ABSTRACT
Epithelia provide a crucial protective barrier for our organs and are also the sites where the majority of carcinomas form. Most studies on epithelia and carcinomas use cell culture or organisms where high-resolution live imaging is inaccessible without invasive techniques. Here, we introduce the developing zebrafish epidermis as an excellent in vivo model system for studying a living epithelium. We developed tools to fluorescently tag specific epithelial cell types and express genes in a mosaic fashion using five Gal4 lines identified from an enhancer trap screen. When crossed to a variety of UAS effector lines, we can now track, ablate or monitor single cells at sub-cellular resolution. Using photo-cleavable morpholino oligonucleotides that target gal4, we can also express genes in a mosaic fashion at specific times during development. Together, this system provides an excellent in vivo alternative to tissue culture cells, without the intrinsic concerns of culture conditions or transformation, and enables the investigation of distinct cell types within living epithelial tissues.
KEY WORDS: Epithelia, Zebrafish, In vivo
Summary: Here we introduce tools to easily track, ablate or monitor subsets of cells in the developing zebrafish epidermis, providing an excellent in vivo model system to study a living epithelium.
INTRODUCTION
Epithelia provide an essential protective barrier for the organs they encase. Barrier function is vital to organ function and identity, and therefore, defects in this barrier can lead to a variety of diseases such as asthma and colitis (Hering et al., 2012; Swindle et al., 2009; Xiao et al., 2011; Zeissig et al., 2007). Furthermore, because cells comprising most epithelia continually turn over by cell death and cell division at some of the highest rates in the body (Blanpain et al., 2007; Hooper, 1956; Pellettieri and Sanchez Alvarado, 2007), defects in cell turnover can lead to the most common solid epithelial tumors, or carcinomas. Additionally, epithelial shape changes are central to morphogenetic movements during embryonic development (Gumbiner, 1992; Montell, 2008; Pilot and Lecuit, 2005). For these reasons, mechanisms driving epithelial morphogenesis and carcinoma development are under intensive study. However, cell lines do not replicate the function and behavior of epithelia in vivo and introduce concerns with respect to the genetic alterations required to immortalize cells in culture. Moreover, in vivo live imaging of simple epithelia in vertebrates is difficult because of their limited accessibility. To surmount this, researchers have cultured tissues ex vivo (Chua et al., 2014; Mahe et al., 2013; Schwank et al., 2013), which might alter the native environment, or have introduced windows for imaging or fiber optic cameras (Flusberg et al., 2008; Ritsma et al., 2013), which cause a wound that can also alter any process under investigation.
The epidermis of developing zebrafish, Danio rerio, overcomes these limitations and provides an experimentally accessible and genetically tractable model for studying epithelia. The optical clarity of developing zebrafish makes it ideal for imaging single cell movements in vivo in real time. The developing epidermis is a bilayered epithelium that sits atop a basement membrane during embryonic and early larval stages (Dane and Tucker, 1985; Le Guellec et al., 2004; Webb and Kimelman, 2005) (Fig. 1) and comprises cells that express defined markers also found in mammalian epithelia and share similar basic structure to those coating human organs such as the breast, lung and prostate (Macias et al., 2011; Pignon et al., 2013; Rock et al., 2010). Additionally, the epidermis of the developing zebrafish contains several specialized cell types that allow for the exchange of oxygen, ions and macromolecules, similar to mammalian bilayered epithelia (Hwang, 2009; Jevtov et al., 2014; Schwerte, 2010). Mammalian genes involved in epithelial morphogenesis, maintenance and disease conditions are conserved in zebrafish (de la Garza et al., 2013; Fukazawa et al., 2010; Lee and Kimelman, 2002; Li et al., 2011; Sabel et al., 2009; Sonawane et al., 2009). Further, gene function within the epidermis can be readily probed using mutations, morpholinos and chemical inhibitors. These attributes make the developing zebrafish an ideal system to study epithelial biology.
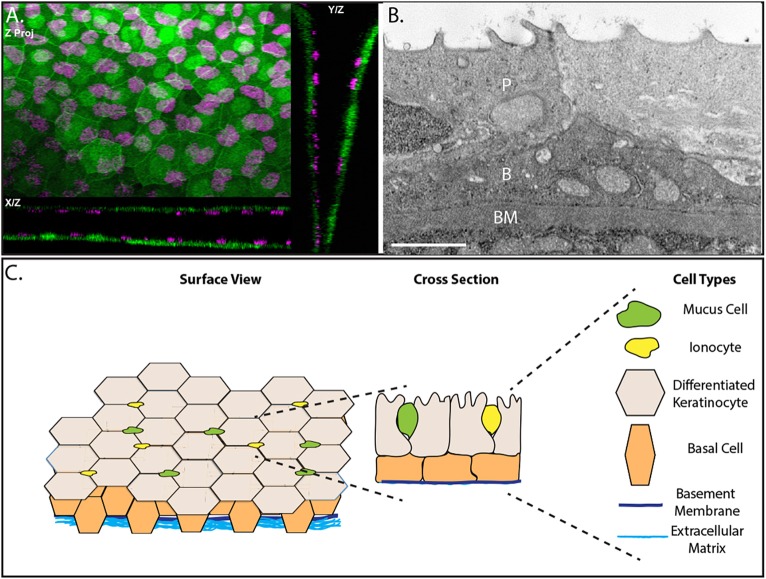
Fig. 1.
The epidermis of developing zebrafish is a bilayer. (A) Confocal maximum intensity projections of an epidermis in 4 day post-fertilization (dpf) Tg(krt4:GFP) (green) transgenic zebrafish with immunostaining for p63 (magenta). (B) A transmission election micrograph of a 4 dpf epidermis. P, periderm; B, basal layer; BM, basement membrane. (C) Schematic of the developing zebrafish epidermis. Scale bar: 2 μm.
Although the zebrafish epidermis is intrinsically easy to image, tools to follow specific cell populations within the epidermis and the ability to perturb gene function in a mosaic manner were previously lacking. Being able to express or disrupt gene function in a subset of epithelial cells has been crucial for discovering numerous fundamental processes in Drosophila (Brand and Dormand, 1995; Fischer et al., 1988; Southall et al., 2008). Using tools established in S. cerevisiae and D. melanogaster (Brand and Dormand, 1995; Giniger et al., 1985), we have created zebrafish epithelial lines using the Gal4 UAS system. Gal4 enhancer trap lines exist in the zebrafish (Davison et al., 2007; Kawakami et al., 2010); however, most screens have focused on the developing nervous system (Asakawa et al., 2008; Satou et al., 2013; Takeuchi et al., 2014). To create an epithelial genetic toolkit, we identified new enhancer trap epithelial lines that can be used to track and ablate different epithelial cell types with spatial and temporal control using gal4 photo-cleavable morpholino oligonucleotides. Together, these tools provide an excellent system to study epithelial development, wound repair, regeneration, pathologies, or any other cell biological process with exquisite molecular and cellular control. Moreover, these tools provide an excellent alternative to cell culture studies that circumvent concerns of transformation and culture conditions.
