Abstract
Although the factors responsible for the recent increase in the prevalence of diabetes worldwide are not entirely known, the morbidity associated with this disease results in substantial health and economic burden on society. Epigenetic modifications, including DNA methylation have been identified as one mechanism by which the environment interacts with the genome and there is evidence that alterations in DNA methylation may contribute to the increased prevalence of both Type 1 and Type 2 diabetes. This review provides a summary of DNA methylation and its role in gene regulation, and includes descriptions of various techniques to measure site-specific and genome wide DNA methylation changes. In addition, we review current literature highlighting the complex relationship between DNA methylation, gene expression, and the development of diabetes and related complications. In studies where both DNA methylation and gene expression changes were reported, DNA methylation status had a strong inverse correlation with gene expression, suggesting that this interaction may be a potential future therapeutic target. We highlight the emerging use of genome wide DNA methylation profiles as a biomarker to predict patients at risk of developing diabetes or specific complications of diabetes. Developing a predictive model that incorporates both genetic information and DNA methylation changes may be an effective diagnostic approach for all types of diabetes and could lead to additional innovative therapies.
Key terms: Islets, insulin secretion, insulin sensitivity, type 2 diabetes, DNA methylation, and epigenetics
Introduction
Over the past several decades the prevalence of diabetes, has increased dramatically from 108 million in 1980 to 422 million in 2014 (1). Type 1 diabetes (T1DM), a T-cell mediated autoimmune process characterized by pancreatic β-cell destruction and insulin deficiency is the most common type of diabetes in children and adolescents accounting for 5–10% of those with diabetes worldwide (2,3). Monogenic forms of diabetes including neonatal diabetes mellitus (NDM) and maturity-onset diabetes of the young (MODY) that result from single gene mutations, account for 1–5% of diabetic cases (4,5). The majority of patients with diabetes worldwide have Type 2 diabetes (T2DM), which accounts for 85–95% of total cases (2) and is characterized by a combination of decreased insulin secretion by the pancreatic β-cells and reduced insulin sensitivity at peripheral target organs(6). T2DM is the most common type of diabetes in adults, but increasingly has also been diagnosed in the youth (7). A recent report by the US National Diabetes Statistics highlighted the estimated annual cost of all forms of diabetes in the United States alone to be $245 billion (8). Although the increased prevalence in both T1DM and T2DM has been well documented throughout the world, the mechanisms responsible for these increases are not well understood.
Epigenetic modifications have been identified as one mechanism by which the environment interacts with the genome and modifies the risk of T1DM (9) and T2DM (10). Epigenetic modifications are often considered as the byproducts of environmental stimuli, which can influence the genetic susceptibility to diabetes. The traditional definition of epigenetic modifications describes changes in DNA methylation, changes in the acetylation, methylation or phosphorylation of histones as well as changes in non-coding RNA expression that are not related to changes in DNA sequence, but are mitotically heritable alterations that can ultimately influence gene expression. However, recent studies have suggested that genetic and epigenetic modes of inheritance may be even more complex than initially thought. For instance, studies suggesting that DNA polymorphisms may result in specific genomic regions that are more or less receptive to epigenetic marks (11–13), thus indicating that epigenetic modifications can be influenced by genetic sequence variations. In addition, environmental factors such as diet, stress, physical activity, and a suboptimal in utero environment can alter the epigenetic states and contribute to changes in gene expression. Overall, there is a complex interplay between genetics, epigenetics and environment, which can influence gene expression. In this review, we will focus on DNA methylation, one of the first epigenetic modifications to be well characterized, especially through genome wide assays, and outline its role in the pathogenesis of diabetes.
I) DNA Methylation and its role in the regulation of gene expression
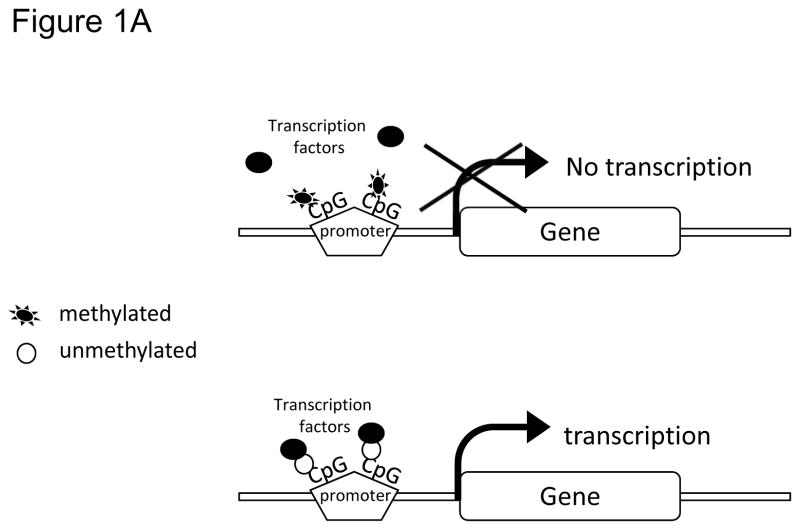
DNA methylation in mammalian cells typically involves covalent addition of a methyl group (-CH3) at the 5′ position of the cytosine ring within the 5′-CpG-3′dinucleotidesto create a 5-methylcytosine (5-mC)(14). CpG sites tend to cluster together as repetitive sequences called as CpG islands (CpGI) found either at promoters of genes, or regions with increased centromeric tandem repeat units (15). In humans, ~70% CpG dinucleotides are methylated, whereas CpG dinucleotides found at CpGI in germ-line tissues and those located near promoters of somatic cells mostly remain unmethylated (15). In order for a gene to be transcribed, the promoter and other regulatory regions of the gene including enhancers must be accessible to transcription factors and other regulatory complexes. DNA methylation decreases accessibility of the DNA and can block the binding of transcription factors, thus affecting gene expression (Figure 1A). In fact, there is evidence that changes in CpG density and methylation status at tissue-specific promoters play an important role in controlling the expression of the associated genes (16).
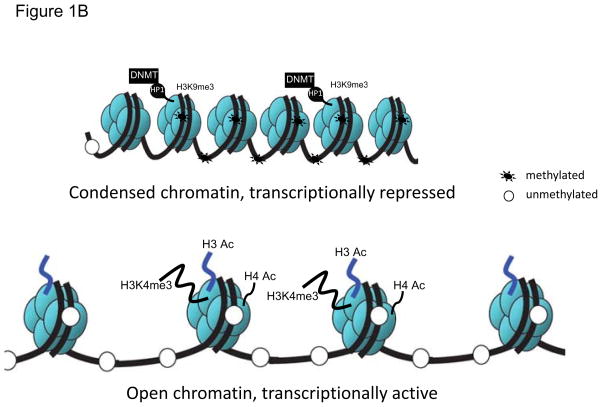
Figure 1.
