Abstract
Rationale: Maintaining optimal symptom control remains the primary objective of asthma treatment. Better understanding of the biologic underpinnings of asthma control may lead to the development of improved clinical and pharmaceutical approaches.
Objectives: To identify molecular pathways and interrelated genes whose differential expression was associated with asthma control.
Methods: We performed gene set enrichment analyses of asthma control in 1,170 adults with asthma, each with gene expression data derived from either whole blood (WB) or unstimulated CD4+ T lymphocytes (CD4), and a self-reported asthma control score representing either the preceding 6 months (chronic) or 7 days (acute). Our study comprised a discovery WB cohort (n = 245, chronic) and three independent, nonoverlapping replication cohorts: a second WB set (n = 448, acute) and two CD4 sets (n = 300, chronic; n = 77, acute).
Measurements and Main Results: In the WB discovery cohort, we found significant overrepresentation of genes associated with asthma control in 1,106 gene sets from the Molecular Signatures Database (false discovery rate, <5%). Of these, 583 (53%) replicated in at least one replication cohort (false discovery rate, <25%). Suboptimal control was associated with signatures of eosinophilic and granulocytic inflammatory signals, whereas optimal control signatures were enriched for immature lymphocytic patterns. These signatures included two related biologic processes related to activation by TREM-1 (triggering receptor expressed on myeloid cells 1) and lipopolysaccharide.
Conclusions: Together, these results demonstrate the existence of specific, reproducible transcriptomic components in blood that vary with degree of asthma control and implicate a novel biologic target (TREM-1).
Keywords: genomics, gene expression, asthma, exacerbation, blood
At a Glance Commentary
Scientific Knowledge on the Subject
Although transcriptomic profiling has been applied to the study of asthma, including differential expression studies comparing patients with moderate and/or severe asthma with healthy control subjects, there is very little known about such genomic underpinnings of asthma control.
What This Study Adds to the Field
This study demonstrates that transcriptomic profiling of peripheral blood and its constituents provides reproducible insights into the underlying determinants of asthma control, including identification of specific immune-related pathways.
Maintaining optimal symptom control remains the primary objective of asthma treatment (1). Despite the availability of several classes of controller medications, including inhaled corticosteroids, leukotriene receptor antagonists, and long-acting β-adrenoceptor agonists, suboptimal asthma control remains common and burdensome. Although insufficient access to healthcare resources and poor patient compliance with medication adherence are major barriers to disease management, a better understanding of the biologic underpinnings of the preexacerbation state is needed to develop targeted clinical and pharmaceutical approaches for maintaining effective asthma control.
One relatively underused strategy for novel pathway identification is to relate differences in asthma phenotypes to gene expression profiles in disease-relevant tissues (2). Three studies of small or modest size have profiled gene expression during acute exacerbation in blood or nasal cells, reporting up-regulation of similar pathways related to innate and adaptive immune responses (3–5). However, these studies focused on expression changes during acute exacerbations and provided no information regarding expression across the broad range of asthma control states that precede exacerbation. Given that cellular and transcriptional responses during acute asthma exacerbation are highly complex (6), the gene pathways perturbed may be quite different from those that destabilize asthma control and trigger exacerbation in the days preceding an acute crisis. Although detection of these earlier alterations may be more informative for the development of preventative controller strategies (7), only one small study of differential gene expression and asthma control has been conducted in circulating leukocytes from children in a broadly uncontrolled severe or controlled persistent disease state (8). Although these profiles related to various kinase signaling pathways are biologically relevant, their modest sample sizes inhibit their broad generalization.
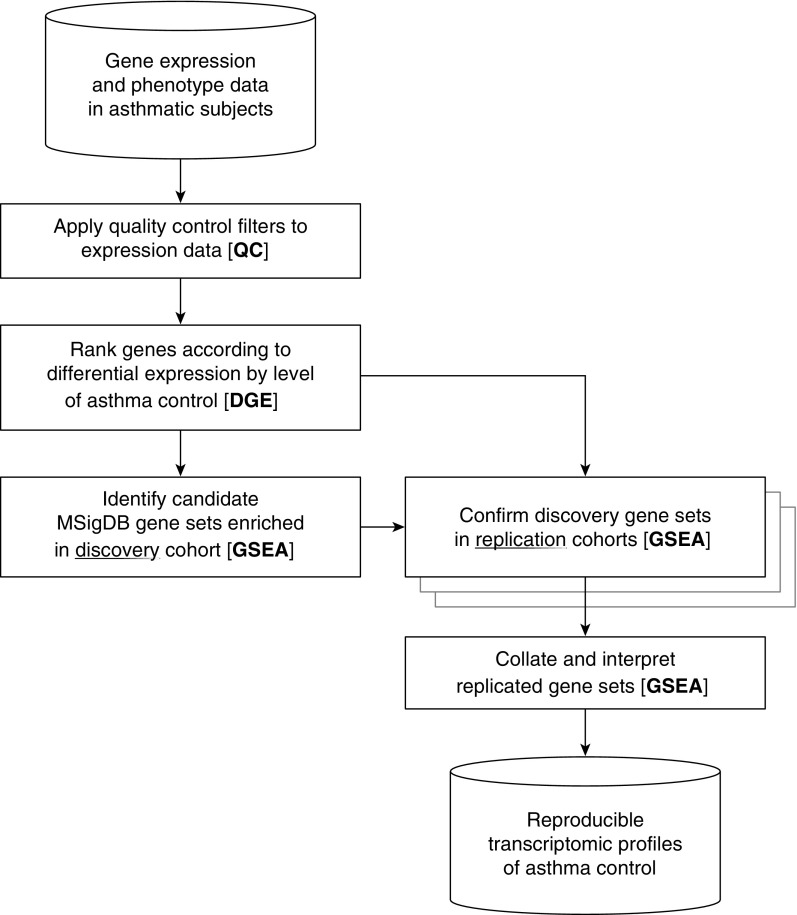
To address these deficiencies and determine the transcriptomic bases of chronic and acute asthma control, we performed a large-scale gene expression-phenotype association analysis of 1,170 adults with asthma (Figure 1), consisting of four nonoverlapping cohorts from Asthma BRIDGE (Asthma BioRepository for Integrative Genomic Exploration) (9) and the CAMP (Childhood Asthma Management Program) study (10). Asthma control was assessed using modified forms of the validated Asthma Control Test (ACT), a composite clinical measure of asthma symptom control (11). Recognizing the polygenic nature of asthma and related phenotypes, such as symptom control, we leveraged gene set enrichment analysis (GSEA) to enable simultaneous assessment of interrelated genes as a unit (12). We identified specific pathway gene sets showing reproducible enrichment in our blood-related datasets across tissue type and phenotypic chronicity. These results help conclusively establish reproducible molecular signatures of asthma control, suggesting pathways possibly amenable to direct therapeutic targeting. Some preliminary results from this study have been previously reported in abstract form (13).
