Abstract
The effects of specific functional groups of pollinators in the diversification of angiosperms are still to be elucidated. We investigated whether the pollination shifts or the specific association with hummingbirds affected the diversification of a highly diverse angiosperm lineage in the Neotropics. We reconstructed a phylogeny of 583 species from the Gesneriaceae family and detected diversification shifts through time, inferred the timing and amount of transitions among pollinator functional groups, and tested the association between hummingbird pollination and speciation and extinction rates. We identified a high frequency of pollinator transitions, including reversals to insect pollination. Diversification rates of the group increased through time since 25 Ma, coinciding with the evolution of hummingbird-adapted flowers and the arrival of hummingbirds in South America. We showed that plants pollinated by hummingbirds have a twofold higher speciation rate compared with plants pollinated by insects, and that transitions among functional groups of pollinators had little impact on the diversification process. We demonstrated that floral specialization on hummingbirds for pollination has triggered rapid diversification in the Gesneriaceae family since the Early Miocene, and that it represents one of the oldest identified plant–hummingbird associations. Biotic drivers of plant diversification in the Neotropics could be more related to this specific type of pollinator (hummingbirds) than to shifts among different functional groups of pollinators.
Keywords: comparative methods, floral traits, ornithophily, pollinator shifts, stochastic mapping
1. Introduction
The current species richness of a group of organisms results from the diversification process that has occurred throughout its evolution. In plants, a variety of intrinsic and extrinsic factors affect the diversification process [1]. Among those factors, changes in climatic conditions [2], the colonization of new geographical areas [3] or the evolution of particular traits might create new possibilities for species diversification [4,5]. In angiosperms, traits such as biotic pollination, floral symmetry and nectar spurs, which are all related to specialized pollination and the ability to generate reproductive isolation, have been proposed as key innovations due to their positive effects on diversification [6,7]. The role of specialized biotic pollination in the diversification of angiosperms is a long-standing question [8], but the mechanisms that led to the apparent association between pollination and species richness are still rather unexplored [9].
One hypothesis is that diversification in angiosperms has been enhanced by the effect of pollinator specialization on reproductive isolation. Spatial and temporal differences in the availability of the most effective pollinator across the species range could produce pollinator shifts, floral divergence, reproductive isolation and, ultimately, speciation in plants [10]. Evidence for pollinator-shift effects in plant speciation have been found for Costus [11], Gladiolus [12] and Lapeirousia [13], and a review of available species-level phylogenies estimated that around 25% of the divergence events could be associated with pollinator shifts in angiosperms [14]. Although these results suggest that frequent pollination shifts have occurred during the speciation events in angiosperms, a large proportion of these events could still occur within specific pollination systems. Indeed, an alternative hypothesis proposes that diversification rates in angiosperms increase with specialization on certain guilds of pollinators, rather than with pollinator shifts per se [12]. For example, vertebrate pollination, and in particular pollination mediated by birds, is associated with plant species richness in various clades [15,16]. The evaluation of the role of ornithophily in the diversification of the whole Gesneriaceae family has recently indicated distinct patterns between Old and New World lineages [17]. The evolution of bird pollination (specifically hummingbird pollination) was associated with an increase in diversification rates in the New World, while no influence was detected for the lineages in the Old World. However, a necessary step to further understand the effects of hummingbird pollination in the New World plant diversity is to evaluate whether the frequent shifts among pollinator groups or the specialization on hummingbird pollination is influencing plant diversification [18]. Surprisingly, the relative contribution of these two processes remains unexplored.
The aim of this study is to evaluate the tempo of evolution of functional groups of pollinators, in particular hummingbirds, and their impact on the diversification rates of the Neotropical lineage of the family Gesneriaceae, hereafter referred to as Gesnerioideae, which is the lineage of this family with hummingbird interactions. Specifically, by expanding the most recent phylogenetic sampling by 129 species, we conducted an accurate evaluation of the pollination syndromes, their evolution and their impact in diversification rate shifts for the subfamily. The Gesnerioideae is a clade of herbaceous plants, shrubs or more rarely small trees. It contains 75 genera and over 1200 species found exclusively in the Neotropics, with the exception of few Southwest Pacific taxa in the tribe Coronanthereae [19,20]. Molecular dating and biogeographic reconstructions have estimated an origin of the Gesnerioideae in South America during the Early Oligocene, with a rapid range expansion into most Neotropical regions [21]. The species in this subfamily exhibit a large diversity of floral morphology associated with repeated adaptations to different pollinators, such as hummingbirds, bees and bats [22–27] (figure 1). Therefore, this clade is particularly interesting to test the mode and tempo with which plant–pollinator interactions have evolved and how they have influenced species diversification.
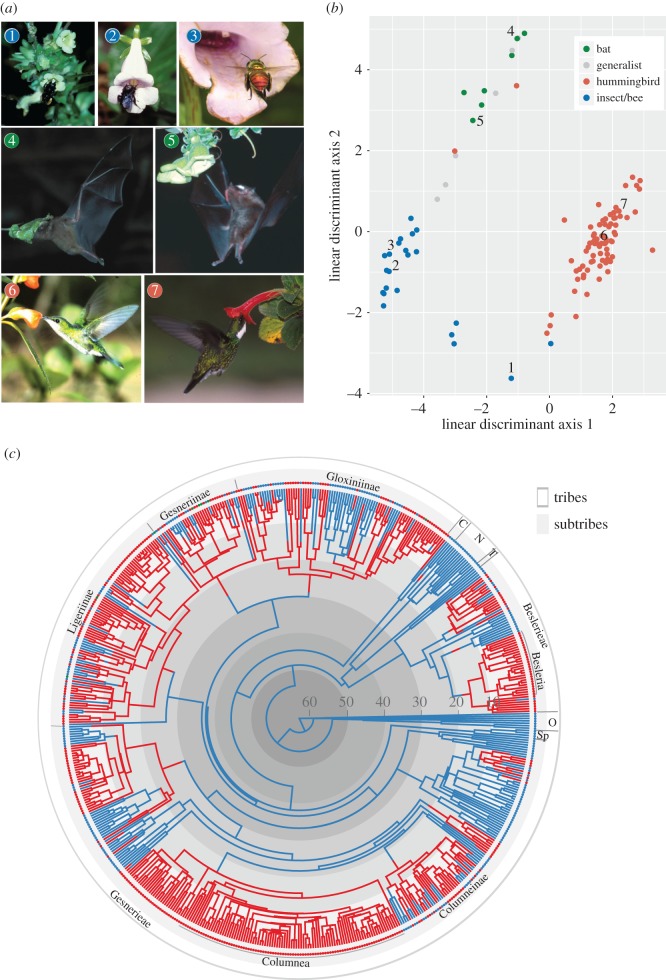
Figure 1.
