To explore how the functional organization of a circuit shapes its emergent behavior, most previous studies have compared experimentally observed patterns of activity to those in shuffled data. However, existing methods for shuffling destroy all correlations between neurons, leaving it ambiguous whether observed patterns are simply a by-product of generic correlations or whether they depend on a specific correlation matrix. Here we describe a nonrandom shuffling procedure that preserves the strengths of observed correlations but rearranges them arbitrarily. We use this procedure to show how the functional organization of prefrontal circuits enables them to generate patterns of activity that have been linked to important aspects of cognition. This procedure may be similarly useful for future studies of emergent circuit behavior.
Keywords: calcium imaging, pattern generation, prefrontal cortex
Abstract
Sequential patterns of prefrontal activity are believed to mediate important behaviors, e.g., working memory, but it remains unclear exactly how they are generated. In accordance with previous studies of cortical circuits, we found that prefrontal microcircuits in young adult mice spontaneously generate many more stereotyped sequences of activity than expected by chance. However, the key question of whether these sequences depend on a specific functional organization within the cortical microcircuit, or emerge simply as a by-product of random interactions between neurons, remains unanswered. We observed that correlations between prefrontal neurons do follow a specific functional organization—they have a small-world topology. However, until now it has not been possible to directly link small-world topologies to specific circuit functions, e.g., sequence generation. Therefore, we developed a novel analysis to address this issue. Specifically, we constructed surrogate data sets that have identical levels of network activity at every point in time but nevertheless represent various network topologies. We call this method shuffling activity to rearrange correlations (SHARC). We found that only surrogate data sets based on the actual small-world functional organization of prefrontal microcircuits were able to reproduce the levels of sequences observed in actual data. As expected, small-world data sets contained many more sequences than surrogate data sets with randomly arranged correlations. Surprisingly, small-world data sets also outperformed data sets in which correlations were maximally clustered. Thus the small-world functional organization of cortical microcircuits, which effectively balances the random and maximally clustered regimes, is optimal for producing stereotyped sequential patterns of activity.
NEW & NOTEWORTHY
To explore how the functional organization of a circuit shapes its emergent behavior, most previous studies have compared experimentally observed patterns of activity to those in shuffled data. However, existing methods for shuffling destroy all correlations between neurons, leaving it ambiguous whether observed patterns are simply a by-product of generic correlations or whether they depend on a specific correlation matrix. Here we describe a nonrandom shuffling procedure that preserves the strengths of observed correlations but rearranges them arbitrarily. We use this procedure to show how the functional organization of prefrontal circuits enables them to generate patterns of activity that have been linked to important aspects of cognition. This procedure may be similarly useful for future studies of emergent circuit behavior.
many in vivo studies have identified sequential patterns of activity within the prefrontal cortex (PFC) and other associational cortices during working memory (Baeg et al. 2003; Fujisawa et al. 2008; Harvey et al. 2012; Seidemann et al. 1996). The visual cortex generates similar patterns both in response to visual stimulation as well as spontaneously (Carrillo-Reid et al. 2015). However, it remains unclear how such types of patterns are generated. Specifically, do these sequences depend on a specific functional organization within cortical microcircuits or emerge simply as a by-product of random interactions between neurons? Many studies have used theory and simulations to show how neural networks resembling local neocortical circuits can generate the sorts of stereotyped trajectories or sequential patterns of activity believed to mediate a variety of functions including motor planning and working memory (Ganguli et al. 2008; Goldman 2009; Hennequin et al. 2014; Laje and Buonomano 2013; Litwin-Kumar and Doiron 2012; Sussillo and Abbott 2009). Nevertheless, it has not yet been possible to demonstrate that the functional organization of an actual neocortical microcircuit contributes to its ability to generate meaningful patterns of multineuron activity.
Recent advances in recording techniques have made it possible to collect large data sets and make important observations, e.g., neurons within isolated cortical networks exhibit correlations that follow a small-world organization (Sadovsky and MacLean 2013) and can fire in a temporally precise manner (Sadovsky and MacLean 2014). However, a fundamental unsolved challenge is developing analytic methods, beyond simulations, that can directly assess how the functional organization of a circuit contributes to the production of emergent patterns of activity. Notably, one study used sophisticated imaging methods to show that isolated neocortical microcircuits in brain slices spontaneously generate sequential patterns of activity (Ikegaya et al. 2004); however, later studies showed that even networks of independently firing neurons or with randomly arranged correlations between neurons could generate similar levels of sequences (Roxin et al. 2008). Most studies to date have compared emergent patterns observed experimentally to those expected based on various models using simulations (Roxin et al. 2008; Sadovsky and MacLean 2013). Using simulations in this way is complicated, because unlike shuffling experimental data, it requires making assumptions so that simulations will approximately, though not exactly, match experimentally observed levels and temporal patterns of activity. Conversely, altering the circuit organization in simulations will invariably alter levels and temporal patterns of activity, making it impossible to disambiguate the individual contributions of these three factors to the observed patterns of activity.
Maximum entropy models have been used to directly assess how the organization of a circuit influences its ability to generate patterns of activity. These models predict instantaneous patterns of network activity by fitting the mean firing rate for each neuron, as well as pairwise functional interactions between neurons, without making additional assumptions (Schneidman et al. 2006; Shlens et al. 2006). Maximum entropy models can reproduce multineuron patterns of activity within the isolated retina and cultures of dissociated cortical neurons and have shown that in these systems emergent patterns of multineuron activity do indeed reflect pairwise functional interactions between neurons. However, these models have been less successful for fitting multineuron patterns of activity from cortical networks in brain slices and in vivo (Ohiorhenuan et al. 2010; Tang et al. 2008). In particular, maximum entropy models do not model sequential patterns of activity observed in cortical networks (Tang et al. 2008), because they predict the distribution of instantaneous patterns of activity. This underscores the need for new methods to assess how sequential patterns of activity in experimental data sets depend on the underlying functional organization of a circuit.
Here we outline one such method, which can dissociate the relative contributions of activity levels, temporal patterns of activity, pairwise correlations, and higher-order structure such as clustering to key features of emergent network-level behavior. We use this method, together with new experimental approaches, to show that the small-world functional organization of local prefrontal microcircuits optimizes the production of stereotyped, sequential patterns of activity believed to mediate important cognitive functions in vivo. This reveals a specific, concrete, and important function for small-world networks, which are found ubiquitously in the nervous system and throughout nature, and demonstrates a new method for addressing the critical question of whether knowing the functional organization underlying a neuronal system helps to understand how it works (Marder 2015).
MATERIALS AND METHODS
All experiments were conducted in accordance with procedures established by the Administrative Panels on Laboratory Animal Care at the University of California, San Francisco.
Subjects.
Initial experiments to characterize GCaMP signals and spontaneous activity in prefrontal slices used wild-type C57BL/6 mice. P26–P33 mice of either sex (Charles River) were injected unilaterally with 500 nl of AAV5/2-hSynapsin1::GCaMP6s (UPenn viral vector core) at the coordinates (in mm) 1.7 anterior-posterior, 0.3 mediolateral, and −2.2 dorsoventral. The virus used is a protein fusion of the GCaMP6s construct with the human synapsin promoter and as such expresses in all neurons both excitatory and inhibitory. Injection rate was 100 nl/min, and the needle was left in the brain for 5 min after injection before retraction. At this injection rate, positive cells could be often be seen several hundred micrometers away from the injection site, resulting in ∼500–750 μm of robust viral expression that corresponded to about two or three useful slices per animal. Neurons expressing GCaMP were generally arranged along the injection tract in a position consistent with expression in deep layers of PFC (cf. Fig. 1A). Mice used in this study were wild-type mice that also served as age-matched or littermate controls for various mouse models of autism and schizophrenia in Luongo et al. (2016). Data presented in Figs. 1–5 are from 29 slices collected from 20 mice all bath-exposed to 2 μM carbachol continuously for 1 h.
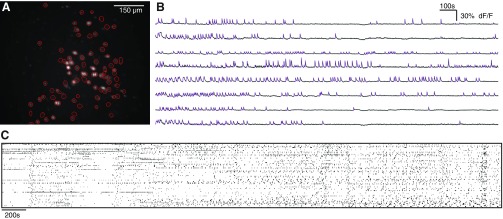
Fig. 1.
Single-photon imaging of GCaMP signals resolves simultaneous activity from many neurons in prefrontal slices. A: regions of interest (ROIs) obtained by an automated algorithm, superimposed on an image showing the maximum standard deviation projection of each pixel during a movie of GCaMP fluorescence. B: sample GCaMP signals from 8 neurons. The overlaid purple lines indicate times at which we detected that each neuron was active. C: example raster of spontaneous network activity for a single 60-min experiment with 82 neurons.
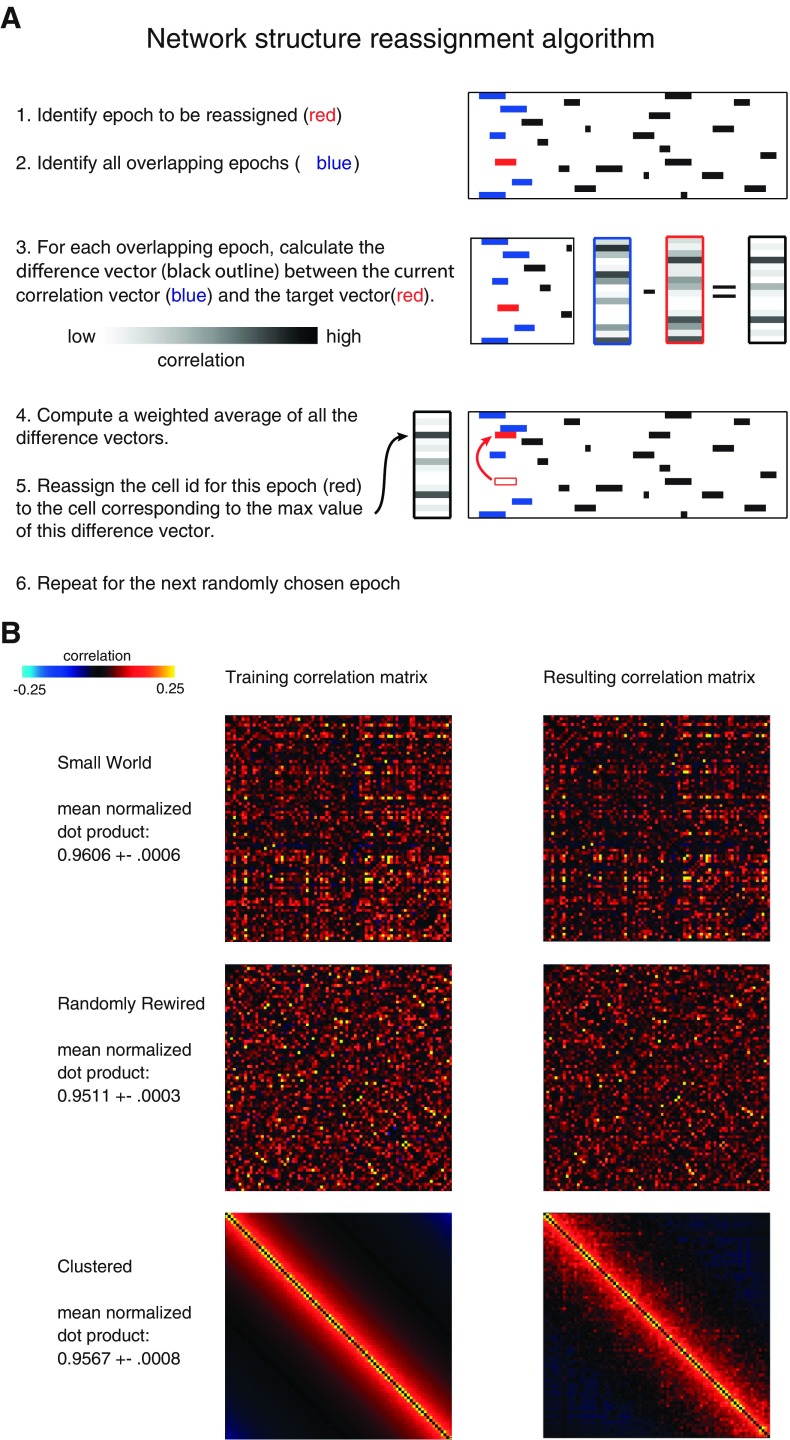
Fig. 5.