Figure 1.
Overview of transcriptomic analyses of asthma control. In this pipeline schematic, cylinders represent input and output data, and boxes represent operations on these data. The discovery cohort is BRIDGE WB and the three replication cohorts are BRIDGE CD4, CAMP WB, and CAMP CD4. BRIDGE = BioRepository for Integrative Genomic Exploration; CAMP = Childhood Asthma Management Program; CD4 = CD4+ T lymphocyte; DGE = differential gene expression; GSEA = gene set enrichment analysis; MSigDB = Molecular Signatures Database, version 4.0; QC = quality control; WB = whole blood.
Methods
The Asthma BRIDGE dataset, which has been described previously (9, 14, 15), comprises a cohort of subjects selected from the EVE asthma genetics consortium (16). Additional detail on subject ascertainment is presented in the Methods section of the online supplement. Briefly, gene expression was measured in two sets of whole blood (WB) samples and one set of unstimulated CD4+ T-lymphocyte (CD4) samples (each sample corresponding to a single subject). An additional CAMP CD4 dataset (distinct from the CAMP WB dataset within Asthma BRIDGE) has also been described previously (17). All gene expression and phenotype data are publicly available to download from the Database of Genotypes and Phenotypes (dbGaP accession number pending) or from the Gene Expression Omnibus (GEO accession number GSE22324), respectively. All 1,170 adult subjects from the four nonoverlapping cohorts had physician-diagnosed asthma. Additional demographic characteristics are summarized in Table 1.
Table 1.
Demographic and Medical Characteristics of Asthma Study Cohorts
| Variable | BRIDGE WB | BRIDGE CD4 | CAMP WB | CAMP CD4 |
|---|---|---|---|---|
| N | 245 | 300 | 448 | 177 |
| Age, yr, mean (SD) | 22.0 (5.2) | 27.1 (15.8) | 20.9 (2.2) | 20.4 (2.2) |
| Female, % | 50.6 | 51.3 | 36.2 | 41.8 |
| Race, % | ||||
| White/European | 13.9 | 31.0 | 57.4 | 100.0 |
| Black/African American | 0.0 | 49.0 | 20.1 | 0.0 |
| Hispanic/Latino | 72.2 | 10.7 | 13.2 | 0.0 |
| Other | 13.9 | 9.3 | 9.4 | 0.0 |
| One or more exacerbations, % | 15.5 | 58.0 | 7.1 | 11.3 |
| History of atopic dermatitis, % | 23.3 | 41.7 | 28.1 | 26.6 |
| History of hay fever, % | 46.5 | 59.3 | 54.5 | 39.5 |
| Ever hospitalized overnight, % | 40.7 | 46.5 | 36.8 | 43.5 |
| Childhood asthma severity, % | ||||
| Mild | — | — | 47.1 | 55.9 |
| Moderate | — | — | 52.9 | 44.1 |
| Body mass index, mean (SD) | — | — | 26.6 (6.1) | 25.6 (5.0) |
| Pre-BD FEV1, % predicted, mean (SD) | — | — | 97.0 (14.5) | 97.4 (12.4) |
| Log10 IgE, ng/ml, mean (SD) | — | — | 2.5 (0.6) | 2.5 (0.7) |
Definition of abbreviations: BD = bronchodilator; BRIDGE = BioRepository for Integrative Genomic Exploration; CAMP = Childhood Asthma Management Program; CD4 = CD4+ T lymphocyte; WB = whole blood.
Age is reported for when the asthma control phenotype and gene expression were measured. A history of atopic dermatitis or hay fever represented a report of having the disease for greater than or equal to 2 years. Overnight hospitalization represents a measure of severity before the preceding 6 months. Asthma-related exacerbation events in the preceding 6 months include visiting a doctor, missing work, visiting an emergency room, or being hospitalized. Childhood asthma severity assessments and measurements of lung function and IgE were only available in the CAMP cohorts. FEV1 is reported before application of a BD.
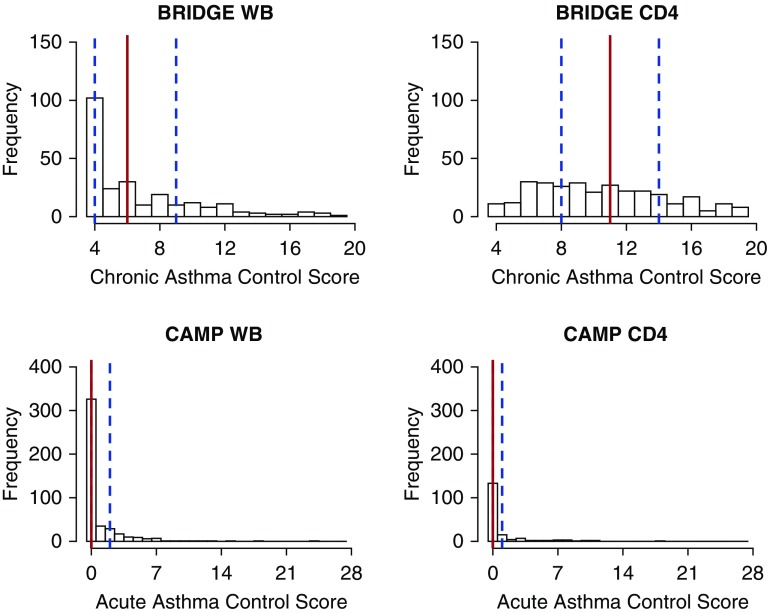
Using clinical phenotype data available in Asthma BRIDGE and in CAMP, we developed two composite scores summarizing self-reported asthma control in the preceding 6 months (chronic, 4–20) and 7 days (acute, 0–28), respectively, each modeled on the ACT questionnaire (11), where higher scores indicate worse asthma control. Based on the medians of the corresponding phenotypic distributions, optimal chronic asthma control was defined as a score less than or equal to six in BRIDGE WB and moderately suboptimal asthma control was defined as a score less than or equal to 11 in BRIDGE CD4 (Figure 2). In CAMP WB and CAMP CD4, optimal acute asthma control was defined as a score of zero. Other separate clinical data, such as medication use, provided further context and validity (see Tables E1 and E2 in the online supplement). Additional detail on the phenotypic data is presented in the Methods section of the online supplement.
Figure 2.
