Abstract
MicroRNA are well-established players in post-transcriptional gene regulation. However, information on the effects of microRNA deregulation mainly relies on bioinformatic prediction of potential targets, whereas proof of the direct physical microRNA/target messenger RNA interaction is mostly lacking. Within the International Cancer Genome Consortium Project “Determining Molecular Mechanisms in Malignant Lymphoma by Sequencing”, we performed miRnome sequencing from 16 Burkitt lymphomas, 19 diffuse large B-cell lymphomas, and 21 follicular lymphomas. Twenty-two miRNA separated Burkitt lymphomas from diffuse large B-cell lymphomas/follicular lymphomas, of which 13 have shown regulation by MYC. Moreover, we found expression of three hitherto unreported microRNA. Additionally, we detected recurrent mutations of hsa-miR-142 in diffuse large B-cell lymphomas and follicular lymphomas, and editing of the hsa-miR-376 cluster, providing evidence for microRNA editing in lymphomagenesis. To interrogate the direct physical interactions of microRNA with messenger RNA, we performed Argonaute-2 photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation experiments. MicroRNA directly targeted 208 messsenger RNA in the Burkitt lymphomas and 328 messenger RNA in the non-Burkitt lymphoma models. This integrative analysis discovered several regulatory pathways of relevance in lymphomagenesis including Ras, PI3K-Akt and MAPK signaling pathways, also recurrently deregulated in lymphomas by mutations. Our dataset reveals that messenger RNA deregulation through microRNA is a highly relevant mechanism in lymphomagenesis.
Introduction
B-cell lymphomas account for approximately 85% of all lymphomas and form a heterogeneous group of lymphoid neoplasms arising at different stages of B-cell development.1 They are classified according to morphological and immunophenotypic features, supplemented by characteristic genomic translocations (WHO 2008). Although these features allow the diagnosis of different histological subtypes of B-cell lymphomas, molecular subtypes remain largely indistinguishable.2 Presumably due to this molecular heterogeneity, many patients do not respond well to common therapy regimens.3 New biomarkers and therapeutic targets need, therefore, to be identified in order to improve the accuracy of lymphoma diagnosis and subsequent therapy selection.
One potential class of biomarkers and/or therapeutic targets is a subset of RNA molecules named microRNA (miRNA). These are small non-coding RNA (17–25 nucleotides in length) that bind mostly to target sequences within the 3’ untranslated region of messenger RNA (mRNA). MiRNA regulate the expression of thousands of mRNA including those with key roles in cell differentiation and cancer pathogenesis.4 MiRNA influence immune cell differentiation and play crucial roles in both early and late B-cell differentiation5 and lymphomagenesis.6 Mechanisms of miRNA dysregulation in lymphomas include copy number alterations (e.g. the miR-17~92 polycistron7), chromosomal translocation (e.g. hsa-miRNA-1258) and mutations (e.g. hsa-miR-1429). Several molecular profiling studies have tried to assess differential miRNA expression in B-cell lymphomas, as recently described.5,6,10 It has been reported that a signature of 38 miRNA containing MYC-regulated and nuclear factor-κB pathway-associated miRNA differentiates Burkitt lymphoma (BL) from diffuse large B-cell lymphoma (DLBCL).11
Available data on miRNA expression profiling in B-cell lymphomas is, however, still preliminary, as published profiles are either mostly not derived from large collections of samples, do not compare subtypes or originate from either quantitative real time polymerase chain reaction (qRT-PCR)-based approaches or microarrays. Next-generation sequencing is able to overcome the disadvantages of previous methods such as probe cross-hybridization12 and the limitations of qRT-PCR, such as restricting the analysis to previously known miRNA. Furthermore, sequencing-based approaches allow for the discovery of novel miRNA and large-scale identification of mutated miRNA.
The present study was performed within the framework of the International Cancer Genome Consortium Project “Determining Molecular Mechanisms in Malignant Lymphoma by Sequencing” (ICGC MMML-Seq). Our aim was to identify, using next-generation sequencing, miRNA signatures in three common subtypes of B-cell lymphomas, BL, DLBCL and follicular lymphoma (FL), and to correlate these to mRNA expression and genomic mutations. Moreover, by performing photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) experiments13 and intersecting the results with the patient-derived mRNA/miRNA expression profiles, we aimed at identifying specific miRNA-mRNA target pairs in BL and DLBCL.
Methods
Patients’ samples
The ICGC MMML-Seq project was approved by the Institutional Review Board of the Medical Faculty of Kiel University (A150/10) and by the recruiting centers. Informed consent was obtained from all patients (or, in the case of children, from their legal guardians). Histopathological, immunophenotypic and genetic characterization of the tumor samples and initial diagnosis (tumor cell content ≥60%) was performed as described recently.14
Next-generation sequencing
Nucleic acids were extracted as previously detailed.14 Libraries for miRNA sequencing were prepared using TruSeq Small RNA sample prep kits (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. with 100 ng – 1 μg total RNA as input. Libraries were size-fractionated on 6 × TBE gels (Life Technologies, Carlsbad, CA, USA). DNA concentration and sizes were analyzed on a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Seven pmol of DNA of each library were loaded onto a flow cell (multiplexing up to four libraries per lane), 50-cycle sequencing was performed using the TruSeq SBS Kit v3 on the HiSeq 2500 (Illumina).
Whole genome sequencing data of tumors and matched controls and transcriptome sequencing data of tumors were generated by the ICGC MMML-Seq project as previously described.14 All sequencing data have been deposited at the European Genomephenome Archive (EGA, http://www.ebi.ac.uk/ega/, accession number EGAS00001001394).
Quantitative real-time polymerase chain reactions
Undiluted reverse transcriptase reactions (20 ng of RNA per sample) were combined with TaqMan Universal Master Mix II (no UNG) (Life Technologies) and amplified (7500HT Real-Time PCR System, Life Technologies) with RNU24 and RNU48 as housekeeping genes. Experiments were performed in triplicate and analyzed using the 2−ΔΔCT method.