A) CpGs at gene promoter sites: methylated and unmethylated. B) Heterochromatin protein 1 (HP1) binds to the trimethylated histone H3 at lysine 9 (H3K9me3), and then recruits DNA methyltransferase (DNMT). DNMTs in turn increase DNA methylation resulting in a compact chromatin and transcription silencing. Whereas, increased histone acetyl transferase activity, increased histone H3 (H3) and histone 4 (H4) acetylation (Ac), and increased trimethylation of histone H3 at lysine 4 (H3K4me3) prevents DNA methylation leading to a relaxed/open chromatin and transcription activation.
In addition, specific histone modifications such as methylation of lysine residues in the amino termini of histone 3 (H3), or removal of acetyl groups from lysine residues on histones H3 and H4 by histone deacetylases can alter the conformation of the chromatin, resulting in a more condensed chromatin form thus restricting access of the transcription factors (15). Regions with condensed inaccessible chromatin structures and methylated CpG sites with associated low rates of mRNA transcription are called heterochromatin (Figure 1B) (15). In contrast, open and active chromatin structures with locally unmethylated CpGs that are associated with high rates of transcription and are referred to as euchromatin (Figure 1B) (15).
DNA methylation and DNA methyl transferases (DNMTs)
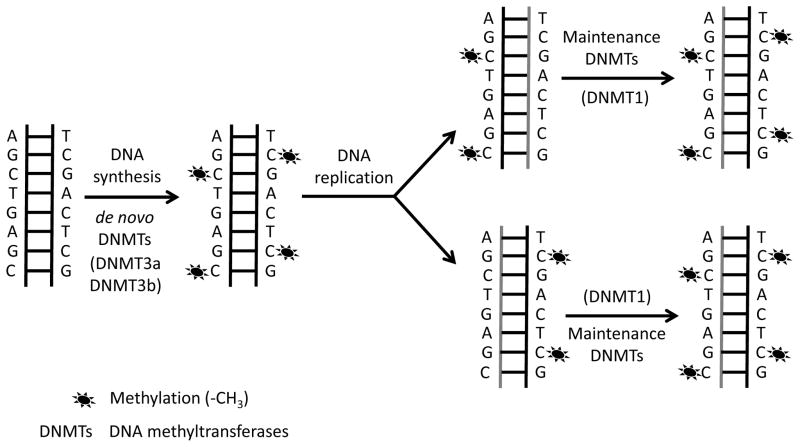
DNA methylation is initiated by enzymes called DNA methyl transferases (DNMTs). DNMT3a and DNMT3b primarily target unmethylated CpGs and establish new methylation marks and therefore, are often characterized as de novo methyl transferases (17,18) (Figure 2). The de novo methylation process occurs during an early embryonic stage in stem cells, and in cancer cells (17,18). Traditionally, DNMT1 has been referred as a maintenance methyl transferase as it recognizes and methylates hemimethylated CpGs during DNA replication, and copies the pre-existing DNA methylation patterns from parental to daughter strands(19) (Figure 2). However more recently, it has been shown that DNMT1, DNMT3a, and DNMT3b could also function co-operatively to methylate DNA (20,21). In fact, DNMT3a and DNMT3b have been shown to function as maintenance methyl transferase in embryonic stem cells (22), and DNMT1 has been proposed to have de novo methyl transferase capacity (23).
Figure 2.
De novo DNA methyltransferases and maintenance DNA methyltransferases.
DNA methylation influences gene expression
DNA methylation is critical during embryonic development in mammalian cells as well as in regulating tissue-specific gene expression and genomic imprinting. Depending on the location of DNA methylation in a genomic sequence, DNA methylation can have varied effects on gene function. For example, usually DNA methylation typically is absent at gene promoters and in intergenic regions such as enhancers, but these regions can become methylated and result in gene silencing (24), ultimately affecting the expression of genes during development and differentiation (25–27). In addition, DNA methylation in CpGI plays a critical role in establishing imprinting marks (28,29), while DNA methylation in the gene body is associated with altered gene expression in dividing cells (30–32). Genomic imprinting is the epigenetic phenomena when genes are expressed based on parent of origin of the allele. Changes in DNA methylation resulting from genomic imprinting haves been linked to 6q24 transient neonatal diabetes (33), where patients with transient neonatal diabetes have decreased methylation of the maternal allele at the 6q24 locus (34,35). In these patients, diabetes resolves after several months of life; however, the underlying mechanism remains to be fully elucidated.
II) Methods for detecting genome-wide and site-specific DNA methylation
Currently various techniques have been developed to determine site-specific and genome wide changes in DNA that associate with changes in gene expression and can be used as markers for risk assessment, early prognosis, and treatment of disease.
In this section, we describe techniques used to measure DNA methylation both at specific sites within the genome and assays that measure DNA methylation on a genome wide scale.
Methods for detecting site-specific DNA methylation
Most site-specific DNA methylation techniques involve bisulfite conversion and are discussed below.
Bisulfite pyrosequencing involves bisulfite treatment of genomic DNA where unmethylated cytosine residues of the DNA are converted to uracil, and methylated cytosines remain unmodified. This is followed by amplification of the bisulfite-converted product through PCR where uracil is converted to thymine using site-targeted biotinylated primers. Finally, successfully converted bisulfite PCR products are sequenced using specifically designed pyrosequencing primers (36). The pyrogram gives percent methylation at each CpG site based on the levels of uncoverted C and converted T. Pyrosequecing offers sequencing of only short stretches of DNA, which limits the number of CpG sites that can be assessed by this technique. However, a potential benefit is that percent methylation at multiple CpG sites (though few in number) can be accurately quantified in one reaction.
Methylation sensitive PCR (MSP) involves use of bisulfite converted DNA followed by PCR with primers specific for methylated versus unmethylated DNA (37). MSP as developed is a non-quantitative technique, but more recently quantitative versions of this technique have become available like MethylLight (38) and quantitative analysis of methylated alleles (QAMA) (39) which use real-time PCR instead of a traditional PCR. A potential drawback of these techniques is that they are based on the assumption that alleles will be methylated or unmethylated and therefore do not take partial allelic methylation due to a heterogeneous cell populations into account.
Bisulfite treatment followed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) uses bisulfite converted DNA, followed by PCR primers using primers with T7 RNA polymerase tag (40) and the products are translated into a single-stranded RNA using T7 polymerase, followed by cleavage by an endoribonuclease such as RNase A. The methylated and unmethylated CpGs generate different cleavage patterns which are then quantitated by mass spectrometry (40). Unlike pyrosequencing, this technique covers wider stretch of DNA and therefore multiple CpG sites can be detected in one reaction. However the technical complexity of this method makes it less user friendly.