Distributions of asthma control scores in study cohorts. In total, there were 245 Asthma BRIDGE WB subjects, 300 Asthma BRIDGE CD4 subjects, 448 CAMP WB subjects, and 177 CAMP CD4 subjects. Within each histogram, the 25th, 50th, and 75th percentiles are marked with dashed blue, solid red, and dashed blue vertical lines, respectively. In BRIDGE WB, optimal chronic asthma control was defined as a score less than or equal to six. In BRIDGE CD4, asthma control was stratified between moderately suboptimal asthma control (score ≤11) and severely suboptimal asthma control. In CAMP WB and CAMP CD4, optimal acute asthma control was defined as a score of zero. BRIDGE = BioRepository for Integrative Genomic Exploration; CAMP = Childhood Asthma Management Program; CD4 = CD4+ T lymphocyte; WB = whole blood.
Subjects were typed on either the HumanHT-12 v4 Expression or HumanRef8 v2 BeadChip platforms (Illumina, Inc., San Diego, CA) (9, 18). Log2-transformed and quantile-normalized data that passed stringent and commonly used quality control metrics were available for 10,456 gene expression probes found across BRIDGE WB, BRIDGE CD4, and CAMP WB and for a further subset of 6,555 probes in CAMP CD4. Additional detail on data management is presented in the Methods section of the online supplement.
To identify biologically meaningful sets of genes differentially expressed between suboptimal and optimal asthma control, we constructed an analytical pipeline in R (version 3.1) and Bioconductor (Figure 1) (19). Differential expression models were adjusted for age, sex, self-reported race, and the first two or three tissue-specific principal components of gene expression to account for batch effects (20). Differential expression results ranked by fold change were inputs for our GSEAs (12) of asthma control, where we tested two human gene set collections (“C7: immunologic signatures” from the Human Immunology Project Consortium, n = 1,012; “C2 CGP: chemical and genetic perturbations,” n = 2,134) from the Molecular Signatures Database (MSigDB, version 4.0) (21). Only gene sets showing strongly significant enrichment (false discovery rate [FDR], <5%) in the discovery cohort (BRIDGE WB) were tested in the three independent replication cohorts (BRIDGE CD4, CAMP WB, CAMP CD4). Gene set replication was defined as a significant enrichment directionally consistent with the discovery cohort (FDR, <25%). Additional analytical detail is presented in the Methods section of the online supplement.
Results
Transcriptomic Analyses of Asthma Control in Discovery Cohort
As the foundation of our transcriptomic analyses of asthma control (Figure 1), we first established a discovery cohort and a definition of optimal and suboptimal control based on clinical phenotypes (Table 1). Among 245 Asthma BRIDGE subjects with WB samples (BRIDGE WB), the chronic asthma control score distribution was heavily left-skewed with a median value of six (Figure 2). Any exacerbation in the preceding 6-month period was reported by only one subject with a control score less than or equal to six and by 37 subjects (31%) with scores greater than six, suggesting this empirical cutoff was reasonable for distinguishing optimal asthma control. Although there was increased prevalence of hay fever and history of asthma-related hospitalizations in subjects with suboptimal asthma control versus those with optimal control (P < 0.05), there was no significant difference in the prevalence of atopic dermatitis between levels of asthma control (see Table E1). There was also no evidence of confounding by indication, because only five subjects (2%) reported using inhaled corticosteroids, leukotriene receptor antagonists, or long-acting β-adrenoceptor agonists in the week preceding their blood draw (see Table E2), with similar proportions of medication use across levels of asthma control.
Recognizing that complex phenotypes, such as asthma control, are likely mediated through the interactions of functionally related biologic processes, we performed a GSEA of 10,456 genes and their differential expressions by level of asthma control in the discovery cohort (see Table E3). As an aside, we found that several of the genes most strongly expressed during suboptimal asthma control had plausible individual connections to asthma biology in the scientific literature (see the Results section in the online supplement). Briefly, GSEA tests whether related genes in a given set are distributed more toward the top or bottom of a ranked list of differential expression results between two biologic states than expected by chance (12). The significance of this enrichment score is determined by repeatedly permuting the observed expression-phenotype relationships to create a null distribution for comparison. The leading edge of an enriched gene set (i.e., the further subset of genes contributing most to the enrichment score) then represents the core biology of interest.
Although numerous biologic gene sets are available as hypotheses to test through GSEA, we focused our analysis on two curated knowledge sets from the MSigDB most likely to provide relevant biologic insights into asthma control: the C7 collection (n = 1,012 sets), related to expression profiling studies comparing human immune cell states (22); and the C2 CGP collection (n = 2,134 sets), related to differential expression in response to chemical and genetic perturbations. With the C7 collection, we hypothesized that groups of transcriptionally coregulated genes in the immune system are differentially expressed by level of asthma control; with the C2 CGP collection, we hypothesized that broader types of perturbing factors influence expression profiles of asthma control. Within BRIDGE WB, we found significant enrichment for 1,106 gene sets (FDR, <5%), including 545 (54%) and 561 (26%) from the C7 and C2 CGP collections, respectively (Table 2; see Table E4). Of these 1,106 enriched gene sets, 784 (71%; 298 C7 and 486 C2 CGP) were positively enriched for suboptimal asthma control (i.e., genes in a given set were distributed toward greater overexpression), whereas 322 (29%; 247 C7 and 75 C2 CGP) were negatively enriched (underexpression). Although these enriched gene C7 and C2 CGP sets comprised 16,975 and 12,601 unique genes, respectively, their leading edges comprised cores of 3,939 and 2,400 unique genes, respectively (see Table E5). This enrichment pattern suggested that genes associated with suboptimal asthma control are localized to specific biologic pathways.
Table 2.
Replication Rates of 1,106 Gene Sets Significantly Enriched for Asthma Control in the Discovery Cohort
| Cohort | C7 (Suboptimal Control) | C7 (Optimal Control) | C2 CGP (Suboptimal Control) | C2 CGP (Optimal Control) |
|---|---|---|---|---|
| BRIDGE WB, n | 298 | 247 | 486 | 75 |
| BRIDGE CD4, n (%) | 7 (2) | 13 (5) | 23 (5) | 12 (16) |
| CAMP WB, n (%) | 177 (59) | 116 (47) | 230 (47) | 17 (23) |
| CAMP CD4, n (%) | 7 (2) | 6 (2) | 24 (5) | 0 (0) |
| 1 replication, n (%) | 180 (60) | 126 (51) | 251 (52) | 26 (35) |
| 2 replications, n (%) | 11 (4) | 9 (4) | 26 (5) | 3 (4) |
| 3 replications, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
Definition of abbreviations: BRIDGE = BioRepository for Integrative Genomic Exploration; C2 CGP = chemical and genetic perturbation signatures; C7 = immunologic signatures from the Human Immunology Project Consortium; CAMP = Childhood Asthma Management Program; CD4 = CD4+ T lymphocyte; MSigDB = Molecular Signatures Database, version 4.0; WB = whole blood.
