Abstract
Modeling of protein structure from amino acid sequence now plays a major role in structural biology. Here we report new developments and progress from the CASP11 community experiment, assessing the state of the art in structure modeling. Notable points include the following: (1) New methods for predicting three dimensional contacts resulted in a few spectacular template free models in this CASP, whereas models based on sequence homology to proteins with experimental structure continue to be the most accurate. (2) Refinement of initial protein models, primarily using molecular dynamics related approaches, has now advanced to the point where the best methods can consistently (though slightly) improve nearly all models. (3) The use of relatively sparse NMR constraints dramatically improves the accuracy of models, and another type of sparse data, chemical crosslinking, introduced in this CASP, also shows promise for producing better models. (4) A new emphasis on modeling protein complexes, in collaboration with CAPRI, has produced interesting results, but also shows the need for more focus on this area. (5) Methods for estimating the accuracy of models have advanced to the point where they are of considerable practical use. (6) A first assessment demonstrates that models can sometimes successfully address biological questions that motivate experimental structure determination. (7) There is continuing progress in accuracy of modeling regions of structure not directly available by comparative modeling, while there is marginal or no progress in some other areas.
Keywords: Protein Structure modeling, Community Wide Experiment, CASP
INTRODUCTION
Experimental protein structures are currently available for less than 1/500th of the proteins with known sequences1. It has long been appreciated that in principle protein structure can be derived from amino acid sequence 1. As a result, many modeling methods have been developed, but it is not always clear how well they perform. This paper describes the 11th CASP community experiment to determine the state of the art in modeling protein three dimensional structure from amino acid sequence and summarizes the most notable developments.
CASP uses blind testing of modeling methods to assess their capabilities: Participants are provided with amino acid sequences of unknown structures and are asked to deposit structure models. These models are then compared with newly determined experimental structures. Results of the CASP experiments are published in special issues of this journal (PROTEINS 2-11). The structure of the CASP11 experiment, participation statistics and number of targets are very similar to those of recent rounds in the series, which have been conducted every two years since 1994, and are described in the Appendix 1 of the Supplementary Material. Altogether almost 60,000 models on 100 prediction targets were collected from 207 modeling groups representing about 100 research labs worldwide. A notable change from recent CASPs is the smaller fraction of targets obtained from large scale Structural Genomics projects (down to 60% this time, compared with 80% in CASP10).
Assessment of modeling performance is divided into a number of categories: models based on homologous templates (template based modeling – TBM, the most useful form of modeling), models produced without detectable homologous templates (free modeling – FM, now often effective for small proteins, but largely stuck for the last decade until new contact prediction methods partially came to the rescue this round), refinement (ability to improve on initial model – critical in template based modeling in order to move away from template bias, and an area with major improvements in the last few rounds), predicting the accuracy of a model (an area that has now advanced to the stage of real usefulness), predicting three dimensional contacts within structures (an area that saw dramatic achievement this round, after 20 years in the doldrums), and exploiting predicted contacts and sparse experimental structure data to build improved models (new in CASP, and so far encompassing NMR and chemical crosslinking). For the first time this CASP, we conducted an experiment for modeling quaternary structure (in collaboration with CAPRI 12) and a pilot assessment of the ability of models to address relevant biological questions. The trouble and expense of determining a structure experimentally is undertaken with specific questions in mind – under what circumstances can models successfully make that effort unnecessary? Full details of targets, participating groups, and all results are available on the CASP web site (http://predictioncenter.org/casp11).
RESULTS
Prediction of residue-residue contacts
The most exciting result in CASP11 was the generation of an accurate three dimensional model of a large (256 residues) protein as a consequence of much more accurate prediction of contacts between protein residues 13. The possibility of predicting three dimensional long range residue-residue contacts from evolutionary information was first recognized about the time CASP started, in 1994 14, and because of its promise, an assessment area on this topic was introduced early in the history of CASP 15. However, until CASP11, results were consistently disappointing, typically with over 80% false positives 16. It is now clear that a major cause of these false positives was a basic theoretical error in the way the data were treated: the methods make use of restrictions on residue type at pairs of positions in the sequence, imposed by three-dimensional proximity. For example, positive and negative charged side chains may be accommodate-able at a pair of contacting positions, but not two positive residues or two negative ones. Similarly, there may be space for a large and small side chain combination, but two large side chains would unavoidably clash, while two small ones may leave a cavity. In the past, measures such as the mutual information between pairs of columns in a multiple sequence alignment have been used to detect such residue type correlations. The flaw in that methodology can be illustrated as follows: suppose two residues at positions A and B are in contact, so the choice of residues in different members of the protein family will be correlated, providing useful information. Similarly if residue B is in contact with residue C, residues at these positions will also be correlated. But these interactions will also generate correlation between residues at A and residues at C, which may not be in contact, leading to a false positive. This kind of knock-on effect is in fact a well-known problem in statistics, when trying to deduce causation 17, and in statistical physics, between sets of interacting particles, for example in Ising models 18. The pre-existing methods for treating this problem have now been adapted for the contact prediction problem by a number of research groups 19-22. Deep, stable alignments are required, but when that condition is satisfied there have been a number of benchmarking studies that suggest good accuracy. In CASP, so far, there have been few targets with suitable alignments, but two such template free-modeling targets in CASP 11 yielded success well beyond that previously seen in CASP, both in protein size and model accuracy (Figure 1 shows one of these, T0806).
