Abstract
Croton yellow vein mosaic virus (CYVMV, genus Begomovirus family Geminiviridae) is a proliferating begomovirus in the Indian sub-continent. The infectious constructs in binary vector was developed against the CYVMV genome and its associated betasatellite. Agroinoculation of the genomic construct of CYVMV produced leaf curl symptoms alone in three species of tobacco, Nicotiana tabacum, N. benthamiana and N. glutinosa. Co-inoculation of betasatellite enhanced the severity of the disease and reduced the incubation time. Based on the infectious clone, a replicon vector pCro, with only the ability to replicate inside the plant was developed. In pCro vector, CP and V2 ORFs from genome of CYVMV was deleted, which resulted localised replication of the molecule with no visible symptoms. Besides the partial CYVMV genome, pCro also has a cassette containing a double 35S promoter, multiple cloning sites and a NOS terminator to overexpress any foreign protein in plant. Episomal release of the replicon from the binary vector backbone after agroinoculation was detected by PCR. A GFP gene was cloned in pCro vector (pCro-GFP) and agroinoculated to N. tabacum resulted in localized expression of GFP at 5 dpi. The CYVMV replicon vector will be a useful tool for studying functional genomics, vaccine expression and gene silencing in plant.
Keywords: Begomovirus , Croton yellow vein mosaic virus, Replicon vector, Gene expression in plant
Introduction
Croton yellow vein mosaic virus (CYVMV) is a distinct virus species of the genus Begomovirus, family Geminiviridae. CYVMV is transmitted by whitefly, Bemisia tabaci, and occurs commonly on Croton bonplandianum causing bright yellow vein mosaic disease. Other than C. Bonplandianum, natural infection of CYVMV has been identified in several weeds and economic plant species e.g., Jatropha sp., Acalypha sp., Cyamopsis sp., Crotalaria juncea, okra, papaya, radish, rapeseed-mustard and tomato [15, 27, 28, 31]. Under the experimental conditions, CYVMV infects as many as 35 plant species of 11 families [27]. CYVMV having broad host range and being efficiently disseminated by B. tabaci is one of the most prevalent begomoviruses in the Indian sub-continent. The complete sequence of the CYVMV genome and the associated croton yellow vein mosaic betasatellite (CroYVMB) are available for several isolates. Biolistic delivery of DNA components generated through rolling-circle amplification (RCA) of monomeric DNA derived from the clones of CYVMV genome and CroYVMB produced typical yellow vein mosaic symptoms in C. bonplandianum [27]. Hence, both the CYVMV and its associated betasatellite are proved to be the etiological agent. Agroinoculation has been widely used for obtaining infectivity of the cloned DNA of begomoviruses [1, 9, 18, 19]. However, agroinfection and the role of CYVMV genome and CroYVMB in disease development have not been studied.
Studies on molecular biology of plant virus led to realize it as a useful genetic resource. Plant viruses are attractive candidates for the molecular farming of pharmaceutical protein in plant system. The idea of plant virus as vector for the production of foreign protein in the plant was first experimented with cauliflower mosaic virus, a dsDNA plant virus [7]. Thereafter, mastreviruses (ssDNA viruses) with monopartite genome, maize streak virus and begomoviruses with bipartite genome, tomato golden mosaic virus and African cassava mosaic virus (ACMV) were attempted as gene expression vectors [10, 32]. The genus Begomovirus currently containing 322 virus species is the largest of all genera of the family Geminiviridae. Begomoviruses have a broad host range, they multiply at a high level in the infected plant cells and they have a small genome, which is easily maneuverable for developing highly infectious clone. These features make begomovirus as an attractive candidate for using as gene expression vector. Based on genomic constituent begomoviruses may be bipartite or monopartite. Initial attempts with few bipartite begomovirus [tomato golden mosaic virus (TGMV) and African cassava mosaic virus (ACMV)] as a gene expression vector were encountered with the structural instability of vector, which undermined the potentiality of geminivirus as an efficient vector for expression of foreign gene in plant. However, the change in the design of vector, where the viral genome was used as replicon by deleting coat protein or/and movement protein encoding sequences and by using the 35S promoter to drive the foreign gene expression brought back geminivirus as a potential candidate for the high level expression of foreign protein in plant [4, 12]. However, geminivirus based efficient gene expression vector is under the developmental stage. Replicon based strategy has been used to design expression/silencing vector for a few mastreviruses [bean yellow dwarf virus (BeYDV), maize streak virus (MSV), wheat dwarf virus (WDV)], curtoviruses [beet curly top virus (BCTV)], bipartite begomoviruses [ACMV, cabbage leaf curl virus (CaLCuV), abuliton mosaic virus (AbMV), tomato leaf curl New Delhi virus (ToLCNDV)] and monopartite begomoviruses [tobacco curly shoot virus (TbCSV), ageratum yellow vein virus (AYVV)] [11, 16, 20, 25, 32]. Of all there geminiviruses, bean yellow dwarf virus (BeYDV) has been extensively used as replicon vector for expressing various foreign proteins in the plant [4]. Besides this, geminiviral replicon vector has been utilized for plant genome engineering [23].
In this study, we have developed an infectious construct of a monopartite begomovirus, CYVMV and shown that CYVMV alone or in the presence of betasatellite, CroYVMB efficiently induce leaf curl disease in the three species of tobacco. The infectious construct of CYVMV was utilized to design an episomal replicon vector by deleting V1 and V2 genes. We demonstrate that the CYVMV replicon vector is useful in expressing green fluorescences protein (GFP) in the tobacco plant.