RESULTS
Identification of GAL4 enhancer lines expressed in distinct cell types of the developing epidermis
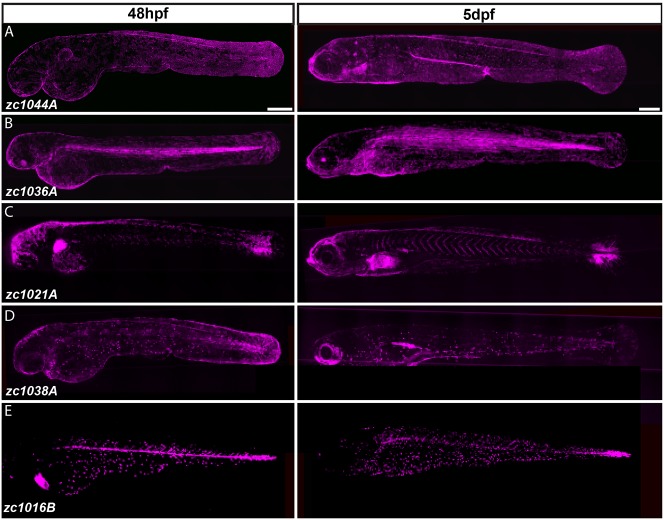
To identify promoters driving expression in diverse cell types of the developing zebrafish epidermis, we screened a collection of Gal4 enhancer trap lines from a large-scale non-biased screen focused on identifying novel neural-specific lines in Danio rerio (Otsuna et al., 2015). The developing zebrafish epidermis is a bilayer during embryonic and larval development that comprises cells similar in function and markers to those comprising mammalian bronchial epithelia (Fig. 1) and acts similarly in oxygen exchange (McLeish et al., 2010; Schwerte, 2010). The basal layer consists of cells that express the epithelial stem cell marker p63 (Bakkers et al., 2002; Lee and Kimelman, 2002), whereas the periderm is an outer superficial layer of differentiated cytokeratin-positive cells (Gong et al., 2002; Wang et al., 2006) that are derived from the enveloping layer (Fukazawa et al., 2010) (Fig. 1A,B). The superficial layer also contains ion transport cells (ionocytes) (Janicke et al., 2007) and secretory mucous-producing cells (Oehlers et al., 2012) that are distributed throughout the tissue (Fig. 1C). We visually screened for expression in these cell types by crossing each Gal4 line to a Tg(UAS-E1b:nfsB-mCherry)c264 line that labels all cells driven by the inserted Gal4 enhancer with red fluorescence (here shown in magenta; Fig. 2A-E). For each line, we screened mCherry expression at 48 hours post-fertilization (hpf), when proliferation is high in both layers of the epidermis and at 5 days post-fertilization (dpf), when proliferation is slow and predominantly in the basal cell layer (Carney et al., 2007). Our analyses identified seven unique lines with reproducible expression in specific epithelial cells within the epidermis. These lines include expression patterns that visually mark the periderm, basal cells, fibroblasts and ionocytes (Table S1). Additionally, two lines express mCherry in multiple epithelial cell and/or tissue types, which we did not characterize further. Some expression patterns also changed during development. For instance, zc1038A showed expression of mCherry within cells at the outer edge of the developing median fin at day 2, but was greatly reduced by day 5 (Fig. 2D).
Fig. 2.
Identification of Gal4 enhancer trap lines expressed in distinct cell types of the developing epidermis. (A-E) Composite images from multiple maximum intensity projections of Gal4 enhancer trap lines driving Tg(UAS-E1b:nfsB-mCherry) (shown in magenta) in 48-hour- and 5-day-old larvae. Scale bars: 250 μm.
To characterize where the different Gal4 lines were expressed, we compared their expression with respect to the superficial keratinocytes of the periderm labeled with GFP by crossing to an existing Tg(krt4:GFP) (Gong et al., 2002) line. The zc1016B line labels a population of superficial cells that reside between the borders of the periderm cells and resemble ionocytes, or cells specific for ion transport across the developing epidermis (Fig. 3A). Analyses of these cells using MitoTracker (Fig. S1) suggest that they represent the ionocytes (Shono et al., 2011), and therefore, we refer to line zc1016B as ‘GET-ionocyte’ (GET, Gal4 enhancer trap). Interestingly, we found that the zc1044A line expresses mCherry in 85% of the krt4:GFP+ periderm cells (98/115 of cells), with variable levels of fluorescence among individual cells (63.2% of the cells expressed mCherry at high levels, 26.3% at an intermediate level, and 10.5% at a low level). Given the pattern of expression, we termed this the ‘GET-periderm’ line (Fig. 3B). The lack of mCherry (Fig. 3B) or Kaede (Fig. 4A) expression in all the periderm cells might reflect that this enhancer expresses in only a subset of periderm cells.
Fig. 3.
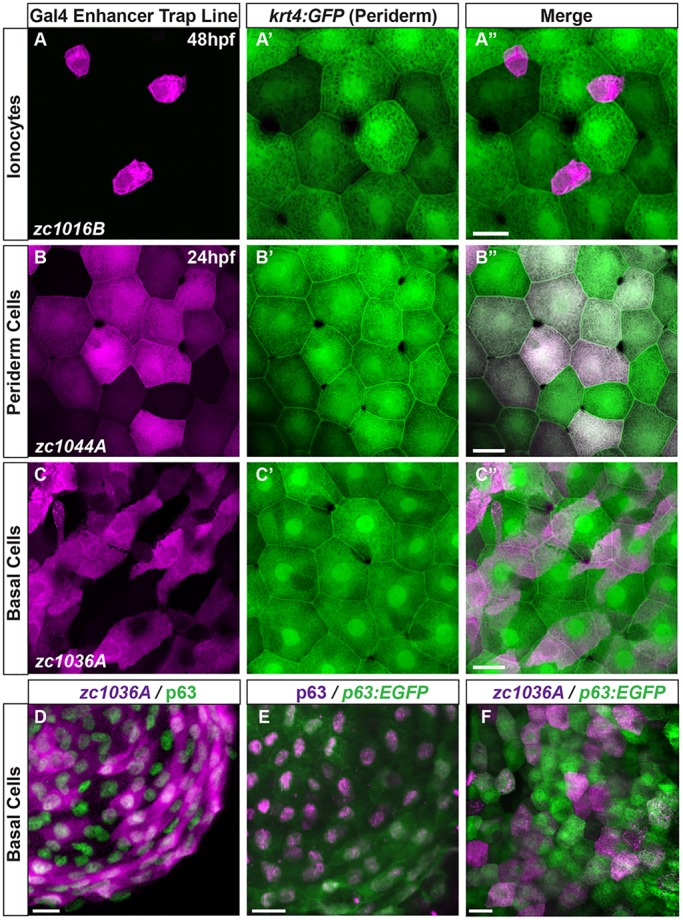
GAL4 enhancer trap lines are expressed in a subset of different epithelial cell types. (A-C) Maximum intensity projections of confocal images demonstrate expression of Gal4 enhancer trap lines (magenta) with respect to the periderm, shown by krt4:GFP expression (green). (D-F) Characterization of the basal cell line with antibody staining for (D) p63 and the GET-basal line (zc1036A), (E) p63 enhancer driving GFP stained with p63 antibody, and (F) p63:EGFP co-localization with the GET-basal line. Scale bars: 25 μm.