Figure 1.
(A) Model TS064_1 of a new-fold CASP11 target T0806 superimposed on the target structure, YaaA from E. coli K-12 (RMSD to target 3.6Å); (B) Structure of the very different best available template (PDB ID 2q07). This is the first CASP example of a high accuracy model for a large (256 residues) new fold target.
Model Refinement
Models based on homologous templates may display high accuracy, but at the same time, contain no new structural information. For example, two members of a protein family with different binding specificity may be similar at the overall structure and sequence levels, but contain local differences, ranging in scale from side chains changes to secondary structure shifts to mini-domains, critical to understanding functional properties 23. Two types of modeling are required to treat these regions. First, differences in main chain and side chain conformations can in principle be obtained by some form of refinement procedure starting from an initial template based model. Second, regions not covered by the closest template can sometimes be modeled using alternative templates or using ab initio based methods. In this section we summarize refinement assessment results, and a later section describes assessment results on non-principal template regions.
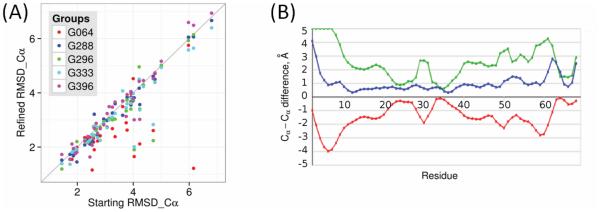
Earlier experience in CASP has shown that it is very challenging to refine an approximate template based model towards the experimental structure. In CASP8 in order to focus attention on this problem, a refinement assessment category was introduced 24. Refinement is a problem where physics based molecular dynamics methods should be effective, and so effort was devoted to encouraging a stronger representation of that community. For each refinement target, participants are provided with one of the best models of that protein obtained in the regular CASP experiment, for use as a starting point, and sometimes also with information identifying problem areas within a model. In the beginning, the results were quite disappointing, but in the last two CASPs, we have seen sustained progress. Figure 2A shows performance on all refinement targets for some of the best-performing groups. There are improvements of up to 2Å RMSD on Cα atoms, and only a small fraction of cases with a minor worsening of RMSD. Figure 2B shows an example illustrating the scale of improvement in local model accuracy that can be achieved.
Figure 2. CASP11 refinement performance.
(A) Cα RMSDs [Å] of the refined vs. original models for some of the best performing groups. Points below the diagonal represent improvement. Where models get worse in refinement, the loss of accuracy is small. In some cases, improvements of 2Å or more are achieved. (B) Best residue-by-residue refinement of the CASP11 target TR829 (Cα-Cα distance to target for the refined (TR829TS064_2, blue) and original (T0829TS499_1, green) models, and the difference between them (improvement, red) [Å]). In this case, there are substantial improvements in the areas that were least accurate (lowest portions of the red line) in the starting structure.
Modeling with the aid of sparse data
Modeling is increasingly used to interpret data obtained with a variety of experimental techniques that provide only sparse information on structure. In general, these combined approaches have the potential to provide information on structure in cases where crystals are too difficult to obtain or structures are too large to be solved by conventional NMR.
In CASP11, an assessment area in sparse data assisted modeling was introduced, with the goal of further spurring interest in developing modeling methods adapted to this task. Data for two experimental techniques were included.
The first is NMR. Modeling has the potential to utilize first pass, sparse NMR data to facilitate studies of larger structures, to provide improved structures of smaller proteins in cases where line broadening and other issues limit data quality, and to accelerate and reduce the cost of structure determination. Solution-state NMR can generally provide accurate 3D structures of small proteins (MW < ~15-20 kDa) 25,26. Larger proteins present a much greater challenge mostly due to the well-known limitations of short transverse spin relaxation rates, broad NMR linewidths, and chemical shift degeneracy. To alleviate these negative effects, sparse NMR data can be obtained using perdeuteration (i.e. replacement of most 1H atoms with 2H), which decreases transverse relaxation rates of the remaining 1H, 15N, and 13C nuclei, and shifts the limits for determining the structure of larger proteins towards the 40-80 kDa range 27-31. However, perdeuteration also reduces the number of 1H-1H NOE-based distance restraints that can be obtained, providing models that are less accurate and precise, and in many cases precluding structure determination by conventional NMR structural modeling techniques altogether. Some previous efforts have shown that combining sophisticated modeling techniques together with sparse NMR can produce higher accuracy structures 28,29,32. In CASP11 we conducted an initial experiment to objectively assess the extent to which current modeling methods can improve the accuracy of structures based on such data.