Materials and methods
Preparation of infectious constructs of CYVMV and CroYVMB
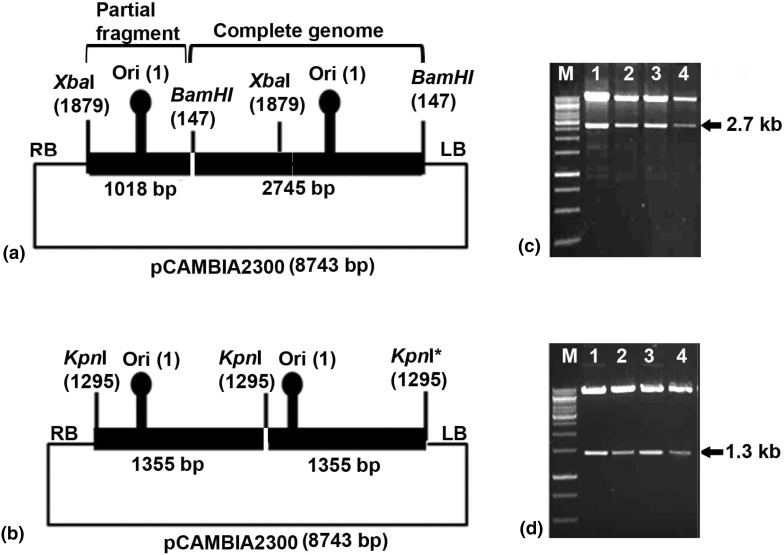
Previously complete genome of an isolate of CYVMV was cloned (M4A, Accession No. JX270684) in the pUC18 vector at BamHI site [28]. A partial tandem repeats (PTR) construct was developed from M4A clone in binary vector pCAMBIA 2300 (Fig. 1a). M4A clone was digested with BamHI and XbaI to generate a 1.0 kb (containing an origin of replication) and 1.7 kb fragments. The 1 kb fragment obtained by digestion with BamHI–XbaI was purified and cloned into the pCAMBIA2300 vector to generate pCAM-M4A-A0.4mer. The full-length (1.0-mer) CYVMV genome was released from M4A clone with BamHI digestion and was re-cloned into BamHI-linearized pCAM-M4A-A0.4mer to generate a partial dimeric construct, designated as pCAM-M4A-A1.4mer (Fig. 1a).
Fig. 1.
Schematic diagram showing design of infectious constructs. a Partial tandem repeat construct to genome of croton yellow vein mosaic virus (CYVMV), b dimeric construct to genome of croton yellow vein mosaic betasatellite (CroYVMB). Confirmation of the constructs by restriction digestion for c CYVMV using XbaI, and d CroYVMB using EcoRI
A CroYVMB molecule (Accession No. JX270685) was earlier cloned (M4β) [28]. The genome of CroYVMB was amplified with a mutated forward primer β-01* (5′tgtaccactacgctacgag3′, T in place of G in restriction site region of KpnI of the universal β-01 primer) and β-02 universal primer [2] using the M4β clone as a template. This amplified product was then cloned into pGEMT easy vector through blunt end ligation to create pGEMT-M4β. The 1.3 kb betasatellite was released from pGEMT-M4β clone through digestion with SacI and KpnI and cloned into pCAMBIA 2300 vector in between the same restriction site, and designated as pCAM-M4β-1.0mer. For inserting another copy of betasatellite, a full-length betasatellite (ca. 1.3 kb) was released as KpnI fragment from M4β and was re-cloned in KpnI-linearized pCAM-M4β-1.0mer. The resulting complete dimeric construct was then designated as pCAM-M4β-2.0mer (Fig. 1b). The tandem orientation of pCAM-M4A-A1.4mer and pCAM-M4β-2.0mer was confirmed by restriction digestion with XbaI, and EcoRI, respectively.
Agroinoculation
The pCAM-M4A-A1.4mer and pCAM-M4β-2.0mer was mobilized into the EHA105 strain of Agrobacterium tumifacience by freeze–thaw method [33]. Similarly, pCAMBIA 2300 were also transformed. Agrobacterium containing these constructs were grown separately in LB broth containing kanamycin and rifampicin at 28 °C for 24 h with 150 rpm. The cells were harvested and re-suspended in agroinfiltration buffer [150 µM Acetosyringone, 10 mM MgCl2, 10 mM 2-(N-morpholino) ethane sulfonic acid (MES, pH 5.7)]. The resuspended culture was further incubated at 28 °C for 1–3 h with 150 rpm to obtain a final O.D of 0.6. The culture containing pCAM-M4A-A1.4mer was infiltrated either alone or co-inoculated with pCAM-M4β-2.0mer into the lower surface of leaves of N. benthamiana, N. glutinosa and N. tabacum plants using 2 ml syringe by pressure infiltration method [13]. All the inoculations were performed in three independent sets and in each case, a mock-inoculation with a culture containing pCAMBIA2300 alone was included as negative control. The agroinoculated seedlings were grown in an insect-free glasshouse for 30 days and observations were recorded periodically.
Detection of viral DNA
Total genomic DNA was extracted by CTAB method [5] from newly emerged leaves of agroinoculated plants of N. benthamiana, N. glutinosa and N. tabacum showing typical symptoms. To detect the presence of CYVMV genome in the agroinoculated plants, a specific primer set [BM90F and BM82R] [27], which is expected to produce ca. 750 bp amplicon, was used. To detect the betasatellite in CroYVMB co-inoculated plants, a CroYVMB specific primer set [BM534F and BM535R] designed by the multiple alignments of full-length betasatellite sequences of CroYVMB isolates and other betasatellite sequences available in the database. The specificity of the primers was validated through online Primer Blast tool [34].