Generation of surrogate data sets with various functional organizations. A: overview of our method for generating each surrogate data set. The method reassigns the cell identities associated with each epoch of activity in the original data set based on a target correlation matrix. B: example of the correlation matrices used for generating surrogate data sets (left) and the correlation matrices for the resulting surrogate data sets (right). Each “training correlation matrix” was the target correlation matrix used as an input to SHARC, and each “resulting correlation matrix” was the correlation matrix associated with a surrogate data set outputted by SHARC. Top: training correlation matrix based on the original pattern of correlations. Middle: training correlation matrix based on a randomly rearranged correlation matrix. Bottom: training correlation matrix based on a maximally clustered correlation matrix.
Slice preparation.
Slices preparation followed our previously described protocol (Gee et al. 2012), with one deviation: immediately after brain slices were prepared, they were transferred to an N-methyl-d-glucamine (NMDG)-based recovery solution for 10 min before being transferred to ACSF for the remainder of their recovery (Zhao et al. 2011). The NMDG-based solution was maintained at 32°C and consisted of the following (in mM): 93 NMDG, 93 HCl, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 25 glucose, 20 HEPES, 5 Na-ascorbate, 5 Na-pyruvate, 2 thiourea, 10 magnesium sulfate, and 0.5 CaCl2. This NMDG preparation method was used to improve the overall health of adult slices to ensure sufficient amounts of activity for analysis. ACSF contained the following (in mM): 126 NaCl, 26 NaHCO3, 2.5 KCl, 1.25 NaH2PO4, 1 MgCl2, 2 CaCl2, and 10 glucose. All recordings were at 32.5 ± 1°C.
Imaging.
GCaMP imaging was performed on a Olympus BX51 upright microscope outfitted with a ×20 1.0 NA water immersion lens with ×0.5 reducer (Olympus) and an ORCA-ER CCD camera (Hamamatsu Photonics). Illumination was delivered with a Lambda DG4 arc lamp (Sutter Instruments). The light path was delivered through a 472/30 excitation filter, a 495-nm single-band dichroic, and a 496-nm long-pass emission filter (Semrock).
All movies that were analyzed consisted of 36,000 frames acquired at 10 Hz (1 h) with 4 × 4 sensor binning yielding a final resolution of 256 × 312 pixels. Light power during imaging was 100–500 μW/mm2. The open-source Micro Manager software suite (v1.4, National Institutes of Health) was used to control all camera parameters and acquire movies. Any movies that had significant drift or movement or lacked significant amounts of activity were excluded from further analysis. Specifically, when movies exhibited drift greater than ∼0.25 soma diameters, the experiment was terminated and any data collected were excluded from analysis. Significant movement could also be detected during independent component analysis by the appearance of elliptical rather than circular segments.
We observed that active, GCaMP-expressing neurons were found within a discrete layer (cf. Fig. 1A) that was consistent with the location of layer 5 of medial PFC.
Signal extraction.
All analyses and signal extraction were performed with MATLAB (MathWorks). Locations of cells were automatically identified with a modified version of the published CellSort 1.1 toolbox (Mukamel et al. 2009). In particular, a factor, μ, specifies the weight between spatial and temporal sparseness: μ = 0 is purely spatial and μ = 1 is purely temporal. For all of our analyses we used μ = 0.2, which was within the optimal range outlined in Mukamel et al. (2009). Signals were extracted from movies, and the baseline fluorescence function, F0, was calculated for every trace using the mode of the kernel density estimate over a 100-s rolling window, implemented via the MATLAB function ksdensity according to the procedure outlined in O'Connor et al. (2010). All signal traces shown represent normalized versions of (F − F0)/F0.
Threshold-based event detection was performed on the traces by detecting increases in (F − F0)/F0 exceeding 2.5σ over 1 s and then further thresholding these events by keeping only those events which exceeded a 4σ increase over 2 s; σ is the standard deviation of (F − F0)/F0, calculated over the entire movie. Thus all detected events have a deviation of at least 4σ from baseline. After these events were identified in the calcium signal from a cell, the cell was considered “active” during the entire period from the beginning to the peak of the event. The beginning of the event was defined as the first point for which (F − F0)/F0 increases by 2.5σ within 1 s. For the variable thresholds used in Fig. 3A, only the second threshold was varied. The peak of the event was defined as the local maximum of the entire event, from the beginning of the event until (F − F0)/F0 returns to the same baseline value. We then created a matrix in which each row corresponds to a neuron and each column corresponds to a frame. Entries in this matrix were 1 if a given neuron was active during a given frame and 0 otherwise. All subsequent analyses were performed on this two-dimensional representation of network activity over time (cf. Fig. 1C).
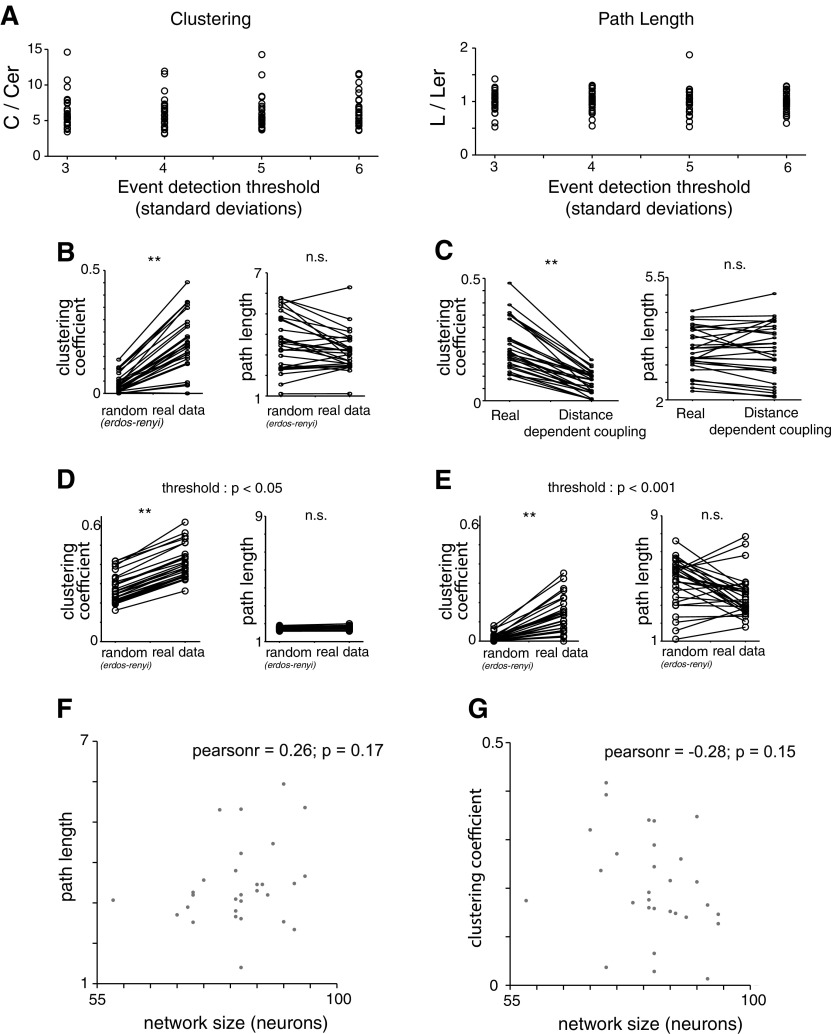
Fig. 3.
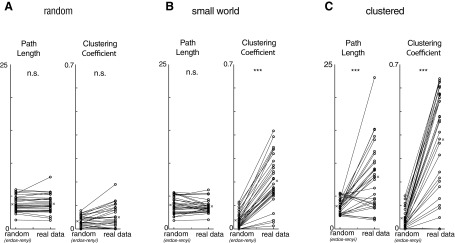
Small-world topology is invariant to event detection threshold, strong correlation threshold, and network size and not due to distance-dependent coupling. A: reanalyzing our original data sets with thresholds of 3, 4, 5, or 6 SDs for event detection and constructing networks based on the pairwise correlation matrix in each case still yields small-world networks. Each dot represents the ratio of either the clustering coefficient, C, or the path length, L, relative to the clustering coefficient or path length for an Erdos-Renyi network (Cer or Ler, respectively), matched to the number of nodes and edge probability. For every threshold, the corresponding network shows nonrandom clustering and random path lengths, characteristic of a small-world topology. B: generating networks based on the pattern of strong correlations between neurons but determining a unique threshold for strong correlations for each cell (rather than using a single threshold for all cells in 1 experiment as was done in our original analysis) still produces a small-world topology showing random path lengths and nonrandom clustering. Each dot represents the ratio of either the clustering coefficient, C, or the path length, L, relative to the clustering coefficient or path length for an Erdos-Renyi network (Cer or Ler, respectively), matched to the number of nodes and edge probability. Networks generated in this way display nonrandom clustering and random path lengths, characteristic of a small-world topology. C: networks generated in according to distance-dependent clustering (i.e., drawn from distribution in Fig. 2C) exhibit much less clustering than networks based on the actual pattern of correlations we observed (“Real”). D: networks generated using a threshold of P < 0.05 to identify “strong correlations” still exhibit small-world characteristics. E: networks generated using a threshold of P < 0.001 to identify “strong correlations” still exhibit small-world characteristics. F: path length plotted as a function of network size illustrates independence (P = 0.17). G: clustering coefficient plotted as a function of network size illustrates independence (P = 0.15). **P < 0.01 by Wilcoxon rank sum test.