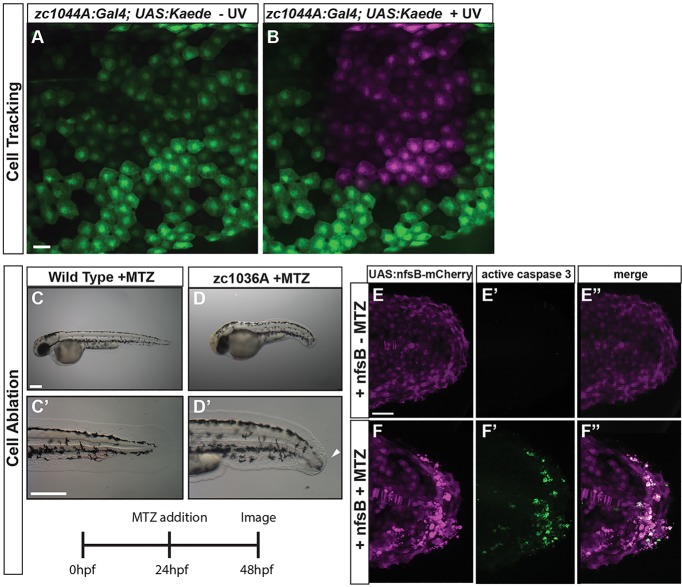
Fig. 4.
Crossing epithelial Gal4 lines to UAS lines that provide tools for developmental studies. (A,B) Cells can be fluorescently labeled and tracked over time with the periderm Gal4 line (zc1044A) driving expression of photo-convertible protein Kaede, Tg(UAS-E1b:Kaede)s1999t. Expression of Kaede is detected as green fluorescence before exposure to blue (UV) light (A), and red (shown here as magenta) after exposure (B). (C-F) The labeled epithelial cells can be readily ablated using the nitroreductase–metronidazole system, Tg(UAS-E1b:nfsB-mCherry). (C-D) Representative bright-field images of wild-type (C) and zc1036A (D) 48 hpf embryos after 24 h treatment with MTZ to ablate the cells. Arrowhead in D′ denotes defects in the developing median fin epidermis. (E-F) The MTZ-exposed zc1036A cells undergo apoptosis (activated caspase-3-positive cells in green) and cause defects in the developing embryo. C-D represent data from a single experiment performed in triplicate. Scale bars: 25 μm in A-B, 250 μm in C-D, 50 μm in E-F.
By contrast, line zc1036A expressed mCherry in a subset of p63-positive basal cells that do not overlap with the krt4:gfp cells (0%, 0/105 cells) but co-label with 78% of cells immunostained with the epithelial stem/progenitor cell marker p63 (Senoo et al., 2007; Yang et al., 1999) (218/277 p63+ cells; Fig. 3C-D). Additionally, we engineered a line that expressed GFP exclusively in all basal epithelial cells using a conserved intron enhancer of the endogenous p63 gene in zebrafish (Antonini et al., 2006, 2015; Pashos et al., 2008) (Fig. S2). Immunohistochemical analyses revealed that 100% (159/159 cells) of the p63:EGFP cells were positive for p63 protein, which localized to the nucleus (Fig. 3E). Crossing the zc1036A Gal4 line to the Tg(p63:EGFP) line showed that UAS:nfsB-mCherry expression colocalized with 65% of p63:EGFP basal cells (77/120 cells; Fig. 3F). Therefore, we named the zc1036A line the ‘GET-basal’ line. Together, these tools allow researchers to specifically label the full population of periderm or basal cell layers or defined subsets of them. To fully characterize the expression pattern for the GET-ionocyte, GET-periderm and GET-basal lines, we also generated a high-resolution atlas across different regions of the animal (Fig. S3A-C). We further characterized our UAS-driven tools using the periderm and basal zebrafish lines, as these cell types are likely to be the most widely used for epithelial cell biology.
UAS lines to study epithelial development
As many developmental studies require the ability to track specific cells and ablate them, we have compiled a number of different UAS effectors to cross to our Gal4 enhancer trap lines to enable cell tracking and ablation. To track cells over time, we crossed the ‘GET-periderm’ line to a Tg(UAS-E1b:Kaede)s1999t line (Davison et al., 2007), to label periderm cells with green fluorescence that can photo-convert upon exposure to blue light and label a subset of cells with red fluorescence (shown as magenta in Fig. 4). By converting the entire larvae, the addition of new cells to the tissue can be followed over time. Cells that are created after photo-conversion will only exhibit green fluorescence within the field of pre-existing cells that express both converted and newly made Kaede protein. Additionally, by limiting the area of exposure to blue light, we can track the fate of a specific population of cells over time (Fig. 4A,B). Using this approach, researchers can track epithelial population dynamics or a targeted subset to study their fates within the developing epidermis.
The Tg(UAS-E1b:nsfB-mCherry)c264 reporter line initially used to screen the Gal4 enhancer trap lines also allows these cells to be targeted for apoptosis upon treatment with metronidazole (MTZ) (Curado et al., 2008; Davison et al., 2007). The Escherichia coli nitroreductase enzyme encoded by the nfsB gene is not normally present in zebrafish and is not toxic on its own, but the addition of MTZ creates a cytotoxic byproduct that causes DNA damage and apoptotic cell death (Curado et al., 2008). Thus, specific cells can be ablated in the developing epidermis. We selectively targeted the basal cells for apoptosis during development, as shown by activated caspase-3 immunostaining that are restricted to mCherry-positive cells, which lead to significant morphological defects in the developing median fin-fold epidermis (Fig. 4D-F). Apoptotic cell death increased in a dose-dependent fashion or with a MTZ single low dose over time (data not shown). By contrast, treating wild-type AB zebrafish with MTZ caused no cell death or other phenotypes (Fig. 4C,E). These two Gal4 lines allow specific epithelial cells to be tracked or ablated to study their roles in development over time.
UAS lines to study cell biology
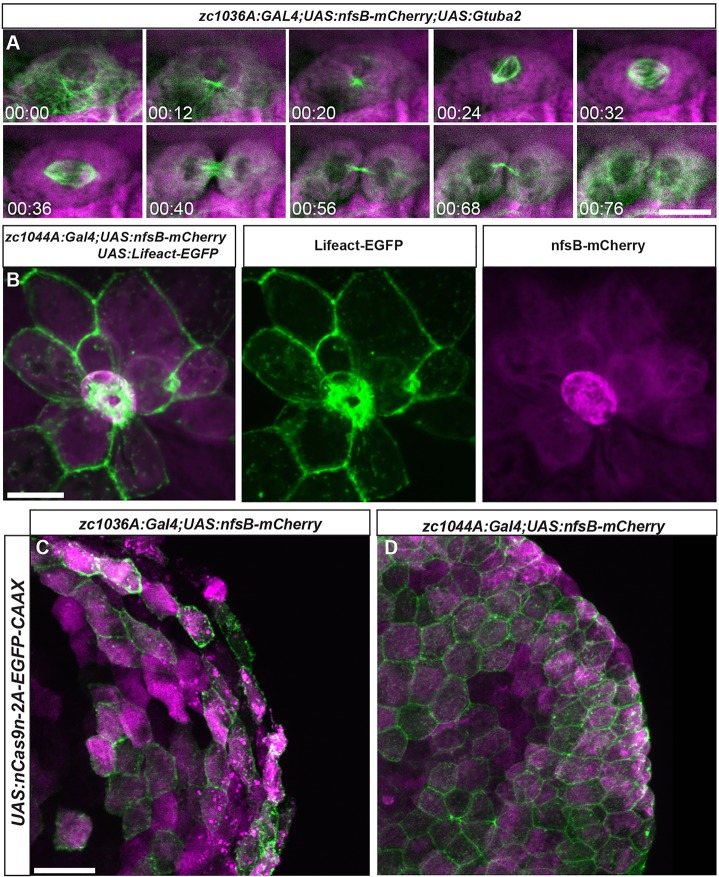
These new epidermal lines, aside from being useful for studying epithelial developmental cell biology, could also be used to replace many cell culture studies by tracking specific fluorescently labeled cytoskeletal proteins. By simply crossing our epidermal zebrafish lines to a growing list of UAS-driven cytoskeletal markers, one can readily follow cell or sub-cellular movements in the periderm or other epidermal cells. For instance, we used spinning-disc confocal microscopy to film basal cells dividing with the GET-basal line crossed to a UAS-GFP-alpha tubulin gene (UAS:Gtuba2) to fluorescently label microtubules (Asakawa and Kawakami, 2010) (Fig. 5A). Basal zebrafish epidermal cell division could be clearly visualized and proceeded with similar kinetics (∼76 min) to those seen in cultured HeLa cells (Mackay et al., 2010). During development this technique could be used to follow mitotic spindle orientation, which can play a crucial role in cell fate decisions (Hernandez and Tirnauer, 2010) and epithelial stratification (Lechler and Fuchs, 2005).