Designing of CYVMV-replicon vector
A multiple cloning sites (MCS) cassette containing the double 35S promoter, restriction enzyme sites (SmaI, KpnI, BamHI, EcoRI) and NOS terminator was amplified from the pRI101 vector (Takara, Japan) with the primers BM 638F and BM 639R. The primers were designed in such a way that the 5′ end of forward primer (BM 638F) contains SpeI and MluI while that of reverse primer (BM 638F) contains HindIII and BsrGI restriction enzyme sites. The PCR mastermix contained 4 µl of 5× reaction buffer, 1.6 µl of 2.5 mM dNTP mix, 0.8 µl each of forward and reverses primers (1.0 micromoles), 1.0 U of Phusion Taq polymerase (NEB, USA) and DNase-free water to make up the volume of 20 µl reaction mixtures. The PCR was performed in a thermocycler, Verti (Invitrogen, USA) with the following thermocycling programme: denaturation at 98 °C for 30 s followed by 40 cycles, each consisting of denaturing at 98 °C for 10 s, primer annealing at 52 °C for 20 s and synthesis at 72 °C for 40 s/kb followed by one cycle of final extension at 72 °C for 10 min. The amplified product was purified with Wizard® SV Gel and PCR Cleanup System (Promega, USA). The purified amplicon was cloned into pGEM®-T Easy Vector Systems (Promega, USA) as per manufacture’s protocol. The clone (pGEM-MCS) was confirmed by restriction digestion and sequencing.
Partial genome of CYVMV [par-CYVMV, containing intergenic region (IR), C1, C2, C3 and C4 ORFs] was amplified from the CYVMV infectious clone using primers BM 640F (with SpeI site) and BM 641R (with MluI site). The IR was also amplified from the CYVMV infectious clone using primers BM 642F (with HindIII site) and BM 643R (with BsrGI site). The amplified products were cloned separately into the pGEM®-T Easy Vector Systems (Promega, USA) as per manufacture’s protocol. Partial genome clone (pGEM-par-CYVMV) and intergenic region clone (pGEM-IR) were confirmed by restriction digestion and sequencing.
To assemble all these clones, first the internal MCS of pCAMBIA 2300 vector was removed by digestion with EcoRI and HindIII and the sites were made blunt end using Quick blunting kit (NEB, USA). The new MCS-cassette was re-amplified from pGEM-MCS using Phusion Taq Polymerase and the amplicon was cloned into the blunt ended pCAMBIA2300 vector to generate pCAM-MCS. The intergenic region was released from pGEM-IR clone with HindIII, BsrGI enzymes and cloned into pCAM-MCS to generate pCAM-MCS-IR. Finally, the partial CYVMV genome was released from the pGEM-par-CYVMV clone with SpeI and MluI digestion and cloned into pCAM-MCS-IR at SpeI and MluI site to generate the replicon vector (pCro). Finally the orientation of different sub-fragments of pCro clone was confirmed by sequencing with AC3F and M13R primer (Table 1). The schematic diagram (Fig. 2) illustrates the detail cloning steps.
Table 1.
Details of the primers used for amplification of different components of CYVMV, CroYVMB, MCS cassette and GFP
| Primer name | Primer sequence (5′-3′) | PCR template | Location | Nucleotide co-ordinates | Amplicon size (kb) |
|---|---|---|---|---|---|
| BM90F | ATGTCGAAGCGTCCAGCAGAT | CYVMV genome | V2 | 302-322 | 0.75 |
| BM82R | TACAGAATCGTAGAAGTAA | Do | V1 | 1045-1063 | |
| BM 638F | CGACGCGTTGTGCGCAATACACTAC | Do | IR | 122-141 | 1.70 |
| BM 639R | GGACTAGTTTAATAAAGATTGAATTTTATTG | Do | C3 | 1075-1094 | |
| BM 640RF | GGTGTACAATTGAATTGGGGACACTCA | Do | IR | 2613-2631 | 0.20 |
| BM 641FR | CCCAAGCTTTGTGCGCAATACACTACTTG | Do | IR | 122-141 | |
| BM 652F | ACTATCACCCTCAATCACTATAC | Do | C1 | 1944-1966 | 3.70 |
| BM 653R | AGAACGGGCAAGACGATG | Do | C1 | 1926-1943 | |
| BM 534F | CARTCATATCCTCCTSYTTGAATTC | CroYVMB | Beta C1 | 312-336 | 0.26 |
| BM 535R | CATATATCAGAATGAGACGGGKTTG | Do | AT rich region | 745-770 | |
| BM 642F | ACGCGTGGACTAGTTTGCATGCCTGCAGGTCC | pRI101 | 35S Promoter | 9009-8991 | 1.30 |
| BM 643R | TGTACACCCAAGCTTCAGGAAACAGCTATGACCAT | pRI101 | NOS terminator | 7701-7682 | |
| BM 650F | CGGGGTACCGTAGATCTGACTAGTAAAGG | pCAMBIA1302 | GFP | 4-23 | 0.77 |
| BM 651F | CGGAATTCGCTAGCTTTGTATAGTTCAT | pCAMBIA1302 | GFP | 713-733 | |
| BM AC3F | CAACCCCTATGTGTTTACAA | CYVMV genome | C3 | 1284-1303 |
Bold and italic indicate position of a restriction enzyme site
Fig. 2.