The first row shows counts stratified by MSigDB collection of gene sets with significant enrichment (false discovery rate, <5%) during suboptimal chronic asthma control or during optimal control in the discovery cohort (BRIDGE WB). The middle three rows show corresponding counts in each of the other three study cohorts of replicated gene sets (consistent direction of enrichment and false discovery rate <25%) and corresponding replication rates. The bottom three rows show the number of discovery gene sets that replicated in at least one, two, or three of the other cohorts, respectively.
Generalizability of Enriched Asthma Control Gene Sets across Replication Cohorts
To assess the generalizability of our discovery GSEA of chronic asthma control, we performed replication analyses in three large independent datasets of nonoverlapping subjects with differential expression and chronic or acute asthma control data (Table 1): a second dataset of WB expression profiles from 448 CAMP subjects participating in Asthma BRIDGE (CAMP WB), and two sets of unstimulated peripheral blood CD4+ T-lymphocyte expression profiles from Asthma BRIDGE (BRIDGE CD4, n = 300) and CAMP (CAMP CD4, n = 177). Gene expression profiles in CAMP CD4 have been used successfully to identify novel genetic susceptibility loci for asthma (23). Notably, the distributions of chronic or acute asthma control scores in the three replication cohorts differed from that observed in the discovery cohort (Figure 2), requiring cohort-specific definitions of optimal asthma control (see the Methods and supplemental Methods sections).
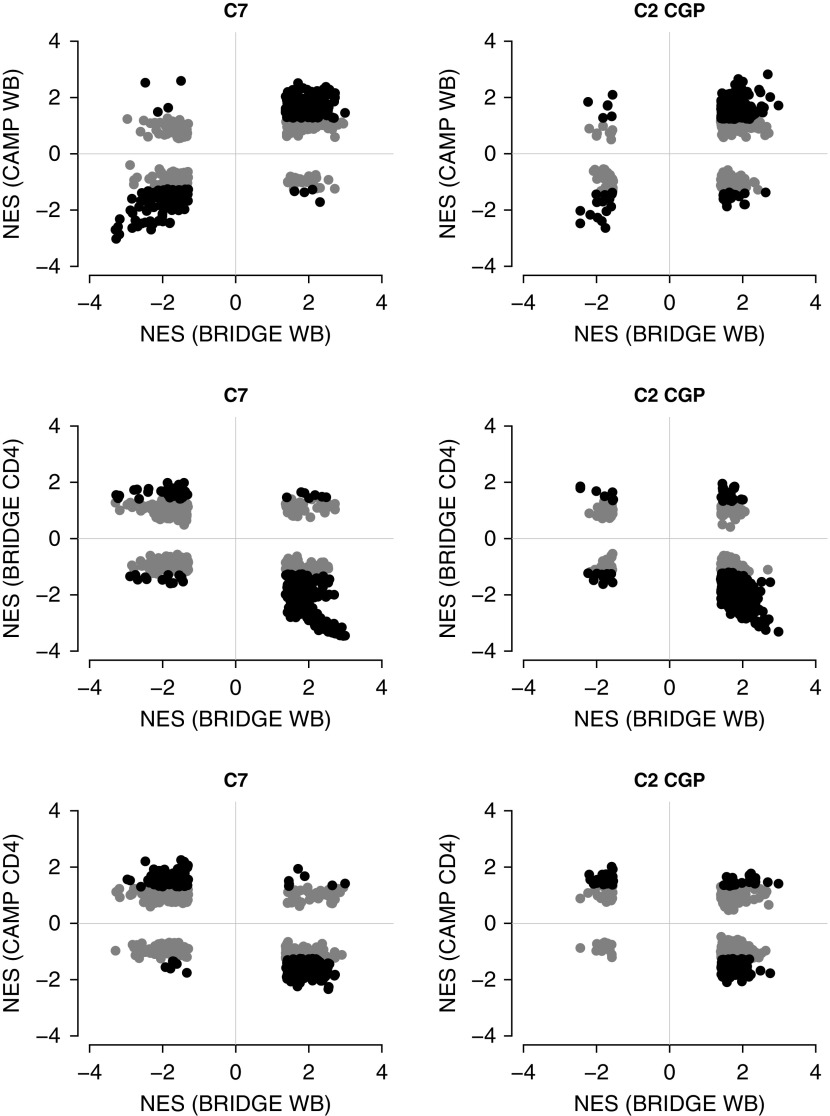
We found strong evidence for replication of our enriched asthma control gene sets across the study datasets. Despite differences in phenotypic distributions and tissue types (WB vs. CD4), 583 (53%) of the 1,106 enriched gene sets identified in BRIDGE WB were moderately replicated in at least one other cohort (i.e., their leading edge was enriched in the same direction with an FDR <25%) and 49 (4%) replicated in at least two cohorts, although none replicated in all three cohorts (Table 2; see Table E4). In general, replication was much stronger within tissue type (BRIDGE WB vs. CAMP WB), than across tissue (BRIDGE WB vs. BRIDGE CD4) (Figure 3; see Figure E1). Imposing more stringent replication criteria magnified these differences: at an FDR less than 5%, a total of 316 (29%) BRIDGE WB gene sets replicated in CAMP WB, but only 12 (1%) and four (<1%) in BRIDGE CD4 and CAMP CD4, respectively. Thus, although substantial consistency in gene set enrichment supported the existence of underlying transcriptomic signatures of asthma control, the robustness of these signals was sensitive to tissue-specific effects.
Figure 3.
Comparisons of gene set enrichment scores for asthma control in discovery cohort with scores in replication cohorts. Each point represents a single gene set from a given MSigDB collection (C7 or C2 CGP) that was enriched in the discovery cohort (FDR, <5%). Position on the x-axis is the NES in the discovery cohort and position on the y-axis is the NES in the replication cohort. Gene set replication occurs when the corresponding point both lies in the lower-left or upper-right quadrants (i.e., shows directional consistency of enrichment) and is marked in black (FDR, <25%) rather than light gray (FDR, ≥25%). BRIDGE = BioRepository for Integrative Genomic Exploration; C2 CGP = chemical and genetic perturbation signatures; C7 = immunologic signatures from the Human Immunology Project Consortium; CAMP = Childhood Asthma Management Program; CD4 = CD4+ T lymphocyte; FDR = false discovery rate; MSigDB = Molecular Signatures Database, version 4.0; NES = normalized enrichment score; WB = whole blood.