(a) Examples of plant–pollinator interactions in the Gesnerioideae (images 1–7); photo information in the electronic supplementary material, table S2. Photo credits: images 1, 2, 4, 5, 7 by I. SanMartin-Gajardo; image 3 by a. Weber; image 6 by L. Freitas. (b) Discriminant analysis conducted for 118 plant species and nine floral traits. Numbers refer to species shown in (a). Electronic supplementary material, table S2, provides the morphological data and the source of pollinator observations for each species. (c) Bayesian common ancestor (CA) phylogenetic reconstruction showing one stochastic mapping of pollination syndromes. White and gray boxes correspond to taxonomic tribes and subtribes. Names following classification by [20]: N, Napeantheae; C, Coronanthereae; T, Titanotricheae; Sp, Sphaerorrhizinae; O, outgroups. Colours on branches correspond to pollination syndromes: blue, insect; red, hummingbird. Trait states: blue, insects; red, hummingbirds; green, bats. Grey concentric circles have 10 Myr span.
Here, we reconstructed one of the largest species-level phylogenies for a group of Neotropical plants based on four DNA loci and a wide sampling of Gesnerioideae species to test for temporal variations and trait-dependent rates of diversification at a continental scale. We specifically investigated (i) the number and timing of transitions among pollination syndromes in the subfamily, and (ii) the temporal match between the evolution of hummingbird pollination and hummingbird diversification in South America [28]. Additionally, we tested (iii) whether the evolution of hummingbird pollination has contributed to the elevated Gesnerioideae diversity in Neotropics, and (iv) whether diversification rates were associated with recurrent shifts of pollinators or the observed species richness was driven specifically by hummingbird-mediated pollination. Addressing these questions in such a large and diverse group of plants will contribute to a better understanding of how ecological factors shape current patterns of species richness in the Neotropics [15].
2. Material and methods
(a). Taxonomic sampling and DNA sequencing
Our taxonomic sampling consisted of 583 species representing all the 75 recognized genera in Gesnerioideae and about 50% of the species in the subfamily [20]. The sampling of each tribe and outgroups is detailed in the electronic supplementary material, methods S1. A total of 475 sequences were amplified from field samples for this study (see ‘Data accessibility’ below) and merged to available Genbank sequences. Sequences were aligned using MAFFT v. 7 [29]) and all sites were scored for accuracy of the alignment using Guidance [30]. We identified the best substitution model for each DNA region using the Akaike information criterion (AIC) as implemented in the phymltest function in R (ape package [31]; see details in the electronic supplementary material, methods S1).
(b). Phylogenetic reconstruction
Relationships among species were reconstructed by Bayesian inference using MrBayes v. 3.2 [32]. Data partitions and details of Bayesian inference are described in the electronic supplementary material, methods S1. Divergence times were estimated using a relaxed clock model with uncorrelated lognormal prior distribution for the rates of substitution and a birth–death prior for the age of each node as implemented in BEAST v. 1.7.0 [33]. Secondary calibration was performed by imposing priors for the divergence times for the clade containing all Gesneriaceae (including Sanango racemosum and members of the Didymocarpoideae family). MCMC settings and tree sampling are described in the electronic supplementary material, methods S1.
(c). Characterization of pollination syndromes
The predictability of pollination syndromes is largely debated [34,35]. However, a recent meta-analysis supported the concept of pollination syndromes, especially for tropical plants [36], and encouraged the use of floral characters as a proxy for pollination interactions in macro-evolutionary studies [16,17]. In Gesnerioideae, several studies combining field observations and multivariate analyses of morphometric data have demonstrated that suites of floral traits could predict specialized pollination by hummingbirds, bees and bats in Drymonia [27], Gesnerieae [25], Nematanthus and Codonanthe [37], and Sinningieae [24].
To further test the validity of pollination syndromes, we assessed thoroughly the correlation among floral traits and functional groups of pollinators among the species of Gesnerioideae with documented pollination systems. An extended bibliographic search was conducted to identify all species with published information about their pollinators (electronic supplementary material, table S2). Flowers of these species were characterized using nine morphological traits reflecting their variation in size, shape and colour (electronic supplementary material, table S2). Trait values were derived from published morphometric datasets, monographic revisions and our own measurements of flowers collected in the field or in living collections, or from scaled images available on J.L.C.'s website (www.gesneriads.ua.edu). Among these traits, the degree of corolla constriction (i.e. tubular versus bell-shaped corolla) and the presence of pouched or urn-shaped corolla have been identified as key traits to discriminate hummingbird- from bee- and bat-pollinated flowers in different groups of Gesnerioidae [24,25,27,37]. Experimental results have also demonstrated the role of flower constriction and anther exsertion in improving the morphological fit between hummingbirds and flowers and/or in deterring less efficient pollinators like bees [38]. We used a discriminant analysis to maximize the differences in each trait among functional groups of pollinators (i.e. hummingbirds, bats, insects and generalists), and to estimate their predictability for the identification of pollination/shape associations. We used the lda and predict functions from the R package MASS [39]. The most discriminant floral traits were then used to predict the functional groups of pollinators for the species included in the phylogeny that lack direct observation of pollinators.
In all subsequent analyses requiring binary states (see below), the bat-pollinated species (8 out of 590 species) were merged into the hummingbird pollination syndrome category. We based this choice on the fact that (i) hummingbirds and nectarivorous bats are both vertebrates with hovering ability, (ii) certain bat-pollinated species are generalists (pollinated also by hummingbirds during late afternoon and at dawn [25]), and (iii) according to a three-state stochastic mapping analysis, most of the bat-pollinated species in Gesnerioideae evolved recently from hummingbird-pollinated species [40] (see the electronic supplementary material, figure S5).
(d). Evolution of pollination syndromes
The study of trait evolution has largely improved by considering evolutionary time in the modelling of a trait change [41], and by including the species diversification process itself in binary-state speciation and extinction (BiSSE) models [42]. Here, we incorporate most of these improvements by jointly modelling the evolution of pollination syndromes and trait-dependent diversification rates (binary-state trait). For this, we used estimates of transition rates between hummingbird and insect pollination syndromes from the BiSSE model that decomposes the evolutionary process into state-specific speciation and extinction rates and two transition rates. We performed an ancestral state reconstruction, accounting for the influence of diversification, using the asr function from the R package diversitree [43] to estimate the marginal probability of each state at each node. This function is only available for the BiSSE model, and not the other extensions of this model, such as the cladogenetic state change speciation and extinction (ClaSSE) model used in the diversification analysis. The temporal assessment of insect and hummingbird-adapted flowers was done by mapping changes in pollination syndromes across the Gesnerioideae phylogenetic tree. We incorporated the BiSSE estimates of ancestral states into the stochastic mapping (modifying the simmap function in the R package phytools [44]; script available from ‘Data accessibility’ below) and ran 200 reconstructions on independent trees. For each stochastic mapping, we divided branch lengths into time bins of 1 Myr and recorded the number of transitions from and to hummingbird pollination syndrome in each bin. We reported the time bin at which 95% of the stochastic mappings have at least one transition event as the onset time for each type of transition. We performed an additional three-state stochastic mapping without considering trait-dependent diversification to explore the evolution among hummingbird, bat and insect pollination syndromes (see the electronic supplementary material, methods S2).