Fig. 5.
Live imaging of cell division and death in the developing epidermis. (A) Still images from a time lapse movie of cell division in the basal Gal4 line (zc1036A) driving expression of fluorescently labeled microtubules Tg(UAS:Gtuba2). (B) Image of a cell being extruded from the epidermis of a live zebrafish in the periderm Gal4 line (zc1044A) driving expression of fluorescently labeled F-actin Tg(UAS:Lifeact-EGFP). (C,D) Expression of Tg(UAS:nCas9n-2A-EGFP-CAAX) in zc1036A (C) and zc0144A (D) GAL4 lines. Scale bars: 20 μm in A, 25 μm in B, 25 μm in C,D.
Additionally, we have developed UAS effector lines to fluorescently label F-actin; Lifeact-EGFP (Riedl et al., 2008) and UtrCH-mCherry (Burkel et al., 2007) to track cell division, extrusion, migration and other epithelial movements during development (Table S2, Fig. S4). We created stable transgenic lines that show fine actin-based microridges in periderm cells and enrichment at cell–cell contacts. For example, we can readily follow extrusion of a cell fated to die, which acts to expel cells without disrupting the barrier function of the epidermis (Fig. 5B) (Eisenhoffer et al., 2012; Rosenblatt et al., 2001). These tools allow real-time imaging of subcellular processes that underlie many epithelial cell shape changes required for morphogenetic movements during development or for any cell biological analysis. Additionally, should researchers want to achieve further mosaic expression of these cytoskeletal reporters, they could simply inject the DNA plasmids expressing UAS:fluorescent tag–cytoskeletal protein into the Gal4 line of interest.
Our new Gal4 lines can also be used to investigate distinct subsets of epithelial cells after genetic manipulation. The CRISPR/Cas9 system (Cong et al., 2013; Haurwitz et al., 2010; Mali et al., 2013) has been shown to reliably insert or delete DNA at precise sites within the zebrafish genome (Hwang et al., 2013; Irion et al., 2014). One strategy for tissue-specific genetic manipulation is to control the expression of the Cas9 protein using a known enhancer element (Ablain et al., 2015). Here we created a UAS-driven nuclear-localized Cas9 that has eGFP-CAAX expression from a 2A peptide to facilitate epithelial cell-type specific loss-of-function studies in the zebrafish (Fig. 5C,D). Additionally, this UAS line can be used with any Gal4 line to facilitate genome editing in a wide variety of cells and tissues with the zebrafish.
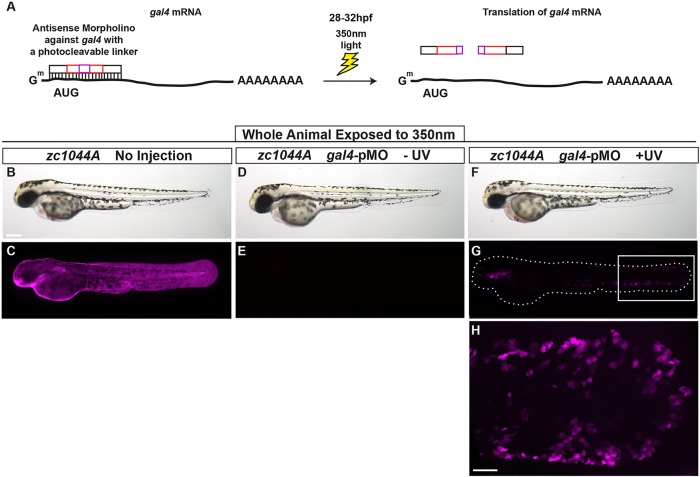
Spatial and temporal control of gene expression using Gal4 photo-cleavable morpholinos
Whereas the Gal4–UAS system can spatially control gene expression of our effector lines, expression of some genes might disrupt embryogenesis. To circumvent this problem and enable expression of genes later in development, we introduced a photo-cleavable morpholino to gal4 (gal4-pMO) (Eisenhoffer et al., 2012; Tallafuss et al., 2012) (Fig. 6A). Using the gal4-pMOs, gene expression can be triggered at later times to entire embryos or to specific cell populations by illuminating only a few cells, as seen in Fig. 4A with Kaede conversion. To drive gene expression at later time points or in specific targeted cells, we injected gal4-pMO into Et(Gal4-VP16)zc1044A;Tg(UAS-E1b:nfsB-mCherry)c264 one-cell-stage embryos and monitored them for fluorescence during development. After 350 nm light exposure to cleave the morpholino and allow Gal4-driven expression, we found that 56% of larvae expressed mCherry within 48 h of light exposure (72 hpf) (Fig. 6B-H). Expression is first seen at the distal edges of the median fin epidermis and in the oral epithelium (Fig. 6G,H). By contrast, >90% of gal4-injected embryos that were not photo-converted had no detectable fluorescence with a standard fluorescence dissecting microscope by 24 hpf (Fig. 6D,E). However, ∼30% of embryos had low levels of fluorescence when examined at higher magnification on a compound microscope, suggesting incomplete knockdown of Gal4-mediated expression. The efficiency of Gal4 activation is dependent on morpholino concentration combined with intensity and duration of the light pulse during conversion. Recovery of Gal4-driven expression in subsets of cells is attributed to the inherent mosaic expression observed in our enhancer trap lines. This approach could also be used in Gal4 lines that show ubiquitous expression throughout the tissue to increase the number of cells that reactivate Gal4 after exposure to UV light. These results show that photo-cleavable morpholinos directed against gal4 can spatially and temporally control gene expression in the epidermis of developing zebrafish using the newly identified lines. These tools will facilitate mosaic and clonal analysis of distinct cell populations within a living epithelial tissue.
Fig. 6.
Spatial and temporal control of Gal4-driven gene expression. (A) The photo-cleavable morpholino directed against gal4 was injected in Et(Gal4-VP16)zc1044A;Tg(UAS-E1b:nsfB-mCherry) transgenic line and embryos were exposed to UV light at 28-32 hpf. (B-H) Representative bright-field (B,D,F) and fluorescent (C,E,G,H) images 48 h after exposure to UV, at 3 dpf. Box in G denotes area of magnification for H. B-H represent data from three independent experiments. Scale bars: 250 μm in B-G, 50 μm in H.