We then examined other possible confounding factors that might influence our interpretations of these replication patterns. There were largely no significant differences in the prevalence of atopic phenotypes between subjects with suboptimal asthma control and those with optimal control, except for increases in hay fever in BRIDGE WB and in high IgE in CAMP WB (P < 0.05). Suboptimal control was also inconsistently associated with several clinical features of asthma severity (see Table E1): in the CAMP WB cohort, but not in CAMP CD4, suboptimal control was associated with more moderate asthma (vs. mild) and with overweight (P < 0.05); in both WB cohorts, but not in the CD4 cohorts, suboptimal control was associated with history of asthma-related hospitalization (P < 0.05). Similarly, inconsistent associations were observed across the cohorts when considering medication usage (see Table E2): in both CAMP cohorts, subjects with optimal control reported higher asthma medication usage (P < 0.05); similar relationships were not observed in either BRIDGE cohort. Thus, although there was some correspondence between asthma control, severity, and medication use in the CAMP cohorts, similar associations were much less pronounced in the BRIDGE cohorts.
To confirm that the gene signatures and enriched gene sets identified in the discovery cohort that replicated in CAMP were not driven by these potential confounders, we performed sensitivity analyses removing CAMP subjects who had been taking any asthma medications in the preceding week before sample procurement. We found that the gene set replication pattern was very similar (see Figure E2), with most of the GSEA results in CAMP (WB, 80%; CD4, 82%) demonstrating directionally consistent associations with the original results (see Figure E3). Together, these data demonstrate that our findings provide robust insights into the molecular underpinnings of asthma control that are not confounded by atopy, severity, or medication.
Biologic Support for Enriched Asthma Control Gene Sets
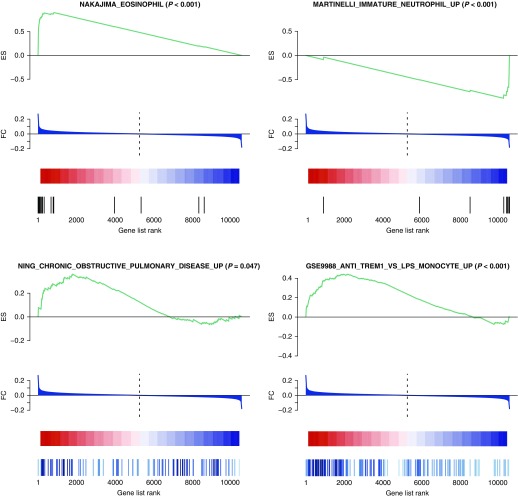
Complementing these replication results and providing initial biologic support for the reliability of our approach was the prominent representation in our results of immunologic processes known to be active in asthma (Figure 4). Among replicated BRIDGE WB gene sets enriched during suboptimal asthma control were signatures of eosinophilic gene expression: nine C7 gene sets derived from a particular gene expression study (GEO DataSeries GSE3982, “Human immune cell transcriptome”) and one C2 CGP gene set (NAKAJIMA_EOSINOPHIL). From the C2 CGP collection, three gene sets related to immature T-lymphocyte-specific transcripts were enriched during optimal asthma control (LEE_DIFFERENTIATING_T_LYMPHOCYTE, LEE_NAIVE_T_LYMPHOCYTE, and LEE_EARLY_T_LYMPHOCYTE_DN), a gene set of transcripts differentially expressed in neutrophils attracted to skin lesions (THEILGAARD_NEUTROPHIL_AT_SKIN_WOUND_DN) was enriched during suboptimal control, and a gene set of transcripts specific to immature neutrophils (MARTINELLI_IMMATURE_NEUTROPHIL_UP) was enriched during suboptimal control. Transcriptional profiles of asthma related to eosinophils and neutrophils have been observed in induced sputum samples (24) and in blood (25).
Figure 4.
Biologic highlights among replicated gene set enrichments for asthma control. In the BRIDGE WB discovery cohort, the differential gene expression comparison was of suboptimal chronic asthma control (score >6) versus optimal control (score ≤6), in red and blue, respectively. All highlighted gene sets were significantly enriched at a false discovery rate less than 5% in BRIDGE WB and replicated in at least one other cohort. Unadjusted enrichment P values are also shown in parentheses next to the gene set name. BRIDGE = BioRepository for Integrative Genomic Exploration; ES = enrichment score; FC = fold change in expression between levels of asthma control; WB = whole blood.
We then examined the replicated GSEA results for the involvement of other types of inflammatory and disease pathways in asthma control (see Table E4). Four C2 CGP gene sets enriched for suboptimal asthma control were programmed cellular responses to specific inflammatory cytokines, including IFN-α1 (IFNA1), IFN-β1 (IFNB1), IL-6 (IL6), and IL-4 (IL4). Notably, enriched during suboptimal asthma control were a pair of gene sets related to chronic obstructive pulmonary disease case status (NING_CHRONIC_OBSTRUCTIVE_PULMONARY_DISEASE_UP and NING_CHRONIC_OBSTRUCTIVE_PULMONARY_DISEASE_DN). These results affirmed that our analytical strategy could identify biologic pathways underlying asthma control.
Novel Biologic Insights into Asthma Control
Encouraged by the high rate of gene set replication across cohorts and the representation of a large number of pathways biologically relevant to asthma, we next examined the 316 gene sets that demonstrated stringent evidence of replication (FDR, <5% in the same direction in CAMP WB) for novel biologic insights (see Table E4). The most striking observation during manual curation of these gene sets was that expression signatures from subjects with optimal asthma control were consistently enriched for transcripts derived from lymphoid cells (e.g., CD4+ and CD8+ T cells and B cells), whereas signatures from subjects with suboptimal control were enriched for transcripts derived from myeloid-derived and monocytic lineages (e.g., neutrophils, basophils, mast cells, and dendritic cells). This pattern was observed across 111 gene sets derived from five independent publicly available GEO datasets that contrasted gene expression profiles between pairs of peripheral blood constituents (GSE3982, GSE22886, GSE29618, GSE10325, and GSE11057). Broadly, these reference profiles demonstrated that specific immune cell types have identifiably distinct sets of transcriptionally coregulated genes relative to one another (e.g., expressed up in B cells vs. myeloid cells), which could explain their contrasting biologic roles in establishing states of asthma control.
To confirm that differences in asthma control did not simply reflect differential blood cell counts, we examined measurements or estimates of those data in the two WB cohorts and found no significant differences in proportions of lymphocytes, monocytes, and neutrophils between subjects with optimal asthma control and those with suboptimal control (see Figure E4). However, suboptimal asthma control was significantly associated with higher measured eosinophil level in CAMP WB (P = 8.5 × 10−6). We then performed a sensitivity analysis in CAMP WB, regressing out eosinophil level in the underlying differential expression model, and found that 81% of the sensitivity GSEA results were consistent with the original results (see Figure E3). Among the 316 strongest results, 99% were consistent, including 11 eosinophil-specific gene sets. In short, the most replicable set of enriched gene sets suggested a strong relationship of expression related to immune cell lineage with asthma control.