Methods for detecting genome-wide DNA methylation
Methylated DNA Immunoprecipitation (MeDIP) and Methyl-CpG binding domain (MBD) protein capture are two commonly used approaches to enrich for methylated DNA regions of the genome which can undergo further analysis with sequencing to detect differentially methylated CpG sites (41). MeDIP uses a monoclonal antibody specific to 5′-methylcytosine to immunoprecipitate single stranded DNA containing one or more CpG sites (42), while the MBD uses the methyl-CpG binding domain of the MBD2 protein to capture double stranded methylated DNA fragments and differentiates variable DNA methylation densities by using different salt concentrations during the elution step (43). One potential drawback of these methods is that the density of methylated cytosines in a particular region of DNA can affect the antibody binding (41). From findings in the literature, it is suggestive that MeDIP is more effective for enriching methylated areas with low CpG density, while MBD protein capture with high, CpG density (43,44).
Luminometric Methylation Assay (LUMA) uses methylation sensitive and insensitive restriction enzymes to digest genomic DNA, followed by quantifying the resulting number of cuts from the restriction enzymes using a pyrosequencer (45,46). As LUMA is unable to identify specific positions in the genome where methylation is located it represents global methylation analysis.
The Infinium Human Methylation 450 Bead Chip (Infinium Methylation 450K; Illumina, Inc. CA, USA) is an array based analysis of 485,000 methylation sites covering approximately 17 CpG sites per gene region distributed across the promoter, 5′UTR, first exon, gene body, and 3′UTR (47). In addition, it also covers most CpGI, CpG shores, flanking regions, and includes some coverage of non-CpG island methylation, and miRNA promoter regions (47). Similar to 450K bead array, Illumina has recently launched the MethylationEPIC bead chip, which offers higher coverage than the 450K array (48). MethylationEPIC covers 850,000 methylation sites across the genome, including 5′-hydroxymethylcytosine patterns and previously unidentifiable, but important, regulatory CpG sites (48). This technique uses low input sample amount (as low as 250 ng) and is reported to be highly reproducible (48). Its moderate cost for genome wide DNA methylation analysis permits for larger number of samples needed for studies of human samples. Thus by offering a comprehensive genome wide DNA methylation analysis, MethylationEPIC is a promising technique to assess genome wide DNA methylation pattern.
Enhanced reduced representation bisulphite sequencing (ERRBS) involves digestion of 10–300 ng of purified genomic DNA using methylation-insensitive restriction enzyme (commonly MspI), followed by end repair, A-tailing, adapter ligation, size selection, bisulfite conversion and PCR amplification (49,50). The prepared libraries are then sequenced and analyzed to determined differentially methylated sites. This assay is designed to target CpG dense promoter regions and repeated elements, which helps to control the high cost of sequencing the entire genome. This technique effectively captures 3 million CpG sites including 85% of CpG islands, and 60% promoters (50). Therefore ERRBS provides a reasonable and cost effective method to measure genome wide DNA methylation.
Whole Genome Bisulfite Sequencing (WGBS) uses 50–100 ng of purified genomic DNA for library preparation that entails fragmentation of the DNA to generate 3′ or 5′ overhangs using an ultrasonicator, followed by end repair, adenylation of 3′ ends, and adaptor ligation (51–54). WGBS aims to profile approximately 15–20 million CpG sites within the whole genome and approximately 1 billion 100 bp end reads are required to obtain 30× average coverage throughout the genome. Once DNA libraries are created, they undergo bisulfite conversion, PCR amplification, followed by next generation sequencing (51–54). Since this technique involves sequencing of the entire genome, it is not very cost effective. However, with declining cost of WGBS, this method will become an critical tool for detecting changes in DNA methylation, especially within intergenic regions and enhancers which have been shown to be key regulatory regions in islets from patients with T2DM and CD34 cells exposed to intrauterine growth restriction (55). Neither the array based genome wide DNA methylation assays, nor ERRBS adequately profile DNA methylation within the intergenic and enhancer regions of the genome. Table 1 provides a summary of few genome-wide DNA methylation techniques that are discussed above.
Table 1.
Genome Wide DNA Methylation Assays.
| WGBS | 450K Infinium Bead Chip Array | MethylationEPICInfinium Bead Chip Array | ERRBS | |
|---|---|---|---|---|
| Regions Sequenced | Whole genome including intergenic and enhancer regions | Predesigned array based | Predesigned array based | Determined by Msp1 digestion to enrich for CpG fragments |
| Genome Coverage | 15–20 million CpG sites | 485,000 methylation sites across the genome | 850,000 methylation sites across the genome | 3 million CpG sites ~85% of CpG islands, and 60% promoters |
| Assay Details | Bisulfite conversion of genomic DNA, followed by next generation sequencing | Bisulfite conversion of genomic DNA followed by annealing bead array | Bisulfite conversion of the genomic DNA, followed by annealing bead array | MspI digestion followed by bisulfite conversion and next generation sequencing |
| Cost per sample | $$$ | $ | $ | $$ |
| Input DNA | 50–100 ng | 500 ng – 1 μg | 250 ng | 10–300 ng |
| Additional coverage information | Comprehensively covers the entire genome including methylated and unmethylated regions. | Most CpG islands, shores, flanking regions, non-CpG island methylation, and miRNA promoter regions | 5′hydroxy-methyl-cytosine patterns and novel CpG regulatory sites | Enrichment in CpG islands, CpG shores, promoters, exons, introns, and intergenic regions |
III) Role of DNA Methylation in Developmental Origins of Diabetes
The period from conception to birth is a time of rapid growth, cellular replication, differentiation, and functional maturation of organ systems. These processes are very sensitive to alterations of the intrauterine metabolic milieu, including pre-eclampsia, maternal hypertension, maternal diabetes and obesity, smoking and exposure to drugs and environmental chemicals, all of which can have long-lasting effects on the offspring. David Barker and colleagues, who developed the developmental origins of health and disease (DOHaD) hypothesis, first described the relationship between low birth weight and the increased risk of cardiovascular disease and T2DM in later life in Hertfordshire population in England (56,57). The results from these epidemiological studies have been replicated in various populations around the world (58).