AGO2 immunoprecipitation
AGO2-PAR-CLIP was carried out as previously described13 with modifications (mostly relating to the washing steps during immunoprecipitation) because of the use of monoclonal anti-AGO2 antibody (#4-642, EMD Millipore, Billerica, MA, USA).15 In brief, following the addition of 4-thiouridine, an immunoprecipitation using a monoclonal anti-AGO2 antibody isolated the RNA-protein complexes. After protein digestion, sequencing adapters were ligated to the purified RNA fragments. Following reverse transcription, PAR-CLIP libraries were sequenced on a HiSeq2500 (Illumina).16
Bioinformatic methods
Genome and transcriptome analysis
Bioinformatic analyses of the genome and transcriptome data were performed as described recently, employing the various pipelines established in the ICGC MMML-Seq14 (additional information is provided in the Online Supplementary Methods).
MicroRNA and immunoprecipitation analysis
Following adapter removal, reads were mapped onto the human genome (1000 genomes project, hs37d5100) using segemehl.17 Novel miRNA prediction was performed using miRanalyzer 0.318 (default parameters), target prediction using miRanda19 (miRsvr-score < −1.2).
After filtering and trimming the PAR-CLIP reads, we obtained a total of 62,281,382 single-end reads, which were aligned with BWA20 with up to two mismatches between a read sequence and the reference sequence (hg19). All reads failing this mapping were aligned against the transcriptome database (Ensembl Genes 75). Aligned reads were piled into clusters by PARA-suite (Kloetgen et al., submitted). As PAR-CLIP reads contain thymidine to cytidine (T-C) conversions at the sites of crosslinks, all identified clusters were filtered to receive the most confident target regions. Excluding clusters containing <5 reads and <25% T-C conversions (excluding 100% T-C conversion sites as these might result from single nucleotide variants) resulted in (prior to pooling) 1,329 clusters for SU-DHL-4, 1,517 clusters for SU-DHL-6, 1,209 clusters for Namalwa and 425 clusters for Raji. Further details (including miRNA-mRNA correlation analyses) are given in the Online Supplementary Methods.
Results
Molecular classification of Burkitt lymphomas versus diffuse large B-cell and follicular lymphomas using a 25 miRNA classifier
We profiled tumor samples from 56 patients including 16 with BL (based on a molecular classifier; all patients ≤18 years), 19 with DLBCL (including 7 with germinal center DLBCL, 10 activated B-cell DLBCL and 2 with type III DLBCL) and 21 with FL (mainly grade 1/2) (Online Supplementary Table S1). We obtained 1,169,752,727 sequencing reads in total (average of 20,888,442 reads per sample, Online Supplementary Table S2). Following normalization of miRNA reads, we performed an unsupervised hierarchical clustering. Unexpectedly (and differently to what we observed at the transcriptomic level, data not shown), no clear distinction between BL, DLBCL and FL was achieved based on miRNA expression profiles (Online Supplementary Figure S1A). We then ranked the miRNA by mean expression and, discarding those that showed little expression variability, chose the top ten miRNA for validating our next-generation sequencing data by qRT-PCR. A correlation analysis showed the consistency of miRNA expression levels regardless of the method of quantification employed [Spearman’s rank correlation test, 10/10, high correlation (R>0.7), P-values for the correlation between qRT-PCR expression and next-generation sequencing expression ≤0.05 in 7/10 cases; details on all P-value calculations are given in the Online Supplementary Bioinformatic Methods)] (Online Supplementary Figure S1B, Online Supplementary Table S3).
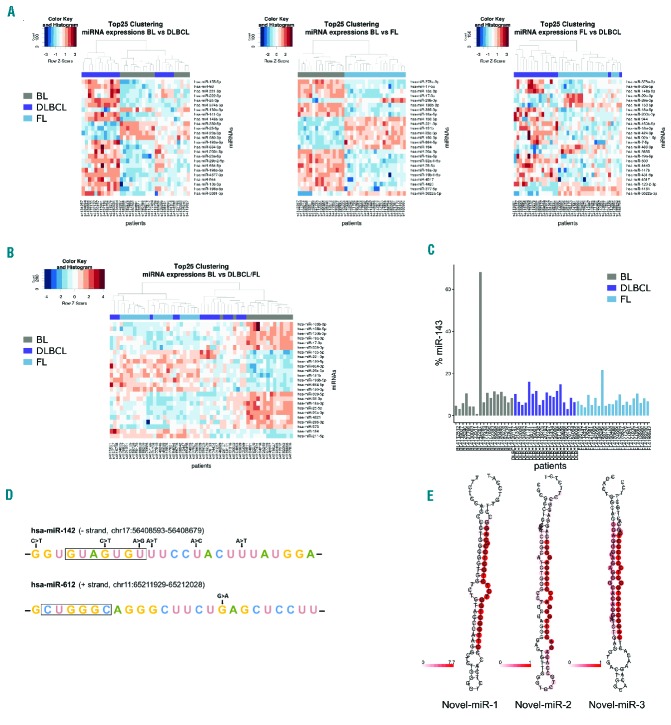
To recognize subtler molecular differences that escape unsupervised clustering approaches, we performed a differential gene expression analysis between BL versus DLBCL, BL versus FL and DLBCL versus FL using edgeR (Online Supplementary Table S4 and Online Supplementary Bioinformatic Methods). Clustering of the top 25 differentially expressed miRNA between each two lymphoma subtypes (BL/DLBCL, BL/FL, and FL/DLBCL) revealed separation according to the subtypes (Figure 1A). Employing this approach, BL and FL separated clearly, whereas the discrimination between BL/DLBCL and FL/DLBCL was less pronounced, most likely due to the molecular heterogeneity of DLBCL.21,22 As there were no patients with dual-hit lymphomas and no DLBCL cases with MYC breaks as single events in our cohort, we were not able to test, whether our classifier was able to single out those cases.
Figure 1.
