Abstract
The human genome produces thousands of long non-coding RNAs (lncRNAs) – transcripts >200 nucleotides long that do not encode proteins. While critical roles in normal biology and disease have been revealed for a subset of lncRNAs, the function of the vast majority remains untested. Here, we developed a CRISPR interference (CRISPRi) platform targeting 16,401 lncRNA loci in 7 diverse cell lines including 6 transformed cell lines and human induced pluripotent stem cells (iPSCs). Large-scale screening identified 499 lncRNA loci required for robust cellular growth, of which 89% showed growth modifying function exclusively in one cell type. We further found that lncRNA knockdown can perturb complex transcriptional networks in a cell type-specific manner. These data underscore the functional importance and cell type-specificity of many lncRNAs.
Introduction
Sequencing efforts have revealed that the human genome produces tens of thousands of long non-coding RNAs (lncRNAs), transcripts over 200 nucleotides that are often spliced and polyadenylated but have no apparent protein coding potential (1–3). Certain lncRNAs play critical roles in cellular function, development, and disease (4, 5). However, of the very large set of lncRNAs – many of which are differentially expressed in tissues and disease states – only a very small fraction have established biological functions, and even fewer are known to function in fundamental aspects of cell biology such as cell proliferation. Currently, it is not possible to predict which lncRNAs are functional, let alone what function they perform. Thus, a large-scale, systematic approach to evaluating the function of the vast population of lncRNAs is critical to understanding the roles that these non-coding transcripts play in cell biology.
A central limitation to systematic efforts to evaluate lncRNA function has been the lack of highly specific, scalable tools for inhibiting lncRNA gene activity (6). Gene deletion studies conducted in mice, flies, and human cells have yielded important biological insights about lncRNAs, but this approach is difficult to scale up (7–10). CRISPR/Cas9 nuclease approaches based on introduction of indels – while both scalable and useful for targeted loss of function studies of protein coding genes by altering the coding frame – are not well suited for the study of lncRNA gene function, as small deletions do not generally disrupt their biological activity (11–13). Nonetheless, larger Cas9-mediated genetic deletions can be effective at eliminating lncRNA genes (6, 14–17). Screens based on RNA interference (RNAi) have been valuable (18, 19) despite challenges with off-target effects (20). However, many lncRNAs localize to the nucleus, where RNAi exhibits variable knockdown efficiency (21).
We previously developed CRISPRi, a technology which can repress transcription of any gene via the targeted recruitment of the nuclease-dead dCas9-KRAB repressor fusion protein to the transcriptional start site (TSS) by a single guide RNA (sgRNA) (22–24). As CRISPRi acts only within a small window (1kb) around the targeted TSS (23), and as dCas9 occludes only 23bp of the targeted DNA strand (25), CRISPRi allows for precise perturbation of any lncRNA gene. By catalyzing repressive chromatin modifications around the TSS and serving as a transcriptional roadblock, CRISPRi tests a broad range of lncRNA gene functions including the production of cis- and trans-acting RNA transcripts (4), cis-mediated regulation related to lncRNA transcription itself (26–29), and enhancer-like function of some lncRNA loci (14, 15, 30). The repressive chromatin modification H3K9me3 catalyzed by CRISPRi is highly specific, with little to no off-target effects due to either spurious dCas9 binding or unintended silencing of distal regulatory elements, as measured by ChIP-seq or RNA-seq (22, 31–34) see also Figure 4C below). To enhance CRISPRi for large-scale screening, we have improved upon the design of CRISPRi sgRNA libraries to optimize on-target activity while further minimizing-off target effects, enabling highly sensitive detection of essential coding genes (35).
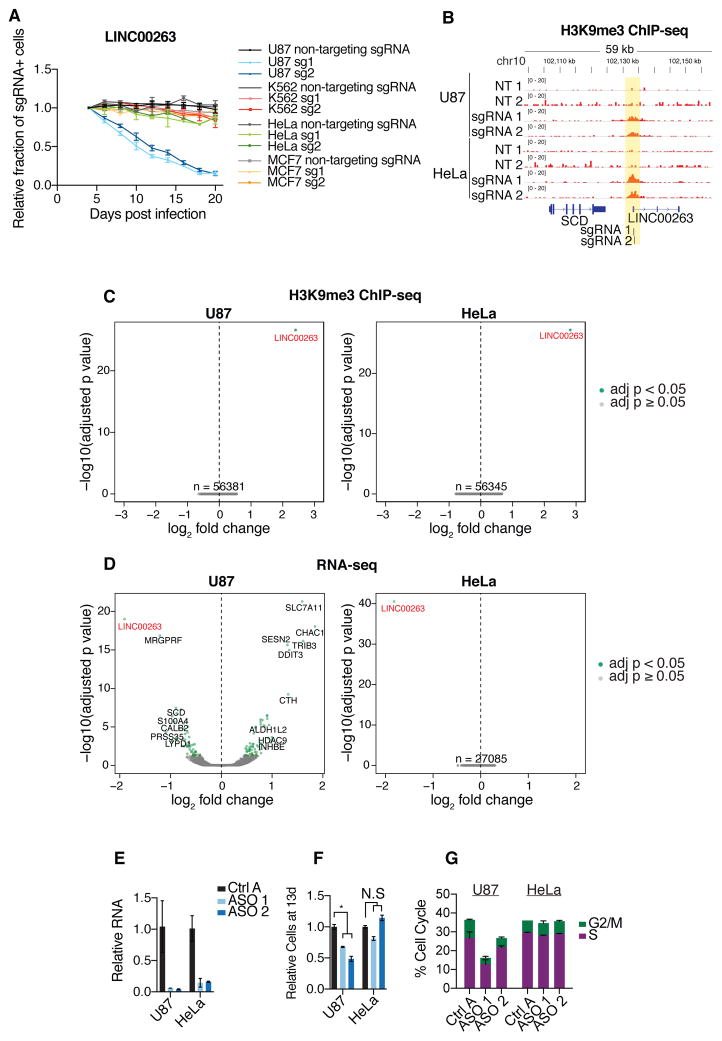
Figure 4. Dissection of cell type-specific growth modifier lncRNA LINC00263.