Although there is an established relationship between early life exposure and the later development of T2DM in human epidemiological studies, animal models are needed to investigate mechanisms underlying these observations. Animal models of fetal programming have been developed in many species including rodents, sheep, guinea pigs and non-human primates. These include various models of intra-uterine growth restriction (IUGR), models of exposure to various environmental chemicals like bisphenol A and phthalates, and models of metabolic abnormalities during pregnancy including gestational diabetes mellitus, and maternal obesity. Although the various models of fetal programming include a wide variety of intrauterine exposures, all of the models described above result in profound effects in the placenta and offspring pancreas resulting in abnormalities in glucose homeostasis. There has been mounting evidence that DNA methylation can affect gene expression in the developing offspring and this is one potential epigenetic mechanism by which early life environment perturbations can affect later life health. In this section we will cover some of the DOHaD models that have indicated alterations in DNA methylation specifically in context of T2DM.
a) Intra-uterine growth restriction (IUGR) models
Exposure to IUGR has profound effects on the expression of genes that control glucose and energy homeostasis in pancreatic islets and peripheral tissues (liver, skeletal muscle and adipose tissue). The following studies indicate that DNA methylation is one mechanism by which an in utero insult can lead to the development of diabetes in adult offspring.
i) Maternal nutrient deprivation (protein restriction) model and DNA methylation
Various studies have shown that maternal diet can induce changes in the heritable epigenome, which would influence the later metabolic health of the offspring. Sandovici et al found reduced expression of a pancreatic transcription factor Hnf4α in rat islets from offspring of mothers who were fed a low protein diet(59). In this model, reduced expression of Hnf4α was associated with increased DNA methylation at P2 promoter, along with reduced activating histone modifications, and increased silencing histone modifications at 3 months of age (59). These studies demonstrate that a maternal sub-optimal nutrient environment influenced the epigenetic state of Hnf4α, which consequently lead to changes in Hnf4α expression. Similarly, maternal low protein diet has been shown to influence locus specific epigenetic changes, including changes at the such as the liver X-receptor alpha (LXRα) promoter and target genes like Abcg5/Abcg8, as well as global DNA methylation in offspring liver (60–62). Other studies have shown that exposure to a maternal low protein diet along with folic acid supplementation result in changes in DNA methylation at the Pparα and glucocorticoid receptor promoter and corresponding changes in gene expression in liver from juvenile and adult rat offspring (63,64). Further, from fetal life to adulthood the offspring of dams fed a low protein diet had altered methylation levels in leptin promoter in adipose tissue, and this was associated with altered feeding behavior in the offspring (65). Therefore, in utero nutritional perturbations can lead to changes in DNA methylation that have life-long effects on the metabolic health of the offspring.
ii) Uterine artery ligation model and DNA methylation
Uterine bilateral artery ligation is another model of inducing IUGR, and animals generated via this technique experience in utero placental insufficiency (UPI) and go on to develop T2DM in adulthood (66). Epigenetic modifications have been shown to play a critical role in regulating the expresison of key genes that are altered in this model. For example, pancreatic homeobox domain 1 (Pdx1), a key pancreatic transcription factor regulating pancreatic differentiation and growth has been shown to have permanently reduced expression in IUGR beta cells (67). This reduction in Pdx1 has been associated with epigenetic changes including an increase in silencing histone modification at the proximal Pdx1 promoter and initiation of de novo DNA methylation leading to permanent reduction of Pdx1 gene expression in adulthood (68). Altered genome wide DNA methylation in the IUGR model was described by Thompson et al who found 1400 differentially methylated loci in isolated islets from adult male rats exposured to IUGR (69). A majority of the differentially methylated CpG sites were found at conserved intergenic regions, and near genes that had been previously described as key regulators of important beta cell processes like cell division and death, vascularization, and insulin secretion (69). Thus, perturbed DNA methylation in UPI model appears to be an important epigenetic mechanism involved in regulating beta cell development and function.
b) Exposure to endocrine disrupting chemicals model and DNA methylation
Unlike the in utero caloric restriction nutritional model where imprinted genes were reported to be unaffected (70), exposure to endocrine disrupting chemicals such as bisphenol A has been shown to affect the expression of imprinted genes. In this context, prenatal exposure to bisphenol A from two weeks prior to mating and throughout gestation, leads to loss of imprinting of Igf2 and gain of Igf2 gene expression in embryos (71). Igf2 is expressed of the paternal allele, and loss of Igf2 imprinting leads to increased expression of Igf2 through both maternal and paternal alleles. This occurs due to an increase on DNA methylation in Igf2 differentially methylated region 1 (DMR1), which is normally unmethylated. A repressor protein, GCF2, binds to unmethylated DMR1, and blocks enhancer access to Igf2 (72). The methylation of DMR1 prevents GCF2 binding and permits enhancer-Igf2 interaction, leading to Igf2 expression (72). In utero exposure to BPA in mice increases Igf2 DMR1 methylation and Igf2 expression from both maternal and paternal allele in embryos (71). While Igf2 has many other roles, it is a key regulator of early beta cell development in islets and aberrant Igf2 imprinting early in life could potentially influence the normal beta cell development. However, whether increased Igf2 expression is the mechanism underlying the impaired glucose tolerance reported in these animals is currently under investigation (73).
c) Gestational Diabetes Mellitus and DNA methylation
Population-based studies demonstrate that the offspring of diabetic mothers have an increased risk for obesity, glucose intolerance, and T2DM (74,75). It has been proposed that early exposure to hyperglycemia and elevated insulin levels may lead to a malprogramming of critical functions related to the development of diabetes and obesity later in life (76). Several studies have sought to identify changes in genome wide DNA methylation resulting from exposure to gestational diabetes mellitus (GDM) as one mechanism contributing to the malprogramming of the offspring(77–79).
Various studies have mapped genome wide DNA methylation using the Infinium 450k bead array or MeDIP in placentae from women with GDM and have found increased number of differentially methylated genes predominantly involved in glucose metabolism pathway and in energy metabolism (80–83). For example, Rong et al found that DNA methylation changes of a subset of tested genes (Rbp4, Glut3, Resistin and Pparα) correlated with gene expression changes determined by qPCR (80). Similarly, Petropoulos and colleagues found correlation between DNA methylation and gene expression of P2rx5, Ccdc15, and Adam12 (81). Alexander et al mapped genome wide DNA methylation using the Infinium 450K methylation array in placentae from Native American and Hispanic women with GDM and correlated the differentially methylated CpG sites with RNA-Seq data and protein levels (under review). They report that placentae from female offspring exposed to GDM were 40% more likely to have significant gains in DNA methylation compared to placentae from male offspring. In addition, changes in DNA methylation corresponded to changes in mRNA expression and protein levels in Piwil3, Cyba, Gstm1, Gstm5, Kcne1 and Nxn. Systems based analysis showed that in utero exposure to GDM lead to significant alterations in DNA methylation in genes related to mitochondrial function, DNA repair, inflammation, and oxidative stress. Together, these studies indicate that in utero exposure to GDM can alter DNA methylation on a genome wide basis and that some of the changes in DNA methylation induced by exposure to GDM are followed by corresponding changes in gene expression.