The consistency of this expression-phenotype pattern may have reflected a specific set of underlying molecular signaling responses that promote a granulocytic inflammatory response activated during suboptimal asthma control. Further inspection of the 316 replicated gene sets revealed two interrelated signaling pathways known to drive granulocytic inflammation: TREM-1, the triggering receptor expressed on myeloid cells 1 (26, 27); and LPS. Expressed on monocytes, macrophages, and neutrophils, TREM-1 is an orphan immunoreceptor that amplifies inflammation induced by pattern-recognition receptors (28) and interacts with the toll-like receptor 4 to promote LPS-mediated granulocytic inflammation (27, 29). These two molecules were represented by six replicated gene sets enriched during suboptimal control that came from the C7 subcollection GSE9988 (“Innate immune responses to TREM-1 activation”) (see Table E4), which consists of profiles derived from a study of monocyte gene expression following treatment with either a TREM-1 activating antibody, LPS, or both (30).
The GSE9988-derived gene sets enriched in our signatures of asthma control support the preferential activation of genes induced by TREM-1 or LPS during suboptimal control. Within this pathway, the gene most differentially expressed in BRIDGE WB and demonstrating strong replication in CAMP WB was CCL23 (Table 3), an IL4/signal transducer and activator of transcription 6–induced chemoattractant of dendritic cells (31, 32). In GSE9988, CCL23 was preferentially up-regulated in LPS-treated monocytes. It is notable that another monocyte-related gene set that similarly included CCL23 was also strongly replicated across the WB datasets: ZHOU_INFLAMMATORY_RESPONSE_LIVE_UP, which characterizes the transcriptomic response in macrophages to a bacterium implicated in periodontal disease (see Table E3). The most strongly replicated of gene sets for suboptimal asthma control was GSE9988_ANTI_TREM1_VS_LPS_MONOCYTE_UP (Figure 4), consisting of genes that were over-expressed in circulating monocytes on activation by TREM-1 but not LPS. Multiple individual leading edge genes from this set demonstrated strong replication in CAMP WB (Table 3), including the transcription factor genes OLIG1, OLIG2, GFOD1, and HSD3B7. Together, these findings suggested that TREM-1 and LPS activation in circulating monocytes contributes to suboptimal asthma control.
Table 3.
Six TREM-1/LPS Signaling Genes Associated with Suboptimal Asthma Control in Whole Blood
| Gene | BRIDGE WB |
CAMP WB |
Fisher Combined P Value | ||||
|---|---|---|---|---|---|---|---|
| Fold Change | Trend P Value | FDR | Fold Change | Trend P Value | FDR | ||
| CCL23 | 1.30 | 3.34 × 10−4 | NS | 1.32 | 8.47 × 10−8 | 8.06 × 10−5 | 3.43 × 10−10 |
| OLIG2 | 1.18 | 2.12 × 10−3 | NS | 1.25 | 8.12 × 10−9 | 1.70 × 10−5 | 2.13 × 10−10 |
| OLIG1 | 1.14 | 1.03 × 10−2 | NS | 1.17 | 4.27 × 10−6 | 1.72 × 10−3 | 3.72 × 10−7 |
| GFOD1 | 1.12 | 6.70 × 10−3 | NS | 1.13 | 1.51 × 10−4 | 2.62 × 10−2 | 6.98 × 10−6 |
| RHOBTB3 | 1.09 | 4.33 × 10−2 | NS | 1.11 | 8.86 × 10−5 | 1.75 × 10−2 | 2.39 × 10−5 |
| HSD3B7 | 1.06 | 1.19 × 10−2 | NS | 1.07 | 2.49 × 10−4 | 3.56 × 10−2 | 1.88 × 10−5 |
Definition of abbreviations: BRIDGE = BioRepository for Integrative Genomic Exploration; CAMP = Childhood Asthma Management Program; FDR = false discovery rate; MSigDB = Molecular Signatures Database, version 4.0; NS = not significant; TREM-1 = triggering receptor expressed on myeloid cells 1; WB = whole blood.
Highlighted here are six TREM-1/LPS signaling genes found in the leading edges of gene sets enriched during suboptimal asthma control in the BRIDGE WB discovery cohort (MSigDB gene set collection GSE9988: “Innate immune responses to TREM-1 activation”), which also demonstrated significant differential expression in the CAMP WB replication cohort (FDR, <5%).
Discussion
Despite a general intuition that expression profiling should provide novel insights into asthma pathobiology, the ability to replicate findings of disease-specific gene expression signatures in independent populations has been challenging, largely because of the lack of large, well-characterized populations with similarly collected phenotypic and molecular data. Our work here demonstrating reproducible gene expression signatures of asthma control was possible because of the availability of 1,170 well-characterized subjects with asthma from the Asthma BRIDGE and CAMP studies. Across the corresponding four study cohorts, there was heterogeneity in the reported duration of asthma control (chronic vs. acute); in the distributions of these asthma control scores; and in subject demographics, disease severity, and asthma medication usage. Nonetheless, we have identified and replicated 583 gene set signatures of asthma control in blood, providing compelling support for specific molecular programs underlying asthma phenotypes.
The strongest, most consistently reproduced gene set enrichments from our discovery analysis in the Asthma BRIDGE WB samples were those also observed in the CAMP WB samples, with substantially less carryover to the CD4+ T-lymphocyte samples. This pattern reflects the much stronger replication rates within (rather than across) tissue type. Importantly, the identified signatures in WB revealed a strong relationship between asthma control and genes expressed by specific immune cell lineages, with optimal asthma control associated with enrichment for lymphoid cell profiles and suboptimal control associated with myeloid and monocytic cell profiles. Notably, these results were robust to adjustments for measured peripheral blood cell counts in our replication cohort, and estimated cell counts in WB using the expression data itself demonstrated minimal differences in major cell population subsets. These observations suggest that the observed differences in expression between asthma control states are not simply caused by differential cell counts in the blood but rather reflect specific immune-related transcriptional pathways regulating the disease state.