The miRnome of B-cell lymphomas. (A) Clustering according to the top 25 differentially expressed miRNA inferred with edgeR between FL (light blue), DLBCL (blue), and BL (gray), in pairwise comparisons. (B) Clustering according to the top 25 differentially expressed miRNA inferred with edgeR between BL and DLBCL/FL. (C) Hsa-miR-143 expression across all patients’ samples. (D) Visualization of the genomic mutations of those miRNA, which show alterations in their mature sequences. The mature sequences of the respective miRNA are shown; the seed sequences are highlighted by black boxes. The positions of the mutations are also indicated. (E) Predicted folding of the three biochemically validated novel miRNA.
Interestingly, 7/25 miRNA differentially expressed between BL/DLBCL (hsa-miRs-23a/29b/130b/146a/155/196b/222) were also part of a recently published, 27-miRNA qRT-PCR-derived classifier for the differentiation of those two subtypes22 (6.9-fold enrichment, one-sided Fisher exact test, P-value for the overlap of the two classifiers 2.322×10−5). In a previous array-based study, we established a classifier consisting of 38 miRNA, which differentiated BL from DLBCL.11 From the top 25 miRNA differentially expressed herein, eight overlapped with those 38 miRNA (hsa-miRs-23a/29b/146a/155/193a/221/222/339) (5.1-fold, P-value for this overlap 5.900×10−5). In summary, five miRNA (hsa-miR-23a/29b/146a/155/222) seem to be robustly able to differentiate BL from DLBCL irrespective of the collection of cases and the method used for analysis. We additionally analyzed previously published microarray data11 for 64 BL cases and 86 DLBCL cases to validate the predictive power of our classifiers on an independent dataset (see the Online Supplementary Methods for further details). We predicted correct class labels for 126/128 cases with a majority vote of at least 80% (recall = 98.44%; 57/58 BL cases and 69/70 DLBCL cases; overall accuracy = 84.0%) on our 25-miRNA-classifier for BL versus DLBCL.
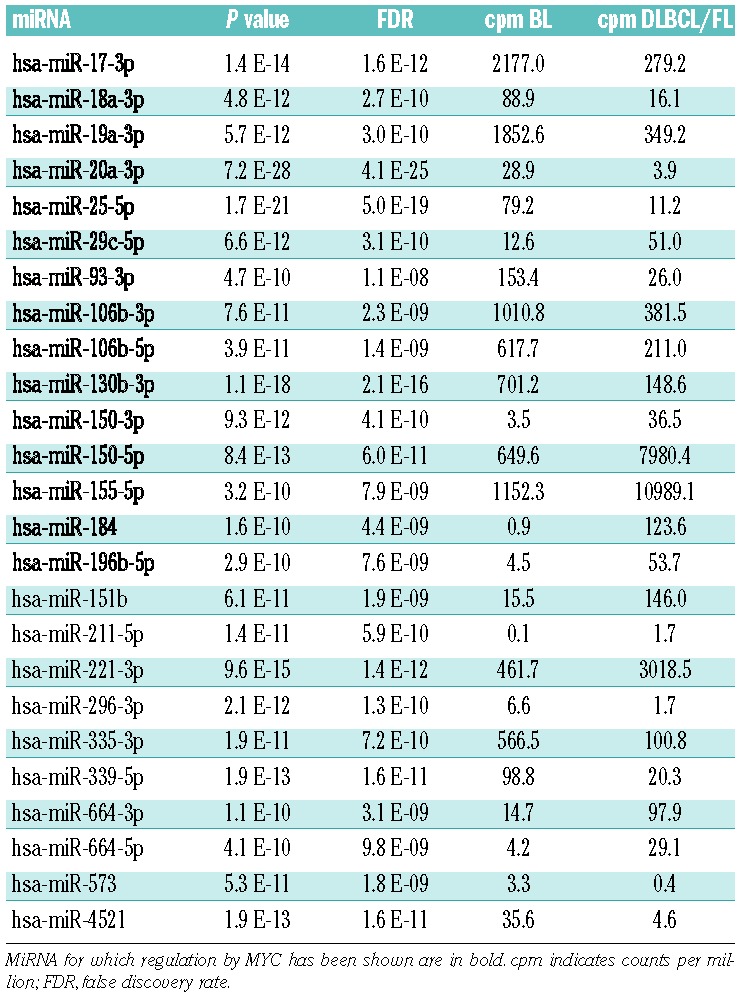
To address the question of how to distinguish BL from the other investigated histological subtypes, we merged DLBCL and FL and clustered the top 25 differentially expressed miRNA between BL and DLBCL/FL inferred with edgeR. This resulted - with the exception of two BL cases - in a clear separation between BL and DLBCL/FL (Figure 1B). Of those top 25 differentially expressed miRNA, 14 were upregulated and 11 were downregulated in BL compared to DLBCL/FL (Table 1). As our analysis takes both “5p” and “3p” versions (previously referred to as mature miRNA and star strand) of each miRNA into account, our classifier consists of 22 unique miRNA. Interestingly, for a total of 13 of these miRNA, regulation by MYC was reported in the literature.23–28
Table 1.
The 25-miRNA classifier separating BL from DLBCL/FL
Hsa-miRNA-143 is highly abundant in germinal center B-cell lymphomas
Contrary to earlier reports,29 hsa-miR-143 showed a very high expression across most lymphoma samples (Figure 1C). Expression ranged from 0.8% to 68.2% (mean 8.9%) of all reads mapping to miRNA for this miRNA alone with no significant differences between subtypes (means 10.8%, 7.4% and 8.9% for BL, FL and DLBCL, respectively). The extremely high expression of this miRNA (68.2%) in patient 4146289 (BL) was confirmed by qRT-PCR, as was the lower expression (0.8%) in patient 4142267 (BL) (Online Supplementary Figure S1B). As hsa-miR-143 forms a bicistronic cluster on chromosomal region 5q33.1 with hsa-miR-145, we also investigated the latter’s expression. MiRNA in bicistronic clusters are transcribed simultaneously and thus show similar expression patterns. The correlation analysis (P-value 0.0034 for the correlation between hsa-miR-143 and hsa-miR-145 expression, R=0.39) confirmed the validity of the hsa-miR-143 expression with similar expression patterns (Online Supplementary Figure S1C). Whole-genome-derived copy number analysis of all patients’ samples did not reveal any relevant alterations in either the promoter or the genomic region of hsa-miR-143/145. The reason for the observed high expression of the hsa-miR-143/145 cluster thus remained unclear.