IV) T2DM, Islets, and DNA methylation
Impaired insulin secretion by the pancreatic islet is a key component to the pathogenesis of T2DM. Therefore, several studies that used a candidate gene approach to study the relationship between changes in DNA methylation and gene expression in islets from donors with T2DM. Increased DNA methylation was measured CpG sites within the promoters of the pancreatic transcription factor Pdx1, the mitochondrial gene regulator Ppargc1a and Insulin gene, which negatively correlated with mRNA expression in all three genes (84–86). Using the genome-wide Infinium450K array, 1,649 CpG sites and 853 genes, including Tcf7l2, Fto, Kcnq1, Irs1, Cdkn1a, and Pde7bwere identified to have significant changes in DNA methylation in human islets from T2DM donors compared to controls (87). Of the 853 genes with differentially methylated CpG sites, 102 genes, including Cdkn1a, Pde7b, Sept9 and Exoc3l2, had significant changes in mRNA expression as measured by microarray (87). Increased DNA methylation at the promoter of Cdkn1a and Pde7b was associated with decreased transcriptional activity in clonal β-cells in vitro as well as impaired glucose stimulated insulin secretion (87). Another genome wide study of DNA methylation using the Infinium27K array found 276 differentially methylated CpG sites of which 96% were hypomethylated in islets of diabetic v.s. non-diabetic donors(88). Changes in differential DNA methylation were correlated with expression changes of 34 genes assessed by microarray (88). The changes in DNA methylation identified in islets from T2DM were not found in DNA from peripheral blood from T2DM donors nor were there induced by exposing a clonal β-cell line to hyperglycemia (89). These studies indicate that alterations in DNA methylation could be a potential mechanism contributing to the pathogenesis of T2DM in the islets.
While changes in islet DNA methylation are linked to T2DM, changes in DNA methylation associated with peripheral insulin resistance may also increase the risk of T2DM. In this review we focused on studies assessing changes in DNA methylation in islets; changes in DNA methylation at peripheral tissues such as liver, muscle and adipose and their association with diabetes are reviewed elsewhere (90,91). Some changes in DNA methylation in peripheral tissues have been shown to be amenable to modification by simple physiological interventions like exercise. For example, a six months exercise intervention induced noticeable alterations in genome wide DNA methylation profile and gene expression in adipose tissue of obese and type 2 diabetic patients (92). Similarly, 3 months exercise program was associated with DNA methylation and gene expression changes in skeletal tissue of non-diabetic male and female volunteers (93). Results from these studies indicate that changes in DNA methylation may not be permanent and in fact may represent key targets of intervention in future studies..
V) T1DM, Islets, Immune Cells and DNA methylation
In monozygotic twin pairs, only 50% co-twins develop type 1 diabetes (94) suggesting that there are both genetic and non-genetic mechanisms responsible for T1DM. With an objective of identifying non-genetic mechanisms influencing the development of T1DM, several studies have measured changes in DNA methylation at CpG sites in immune cells including peripheral lymphocytes (95) and monocytes(96). Stefan et al used the Infinium 27K bead array to measure changes in DNA methylation in DNA isolated from lymphoblast cell lines from 3 pairs of monozygotic twins discordant for T1DM and 6 pairs of monozygotic twins concordant for T1DM. They identified 88 CpG sites with significant changes in DNA methylation. Functional genomics analysis indicated that the affected genes were clustered in the immune response and defense response pathways and included genes that had previously been associated with T1DM pathogenesis including Hla, Ins, Il2rb and Cd226 (95). No data on gene expression was reported in this dataset. A similar study of monozygotic twin pairs discordant for T1DM profiled changes in DNA methylation in DNA isolated from whole blood with the Infinium 450K array. The authors report modest methylation differences for the MHC region and T1D-associated CpG sites in Bach2, Ins, Igf2, and Clec16a (DNA methylation difference range: 2.2%–5.0%). Other genes reported to have significant differences in DNA methylation were Magi2, Fancc, and Pcdhb16 (DNA methylation difference range: 6.9%–16.1%). No gene expression data was reported and it is not clear what changes in DNA methylation from peripheral blood samples represent in this population (97). Epigenetic changes are challenging to interpret in a mixed cell population such as whole blood because these changes may reflect differences in white blood cell populations, which may or may not be related to a disease phenotype.
Recently a study highlighted the cross talk between immune responses and β-cell specific DNA methylation changes at Ins1 and Ins2 in islets from NOD mice, and in human beta cells in vitro (98). In the NOD mouse model of T1DM, inflammatory cytokines including TNF, IFNγ, IL6 and IL1B increase with age (98). Rui et al showed reduced insulin gene expression and increased percent DNA methylation at exon 2 of Ins1 and exon1 of Ins2 genes in sorted beta cells from 4 week-old NOD mice cultured in media with cytokines (98). Moreover, increased cytokines induced mRNA expression levels of DNMTs in sorted β-cells of cultured islets from NOD mice and from human non-diabetic donors (98). This study suggests that increased cytokine levels associated with T1DM induce increased DNA methylation and decreased insulin mRNA levels in islets. Recently there has been interest in quantifying the amount unmethylated preproinsulin DNA in the circulation as a biomarker of beta cells death. Beta cells have a much higher frequency of unmethylated CpG sites within the preproinsulin DNA sequence than other cells, and upon β-cell death these DNA sequences are released into the circulation (99). Studies have found increased levels of unmethylated preproinsulin DNA in peripheral blood samples of patients with new-onset type 1 diabetes compared with controls (100,101).
VI) Diabetic Nephropathy and DNA methylation
The studies highlighted above focus on evidence that DNA methylation is associated with the pathogenesis of diabetes, but there is also evidence that changes in DNA methylation can be induced by hyperglycemia and other metabolic byproducts of diabetes which contribute to the development of diabetic complications in peripheral organs (102). In addition, DNA methylation assays in peripherally accessible cells have been used as a biomarker to predict which patients are at increased risk of developing particular complications of both T1DM and T2DM. Here we focus on studies examining the relationship between DNA methylation and diabetic nephropathy.