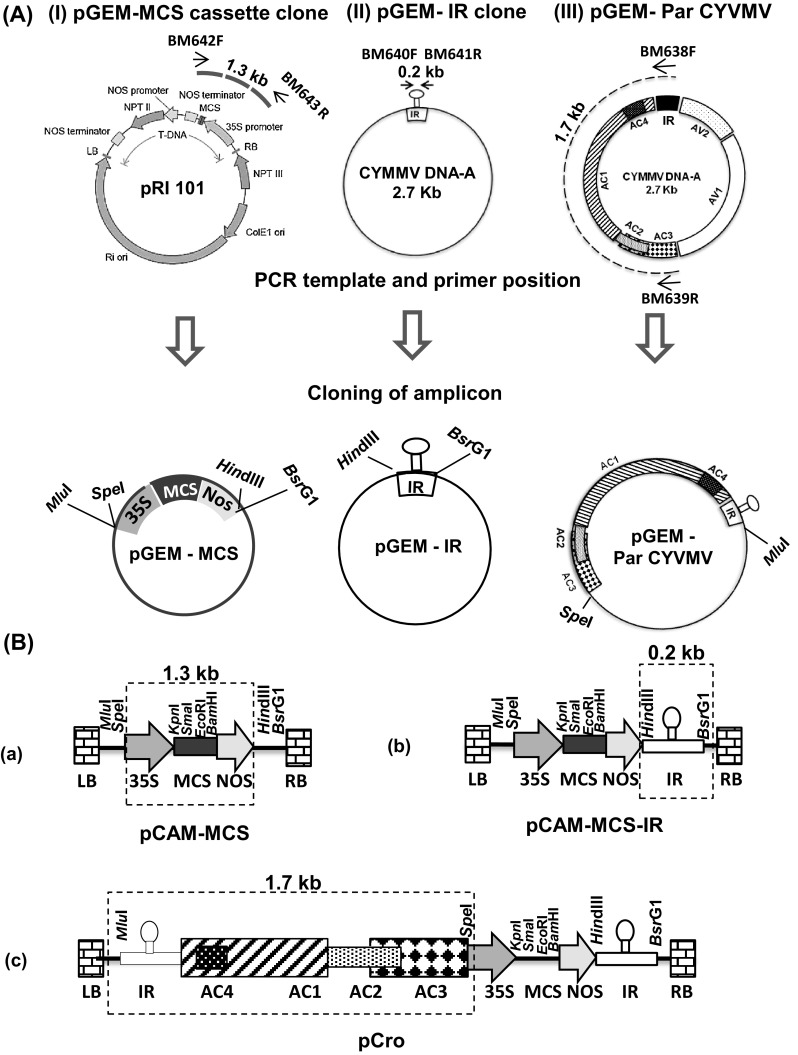
Schematic representation of the design of pCro replicon vector based on genome of croton yellow vein mosaic virus (CYVMV). a Amplification and cloning of i MCS cassette, ii intergenic region (IR) and iii partial genome of CYVMV (par-CYVMV) in pGEMT-easy vector. Template for PCR, primer name, their positions and expected amplicon sizes were depicted in the upper panel. b Sequential sub-cloning of i MCS cassette, ii IR and iii par-CYVMV from pGEMT vector to a blunted pCAMBIA2300 vector to develop pCro vector
Cloning of GFP in CYVMV replicon vector
The GFP gene was amplified from the pCAMBIA1302 using primers BM 650F (with KpnI site) and BM 651R (with EcoRI site) following the PCR conditions mentioned earlier. The amplified GFP gene was cloned into pCro replicon vector at KpnI and EcoRI sites to generate pCro-GFP.
Plant inoculation and GFP expression analysis
The pCro and pCro-GFP were mobilized separately into the EHA105 strain of A. tumifacience and agroinfiltration was carried out in N. tabacum as described earlier. The plant genomic DNA was isolated from the pCro and pCro-GFP-infiltrated N. tabacum leaf using DNeasy Plant Mini Kit (Qiagen, USA) as per manufacturer’s protocol. After plant inoculation, episome formation from the agro-constructs of pCro and pCro-GFP was assessed through inverse PCR using outward primer pairs BM 640R/BM 642F and BM 652F/BM 653R, respectively (outward arrows in Fig. 5a). 200 ng of total plant DNA from the agroinfiltrated plants was used in inverse PCR keeping all other PCR parameters same as described earlier. Expression of green fluorescent protein (GFP) was observed through the epi-fluorescence analysis of infiltrated leaf at 5 days post inoculation (dpi) under the UVL-56 illuminating ultraviolet (UV) light at 365 nm (UV product, Upland, CA) in the dark room and photographed with a digital camera.
Fig. 5.
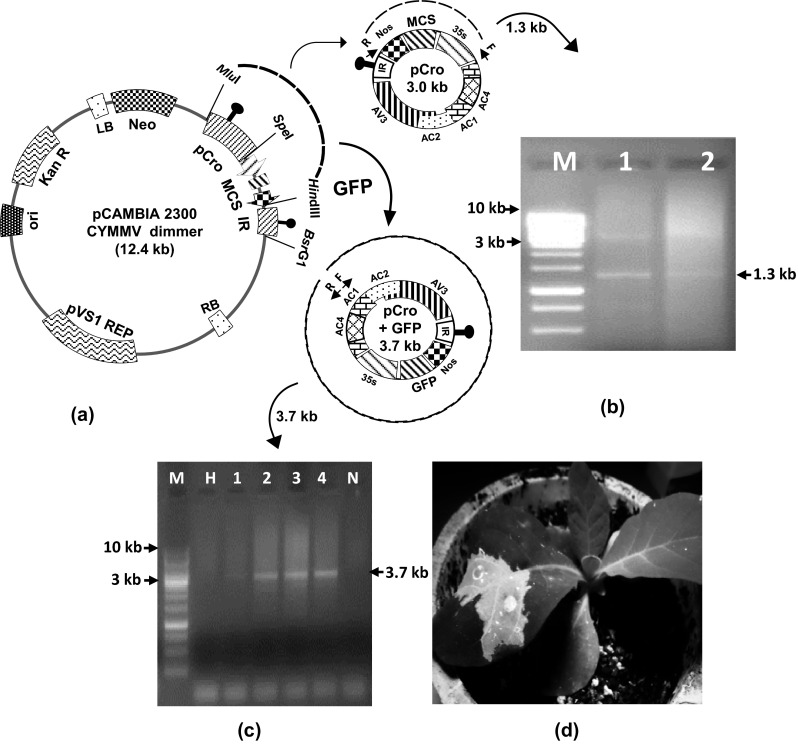
Expression of GFP in N. tabacum plant using pCro vector. a Schematic representation of episomal release of pCro and pCro-GFP from binary vector backbone. Inverse primer positions are indicated as small arrows within the episomal fragments, b detection of pCro episome from agroinoculated plants by inverse-PCR which yielded a 1.3 kb amplicon, c detection of pCro-GFP episome from agroinoculated plants by PCR which yielded a 3.7 kb amplicon. M 1 kb ladder, N negative control, H healthy control; d epifluorescence of GFP from N. tabacum under UV light