(e). Diversification analysis
First, we tested whether diversification rates were constant or varied through time using the R package TreePar [45]. The model settings are described in the electronic supplementary material, methods S3. Second, we tested a range of trait-dependent diversification models to assess correlations between evolution of pollination syndromes in Gesnerioideae and changes in speciation and extinction rates. Those models included the BiSSE [42] and ClaSSE [46] classes. These models allow us to distinguish whether diversification rates are associated with any particular pollination syndrome (i.e. BiSSE) or whether they change in response to shifts in pollination syndromes (switches between insect and hummingbird syndromes, i.e. ClaSSE). We compared the BiSSE and ClaSSE models with a recently proposed trait-independent model where unobserved states, which are independent of our pollination syndromes, account for differences in the diversification process (called CID2 [47]). A binary trait was used to represent the pollination syndromes (insect as state 0; hummingbird as state 1). Sampling fraction was accounted in all trait-dependent and CID2 models as 0.53 and 0.48 for insect and hummingbird pollination, respectively. We estimated different speciation rates in BiSSE to obtain the parameters λ0 and λ1 for the speciation associated with insect and hummingbird pollination syndromes, respectively. For the ClaSSE model, we denoted the speciation within pollination syndromes (λ000, λ111), the speciation associated with a switch in trait for one of the descendant species, namely from insect to hummingbird pollination syndrome (λ001, λ101), and the speciation rates associated with switch in both descendant species (λ011, λ100). Each model also included two state-specific extinction rates (μ0, μ1) and two transition rates (q01, q10). We compared eight BiSSE and 13 ClaSSE models using maximum-likelihood estimates in the R package diversitree [43]. The best model was selected based on AICc, and we estimated the posterior distributions of each parameter for the best model in a Bayesian framework [5]. Priors and MCMC parameters are described in the electronic supplementary material, methods S3.
Methods associating traits and diversification should be taken with caution [48]; these issues were minimized in our dataset and analyses (see the electronic supplementary material, methods S4), rejecting that rates may vary over the tree or through time [49].
3. Results
(a). Phylogenetic reconstruction
The best models of molecular evolution were GTR + Γ and GTR + Γ + I for the nuclear and chloroplast DNA partition, respectively (electronic supplementary material, figure S1). The MrBayes and BEAST analyses resulted in congruent topologies. Our phylogenetic reconstruction (figure 1c) constitutes one of the largest species-level phylogenetic analysis for Neotropical plants. The topology corroborates the formal previous classifications [20], namely that Gesnerioideae comprises five tribes and 12 subtribes (posterior probabilities > 0.99; electronic supplementary material, figure S4). Relationships among tribes had a high support (posterior probabilities > 0.99), except that Titanotricheae, Napeantheae and Beslerieae (composed of the genera Besleria, Gasteranthus, Reldia, Cremosperma, Shuaria, Anetanthus and Tylopsacas) formed a clade (PP = 0.508 in the BEAST MCC tree) sister to the rest of the Gesnerioideae. The tribe Coronanthereae was sister to the Gesnerieae in agreement with prior results [19]. Five highly supported clades were resolved in the Gesnerieae, corresponding to the subtribes Gesneriinae, Gloxiniinae, Columneinae, Sphaerorrhizinae and Ligeriinae. Generic and infrageneric relationships largely agree with previous phylogenetic results obtained for these lineages [37,50–53]. Out of 74 genera of Gesnerioideae, eight appeared non-monophyletic and are still in need of further taxonomical revision (Achimenes, Diastema, Gesneria, Mandirola, Paliavana, Phinaea, Sinningia and Vanhouttea).
(b). Characterization and evolution of pollination syndromes
Overall, 118 species with documented pollination systems were recorded from the literature (electronic supplementary material, table S2). Among them, 82 species were pollinated by hummingbirds, 19 species pollinated by bees, three species pollinated by other insects (butterfly, diptera and moth), and seven species pollinated by bats (electronic supplementary material, table S2). Seven other species are pollinated by a mix of nocturnal and diurnal visitors (e.g. hummingbird, bat and moth). These generalist species of Gesnerioideae have so far been recorded only on the Caribbean islands in pollinator-depauperate environments [26,54]. The discriminant analyses explained a large proportion of the floral trait variability (axes 1 and 2 with a 76.35% and 22.34% of explained variance, respectively). Linear discriminant axis 1 had a positive loading for corolla tube shape and lateral compression, and a negative loading for corolla length and the corolla width at mouth. Linear discriminant axis 2 had a positive loading for most of the traits except corolla length and tube shape (electronic supplementary material, table S3 and figure S2). The predictability of each group of functional pollinators was high (hummingbirds = 0.974, insects = 0.954, bats = 1.00, generalists = 0.66), and their separation in the morphological space was clear (figure 1b). Only five species of 118 (approx. 5%) have a group predictability lower than 0.8; these are one bee-pollinated (S. villosa), three generalists (G. viridiflora, R. leucomallon, R. vernicosum) and one hummingbird-pollinated (P. sericiflora, a species with flower morphology related to the bat syndrome but effectively pollinated by hummingbirds [22]). The standardized coefficients of each trait determine the contribution of the respective trait to the discriminant function among the groups. Based on these values (electronic supplementary material, table S3), we selected tube shape and lobe symmetry (electronic supplementary material, figures S2 and S3) as a proxy to assign pollination syndrome for species in the phylogeny whose pollination biology is unstudied in the field. Using this approach, and the information listed in the electronic supplementary material, table S2, we inferred 351 species with hummingbird pollination syndrome, eight species pollinated by bats and 231 species with insect pollination syndrome among the 590 taxa included in our phylogenetic tree (electronic supplementary material, table S1).
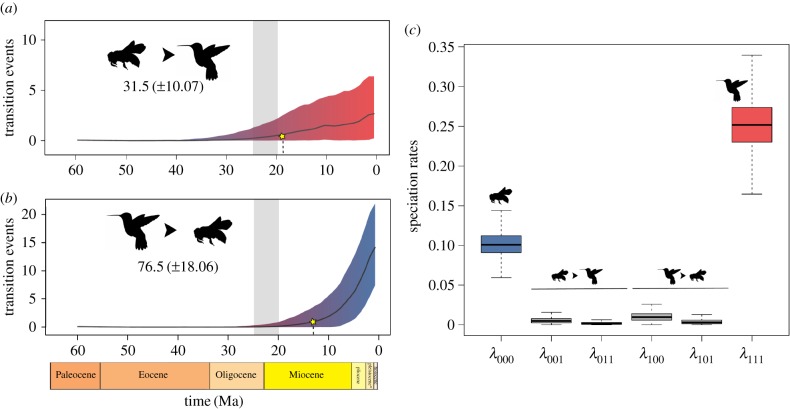
The BiSSE estimates of transition rates between pollination syndromes indicated a median rate from insect to hummingbird pollination syndrome of 0.009, and from hummingbird to insect pollination syndrome of 0.044. Our stochastic mapping showed that pollination syndromes evolved on average from insect to hummingbird 31.50 (±10.07) times. Transitions to hummingbird pollination syndromes first occurred around 18.5 Ma and then increased in frequency over time (figure 2a). These transitions were reconstructed at or near the crown of large clades of Gesnerioideae, such as Besleria, Ligeriineae, Gesneriinae, Gloxiniinae and Columnea + Glossoloma genera (figure 1c). Reversions from hummingbird to insect pollination were highly frequent (on average 76.50 ± 18.06 times). These reversions to insect pollination started around 12.5 Ma and were mainly reconstructed on terminal branches or within clades including few species (figures 1c and 2b). Our three-state reconstruction treating bat and hummingbird syndromes separately showed that transitions to bat-pollinated flowers (all bat-pollinated species in Gesnerioideae have been observed in the field) have occurred at least seven times, since around 9.5 Ma, and mainly from hummingbird-adapted flowers (electronic supplementary material, figures S4 and S5).