19 targets were included, ranging from 110 to 544 amino acids in size, all with no identifiable structure templates to allow modeling by homology. The Montelione group (Rutgers) provided simulated restraints from ambiguous NOESY cross peak assignments similar to those that could routinely be obtained from true sparse NMR experiments. Using crystal structures of the targets, the Montelione lab simulated NOESY peak lists and chemical shifts assuming uniformly 2H,13C,15N-enriched protein samples prepared with 1H-13C labeling of sidechain Ala, Leu, Val, and Ile(δ) methyl groups 29,33-36. The ASDP program 37-39 was then used to generate an initial set of both unique and ambiguous distance atom pair restraints from these NOESY peak lists. These distance-restraint data were distributed to the CASP community after the initial structure predictions were collected, but prior to release of the experimental coordinates. There was a wide range of performance by CASP groups, with the best consistently generating more accurate models than obtained using conventional NMR structure generation methods on the same data (benchmarked with CNS 40,41 and partially with ASDP, by the Montelione group and the CASP assessor) (Figure 3). Compared to the CNS results on the 19 target set, best CASP models were on average better by more than 17 GDT_TS units (Ca RMSDs lower by more than 1.1 Å), suggesting considerable potential in for the techniques developed by CASP participants.
Figure 3. Improvement in model accuracy using NMR simulated sparse data.
(A) Improvement in terms of the GDT_TS score for all the CASP11 modeling targets for which sparse data were available. Scores for the best original model submitted in CASP (“unassisted”, blue), results obtained with CNS (green), and obtained by CASP participants (red) utilizing the sparse data are shown. For most targets, CASP data assisted results show a dramatic improvement in model accuracy. For example, for the first target, Ts814, GDT_TS improves from 16 to 70, corresponding to a change in Cα RMSD from 21.1 Å to 2.6 Å. Improvements using conventional CNS procedures are considerably smaller. (B) Best models obtained for CASP11 target T0777. Left: Best unassisted model (RMSD to the experimental structure is 14.7 Å); Center: Experimental structure; Right: Best model obtained with sparse data (3.7 Å RMSD).
The second sparse data area is modeling using mass spectrometry data on residue-residue cross-links (XL-MS). Typically X-ray crystallography and NMR are the methods of choice for determining protein structure. In cases where these options are not viable, because of sample amount, or the absence of crystals and/or high quality NMR spectra, CX-MS combined with modeling can offer a possible alternative requiring smaller samples (nano to micromolar), simpler protocols, in many cases shorter time to obtain the structure, and lower costs 42,43. Residue-residue distance restraints, obtained from analysis of cross linked peptides, allow the generation of protein-protein interaction maps and potentially provide information on the structure of component domains. The usefulness of this approach has been demonstrated most strongly in the case of complexes, where comparatively few cross links are needed to determine relative subunit orientation, while solving the structure of the complete multidomain complex continues to present a challenge. For single chain proteins, the relatively small numbers of restraints, and their length (20-25Å), make development of specialized improved modeling techniques critical to success.
In CASP11 we focused on single proteins as targets for CX-MS assisted modeling, in collaboration with the Rappsilber lab (U. of Edinburgh/Technical U. Berlin). The Rappsilber group are using novel photo-crosslinking methodology, which effectively increases the density of generated restraints (from about 0.15 for standard Lys-Lys chemistry to > 2.5 contacts/residue) 44. Four protein samples, ranging from 204 to 420 amino acids in length, were obtained through CASP relations with target providers (two from individual researchers and two from the JCSG PSI structural genomics center). The Rappsilber group cross-linked these proteins, and used mass spectrometry to obtain distance restraints. These data were then provided to the CASP community and were used by 19 groups to model the structures. Little or no improvement was found in the models produced using the cross-link data. There are a number of reasons for this. Because of the very short data acquisition time necessitated by the CASP target release schedule, the experimental datasets had an imbalance in structure coverage and a comparatively small number of generated restraints (0.6-1.2 versus a potential >2.5 contacts/residue). A higher lysine content (lysines are the XL-MS reagent primary targets) and a more uniform distribution of digestion sites in proteins selected for the experiment would also have enhanced the experimental data quality. The apparent lack of improvement in model accuracy when the XL-MS constraints are used may also stem from the relatively early stage of development of the modeling techniques adapted for this task. For example, the molecular nature of the cross-links, including their position on the surface the protein, rather than simple Cartesian distance restraints should be taken into account. We expect to expand this segment of CASP in the next experiment, with more accurate single protein data sets as well as data for complexes.
Modeling of molecular assemblies
The structure of protein complexes is of increasing importance in biology. At the beginning of CASP, we also included some complexes as targets, until the CAPRI community experiments began specializing in this area 12. CASP11 included a joint CAPRI/CASP section for protein complexes, with participants from both communities and assessment based on well-established CAPRI procedures 12. CAPRI and CASP participants in this category were asked to model the quaternary structure of appropriate CASP oligomeric targets (dimers and tetramers). Modeling of three CASP targets that are transitory hetero-complexes was also included, giving a total of 27 targets. Twenty-nine CAPRI and fifteen CASP groups participated in this experiment. CASP and CAPRI participant best performances were similar. For example, the best CASP group submitted “acceptable” (by the CAPRI evaluation standards 12) models on 15 targets while the best CAPRI group did so on 16 (out of the 25 assessed targets). Of 12 participating servers, the best CASP server ranked 4th overall, submitting 9 acceptable models vs. 15 from the best CAPRI server. An important part of this test was sharing component modeling results across the communities, for use in docking. Generally, interface accuracy decreased with the accuracy of the monomer structure models. Overall, where homologous structures of complexes were available, results were encouraging. But it is clear that docking without the use of an interface template still presents major challenges. Detailed results of this experiment are presented in a separate paper in this issue (Lensink et al).