Correlations between cells were calculated between the binary event trains corresponding to those two cells after subtraction of the mean level of activity from each event train. The mean level of activity at a given point in time was calculated as a unitary Gaussian filter (sigma: 50 frames) applied to the event train yielding a slow-varying estimate of the mean rate.
Figure 2C was generated by computing the mean correlation of data sets in which each event was randomly reassigned (within each cell) by shifting a range of ±X frames ranging from 1 frame (100 ms) to 80 frames (8 s). Each individual “epoch,” i.e., each continuous period of activity within one neuron's activity raster, was shifted by a unique random offset, as opposed to shuffled data sets in which large segments of an activity raster (including many periods of activity and inactivity) were shifted together.
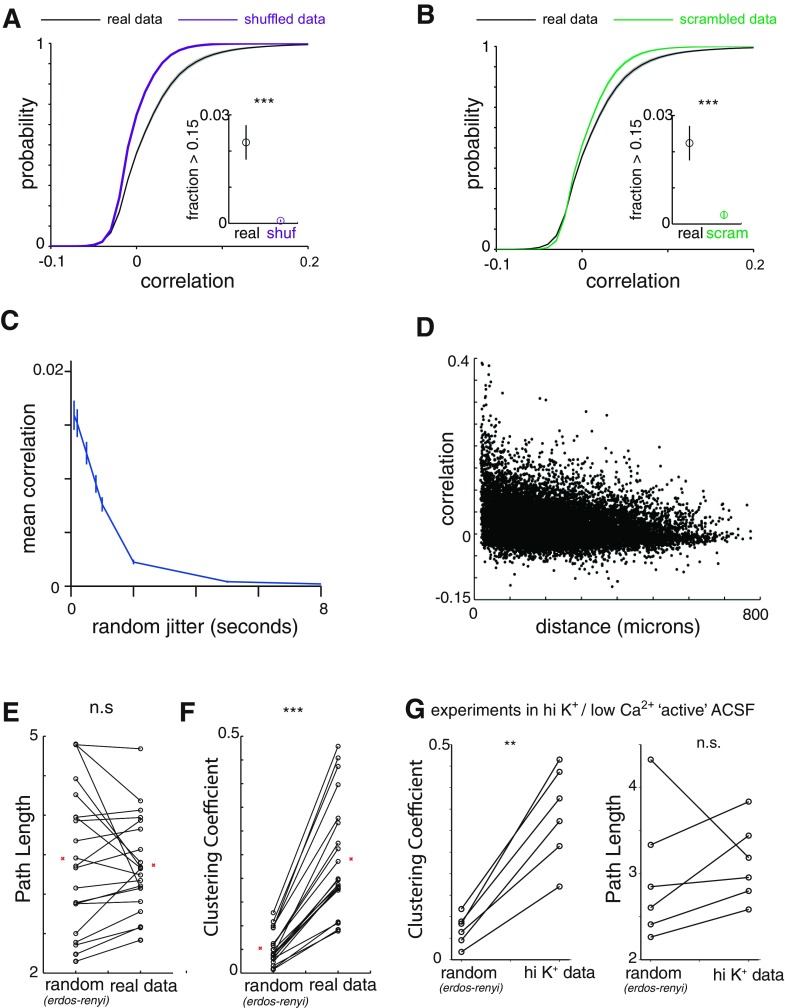
Fig. 2.
Spontaneous prefrontal network activity is enriched in positive correlations and events in which multiple neurons are coactive. A: cumulative probability distributions for correlations in original data sets (“real”; n = 29 experiments) vs. those observed in data shuffled by shifting each neuron's event train by a different random amount (“shuffled”). Inset: fraction of correlations > 0.15, a level of correlation that is extremely rarely observed in shuffled data sets. ***P < 0.001 by Wilcoxon rank sum test. B: cumulative probability distributions for correlations in original data sets (n = 29 experiments) vs. randomly reassigning the neuronal identity associated with each event (“scrambled”). Inset: fraction of correlations > 0.15, a level of correlation that is extremely rare in shuffled data sets. ***P < 0.001 by Wilcoxon rank sum test. C: mean correlation of data sets when each event is randomly reassigned to within a plus or minus range of jitter ranging from 100 ms (1 frame) to 8 s (80 frames). Most correlation is abolished by jitter of 1–2 s. D: correlations between neurons plotted as a function of distance between those neurons. The correlation is weakly related to the distance between 2 neurons, but high correlations occur even at large distances. E: average path length (right) and average path length of a random Erdos-Renyi connected network of the same number of nodes and edge probability (left). n.s., Not significant. F: average clustering coefficient (right) and average clustering coefficient of a random Erdos-Renyi connected network of the same number of nodes and edge probability (left). ***P < 0.001 by Wilcoxon rank sum test. G: we constructed networks based on the pattern of strong correlations in data sets recorded with a high K+, low Ca2+ “active” ACSF instead of carbachol to elicit activity. These “active ACSF” data sets also display random path lengths and nonrandom clustering, characteristic of a small-world topology. **P < 0.01 by Wilcoxon rank sum test.
The standard deviation projection in Fig. 1 was obtained as follows. For each pixel, we computed the standard deviation of (F − F0)/F0 over 100-s intervals throughout the movie then plotted the maximum value of these standard deviations.
Simulations of a random, distance-dependent coupling rule.
The networks based on a random, distance-dependent coupling rule (Fig. 3) were constructed by first measuring correlations as a function of distance across all experiments, as shown in Fig. 2D. A distribution of correlation as a function of distance was then constructed by binning into 50-μm bins. Then, to construct simulated networks, for each pair of neurons we made a random draw from this distribution of correlations, based on the physical distance between this pair of neurons. In this way, we obtained simulated correlation matrices, based on a random, distance-dependent rule for correlations.
Creating and analyzing functional networks.
Edges or functional connections between neurons were determined on the basis of significant correlation. First, all data sets were shuffled 100 times, and from this a null distribution of correlations was estimated and the threshold for an edge, θ, was set as any correlation above the 99% threshold of the null distribution. While the correlation distributions were mean subtracted and thus two tailed, rarely were there any significant negative correlations, and thus only positive correlations were considered. In the case of Fig. 3B, where the threshold was done on a per cell basis, networks were shuffled 500 times and the 99% threshold was used for each individual interaction to determine significance.
The clustering coefficient for each node was calculated by considering the subgraph of the k nodes connected to a given node and then calculating , where e is the total number of edges between the k nodes connected to the principal node divided by the total possible number of edges between all k nodes, which is kC2. So a clustering coefficient of 1 would mean that all possible edges between k neighbors exist, whereas a clustering coefficient of 0 would indicate that none of the k neighbors share an edge.
To compare real, experimentally observed, networks to random ones, we first generated random networks with an Erdos-Renyi model in which all possible edges are equally likely. Specifically, if the real network has an edge probability of p, then we generated a random network in which each possible edge was randomly added with probability p. That is, for each pair of nodes we generated a random number between 0 and 1 (with the MATLAB function rand) and then added an edge between those two nodes if this random number was <p. Random path length/clustering coefficients denoted in all figures represent the average of 100 random networks.
Detecting and quantifying temporal sequences of activity.
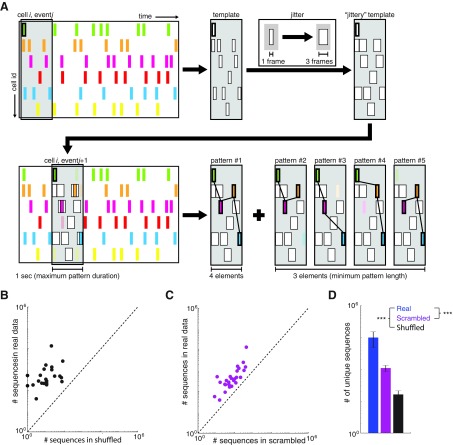
We implemented a template-matching algorithm to identify stereotyped patterns of sequential activity (Ikegaya et al. 2004; Roxin et al. 2008). Briefly, we began with a reference event in a given cell i and then identified all of the other cells that became active in a 1-s (10 frames) window following the reference event. This was stored as a “template vector” of cell IDs and activation times relative to the reference event (i.e., offset times). This template was then shifted to each subsequent event of cell i. Any events that align with the template constitute a pattern. One frame of jitter was allowed when matching event times. We identified all possible patterns of sequential activation that begin with the reference event. For example, if a reference event in cell 1 was followed sequentially by events in cells 5, 17, and 37, then we would identify both a four-cell sequence (1 → 5 → 17 → 37) and three three-cell sequences (1 → 5 → 17, 1 → 5 → 37, 1 → 17 → 37) as illustrated in Fig. 3. A “pattern vector” containing the cell IDs and offset times of each matched event was stored for each identified sequence. If this pattern vector matched an existing pattern vector—again allowing one frame of jitter—then it was counted as an additional incidence of that pattern; otherwise, it was stored as a new pattern. For the purpose of defining “unique patterns,” patterns had to repeat at least three times in data to be counted. This process was repeated iteratively, and every active state in every cell was used as a reference event. The algorithm was not parallelized and required ∼4 h per data set running on a 2.0-GHz dual-core processor.
Types of simulated data sets.
We compared the numbers of sequential patterns of activity (quantified as described above) in our actual, experimentally observed data sets to those in various forms of simulated data sets. First, we generated “shuffled data sets” simply by shifting large chunks of each neuron's event train in time. Specifically, we randomly subdivided each even train into six segments of random length, circularly shuffled each of these by a random amount, and then recomposed them together to construct the full event train. For example, a cell's event train might be separated into segments of 4,000, 6,000, 5,000, 8,000, 10,000, and 3,000 frames, which were individually shuffled and then recombined to form the shuffled data set. Second, we generated “scrambled” versions of experimentally observed data sets in which we preserved the number of neurons active at each point in time but randomly reassigned the identities of the specific neurons that are active or inactive. Specifically, we identified all of the “epochs” of activity within each data set, i.e., all the periods of time during which a neuron was continuously active. Each epoch is defined by a start time, duration, and associated neuron, and a data set is fully specified by the corresponding list of epochs. Thus we created each scrambled data set using a new list of epochs, created by combining an original list of epoch times and durations with a randomly permuted list of associated neurons.
Third, as described in detail below, we created surrogate data sets in which we nonrandomly shuffled which neurons were active at each point in time, in order to achieve a desired correlation matrix. For this, we developed a new method we call shuffling activity to rearrange correlations (SHARC). We used SHARC to create surrogate data sets that simulate three different network topologies (described in more detail in results): small world, random, and clustered. Fourth, we simulated independent homogeneous or inhomogeneous Poisson neurons (with firing rates matched to the experimentally observed ones). Finally, in one case we used SHARC to shuffle the simulated activity of Poisson neurons, in order to create surrogate data sets that match the pattern of experimentally observed correlations.
Creating surrogate data sets by shuffling activity to rearrange correlations.