DISCUSSION
A major challenge for epithelial biology studies is analyzing cell behaviors in vivo. The developing zebrafish epidermis provides an ideal system for investigating epithelia, yet tools to mark and track specific cell types were previously lacking. Here we describe novel Gal4 enhancer trap lines that can be used to visualize specific epidermal cells in developing zebrafish to enable time lapse imaging, overexpress genes of interest, and target cells for ablation. Importantly, we identified new lines with expression restricted to the outer periderm or basal cells, two commonly studied cell types in epithelial cell biology. Our methods allow researchers to easily study specific cell populations within a living epithelial bilayer.
By combining new and existing UAS effectors with our Gal4 lines, any biological process in epithelia can be studied at subcellular resolution. For example, using our system, one can now follow or ablate specific epithelial cells during development. Additionally, because of the exquisite subcellular resolution of processes visualized by our UAS-driven fluorescent cytoskeletal lines, we believe that these lines could supplement studies using epithelial cell culture lines and eliminate various concerns about their intrinsic transformed states. Finally, zebrafish epidermis could provide an excellent model for epithelial bilayers, a primary site of malignancy formation in the human body.
An important, yet unexpected, aspect of our Gal4 lines is that they are expressed in subsets of both the periderm and basal layers of the developing zebrafish epidermis. Many studies in epithelia rely on expression or knockdown in a mosaic fashion when trying to resolve whether a protein functions in a cell-autonomous manner, or so that function can be probed in genes that would be lethal if expressed in all cells. Therefore, having these lines express intrinsically in a mosaic fashion could be an additional asset for many epithelial studies. In cases where mosaic expression is not desired, the krt4 and our novel p63 enhancer elements express ubiquitously in the periderm and basal layers, respectively.
The tools for the zebrafish epidermis presented here provide an excellent system to investigate pathologies in epithelial bilayers. For example, our Gal4 enhancer trap lines can be used to manipulate gene expression within specific epithelial cell types, as well as follow their movements and behaviors under physiological conditions and during pathogenesis or carcinogenesis. Recent studies have demonstrated that mutations in conserved tumor suppressor genes or the sphingosine-1-phosphate signaling pathway lead to altered epithelial cell behaviors and hallmarks seen in aggressive tumors (Gu et al., 2015; Marshall et al., 2011; Reischauer et al., 2009), highlighting the utility of the zebrafish epidermis to model the earliest events of carcinoma formation and progression. Our toolset now provides new strategies for loss-of-function studies for the zebrafish epidermis (Fig. 5C,D), and combined with existing mutant lines that exhibit altered epithelial morphology or function (Amsterdam et al., 2004; Carney et al., 2007; van Eeden et al., 1996; Webb et al., 2008), will enable live imaging and in vivo perturbation studies to dissect the cell and molecular changes that promote transition to a disease state in bilayered epithelia.
In conclusion, we have identified five novel Gal4 enhancer trap lines that can drive expression of any gene in different cells of the developing zebrafish epidermis. Using UAS effector lines, we developed methods to track or ablate cells in the periderm, and observe subcellular microtubule and actin dynamics in either cell layer. Finally, we showed that photo-cleavable morpholinos can be used to spatially and temporally control expression of any gene driven by a UAS element in specific epithelial cells. The Gal4 enhancer trap lines described here have been deposited in the Zebrafish International Resource Center (ZIRC) for public distribution (Table S1). Additionally, the UAS effector lines are available for distribution as plasmids (Table S2). Together, these tools provide a set of reagents that will allow researchers to study specific subsets of epithelial cells and their dynamics during development, repair or carcinogenesis and will provide an in vivo alternative to study processes at resolutions typically found in cell culture.
MATERIALS AND METHODS
Zebrafish husbandry and maintenance
Zebrafish, Danio rerio, were maintained at 28.5°C in water with a pH of 7.5, and kept on a 14 h/10 h light/dark cycle according to Westerfield (2007). The zebrafish used in this study were handled in accordance with the guidelines of the University of Utah Institutional Animal Care and Use Committee.
Gal4 enhancer trap screen
To generate the enhancer-trap lines, the Et(Gal4-VP16;myl7:GFP) plasmid in the Tol2 backbone and transposase mRNA was injected into 1-cell-stage developing zebrafish embryos, as described in Otsuna et al. (2015). Briefly, F0 embryos were raised to maturity and then crossed to a Tg(UAS-E1b:nfsB-mCherry)c264 reporter line. The F1 transgenic embryos were identified based on epithelial fluorescent mCherry expression patterns and imaged at 2 and 5 days post-fertilization. Embryos positive for epithelial mCherry expression were raised and then outcrossed to the F3 generation for further characterization.
p63 enhancer cloning and transgene construction
Multiz tracks on the University of California Santa Cruz Genome Browser revealed an intronic region in the zebrafish p63 gene with multiple regions of homology in intron 4 of mouse p63, which contains multiple enhancers that direct tissue-specific expression in transgenic mice (Antonini et al., 2006, 2015; Pashos et al., 2008). The 5564 bp region containing these sequences was amplified with the primers: For, TATTGAACTTGAGCGCAACT; Rev, TCGATATGCCTCTGTACGC. The resulting PCR product was cloned into pDONR221 and subsequently into pGW_cfosEGFP universal expression plasmid (Fisher et al., 2006). The PCR product was also cloned into the Tol2 p5E vector (Kwan et al., 2007).
Generation of UAS reporters
Lifeact-EGFP, Lifeact-mCherry, UtrCH-mCherry and nCas9n were all cloned into the middle entry vector of the Tol2 kit (Tol2 kit plasmid #218) (Kwan et al., 2007). They were then combined with E1b-UAS 5′ and poly A 3′ entry clones into the Tol2 pDEST_CG2 (Tol2 kit plasmid #395) destination vector. The Cas9 middle entry clone was combined with the E1b-UAS 5′ and p3E-2A-EGFP-CAAX-poly A (Tol2 kit plasmid #458) into the pDESTTol2pA2 (Tol2 kit plasmid #394) destination vector. Injections were performed with 25 pg of the purified plasmids along with 50 pg of transposase mRNA into wild-type AB strain, Et(Gal4-VP16)zc1044A;Tg(UAS-E1b:nsfB-mCherry)c264 or Et(Gal4-VP16)zc1036A;Tg(UAS-E1b:nsfB-mCherry)c264 developing zebrafish embryos at the 1-cell-stage. Potential carriers were identified by fluorescence expression and raised to adulthood.
Kaede photo-conversion
Et(Gal4-VP16)zc1044A; Tg(UAS-E1b:Kaede)s1999t embryos were anesthetized and mounted in 1% low melt agarose in E3 medium. Initial images were taken of both the 488 nm (green) and 568 nm (red) channels using a 20× objective. Embryos were then exposed to 350 nm light, with the area of exposure restricted by using a 40× objective. Post-conversion images were then taken of the embryo at 20× using the previous exposure times.
Immunohistochemistry
Zebrafish embryos and/or larvae were fixed and stained according to Eisenhoffer et al. (2012) with primary antibodies against p63 (Ab11149; Abcam, Cambridge, MA; 1:500) or activated caspase-3 (559565; BD Pharmigen, San Jose, CA; 1:700). For staining of mitochondria, embryos were incubated with 500 nM of Mitotracker 488 (M7514; Thermo Fisher Scientific, Waltham, MA) for 30 min in the dark, rinsed and mounted for imaging.