We identified a further subset of 316 asthma control gene sets demonstrating very stringent replication (FDR, <5%) in our WB analyses, revealing many plausible candidate pathways for additional investigation. One of the most compelling was the apparent activation of the TREM-1 (and related LPS) signaling pathway in subjects with suboptimal asthma control. Enrichment was noted for six gene sets that were differentially expressed in circulating murine monocytes in response to stimulation with TREM-1, LPS, or both (30). TREM-1 is a critical amplifier of inflammatory responses induced by toll-like receptor 4 (27) and other pattern-recognition receptors (28, 29). Figure 2 in a review by Colonna (29) on TREM-related immunology illustrates schematically its influence on downstream inflammatory response. Murine models also support a role for TREM-1 in the modulation of fungus-induced allergic asthma (33), and increased expression of TREM1 was observed in transcriptomic profiles of children with nonallergic asthma (25). Moreover, Bucova and coworkers (34) observed increased protein plasma levels of TREM-1 among adults with more severe allergic asthma, especially among those experiencing acute exacerbations. Relatedly, indoor endotoxin exposure is also a strong risk factor for asthma exacerbation (35). Although the TREM1 gene itself was not differentially expressed in our data, the body of transcriptomic evidence presented here implicates increased TREM-1 signaling as a potential early molecular indicator of worsening asthma control. Our work supports further investigation of the TREM-1 signaling pathway as potential therapeutic target for disrupting asthma-related inflammatory signaling.
We recognize that there are limitations to our transcriptomic study of asthma control. The compositions of our chronic and acute scores, representing the preceding 6 months and 7 days, respectively, do not correspond exactly to the 4-week ACT score widely used in clinical practice (11), although our findings strongly suggest that our scores do provide meaningful biologic information. We stress that the profiles identified provide insights into the biology of asthma control, but should not be considered for use in the clinical setting as prognostic biomarkers. Validation of these signatures in prospective cohorts designed for assessment of real-world practice would be required. Another consideration is temporality: the cross-sectional study design precludes definitive conclusions regarding causal relationships, namely whether the observed signatures are a determinant or a consequence of asthma control status. Although prospective studies would provide clarity in this regard, they are technically challenging. A potential alternative strategy could include genetic studies of asthma control, using the genes identified herein as candidates for prioritization.
In summary, we have leveraged a unique and sizable collection of phenotypic and transcriptomic data from four large independent cohorts to identify important biologic pathways of asthma control in adults. The reproducibility and specificity of these results strongly motivates embarking on functional and mechanistic investigation of these candidate asthma pathways.
Acknowledgments
Acknowledgment
The authors thank the Asthma BRIDGE Consortium study staff for their assistance and contributions, and all of the study subjects for their participation in the Asthma BRIDGE initiative.
Footnotes
Supported by extramural grants from the NHLBI at the National Institutes of Health (RC2 HL101543 [America Recovery and Reinvestment Act “Grand Opportunity”], R01 HL086601, T32 HL007427, and K01 HL127265); and by the National Institutes of Health Intramural Research Program at the National Institute of Environmental Health Sciences (S.J.L.).
Author Contributions: D.C.C.-C., V.J.C., S.T.W., and B.A.R. conceived the analyses. F.D.M., R.C.S., R.F.L., A.H.L., F.D.G., J.M., W.J.G., C.O., J.A.K., S.R.W., E.T.N., D.L.N., K.C.B., S.J.L., A.B.-V., S.T.W., and B.A.R. supervised the data generation. D.C.C.-C. and W.Q. developed and ran the data quality control pipeline. D.C.C.-C. developed and ran the analytical pipeline. D.C.C.-C. and B.A.R. wrote the manuscript. All authors read and approved the final manuscript.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201601-0107OC on August 5, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.National Heart Lung and Blood Institute, National Asthma Education and Prevention ProgramExpert Panel Report 3 (EPR-3): guidelines for the diagnosis and management of asthma, summary report 2007. Bethesda, MA: U.S. Department of Health and Human Services, National Institutes of Health2007
- 2.Hansel NN, Diette GB. Gene expression profiling in human asthma. Proc Am Thorac Soc. 2007;4:32–36. doi: 10.1513/pats.200606-132JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guajardo JR, Schleifer KW, Daines MO, Ruddy RM, Aronow BJ, Wills-Karp M, Hershey GK. Altered gene expression profiles in nasal respiratory epithelium reflect stable versus acute childhood asthma. J Allergy Clin Immunol. 2005;115:243–251. doi: 10.1016/j.jaci.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 4.Aoki T, Matsumoto Y, Hirata K, Ochiai K, Okada M, Ichikawa K, Shibasaki M, Arinami T, Sumazaki R, Noguchi E. Expression profiling of genes related to asthma exacerbations. Clin Exp Allergy. 2009;39:213–221. doi: 10.1111/j.1365-2222.2008.03186.x. [DOI] [PubMed] [Google Scholar]
- 5.Bjornsdottir US, Holgate ST, Reddy PS, Hill AA, McKee CM, Csimma CI, Weaver AA, Legault HM, Small CG, Ramsey RC, et al. Pathways activated during human asthma exacerbation as revealed by gene expression patterns in blood. PLoS One. 2011;6:e21902. doi: 10.1371/journal.pone.0021902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McErlean P, Berdnikovs S, Favoreto S, Jr, Shen J, Biyasheva A, Barbeau R, Eisley C, Barczak A, Ward T, Schleimer RP, et al. Asthmatics with exacerbation during acute respiratory illness exhibit unique transcriptional signatures within the nasal mucosa. Genome Med. 2014;6:1. doi: 10.1186/gm520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman ED, Reddel HK, Eriksson G, Peterson S, Ostlund O, Sears MR, Jenkins C, Humbert M, Buhl R, Harrison TW, et al. Overall asthma control: the relationship between current control and future risk J Allergy Clin Immunol 2010. 125:600–608 [DOI] [PubMed]