A) Internally-controlled growth assays for 2 independent sgRNAs targeting the TSS of LINC00263 and non-targeting sgRNA in U87, K562, HeLa, and MCF7 cells. B) ChIP-seq against H3K9me3 in replicates of U87 and HeLa cells infected with non-targeting sgRNAs or LINC00263 sgRNAs. Values represent normalized reads. C) Volcano plots for ChIP-seq samples in (B), representing genome-wide differential enrichment of H3K9me3 at promoter regions. Fold changes are between LINC00263 sgRNAs over non-targeting sgRNAs. D) Volcano plots for RNA-seq differential expression following infection of LINC00263 sgRNAs compared to infection of non-targeting sgRNAs. E) qPCR of ASO knockdown of LINC00263 in U87 and HeLa cells. F) Proportion of cells at 13 days post ASO transfection, relative to control ASO. G) Percentage of cells in S or G2/M phases following ASO knockdown of LINC00263. * indicates p = 0.0029.
Here, we developed CRISPRi libraries targeting 16,401 lncRNA loci (with 10 sgRNAs per TSS), and conducted screens for genes that are required for robust growth in 7 human cell types—6 transformed cell lines and induced pluripotent stem cells (iPSCs)(36). These large-scale screens, coupled with extensive validation studies, greatly increased the number of lncRNA genes known to have biological function and revealed lncRNA function to be highly cell type-specific. Our studies thus help elucidate the biology contained within the lncRNA genome, and provide a tool for both large-scale and targeted investigations of lncRNA function.
Results
CRISPRi screens identify lncRNA loci that modify cell growth
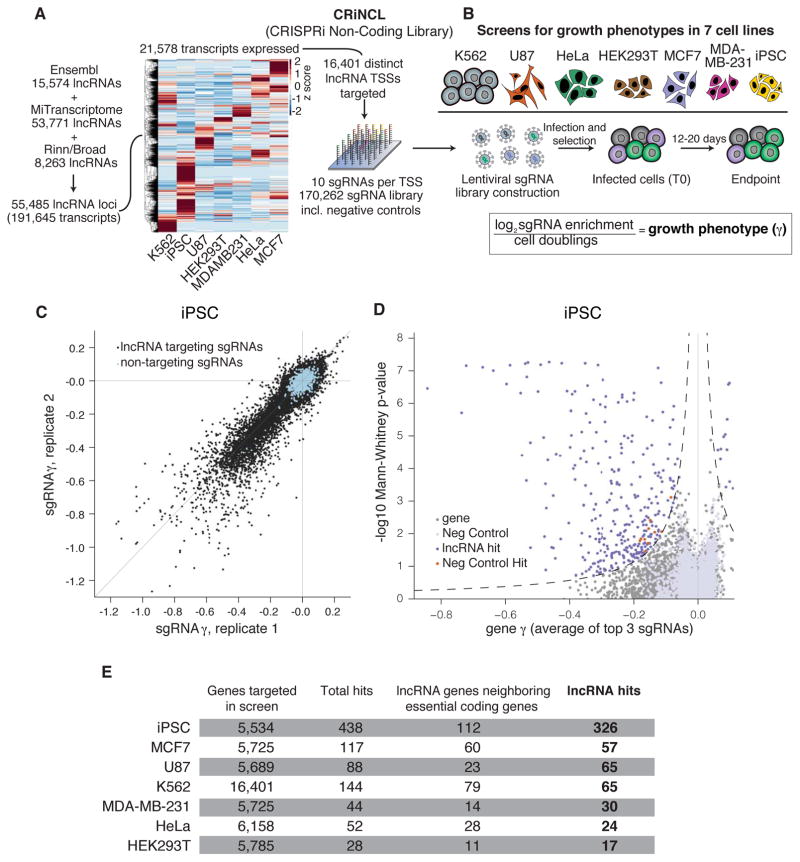
We first designed an sgRNA library to enable genome-scale CRISPRi screening of lncRNA gene function. We generated a comprehensive lncRNA gene set by merging three major non-coding transcriptome annotations (37–39), prioritized ~1/3 of these genes based on expression in any of a panel of cancer and non-transformed cell lines (Table S1), and designed 10 sgRNAs targeting each lncRNA transcription start site (TSS) using the hCRISPRi-v2.1 algorithm (35) (Figure 1A and S1). The cell lines represent a broad range of cell types studied by the ENCODE project (40), including a chronic myeloid leukemia cell line (K562), the cervical cancer line HeLa, a glioblastoma line (U87), and two mammary adenocarcinoma lines (MCF7 and MDA-MB-231). We also chose an iPSC line that inducibly expresses CRISPRi components (33, 41). The library, termed “CRiNCL” for CRISPRi Non-Coding Library, is available as pooled lentiviral plasmid libraries on Addgene and in silico as Table S2.
Figure 1. CRISPRi screens identify lncRNA genes that modify cell growth.