Transmission electron microscopy
Samples were fixed in 3% glutaraldehyde+2% paraformaldehyde+0.1 M sodium cacodylate buffer and treated with 0.1% Millipore-filtered cacodylate buffer tannic acid. They were post-fixed with 1% buffered osmium tetroxide for 30 min, stained in block with 1% Millipore-filtered uranyl acetate. Samples were dehydrated in increasing concentrations of ethanol, infiltrated, embedded in LX-112 medium and polymerized in an oven at 60°C for about 3 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica, Deerfield, IL), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL, Peabody, MA) at an accelerating voltage of 80 kV. Digital images were obtained using AMT Imaging system (Advanced Microscopy Techniques, Danvers, MA).
Live imaging
Embryos or larvae were anesthetized with 0.04% tricaine, mounted in a 10 mm MatTek culture dish with 1% low-melt agarose in E3 embryo medium, and images were then acquired on an inverted Nikon microscope equipped with a Yokugawa spinning disc head, three Coherent solid-state lasers, and an Andor electron-multiplied, cooled CCD camera as in Eisenhoffer and Rosenblatt (2011). Image acquisition and post-processing were performed using Andor iQ software. For high-resolution images of the whole animal at different developmental stages, individual images were acquired that spanned the entirety of the animals in the x, y and z planes using a 10× objective and the tiling/montage feature within the Andor iQ software. Maximum intensity projections were generated from the resulting data and a single composite image was created from the individual panels using the montage feature in Andor iQ.
Photo-morpholino injections and photo-conversion
The photo-cleavable morpholino antisense oligonucleotides used in this study were acquired from Gene Tools, LLC. Et(Gal4-VP16)zc1044A;Tg(UAS-E1b:nsfB-mCherry)c264 one-cell stage zebrafish embryos were injected with 0.2 mM photo-cleavable morpholino oligonucleotides directed against gal4 and then allowed to develop at 28.5°C. At 24 hpf, the embryos were then converted on a Nikon 90iEclipse compound fluorescent microscope using a 10× objective with exposure to 305 nm light for 60 s. Alternatively, embryos were exposed to 350 nm light using the GeneTools lightbox on the highest intensity setting with exposure of 5 min. Either condition resulted in very little toxicity to wild-type uninjected embryos. Sequence for the gal4 antisense photo-cleavable morpholino oligonucleotide: GTTCGATAGATACATGTAGCTTCAT.
Acknowledgements
We thank members of the Rosenblatt laboratory for scientific discussions, suggestions, and comments. We would also like to thank Maurine Hobbs and the staff of the Centralized Zebrafish Resource at the University of Utah for excellent maintenance and care of the zebrafish. We thank Kenneth Dunner Jr. from the High Resolution Electron Microscopy Facility for support with the electron microscopy and associated sample preparation, partially supported by the MD Anderson Cancer Center CCSG P30CA016672.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
G.T.E. and J.R. designed the research. G.T.E., O.E.R., and G.S. performed experiments and G.T.E. analyzed the data. C.D.B, J.L.B., J.L., and D.S.W. generated key reagents and transgenic lines, and provided analysis of the resulting data. H.O., C.-B.C., R.I.D. carried out the initial GAL4 enhancer trap screen. G.T.E. and J.R. wrote the paper, and all authors provided edits.
Funding
This work was supported by National Institutes of Health-National Institute of General Medical Sciences [1R01GM102169-02 and 1DP2OD002056-01 to J.R.]. G.T.E. was supported by American Cancer Society Post Doctoral Fellowship [120464-PF-11-095-01-CSM] and Cancer Prevention Research Institute of Texas (CPRIT) First-Time Tenure-Track Faculty Recruitment Award [RR140077]. Deposited in PMC for release after 12 months.
Data availability
The Gal4 enhancer trap lines described here have been deposited in the Zebrafish International Resource Center (ZIRC) for public distribution (see Table S1)
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.184341.supplemental
References
- Ablain J., Durand E. M., Yang S., Zhou Y. and Zon L. I. (2015). A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 32, 756-764. 10.1016/j.devcel.2015.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A., Nissen R. M., Sun Z., Swindell E. C., Farrington S. and Hopkins N. (2004). Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 101, 12792-12797. 10.1073/pnas.0403929101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonini D., Rossi B., Han R., Minichiello A., Di Palma T., Corrado M., Banfi S., Zannini M., Brissette J. L. and Missero C. (2006). An autoregulatory loop directs the tissue-specific expression of p63 through a long-range evolutionarily conserved enhancer. Mol. Cell. Biol. 26, 3308-3318. 10.1128/MCB.26.8.3308-3318.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonini D., Sirico A., Aberdam E., Ambrosio R., Campanile C., Fagoonee S., Altruda F., Aberdam D., Brissette J. L. and Missero C. (2015). A composite enhancer regulates p63 gene expression in epidermal morphogenesis and in keratinocyte differentiation by multiple mechanisms. Nucleic Acids Res. 43, 862-874. 10.1093/nar/gku1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K. and Kawakami K. (2010). A transgenic zebrafish for monitoring in vivo microtubule structures. Dev. Dyn. 239, 2695-2699. 10.1002/dvdy.22400 [DOI] [PubMed] [Google Scholar]
- Asakawa K., Suster M. L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M. and Kawakami K. (2008). Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA 105, 1255-1260. 10.1073/pnas.0704963105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkers J., Hild M., Kramer C., Furutani-Seiki M. and Hammerschmidt M. (2002). Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev. Cell 2, 617-627. 10.1016/S1534-5807(02)00163-6 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Horsley V. and Fuchs E. (2007). Epithelial stem cells: turning over new leaves. Cell 128, 445-458. 10.1016/j.cell.2007.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H. and Dormand E.-L. (1995). The GAL4 system as a tool for unravelling the mysteries of the Drosophila nervous system. Curr. Opin. Neurobiol. 5, 572-578. 10.1016/0959-4388(95)80061-1 [DOI] [PubMed] [Google Scholar]
- Burkel B. M., von Dassow G. and Bement W. M. (2007). Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil. Cytoskelet. 64, 822-832. 10.1002/cm.20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney T. J., von der Hardt S., Sonntag C., Amsterdam A., Topczewski J., Hopkins N. and Hammerschmidt M. (2007). Inactivation of serine protease Matriptase1a by its inhibitor Hai1 is required for epithelial integrity of the zebrafish epidermis. Development 134, 3461-3471. 10.1242/dev.004556 [DOI] [PubMed] [Google Scholar]
- Chua C. W., Shibata M., Lei M., Toivanen R., Barlow L. J., Bergren S. K., Badani K. K., McKiernan J. M., Benson M. C., Hibshoosh H. et al. (2014). Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 16, 951-961, 951-954 10.1038/ncb3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A. et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado S., Stainier D. Y. R. and Anderson R. M. (2008). Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 3, 948-954. 10.1038/nprot.2008.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dane P. J. and Tucker J. B. (1985). Modulation of epidermal cell shaping and extracellular matrix during caudal fin morphogenesis in the zebra fish Brachydanio rerio. J. Embryol. Exp. Morphol. 87, 145-161. [PubMed] [Google Scholar]