Results
Infectivity analysis of the agro-constructs
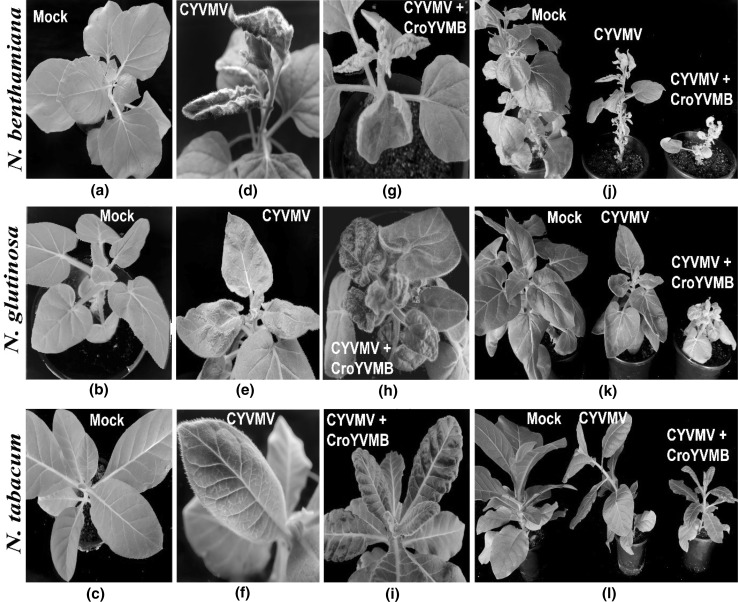
Restriction digestion of pCAM-M4A-A1.4mer with XbaI and pCAM-M4β-2.0mer with EcoRI, released 2.7 and 1.3 kb fragments, respectively (Fig. 1c, d) and thus confirmed the tandem orientation of the constructs. The result of the infectivity experiment was summarized in Table 2. In every set of experiment, more number of plants of N. benthamiana developed symptom compared to other two species of Nicotiana. Inoculation of pCAM-M4A-A1.4mer alone produced leaf curling, leaf rolling and vein thickening symptoms on N. benthamiana at 10–13 dpi (Fig. 3d). In N. glutinosa and N. tabacum it produced mild puckering and vein clearing symptoms, respectively (Fig. 3e, f). The incubation time in N. tabacum is more (30–35 dpi) than that in N. glutinosa (15–20 dpi) (Table 2). Co-inoculation of pCAM-M4A-A1.4mer with pCAM-M4β-2.0mer on N. benthamiana, N. glutinosa and N. tabacum resulted in severe leaf curl and stunting symptoms (Fig. 3j–l). The incubation time was 8–10, 10–12 and 25–30 dpi for N. benthamiana, N. glutinosa and N. tabacum, respectively when both the constructs were co-inoculated. Plants inoculated with pCAMBIA2300 alone, however, did not produce any symptoms.
Table 2.
Agroinfectivity analysis of the cloned DNA of croton yellow vein mosaic virus (CYVMV) and croton yellow vein mosaic betasatellite (CroYVMB) on different Nicotiana spp.
| Constructa | Plant | No. of symptomatic plants out of ten no. inoculated | dpi | Systemic symptoms | ||
|---|---|---|---|---|---|---|
| E1 | E2 | E3 | ||||
| CYVMV | NB | 10 | 8 | 8 | 10–13 | Leaf curling, leaf rolling and vein thickening |
| NG | 6 | 7 | 5 | 15–20 | Mild puckering on leaves | |
| NT | 2 | 1 | 4 | 30–35 | Vein clearing | |
| CYVMV + CroYVMB | NB | 10 | 10 | 9 | 8–10 | Severe leaf curling, leaf rolling, vein thickening and stunting |
| NG | 4 | 8 | 7 | 10–12 | Severe leaf curling with puckering, smalling of leaves, stunting | |
| NT | 4 | 7 | 6 | 25–30 | Puckering on leaves and stunting | |
| pC2300 (mock inoculation) | NB | 0 | 0 | 0 | – | No symptoms |
| NG | 0 | 0 | 0 | – | No symptoms | |
| NT | 0 | 0 | 0 | – | No symptoms | |
NB, Nicotiana benthamiana; NG, N. glutinosa; NT, N. tabacum; dpi, days post inoculation; E, experiment number
aCYVMV: a partial dimer was used, CroYVMB: croton yellow vein mosaic betasatellite dimer was used. Constructs were mobilized into Agrobacterium tumefaciens EHA105 strain
Fig. 3.