Estimation of model accuracy
Reliable a priori estimates of global and local accuracy of models are critical in determining the usefulness of a model to address a specific problem. Indeed one primary reason for the limited utilization of model structures by the broader biological community is the lack of information on what to believe and what not to believe. For these reasons, in CASP7 (2006) an assessment area on methods for estimating model accuracy was created 45.
This focus has resulted in a steady improvement in the capabilities of methods in this area 46-49 and has contributed to the development of new methods and improvement of the existing methods that are capable of estimating accuracy on the basis of a single model (as opposed to methods that require some form of clustering using large sets of models for the same target, a rather artificial scenario). The best single-model accuracy estimate methods are now as effective as clustering methods in recognizing best models in a set of candidates with an average accuracy error of 7% 49. These methods were shown to be particularly good in picking relatively good models for difficult targets, when the model pool is dominated by models of poor quality. On a finer resolution scale, the methods are able to distinguish between residues that are reasonably accurately modeled at the main chain level and those that are not (binary accuracy of 0.88, ROC-curve based accuracy of 0.92 (area under the curve) using the 3.8 Å Cα error threshold) 49. An example of a successful prediction of inaccurate regions in a model is shown in Figure 4. Overall, this focus area has established that methods of estimating accuracy, while not perfect, are already very useful.
Figure 4. Example of successfully identifying an inaccurate region in a model.
(A) Structural superposition of the model T0766TS160_2 (magenta) and CASP11 target T0766 (cyan); (B) The poorly modeled strand-turn-strand-helix motif (residues 52-64) is detected by the accuracy estimator ModFOLD-single59 (green), with predicted main chain accuracy closely tracking the actual error curve (blue) (red – difference between the estimated and actual error).
Suitability of models for answering relevant functional questions
In CASP, models are primarily evaluated by comparison of their coordinates with those of the corresponding experimental structure, and this remains the gold standard. However, the trouble and expense of determining an experimental structure is almost always undertaken with the goal of answering a specific biological question. In practice therefore, the key question is not whether the model is as accurate as an experimental structure would be – it often is not – but whether it is accurate enough to answer the relevant biological question. At one extreme, some questions can be addressed with very low accuracy models – for example the location of likely epitopes. At the other extreme, very high accuracy may be required, for, say, drug design purposes 50. Thus, one important criterion on which to judge a model is the degree to which it answers the relevant questions. In CASP 11, we included a pilot experiment on assessing models in these terms. The analysis was done by Roland Dunbrack, assessor in the template-based category. For 39 target proteins, at least some information on function was available from the target contributors. In some cases, the target provider supplied specific functional questions, in others, the assessor judged likely motivation for getting the structure. Some examples: (1) For the complex of the protein kinase II leucine zipper and the ab11b GTPase (targets 797 and 798), the question of interest was the relative orientation of the components of the heterodimer. A number of groups successfully modeled the full structure. (2) For target 792, the LOTUS domain of Drosophila Oskar, the question was the structure of the homodimer. Assessment showed that docking using the best CASP models of the monomer produced the correct structure. (3) A number of targets have disease relevant mutations, and the assessor evaluated whether the impact of these on molecular function could be deduced from the models. In some cases, such as target 783, Human Isoprenoid Synthase, where mutations occurring in the interior of a domain cause a monogenic disease, dystroglycanopathy, this was judged possible. In others, such as target 794, VNN1, containing mutations involved in cancer, results were mixed.
Backbone and Alignment Accuracy
Figure 5 compares the backbone accuracy of models for each of the 11 CASPs as a function of target difficulty (Appendix 2 in Supplementary Material), in terms of the standard CASP measure, GDT_TS 51,52. While progress from CASP to CASP was marked in the earlier experiments, recently, by this measure, there is little progress. Comparison of alignment accuracy as a function of target difficulty also shows no apparent progress in recent CASPs (Appendix 3, Supplementary Figure S4). As noted earlier, the majority of targets in each CASP are suitable for comparative modeling, starting with alignment of the target sequence to those of possible structure templates so that a lack of progress in alignment is consistent with the results for backbone accuracy.
Figure 5. Best GDT_TS scores of submitted models for targets in all CASPs, as a function of target difficulty.
For recent CASPs, human/server targets only are included, and in earlier CASPs - all targets. Trend line for CASP11 runs similar to other CASPs (starting from CASP5) in the mid- and hard- sections of the difficulty range and is shorter and lower at the easy end (as there were no very easy human/server targets in CASP11, and a few short non-globular domains marked on the graph pull the curve down in that area).
It is possible that the approximate method of determining target difficulty masks progress in model accuracy. To see if this is the case, we consider an alternative way of estimating progress between CASPs, rather than attempting to normalize for target difficulty. That is to compare performance of current methods with the earlier ones on the same targets. There are two modeling servers in CASP where this is possible – the methodology has not been altered, but it is possible to use contemporary structure and sequence databases. One of these is SAM-T08 53, unaltered since CASP8. Figure 6 shows a comparison of the GDT_TS scores for the best models for each target in CASPs 8, 9, 10 and 11 with the corresponding model from SAM-T08. The SAM-T08 results were obtained using the current sequence and structure databases available at the time of each CASP. Results here also suggest no substantial progress in overall model accuracy. Similar calculations were performed using another frozen server method (FFAS03) and for the human-expert groups (Baker and Zhang) rather than best models. All results are similar, and so not included in here.