- Davison J. M., Akitake C. M., Goll M. G., Rhee J. M., Gosse N., Baier H., Halpern M. E., Leach S. D. and Parsons M. J. (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304, 811-824. 10.1016/j.ydbio.2007.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Garza G., Schleiffarth J. R., Dunnwald M., Mankad A., Weirather J. L., Bonde G., Butcher S., Mansour T. A., Kousa Y. A., Fukazawa C. F. et al. (2013). Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J. Invest. Dermatol. 133, 68-77. 10.1038/jid.2012.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer G. T. and Rosenblatt J. (2011). Live imaging of cell extrusion from the epidermis of developing zebrafish. J. Vis. Exp. 52, e2689 10.3791/2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer G. T., Loftus P. D., Yoshigi M., Otsuna H., Chien C.-B., Morcos P. A. and Rosenblatt J. (2012). Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 484, 546-549. 10.1038/nature10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J. A., Giniger E., Maniatis T. and Ptashne M. (1988). GAL4 activates transcription in Drosophila. Nature 332, 853-856. 10.1038/332853a0 [DOI] [PubMed] [Google Scholar]
- Fisher S., Grice E. A., Vinton R. M., Bessling S. L., Urasaki A., Kawakami K. and McCallion A. S. (2006). Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 1, 1297-1305. 10.1038/nprot.2006.230 [DOI] [PubMed] [Google Scholar]
- Flusberg B. A., Nimmerjahn A., Cocker E. D., Mukamel E. A., Barretto R. P. J., Ko T. H., Burns L. D., Jung J. C. and Schnitzer M. J. (2008). High-speed, miniaturized fluorescence microscopy in freely moving mice. Nat. Methods 5, 935-938. 10.1038/nmeth.1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa C., Santiago C., Park K. M., Deery W. J., Gomez de la Torre Canny S., Holterhoff C. K. and Wagner D. S. (2010). poky/chuk/ikk1 is required for differentiation of the zebrafish embryonic epidermis. Dev. Biol. 346, 272-283. 10.1016/j.ydbio.2010.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M. and Ptashne M. (1985). Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40, 767-774. 10.1016/0092-8674(85)90336-8 [DOI] [PubMed] [Google Scholar]
- Gong Z., Ju B., Wang X., He J., Wan H., Sudha P. M. and Yan T. (2002). Green fluorescent protein expression in germ-line transmitted transgenic zebrafish under a stratified epithelial promoter from keratin8. Dev. Dyn. 223, 204-215. 10.1002/dvdy.10051 [DOI] [PubMed] [Google Scholar]
- Gu Y., Shea J., Slattum G., Firpo M. A., Alexander M., Mulvihill S. J., Golubovskaya V. M. and Rosenblatt J. (2015). Defective apical extrusion signaling contributes to aggressive tumor hallmarks. Elife 4, e04069 10.7554/elife.04069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B. M. (1992). Epithelial morphogenesis. Cell 69, 385-387. 10.1016/0092-8674(92)90440-N [DOI] [PubMed] [Google Scholar]
- Haurwitz R. E., Jinek M., Wiedenheft B., Zhou K. and Doudna J. A. (2010). Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355-1358. 10.1126/science.1192272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering N. A., Fromm M. and Schulzke J.-D. (2012). Determinants of colonic barrier function in inflammatory bowel disease and potential therapeutics. J. Physiol. 590, 1035-1044. 10.1113/jphysiol.2011.224568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez P. and Tirnauer J. S. (2010). Tumor suppressor interactions with microtubules: keeping cell polarity and cell division on track. Dis. Model. Mech. 3, 304-315. 10.1242/dmm.004507 [DOI] [PubMed] [Google Scholar]
- Hooper C. E. S. (1956). Cell turnover in epithelial populations. J. Histochem. Cytochem. 4, 531-540. 10.1177/4.6.531 [DOI] [PubMed] [Google Scholar]
- Hwang P.-P. (2009). Ion uptake and acid secretion in zebrafish (Danio rerio). J. Exp. Biol. 212, 1745-1752. 10.1242/jeb.026054 [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J.-R. J. and Joung J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U., Krauss J. and Nusslein-Volhard C. (2014). Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141, 4827-4830. 10.1242/dev.115584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke M., Carney T. J. and Hammerschmidt M. (2007). Foxi3 transcription factors and Notch signaling control the formation of skin ionocytes from epidermal precursors of the zebrafish embryo. Dev. Biol. 307, 258-271. 10.1016/j.ydbio.2007.04.044 [DOI] [PubMed] [Google Scholar]
- Jevtov I., Samuelsson T., Yao G., Amsterdam A. and Ribbeck K. (2014). Zebrafish as a model to study live mucus physiology. Sci. Rep. 4, 6653 10.1038/srep06653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Abe G., Asada T., Asakawa K., Fukuda R., Ito A., Lal P., Mouri N., Muto A., Suster M. L. et al. (2010). zTrap: zebrafish gene trap and enhancer trap database. BMC Dev. Biol. 10, 105 10.1186/1471-213X-10-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P. and Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Le Guellec D., Morvan-Dubois G. and Sire J.-Y. (2004). Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio). Int. J. Dev. Biol. 48, 217-231. 10.1387/ijdb.15272388 [DOI] [PubMed] [Google Scholar]
- Lechler T. and Fuchs E. (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275-280. 10.1038/nature03922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. and Kimelman D. (2002). A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell 2, 607-616. 10.1016/S1534-5807(02)00166-1 [DOI] [PubMed] [Google Scholar]
- Li Q., Frank M., Akiyama M., Shimizu H., Ho S.-Y., Thisse C., Thisse B., Sprecher E. and Uitto J. (2011). Abca12-mediated lipid transport and Snap29-dependent trafficking of lamellar granules are crucial for epidermal morphogenesis in a zebrafish model of ichthyosis. Dis. Model. Mech. 4, 777-785. 10.1242/dmm.007146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias H., Moran A., Samara Y., Moreno M., Compton J. E., Harburg G., Strickland P. and Hinck L. (2011). SLIT/ROBO1 signaling suppresses mammary branching morphogenesis by limiting basal cell number. Dev. Cell 20, 827-840. 10.1016/j.devcel.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D. R., Ullman K. S. and Rodesch C. K. (2010). Time-lapse imaging of mitosis after siRNA transfection. J. Vis. Exp. 40, e1878 10.3791/1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe M. M., Aihara E., Schumacher M. A., Zavros Y., Montrose M. H., Helmrath M. A., Sato T. and Shroyer N. F. (2013). Establishment of gastrointestinal epithelial organoids. Curr. Protoc. Mouse Biol. 3, 217-240. 10.1002/9780470942390.mo130179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E. and Church G. M. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823-826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall T. W., Lloyd I. E., Delalande J. M., Nathke I. and Rosenblatt J. (2011). The tumor suppressor adenomatous polyposis coli controls the direction in which a cell extrudes from an epithelium. Mol. Biol. Cell 22, 3962-3970. 10.1091/mbc.E11-05-0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeish J. A., Chico T. J. A., Taylor H. B., Tucker C., Donaldson K. and Brown S. B. (2010). Skin exposure to micro- and nano-particles can cause haemostasis in zebrafish larvae. Thromb. Haemost. 103, 797-807. 10.1160/TH09-06-0413 [DOI] [PubMed] [Google Scholar]