Agroinfectivity of croton yellow vein mosaic virus (CYVMV) and croton yellow vein mosaic betasatellite (CroYVMB) on Nicotiana benthamiana, N. glutinosa and N. tabacum. a–c Mock inoculated plants showing no symptom, d–f inoculated plants with CYVMV construct alone showing different symptoms in the different species of tobacco, g–i inoculated plants with CYVMV DNA-A and betasatellite CroYVMB showing severe symptoms, j–l comparison of overall plant growth and stunting symptoms in the different tobacco species
Detection of CYVMV and CroYVMB in agroinoculated plants
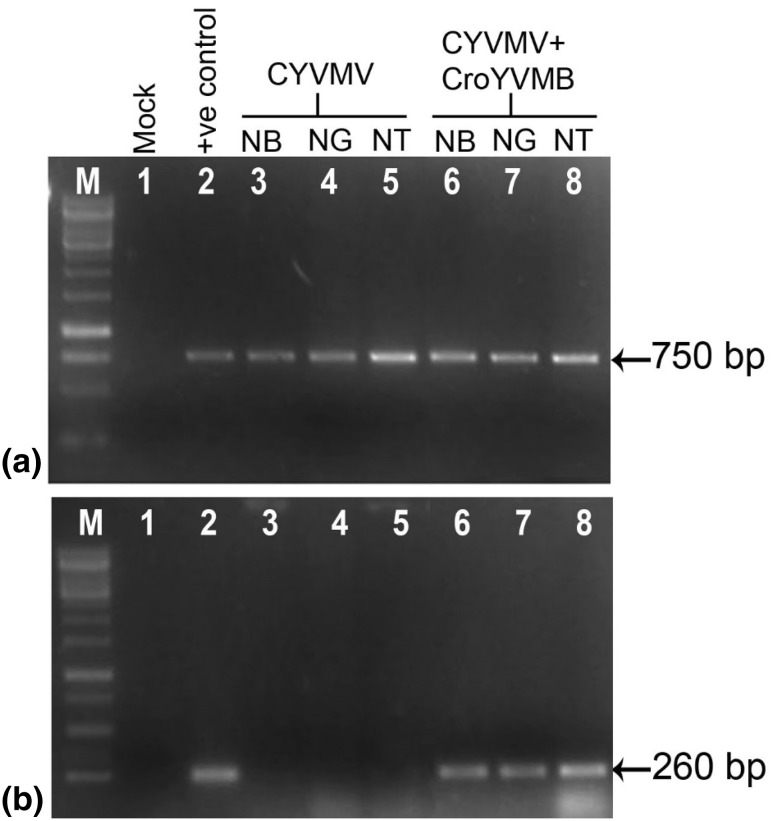
Newly developed symptomatic leaves of plants agroinoculated either with the construct of CYVMV alone or in combination with CroYVMB showed expected 750 bp amplicon specific to CYVMV (Fig. 4a), indicating replication and systemic distribution of the virus. Plants, which were co-inoculated with betasatellite produced CroYVMB specific 260 bp amplicon (Fig. 4b). However, no amplification was observed in the plants inoculated either with CroYVMB alone (data not shown) or plants inoculated with binary vector, pC2300.
Fig. 4.
PCR confirmation of the presence of genome of a CYVMV and b CroYVMB in agroinoculated Nicotiana benthamiana (NB), N. glutinosa (NG) and N. tabacum (NT) plants. M marker; lane 1 mock inoculated plant; lane 2 positive control for CYVMV and CroYVMB; lane 3–5 amplification of CYVMV (for a) or CroYVMB (for b) from NB, NG and NT plants, respectively inoculated with only CYVMV; lane 6–8 amplification of CYVMV (for a) or CroYVMB (for b) from NB, NG and NT plants, respectively inoculated with CYVMV and CroYVMB
CYVMV-based replicon vector
PCR amplification of MCS cassette, IR and partial CYVMV genome yielded 1.3, 0.2 and 1.7 kb amplicons, respectively. The strategy adopted to construct CYVMV replicon has been described in detail in the “Materials and methods” section. After sequential cloning of these components into a blunt pCAMBIA2300 vector, orientations of these fragments were confirmed by appropriate restriction digestion and by PCR using appropriate primers (data not shown). The final resulting plasmid, designated as pCro, is depicted in Fig. 2b(iii), showing the various restriction sites and the positions of the cloned fragments.
In-planta expression of GFP through CYVMV replicon
Amplification of GFP gene resulted in an amplicon of 0.77 kb, which was cloned in the pCro vector. After agroinoculation of pCro plasmid either alone or with GFP (pCro-GFP) into plants, no visible symptom of the virus is produced. However, a circular episome of 3.0 kb for pCro and 3.7 kb for pCro-GFP were excised out (Fig. 5a) and replicates independently in leaves for a period of time. These replicon episomes were detected by inverse PCR, which resulted in 1.3 and 3.7 kb amplicons for pCro and pCro-GFP, respectively in only the inoculated portion of the leaf at 5 dpi and no such amplicon was observed in healthy control (Fig. 5b, c). The epifluorescence of GFP was observed in the pCro-GFP inoculated leaves under the UV lamp at 5 dpi (Fig. 5d).
Discussion
Highly infectious binary vector based constructs to genome of CYVMV and CroYVMB were developed and agro infectivity of these constructs was demonstrated in this study. Infectivity of the agro-mobilized constructs of CYVMV and CroYVMB was assessed on N. benthamiana, N. glutinosa and N. tabacum based on symptom production and PCR analysis. Partial tandem repeat construct of genome of CYVMV is infectious and could produce leaf curl symptoms alone. Co-inoculation with the betasatellite reduced the incubation time for symptom development and enhanced the symptom severity. This study indicated that CYVMV was a typical monopartite begomovirus and the betasatellite was involved only to modulate the symptom severity. However, earlier studies indicated that both CYVMV and CroYVMB are essential for production of yellow vein mosaic symptom in C. bonplandianum [27]. There are two types of betasatellite associated monopartite begomoviruses depending upon whether or not the cognate betasatellite molecules are essential for inducing symptoms [8]. In the majority of these begomoviruses, betasatellite molecules are necessary for symptom development [3, 8, 14, 29]. In the second group, betasatellite is dispensable [17, 30]. The present study and earlier observation suggested that in the case of CYVMV, the requirement of betasatellite for induction of symptom is host dependent and CYVMV is capable of inducing symptom alone in at least three different tobacco species.