To identify molecular pathways associated with the high expression of hsa-miR-143, we performed a target prediction and investigated which of the predicted targets were downregulated in the respective RNASeq data. This resulted in 186 predicted hsa-miR-143/mRNA interaction pairs (Online Supplementary Table S5). Gene Ontology (GO) analysis employing Gorilla30 revealed that the GO term “ubiquitin-protein transferase activity” (GO:0004842) showed the highest enrichment (5.33-fold, P-value <0.001). The associated target genes are listed in Online Supplementary Table S6, the entire GO output in Online Supplementary Table S7.
Hsa-miR-142 is recurrently mutated in its mature sequence in diffuse large B-cell lymphomas and follicular lymphomas
Next, we searched for mutated miRNA, which were detectable at both DNA and RNA levels. Mutations in miRNA located in the IGH gene locus were excluded. We identified ten mutations (Table 2) in eight patients (6 mutations in 5 DLBCL patients, 4 mutations in 3 FL patients) with a total of four miRNA affected (hsa-miR-142/-612/-3655/-4322). In two miRNA (hsa-miR-142/-612), the mutations were within the mature sequence (Figure 1D).
Table 2.
Genomically mutated miRNA.
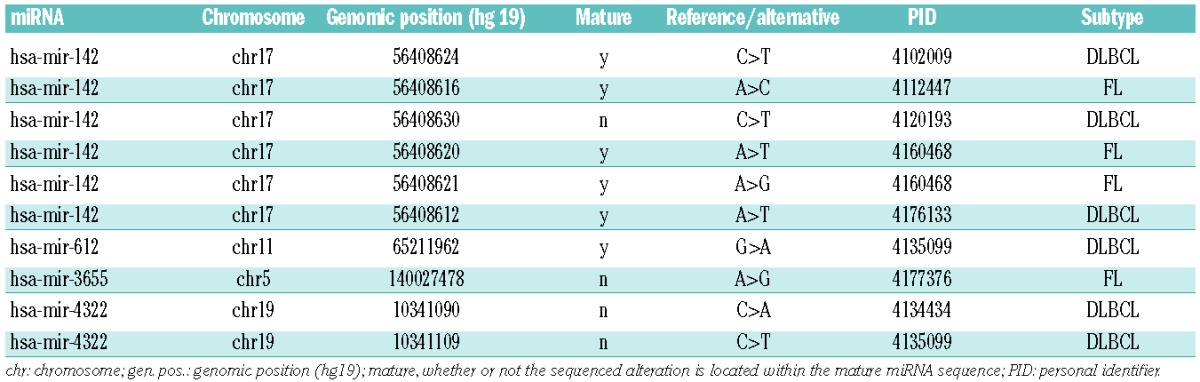
Hsa-miR-142 was the most frequently mutated miRNA with six different mutations in 5/40 DLBCL/FL patients. Two of those were located within the seed sequence. Looking at the subgroups, this broke up into 3/19 in DLBCL and 2/21 in FL. A recent publication9 reported mutation of hsa-miR-142 in 11/56 DLBCL cases. Our data therefore confirm the mutation frequency in DLBCL and extend this finding to FL.
The hsa-miR-376 cluster is recurrently edited in germinal center B-cell lymphoma subtypes
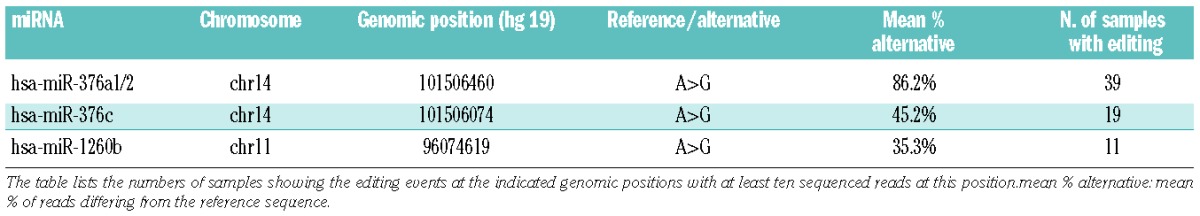
RNA editing is a process in which (most commonly) adenosine deaminases perform the site-specific hydrolytic deamination of adenosine to inosine.31 When an RNA molecule contains an inosine, the sequence change is usually A-to-G. We searched for mutations exclusive to the miRNA data (not seen at the genomic level), which thus represented bona fide miRNA editing events. Starting with all single nucleotide variants, we restricted our search to those in the seed regions and discarded known single nucleotide variants as reported in dbSNP_135 including rare variants. The remaining 40 candidates were manually evaluated (correct position of single nucleotide variants in sequence reads, A-to-G change, sequencing quality of errors), narrowing the list to four single nucleotide variants (Table 3). These mapped to hsa-miR-1260b, hsa-miR-376a1/2, and hsa-miR-376c, with the hsa-miR-376 family belonging to the same genomic cluster on 14q32. Editing frequencies (edited reads versus all reads) ranged from 35–86% across miRNA in the lymphoma samples showing this phenomenon. The editing “efficiency” (percent alternative base) and the expression of ADAR, one of the main enzymes responsible for RNA editing,31 per case (with observed editing) showed a weak correlation (P-value 0.044; R=0.30), possibly pointing to the mechanism behind the observed miRNA editing.
Table 3.
RNA editing events across lymphoma subtypes.