A) Schematic of CRISPRi library design strategy. Three lncRNA annotation sets were merged, prioritized by expression in the indicated cell lines, and targeted by 10 sgRNAs per TSS using the hCRISPRi-v2.1 algorithm. Heatmap represents expression as z-score of fragments per kilobase million (FPKM) within each cell line (see Figure S1 for TPM values). B) Schematic of growth screens performed in 7 different cell lines, and formula for calculation of the growth phenotype (γ). C) Scatter plot of sgRNA phenotypes from two independent replicates of a CRISPRi screen performed in iPSCs. D) Volcano plot of gene γ and p-value. Screen replicates were averaged, and sgRNAs targeting the same gene were collapsed into a growth phenotype for each gene by the average of the 3 top scoring sgRNAs by absolute value, and assigned a p-value by the Mann-Whitney test of all 10 sgRNAs compared to the non-targeting controls. Negative control genes were randomly generated from the set of non-targeting sgRNAs, and dashed lines represents a threshold for calling hits by screen score (see Methods). Neighbor hits are not displayed for clarity (see Figure S3A,B). E) Summary table of all CRISPRi growth screens performed.
We used this library to conduct screens for lncRNA loci that increase or decrease cell growth in each of 7 cell lines. We infected the full lentiviral library or targeted sublibraries (Figure S2A) into each cell line engineered to express dCas9-KRAB (22, 23, 33, 42), selected for infected cells by puromycin selection, and cultured for between 12 and 20 days, measuring sgRNA enrichment by Illumina sequencing (Figure 1B and Table S3). The fraction of cells infected with the sgRNA library remained stable over the course of the screen (23), indicating that CRISPRi targeting of lncRNA loci does not exhibit non-specific toxicity (Figure S2B). To facilitate comparisons between screens conducted for different durations and in cell lines with different growth rates, we normalized sgRNA enrichment by total cell doublings to obtain the quantitative growth phenotype γ, which reflects the positive or negative impact on cell growth caused by knockdown of a given gene (43) (Figure 1B).
Analysis of biological replicates revealed that the γ for targeting sgRNAs showed strong and reproducible phenotypes (Pearson r = 0.34–0.90) while non-targeting control sgRNAs were tightly distributed around 0 (Figure 1C and S2C, Table S3). We averaged replicate sgRNA phenotypes and used these to score lncRNA genes (23, 35), calculating gene phenotypes from the mean of the top 3 sgRNAs targeting the gene and Mann-Whitney p-values from all 10 sgRNAs compared to non-targeting control sgRNAs (Figure 1D and S3A, Table S4). Within each screen, we also randomly sampled non-targeting sgRNA phenotypes to generate “negative control genes” and analyzed them as with lncRNA genes (see Methods), enabling us to estimate an empirical false discovery rate for each screen as well as the combined screen dataset (Figure S2D). LncRNA genes were considered to be hits if their combined phenotype effect size and p-value (referred to here as “screen score”) exceeded a consistent threshold applied to each screen corresponding to an empirical false discovery rate of 5% (Figure S3C). Overall, we found between 28 and 438 lncRNA loci hits in each cell line (Figure 1E and S3A, Table S4).
We observed that for 169 of these lncRNA hits, the TSS of the non-coding gene was within 1kb of the TSS of a coding gene previously found to be essential in a CRISPRi screen (23), making it difficult to determine whether the observed phenotypes were due to knockdown of the target lncRNA or direct inhibition of the neighboring coding gene (Figure S3B). We thus removed these hits from the total set of hit genes for downstream analyses (Figure 1E, S3A, S3D), resulting in 169 “neighbor hits” and 499 “lncRNA hits,” 299 of which are distal from any protein coding gene (~90% of which would not measurably impact growth upon knockdown). The 1kb threshold was chosen based on the maximum distance at which CRISPRi is effective as revealed by analysis of dense sgRNA tiling and genome-scale screens (Figure S4) (23); increasing this threshold to 10kb classifies only an additional 19 genes as neighbor hits (Figure S3D).
A larger fraction of lncRNAs hits were observed in the iPSC screen, suggesting that this cell line is either more susceptible to growth perturbations or that iPSCs were differentiating to other cell types with lower growth rates. We therefore investigated iPSC differentiation in a secondary fluorescence activated cell sorting (FACS)-based screen by assessing loss of pluripotency as indicated by decreased POU5F1/OCT4 expression. CRISPRi targeting of only 9 lncRNA loci reduced POU5F1/OCT4 expression (Figure S5, Tables S5–6), suggesting that the majority of lncRNA hits identified in iPSCs primarily affect cell growth. To confirm that the increased fraction of lncRNA hits in iPSCs was not due to technical differences in CRISPRi function between cell lines, we performed a CRISPRi screen for protein-coding genes required for cell growth in iPSCs (Figure S6A, Table S7). These results corresponded well with our previously published K562 growth screen (35) in both the number of genes found to have function and in the ability to specifically identify known essential genes (Figure S6B–C) (44). Taken together, our screens identified 499 lncRNA genes that modify cell growth and have no essential coding gene neighbors, representing a large set of unstudied non-protein-coding genes serving important functions in cell biology.