Diabetic nephropathy (DN) characterized by persistent proteinuria, hypertension and a persistent decline in the glomerular filtration rate, is the leading cause of end stage renal disease in the US and Europe. In diabetic nephropathy, prolonged exposure to hyperglycemia induces production of cytokines, chemokines and growth factors including transforming growth factors beta 1 (TGFβ) and connective tissue growth factors (CTGF), which leads to abnormal glomerular pathology. Brennan et al measured DNA methylation with MALDI-TOF mass spectroscopy at the 5′promoter region of 192 candidate genes previously identified to be differentially expressed in in vitro models of DN (human mesangial cells and proximal tubular epithelial cells treated with stimuli including glucose, TGFβ and CTGF) and in renal biopsies from individual with DN (103). They report that 301 CpGs in 38 out of 192 genes were differentially methylated (defined as a >20% difference in methylation). GO analysis of the differentially methylated genes revealed that the predominant biological function of the affected genes was organism development. In a separate study, DNA methylation profiling with the Infinium 27K bead array from samples of whole blood was used to identify 19 prospective CpG sites associated with risk of DN in T1Y1 compared to those without nephropathy (104). Additional studies using various DNA methylation assays and DNA collected from peripheral blood samples or saliva identified specific DNA methylation profiles for diabetic patients with and without nephropathy (105–107). Most of these studies proposed using DNA methylation profiles as biomarkers to help predict disease status and progression and did not report associated gene expression data.
In summary, there is some evidence suggesting that exposure to cytokines and chemokines as well as hyperglycemia can alter gene expression and DNA methylation in the diabetic kidney. In addition, DNA methylation profiles may provide insight about which patients with diabetes are at particular risks for developing DN, but this remains to be tested prospectively.
VII) Conclusion and Future Directions
Diabetes is an increasing health concern worldwide and a substantial economic burden to individuals and society. It is becoming increasingly important to identify improved biomarkers that can help characterize populations that are at risk o and also can help identify at characterize populations at risk of developing diabetes. In addition, these biomarkers can assist wtih, improvement of the diabetes clinical diagnostic processes, and develop targeted treatment options for patients with both T1 and T2DM. Recently our knowledge of epigenetic contributions to the pathogenesis of diabetes has expanded phenomenally, with DNA methylation being at the forefront of this advancement. Throughout this review, we discussed various studies showing a complex relationship between DNA methylation, genes expression, and the development of diabetes and related complications (Figure 3). The use of genome wide DNA methylation profiles as a biomarker to predict at-risk patients is in its infancy and needs additional study. In studies where both DNA methylation and gene expression changes were reported, DNA methylation correlated well with gene expression changes, indicating a possible future therapeutic target. However, mechanistic insights into environmental exposures and genetic predisposition to changes in DNA methylation leading to diabetes continue to be an area of active research. One particularly interesting area of this study aims to understand how byproducts of metabolism could act as co-factors to influence epigenetic programming of gene expression (108). Developing a model incorporating genetic and DNA methylation changes together could be useful for development of effective diagnostic approaches and innovative therapeutic targets for diabetes.
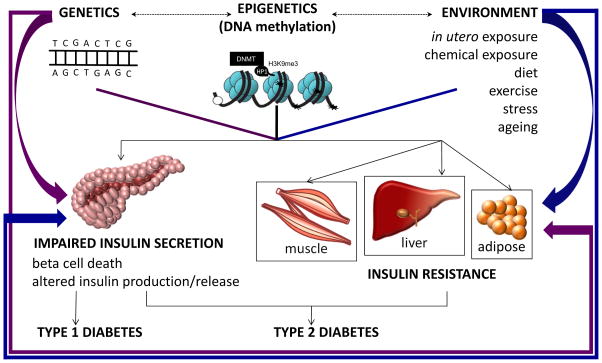
Figure 3.
DNA methylation and its relationship to Type 1 and Type 2 diabetes. There is a complex interaction between genetics, epigenetics and environment (shown by dashed arrows). Epigenetics (DNA methylation) in combination with genetic and environmental stimuli could either impair pancreatic development and insulin secretion, or lead to insulin resistance at peripheral tissues such as liver, muscle and adipose. A combination of impaired insulin secretion and insulin resistance underlie Type 2 diabetes, whereas impaired pancreatic development and insulin secretion underlie Type 1 diabetes. Genetic inheritance (purple arrows) or abnormal environmental stimuli (blue arrows) alone could either impair pancreatic function, or lead to insulin resistance independently.
Acknowledgments
This publication was supported by the National Institute of Environmental Health Sciences and the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award numbers SEP: K08 DK090302, P30 ES013508. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- T1DM
Type 1 diabetes
- T2DM
Type 2 diabetes
- CpGI
CpG islands
- DNMTs
DNA methyl transferases
- DOHaD
Developmental Origins of Health and Disease
- IUGR
Intra-uterine growth restriction
- BPA
Bisphenol A
- GDM
Gestation Diabetes Mellitus
- NDM
Neonatal Diabetes
- DN
Diabetic nephropathy
Footnotes
Disclosure statement: The authors have nothing to disclose.