Discovery of three hitherto unreported microRNA expressed in germinal center B-cell lymphomas
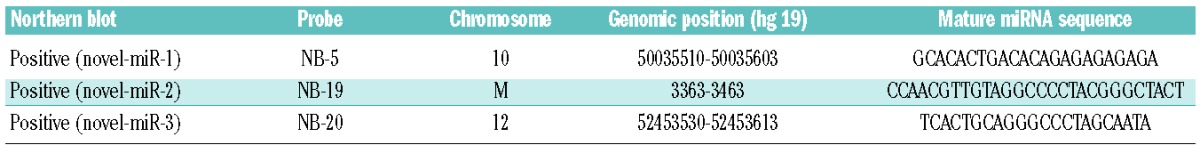
We employed miRanalyzer to predict hitherto unreported miRNA,18 then choosing a subset of 20 (Online Supplementary Table S8), and observed the correct processing of three candidates (Table 4) by northern blotting (Online Supplementary Figure S2A). Secondary structures of these three hitherto unreported miRNA as predicted by RNAfold32 are depicted in Figure 1E.
Table 4.
Novel miRNA in B-cell lymphomas.
Novel-miR-1 was moderately expressed in SU-DHL-4 and weakly expressed in Namalwa and Raji. Novel-miR-2 was expressed in Raji and SU-DHL-4, and novel-miR-3 was expressed in Raji and Namalwa (Online Supplementary Figure S2A). We next assessed publicly available RNASeq data of 16 cell lines (details in the Online Supplementary Material) across a variety of tissues/diseases for expression of our three novel miRNA. Novel-miR-2 and novel-miR-3 were broadly expressed (in 16/16 cell lines and 12/16 cell lines, respectively), whereas novel-miR-1 showed restricted expression and was only detected in the B-lymphoblastoid cell line GM12878 (data not shown). We then focused on novel-miR-1 (restricted expression) and novel-miR-2 (broad expression) for further experiments.
We performed overexpression/knockdown studies in SU-DHL-4 (novel-miR-1) and Raji (novel-miR-2) followed by RNASeq (Online Supplementary Figure S2B). In order to identify only those mRNA whose differential expression was due to direct targeting effects, we searched for mRNA that carried the respective seed sequence, had a significant miRanda score and were inversely regulated (false discovery rate for all further calculations <0.05).
Downregulation of novel-miR-1 and novel-miR-2 resulted in two (EIF3C, MPEG1) and three (HLA-DRB5, PFKFB4, PPP1R35) upregulated mRNA, respectively. Upregulation of novel-miR-1 led to the downregulation of 55 coding mRNA (Online Supplementary Table S9), whereas overexpression of novel-miR-2 only resulted in two downregulated mRNA (SLCO2B1, UPP1). Interestingly, there were many genes previously reported in the context of lymphomagenesis among those mRNA, which carried novel-miR-1 seed sequences. These genes represent its bona fide direct targets and included CARD11, E2F1, MCM2 and MCM7. Novel-miR-1 thus potentially represents a new player in lymphomagenesis. Sequences of novel-miR-1/-2/-3 have been submitted to miRBase.
AGO2 immunoprecipitation identifies direct mRNA-miRNA interactions in lymphoma subtypes
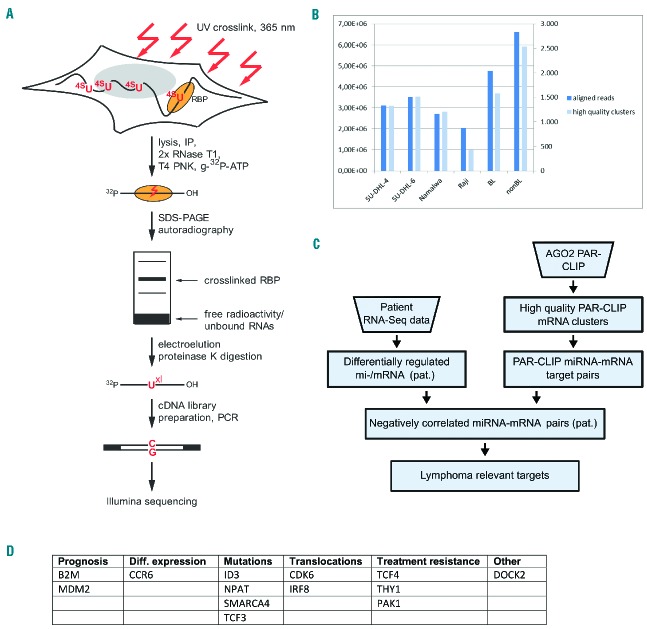
To identify those mRNA that were physically targeted by miRNA in Argonaute-miRNA-mRNA complexes (rather than using bioinformatic predictions to identify putative interactions only), we performed PAR-CLIP experiments of endogenous AGO213 (Figure 2A) in two BL cell lines (Namalwa, Raji) and two non-BL cell lines (SU-DHL-4, SU-DHL-6; both t(14;18)-positive). Merging the BL and the non-BL sequencing reads resulted in 1,587 and 2,532 clusters, respectively (individual read numbers are shown in Figure 2B). Combining these miRNA-target sites with the transcriptome data also available for each patient (Figure 2C) led to 302 (BL)/540 (non-BL) miRNA-mRNA interactions with negative correlations, with several genes being targeted by more than one miRNA (Online Supplementary Table S10). At the individual gene level the numbers were 208 (BL) and 328 (non-BL).
Figure 2.
Direct miRNA-mRNA regulation in B-cell lymphomas. (A) PAR-CLIP principle. Following the addition of 4-thiouridine, an immunoprecipitation with subsequent protein digestion is performed. Purified RNA fragments are reverse transcribed and cDNA libraries are sequenced on a HiSeq2500 followed by bioinformatic analysis (adapted from Hafner et al.13). (B) PAR-CLIP library statistics. The left y-axis shows the number of aligned reads, the right y-axis the number of high quality PAR-CLIP clusters. The cell lines employed are indicated. (C) Flow chart of the integrative miRNA-mRNA regulation analysis (adapted from Farazi et al.15). (D) List of lymphoma relevant genes for which regulation by distinct miRNA could be elucidated.
