175 (Date of assess: 10/05/2016) (2012).
- While G. M. et al. Sexual selection drives asymmetric introgression in wall lizards. Ecol. Lett. 18, 1366–1375 (2015). [DOI] [PubMed] [Google Scholar]
- Donnelly M. J., Cuamba N., Charlwood J. D., Collins F. H. & Townson H. Population structure in the malaria vector, Anopheles arabiensis Patton, in East Africa. Heredity 83, 408–417 (1999). [DOI] [PubMed] [Google Scholar]
- Fanello C., Santolamazza F. & della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med. Vet. Entomol. 16, 461–464 (2002). [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 86, 485–486 (1995). [Google Scholar]
- Excoffier L., Laval G. & Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinf. Online 1, 47–50 (2005). [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C., Hutchinson W. F., Wills D. P. M. & Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538 (2004). [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D. A. & VonHoldt B. M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (2012). [Google Scholar]
- Evanno G., Regnaut S. & Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- Jakobsson M. & Rosenberg N. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007). [DOI] [PubMed] [Google Scholar]
- Chen C., Durand E., Forbes F. & Francois O. Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol. Ecol. Notes 7, 747–756 (2007). [Google Scholar]
- Jay F. et al. Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol. Ecol. 21, 2354–2368 (2012). [DOI] [PubMed] [Google Scholar]
- The Anopheles gambiae 1000 Genomes Consortium. Ag1000G phase 1 AR2 data release. MalariaGEN. http://www.malariagen.net/data/ag1000g-phase1-AR2 (Date of Access: 19/12/2014) (2014).
- DePristo M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. S. qqman: An R package for visualizing GWAS results using QQ and Manhattan plots. BioRxiv, doi: 10.1101/005165 (2014). [DOI] [Google Scholar]
- Chang C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7, doi: 10.1186/s13742-015-0047-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A. L. & Reich D. Population structure and eigenanalysis. PLoS Genet. 2, e190, doi: 10.1371/journal.pgen.0020190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Torres D. et al. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol. Biol. 7, 179–184 (1998). [DOI] [PubMed] [Google Scholar]
- Ranson H. et al. Identification of a point mutation in the voltage-gated sodium channel gene of Kenyan Anopheles gambiae associated with resistance to DDT and pyrethroids. Insect Mol. Biol. 9, 491–497 (2000). [DOI] [PubMed] [Google Scholar]
- Janeira F. et al. A primer-introduced restriction analysis-polymerase chain reaction method to detect knockdown resistance mutations in Anopheles gambiae. J. Med. Entomol. 45, 237–241 (2008). [DOI] [PubMed] [Google Scholar]
- Weill M. et al. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol. Biol. 13, 1–7 (2004). [DOI] [PubMed] [Google Scholar]
- Simões M. J., Próspero M. I. & Ribeiro H. Optimização Da Técnica ELISA ‘Two Sites’ Utilizada Na Identificação de Refeições Sanguíneas de Mosquitos. Rev. Port. Doenc. Infec. 18, 225–229 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.