Figure 2.
(a,b) Estimated number of transitions through time for pollination syndromes. Numbers below the pictograms correspond to the mean total number of transitions between the states and the standard deviation. Stars denote the starting point in time where at least one transition is recorded in 95% of the reconstructions. Grey bar is the age of the most recent CA of extant hummingbirds (20.3–24.7 Ma [28]). (c) State-dependent speciation rate estimates from ClaSSE model. λ = speciation rate (specific parameters in Diversification analysis section), 0 = insect, 1 = hummingbird. Colours and pictograms correspond to the binary pollination syndrome: blue for insect, and red for hummingbird states.
(c). Diversification analysis
Our analyses of temporal shifts during the diversification of Gesnerioideae detected a single shift in diversification rate (p-value < 0.001; electronic supplementary material, figure S6) that most probably occurred around 18.5 Ma (95% confidence interval = 5.0–25.5 Ma). The mean net diversification rates were 0.067 and 0.177 Ma−1 for the periods before and after the shift, respectively. The comparison of models for trait-dependent diversification indicates that the model with the best AICc is the ClaSSE with all different speciation rates (though λ011 and λ101 constrained to be zero), extinction rates constrained to be zero and equal transition rates (table 1; see complete electronic supplementary material, table S7). We clearly rejected a trait-independent process shaping the diversification of the Gesnerioideae (CID2 null model with a δAICc = 25.0 from the best model). The MCMC parameter estimates suggested that speciation rates within pollination syndromes were higher than those associated with shifts between them (figure 2c). Furthermore, species within the hummingbird pollination syndrome have at least a twofold higher rate (mean λ111 = 0.252 Myr−1, 95% HPD = 0.193–0.314) than species within the insect pollination syndrome (mean λ000 = 0.102 Myr−1, 95% HPD = 0.071–0.133). All speciation rates associated with a shift in pollination syndrome, regardless of the direction of the shifts, are lower and close to 0.01, and thus an order of magnitude lower than the rates within pollination syndrome (λ001 = 0.006, λ011 = 0.003, λ101 = 0.004, λ100 = 0.010; figure 2c). Posterior distributions of extinction rate showed a higher extinction rate for hummingbird pollination syndrome species (mean μ0 = 0.015, 95% HPD = 0.000–0.041, and μ1 = 0.027, 95% HPD = 0.000–0.077). Transition rates between pollination syndrome states supported a higher rate of reversals to insect pollination syndrome (mean q01 = 0.006, HPD = 0.000–0.009, and mean q10 = 0.023, HPD = 0.000–0.040), and were of similar magnitude to the rates estimated by BiSSE (electronic supplementary material, table S4).
Table 1.
Summary of the best models for each class (based on AICc) of trait-dependent diversification models (BiSSE and ClaSSE), and the null trait-independent model (CID2). Italics indicate best model AICc.
| description | speciation | extinction | transition | np | LogL | AICc | δAICc |
|---|---|---|---|---|---|---|---|
| BiSSE | |||||||
| different λ and q | λ0 ≠ λ1 | μ0 = μ1 | q0_1 ≠ q1_0 | 5 | −2089.00 | 4188.10 | 10.10 |
| ClaSSE | |||||||
| different λa,b,c | λ000 ≠ λ111 ≠ λ001 ≠ λ100, λ011 = zero, λ101 = zero | μ0 = μ1 = 0 | q0_1 = q1_0 | 5 | −2083.94 | 4177.99 | 0.00 |
| null model | |||||||
| CID2 null modeld | τ0A = τ1A ≠ τ0B = τ1B | ε0A = ε1A ≠ ε0B = ε1B | all q are equal | 5 | −2096.44 | 4202.98 | 25.00 |
aParameters λ011 and λ100 correspond to an insect-pollinated species that gives rise to two hummingbird-pollinated species, and vice versa.
bParameters λ000 and λ111 correspond to an insect-pollinated species that gives rise to two insect-pollinated species, and vice versa for hummingbird-pollinated.
cParameters λ010 and λ101 correspond to an insect-pollinated species that gives rise to one insect, and one hummingbird-pollinated species.
dCID2 is the described null model for trait-dependent diversification models in [47]. τ and ε correspond to turnover and extinction fraction parameters.
We found that diversification results are robust to the misidentification of functional groups of pollinators at the tips of the phylogenetic tree. First, the difference in rates of speciation between the two pollination syndromes (λ000 and λ111) is persistent if we remove the species-rich genus Columnea, which includes exclusively hummingbird pollination syndrome species (electronic supplementary material, figure S7b), and the few bat-pollinated species (electronic supplementary material, table S6). Second, the test for possible misidentification of functional groups of pollinators indicated that our estimation of speciation and extinction rates are extremely robust to up to 10% of equivocal states (for both insect and hummingbird) and that even 15% of misidentification leads to qualitatively similar results (electronic supplementary material, figures S8–S10). Finally, the simulations of traits, whose evolution is independent from the diversification process, showed that the estimated speciation rates within pollination syndromes (i.e. λ000 and λ111) are equal, under the null hypothesis, as well as the extinction and transition rates (electronic supplementary material, figure S7a). The effect of hummingbird pollination syndrome on diversification that we detected is thus not likely to be due to a particular shape of the phylogeny that could lead to a false detection of an association between traits and speciation [49].
4. Discussion
We showed that hummingbird pollination probably played a role in the diversification dynamics of Gesnerioideae in the Neotropics. Two lines of evidence support this result. First, the diversification of this subfamily increased substantially around 20 Ma. This period corresponds closely to the dating for the common ancestor of hummingbirds into South America (22.4 Ma, 95% HPD: 20.3–24.7 Ma in [28]; 24–25 Ma in [55]) and the first appearance of plant species with hummingbird pollination syndrome in the Gesnerioideae (figure 2a). Second, we clearly show that species with hummingbird pollination syndrome have higher rates of speciation compared with species with insect pollination syndrome. On the other hand, we did not find high speciation rates associated with transitions between pollinator syndromes. These results indicate that species richness in this plant group has been driven by speciation within hummingbird-pollinated lineages, without involving shifts among functional groups of pollinators, contradicting the classical pollinator-shift hypothesis [14].