Many of the genes showing direct regulation by miRNA have well-known roles in lymphomagenesis (Figure 2D). These genes fell into different functional categories, some for which expression was correlated to prognosis33 [B2M33 (targeted by hsa-miR-106b), MDM234 (hsa-miR-361)], for which differential expression was shown [CCR635 (hsa-miR-296)] or were correlated to treatment resistance [e.g. THY136 (hsa-miR-149)]. For other targeted genes, mutations [ID314 (hsa-miR-4424), NPAT37 (hsa-miR-4518), SMARCA438 (hsa-miR-2467), TCF339 (hsa-miR-184)] or translocations [e.g. CDK640 (hsa-miR-148b)] have been described in several types of lymphomas.
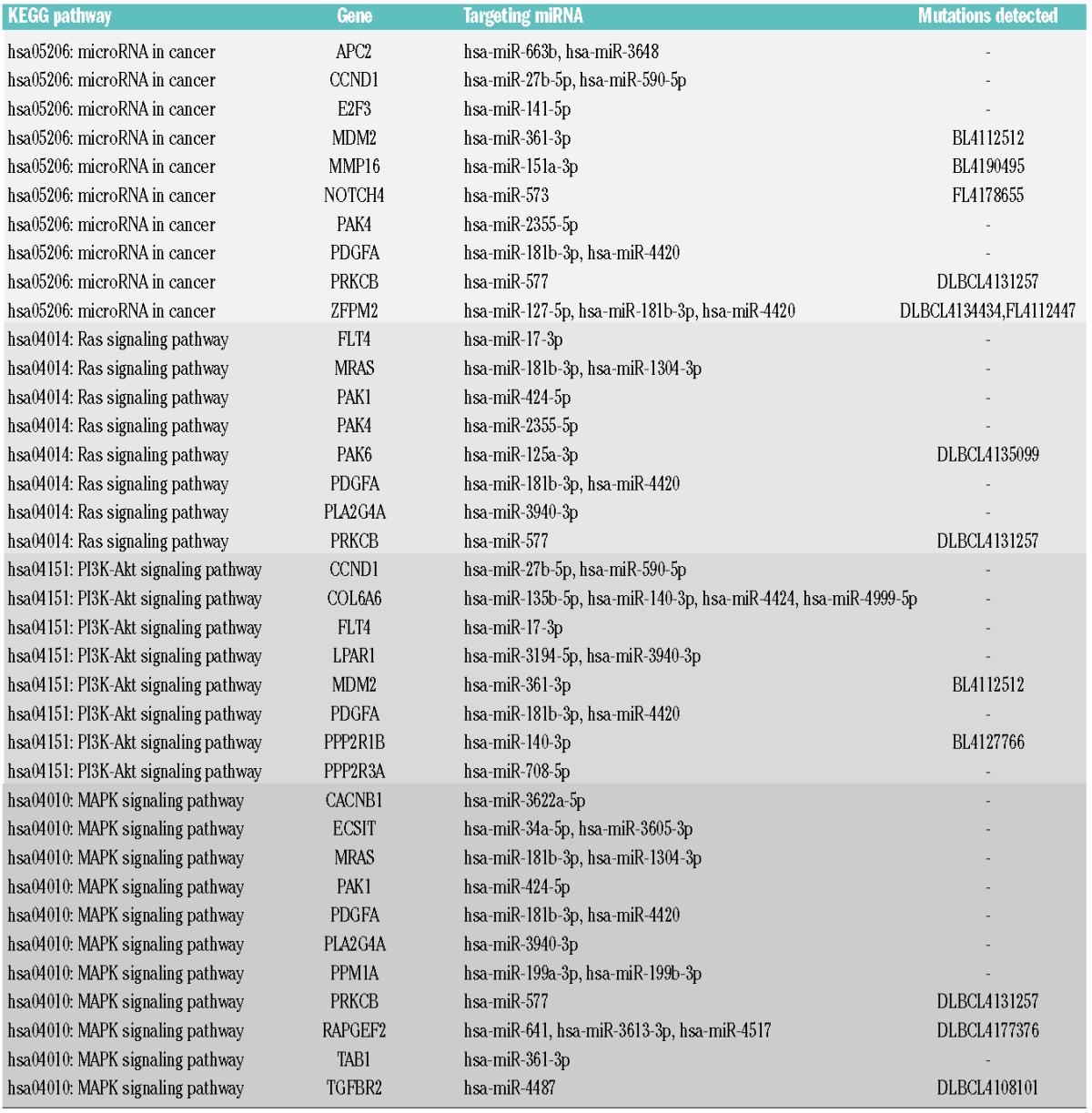
Significantly enriched and lymphoma-relevant targeted KEGG pathways (Table 5) showing a differential expression between BL and non-BL included “miRNA in cancer” (hsa05206, 10 genes, P-value 7.56×10−7, enrichment 7.7), “MAPK signaling” (hsa04010, 11 genes, P-value 1.79×10−8, enrichment 9.7), “Ras signaling” (hsa04014, 8 genes, P-value 7.56×10−6, enrichment 8.0), and “PI3K-Akt signaling” (hsa04151, 8 genes, P-value 1.48×10−4, enrichment 5.2). Total numbers of genomically detected mutations in the four mentioned pathways were in that order 122 (297 genes in the pathway), 111 (257 genes), 83 (228 genes) and 124 (347 genes). As these overlaps were not statistically significant (P-values 0.221 to 0.409), this suggests that the respective pathways are targeted and deregulated either by virtue of miRNA interference or by mutations.
Table 5.
Targeted KEGG pathways and associated miRNA-mRNA regulation pairs.
Discussion
We here report a deep sequencing analysis to identify differences in miRNA expression in samples from patients with BL, FL and DLBCL collected within the ICGC MMML-Seq Consortium. Comparing our miRNA classifiers separating the three entities to previous array- and qRT-PCR based studies, five miRNA (hsa-miRs-23a/29b/146a/155/222) were recurrently identified to differentiate BL from DLBCL11,22 and two miRNA (hsa-miR-92/150) to robustly separate FL from DLBCL.41,42 Of note, 13 of those miRNA differentiating BL/DLBCL were previously reported to be regulated by MYC,23–28 emphasizing the role of MYC in the pathogenesis of BL.