As described in more detail in results, we created surrogate data sets using the times and durations of active states observed in the original data set but reassigned the cell identities associated with each active state. The cell identities were reassigned based on an iterative process and a target correlation matrix, C. To create a surrogate data set based on the original pattern of correlations, we simply used the correlation matrix obtained from the original data set, Ci,orig. To obtain Ci,rand we randomly rearranged the values in Ci,orig (while maintaining the symmetry of the correlation matrix).
Based on the target correlation matrices, defined above, we reassigned the cell identities associated with each active state. Specifically, we randomly selected an active state, i. We then found all the active states that overlapped with this active state. Next, we selected the subset of these active states for which new cell identities had already been assigned; call this set X. Let rj represent the number of time points over which active state j ∈ X overlaps with active state i, and let nj represent the identity of the cell assigned to active state j ∈ X. Li and Lj are the lengths of active states i and j, respectively. Then we constructed a vector,
where represents row j of the target correlation matrix, i.e., the target correlations between neuron nj and the other neurons, and contains the current values of the correlations between neuron nj and the other neurons based on the reassigned portion of the surrogate data set. This step can be thought of as “guessing” which cell should be assigned to a particular epoch of activity by first figuring out what other cells are active at the same time and then choosing cells that are strongly correlated with these known active cells. Note that we assign values of (i.e., construct “guesses” about which cell should be active), using the difference between the current correlation matrix () and the target correlation matrix (), in order to identify cell pairs for which the current correlation deviates from the target value, and force the new correlation matrix to progressively approximately the target correlation matrix.
We set elements of to zero if the corresponding neuron had already been assigned to an active state that overlaps with active state i, i.e., element nj of was set to zero . Finally, we assigned active state i to the neuron corresponding to the maximum value of . This can be thought of as choosing the cell that represents the “consensus” based on tallying up all of the “guesses” about which cells “should” be assigned to the epoch of activity being considered. If all the elements of were zero, e.g., because there are no overlapping active states that have had new cell identities assigned, then we simply used the cell identity of this active state from the original data set. We repeated this procedure for every epoch of activity.
For surrogate data sets that were fit to correlation matrices from other experiments, if there was a mismatch in the number of cells in the two experiments, the experiment with more cells was downsampled by randomly choosing a subset of those neurons. For example if activity from data set A with 80 neurons was going to be fit to a matrix from data set B with 70 neurons, only 70 randomly chosen neurons from data set A would be fit to data set B. A MATLAB implementation of SHARC is available at https://github.com/fluongo/SHARC.
To simulate randomly rewired networks, we randomly rearranged all elements of an experimentally observed correlation matrix (maintaining symmetry). This yielded a randomly rearranged correlation matrix that was used as input to SHARC. To simulate maximally clustered networks, we started with a set of experimentally observed correlations. Then we arranged all neurons along an imaginary line (in order of their indexes, i.e., neuron 1 followed by neuron 2, etc.) and then reassigned the experimentally observed correlations based on a distance-dependent rule. That is, the strongest correlations were assigned to pairs of “neighboring” neurons, and the weakest correlations were assigned to neurons on opposite ends of the imaginary line. Examples of the randomly rewired and maximally clustered correlation matrices used as inputs to SHARC are shown in Fig. 5B.
Simulating networks of independent Poisson neurons.
We simulated networks of independent Poisson neurons that were rate-matched to neurons in our actual experiments. For each simulated neuron, we drew event times from a Poisson distribution with either a constant rate parameter, equivalent to the rate observed in the corresponding actual neuron over the entire experiment (homogeneous), or a time-varying rate parameter, calculated with a 60-s sliding average of the corresponding actual neuron's event rate (inhomogeneous). In this way, for each experimental data set, we simulated a network of independent, rate-matched homogeneous or inhomogeneous Poisson neurons. As for our other data sets, we then detected stereotyped sequential patterns of activity in these networks of Poisson neurons. The results of this analysis are shown in Fig. 9.
Fig. 9.
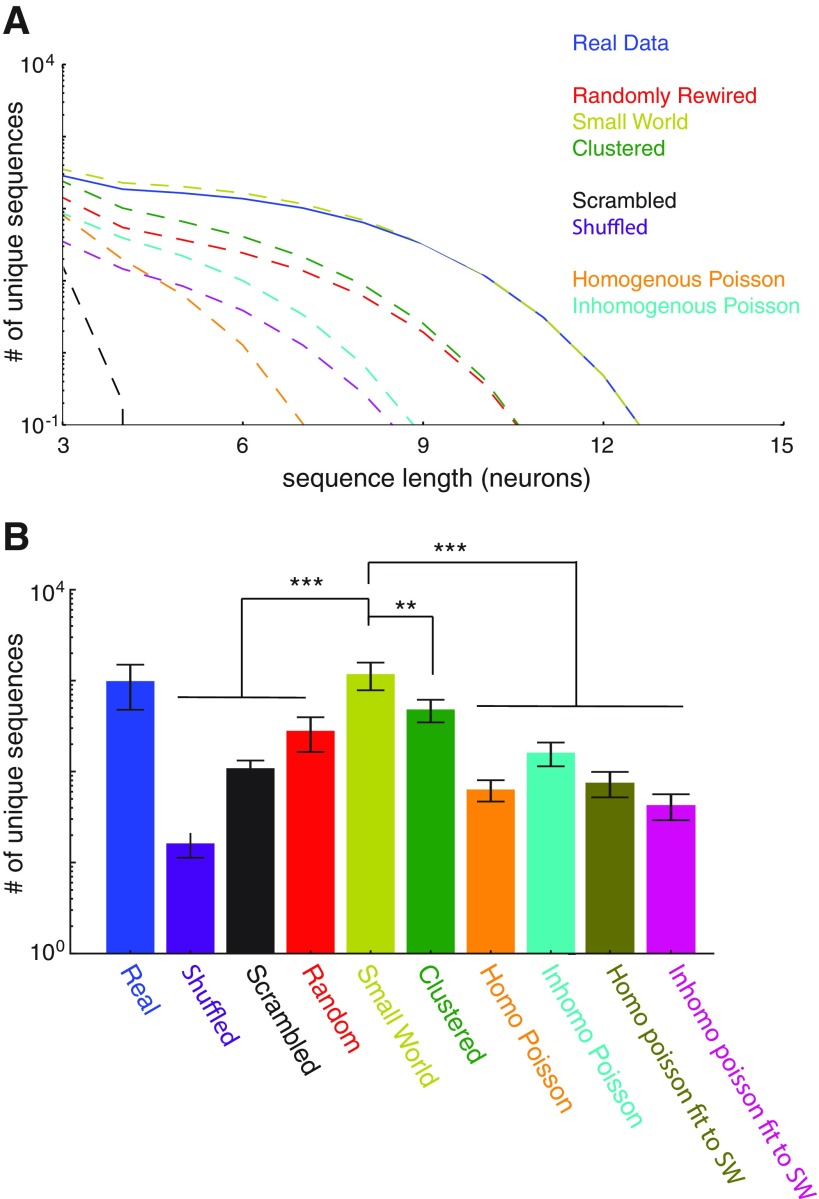
Small-world networks outperform clustered networks, random networks, and independent homogeneous/inhomogeneous Poisson neurons and reproduce the levels of sequences observed in actual data. A: the average number of unique temporal sequences is plotted as a function of the sequence length (in neurons) for surrogate data sets representing various types of networks, as well as our experimentally recorded data sets. For each experimental data set (n = 29), we generated 1 surrogate data set of each type, such that all of these surrogate data sets conformed to the temporal pattern of activity in the experimental data set. B: bar graph summarizing the total number of unique temporal sequences within various types of data sets. Real data contain more sequences than shuffled and scrambled data. Only SHARC-generated surrogate data sets based on the observed small-world correlations recapitulate the experimentally observed levels of sequences, outperforming SHARC-generated surrogate data sets based on either random or clustered correlations. Neither independent homogeneous Poisson nor inhomogeneous Poisson neurons produce high levels of sequences. Using SHARC to fit the activity of either homogeneous or inhomogeneous Poisson neurons to the experimentally observed small-world pattern of correlations also fails to recapitulate the levels of sequences observed in real data. **P < 0.05, ***P < 0.001 by Wilcoxon rank sum test.
Statistical analysis.
Unless otherwise noted, we used the Mann-Whitney U-test to compare pairs of groups, repeated-measures ANOVA to compare multiple groups, and the two-tailed Kolmogorov-Smirnov test to compare pairs of distributions. Error bars where shown indicate standard error unless otherwise noted. Cell identification, signal extraction and normalization, event detection, and all other data analysis was done with fully automated routines that were independent of the investigator and thus blinded.
RESULTS
We initially used single-photon, wide-field imaging to capture fluorescent signals from the genetically encoded Ca2+ indicator GCaMP6s (Chen et al. 2013) in acute brain slices from late adolescent (P41–P57) mice (n = 29). We recorded sparse, robust GCaMP signals from neurons in deep layers of the medial PFC, where virus had been injected to drive GCaMP expression (Fig. 1). GCaMP6s has a rise time of ∼100 ms, making it difficult to resolve signals faster than this timescale (Chen et al. 2013). Therefore, we imaged GCaMP signals at 10 Hz. Thus, while GCaMP sensors are in principle capable of resolving single spikes (Chen et al. 2013), the events detected here reflect increases in spike rate rather than single spikes. We used a low concentration of the cholinergic agonist carbachol (2 μM) in the bath to model basal cholinergic tone in vivo and promote spontaneous network activity (Fellous and Sejnowski 2000). Using a combination of independent component analysis and image segmentation (Mukamel et al. 2009), we identified the locations of neurons, measured their GCaMP signals, and detected events corresponding to increased activity in these neurons (Fig. 1). Each individual experiment recorded from 58–94 active neurons over ∼1 h. Some of these represent control experiments from a recent study of neuronal correlations in mice that model autism (Luongo et al. 2016); however, that study only looked at pairwise correlations and did not examine the sequential patterns of activity that are the subject of this study.
Prefrontal microcircuit activity contains more correlations than expected by chance.
First, we determined whether prefrontal microcircuits generate spontaneous activity that is randomly distributed vs. organized according to some structure, by computing correlations between activity in different neurons. We found that the distribution of correlations included more strong correlations than would be expected by chance (Fig. 2A). We compared these correlations to those obtained after shuffling our data. Notably, neurons exhibit long-lasting periods of increased activity interspersed with periods of quiescence. To preserve the autocorrelated temporal structure in shuffled data, we first simply shifted large chunks of each neuron's event train in time (materials and methods). Shuffling data in this way preserves the interevent interval distribution of each individual neuron (autocorrelation) but should destroy any structure between neurons. As shown in Fig. 2A, we found that real data contained an excess of strong correlations between different neurons compared with these shuffled data.
To further disentangle the effect of this temporal structure, we also compared the correlations observed to those found in a “scrambled” version of each data set. Unlike the “shuffled” data sets, these scrambled data sets account for any network-wide nonstationarity that may be present. These represent an important control, as correlation measures have been shown to be particularly sensitive to violations of stationarity (Brody 1999).