(a). Evolution of hummingbird pollination syndrome in Gesnerioideae
Our study suggests that Gesnerioideae was ancestrally pollinated by insects and that at least 31 transitions to hummingbird and bat pollination syndromes occurred during its evolution (figure 1c; electronic supplementary material, figure S4). The repeated evolution of hummingbird pollination syndrome in independent Gesnerioideae lineages centred in different geographical areas, such as the Brazilian Atlantic forest, Andes, Caribbean islands and Central America [21], is indicative of the success of this ecological interaction in multiple biomes of the Neotropics. We found also frequent state reversals from hummingbird to insect pollination syndrome providing evidence against the hypothesis that the evolution of hummingbird pollination could act as a dead end from which reversals to insect pollination are rare or no longer possible [14,56–58]. The convergent evolution of distinct floral morphologies and pigmentation in relatively short periods of time, as well as the reversibility of this system, are striking and encourage the investigation of whether a shared molecular basis might control these phenotypic transitions [59].
Flowers corresponding to the hummingbird pollination syndrome appeared in Gesnerioideae around 18.5 Ma (figure 2a), when hummingbirds were already present and diversifying in South America [28,55]. This rather early origin of hummingbird flowers, and the inferred south American origin of the Gesnerioideae [21], indicate that this plant group could be one of the oldest to have established interactions with the first hummingbirds living in the tropical regions of South America, unlike more recent hummingbird-adapted plant lineages [15,16,55,60]. This old interaction might have given the Gesnerioideae species enough time for the evolution of hummingbird pollination in separate lineages, and a substantial amount of transitions back to insect pollination.
(b). Effect of hummingbird pollination on plant diversification
Our finding of a preferred trait-dependent model indicates an effect of pollination syndromes in the diversification of the Gesnerioideae. A twofold increase in speciation rates for species with a hummingbird pollination compared with insect pollination syndrome suggests that floral morphologies associated with hummingbird pollinators may promote mechanisms that lead to the generation of new species. By contrast, speciation rates associated with shifts in pollination syndromes (i.e. between insect and hummingbird pollination syndromes) were between 20 and 80 times lower than those within pollination syndromes (electronic supplementary material, table S5). These results indicate that the classical pollinator-shift hypothesis driving plant speciation does not explain the Gesnerioideae diversification. Instead, species richness in this plant group has been driven by speciation within hummingbird-pollinated lineages, without involving shifts among functional groups of pollinators.
Why hummingbird pollination promotes plant speciation remains unclear, but we propose plausible mechanisms as a starting point for future research. First, the evolution of tubular or gullet-like flowers characterizing most hummingbird flowers may have directly accelerated speciation by promoting specialized relationships with the different categories of hummingbird species and the evolution of rapid prezygotic reproductive barriers [61,62]. Hummingbirds vary dramatically in bill size and shape, and these characteristics largely match with the flower morphology of the species they feed on [63–65]. Although most plant species are visited by several hummingbird species, specialization in the plant–pollinator network was found to increase at low and medium elevations [66], and in species-rich communities in which closely related hummingbirds visited distinct sets of flowering species [67]. Thus, floral specialization in hummingbird-pollinated species is frequently more specialized than initially assumed, which may result in greater potential for pollen segregation and, eventually, speciation. Second, flower specialization and specific pollen placement on the hummingbird body may limit interspecific pollen transfer among species sharing pollinators, thereby increasing the number of plant species that can co-occur in the same community [37,68,69]. It has been suggested that this process could decrease extinction rates [9], but also potentially increase the carrying capacity of hummingbird-pollinated lineages per unit of area, a factor that can limit the decline of diversification rates over time [70]. Finally, beyond floral specialization, bird pollination could have a direct impact on gene flow and the geography of plant speciation. Compared with insects, bird pollination increases the efficiency of pollen transfer and deposition [71], potentially affecting the connectivity among natural populations even in the context of a patchy distribution of suitable habitat [72]. Such improved transfer of pollen over long distances could enable the maintenance of isolated population providing the precondition for allopatric speciation. This has been suggested for the Andean species of Passiflora [73] and other hummingbird-pollinated lineages in the Gesneriaceae like Columnea and Dircaea [24,74]. Finally, and consistent with the last argument, hummingbird pollination is considered to be more efficient than insect pollination in Neotropical cloud forests at middle to high elevations, because insects are less active in cool, foggy, and wet conditions [75,76]. Altogether these patterns suggest that hummingbird-pollinated species could have more opportunities to colonize a wider geographical and climatic range, and to establish complex plant–pollinator interactions compared with insect-pollinated lineages. Although these hypotheses remain largely untested, such factors could potentially trigger diversification in angiosperm. Testing these hypotheses requires more complete morphological characterization of the plant species and plant–pollinator interaction data, to better understand how biotic interactions have shaped biodiversity and macro-evolutionary patterns in the Neotropical region.
Our study provides new insights into one of the most intriguing factors influencing the diversification of Neotropical plant lineages, namely the impact of hummingbird pollination. However, additional ecological factors should not be excluded to conduct holistic examinations as encouraged by recent studies [15,16]. The evolution of epiphytism and different growth forms, the colonization of new biomes, and the geological history have potentially influenced the diversification and distribution of Gesnerioideae [17]. Currently, factors are usually evaluated independently, making it difficult to test their joint effects [77], and they should be taken with precaution, for instance, in traits with very few evolutionary transitions (such as epiphytism in the species-rich Columneinae clade). An optimal methodology should consider multiple factors simultaneously and allow a particular combination of those (i.e. colonization of a new area, with an in situ new trait state) to affect the diversification process (as discussed in [77,78]). Such approaches are, however, not yet available, and there is a current need to develop thoughtful tests for modelling simultaneously the success of multiple ecological interactions, considering the caveats of the methods and data, while integrating global biodiversity patterns.
5. Conclusion
We identified a strong and positive effect of hummingbird pollination syndrome on the process of species diversification in the subfamily Gesnerioideae. This effect was probably triggered by the repeated acquisition of hummingbird pollination when this pollination niche became available in South America during the Early Miocene. Plants with a hummingbird pollination syndrome have a twofold increase in the rate of speciation, suggesting a positive effect of hummingbird pollination on the establishment of reproductive isolation. Our findings complement the global understanding of the diversification processes leading to the exceptional diversity of flowering plants in the Neotropics, and provide new directions towards further testing the role played by plant–pollinator relationships in the build-up of plant diversity.
Supplementary Material
Acknowledgements
We thank Alain Chautems, Lianka Cairampoma, Andrea O. De Araujo, Valquiria Ferreira Dutra, Mauro Peixoto, Angela Cano, Harri Lorenzi, Gabriel E. Ferreira, Juvenal E. Batista, Mireya Correa, Marina Wolowski and the staff of the Smithsonian Tropical Research Institute for their contribution to the fieldwork; Régine Niba and Fadil Avdija for their contribution to the laboratory work; Yvonne Menneret, Marina Magnette, Matthieu Grillet, Alexandre Chappuis, Bertrand Guigon and Vincent Goldschmid for the propagation and maintenance of the Gesneriaceae collection at Geneva. Collection permits were granted by the CNPq in Brazil (CMC 038/03) and in the ANAM in Panama (SC/P-43–10). Images in figure 1a were kindly provided by Ivonne SanMartin-Gajardo, Anton Weber and Leandro Freitas. We thank Daniele Silvestro for the helpful discussions and support for the analyses, and Catherine Graham and two anonymous reviewers for their advice on the manuscript. All analyses were run at the High-performance Computing Center (Vital-IT) from the Swiss Institute of Bioinformatics.