The greater discriminative power between BL, DLBCL and FL based on unsupervised analysis of the RNA-Seq data likely comes from less variation among the patients, which might be partly due to the higher number of analyzed genes when compared to miRNA-Seq as well as overlapping effects of some miRNA. Supervised analysis based on differentially expressed miRNA did, however, have a similar discriminative power as the supervised analysis of differentially expressed mRNA.
We identified hsa-miR-143 as highly expressed (compared to all other miRNA) across all three subtypes. This miRNA has hitherto mostly been discussed as a tumor suppressor in (mainly) epithelial malignancies.43 However, a recent study in colorectal cancer found hsa-miR-143 overexpression correlated to short overall survival.44 Earlier publications also reported a downregulation (mostly associated with its deletion) of the hsa-miR143/145 cluster in some leukemias and lymphomas.45,46 Examples of other miRNA, which have - depending on the tumor type - been described as both tumor suppressors and oncogenes include hsa-miR-26a, and the hsa-miR-141/200a-cluster.4 The high expression of hsa-miR-143 raises the possibility of a new and more general role for this miRNA in lymphomagenesis.
We describe recurrent mutations in hsa-miR-142 in FL at a frequency of 9.5%. Additionally, we confirm recurrent mutations of hsa-miR-142, at a frequency of 12.5% in DLBCL compared to 19.6% as previously published.9 Hsa-miR-142 mutations lead to the generation of new target sites as well as abolishing originally canonical ones in lymphoma-relevant genes, suggesting that hsa-miR-142 mutations act as a pathogenic mechanism across lymphoma subtypes. Other - albeit non-recurrent - seed sequence mutations affected hsa-miR-612, which was previously shown to suppress local invasion and distant colonization of hepatocellular carcinoma47 but has not been linked to lymphoid malignancies yet.
RNA editing as a post-transcriptional modification is the site-specific alteration of an RNA transcript. The most frequently observed form is adenosine to inosine (A-to-I) editing, catalyzed by ADAR enzymes. Both the splicing and the translation machinery recognize inosines as guanosines. RNA editing occurs in a tissue-specific manner and increases the diversity of protein products in the case of mRNA editing. The specific deamination of miRNA affects the stability of their precursors and thus the processing efficacy as well as results in the generation of novel mRNA targets sites in addition to altering existing ones.48 The hsa-mir-376 family was previously shown to be subject to miRNA editing in different cancer types, although not yet in lymphoma. This resulted in an altered mRNA target profile with both the loss of regulation of previous targets as well as the gain of new targets.49,50 Both aspects promoted the respective cancers. We provide here evidence of miRNA editing (hsa-miR-1260b, hsa-miR-376a1/2, and hsa-miR-376c) in lymphomas.
Only sequencing data allow larger scale identification of novel miRNA. The current release 21 of miRBase lists 1,881 human miRNA. Similar to previous studies,10 we identified hundreds of putative novel miRNA candidates. Through northern blot experiments, we provide experimental evidence of the correct processing of three novel miRNA. Novel-miR-1 emerged as the most interesting candidate, only being detectable in SU-DHL-4, Namalwa and a B-lymphoblastoid cell lines. Our analysis showed that it regulates many well-known lymphoma genes including CARD11, E2F1, MCM2 and MCM7, thus presenting itself as a potential novel player in lymphomagenesis.
Through our integrative analysis of miRNA and mRNA profiles in patients’ samples in combination with AGO2 PAR-CLIP data, for the first time it is possible to pinpoint individual, biochemically defined miRNA/mRNA target interactions in lymphomas as well as functional consequences of miRNA dysregulation. We focused our analysis on those target pairs (208 in BL, 328 in DLBCL/FL) with consistent expression changes (presumably due to aberrant miRNA expression) in the respective patients’ RNASeq data. Just performing a correlation analysis between differentially expressed miRNA and mRNA in patients’ samples coupled with a miRanda target prediction would have resulted in a much greater number of predicted interaction pairs (2,151 predicted pairs, data not shown). We described associated regulatory pathways including “Ras signaling”, “PI3K-Akt signaling”, and “MAPK signaling”. As there was very little overlap between those mRNA that are targeted by miRNA and those genes for which genomic mutations were detected (in those pathways), we suggest miRNA-mRNA targeting with subsequent deregulation as an additional oncogenic mechanism. We also provide evidence of miRNA regulation of many genes with already established roles in lymphomagenesis, including ID3, CDK6, MDM2, SMARCA4, and TCF3.
Our miRNA expression profiles uncovered subtype-specific differences in miRNA expression, evidence of recurrent hsa-miR-142 mutations in FL and DLBCL as well as miRNA editing and revealed distinct miRNA/mRNA target interaction pairs with roles in lymphomagenesis. Thus, we confirm and extend the important role that miRNA play in lymphomagenesis.
Supplementary Material
Acknowledgments
The project was funded by the Federal Ministry of Education and Research in Germany (BMBF) within the Program for Medical Genome Research (01KU1002A through 01KU1002J). The authors also acknowledge funding from Heinrich-Heine University Duesseldorf (Forschungskommission 17/2013), the Deutsche Forschungsgemeinschaft (DFG, HO 5456/3-1) and the Duesseldorf School of Oncology (funded by the Comprehensive Cancer Center Düsseldorf/Deutsche Krebshilfe and the Medical Faculty HHU Duesseldorf).