lncRNA CRISPRi phenotypes are reproducible with robust knockdown
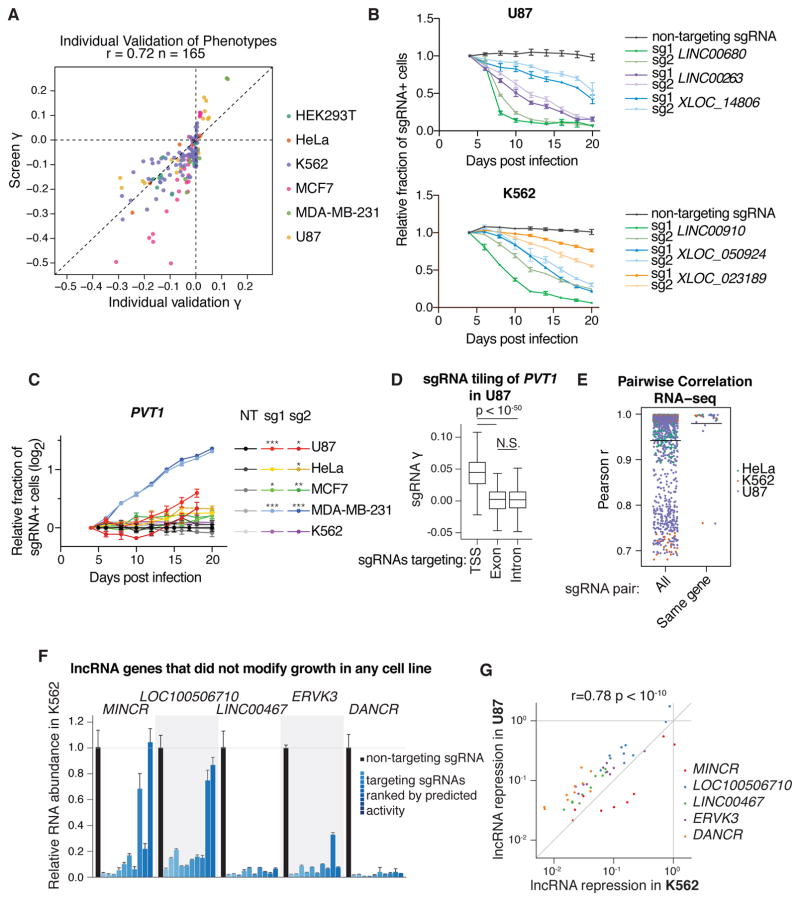
Extensive validation studies argue for the low false-positive and -negative rates of our studies. First, we individually cloned the top two sgRNAs targeting 65 representative lncRNA hit loci, 41 of which were hits in only one cell line. We tested whether the observed phenotypes from the screens were reproducible using internally-controlled growth assays, in which the fraction of cells infected with an sgRNA were measured over time by flow cytometry. We monitored the growth effects of sgRNAs in the cell lines in which they exhibited a phenotype in the screen, as well as several sgRNAs in cell lines where they showed no effect, and found that the individual sgRNA growth phenotypes (γ) correlated well with the screen γ (Pearson r = 0.72, Figure 2A). This confirmed both that lncRNA knockdown phenotypes were reproducible and that the difference in lncRNA phenotype between cell lines was not due to technical differences between genome-scale screens. Analyzing these phenotypes over time further revealed distinct kinetics of cell depletion mediated by lncRNA knockdown (Figure 2B). For 12 lncRNA hits, we measured the levels of knockdown by qPCR and found over 70–95% knockdown for most of the targeted transcripts (14/14 sgRNAs in U87; 10/16 sgRNAs in MCF7) despite the effect of cellular depletion (Figure S7A).
Figure 2. Validation of screen results shows reproducible phenotypes, correlated transcriptome responses, and robust knockdown of target transcripts.
A) Individual sgRNA phenotypes from internally-controlled growth assays (B,C) compared to sgRNA phenotypes from screens. Individual growth phenotypes were calculated from relative fraction of sgRNA-containing cells at the endpoint, divided by the number of doublings from 4 days post-infection. Screen growth phenotypes represent the replicate average phenotype from the indicated cell line. B) Internally-controlled growth assays performed with sgRNAs targeting lncRNA hit genes in U87 and K562. Cells were infected with lentivirus of the sgRNA expression vector (including a BFP marker gene) and passaged for 20 days. The fraction of sgRNA-containing cells was measured as the fraction of high-BFP-expressing cells by flow cytometry, and expressed relative to the fraction at 4 days post infection. Points represent the mean and standard deviation of 3 biological replicates. C) Internally-controlled growth assays of PVT1-targeting sgRNAs in 5 cell lines. Assays were performed as in (B). Asterisks represent t-test p-values compared to the non-targeting (NT) sgRNA at the assay endpoint (* < 0.05, ** < 0.01, *** < 0.001). D) Boxplot of sgRNA growth phenotypes from tiling screen of PVT1 in U87 cells. TSS represents all sgRNAs within 1kb of the PVT1 “p1” and “p2” TSSs as annotated by FANTOM, exon represents sgRNAs targeting any PVT1 exon annotated by Ensembl, and intron represents all other sgRNAs (see Figure S7B). sgRNA γs are the average of two replicates. E) Pairwise correlation of gene expression profiles for independent sgRNAs. Expression profiles were measured by RNA-seq and correlations were calculated from transcripts per million (TPM) of genes with significant variation of expression (see Methods). “All” represents every sgRNA pair from the same cell line with the same phenotype direction, except same-sgRNA and same-gene pairs. F) Relative RNA abundance in K562 of lncRNA genes that were not hits in any cell line. RNA abundance for all 10 sgRNAs targeting the indicated genes in the CRiNCL library was measured by qPCR. Each bar represents the mean and standard deviation of 3 biological replicates, and is ordered by decreasing activity as predicted by the hCRISPRi-v2.1 algorithm. G) Correlation of lncRNA repression in K562 and U87. Points represent mean values from (F) and Figure S7C.
In four cell lines, knockdown of lncRNA PVT1 had a pro-growth phenotype. As PVT1 had previously been characterized as a proto-oncogene (45) and pro-growth phenotypes in cancer cell lines are uncommon (23, 46), we validated the pro-growth phenotype (Figure 2C and S7A) and investigated this complex locus further by conducting a CRISPRi screen in U87 cells with an sgRNA library tiling every possible site along the locus (17,469 sgRNAs). We found that only sgRNAs within 1kb of the most upstream TSSs, which is distal to any mapped enhancers, caused a consistent pro-growth phenotype (Figure 2D and S7B, Table S8). Within this TSS region, the majority of sgRNAs promoted cell growth, and knockdown of the major isoform was confirmed by qPCR (Figure S7A). sgRNAs outside of this 1 kb window around the TSS, which would not be expected to affect transcription of the major PVT1 isoform (23), showed no consistent impact on growth, arguing that the observed pro-growth phenotype is mediated by transcriptional interference.