Data accessibility
The data and additional details of the approaches used in this paper are available in the electronic supplementary material and at Dryad Digital Repository [79]. R script available from www2.unil.ch/phylo/files/software/make.simmap.BiSSE.R.
Authors' contributions
M.L.S.-S., N.S. and M.P. planned and designed the research and performed the analyses. M.P. and J.L.C. conducted fieldwork and gathered data. M.L.S.-S., J.R., J.L.C., N.S. and M.P. wrote the manuscript. All authors gave final approval for publication.
Competing interests
The authors have declared that no competing interests exist.
Funding
This study was funded by the Ville de Genève, the Faculty of Biology and Medicine at the University of Lausanne and the Swiss National Science foundation (grant no. CRSII3_147630).
References
- 1.Vamosi JC, Vamosi SM. 2011. Factors influencing diversification in angiosperms: at the crossroads of intrinsic and extrinsic traits. Am. J. Bot. 98, 460–471. ( 10.3732/ajb.1000311) [DOI] [PubMed] [Google Scholar]
- 2.Fiz-Palacios O, Schneider H, Heinrichs J, Savolainen V. 2011. Diversification of land plants: insights from a family-level phylogenetic analysis. BMC Evol. Biol. 11, 1 ( 10.1186/1471-2148-11-341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes C, Eastwood R. 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl Acad. Sci. USA 103, 10 334–10 339. ( 10.1073/pnas.0601928103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litsios G, Wuest RO, Kostikova A, Forest F, Lexer C, Linder HP, Pearman PB, Zimmermann NE, Salamin N. 2014. Effects of a fire response trait on diversification in replicated radiations. Evolution 68, 453–465. ( 10.1111/evo.12273) [DOI] [PubMed] [Google Scholar]
- 5.Silvestro D, Zizka G, Schulte K. 2014. Disentangling the effects of key innovations on the diversification of Bromelioideae (Bromeliaceae). Evolution 68, 163–175. ( 10.1111/evo.12236) [DOI] [PubMed] [Google Scholar]
- 6.Hodges SA, Arnold ML. 1995. Spurring plant diversification: are floral nectar spurs a key innovation? Proc. R. Soc. Lond. B 262, 343–348. ( 10.1098/rspb.1995.0215) [DOI] [Google Scholar]
- 7.Sargent RD. 2004. Floral symmetry affects speciation rates in angiosperms. Proc. R. Soc. Lond. B 271, 603–608. ( 10.1098/rspb.2003.2644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stebbins GL. 1974. Flowering plants: evolution above the species level. Cambridge, MA: The Belknap Press of Harvard University Press. [Google Scholar]
- 9.Armbruster WS, Muchhala N. 2009. Associations between floral specialization and species diversity: cause, effect, or correlation? Evol. Ecol. 23, 159–179. ( 10.1007/s10682-008-9259-z) [DOI] [Google Scholar]
- 10.Van der Niet T, Peakall R, Johnson SD. 2014. Pollinator-driven ecological speciation in plants: new evidence and future perspectives. Ann. Bot. 113, 199–211. ( 10.1093/aob/mct290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kay KM, Reeves PA, Olmstead RG, Schemske DW. 2005. Rapid speciation and the evolution of hummingbird pollination in neotropical Costus subgenus Costus (Costaceae): evidence from nrDNA ITS and ETS sequences. Am. J. Bot. 92, 1899–1910. ( 10.3732/ajb.92.11.1899) [DOI] [PubMed] [Google Scholar]
- 12.Valente LM, Manning JC, Goldblatt P, Vargas P. 2012. Did pollination shifts drive diversification in southern African Gladiolus? Evaluating the model of pollinator-driven speciation. Am. Nat. 180, 83–98. ( 10.1086/666003) [DOI] [PubMed] [Google Scholar]
- 13.Forest F, Goldblatt P, Manning JC, Baker D, Colville JF, Devey DS, Jose S, Kaye M, Buerki S. 2014. Pollinator shifts as triggers of speciation in painted petal irises (Lapeirousia: Iridaceae). Ann. Bot. 113, 357–371. ( 10.1093/aob/mct248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Niet T, Johnson SD. 2012. Phylogenetic evidence for pollinator-driven diversification of angiosperms. Trends Ecol. Evol. 27, 353–361. ( 10.1016/j.tree.2012.02.002) [DOI] [PubMed] [Google Scholar]
- 15.Givnish TJ, et al. 2014. Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Mol. Phylogenet. Evol. 71, 55–78. ( 10.1016/j.ympev.2013.10.010) [DOI] [PubMed] [Google Scholar]
- 16.Lagomarsino LP, Condamine FL, Antonelli A, Mulch A, Davis CC. 2016. The abiotic and biotic drivers of rapid diversification in Andean bellflowers (Campanulaceae). New Phytol. 210, 1430–1442. ( 10.1111/nph.13920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roalson EH, Roberts WR. 2016. Distinct processes drive diversification in different clades of Gesneriaceae. Syst. Biol. 65, 662–684. ( 10.1093/sysbio/syw012) [DOI] [PubMed] [Google Scholar]
- 18.Schmidt-Lebuhn AN, Kessler M, Hensen I. 2007. Hummingbirds as drivers of plant speciation? Trends Plant Sci. 12, 329–331. ( 10.1016/j.tplants.2007.06.009) [DOI] [PubMed] [Google Scholar]
- 19.Woo VL, Funke MM, Smith JF, Lockhart PJ, Garnock-Jones PJ. 2011. New World origins of southwest Pacific Gesneriaceae: multiple movements across and within the South Pacific. Int. J. Plant Sci. 172, 434–457. ( 10.1086/658183) [DOI] [Google Scholar]
- 20.Weber A, Clark JL, Möller M. 2013. A new formal classification of Gesneriaceae. Selbyana 31, 68–94. [Google Scholar]
- 21.Perret M, Chautems A, Araujo AO, Salamin N. 2013. Temporal and spatial origin of Gesneriaceae in the New World inferred from plastid DNA sequences. Bot. J. Linn. Soc. 171, 61–79. ( 10.1111/j.1095-8339.2012.01303.x) [DOI] [Google Scholar]
- 22.SanMartin-Gajardo I, Sazima M. 2005. Chiropterophily in Sinningieae (Gesneriaceae): Sinningia brasiliensis and Paliavana prasinata are bat-pollinated, but P. sericiflora is not. Not yet? Ann. Bot. 95, 1097–1103. ( 10.1093/aob/mci124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.SanMartin-Gajardo I, Sazima M. 2005. Espécies de Vanhouttea Lem. e Sinningia Nees (Gesneriaceae) polinizadas por beija-flores: interações relacionadas ao hábitat da planta e ao néctar. Revista Brasil. Bot. 28, 441–450. [Google Scholar]