In this study, a CYVMV genome-based replicating shuttle vector, pCro, was constructed and tested for its ability to express GFP in the plant. The addition of an MCS under the control of a double 35S promoter increases the opportunity of cloning different genes and directs their translation. Efforts to develop geminiviral replicons for gene expression vector began long back, but the genome size constraints have hindered the use of full viruses for heterologous protein expression. Virus genome size constraints are imposed by cell-to-cell movement through plasmodesmata and not by replication [6]. Therefore, virus genes involved in encapsidation and cell-to-cell movement can be deleted to increase the cargo capacity for large heterologous sequences. Deletion of coat protein in some bipartite begomoviruses has been employed to insert foreign sequence up to 800 nt [22]. Geminivirus replicon vector was also developed by deleting AV2/V2 and CP genes of other geminiviruses [11, 21, 24, 25]. In this study, we deleted the V2 and V1 gene of CYVMV and retained the elements required for rolling-circle replication. This replicon based vector enabled us to simultaneously amplify and expresses foreign gene in high-copy number by rolling-circle replication. Due to the absence of the V2 and V1 genes, the vector is incapable of spreading and developing disease throughout the host plant. The V2 gene has been shown to be involved in movement and it acts as a potent silencing suppressor in other begomoviruses [26, 35]. Deletion of V2 may hinder the rate of virus multiplication which needs to be studied in future. Additionally, lack of CP eliminates any potential for insect-based transfer of the geminivirus replicon between plants, and thus ensures the biosafety of such replicon vector.
In summary, we have established that agro construct of CYVMV genome alone is highly infectious in the different tobacco species. Using this infectious construct, a CYVMV-based replicon vector was developed for transient expression of GFP in plants. Such monopartite begomovirus based vector is rare and will help in the production of vaccine and other pharmaceuticals in the plant. As the CYVMV has a large host range, the system that we are presenting here will be suitable for expressing proteins in many plant species. This replicon vector has obvious applications in the area of functional genomics either as an overexpression or as a silencing system. Based on this basic replicon vector, in future, improvement can be made to increase its efficiency and distribution throughout the plants.
Acknowledgments
The financial support from National Agricultural Science Fund, ICAR is thankfully acknowledged.
References
- 1.Bi H, Aileni M, Zhang P. Evaluation of cassava varieties for cassava mosaic disease resistance jointly by agroinoculation screening and molecular markers. Afr J Plant Sci. 2010;4:330–338. [Google Scholar]
- 2.Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. Universal primers for the PCR-mediated amplification of DNA-β: a molecule associated with some monopartite begomoviruses. Mol Bio-Technol. 2002;20:315–318. doi: 10.1385/MB:20:3:315. [DOI] [PubMed] [Google Scholar]
- 3.Briddon RW, Mansoor S, Bedford ID, Pinner MS, Saunders K, Stanley J, Zafar Y, Malik KA, Markham PG. Identification of DNA components required for induction of cotton leaf curl disease. Virology. 2001;285:234–243. doi: 10.1006/viro.2001.0949. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q, He J, Phoolcharoen W, Mason HS. Geminiviral vectors based on bean yellow dwarf virus for production of vaccine antigens and monoclonal antibodies in plants. Hum Vaccines. 2011;7:331–338. doi: 10.4161/hv.7.3.14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 6.Gilbertson RL, Sudarshana M, Jiang H, Rojas MR, Lucas WJ. Limitations on geminivirus genome size imposed by plasmodesmata and virus-encoded movement protein: insights into DNA trafficking. Plant Cell. 2003;15:2578–2591. doi: 10.1105/tpc.015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gronenborn B, Gardner RC, Schaefer S, Shepherd RJ. Propagation of foreign DNA in plants using cauliflower mosaic virus as vector. Nature. 1981;294:773–776. doi: 10.1038/294773a0. [DOI] [PubMed] [Google Scholar]
- 8.Guo W, Jiang T, Zhang X, Li G, Zhou X. Molecular variation of satellite DNA beta molecules associated with Malvastrum yellow vein virus and their role in pathogenicity. Appl Environ Microbiol. 2008;74:1909–1913. doi: 10.1128/AEM.02461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagen C, Rojas MR. Biology and molecular characterization of cucurbit leaf crumple virus, an emergent cucurbit-infecting begomovirus in the Imperial Valley of California. Plant Dis. 2008;92(5):781–793. doi: 10.1094/PDIS-92-5-0781. [DOI] [PubMed] [Google Scholar]
- 10.Hayes RJ, Coutts RHA, Buck KW. Stability and expression of bacterial genes in replicating geminivirus vectors in plants. Nucl Acids Res. 1989;17:2391–2403. doi: 10.1093/nar/17.7.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Z, Chen Q, Hjelm B, Arntzen C, Mason H. A DNA replicon system for rapid high-level production of virus-like particles in plants. Biotechnol Bioeng. 2009;103:706–714. doi: 10.1002/bit.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Z, Phoolcharoen W, Lai H, Piensook K, Cardineau G, Zeitlin L, Whaley KJ, Arntzen CJ, Mason HS, Chen Q. High-level rapid production of full-size monoclonal antibodies in plants by a single-vector DNA replicon system. Biotechnol Bioeng. 2010;106:9–17. doi: 10.1002/bit.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia H, Pang Y, Fang R. Agroinoculation as a simple way to deliver a tobacco mosaic virus-based expression vector. Acta Bot Sin. 2003;45(7):770–773. [Google Scholar]