To understand how we generated these scrambled data sets, first note that each original data set is composed of a set of epochs of activity, where each epoch is defined by which neuron was active, when it became active, and how long it remained active. To “scramble” a data set, we simply shuffled the list of neuron identities associated with each epoch. As a result, each neuron has exactly the same number of epochs of activity in the original and scrambled data sets, i.e., the scrambled data set is effectively “rate-matched.” These scrambled data sets, which maintain both the temporal structure of total network activity and the distribution of neuronal rates, nevertheless contain significantly fewer correlations than the original data sets (Fig. 2B).
We also measured how correlations change when temporal jitter is introduced into the activity raster for each neuron. The results are shown in Fig. 2C. The mean correlation remains relatively high when each activity raster is jittered by 1, 2, or 4 frames (0.1–0.4 s) and then drops off sharply when the jitter is >10 frames (1 s). Thus, consistent with the kinetics of GCaMP and our observation that increases in GCaMP signals tend to last for 0.5–1 s, the correlations we calculated seem to measure increases and decreases in activity occurring on timescales of ∼0.5 s. Importantly, our sampling rate (10 Hz) should be appropriate for measuring correlations on this timescale (as opposed to say, correlations between single spikes).
Importantly, this timescale is much slower than our 10 Hz framerate, and thus calcium imaging represents a viable technique for measuring these interactions. Notably, in the original data sets, strong correlations were present even for pairs of neurons separated by large distances (Fig. 2D).
Prefrontal microcircuit activity has a small-world organization.
The previous analysis shows that the strengths of correlations exceed those expected by chance. Next, we asked whether the arrangement of correlations between neurons is random or follows a specific pattern. To address this question, we examined the structure of a network in which each neuron represents a node and the strong correlations between neurons represent edges. We assumed that an edge is present between two neurons if the correlation between them exceeds the noise level and absent otherwise. Networks constructed in this way, using the correlations between neurons, describe the functional organization of a neural circuit rather than the organization of physical, synaptic connections. Such functional networks have been widely studied and shown to be powerful tools for exploring the emergent behavior of neuronal circuits (Downes et al. 2012; Gururangan et al. 2014; Sadovsky and MacLean 2013; Schneidman et al. 2006; Shlens et al. 2006; Tkacik et al. 2014) and in many cases reflect important aspects of physical connectivity (Cossell et al. 2015; Ko et al. 2011; Stafford et al. 2014).
We next calculated clustering coefficients and path lengths for each functional network. These measures are used to distinguish between randomly wired networks, small-world networks, and clustered networks (Watts and Strogatz 1998). These three classes of networks can all have the same number of edges (strong correlations) but differ in how those edges are arranged. In randomly wired networks edges (correlations) between neurons are distributed randomly, whereas in clustered networks edges are distributed topographically, such that each neuron is connected (strongly correlated) with other “nearby” neurons, which also connect to each other, forming clusters. Small-world networks are formed when a small fraction of the edges in a clustered network are randomly rewired, and they are thought to exhibit desirable features of both clustered and random networks. For example, clustered, but not random, networks have distributed structure, which can be useful, e.g., for generating stereotyped patterns of activity as we show below. By contrast, random networks have a short path length, i.e., the average distance (along edges) from one node to another is relatively short, facilitating information flow through the network. Real-world networks, e.g., social networks, often possess a small-world organization that maintains some structure while also having short path lengths typical of random networks.
Clustering coefficients are defined as follows. Suppose node A is connected to nodes B and C—then the clustering coefficient is just the probability that B and C are themselves connected. We find that our data sets all contain 1) clustering coefficients that are markedly higher than those found in random networks (Fig. 2F; actual clustering coefficients: 0.30 ± 0.02 vs. 0.07 ± 0.01 for random networks, P < 0.001) and 2) low path lengths, similar to those in random networks (Fig. 2E; actual path lengths: 3.0 ± 0.2 vs. 3.1 ± 0.4 for random networks, P = 0.78). Random networks were constructed with the Erdos-Renyi model. This yields a random graph that has the same number of nodes (N) and edge probability (p) as the original, experimentally observed network. Specifically, each possible edge was included with a probability p. Finally, a similar small-world organization was observed when we elicited activity by exposing prefrontal slices to a high-K+, low-Ca2+ ACSF instead of 2 μM carbachol, highlighting that this organization was not simply a by-product of cholinergic activation (Fig. 2G). These results do not change if we vary the threshold for detecting events within Ca2+ signals (Fig. 3A) or if we determine the threshold for defining “strong correlations” on an individual cell basis, instead of using a single threshold for the entire network (Fig. 3B).
Our data sets also contain substantially more clustering than is expected simply based on a random, distance-dependent rule for coupling (Fig. 3C). These small-world characteristics were also present when we constructed networks with either a less stringent (P < 0.05) or more stringent (P < 0.001) threshold to identify strong correlations in our experimental data sets (Fig. 3, D and E). Finally, both path length and clustering were independent of the number of neurons (nodes) in the network (Fig. 3, F and G), which is, again, a canonical feature of small-world networks (Humphries and Gurney 2008). Thus, within prefrontal microcircuits, correlations are arranged nonrandomly, forming small-world networks.
Prefrontal microcircuits spontaneously generate sequential patterns of activity.
Given the bevy of modeling studies that have described ways in which the functional organization of recurrent networks can shape patterns of emergent activity, we decided to measure the capacity of prefrontal microcircuits to generate sequential patterns of activity (Abeles et al. 1993; Aviel et al. 2003; Diesmann et al. 1999). We used a template-matching algorithm similar to that used previously (Ikegaya et al. 2004) to count the number of occurrences of each potential sequence of activity. As diagrammed in Fig. 4A, this algorithm identifies every pattern of activation within a 1-s window following the onset of activity in a neuron, counts the number of times the same pattern recurs (allowing for a jitter of ±1 frames, i.e., 0.1 s), and counts all subpatterns within the original pattern. To determine whether these data sets contain more sequential patterns of activity than expected by chance, we initially made comparisons to the two types of control data sets described above. First, we calculated the number of sequential patterns in data sets that had been shuffled by shifting each neuron's activity train in time. Second, we calculated the number of sequential patterns in a scrambled version of each data set, in which the cell identities corresponding to each epoch of activity had been randomly reassigned, while maintaining the temporal pattern of total network activity and the distribution of activity levels across cells. Every one of our original data sets contained more sequential patterns of activity than the corresponding shuffled (Fig. 4, B and D) or scrambled (Fig. 4, C and D) data sets. Thus our original data sets generate more stereotyped sequential patterns of activity (i.e., sequences that were observed multiple times) than expected simply by chance.
Fig. 4.
Prefrontal microcircuits spontaneously generate many more stereotyped sequences of activity than expected by chance. A: schematic outlining the detection of temporal patterns. Briefly, a reference event was chosen, and from that event a template was constructed based on all the active events that occurred during the following 1 s. This template was counted as an existing pattern if it matched a previously generated template (allowing for up to 1 frame of jitter) or as a new pattern otherwise. Each subpattern contained within the original template was similarly compared. This process was repeated iteratively using all events in all cells as the reference event. B: for each data set (n = 29), the number of unique temporal patterns is plotted against the number of patterns in a shuffled version of the same data set. Each data set was shuffled by shifting the activity raster for each neuron over in time by a random amount (different for each neuron). All 29 points lie above the unity line, indicating that actual data sets always contain more unique temporal patterns than expected by chance. C: for each data set (n = 29), the number of unique temporal patterns is plotted against the number of patterns in a “scrambled” version of the same data set. Each scrambled data set was generated from the original data set by randomly reassigning the identity of the cell associated with each epoch of activity. Again, all 29 points lie above the unity line, indicating that actual data sets always contain more unique temporal patterns than expected by chance. D: bar plot summarizing the data plotted in B and C. ***P < 0.001 by Wilcoxon rank sum test.
Understanding how the small-world organization of prefrontal microcircuits contributes to sequence generation.
Of course, the critical question we set out to address is whether the functional organization of prefrontal microcircuits facilitates sequence generation. As described in the introduction, previous studies have, in most cases, failed to observe sequential patterns of microcircuit activity exceeding the number expected based on randomly arranged correlations between neurons. Naively, one would expect the small-world organization we found to facilitate sequence generation, because the presence of clustering in a network implies that specific groups of neurons are repeatedly coactive, which should lead to the repeated occurrence of sequences composed of these neurons. Thus we would first like to confirm this intuition, that small-world networks generate more sequences than networks in which correlations are randomly arranged.
We would also like to address two additional questions whose answers are less obvious. First, does increasing clustering beyond the levels observed in our original data sets further increase sequence generation, i.e., would maximally clustered networks, in which correlations are arranged topographically, generate even more sequential patterns of activity than small-world networks? The previously laid out intuition would suggest that more clustered networks should generate more sequences. On the other hand, stronger clustering implies that once one or two cells are known to be coactive, the remaining coactive cells should become more predictable. This might reduce the number of unique sequences that occur in more clustered networks.
A second question is whether the temporal pattern of total network activity (i.e., the number of neurons active as a function of time) together with the functional organization of the network [i.e., the (undirected) pairwise correlation matrix] are sufficient to explain the number of sequential patterns of activity we observed. That is, does a model that includes the temporal pattern of total network activity and the pairwise correlation matrix, but makes no additional assumptions, reproduce the levels of sequences observed in our original data sets? Or do additional aspects of network activity, e.g., the detailed statistics of activity within individual neurons or knowledge about whether one neuron tends to be active before vs. after another, contribute to sequence generation?
To address the preceding questions, we would ideally be able to compare patterns of network activity that are similar overall but reflect specific changes in the arrangement of correlations. Therefore, we devised a novel method to create surrogate data sets that all have exactly the same amount and temporal pattern of activity as each original data set, i.e., if N neurons were active at time t in the original data set, then N neurons would be active at time t in each surrogate data set as well. However, these surrogate data sets either preserve or alter the correlations between neurons and can thereby evaluate how specific changes in the organization of pairwise correlations would affect the occurrence of sequential patterns of activity. Note that maximum entropy models can be used in a similar way, to test whether knowing the mean activity level for each neuron and the pairwise functional interactions between neurons is sufficient to predict patterns of network activity. However, as noted above, maximum entropy models only predict instantaneous patterns of activity, whereas we are interested in sequential patterns of network activity. Therefore, our method (described below) generates surrogate data sets that capture the evolution of network activity over time. We refer to this method as shuffling activity to rearrange correlations, or SHARC.
To understand how we generated these surrogate data sets, again consider that each original data set can be defined as a set of epochs of activity, where each epoch is defined by which neuron was active, when it became active, and how long it remained active. Each of our original data sets is also associated with a set of correlations. As described below, to generate each surrogate data set we simply reassigned the neuron associated with each epoch of activity, in order to achieve a new, target set of correlations (this can be thought of as nonrandomly shuffling the cell identities associated with each epoch of activity). In this way, each surrogate data set preserved the overall temporal structure of network activity because the timing and duration of every epoch of activity were the same in each original data set and all the corresponding surrogate data sets. Thus the total number of neurons active at any given point in time would be the same for all of these data sets. However, the particular combination of cells active at a given point in time would differ across data sets, and, as a result, each surrogate data set also had a different set of correlations.