Repression of lncRNA loci elicits lncRNA-specific transcriptome responses
To better understand the consequences of lncRNA CRISPRi, we performed RNA-seq following CRISPRi knockdown of 42 lncRNA hits in 3 cell types. 32 of these lncRNA loci were hits in only one cell type. Selected lncRNA loci did not have essential coding gene neighbors, and 2 or more sgRNAs per gene were tested individually. Distinct sgRNAs targeting the same lncRNA TSS resulted in highly correlated transcriptome responses (mean Pearson r = 0.980; Figure 2E) that were generally proximal to each other in hierarchical clustering analysis (Figure S8A–D). By contrast, pairs of sgRNAs targeting different hit lncRNA loci with the same phenotype direction had transcriptome responses that were more dissimilar (mean Pearson r = 0.942, Mann-Whitney p-value compared to same-gene pairs = 6.4×10−08;), suggesting distinct molecular mechanisms of the lncRNAs despite having similar phenotypes (Figure 2E).
RNA-seq analysis of differential gene expression also revealed several clusters of co-expressed genes, suggesting that growth modifier lncRNA loci regulate critical pathways (Figure S8A–D and Table S9). For instance, 2 lncRNA knockdowns that caused increased growth in U87 cells clustered by upregulation of translation genes (p = 3.2×10−37), while other pro-growth sgRNAs showed correlated changes in expression of DNA replication (p = 2.0×10−10) and post-transcriptional regulation (p = 3.0×10−08). Clusters enriched for genes in the p53 pathway (e.g. ATF3) were upregulated by many anti-growth sgRNAs in both U87 and HeLa cells. Interestingly, K562 cells showed clusters of genes enriched for platelet degranulation (p = 1.6×10−05) and response to decreasing oxygen levels (p = 5.0×10−05). The median magnitude of log2 fold changes for differentially expressed genes in U87, HeLa, and K562 were 0.67, 0.86, and 1.17, respectively (Figure S8E), with several genes exhibiting > 2 fold up- or down-regulation consistently across many samples (Figure S8F). These results indicate that different lncRNAs can regulate distinct biological pathways that affect cell growth and proliferation.
Analysis of the chromosomal location of differentially expressed genes did not reveal a global trend toward transcriptional changes on the targeted chromosome (Figure S9). We did however find that knockdown of 14 lncRNA loci resulted in local transcriptional changes within a 20 gene window (Figure S10), suggesting that certain lncRNAs may preferentially act locally.
CRISPRi robustly inhibits lncRNA transcription
The fraction of growth modifier lncRNA loci identified in our screens (1–8% per cell line) was less than the fraction of essential protein-coding genes in previous reports (10–11%) (35, 46). We therefore wanted to assess whether lncRNA genes that did not appear as a hit in any screen were true negatives or simply a result of ineffective repression by CRISPRi. To this end, using all 10 sgRNAs per gene, we measured the knockdown of five arbitrarily selected lncRNA genes that had no observed phenotype in any cells and were expressed in both K562 and U87 cells (Figure 2F and S7C). Of these 100 knockdown measurements, 61 showed over 90% repression of the targeted lncRNA. Furthermore, with the exception of LOC100506710 in U87 cells, all lncRNAs were repressed by at least 90% by at least three different sgRNAs. For all sgRNAs, lncRNA knockdown efficiency correlated with their predicted CRISPRi activity, and the efficiency of knockdown was highly correlated between K562 and U87 cells (Pearson r = 0.78; Figure 2G). Based on these findings, with the exception of cases where a small amount of residual transcript is sufficient for lncRNA function, we infer that the majority of lncRNA loci that did not appear as a screen hit produce transcripts that are not essential for robust growth of the cell line screened.
Growth modifier lncRNA function is highly cell type-specific
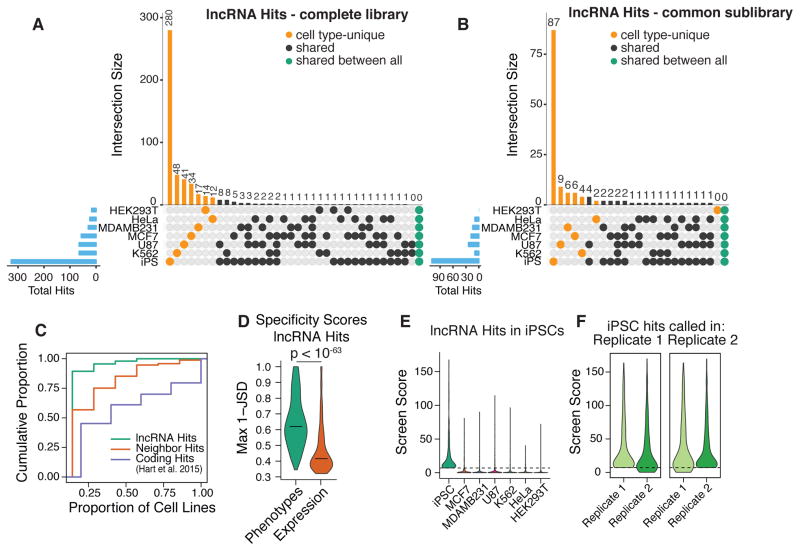
We next determined the number of lncRNA hits that were unique to a specific cell type or common to any combination of two or more of the cell types screened. The vast majority (89.4%) of lncRNA hits were unique to only one cell type, with none being a hit in 5 or more cell types (Figure 3A–C). Even when we restricted this analysis to the 1,329 lncRNAs expressed in all 7 cell types, 82.6% of the lncRNA hits modified growth in only one cell type (Figure 3B). Analysis of cell type-specificity scores based on the Jensen-Shannon distance, which quantifies how closely a given distribution resembles “perfect” specificity (37), revealed that the specificity of lncRNA screen scores was far greater than the specificity of lncRNA expression, for lncRNA hits (Figure 3D). Therefore, differential expression patterns alone are not sufficient to predict functional lncRNAs. Cross comparison of screen score distributions for lncRNAs that scored as hits in each cell type revealed that the threshold used for calling hits did not account for the cell type specificity (Figure 3E, S11D–E). Furthermore, cross-comparison of screen scores between replicates does not support technical variation as the source of the apparent cell type-specific function (Figure 3F and S11F).