- 14.Jose J, Usha R. Bhendi yellow vein mosaic disease in India is caused by association of a DNA Beta satellite with a begomovirus. Virology. 2003;305:310–317. doi: 10.1006/viro.2002.1768. [DOI] [PubMed] [Google Scholar]
- 15.Khan MS, Tiwari AK, Ji SH, Chun SC. First report of a croton yellow vein mosaic virus (CYVMV) associated with tomato leaf curl disease in India. J Phytopathol. 2015;163:777–779. doi: 10.1111/jph.12319. [DOI] [Google Scholar]
- 16.Kim KI, Sunter G, Bisaro DM, Chung IS. Improved expression of recombinant GFP using a replicating vector based on Beet curly top virus in leaf-disks and infiltrated Nicotiana benthamiana leaves. Plant Mol Biol. 2007;64:103–112. doi: 10.1007/s11103-007-9137-z. [DOI] [PubMed] [Google Scholar]
- 17.Li ZH, Xie Y, Zhou XP. Tobacco curly shoot virus DNAβ is not necessary for infection but intensifies symptoms in a host dependent manner. Phytopathology. 2005;95:902–908. doi: 10.1094/PHYTO-95-0902. [DOI] [PubMed] [Google Scholar]
- 18.Malathi VG, Surendranath B, Nagma A, Roy A. Adaptation of new hosts shown by the cloned components of Mungbean yellow mosaic India virus causing cowpea golden mosaic in Northern India. Can J Plant Pathol. 2005;27:439–447. doi: 10.1080/07060660509507243. [DOI] [Google Scholar]
- 19.Mandal B, Varma A, Malathi VG. Systemic infection of Vigna mungo using the cloned DNAs of the black gram isolate mungbean yellow mosaic geminivirus through agroinoculation and transmission of the progeny virus by whiteflies. J Phytopathol. 1997;145:503–510. doi: 10.1111/j.1439-0434.1997.tb00358.x. [DOI] [Google Scholar]
- 20.Matzeit V, Schaefer S, Kammann M, Schalk HJ, Schell J, Gronenborn B. Wheat dwarf virus vectors replicate and express foreign genes in cells of monocotyledonous plants. Plant Cell. 1991;3:247–258. doi: 10.1105/tpc.3.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mor TS, Moon YS, Palmer KE, Mason HS. Geminivirus vectors for high-level expression of foreign proteins in plant cells. Biotechnol Bioeng. 2003;81:430–437. doi: 10.1002/bit.10483. [DOI] [PubMed] [Google Scholar]
- 22.Muangsan N, Robertson D. Geminivirus vectors for transient gene silencing in plants. Methods Mol Biol. 2004;265:101–115. doi: 10.1385/1-59259-775-0:101. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas J, Baltes JG, Tomas C, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26:151–163. doi: 10.1105/tpc.113.119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer KE, Thomson JA, Rybicki EP. Generation of maize cell lines containing autonomously replicating maize streak virus-based gene vectors. Arch Virol. 1999;144:1345–1360. doi: 10.1007/s007050050591. [DOI] [PubMed] [Google Scholar]
- 25.Pandey P, Choudhury NR, Mukherjee SK. A geminiviral amplicon (VA) derived from Tomato leaf curl virus (ToLCV) can replicate in a wide variety of plant species and also acts as a VIGS vector. Virol J. 2009;6:152. doi: 10.1186/1743-422X-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priyadarshini CGP, Ambika MV, Tippeswamy R, Savithri HS. Functional characterization of coat protein and V2 involved in cell to cell movement of cotton leaf curl kokhran virus-Dabawali. PLoS One. 2011;6(11):e26929. doi: 10.1371/journal.pone.0026929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pramesh D, Mandal B, Phaneendra C, Muniyappa V. Host range and genetic diversity of croton yellow vein mosaic virus, a weed-infecting monopartite begomovirus causing leaf curl disease in tomato. ArchVirol. 2013;158:531–542. doi: 10.1007/s00705-012-1511-8. [DOI] [PubMed] [Google Scholar]
- 28.Roy A, Spoorthi P, Bag MK, Prasad TV, Singh R, Dutta M, Mandal B. A leaf curl disease in germplasm of rapeseed-mustard in India: molecular evidence of a weed-infecting begomovirus–betasatellite complex emerging in a new crop. J Phytopathol. 2013;161:522–535. doi: 10.1111/jph.12102. [DOI] [Google Scholar]
- 29.Saunders K, Bedford ID, Briddon RW, Markham PG, Wong SM, Stanley J. A unique virus complex causes Ageratum yellow vein disease. Proc Natl Acad Sci USA. 2000;97:6890–6895. doi: 10.1073/pnas.97.12.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shilpi S, Kumar A, Biswas S, Roy A, Mandal B. A recombinant Tobacco curly shoot virus causes leaf curl disease in tomato in a north-eastern state of India and has potentiality to trans-replicate a non-cognate betasatellite. Virus Genes. 2015;50:87–96. doi: 10.1007/s11262-014-1141-1. [DOI] [PubMed] [Google Scholar]
- 31.Snehi SK, Raj SK, Khan MS, Prasad V. Molecular identification of a new begomovirus associated with yellow mosaic disease of Jatropha gossypifolia in India. Arch Virol. 2011;156:2303–2307. doi: 10.1007/s00705-011-1118-5. [DOI] [PubMed] [Google Scholar]
- 32.Ward A, Etessami P, Stanley J. Expression of a bacterial gene in plants mediated by infectious geminivirus DNA. EMBO J. 1988;7:1583–1587. doi: 10.1002/j.1460-2075.1988.tb02983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weigel D, Glazebrook J. Transformation of Agrobacterium using the freeze–thaw method. CSH Protoc. 2006;2006(7). doi:10.1101/pdb.prot4666. [DOI] [PubMed]
- 34.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Dong J, Xu Y, Wu J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2012;163:51–58. doi: 10.1016/j.virusres.2011.08.009. [DOI] [PubMed] [Google Scholar]