The target correlation matrix could be chosen to preserve the pattern of strong correlations present in our original data sets. Alternatively, the target correlation matrix could randomly rearrange these correlations, preserving the number of strong correlations but disrupting their organization. The iterative optimization procedure (SHARC) by which we reassigned the cell identities in order to achieve the target correlation matrix is diagrammed in Fig. 5A. First, we randomly selected an epoch of activity, i, whose cell identity had not yet been reassigned. Second, we found all the other epochs of activity j that 1) overlap in time with epoch i and 2) already have had a new cell identity assigned. Third, for each cell assigned to an epoch j that overlaps with epoch i, we computed the difference between the target correlation vector corresponding to that cell and the current value of that correlation vector based on the partially constructed surrogate data set. (This step can be thought of as “guessing” which cell should be assigned to a particular epoch of activity by first figuring out what other cells are active at the same time, then choosing cells that are strongly correlated with these known active cells). Next, we multiplied each difference vector by the amount of overlap between epoch j and epoch i and computed a weighted sum of all the difference vectors. (This step can be thought of as tallying up all of the “guesses” about which cells “should” be assigned to the epoch of activity being considered.) Finally, we selected a new cell identity for epoch i by simply choosing the cell corresponding to the maximum value of this weighted difference vector, excluding all cells that were already active during any part of epoch i (i.e., we chose the cell that represents the “consensus” based on tallying up all of the “guesses”). If the difference vector was empty, e.g., because there were no overlapping epochs of activity that have had new cell identities assigned, then we simply used the cell identity of this epoch of activity from the original data set.
Of course, although the target matrices were used to generate the surrogate data sets, the resulting surrogate data sets had slightly different correlation matrices (this reflects both the stochastic nature of the algorithm and the constraint that the temporal pattern of activity is fixed). Examples of correlation matrices for surrogate data sets are shown alongside their target correlation matrices in Fig. 5B. As Fig. 5B shows, the surrogate data sets had the desired properties, i.e., correlation matrices for the surrogate data sets were very similar to the target matrices. For example, the correlation matrices for surrogate data sets designed to preserve the arrangement of correlations present in the original data sets were very similar to the original correlation matrices, as evidenced by a normalized dot product of 0.96 ± 0.02 (mean ± SD). This original arrangement of correlations was lost in the surrogate data sets generated from correlation matrices that had been randomly rearranged, as the normalized dot product in this case was markedly lower: 0.21 ± 0.14 (P < 0.001). Notably, for the randomly rearranged case, the surrogate data sets did have correlation matrices very similar to their (randomly rearranged) target matrices, as evidenced by a normalized dot product of 0.95 ± 0.01. Thus SHARC creates surrogate data sets that conform to the training correlation matrices.
Small-world networks generate more sequences than random, clustered, or Poisson networks.
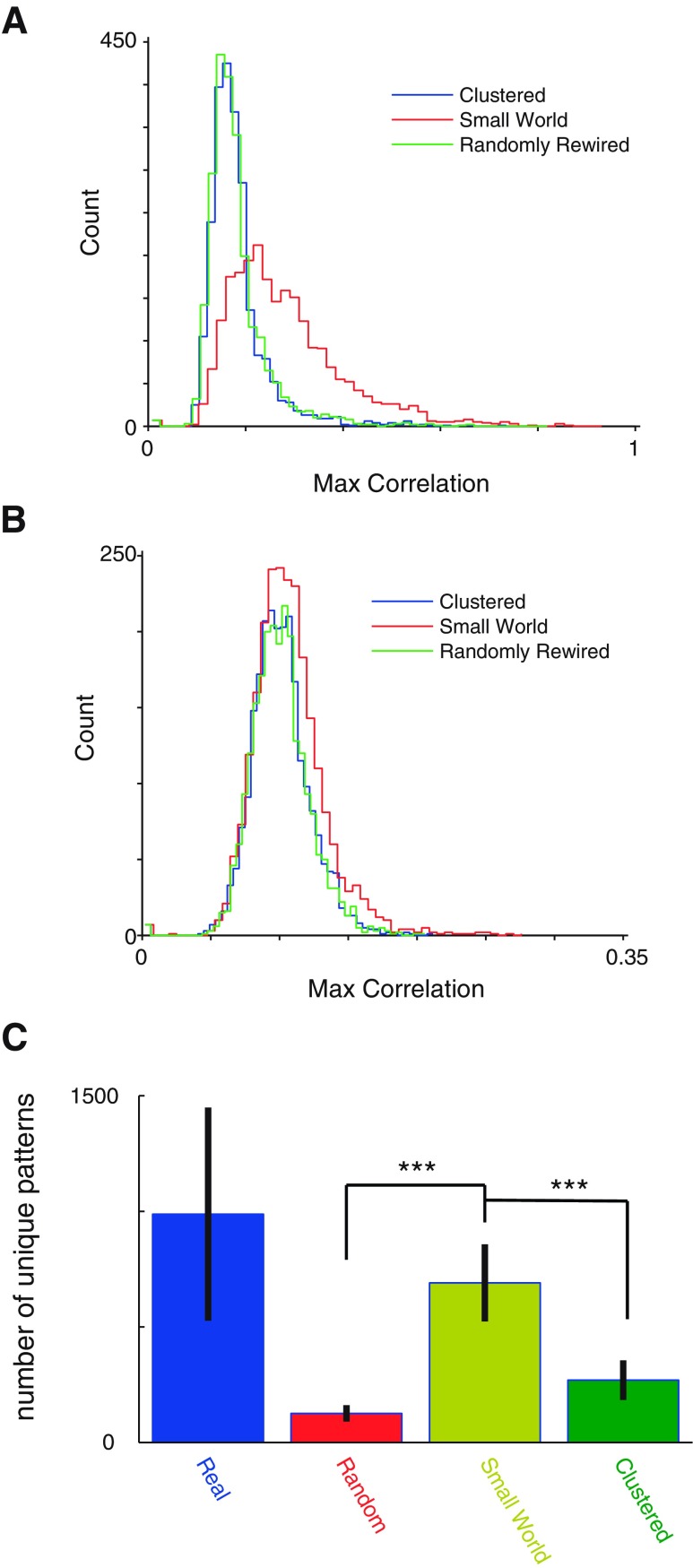
For each of our 29 data sets, we used SHARC to generate surrogate data sets that preserved the original small-world organization of pairwise correlations, randomly rearranged these correlations, or rearranged these correlations topographically, resulting in a maximally clustered network (cf. Fig. 5B). Examples of rasters and correlation matrices generated with SHARC are illustrated in Fig. 6 and Fig. 7, respectively. We confirmed that the network properties of each surrogate data set matched the respective network type—clustered, random, or small world (Fig. 8). Then we counted the number of sequential patterns of activity (as a function of sequence length) for each original data set, shuffled data set, scrambled data set, surrogate data set based on randomly rewiring the correlation matrix, surrogate data set that preserved the original small-world correlation matrix, and surrogate data set based on a maximally clustered correlation matrix (Fig. 9A). Figure 9B summarizes the total number of sequential patterns of activity for each case. As shown in Fig. 9, surrogate data sets that preserve the original small-world pattern of correlations contain the same number of sequences as our original data sets and generate significantly more sequential patterns of activity than either random or maximally clustered networks. In addition, we generated additional surrogates of either rate-matched homogeneous or inhomogeneous Poisson neurons and found that our data sets contained many more sequences than expected by networks of independent Poisson neurons (Fig. 9). This helps resolve conflicts from earlier studies that found that numbers of sequences observed in shorter data sets (∼2 min) could be accounted for using independently rate-matched Poisson neurons (Roxin et al. 2008). Finally, we wondered whether we could utilize SHARC to test whether the small-world pattern of correlations alone is sufficient to reproduce sequential patterns of activity. Specifically, we started with simulated activity from independently firing Poisson neurons, with either homogeneous or inhomogeneous rates matched to the experimentally observed firing rates. Then we used SHARC to rearrange these firing patterns so that their correlations matched the small-world pattern of the experimentally observed correlations. However, surrogate data sets constructed in this manner failed to recapitulate the high levels of sequences observed in either the original data sets or the small-world surrogate data sets (Fig. 9B). This suggests that additional features of the original data sets, specifically the number of neurons firing at each point in time and/or the duration of each period of activity, also contribute to sequence generation.
Fig. 6.
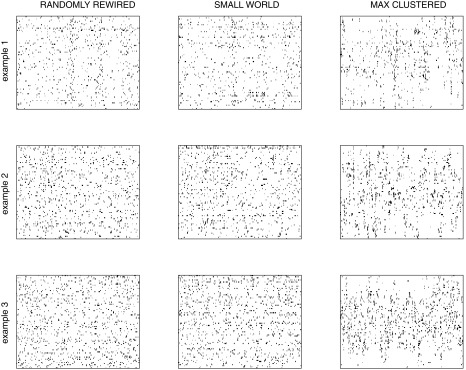
Sample activity rasters from surrogate data sets: examples of the 3 different types of surrogate data sets, each generated for 3 different experiments. For each experiment, we generated surrogate data sets based on the original pattern of activity and a randomly rewired correlation matrix (left), the original small-world correlation matrix (center), or a maximally clustered correlation matrix (right). Thus all 3 of the data sets in each row, which correspond to different network topologies, nevertheless have the same temporal level of total network activity, i.e., at each point in time, the same number of neurons will be active in all three networks. Each raster represents 600 s of data.
Fig. 7.
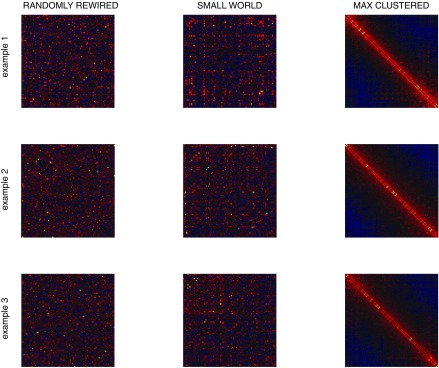
Sample correlation matrices from surrogate data sets: correlation matrices corresponding to each of the activity rasters presented in Fig. 6.
Fig. 8.
Path lengths and clustering coefficients for surrogate data sets. A: path lengths (left) and clustering coefficients (right) for surrogate data sets fit to randomly rewired correlation matrices compared with Erdos-Renyi random networks. Randomly rewired networks match the path lengths and clustering coefficients of Erdos-Renyi networks. B: path lengths (left) and clustering coefficients (right) for surrogate data sets fit to small-world correlation matrices compared with Erdos-Renyi random networks. Small-world networks have similar path lengths and greater clustering compared with Erdos-Renyi random networks. C: path lengths (left) and clustering coefficients (right) for surrogate data sets fit to maximally clustered correlation matrices compared with Erdos-Renyi random networks. Clustered networks have greater path lengths and clustering than Erdos-Renyi random networks. ***P < 0.001 by Wilcoxon rank sum test.