- 24.Perret M, Chautems A, Spichiger R, Barraclough TG, Savolainen V. 2007. The geographical pattern of speciation and floral diversification in the Neotropics: the tribe Sinningieae (Gesneriaceae) as a case study. Evolution. 61, 1641–1660. ( 10.1111/j.1558-5646.2007.00136.x) [DOI] [PubMed] [Google Scholar]
- 25.Martén-Rodríguez S, Almarales-Castro A, Fenster CB. 2009. Evaluation of pollination syndromes in Antillean Gesneriaceae: evidence for bat, hummingbird and generalized flowers. J. Ecol. 97, 348–359. ( 10.1111/j.1365-2745.2008.01465.x) [DOI] [Google Scholar]
- 26.Martén-Rodríguez S, Quesada M, Castro A-A, Lopezaraiza-Mikel M, Fenster CB, Phillips R. 2015. A comparison of reproductive strategies between island and mainland Caribbean Gesneriaceae. J. Ecol. 103, 1190–1204. ( 10.1111/1365-2745.12457) [DOI] [Google Scholar]
- 27.Clark JL, Clavijo L, Muchhala N. 2015. Convergence of anti-bee pollination mechanisms in the Neotropical plant genus Drymonia (Gesneriaceae). Evol. Ecol. 29, 355–377. ( 10.1007/s10682-014-9729-4) [DOI] [Google Scholar]
- 28.McGuire JA, Witt CC, Remsen JV Jr, Corl A, Rabosky DL, Altshuler DL, Dudley R. 2014. Molecular phylogenetics and the diversification of hummingbirds. Curr. Biol. 24, 910–916. ( 10.1016/j.cub.2014.03.016) [DOI] [PubMed] [Google Scholar]
- 29.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. 2010. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38, W23–W28. ( 10.1093/nar/gkq443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 32.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. 2004. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 35, 375–403. ( 10.1146/annurev.ecolsys.34.011802.132347) [DOI] [Google Scholar]
- 35.Ollerton J, Alarcon R, Waser NM, Price MV, Watts S, Cranmer L, Hingston A, Peter CI, Rotenberry J. 2009. A global test of the pollination syndrome hypothesis. Ann. Bot. 103, 1471–1480. ( 10.1093/aob/mcp031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosas-Guerrero V, Aguilar R, Marten-Rodriguez S, Ashworth L, Lopezaraiza-Mikel M, Bastida JM, Quesada M. 2014. A quantitative review of pollination syndromes: do floral traits predict effective pollinators? Ecol. Lett. 17, 388–400. ( 10.1111/ele.12224) [DOI] [PubMed] [Google Scholar]
- 37.Serrano-Serrano ML, Perret M, Guignard M, Chautems A, Silvestro D, Salamin N. 2015. Decoupled evolution of floral traits and climatic preferences in a clade of Neotropical Gesneriaceae. BMC Evol. Biol. 15, 247 ( 10.1186/s12862-015-0527-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castellanos MC, Wilson P, Thomson JD. 2004. 'Anti-bee' and 'pro-bird' changes during the evolution of hummingbird pollination in Penstemon flowers. J. Evol. Biol. 17, 876–885. ( 10.1111/j.1420-9101.2004.00729.x) [DOI] [PubMed] [Google Scholar]
- 39.Ripley B, Venables B, Bates DM, Hornik K, Gebhardt A, Firth D, Ripley MB. 2013. Package ‘MASS’. CRAN Repository. See http://cran.r-projectorg/web/packages/MASS/MASS.pdf.
- 40.Fleming TH, Geiselman C, Kress WJ. 2009. The evolution of bat pollination: a phylogenetic perspective. Ann. Bot. 104, 1017–1043. ( 10.1093/aob/mcp197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huelsenbeck JP, Nielsen R, Bollback JP. 2003. Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158. ( 10.1080/10635150390192780) [DOI] [PubMed] [Google Scholar]
- 42.Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710. ( 10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 43.FitzJohn RG. 2012. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092. ( 10.1111/j.2041-210X.2012.00234.x) [DOI] [Google Scholar]
- 44.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 45.Stadler T. 2011. Mammalian phylogeny reveals recent diversification rate shifts. Proc. Natl Acad. Sci. USA 108, 6187–6192. ( 10.1073/pnas.1016876108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldberg EE, Igic B. 2012. Tempo and mode in plant breeding system evolution. Evolution 66, 3701–3709. ( 10.1111/j.1558-5646.2012.01730.x) [DOI] [PubMed] [Google Scholar]
- 47.Beaulieu JM, O'Meara BC. 2016. Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst. Biol. 65, 583–601. ( 10.1093/sysbio/syw022) [DOI] [PubMed] [Google Scholar]
- 48.Maddison WP, FitzJohn RG. 2015. The unsolved challenge to phylogenetic correlation tests for categorical characters. Syst. Biol. 64, 127–136. ( 10.1093/sysbio/syu070) [DOI] [PubMed] [Google Scholar]
- 49.Rabosky DL, Goldberg EE. 2015. Model inadequacy and mistaken inferences of trait-dependent speciation. Syst. Biol. 64, 340–355. ( 10.1093/sysbio/syu131) [DOI] [PubMed] [Google Scholar]
- 50.Clark JL, Funke MM, Duffy AM, Smith JF. 2012. Phylogeny of a Neotropical clade in the Gesneriaceae: more tales of convergent evolution. Int. J. Plant Sci. 173, 894–916. ( 10.1086/667229) [DOI] [Google Scholar]
- 51.Mora MM, Clark JL. 2016. Molecular phylogeny of the Neotropical genus Paradrymonia (Gesneriaceae), reexamination of generic concepts and the resurrection of Trichodrymonia and Centrosolenia. Syst. Bot. 41, 82–104. ( 10.1600/036364416X690561) [DOI] [Google Scholar]
- 52.Ferreira GE, Chautems A, Hopkins MJ, Perret M. 2016. Independent evolution of pouched flowers in the Amazon is supported by the discovery of a new species of Lesia (Gesneriaceae) from Serra do Aracá tepui in Brazil. Plant Syst. Evol. 302, 1109–1119. ( 10.1007/s00606-016-1320-8) [DOI] [Google Scholar]
- 53.Araujo AO, Chautems A, Cardoso-Gustavson P, Souza VC, Perret M. 2016. Taxonomic revision and phylogenetic position of the Brazilian endemic genus Sphaerorrhiza (Sphaerorrhizinae, Gesneriaceae) including two new species. Syst. Bot. 41, 651–664. ( 10.1600/036364416X692352) [DOI] [Google Scholar]
- 54.Marten-Rodriguez S, Fenster CB, Agnarsson I, Skog LE, Zimmer EA. 2010. Evolutionary breakdown of pollination specialization in a Caribbean plant radiation. New Phytol. 188, 403–417. ( 10.1111/j.1469-8137.2010.03330.x) [DOI] [PubMed] [Google Scholar]