Figure 6. Comparison of backbone accuracy of the best CASP models (CASP8-11) with the results of the frozen-in-time prediction method (SAM-T08).
Trend lines are very similar, suggesting no substantial progress.
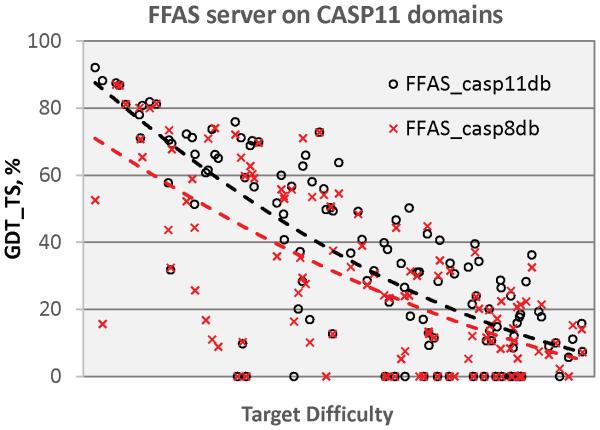
Although overall model accuracy has not improved much when the increased size of sequence and structure databases is taken into account (Figures 5 and 6), examination of improvement in accuracy without correcting for database changes shows impressive improvement (Figure 7). This figure compares performance of the FFA03 server on CASP11 targets using then contemporary databases and using databases from the time of CASP8, six years earlier. A substantial fraction of CASP11 targets now have much improved templates available resulting in improved accuracy, in some cases dramatically so, with improvements in model accuracy of 60 GDT_TS units.
Figure 7.
Comparison of GDT_TS scores for models of CASP11 targets generated with a reference CASP server (FFAS03) using sequence and structure databases available during CASP11 (black) and using the databases available during the CASP8 experiment (red). Quadratic trend lines show that FFAS models using contemporary databases are often substantially improved over those possible six years earlier, because of increased database size, particularly for the now less difficult targets. Most of the improvement comes from the increased availability of suitable structure templates.
Accuracy of regions structurally divergent from a principal template
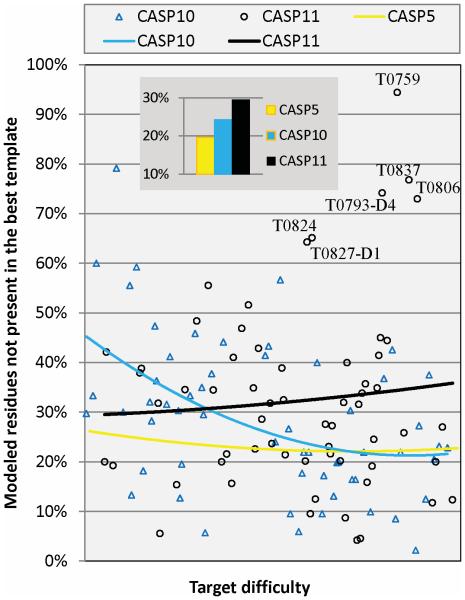
In contrast to the rather discouraging picture of no progress presented by the overall backbone and alignment accuracy measures, examination of the accuracy of regions not model-able from the closest template is more encouraging. A single template will usually not provide a structural scaffold for all of the residues in a target protein, and in the middle range of target difficulty, 50% or more of target may not be covered. Modeling of these non-principal template covered regions (sometimes misleadingly called ‘loops’) will often be key to correctly characterizing functional differences between the template protein and the target, so that progress in this area is important. Figure 8 shows the % of ‘loop’ residues correctly modeled (Cα error less than 3.8Å in a global superposition) for each target with at least 15 of such residues. As we have noted previously, there was a substantial improvement between CASP5 (yellow) and CASP10 (blue) over most of the target difficulty range, except for the most difficult, mostly non-template, targets. CASP11 (black) shows a further improvement from CASP10, with the most gain towards the difficult end of the target range. The average coverage histogram shows the overall improvement at about 5%, from 25 to 30, about the same as the whole gain from CASP5 to CASP10. Examination of the individual targets shows that the improvement is driven by outstanding performance for six targets, two of which (T0806 and T0824) are the result of the use of new contact prediction methods (discussed above).
Figure 8. Percentage of residues successfully modeled that were not available from the single best template.
Only targets in which at least 15 residues could not be aligned to the best template are included. Each point represents the best model for a target in CASP10 and 11. Quadratic fit lines are threaded through the data. The trend line for CASP5 is shown for comparison. The insert shows average improvement percentage over all targets in CASP5, 10 and 11. Clearly, CASP11 performance in this aspect improved over that of CASP5 and CASP10.
DISCUSSION
Results from this and other recent CASP experiments are somewhat paradoxical. On the one hand, particular areas of modeling show impressive progress. As documented above, this time these were contact prediction, refinement, assignment of model accuracy, and modeling of non-principal templates regions. Not discussed here, the stereo-chemical quality of models also continues to improve. These are all important aspects of modeling, and the progress is heartening to see. Also, comparison of performance on CASP 11 targets using then contemporary databases and those available only six years earlier (figure 7) shows dramatic improvement in backbone accuracy for many targets. In practical terms, then, the fraction of protein domains for which good models can be built is increasing rapidly.