These findings directly address the two questions raised earlier. First, an analytical method that preserves the overall pattern of network activity (i.e., the timing and duration of each epoch of activity) and the pairwise correlation matrix but makes no additional assumptions is sufficient to reproduce the levels of sequential patterns of activity observed in our original data sets. Second, the small-world organization of pairwise correlations we found in prefrontal microcircuits optimizes the production of these sequential patterns.
An important question is whether SHARC simply “renames” the cells. That is, perhaps small-world surrogate data sets match the performance of the original data sets because each neuron in a surrogate data set effectively mimics the activity pattern of a neuron in the original data set? To address this question, we calculated the correlation between the event train of each surrogate data set neuron and the most similar event train in the original data set. This correlation was ∼0.2 for random/clustered surrogate data sets and ∼0.3 for small-world surrogate data sets (Fig. 10A). Thus cells in the small-world surrogate data sets are not simply “renamed” versions of cells in the original data sets. Furthermore, this slight increase in correlation for small-world surrogate data sets relative to random/clustered surrogate data sets could be eliminated simply by generating surrogate data sets using an activity pattern from one experiment together with a correlation matrix from a different experiment. In this case, the correlation between each surrogate neuron and the most similar neuron in each original data set was ∼0.1 for random, clustered, and small-world surrogate data sets (Fig. 10B). Furthermore, even when we generated surrogate data sets in this way, using the activity pattern from one experiment together with the correlation matrix from a different experiment, small-world surrogate data sets continued to reproduce the number of sequences observed in actual data sets and to outperform random and clustered data sets (Fig. 10C).
Fig. 10.
A bias exists toward less relabeling of neurons in small-world surrogates that can be accounted for by fitting to correlation matrices from other data sets, and in this case small-world surrogates still outperform random and clustered networks. A: for each neuron in a surrogate data set, we found the maximum correlation between its event train and the event train of a neuron in the corresponding original (actual) data set, i.e., the most similar actual neuron. The distribution of these correlations is plotted for each type of surrogate data set. In all cases, these correlations tend to be substantially <1, indicating that neurons in the surrogate data sets are not just “renamed” versions of the original neurons. Note that neurons in small-world surrogate data sets tend to have slightly higher correlations with actual neurons than neurons in either maximally clustered or randomly rewired data sets. B: same as A, except that surrogate data sets have now been generated with the activity raster from 1 experiment and the correlation matrix from a different experiment. We computed correlations between each neuron in a surrogate data set and neurons within the activity raster that was used to generate that surrogate data set. This procedure eliminates the slightly higher correlations between simulated neurons and real neurons observed for the small-world surrogate data sets shown in A. C: for each activity raster, we generated 3 surrogate data sets of each type, each based on the correlation matrix from a different experiment. We used the original correlation matrix (from a different experiment) to generate small-world surrogate data sets or correlation matrices that had been reorganized topographically or randomly shuffled to generated maximally clustered or randomly rewired data sets, respectively. As in our original analysis, only small-world surrogate data sets contained a similar number of stereotyped sequential patterns of activity, compared with the original data sets. Maximally clustered and randomly rewired surrogate data sets both contained significantly fewer stereotyped sequential patterns of activity than the small-world surrogate data sets. ***P < 0.001.
DISCUSSION
This study evaluated how the functional organization of prefrontal microcircuits, i.e., the small-world correlation matrix, contributes to an important function of these circuits: their ability to generate multineuron sequences of activity. We found that pairwise correlations between prefrontal neurons form a small-world network that optimizes the production of a diverse set of such sequences. Conversely, this small-world organization is sufficient to explain the occurrence of sequential patterns of activity in these circuits. We also outline a general method (shuffling activity to rearrange correlations, or SHARC) for generating surrogate data sets that makes minimal assumptions about the structure of the underlying data, maintains the temporal structure of total network activity, and either preserves the pattern of pairwise correlations or alters it in specific ways. SHARC will complement other approaches, e.g., theory, simulations, and maximum entropy models, for linking the functional organization of a circuit to the emergence of complex multineuronal patterns of activity.
Small-world neural networks.
Small-world networks were first formalized to describe diverse real-world networks that combine desirable features of both highly structured and randomly connected networks—clustering and short path lengths, respectively (Watts and Strogatz 1998). These features have been hypothesized to reflect an ability of small-world networks to process information on both local and global scales. Small-world topologies have been described for inter- and intra-areal functional connectivity in cat, monkey, and human cortex (Bassett and Bullmore 2006; Sporns and Zwi 2004). Small-world topologies have also been identified for microcircuits in neuronal culture (Downes et al. 2012), rat (Song et al. 2005) and cat (Yu et al. 2008) visual cortex, deep layers of monkey cortex (Gerhard et al. 2011), and various cortical microcircuits of mice (Sadovsky and MacLean 2013). In addition, models of small-world topology have been shown to enable network-wide reverberations of activity (Volman and Gerkin 2011). Introducing higher-order interactions such as clustering into a model of otherwise randomly connected cortical neurons nicely recapitulates the doubly stochastic nature of cortical spike trains, further supporting the presence of small-world topologies within cortical microcircuits (Litwin-Kumar and Doiron 2012).
In this context, our results support the idea that small-world networks represent a common organizing principle for neural systems (rather than a PFC-specific feature). More importantly, they demonstrate a specific and concrete way in which such an organization actually impacts neural circuit function. Small-world networks have been hypothesized to effectively balance local and global modes of computation, but until now applications of these ideas to neural networks have been largely abstract. Here we show exactly how this balance enables prefrontal microcircuits to generate stereotyped temporal patterns of activity, while also maximizing the diversity of such patterns. This solves a challenging trade-off between the tendencies of random networks to produce multitudes of patterns that fail to repeat and highly clustered networks to produce a small number of extremely repetitive patterns. Neural circuits that need to generate these patterns in order to reliably and flexibly support a diverse repertoire of behaviors must balance these two regimes, and here we show that the small-world functional organization of actual prefrontal microcircuits achieves this balance. In this way, prefrontal microcircuits can contribute to the stereotyped patterns of activity that have been associated with flexible behavior in vivo (Baeg et al. 2003; Fujisawa et al. 2008; Harvey et al. 2012; Seidemann et al. 1996).
Remarkably, surrogate data sets based on a small-world network of pairwise correlations between neurons and the temporal pattern of total network activity we observed are sufficient to largely reproduce the levels of sequential patterns observed in our original recordings. Naively, one might have expected undirected pairwise correlations to be extremely limited in their ability to capture temporally structured activity, and that adequately describing sequential patterns of activity would require knowledge about the underlying, asymmetric pattern of neural connections, or at least which cell in a correlated pair tends to be active first. However, we find that at the level of cortical microcircuits the undirected pairwise correlation matrix, with its small-world topology, captures the key features of network organization involved in sequence generation. This is reminiscent of other studies that have used coupling between neurons to explain complex features of activity in actual neural circuits (Stevenson et al. 2012).
Interestingly, a distance-dependent, random coupling rule was not sufficient to reproduce the level of “small-worldness” we found in prefrontal microcircuits. Consistent with previous studies of cultured neurons (Downes et al. 2012), this implies that additional mechanisms, besides a local random connection rule, give rise to the small-world organization present in actual neural networks.
Finally, to be clear: we have shown that surrogate data sets based on the experimentally observed small-world correlation matrices outperform surrogate data sets based on random or clustered correlation matrices, as well as those based on Poisson neurons. Thus we have shown that the functional organization of the local circuit contributes to the production of stereotyped, sequential patterns of activity. That being said, there are other features of the experimentally observed correlation matrices (e.g., the degree distribution) that contribute to their overall “small-worldness,” and we have not tried to dissociate the contributions of each of these. Future studies exploring a wider range of network topologies could potentially disentangle the relative contributions of various graph features for producing specific temporal patterns of neural activity.
Relationship to in vivo studies.
Of course, it will be important to compare our findings to features of activity occurring in vivo. That being said, in vivo recordings would address questions fundamentally different from those addressed here. Activity in vivo is strongly influenced by synaptic inputs originating outside the PFC. Thus, if we observed a small-world organization and/or sequential patterns of activity in vivo, it would be unclear whether these phenomena reflect local features of prefrontal circuits or are imposed by external inputs. Two other recent studies similarly used brain slices to establish the presence of small-world topologies and temporally precise firing in cortical microcircuits (Sadovsky and MacLean 2013, 2014).
Physiological relevance of activity sequences during spontaneous UP/DOWN states in brain slices.
The spontaneous activity we observed in prefrontal brain slices consists of population-wide increases in activity interspersed with periods of quiescence (Fig. 1B). This pattern of alternating “UP” and “DOWN” states conforms to a slow oscillation that has been well described and dominates spontaneous cortical activity both in vitro and in vivo (Luczak et al. 2007, 2009; MacLean et al. 2005; Sanchez-Vives and McCormick 2000; Steriade et al. 1993). Notably, spatiotemporal patterns observed within spontaneous UP and DOWN states recapitulate the patterns observed in response to sensory stimulation both in vitro (MacLean et al. 2005) and in vivo (Carrillo-Reid et al. 2015; Luczak et al. 2009). Importantly, the sensory stimulation-evoked patterns comprise a subset of the spontaneously occurring ones (Luczak et al. 2009), suggesting that spontaneous activity defines a space of possible patterns that is sampled during behavior. Of course, not all spontaneously occurring patterns will be behaviorally useful—many might correspond to degraded versions of specific behavioral programs or nonsensical combinations of multiple programs, etc. Nevertheless, in this framework, our findings suggest that small-world architectures enlarge the space of activity patterns that could potentially drive behavior, and could thus be thought of as creating network dynamics that facilitate potential behavioral flexibility. It will be critical for future studies to elucidate how factors such as external input select among this set of spontaneously occurring patterns to recruit specific, behaviorally relevant ones.
Using SHARC to reveal how multineuron patterns of activity depend on the functional organization of neural networks.
Neural representations involving populations of cortical neurons are more informative than individual spike trains (Averbeck et al. 2006), and several recent studies have shown how neuronal ensembles can carry information in vivo (Carrillo-Reid et al. 2015; Fujisawa et al. 2008; Harvey et al. 2012). As more studies identify patterns of activity within neuronal ensembles either in reduced preparations or in vivo, a critical question will be whether those patterns are simply a by-product of randomly arranged correlations between neurons or depend on more complex patterns of functional interactions. A closely related question is whether knowing the functional organization of a network helps to understand how that circuit actually works (Marder 2015). The computational method presented here, SHARC, represents a powerful new tool for answering both of these questions. SHARC reassigns the cell identities associated with activity in the original data set, rather than simulating new patterns of activity. This is critical, because this method preserves the exact pattern of total network activity present in the original data set, making it possible to directly probe and dissociate the relative contribution of activity levels, temporal patterns of activity, pairwise correlations, and higher-order structure such as clustering to key features of emergent network-level behavior. Here we used SHARC to investigate the relationship between organization and function in prefrontal microcircuits. We found that correlations between prefrontal neurons form a small-world network that optimizes the ability of prefrontal microcircuits to generate sequential patterns of activity that have been linked to important behavioral functions. This reveals an important function for small-world neural networks and illustrates a novel approach for linking the functional organization of a neural network to the patterns of activity it generates.