- 55.Abrahamczyk S, Renner SS. 2015. The temporal build-up of hummingbird/plant mutualisms in North America and temperate South America. BMC Evol. Biol. 15, 104 ( 10.1186/s12862-015-0388-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson P, Wolfe AD, Armbruster WS, Thomson JD. 2007. Constrained lability in floral evolution: counting convergent origins of hummingbird pollination in Penstemon and Keckiella. New Phytol. 176, 883–890. ( 10.1111/j.1469-8137.2007.02219.x) [DOI] [PubMed] [Google Scholar]
- 57.Tripp EA, Manos PS. 2008. Is floral specialization an evolutionary dead-end? Pollination system transitions in Ruellia (Acanthaceae). Evolution 62, 1712–1737. ( 10.1111/j.1558-5646.2008.00398.x) [DOI] [PubMed] [Google Scholar]
- 58.Barrett SC. 2013. The evolution of plant reproductive systems: how often are transitions irreversible? Proc. R. Soc. B 280, 20130913 ( 10.1098/rspb.2013.0913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cronk Q, Ojeda I. 2008. Bird-pollinated flowers in an evolutionary and molecular context. J. Exp. Bot. 59, 715–727. ( 10.1093/jxb/ern009) [DOI] [PubMed] [Google Scholar]
- 60.Tripp EA, McDade LA. 2013. Time-calibrated phylogenies of hummingbirds and hummingbird-pollinated plants reject a hypothesis of diffuse co-evolution. Aliso 31, 89–103. ( 10.5642/aliso.20133102.05) [DOI] [Google Scholar]
- 61.Givnish TJ. 2010. Ecology of plant speciation. Taxon 59, 1326–1366. [Google Scholar]
- 62.Betts MG, Hadley AS, Kress WJ. 2015. Pollinator recognition by a keystone tropical plant. Proc. Natl Acad. Sci. USA 112, 3433–3438. ( 10.1073/pnas.1419522112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stiles FG. 1985. On the role of birds in the dynamics of Neotropical forests. In Conservation of tropical forest birds (eds Diamond AW, Lovejoy J), pp. 49–59. Cambridge, UK: International Council of Bird Preservation. [Google Scholar]
- 64.Temeles EJ, Koulouris CR, Sander SE, Kress WJ. 2009. Effect of flower shape and size on foraging performance and trade-offs in a tropical hummingbird. Ecology 90, 1147–1161. ( 10.1890/08-0695.1) [DOI] [PubMed] [Google Scholar]
- 65.Maglianesi MA, Blüthgen N, Böhning-Gaese K, Schleuning M. 2014. Morphological traits determine specialization and resource use in plant–hummingbird networks in the Neotropics. Ecology 95, 3325–3334. ( 10.1890/13-2261.1) [DOI] [Google Scholar]
- 66.Maglianesi MA, Böhning-Gaese K, Schleuning M. 2015. Different foraging preferences of hummingbirds on artificial and natural flowers reveal mechanisms structuring plant–pollinator interactions. J. Anim. Ecol. 84, 655–664. ( 10.1111/1365-2656.12319) [DOI] [PubMed] [Google Scholar]
- 67.Martín González AM, et al. 2015. The macroecology of phylogenetically structured hummingbird–plant networks. Glob. Ecol. Biogeogr. 24, 1212–1224. ( 10.1111/geb.12355) [DOI] [Google Scholar]
- 68.Brown JH, Kodric-Brown A. 1979. Convergence, competition, and mimicry in a temperate community of hummingbird-pollinated flowers. Ecology 60, 1022–1035. ( 10.2307/1936870) [DOI] [Google Scholar]
- 69.Sazima I, Buzato S, Sazima M. 1996. An assemblage of hummingbird-pollinated flowers in a montane forest in southeastern Brazil. Botanica Acta 109, 149–160. ( 10.1111/j.1438-8677.1996.tb00555.x) [DOI] [Google Scholar]
- 70.Vamosi JC, Vamosi SM. 2010. Key innovations within a geographical context in flowering plants: towards resolving Darwin's abominable mystery. Ecol. Lett. 13, 1270–1279. ( 10.1111/j.1461-0248.2010.01521.x) [DOI] [PubMed] [Google Scholar]
- 71.Castellanos MC, Wilson P, Thomson JD. 2003. Pollen transfer by hummingbirds and bumblebees, and the divergence of pollination modes in Penstemon. Evolution 57, 2742–2752. ( 10.1111/j.0014-3820.2003.tb01516.x) [DOI] [PubMed] [Google Scholar]
- 72.Hughes M, Möller M, Edwards TJ, Bellstedt DU, De Villiers M. 2007. The impact of pollination syndrome and habitat on gene flow: a comparative study of two Streptocarpus (Gesneriaceae) species. Am. J. Bot. 94, 1688–1695. ( 10.3732/ajb.94.10.1688) [DOI] [PubMed] [Google Scholar]
- 73.Abrahamczyk S, Souto-Vilaros D, Renner SS. 2014. Escape from extreme specialization: passionflowers, bats and the sword-billed hummingbird. Proc. R. Soc. B 281, 20140888 ( 10.1098/rspb.2014.0888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schulte LJ, Clark JL, Novak SJ, Jeffries SK, Smith JF. 2015. Speciation within Columnea section angustiflora (Gesneriaceae): islands, pollinators and climate. Mol. Phylogenet. Evol. 84, 125–144. ( 10.1016/j.ympev.2014.12.008) [DOI] [PubMed] [Google Scholar]
- 75.Armbruster WS, Berg EE. 1994. Thermal ecology of male euglossine bees in a tropical wet forest: fragrance foraging in relation to operative temperature. Biotropica 26, 50–60. ( 10.2307/2389110) [DOI] [Google Scholar]
- 76.Cruden RW. 1972. Pollinators in high-elevation ecosystems: relative effectiveness of birds and bees. Science 176, 1439–1440. ( 10.1126/science.176.4042.1439) [DOI] [PubMed] [Google Scholar]
- 77.Donoghue MJ, Sanderson MJ. 2015. Confluence, synnovation, and depauperons in plant diversification. New Phytol. 207, 260–274. ( 10.1111/nph.13367) [DOI] [PubMed] [Google Scholar]
- 78.Etienne RS, Haegeman B. 2012. A conceptual and statistical framework for adaptive radiations with a key role for diversity dependence. Am. Nat. 180, E75–E89. ( 10.1086/667574) [DOI] [PubMed] [Google Scholar]
- 79.Serrano-Serrano ML, Rolland J, Clark JL, Salamin N, Perret M. 2017. Data from: Hummingbird pollination and the diversification of angiosperms: an old and successful association in Gesneriaceae. Dryad Digital Repository. ( 10.5061/dryad.m7589) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Serrano-Serrano ML, Rolland J, Clark JL, Salamin N, Perret M. 2017. Data from: Hummingbird pollination and the diversification of angiosperms: an old and successful association in Gesneriaceae. Dryad Digital Repository. ( 10.5061/dryad.m7589) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The data and additional details of the approaches used in this paper are available in the electronic supplementary material and at Dryad Digital Repository [79]. R script available from www2.unil.ch/phylo/files/software/make.simmap.BiSSE.R.