- 8.Persson H, Kwon AT, Ramilowski JA, Silberberg G, Söderhäll C, Orsmark-Pietras C, Nordlund B, Konradsen JR, de Hoon MJ, Melén E, et al. Transcriptome analysis of controlled and therapy-resistant childhood asthma reveals distinct gene expression profiles. J Allergy Clin Immunol. 2015;136:638–648. doi: 10.1016/j.jaci.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 9.Raby B, Barnes K, Beaty TH, Bosco A, Carey VJ, Castro M, Cheadle C, Gilliland FD, Talat K, Islam S, et al. Asthma BRIDGE: the Asthma Biorepository for Integrative Genomic Exploration [abstract]. Am J Respir Crit Care Med 20111836189 [Google Scholar]
- 10.Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials. 1999;20:91–120. [PubMed] [Google Scholar]
- 11.Cloutier MM, Schatz M, Castro M, Clark N, Kelly HW, Mangione-Smith R, Sheller J, Sorkness C, Stoloff S, Gergen P. Asthma outcomes: composite scores of asthma control. J Allergy Clin Immunol. 2012;129(Suppl. 3):S24–S33. doi: 10.1016/j.jaci.2011.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croteau-Chonka DC, Qiu W, Carey VJ, Weiss ST, Raby BA. Asthma BioRepository for Integrative Genomic Exploration (BRIDGE) Consortium. Gene set enrichment analyses of gene expression associations with asthma control hint at candidate drug pathways [abstract] Am J Respir Crit Care Med. 2014;189 [Google Scholar]
- 14.Yan X, Chu JH, Gomez J, Koenigs M, Holm C, He X, Perez MF, Zhao H, Mane S, Martinez FD, et al. Noninvasive analysis of the sputum transcriptome discriminates clinical phenotypes of asthma. Am J Respir Crit Care Med. 2015;191:1116–1125. doi: 10.1164/rccm.201408-1440OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brehm JM, Ramratnam SK, Tse SM, Croteau-Chonka DC, Pino-Yanes M, Rosas-Salazar C, Litonjua AA, Raby BA, Boutaoui N, Han YY, et al. Stress and bronchodilator response in children with asthma. Am J Respir Crit Care Med. 2015;192:47–56. doi: 10.1164/rccm.201501-0037OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, Himes BE, Levin AM, Mathias RA, Hancock DB, et al. Mexico City Childhood Asthma Study (MCAAS); Children’s Health Study (CHS) and HARBORS study; Genetics of Asthma in Latino Americans (GALA) Study, Study of Genes-Environment and Admixture in Latino Americans (GALA2) and Study of African Americans, Asthma, Genes & Environments (SAGE); Childhood Asthma Research and Education (CARE) Network; Childhood Asthma Management Program (CAMP); Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity (SAPPHIRE); Genetic Research on Asthma in African Diaspora (GRAAD) Study. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunninghake GM, Chu JH, Sharma SS, Cho MH, Himes BE, Rogers AJ, Murphy A, Carey VJ, Raby BA. The CD4+ T-cell transcriptome and serum IgE in asthma: IL17RB and the role of sex. BMC Pulm Med. 2011;11:17. doi: 10.1186/1471-2466-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy A, Chu JH, Xu M, Carey VJ, Lazarus R, Liu A, Szefler SJ, Strunk R, Demuth K, Castro M, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19:4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma S, Murphy A, Howrylak J, Himes B, Cho MH, Chu JH, Hunninghake GM, Fuhlbrigge A, Klanderman B, Ziniti J, et al. The impact of self-identified race on epidemiologic studies of gene expression. Genet Epidemiol. 2011;35:93–101. doi: 10.1002/gepi.20560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, Mesirov JP, Haining WN. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. 2016;44:194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma S, Zhou X, Thibault DM, Himes BE, Liu A, Szefler SJ, Strunk R, Castro M, Hansel NN, Diette GB, et al. A genome-wide survey of CD4(+) lymphocyte regulatory genetic variants identifies novel asthma genes. J Allergy Clin Immunol. 2014;134:1153–1162. doi: 10.1016/j.jaci.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127:153–160. doi: 10.1016/j.jaci.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 25.Raedler D, Ballenberger N, Klucker E, Böck A, Otto R, Prazeres da Costa O, Holst O, Illig T, Buch T, von Mutius E, et al. Identification of novel immune phenotypes for allergic and nonallergic childhood asthma. J Allergy Clin Immunol. 2015;135:81–91. doi: 10.1016/j.jaci.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 26.Lucas K, Maes M. Role of the toll like receptor (TLR) radical cycle in chronic inflammation: possible treatments targeting the TLR4 pathway. Mol Neurobiol. 2013;48:190–204. doi: 10.1007/s12035-013-8425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arts RJ, Joosten LA, Dinarello CA, Kullberg BJ, van der Meer JW, Netea MG. TREM-1 interaction with the LPS/TLR4 receptor complex. Eur Cytokine Netw. 2011;22:11–14. doi: 10.1684/ecn.2011.0274. [DOI] [PubMed] [Google Scholar]
- 28.Colonna M, Facchetti F. TREM-1 (triggering receptor expressed on myeloid cells): a new player in acute inflammatory responses. J Infect Dis. 2003;187:S397–S401. doi: 10.1086/374754. [DOI] [PubMed] [Google Scholar]
- 29.Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003;3:445–453. doi: 10.1038/nri1106. [DOI] [PubMed] [Google Scholar]
- 30.Dower K, Ellis DK, Saraf K, Jelinsky SA, Lin LL. Innate immune responses to TREM-1 activation: overlap, divergence, and positive and negative cross-talk with bacterial lipopolysaccharide. J Immunol. 2008;180:3520–3534. doi: 10.4049/jimmunol.180.5.3520. [DOI] [PubMed] [Google Scholar]
- 31.Nardelli B, Tiffany HL, Bong GW, Yourey PA, Morahan DK, Li Y, Murphy PM, Alderson RF. Characterization of the signal transduction pathway activated in human monocytes and dendritic cells by MPIF-1, a specific ligand for CC chemokine receptor 1. J Immunol. 1999;162:435–444. [PubMed] [Google Scholar]
- 32.Novak H, Müller A, Harrer N, Günther C, Carballido JM, Woisetschläger M. CCL23 expression is induced by IL-4 in a STAT6-dependent fashion. J Immunol. 2007;178:4335–4341. doi: 10.4049/jimmunol.178.7.4335. [DOI] [PubMed] [Google Scholar]
- 33.Buckland KF, Ramaprakash H, Murray LA, Carpenter KJ, Choi ES, Kunkel SL, Lukacs NW, Xing Z, Aoki N, Hartl D, et al. Triggering receptor expressed on myeloid cells-1 (TREM-1) modulates immune responses to Aspergillus fumigatus during fungal asthma in mice. Immunol Invest. 2011;40:692–722. doi: 10.3109/08820139.2011.578270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bucova M, Suchankova M, Dzurilla M, Vrlik M, Novosadova H, Tedlova E, Urban S, Hornakova E, Seligova M, Durmanova V, et al. Inflammatory marker sTREM-1 reflects the clinical stage and respiratory tract obstruction in allergic asthma bronchiale patients and correlates with number of neutrophils. Mediators Inflamm. 2012;2012:628754. doi: 10.1155/2012/628754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanchongkittiphon W, Mendell MJ, Gaffin JM, Wang G, Phipatanakul W. Indoor environmental exposures and exacerbation of asthma: an update to the 2000 review by the Institute of Medicine. Environ Health Perspect. 2015;123:6–20. doi: 10.1289/ehp.1307922. [DOI] [PMC free article] [PubMed] [Google Scholar]