GRANTS
This work was supported by the Staglin Family and International Mental Health Research Organization (IMHRO), the Simons Foundation for Autism Research, the Alfred P. Sloan Foundation, National Institute of Mental Health (R00 MH-085946), and the National Institutes of Health Office of the Director (DP2MH10001101). F. J. Luongo is additionally supported by an IMSD fellowship from National Institute of General Medical Sciences (R25 GM-56847).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
F.J.L. and V.S.S. conception and design of research; F.J.L. and M.E.H. performed experiments; F.J.L., C.A.Z., and V.S.S. analyzed data; F.J.L. and V.S.S. interpreted results of experiments; F.J.L. prepared figures; F.J.L. and V.S.S. drafted manuscript; F.J.L. and V.S.S. edited and revised manuscript; F.J.L. and V.S.S. approved final version of manuscript.
ACKNOWLEDGMENTS
Members of the Sohal laboratory provided helpful discussions. We also acknowledge the use of GCaMP constructs developed by Vivek Jayaraman, Rex Kerr, Douglas Kim, Loren Looger, and Karel Svoboda as part of the GENIE Project at the Janelia Farm Research Campus, Howard Hughes Medical Institute.
REFERENCES
- Abeles M, Bergman H, Margalit E, Vaadia E. Spatiotemporal firing patterns in the frontal cortex of behaving monkeys. J Neurophysiol 70: 1629–1638, 1993. [DOI] [PubMed] [Google Scholar]
- Averbeck BB, Latham PE, Pouget A. Neural correlations, population coding and computation. Nat Rev Neurosci 7: 358–366, 2006. [DOI] [PubMed] [Google Scholar]
- Aviel Y, Mehring C, Abeles M, Horn D. On embedding synfire chains in a balanced network. Neural Comput 15: 1321–1340, 2003. [DOI] [PubMed] [Google Scholar]
- Baeg EH, Kim YB, Huh K, Mook-Jung I, Kim HT, Jung MW. Dynamics of population code for working memory in the prefrontal cortex. Neuron 40: 177–188, 2003. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist 12: 512–523, 2006. [DOI] [PubMed] [Google Scholar]
- Brody CD. Correlations without synchrony. Neural Comput 11: 1537–1551, 1999. [DOI] [PubMed] [Google Scholar]
- Carrillo-Reid L, Miller JE, Hamm JP, Jackson J, Yuste R. Endogenous sequential cortical activity evoked by visual stimuli. J Neurosci 35: 8813–8828, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499: 295–300, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossell L, Iacaruso MF, Muir DR, Houlton R, Sader EN, Ko H, Hofer SB, Mrsic-Flogel TD. Functional organization of excitatory synaptic strength in primary visual cortex. Nature 518: 399–403, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diesmann M, Gewaltig MO, Aertsen A. Stable propagation of synchronous spiking in cortical neural networks. Nature 402: 529–533, 1999. [DOI] [PubMed] [Google Scholar]
- Downes JH, Hammond MW, Xydas D, Spencer MC, Becerra VM, Warwick K, Whalley BJ, Nasuto SJ. Emergence of a small-world functional network in cultured neurons. PLoS Comput Biol 8: e1002522, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellous JM, Sejnowski TJ. Cholinergic induction of oscillations in the hippocampal slice in the slow (0.5–2 Hz), theta (5–12 Hz), and gamma (35–70 Hz) bands. Hippocampus 10: 187–197, 2000. [DOI] [PubMed] [Google Scholar]
- Fujisawa S, Amarasingham A, Harrison MT, Buzsaki G. Behavior-dependent short-term assembly dynamics in the medial prefrontal cortex. Nat Neurosci 11: 823–833, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli S, Huh D, Sompolinsky H. Memory traces in dynamical systems. Proc Natl Acad Sci USA 105: 18970–18975, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee S, Ellwood I, Patel T, Luongo F, Deisseroth K, Sohal VS. Synaptic activity unmasks dopamine D2 receptor modulation of a specific class of layer V pyramidal neurons in prefrontal cortex. J Neurosci 32: 4959–4971, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard F, Pipa G, Lima B, Neuenschwander S, Gerstner W. Extraction of network topology from multi-electrode recordings: is there a small-world effect? Front Comput Neurosci 5: 4, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MS. Memory without feedback in a neural network. Neuron 61: 621–634, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururangan SS, Sadovsky AJ, MacLean JN. Analysis of graph invariants in functional neocortical circuitry reveals generalized features common to three areas of sensory cortex. PLoS Comput Biol 10: e1003710, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Coen P, Tank DW. Choice-specific sequences in parietal cortex during a virtual-navigation decision task. Nature 484: 62–68, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennequin G, Vogels TP, Gerstner W. Optimal control of transient dynamics in balanced networks supports generation of complex movements. Neuron 82: 1394–1406, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MD, Gurney K. Network “small-world-ness”: a quantitative method for determining canonical network equivalence. PLoS One 3: e0002051, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegaya Y, Aaron G, Cossart R, Aronov D, Lampl I, Ferster D, Yuste R. Synfire chains and cortical songs: temporal modules of cortical activity. Science 304: 559–564, 2004. [DOI] [PubMed] [Google Scholar]
- Ko H, Hofer SB, Pichler B, Buchanan KA, Sjostrom PJ, Mrsic-Flogel TD. Functional specificity of local synaptic connections in neocortical networks. Nature 473: 87–91, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laje R, Buonomano DV. Robust timing and motor patterns by taming chaos in recurrent neural networks. Nat Neurosci 16: 925–933, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin-Kumar A, Doiron B. Slow dynamics and high variability in balanced cortical networks with clustered connections. Nat Neurosci 15: 1498–1505, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak A, Bartho P, Harris KD. Spontaneous events outline the realm of possible sensory responses in neocortical populations. Neuron 62: 413–425, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak A, Bartho P, Marguet SL, Buzsaki G, Harris KD. Sequential structure of neocortical spontaneous activity in vivo. Proc Natl Acad Sci USA 104: 347–352, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo FJ, Horn ME, Sohal VS. Putative microcircuit-level substrates for attention are disrupted in mouse models of autism. Biol Psychiatry 79: 667–675, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JN, Watson BO, Aaron GB, Yuste R. Internal dynamics determine the cortical response to thalamic stimulation. Neuron 48: 811–823, 2005. [DOI] [PubMed] [Google Scholar]
- Marder E. Understanding brains: details, intuition, and big data. PLoS Biol 13: e1002147, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63: 747–760, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor DH, Peron SP, Huber D, Svoboda K. Neural activity in barrel cortex underlying vibrissa-based object localization in mice. Neuron 67: 1048–1061, 2010. [DOI] [PubMed] [Google Scholar]
- Ohiorhenuan IE, Mechler F, Purpura KP, Schmid AM, Hu Q, Victor JD. Sparse coding and high-order correlations in fine-scale cortical networks. Nature 466: 617–621, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxin A, Hakim V, Brunel N. The statistics of repeating patterns of cortical activity can be reproduced by a model network of stochastic binary neurons. J Neurosci 28: 10734–10745, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovsky AJ, MacLean JN. Scaling of topologically similar functional modules defines mouse primary auditory and somatosensory microcircuitry. J Neurosci 33: 14048–14060, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovsky AJ, MacLean JN. Mouse visual neocortex supports multiple stereotyped patterns of microcircuit activity. J Neurosci 34: 7769–7777, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vives MV, McCormick DA. Cellular and network mechanisms of rhythmic recurrent activity in neocortex. Nat Neurosci 3: 1027–1034, 2000. [DOI] [PubMed] [Google Scholar]
- Schneidman E, Berry MJ 2nd, Segev R, Bialek W. Weak pairwise correlations imply strongly correlated network states in a neural population. Nature 440: 1007–1012, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidemann E, Meilijson I, Abeles M, Bergman H, Vaadia E. Simultaneously recorded single units in the frontal cortex go through sequences of discrete and stable states in monkeys performing a delayed localization task. J Neurosci 16: 752–768, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlens J, Field GD, Gauthier JL, Grivich MI, Petrusca D, Sher A, Litke AM, Chichilnisky EJ. The structure of multi-neuron firing patterns in primate retina. J Neurosci 26: 8254–8266, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Sjostrom PJ, Reigl M, Nelson S, Chklovskii DB. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol 3: e68, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Zwi JD. The small world of the cerebral cortex. Neuroinformatics 2: 145–162, 2004. [DOI] [PubMed] [Google Scholar]
- Stafford JM, Jarrett BR, Miranda-Dominguez O, Mills BD, Cain N, Mihalas S, Lahvis GP, Lattal KM, Mitchell SH, David SV, Fryer JD, Nigg JT, Fair DA. Large-scale topology and the default mode network in the mouse connectome. Proc Natl Acad Sci USA 111: 18745–18750, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steriade M, Nunez A, Amzica F. A novel slow (<1 Hz) oscillation of neocortical neurons in vivo: depolarizing and hyperpolarizing components. J Neurosci 13: 3252–3265, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson IH, London BM, Oby ER, Sachs NA, Reimer J, Englitz B, David SV, Shamma SA, Blanche TJ, Mizuseki K, Zandvakili A, Hatsopoulos NG, Miller LE, Kording KP. Functional connectivity and tuning curves in populations of simultaneously recorded neurons. PLoS Comput Biol 8: e1002775, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussillo D, Abbott LF. Generating coherent patterns of activity from chaotic neural networks. Neuron 63: 544–557, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A, Jackson D, Hobbs J, Chen W, Smith JL, Patel H, Prieto A, Petrusca D, Grivich MI, Sher A, Hottowy P, Dabrowski W, Litke AM, Beggs JM. A maximum entropy model applied to spatial and temporal correlations from cortical networks in vitro. J Neurosci 28: 505–518, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacik G, Marre O, Amodei D, Schneidman E, Bialek W, Berry MJ 2nd. Searching for collective behavior in a large network of sensory neurons. PLoS Comput Biol 10: e1003408, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volman V, Gerkin RC. Synaptic scaling stabilizes persistent activity driven by asynchronous neurotransmitter release. Neural Comput 23: 927–957, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of “small-world” networks. Nature 393: 440–442, 1998. [DOI] [PubMed] [Google Scholar]
- Yu S, Huang D, Singer W, Nikolic D. A small world of neuronal synchrony. Cereb Cortex 18: 2891–2901, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Ting JT, Atallah HE, Qiu L, Tan J, Gloss B, Augustine GJ, Deisseroth K, Luo M, Graybiel AM, Feng G. Cell type-specific channelrhodopsin-2 transgenic mice for optogenetic dissection of neural circuitry function. Nat Methods 8: 745–752, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]