Figure 3. Growth modifier lncRNA function is highly cell type-specific.
A) Numbers of lncRNA hits for each set of cell types in the complete library and (B) common sublibrary (lncRNAs that were expressed and screened in all cell types). Blue bars indicate total number of lncRNA hits in each cell type. C) Cumulative distribution function for the proportion of cell types in which each gene is a hit. Protein coding hits were obtained from Hart et al. 2015 using the authors’ 5% FDR Bayes Factor threshold. D) Distributions of the maximum 1 - Jenson Shannon distance (JSD) metric of cell type-specificity for lncRNA hit screen scores and expression values. Horizontal lines – median. E) Distributions of screen scores across all cell types for lncRNAs that were hits in iPSCs. Dashed line represents screen score threshold for calling hit genes. F) Distributions of screen scores across both replicates of iPS cells, for lncRNAs that would be called as hits in replicate 1 (left) and in replicate 2 (right).
In contrast to the sparse cell type overlap of lncRNA hits, analysis of published protein coding screens across similar numbers of cell types (46, 47) revealed that the majority (54.8% in (47), 67.3% in (46)) of essential protein coding genes are hits in 2 or more cell types, with 20.4% and 30.8% being essential to all cell types screened in (47) and (46), respectively (Figure 3C, S11A–B). In addition, “neighbor hits” (lncRNA loci that are within 1kb of an essential protein coding gene), were more likely to modify growth in multiple cell types, suggesting that CRISPRi targeted to these loci represses the adjacent essential coding gene, at least in some cases (Figure 3C, S11C,E).
Cell type-specific lncRNAs elicit highly divergent phenotypes
We sought to better understand the cell type-specific function of specific lncRNAs. We focused on LINC00263, which despite being expressed in all 7 cell lines screened, had a much stronger negative growth phenotype in U87 than in any other cell line (Figure S12A). The abundance of LINC00263 transcript in a given cell line was also poorly correlated with the corresponding screen phenotype (Pearson r = 0.266). Validating these screen results, in internally controlled growth assays, two distinct sgRNAs to the TSS of LINC00263 reduced the propagation of only U87 cells and not K562, MCF7 or HeLa cells (Figure 4A). H3K9me3 is a chromatin modification that is a result of local dCas9-KRAB activity (31), and in both U87 and HeLa cells with LINC00263 CRISPRi targeting, ChIP-seq analysis demonstrated equal enrichment of H3K9me3 specifically at the LINC00263 promoter for two independent sgRNAs (Figure 4B,C, S12B,C). However, despite such evidence of equivalent and specific CRISPRi targeting, U87 and HeLa cells had substantially different transcriptome changes after LINC00263 knockdown. While U87 cells upregulated genes related to ER stress (e.g. ATF4, CHAC1; GO term p = 4.51×10−09) and apoptosis (e.g. DDIT3, SOD2; GO term p = 3.39×10−08), only LINC00263 itself was differentially expressed in HeLa cells (adj. p < 0.05; Figure 4D). In K562 cells, these same 2 sgRNAs also produced very little transcriptional change (Figure S12D). Of note, in all three cell lines, knockdown efficiency of LINC00263 was equivalent (Figure 4D, S12D). Consistent with our observations for LINC00263, knockdown of PVT1 and LINC00909, which were hits in U87 but not in HeLa, produced many more differentially expressed genes in U87 (Figure S12E). By contrast, depletion of LINC00680, which was a hit in both U87 and HeLa cells, resulted in comparable numbers of differentially expressed genes in U87 and HeLa cells (Figure S12E). Our results suggest that the specificity of lncRNA function is not due to differences in CRISPRi activity, but is related to differences in transcriptional networks across cell types.
We then targeted the LINC00263 lncRNA transcript with antisense oligonucleotides (ASOs) that degrade RNA via an RNaseH-based mechanism. In both U87 and HeLa cells, ASOs reduced LINC00263 transcript levels by 85–95% (Figure 4E). However, LINC00263 ASOs decreased proliferation in U87 cells but not in HeLa cells (Figure 4F,G). The magnitude of proliferation decrease was also comparable to CRISPRi (Figure S12F,G), further supporting the cell type-specific function of this lncRNA. ASO knockdown of three other U87 lncRNA hits also reduced cell proliferation (Figure S12H,I), providing additional evidence for the functional contribution of the lncRNA molecule in these examples.