On the other hand, overall backbone accuracy, after taking into account improvement in database coverage of sequence and structure has changed little in a decade (figures 5 and 6). Why is that? We offer the following explanation. For comparative models, overall accuracy is dominated by the accuracy of sequence alignment with a primary template. Supplementary figure S3 shows that overall alignment accuracy as a function of target difficulty has a very similar form to that for overall backbone accuracy (figure 5). Alignment accuracy increased dramatically in the first decade of CASP, and the fraction of misaligned residues is typically not more than 10 to 15% of the alignable regions. At that level, it is doubtful that sequence based methods can yield much further improvement – alternative alignments must be explored and evaluated structurally, and that is still not viable.
Improvement in the accuracy of non-principal template regions in the last ten years is substantial – from 20% to 30% modeled correctly by the criteria used. There is a long way to go still, but improvements continue. Similarly, in the last few CASPs, refinement has advanced from totally ineffective to typically making small gains in accuracy, a major achievement.
As noted in previous papers in this series54,55, in template free (FM) modeling, over the last decade there has been substantial progress in accuracy for small (less than 100 residues) targets, building on earlier methodological advances but the techniques used have not appeared to scale to larger structures. But encouragingly, in this CASP, the FM assessor found evidence of substantial progress across the size spectrum56. Very significant issues are still to be overcome however.
As noted earlier, the most dramatic result in this CASP was for two template free targets where the improved methods for predicting three-dimensional contacts could be applied (T0826 and T0824-D1). This is the first objective evidence that these methods do work under appropriate circumstances. The primary requirement is a high quality deep alignment. The very rapid rate of sequencing now occurring is likely to bring many targets within range of this approach in the next few years, and has the potential to transform the field.
While this prospect is encouraging, it should be noted that contact information will not be as strong for regions of structure that reflect varying function within a family, similar in nature to those regions not covered by a template in current modeling. Improved methods will still be needed there (and of course in refinement). The improvement to date has come from two sources. The first is that some of these regions are covered by alternative templates. A great deal of effort has been put into developing methods that utilize information from multiple templates (see for example 57), but either the data are still too limited or the methods are not yet optimal. The other approach to modeling these regions is more ab initio in nature. Given the progress in template free modeling of small proteins, it is puzzling that such techniques have not had a greater impact on this problem. It is not clear whether that is because it is harder than it looks (although the regions themselves are often relatively small, models rely on an accurate environment provided by the rest of the structure) or the problem has not yet received enough attention. CASP will continue to particularly encourage efforts in this area.
Perhaps the area where progress is of most practical significance (as opposed to dramatic) is that of estimating the accuracy of models, both overall and at the residue level. Models will never be perfect, and have a wide range of accuracy, so that if no information on error levels is provided, they are almost useless. Reliability of the accuracy estimators is already high enough to be of considerable practical value 49,58. For example, if a structure model is the basis for interpreting the impact of a genetic mutation, it may be possible to determine whether the local model features are accurate enough for the purpose. Current methods are broadly of two types – those that use consensus information across a number of models – where the models agree, the structure is likely more accurate; and those based on some kind of evaluation of the atomic interactions in a model, including methods that use potentials of mean force and other energy related approaches. Consensus methods work surprisingly well, but have obvious limitations. For example, the exceptional models for two free modeling targets discussed earlier (T0806 and T0824-D1) would appear very inaccurate with that approach, because no other models come close. An encouraging development this CASP was improved performance of the energy related approaches, so that by some measures they are now competitive with consensus ones.
CASP continues to explore new areas of modeling. This time, tests of exploiting sparse NMR data were more extensive, and chemical crosslinking data were also considered. Results from NMR are encouraging, but these were still simulated data, and did not include one of the most useful data types, chemical shifts. Thus it is hard yet to judge the significance of the results. By and large, the efforts with crosslinking data were not very effective, probably because of limited and poor data quality. This is an area CASP plans to develop further next time. The collaboration with CAPRI on testing methods for modeling protein complexes reflects recognition that many biological questions of interest involve protein-protein interaction of some form. In spite of the many years of effort the docking community has made in this area, overall, results were not impressive. We expect that working together will speed progress in future. As noted earlier, the real test of a model is not whether it is similar to the experimental structure but whether it can answer some relevant biological question or be used to generate useful hypotheses. For that reason, we plan to continue to emphasize this area.
Further information
The other papers in this PROTEINS virtual special issue provide reports by some of the better performing prediction teams, a description of the targets as well as an analysis of some of them by the target providers, and the assessments. A list of the papers is provided in Supplementary table 1. All the modeling and assessment papers in this issue have been peer reviewed. The CASP web site (http://predictioncenter.org) provides extensive details of the targets, the predictions, and the numerical analyses. A CASP12 experiment is planned, beginning in the spring of 2016, and culminating in a meeting in December of that year. Those interested should check the CASP web site for further announcements.
Supplementary Material
Acknowledgments
The organizers greatly appreciate the dedication, creativity, and hard work of the assessment teams, led by Roland Dunbrack (Fox Chase Cancer Institute, responsible for Template based modeling, refinement, and biological relevance assessments) and Nick Grishin (UT Southwestern medical school and HHMI, responsible for Template free Contact assisted and CASP ROLL assessments). As always, we are grateful to the members of the experimental community, particularly the structural genomics centers, who provided targets. Taking part required courage and commitment on the part of all the modeling groups. Once again the assessment teams worked extremely hard and effectively to extract major insights from the results. Many thanks to L. Jaroszewski and Z. Li for re-configuration of the FFAS server so that it could use old CASP8 databases for building models. We again thank PROTEINS for providing a mechanism for peer reviewed publication of the outcome of the experiment.