- Montell D. J. (2008). Morphogenetic cell movements: diversity from modular mechanical properties. Science 322, 1502-1505. 10.1126/science.1164073 [DOI] [PubMed] [Google Scholar]
- Oehlers S. H., Flores M. V., Hall C. J., Crosier K. E. and Crosier P. S. (2012). Retinoic acid suppresses intestinal mucus production and exacerbates experimental enterocolitis. Dis. Model. Mech. 5, 457-467. 10.1242/dmm.009365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuna H., Hutcheson D. A., Duncan R. N., McPherson A. D., Scoresby A. N., Gaynes B. F., Tong Z., Fujimoto E., Kwan K. M., Chien C. B. et al. (2015). High-resolution analysis of CNS expression patterns in zebrafish GAL4 enhancer-trap lines. Dev. Dyn. 244, 785-796. 10.1002/dvdy.24260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashos E. E., Kague E. and Fisher S. (2008). Evaluation of cis-regulatory function in zebrafish. Brief. Funct. Genomics Proteomics 7, 465-473. 10.1093/bfgp/eln045 [DOI] [PubMed] [Google Scholar]
- Pellettieri J. and Sanchez Alvarado A. (2007). Cell turnover and adult tissue homeostasis: from humans to planarians. Annu. Rev. Genet. 41, 83-105. 10.1146/annurev.genet.41.110306.130244 [DOI] [PubMed] [Google Scholar]
- Pignon J.-C., Grisanzio C., Geng Y., Song J., Shivdasani R. A. and Signoretti S. (2013). p63-expressing cells are the stem cells of developing prostate, bladder, and colorectal epithelia. Proc. Natl. Acad. Sci. USA 110, 8105-8110. 10.1073/pnas.1221216110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot F. and Lecuit T. (2005). Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev. Dyn. 232, 685-694. 10.1002/dvdy.20334 [DOI] [PubMed] [Google Scholar]
- Reischauer S., Levesque M. P., Nusslein-Volhard C. and Sonawane M. (2009). Lgl2 executes its function as a tumor suppressor by regulating ErbB signaling in the zebrafish epidermis. PLoS Genet. 5, e1000720 10.1371/journal.pgen.1000720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A. H., Kessenbrock K., Yu J. H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T. A., Werb Z. et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605-607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsma L., Steller E. J. A., Ellenbroek S. I. J., Kranenburg O., Borel Rinkes I. H. M. and van Rheenen J. (2013). Surgical implantation of an abdominal imaging window for intravital microscopy. Nat. Protoc. 8, 583-594. 10.1038/nprot.2013.026 [DOI] [PubMed] [Google Scholar]
- Rock J. R., Randell S. H. and Hogan B. L. M. (2010). Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 3, 545-556. 10.1242/dmm.006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J., Raff M. C. and Cramer L. P. (2001). An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 11, 1847-1857. 10.1016/S0960-9822(01)00587-5 [DOI] [PubMed] [Google Scholar]
- Sabel J. L., d'Alencon C., O'Brien E. K., Van Otterloo E., Lutz K., Cuykendall T. N., Schutte B. C., Houston D. W. and Cornell R. A. (2009). Maternal Interferon Regulatory Factor 6 is required for the differentiation of primary superficial epithelia in Danio and Xenopus embryos. Dev. Biol. 325, 249-262. 10.1016/j.ydbio.2008.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C., Kimura Y., Hirata H., Suster M. L., Kawakami K. and Higashijima S.-I. (2013). Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development 140, 3927-3931. 10.1242/dev.099531 [DOI] [PubMed] [Google Scholar]
- Schwank G., Andersson-Rolf A., Koo B.-K., Sasaki N. and Clevers H. (2013). Generation of BAC transgenic epithelial organoids. PLoS ONE 8, e76871 10.1371/journal.pone.0076871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerte T. (2010). Skin epithelium of zebrafish may work as an airway epithelia analogue model to evaluate systemic effects of micro- and nano-particles. Thromb. Haemost. 103, 692-693. 10.1160/TH10-01-0042 [DOI] [PubMed] [Google Scholar]
- Senoo M., Pinto F., Crum C. P. and McKeon F. (2007). p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129, 523-536. 10.1016/j.cell.2007.02.045 [DOI] [PubMed] [Google Scholar]
- Shono T., Kurokawa D., Miyake T. and Okabe M. (2011). Acquisition of glial cells missing 2 enhancers contributes to a diversity of ionocytes in zebrafish. PLoS ONE 6, e23746 10.1371/journal.pone.0023746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane M., Martin-Maischein H., Schwarz H. and Nusslein-Volhard C. (2009). Lgl2 and E-cadherin act antagonistically to regulate hemidesmosome formation during epidermal development in zebrafish. Development 136, 1231-1240. 10.1242/dev.032508 [DOI] [PubMed] [Google Scholar]
- Southall T. D., Elliott D. A. and Brand A. H. (2008). The GAL4 system: a versatile toolkit for gene expression in Drosophila. CSH Protoc. 2008, pdb top49 10.1101/pdb.top49 [DOI] [PubMed] [Google Scholar]
- Swindle E. J., Collins J. E. and Davies D. E. (2009). Breakdown in epithelial barrier function in patients with asthma: identification of novel therapeutic approaches. J. Allergy Clin, Immunol. 124, 23-34; quiz 35-26 10.1016/j.jaci.2009.05.037 [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Matsuda K., Yamaguchi S., Asakawa K., Miyasaka N., Lal P., Yoshihara Y., Koga A., Kawakami K., Shimizu T. et al. (2014). Establishment of Gal4 transgenic zebrafish lines for analysis of development of cerebellar neural circuitry. Dev. Biol. 397, 1-17. 10.1016/j.ydbio.2014.09.030 [DOI] [PubMed] [Google Scholar]
- Tallafuss A., Gibson D., Morcos P., Li Y., Seredick S., Eisen J. and Washbourne P. (2012). Turning gene function ON and OFF using sense and antisense photo-morpholinos in zebrafish. Development 139, 1691-1699. 10.1242/dev.072702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden F. J., Granato M., Schach U., Brand M., Furutani-Seiki M., Haffter P., Hammerschmidt M., Heisenberg C. P., Jiang Y. J., Kane D. A. et al. (1996). Genetic analysis of fin formation in the zebrafish, Danio rerio. Development 123, 255-262. [DOI] [PubMed] [Google Scholar]
- Wang Y.-H., Chen Y.-H., Lin Y.-J. and Tsai H.-J. (2006). Spatiotemporal expression of zebrafish keratin 18 during early embryogenesis and the establishment of a keratin 18:RFP transgenic line. Gene Expr. Patterns 6, 335-339. 10.1016/j.modgep.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Webb A. E. and Kimelman D. (2005). Analysis of early epidermal development in zebrafish. Methods Mol. Biol. 289, 137-146. [DOI] [PubMed] [Google Scholar]
- Webb A. E., Driever W. and Kimelman D. (2008). psoriasis regulates epidermal development in zebrafish. Dev. Dyn. 237, 1153-1164. 10.1002/dvdy.21509 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2007). The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio*), 5th edition Eugene, OR USA: University of Oregon Press. [Google Scholar]
- Xiao C., Puddicombe S. M., Field S., Haywood J., Broughton-Head V., Puxeddu I., Haitchi H. M., Vernon-Wilson E., Sammut D., Bedke N. et al. (2011). Defective epithelial barrier function in asthma. J. Allergy Clin. Immunol. 128, 549-556.e541-512. 10.1016/j.jaci.2011.05.038 [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C. et al. (1999). p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714-718. 10.1038/19539 [DOI] [PubMed] [Google Scholar]
- Zeissig S., Burgel N., Gunzel D., Richter J., Mankertz J., Wahnschaffe U., Kroesen A. J., Zeitz M., Fromm M. and Schulzke J.-D. (2007). Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut 56, 61-72. 10.1136/gut.2006.094375 [DOI] [PMC free article] [PubMed] [Google Scholar]