Machine learning identifies features predictive of growth modifier lncRNAs
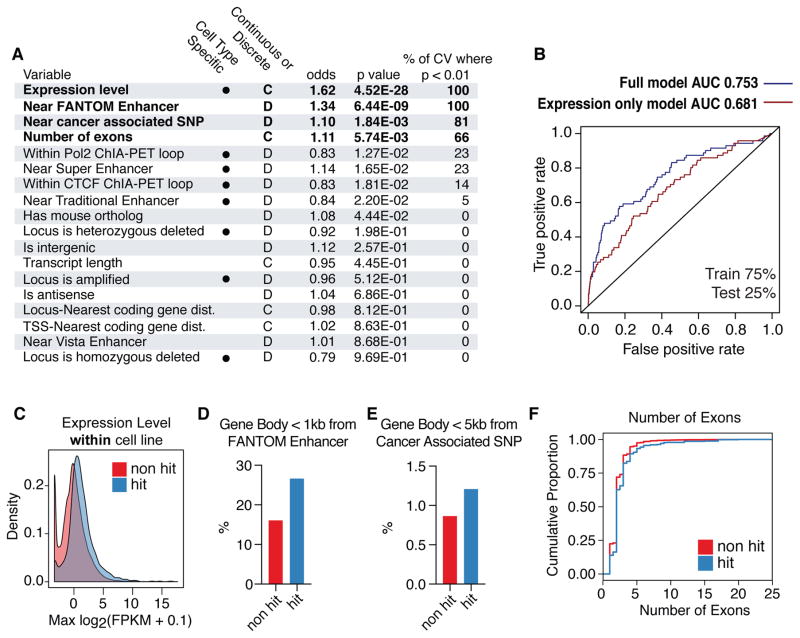
Using data from our genome-scale screens, we sought to identify properties of the lncRNA hits that can distinguish them from non-hit lncRNAs. 18 classes of genomic data such as enhancer maps, expression levels, chromosomal looping data, conservation, and copy number variation from ENCODE (40), FANTOM (48), Vista (49), and other sources (50–52) were compared with all lncRNA loci screened in this study. 8 of these properties (expression, Pol2/CTCF looping by ChIA-PET, enhancers and super enhancers from (51), copy number variation) were cell type-dependent. Generalized linear models were constructed to assess which genomic properties are predictive of lncRNA function (see Methods). Expression levels within each cell line, lncRNA gene body within 1kb of a mapped FANTOM Enhancer, lncRNA gene body within 5kb of a cancer-associated single nucleotide polymorphism (SNP) (50), and the number of exons were significant predictors of lncRNAs hits (p < 0.01) in repeated 10-fold cross validation (Figure 5, Table S10). 99.6% of lncRNA genes that were screened but not apparently expressed were not called as hits (Figure 5C). Whether the 11 growth modifier hits of such “non-expressed” lncRNA loci represent non-lncRNA mediated effects, inaccurate quantitation of the transcript levels, or effects mediated by lncRNAs acting at low expression remains to be determined. In support of the latter possibility, HOTTIP has been reported to function despite being expressed at ~0.3 copies per cell (53). Nonetheless, many highly expressed lncRNAs were not hits (e.g. 154 non-hit lncRNAs were detected at FPKM > 100), and the accuracy for predicting lncRNA hits was greater for a model using all variables as compared to a model that relied only on expression levels (Figure 5B).
Figure 5. Machine learning identifies genomic features of growth modifier lncRNAs.
A) Results from logistic regression model using 18 classes of genomic data as possible predictors of growth modifier lncRNAs. Cell type dependent variables are marked. Odds ratios represent relative impact of 1 standard deviation increase of given variable. Significant variables (p < 0.01) are bolded. Results of 10-fold cross validation are represented as the % of cross validation iterations where the given variable is significant. B) ROC curves for full model compared to model using only expression data. C) Density plot of expression levels for lncRNAs that scored as hits and non-hits, aggregated across all cell types. D) Percentage of non-hit (red) and hit (blue) lncRNAs whose gene bodies resided < 1 kb from an annotated FANTOM enhancer. E) Percentage of non-hit (red) and hit (blue) lncRNAs whose gene bodies resided < 5 kb from a cancer associated SNP. F) Cumulative distribution function of number of exons for non-hit (red) and hit (blue) lncRNAs transcripts.
Compared to non-hit lncRNAs, hit lncRNA gene bodies were 1.66 times more likely to be within 1kb of a mapped enhancer (Figure 5D). This represented 127 of the lncRNA hit loci identified in our screens. However, the FANTOM enhancer annotations used for our analyses were derived from hundreds of different cell types, and thus only a fraction of these enhancers are active in any given cell type in our screen (48, 49). Hit loci were also 1.4 times more likely to be within 5kb of a cancer-associated SNP (Figure 5E). That our hits were enriched for multi-exonic lncRNAs is consistent with the concept that lncRNA splicing can be an aspect of lncRNA function (26) (Figure 5F). However, the explanatory power of exon number was relatively low, and our screen did identify several single exon hits such as NEAT1. However, no genomic property analyzed, alone or in aggregate, fully predicted growth modifier lncRNAs in a given cell type, underscoring the importance of performing loss-of-function screens for defining sets of functional genes.
Discussion
By employing CRISPRi for systematic, large-scale screens for lncRNA function in multiple cell lines, we identified 499 lncRNA loci that are required for robust cell growth. This work increases considerably the number of known functional lncRNAs and revealed that the large majority (89%) of lncRNA genes identified modified growth in just one cell type. Studies of the protein-coding genome with similar large-scale screening efforts showed that an essential gene in one cell type is highly likely to be essential in the other cell types tested (46, 47). In contrast to protein coding genes, of the 1,329 lncRNA genes expressed in all of the seven different cell lines tested, not one lncRNA gene was required for robust cell growth in all cell types, with the large majority of lncRNA gene hits being specific to just one cell line. Our results thus reveal a critical role of cellular context in determining lncRNA function.