We would like to thank H. Lammert for excellent technical assistance and M. Gombert for assistance with sequencing and data handling.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/101/11/1380
References
- 1.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362(15):1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campo E, Swerdlow SH, Harris NL, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117(19):5019–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinha R, Nastoupil L, Flowers CR. Treatment strategies for patients with diffuse large B-cell lymphoma: past, present, and future. Blood Lymphat Cancer. 2012;2012(2):87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011; 223(2):102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Lisio L, Martinez N, Montes-Moreno S, et al. The role of miRNAs in the pathogenesis and diagnosis of B-cell lymphomas. Blood. 2012;120(9):1782–1790. [DOI] [PubMed] [Google Scholar]
- 6.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: how a complex biology gets more complex. Leukemia. 2015;29(5):1004–1017. [DOI] [PubMed] [Google Scholar]
- 7.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enomoto Y, Kitaura J, Hatakeyama K, et al. Emu/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia. 2011;25(12):1849–1856. [DOI] [PubMed] [Google Scholar]
- 9.Kwanhian W, Lenze D, Alles J, et al. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 2012;1(2):141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim EL, Trinh DL, Scott DW, et al. Comprehensive miRNA sequence analysis reveals survival differences in diffuse large B-cell lymphoma patients. Genome Biol. 2015;16:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenze D, Leoncini L, Hummel M, et al. The different epidemiologic subtypes of Burkitt lymphoma share a homogenous micro RNA profile distinct from diffuse large B-cell lymphoma. Leukemia. 2011;25(12):1869–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009;10(5):490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter J, Schlesner M, Hoffmann S, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. [DOI] [PubMed] [Google Scholar]
- 15.Farazi TA, Ten Hoeve JJ, Brown M, et al. Identification of distinct miRNA target regulation between breast cancer molecular subtypes using AGO2-PAR-CLIP and patient datasets. Genome Biol. 2014;15(1):R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spitzer J, Hafner M, Landthaler M, et al. PAR-CLIP (Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation): a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol. 2014;539:113–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann S, Otto C, Kurtz S, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol. 2009;5(9):e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackenberg M, Sturm M, Langenberger D, Falcon-Perez JM, Aransay AM. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37(Web Server issue):W68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biol. 2004; 5(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. [DOI] [PubMed] [Google Scholar]
- 22.Iqbal J, Shen Y, Huang X, et al. Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. Blood. 2015;125(7):1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Lisio L, Sanchez-Beato M, Gomez-Lopez G, et al. MicroRNA signatures in B-cell lymphomas. Blood Cancer J. 2012;2(2):e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Mai C, Yang H, et al. Candidate tumour suppressor CCDC19 regulates miR-184 direct targeting of C-Myc thereby suppressing cell growth in non-small cell lung cancers. J Cell Mol Med. 2014;18(8):1667–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao J, Zhao X. c-MYC-miRNA circuitry: a central regulator of aggressive B-cell malignancies. Cell Cycle. 2014;13(2):191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong L, Jiang W, Zhou R, Mao C, Guo Z. Identification and analysis of the regulatory network of Myc and microRNAs from high-throughput experimental data. Comput Biol Med. 2013;43(9):1252–1260. [DOI] [PubMed] [Google Scholar]
- 28.Zhao ZN, Bai JX, Zhou Q, et al. TSA suppresses miR-106b-93-25 cluster expression through downregulation of MYC and inhibits proliferation and induces apoptosis in human EMC. PLoS One. 2012;7(9):e45133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98(12):1914–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomaselli S, Locatelli F, Gallo A. The RNA editing enzymes ADARs: mechanism of action and human disease. Cell Tissue Res. 2014;356(3):527–532. [DOI] [PubMed] [Google Scholar]
- 32.Denman RB. Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques. 1993;15(6):1090–1095. [PubMed] [Google Scholar]
- 33.Hagberg H, Killander A, Simonsson B. Serum beta 2-microglobulin in malignant lymphoma. Cancer. 1983;51(12):2220–2225. [DOI] [PubMed] [Google Scholar]
- 34.Solenthaler M, Matutes E, Brito-Babapulle V, Morilla R, Catovsky D. p53 and mdm2 in mantle cell lymphoma in leukemic phase. Haematologica. 2002;87(11):1141–1150. [PubMed] [Google Scholar]
- 35.Durig J, Schmucker U, Duhrsen U. Differential expression of chemokine receptors in B cell malignancies. Leukemia. 2001;15(5):752–756. [DOI] [PubMed] [Google Scholar]
- 36.Ishiura Y, Kotani N, Yamashita R, et al. Anomalous expression of Thy1 (CD90) in B-cell lymphoma cells and proliferation inhibition by anti-Thy1 antibody treatment. Biochem Biophys Res Commun. 2010;396(2):329–334. [DOI] [PubMed] [Google Scholar]
- 37.Kuppers R. NPAT mutations in Hodgkin lymphoma. Blood. 2011;118(3):484–485. [DOI] [PubMed] [Google Scholar]
- 38.Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen D, Law ME, Theis JD, et al. Clinicopathologic features of CDK6 translocation-associated B-cell lymphoproliferative disorders. Am J Surg Pathol. 2009;33(5):720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrie CH, Chi J, Taylor S, et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. J Cell Mol Med. 2009;13(7):1248–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roehle A, Hoefig KP, Repsilber D, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142(5):732–744. [DOI] [PubMed] [Google Scholar]
- 43.Kent OA, McCall MN, Cornish TC, Halushka MK. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014;42(12):7528–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schou JV, Rossi S, Jensen BV, et al. miR-345 in Metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PLoS One. 2014; 9(6):e99886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dou L, Zheng D, Li J, et al. Methylation-mediated repression of microRNA-143 enhances MLL-AF4 oncogene expression. Oncogene. 2012;31(4):507–517. [DOI] [PubMed] [Google Scholar]
- 46.Liu C, Iqbal J, Teruya-Feldstein J, et al. MicroRNA expression profiling identifies molecular signatures associated with anaplastic large cell lymphoma. Blood. 2013;122(12):2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao ZH, Wan JL, Zeng LY, et al. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210(4):789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blow MJ, Grocock RJ, van Dongen S, et al. RNA editing of human microRNAs. Genome Biol. 2006;7(4):R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choudhury Y, Tay FC, Lam DH, et al. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest. 2012;122(11):4059–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizuguchi Y, Mishima T, Yokomuro S, et al. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS One. 2011;6(1):e15304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.