The Prediction Center is supported by a grant from the US National Institute of General Medical Sciences (NIGMS/NIH) – grant R01GM100482 to KF. Partial meeting support was provided by the grant R13GM109649 to KF.
Footnotes
As of December 2015, 106K structures in the PDB vs 55M sequences in the UniProtKB.
References
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(96):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23(3):ii–v. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 3.Moult J, Hubbard T, Bryant SH, Fidelis K, Pedersen JT. Critical assessment of methods of protein structure prediction (CASP): round II. Proteins. 1997;(Suppl 1):2–6. [PubMed] [Google Scholar]
- 4.Moult J, Hubbard T, Fidelis K, Pedersen JT. Critical assessment of methods of protein structure prediction (CASP): round III. Proteins. 1999;(Suppl 3):2–6. [PubMed] [Google Scholar]
- 5.Moult J, Fidelis K, Zemla A, Hubbard T. Critical assessment of methods of protein structure prediction (CASP): round IV. Proteins. 2001;(Suppl 5):2–7. [PubMed] [Google Scholar]
- 6.Moult J, Fidelis K, Zemla A, Hubbard T. Critical assessment of methods of protein structure prediction (CASP)-round V. Proteins. 2003;53(Suppl 6):334–339. doi: 10.1002/prot.10556. [DOI] [PubMed] [Google Scholar]
- 7.Moult J, Fidelis K, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)--round 6. Proteins. 2005;61(Suppl 7):3–7. doi: 10.1002/prot.20716. [DOI] [PubMed] [Google Scholar]
- 8.Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69(Suppl 8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moult J, Fidelis K, Kryshtafovych A, Rost B, Tramontano A. Critical assessment of methods of protein structure prediction - Round VIII. Proteins. 2009;77(Suppl 9):1–4. doi: 10.1002/prot.22589. [DOI] [PubMed] [Google Scholar]
- 10.Moult J, Fidelis K, Kryshtafovych A, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)--round IX. Proteins. 2011;79(Suppl 10):1–5. doi: 10.1002/prot.23200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)--round x. Proteins. 2014;82(Suppl 2):1–6. doi: 10.1002/prot.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lensink MF, Wodak SJ. Docking, scoring, and affinity prediction in CAPRI. Proteins. 2013;81(12):2082–2095. doi: 10.1002/prot.24428. [DOI] [PubMed] [Google Scholar]
- 13.Monastyrskyy B, D'Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. New encouraging developments in contact prediction: Assessment of the CASP11 results. Proteins. 2015 doi: 10.1002/prot.24943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gobel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 1994;18(4):309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 15.Lesk AM. CASP2: report on ab initio predictions. Proteins. 1997;(Suppl 1):151–166. doi: 10.1002/(sici)1097-0134(1997)1+<151::aid-prot20>3.3.co;2-j. [DOI] [PubMed] [Google Scholar]
- 16.Monastyrskyy B, D'Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue-residue contact prediction in CASP10. Proteins. 2014;82(Suppl 2):138–153. doi: 10.1002/prot.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baba K, Shibata R, Sibuya M. Partial correlation and conditional correlation as measures of conditional independence. Aust Nz J Stat. 2004;46(4):657–664. [Google Scholar]
- 18.Binney JJ, Dowrick NJ, Fisher AJ, Newman MEJ. The Theory of Critical Phenomena: An Introduction to the Renormalization Group. Clarendon Press; Oxford: 1992. [Google Scholar]
- 19.Burger L, van Nimwegen E. Disentangling direct from indirect co-evolution of residues in protein alignments. PLoS Comput Biol. 2010;6(1):e1000633. doi: 10.1371/journal.pcbi.1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A. 2011;108(49):E1293–1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones DT, Buchan DW, Cozzetto D, Pontil M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28(2):184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- 22.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci U S A. 2013;110(39):15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim K, Zhang H, Tempczyk A, Bonander N, Toedt J, Howard A, Eisenstein E, Herzberg O. Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine. Proteins. 2001;45(4):397–407. doi: 10.1002/prot.10004. [DOI] [PubMed] [Google Scholar]
- 24.MacCallum JL, Hua L, Schnieders MJ, Pande VS, Jacobson MP, Dill KA. Assessment of the protein-structure refinement category in CASP8. Proteins. 2009;77(Suppl 9):66–80. doi: 10.1002/prot.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao B, Guan R, Montelione GT. Improved technologies now routinely provide protein NMR structures useful for molecular replacement. Structure. 2011;19(6):757–766. doi: 10.1016/j.str.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao B, Tejero R, Baker D, Montelione GT. Protein NMR structures refined with Rosetta have higher accuracy relative to corresponding X-ray crystal structures. Journal of the American Chemical Society. 2014;136(5):1893–1906. doi: 10.1021/ja409845w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321(5893):1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, Kennedy MA, Prestegard J, Montelione GT, Baker D. NMR structure determination for larger proteins using backbone-only data. Science. 2010;327(5968):1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(27):10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tugarinov V, Choy WY, Orekhov VY, Kay LE. Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(3):622–627. doi: 10.1073/pnas.0407792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grishaev A, Tugarinov V, Kay LE, Trewhella J, Bax A. Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints. Journal of Biomolecular Nmr. 2008;40(2):95–106. doi: 10.1007/s10858-007-9211-5. [DOI] [PubMed] [Google Scholar]