Several clues to this specificity of lncRNA function emerge from our analyses. First, although cell type-specific expression of lncRNAs was the strongest predictor of lncRNA hits in our machine learning model (Figure 5A,C), it did not fully explain this functional specificity (Figure 3, 5B). For example, RNA-seq analysis points to LINC00263 playing a role in a complex transcriptional network required for U87 cells, but that despite being expressed in other cell types, LINC00263 appears dispensable for the normal expression of nearly all genes in these other cells (Figure 4D and S12D,E). Taking advantage of the scale of our dataset, we have also begun to discover genomic features that predict growth modifying function. Our finding that enhancer proximity and chromosome contacts correlate with lncRNA function suggests that higher-order chromatin structure can play a role in such specificity of lncRNA function (28, 29) (30). The extent to which cell type-specific function of enhancer-templated lncRNAs results from repression of the transcript itself or its genomic locus remains an important open question. In any case, the association of lncRNA function with higher order chromatin structure is consistent with the emerging view that chromosomal looping between lncRNA promoters and target genes differs between cell types (54) and is critical to lncRNA function (55). Finally, our finding that genomic regions containing growth modifier lncRNAs are enriched for cancer risk SNPs suggests that these lncRNAs may contribute to the pathogenesis of cancer.
Regardless of the mechanism(s) of the observed cell type-specificity of lncRNAs, this finding has implications for understanding the biological roles of lncRNAs. LncRNAs appear to have originated much later than protein coding genes, consistent with their not playing generic housekeeping roles (3, 56). Our study, which focused on lncRNAs required for robust cell growth, underestimates the true number of functional lncRNAs in these cell types, as lncRNAs have been shown to regulate more evolutionarily complex cellular decisions such as cell fate (7, 19, 57, 58), cancer metastasis (59, 60), and perhaps neuronal function (61). The CRISPRi tools developed here can now be applied to the study of such higher order cellular processes, where lncRNAs might exhibit even greater richness of function. Finally, the exquisite cell type-specificity of lncRNA gene function has clear implications for targeted therapy.
Supplementary Material
Acknowledgments
We thank the members of the Lim and Weissman labs, particularly Alex Fields, Joshua Dunn, Manny DeVera, Miao Cui, and David Wu for helpful discussions and assistance. We thank Annie Truong for assistance with iPS cell culturing and Nathan Salomonis for iPSC RNA-seq data. We also thank Eric Chow and Derek Bogdanoff of the UCSF Center for Advanced Technology for sequencing assistance, and Laurakay Bruhn, Daniel Ryan, Luke Fairbairn, and Peter Tsang of Agilent Technologies for their assistance on the design and synthesis of oligonucleotide pools.
This project was supported by NIH 1R01NS091544-01A1, VA 5I01 BX000252-07, NIH SPORE DRP, the Shurl and Kay Curci Foundation, the LoGlio Foundation, and the Hana Jabsheh Initiative (to D.A.L.). S.J.L. is supported by NIH F30 NS092319-01. M.A.H., J.E.V., M.Y.C., Y.C., L.A.G., and J.S.W. were supported by the Howard Hughes Medical Institutes and the National Institutes of Health (P50 GM102706, U01 CA168370, R01 DA036858). S.W.C. and H.Y.C. are supported by National Institutes of Health (R35-CA209919, P50-HG007735). B.R.C and M.A.M are supported by the Gladstone Institutes and National Institutes of Health (U01HL100406, P01HL089707, R01HL130533). L.A.G. is supported by the NIH/NCI Pathway to Independence Award (K99CA204602). Oligonucleotide pools were provided courtesy of the Innovative Genomics Initiative. Sequencing data are deposited in GSE85011.
Footnotes
References
- 1.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Bassett AR, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. The EMBO Journal. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aparicio-Prat E, et al. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genomics. 2015;16:846. doi: 10.1186/s12864-015-2086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho TT, et al. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2014;43:gku1198–e17. doi: 10.1093/nar/gku1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin Y, et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2015;16:504–516. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Paralkar VR, et al. Unlinking an lncRNA from Its Associated cis Element. Molecular Cell. 2016;62:104–110. doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groff AF, et al. In Vivo Characterization of Linc-p21 Reveals Functional cis-Regulatory DNA Elements. Cell Reports. 2016;16:2178–2186. doi: 10.1016/j.celrep.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu S, et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016 doi: 10.1038/nbt.3715.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin N, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Molecular Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Y, Cullen BR. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8:855–860. doi: 10.1017/s1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimasu H, et al. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engreitz JM, Haines JE, Munson G, Chen J, Perez EM. Neighborhood regulation by lncRNA promoters, transcription, and splicing. 2016 doi: 10.1101/050948.. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ørom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fulco CP, et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016:1–13. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakore PI, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Meth. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amabile A, et al. Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell. 2016;167:219–232.e14. doi: 10.1016/j.cell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandegar MA, et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun CJ, et al. Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proceedings of the National Academy of Sciences. 2016;113:E3892–900. doi: 10.1073/pnas.1600582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horlbeck MA, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 2016;5 doi: 10.7554/eLife.19760.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K, et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Stem Cells. 2007;131:1–12. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature Genetics. 2015 doi: 10.1038/ng.3192.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium TEP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 41.Kreitzer FR, et al. A robust method to derive functional neural crest cells from human pluripotent stem cells. Am J Stem Cells. 2013;2:119–131. [PMC free article] [PubMed] [Google Scholar]
- 42.Liu SJ, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kampmann M, Bassik MC, Weissman JS. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proceedings of the National Academy of Sciences. 2013;110:E2317–26. doi: 10.1073/pnas.1307002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Molecular Systems Biology. 2014;10:733–733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tseng YY, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 48.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan X, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, et al. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol. 2016;17:19. doi: 10.1186/s13059-016-0880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma W, et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Meth. 2015;12:71–78. doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973–1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 57.Ramos AD, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Research. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatinstate to promote cancer metastasis. Nature. 2011;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 62.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salomonis N, et al. Integrated Genomic Analysis of Diverse Induced Pluripotent Stem Cells from the Progenitor Cell Biology Consortium. Stem Cell Reports. 2016;7:110–125. doi: 10.1016/j.stemcr.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Meth. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 67.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Geen H, Echipare L, Farnham PJ. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. Methods Mol Biol. 2011;791:265–286. doi: 10.1007/978-1-61779-316-5_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.