References
- 1.Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet. 2016;387:1513–1530. doi: 10.1016/S0140-6736(16)00618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diagnosis and classification of diabetes mellitus. Diabetes Care. 2009;32:S62–67. doi: 10.2337/dc2309-S2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rewers M, Norris J, Dabelea D. Epidemiology of type 1 Diabetes Mellitus. Adv Exp Med Biol. 2004;552:219–246. [PubMed] [Google Scholar]
- 4.Kanakatti Shankar R, Pihoker C, Dolan LM, Standiford D, Badaru A, Dabelea D, Rodriguez B, Black MH, Imperatore G, Hattersley A, Ellard S, Gilliam LK. Permanent neonatal diabetes mellitus: prevalence and genetic diagnosis in the SEARCH for Diabetes in Youth Study. Pediatr Diabetes. 2013;14:174–180. doi: 10.1111/pedi.12003. doi:110.1111/pedi.12003. Epub 12012 Oct 12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter WE. Molecular and biochemical analysis of the MODY syndromes. Pediatr Diabetes. 2000;1:88–117. doi: 10.1034/j.1399-5448.2000.010206.x. [DOI] [PubMed] [Google Scholar]
- 6.Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003;46:3–19. doi: 10.1007/s00125-002-1009-0. Epub 2003 Jan 2011. [DOI] [PubMed] [Google Scholar]
- 7.D’Adamo E, Caprio S. Type 2 diabetes in youth: epidemiology and pathophysiology. Diabetes Care. 2011;34:S161–165. doi: 10.2337/dc11-s212. doi:110.2337/dc2311-s2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Economic costs of diabetes in the U S in 2012. Diabetes Care. 2013;36:1033–1046. doi: 10.2337/dc12-2625. doi:1010.2337/dc1012-2625. Epub 2013 Mar 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang MN, Buzzetti R, Pozzilli P. Epigenetics in autoimmune diseases with focus on type 1 diabetes. Diabetes Metab Res Rev. 2013;29:8–18. doi: 10.1002/dmrr.2375. [DOI] [PubMed] [Google Scholar]
- 10.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. doi:2710.2337/db2709-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Craig DW, Redman M, Gershon ES, Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–419. doi: 10.1016/j.ajhg.2010.02.005. doi:410.1016/j.ajhg.2010.1002.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dayeh TA, Olsson AH, Volkov P, Almgren P, Ronn T, Ling C. Identification of CpG-SNPs associated with type 2 diabetes and differential DNA methylation in human pancreatic islets. Diabetologia. 2013;56:1036–1046. doi: 10.1007/s00125-012-2815-7. doi:1010.1007/s00125-00012-02815-00127. Epub 02013 Mar 00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsson AH, Volkov P, Bacos K, Dayeh T, Hall E, Nilsson EA, Ladenvall C, Ronn T, Ling C. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 2014;10:e1004735. doi: 10.1371/journal.pgen.1004735. doi:1004710.1001371/journal.pgen.1004735. eCollection 1002014 Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. doi:210.1038/nrg3354. Epub 2013 Feb 1012. [DOI] [PubMed] [Google Scholar]
- 15.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. doi:410.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 16.Ponnaluri VK, Ehrlich KC, Zhang G, Lacey M, Johnston D, Pradhan S, Ehrlich M. Association of 5-hydroxymethylation and 5-methylation of DNA cytosine with tissue-specific gene expression. Epigenetics. 2016;2:0. doi: 10.1080/15592294.2016.1265713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 18.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. Epub 12005 Jan 13324. [DOI] [PubMed] [Google Scholar]
- 19.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. Epub 42004 Aug 48331. [DOI] [PubMed] [Google Scholar]
- 20.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. doi:810.1038/nrg2651. Epub 2009 Sep 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vertino PM, Yen RW, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-1114-1183-r1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. Epub 2007 Sep 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. doi:710.1038/nature07107. Epub 02008 Jul 07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. doi:1010.1038/ng.1708. Epub 2010 Nov 1097. [DOI] [PubMed] [Google Scholar]
- 28.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 29.Choi JD, Underkoffler LA, Wood AJ, Collins JN, Williams PT, Golden JA, Schuster EF, Jr, Loomes KM, Oakey RJ. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol Cell Biol. 2005;25:5514–5522. doi: 10.1128/MCB.25.13.5514-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 31.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. doi:310.1038/nbt.1533. Epub 2009 Mar 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20:670–680. doi: 10.1093/hmg/ddq513. doi:610.1093/hmg/ddq1513. Epub 2010 Nov 1026. [DOI] [PubMed] [Google Scholar]
- 33.Temple IK, Shield JP. Transient neonatal diabetes, a disorder of imprinting. J Med Genet. 2002;39:872–875. doi: 10.1136/jmg.39.12.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Docherty LE, Kabwama S, Lehmann A, Hawke E, Harrison L, Flanagan SE, Ellard S, Hattersley AT, Shield JP, Ennis S, Mackay DJ, Temple IK. Clinical presentation of 6q24 transient neonatal diabetes mellitus (6q24 TNDM) and genotype-phenotype correlation in an international cohort of patients. Diabetologia. 2013;56:758–762. doi: 10.1007/s00125-013-2832-1. doi:710.1007/s00125-00013-02832-00121. Epub 02013 Feb 00126. [DOI] [PubMed] [Google Scholar]
- 35.Flanagan SE, Mackay DJ, Greeley SA, McDonald TJ, Mericq V, Hassing J, Richmond EJ, Martin WR, Acerini C, Kaulfers AM, Flynn DP, Popovic J, Sperling MA, Hussain K, Ellard S, Hattersley AT. Hypoglycaemia following diabetes remission in patients with 6q24 methylation defects: expanding the clinical phenotype. Diabetologia. 2013;56:218–221. doi: 10.1007/s00125-012-2766-z. doi:210.1007/s00125-00012-02766-z. Epub 02012 Oct 00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 37.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeschnigk M, Bohringer S, Price EA, Onadim Z, Masshofer L, Lohmann DR. A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus. Nucleic Acids Res. 2004;32:e125. doi: 10.1093/nar/gnh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. Epub 12005 Oct 15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouse SD, Nagarajan RO, Costello JF. Genome-scale DNA methylation analysis. Epigenomics. 2010;2:105–117. doi: 10.2217/epi.09.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. Epub 2005 Jul 2010. [DOI] [PubMed] [Google Scholar]
- 43.Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, Robinson MD, Clark SJ. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics : official journal of the DNA Methylation Society. 2011;6:34–44. doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- 44.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic acids research. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–48. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- 46.Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. doi:710.2217/epi.2211.2105. [DOI] [PubMed] [Google Scholar]
- 48.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–399. doi: 10.2217/epi.15.114. doi:310.2217/epi.2215.2114. Epub 2015 Dec 2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. Print 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. doi:410.1038/nprot.2010.1190. Epub 2011 Mar 1018. [DOI] [PubMed] [Google Scholar]