- 32.Sgourakis NG, Natarajan K, Ying J, Vogeli B, Boyd LF, Margulies DH, Bax A. The Structure of Mouse Cytomegalovirus m04 Protein Obtained from Sparse NMR Data Reveals a Conserved Fold of the m02-m06 Viral Immune Modulator Family. Structure. 2014;22(9):1263–1273. doi: 10.1016/j.str.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner KH, Rosen MK, Kay LE. Global folds of highly deuterated, methyl-protonated proteins by multidimensional NMR. Biochemistry. 1997;36(6):1389–1401. doi: 10.1021/bi9624806. [DOI] [PubMed] [Google Scholar]
- 34.Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE. Selective methyl group protonation of perdeuterated proteins. Journal of Molecular Biology. 1996;263(5):627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 35.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nature Protocols. 2006;1(2):749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 36.Zheng D, Huang YJ, Moseley HN, Xiao R, Aramini J, Swapna GV, Montelione GT. Automated protein fold determination using a minimal NMR constraint strategy. Protein science : a publication of the Protein Society. 2003;12(6):1232–1246. doi: 10.1110/ps.0300203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang YJ, Moseley HN, Baran MC, Arrowsmith C, Powers R, Tejero R, Szyperski T, Montelione GT. An integrated platform for automated analysis of protein NMR structures. Methods in enzymology. 2005;394:111–141. doi: 10.1016/S0076-6879(05)94005-6. [DOI] [PubMed] [Google Scholar]
- 38.Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. Journal of the American Chemical Society. 2005;127(6):1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- 39.Huang YJ, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 2006;62(3):587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. Pt 5. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2(11):2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 42.Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173(3):530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walzthoeni T, Leitner A, Stengel F, Aebersold R. Mass spectrometry supported determination of protein complex structure. Curr Opin Struct Biol. 2013;23(2):252–260. doi: 10.1016/j.sbi.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Belsom A, Schneider M, Fischer L, Brock O, Rappsilber J. Serum Albumin Domain Structures in Human Blood Serum by Mass Spectrometry and Computational Biology. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.M115.048504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cozzetto D, Kryshtafovych A, Ceriani M, Tramontano A. Assessment of predictions in the model quality assessment category. Proteins. 2007;69(Suppl 8):175–183. doi: 10.1002/prot.21669. [DOI] [PubMed] [Google Scholar]
- 46.Cozzetto D, Kryshtafovych A, Tramontano A. Evaluation of CASP8 model quality predictions. Proteins. 2009;77(Suppl 9):157–166. doi: 10.1002/prot.22534. [DOI] [PubMed] [Google Scholar]
- 47.Kryshtafovych A, Fidelis K, Tramontano A. Evaluation of model quality predictions in CASP9. Proteins. 2011;79(Suppl 10):91–106. doi: 10.1002/prot.23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kryshtafovych A, Barbato A, Fidelis K, Monastyrskyy B, Schwede T, Tramontano A. Assessment of the assessment: evaluation of the model quality estimates in CASP10. Proteins. 2014;82(Suppl 2):112–126. doi: 10.1002/prot.24347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kryshtafovych A, Barbato A, Monastyrskyy B, Fidelis K, Schwede T, Tramontano A. Methods of model accuracy estimation can help selecting the best models from decoy sets: Assessment of model accuracy estimations in CASP11. Proteins. 2015 doi: 10.1002/prot.24919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294(5540):93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 51.Zemla A, Venclovas Moult J, Fidelis K. Processing and evaluation of predictions in CASP4. Proteins. 2001;(Suppl 5):13–21. doi: 10.1002/prot.10052. [DOI] [PubMed] [Google Scholar]
- 52.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karplus K. SAM-T08, HMM-based protein structure prediction. Nucleic Acids Res. 2009;37:W492–497. doi: 10.1093/nar/gkp403. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kryshtafovych A, Fidelis K, Moult J. CASP9 results compared to those of previous CASP experiments. Proteins. 2011;79(Suppl 10):196–207. doi: 10.1002/prot.23182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kryshtafovych A, Fidelis K, Moult J. CASP10 results compared to those of previous CASP experiments. Proteins. 2014;82(Suppl 2):164–174. doi: 10.1002/prot.24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinch LN, Li W, Monastyrskyy B, Kryshtafovych A, Grishin NV. Evaluation of free modeling targets in CASP11 and ROLL. Proteins. 2015 doi: 10.1002/prot.24973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7(8):1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kryshtafovych A, Fidelis K. Protein structure prediction and model quality assessment. Drug Discov Today. 2009;14(7/8):386–393. doi: 10.1016/j.drudis.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGuffin LJ, Buenavista MT, Roche DB. The ModFOLD4 server for the quality assessment of 3D protein models. Nucleic Acids Res. 2013;41:W368–372. doi: 10.1093/nar/gkt294. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.