- 51.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. doi:210.1038/nature06745. Epub 02008 Feb 06717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. doi:310.1038/nature08514. Epub 02009 Oct 08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. doi:210.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Busche S, Shao X, Caron M, Kwan T, Allum F, Cheung WA, Ge B, Westfall S, Simon MM, Barrett A, Bell JT, McCarthy MI, Deloukas P, Blanchette M, Bourque G, Spector TD, Lathrop M, Pastinen T, Grundberg E. Population whole-genome bisulfite sequencing across two tissues highlights the environment as the principal source of human methylome variation. Genome Biol. 2015;16:290. doi: 10.1186/s13059-13015-10856-13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasquali L, Gaulton KJ, Rodriguez-Segui SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Moran I, Gomez-Marin C, van de Bunt M, Ponsa-Cobas J, Castro N, Nammo T, Cebola I, Garcia-Hurtado J, Maestro MA, Pattou F, Piemonti L, Berney T, Gloyn AL, Ravassard P, Gomez-Skarmeta JL, Muller F, McCarthy MI, Ferrer J. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–143. doi: 10.1038/ng.2870. doi:110.1038/ng.2870. Epub 2014 Jan 1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet. 1989;2:577–580. doi: 10.1016/s0140-6736(89)90710-1. [DOI] [PubMed] [Google Scholar]
- 57.Hales CN, Barker DJ, Clark PM, Cox LJ, Fall C, Osmond C, Winter PD. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ. 1991;303:1019–1022. doi: 10.1136/bmj.303.6809.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rich-Edwards JW, Colditz GA, Stampfer MJ, Willett WC, Gillman MW, Hennekens CH, Speizer FE, Manson JE. Birthweight and the risk for type 2 diabetes mellitus in adult women. Ann Intern Med. 1999;130:278–284. doi: 10.7326/0003-4819-130-4_part_1-199902160-00005. [DOI] [PubMed] [Google Scholar]
- 59.Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y, Jones RH, Marquez VE, Cairns W, Tadayyon M, O’Neill LP, Murrell A, Ling C, Constancia M, Ozanne SE. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci U S A. 2011;108:5449–5454. doi: 10.1073/pnas.1019007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Straten EM, Bloks VW, Huijkman NC, Baller JF, van Meer H, Lutjohann D, Kuipers F, Plosch T. The liver X-receptor gene promoter is hypermethylated in a mouse model of prenatal protein restriction. Am J Physiol Regul Integr Comp Physiol. 2010;298:R275–282. doi: 10.1152/ajpregu.00413.2009. doi:210.1152/ajpregu.00413.02009. Epub 02009 Nov 00414. [DOI] [PubMed] [Google Scholar]
- 61.Rees WD, Hay SM, Brown DS, Antipatis C, Palmer RM. Maternal protein deficiency causes hypermethylation of DNA in the livers of rat fetuses. J Nutr. 2000;130:1821–1826. doi: 10.1093/jn/130.7.1821. [DOI] [PubMed] [Google Scholar]
- 62.Altmann S, Murani E, Schwerin M, Metges CC, Wimmers K, Ponsuksili S. Dietary protein restriction and excess of pregnant German Landrace sows induce changes in hepatic gene expression and promoter methylation of key metabolic genes in the offspring. J Nutr Biochem. 2013;24:484–495. doi: 10.1016/j.jnutbio.2012.01.011. doi:410.1016/j.jnutbio.2012.1001.1011. Epub 2012 Jun 1027. [DOI] [PubMed] [Google Scholar]
- 63.Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr. 2008;100:278–282. doi: 10.1017/S0007114507894438. doi:210.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 65.Jousse C, Parry L, Lambert-Langlais S, Maurin AC, Averous J, Bruhat A, Carraro V, Tost J, Letteron P, Chen P, Jockers R, Launay JM, Mallet J, Fafournoux P. Perinatal undernutrition affects the methylation and expression of the leptin gene in adults: implication for the understanding of metabolic syndrome. FASEB J. 2011;25:3271–3278. doi: 10.1096/fj.11-181792. doi:3210.1096/fj.3211-181792. Epub 182011 Jun 181713. [DOI] [PubMed] [Google Scholar]
- 66.Simmons RA, Templeton LJ, Gertz SJ. Intrauterine growth retardation leads to the development of type 2 diabetes in the rat. Diabetes. 2001;50:2279–2286. doi: 10.2337/diabetes.50.10.2279. [DOI] [PubMed] [Google Scholar]
- 67.Stoffers DA, Desai BM, DeLeon DD, Simmons RA. Neonatal exendin-4 prevents the development of diabetes in the intrauterine growth retarded rat. Diabetes. 2003;52:734–740. doi: 10.2337/diabetes.52.3.734. [DOI] [PubMed] [Google Scholar]
- 68.Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson RF, Fazzari MJ, Niu H, Barzilai N, Simmons RA, Greally JM. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. J Biol Chem. 2010;285:15111–15118. doi: 10.1074/jbc.M109.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla M, Andrews S, Didier N, Charalambous M, McEwen K, Marazzi G, Sassoon D, Patti ME, Ferguson-Smith AC. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet. 2012;8:e1002605. doi: 10.1371/journal.pgen.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9:e1003401. doi: 10.1371/journal.pgen.1003401. doi:1003410.1001371/journal.pgen.1003401. Epub 1002013 Apr 1003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eden S, Constancia M, Hashimshony T, Dean W, Goldstein B, Johnson AC, Keshet I, Reik W, Cedar H. An upstream repressor element plays a role in Igf2 imprinting. EMBO J. 2001;20:3518–3525. doi: 10.1093/emboj/20.13.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Susiarjo M, Xin F, Bansal A, Stefaniak M, Li C, Simmons RA, Bartolomei MS. Bisphenol a exposure disrupts metabolic health across multiple generations in the mouse. Endocrinology. 2015;156:2049–2058. doi: 10.1210/en.2014-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pettitt DJ, Aleck KA, Baird HR, Carraher MJ, Bennett PH, Knowler WC. Congenital susceptibility to NIDDM. Role of intrauterine environment Diabetes. 1988;37:622–628. doi: 10.2337/diab.37.5.622. [DOI] [PubMed] [Google Scholar]
- 75.Silverman BL, Metzger BE, Cho NH, Loeb CA. Impaired glucose tolerance in adolescent offspring of diabetic mothers. Relationship to fetal hyperinsulinism Diabetes care. 1995;18:611–617. doi: 10.2337/diacare.18.5.611. [DOI] [PubMed] [Google Scholar]
- 76.Plagemann A. Perinatal programming and functional teratogenesis: impact on body weight regulation and obesity. Physiol Behav. 2005;86:661–668. doi: 10.1016/j.physbeh.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 77.Pettitt DJ, Aleck KA, Baird HR, Carraher MJ, Bennett PH, Knowler WC. Congenital susceptibility to NIDDM. Role of intrauterine environment Diabetes. 1988;37:622–628. doi: 10.2337/diab.37.5.622. [DOI] [PubMed] [Google Scholar]
- 78.Silverman BL, Metzger BE, Cho NH, Loeb CA. Impaired glucose tolerance in adolescent offspring of diabetic mothers. Relationship to fetal hyperinsulinism Diabetes Care. 1995;18:611–617. doi: 10.2337/diacare.18.5.611. [DOI] [PubMed] [Google Scholar]
- 79.Plagemann A. Perinatal programming and functional teratogenesis: impact on body weight regulation and obesity. Physiol Behav. 2005;86:661–668. doi: 10.1016/j.physbeh.2005.08.065. Epub 2005 Nov 2008. [DOI] [PubMed] [Google Scholar]
- 80.Rong C, Cui X, Chen J, Qian Y, Jia R, Hu Y. DNA methylation profiles in placenta and its association with gestational diabetes mellitus. Exp Clin Endocrinol Diabetes. 2015;123:282–288. doi: 10.1055/s-0034-1398666. doi:210.1055/s-0034-1398666. Epub 1392015 Apr 1398621. [DOI] [PubMed] [Google Scholar]
- 81.Petropoulos S, Guillemin C, Ergaz Z, Dimov S, Suderman M, Weinstein-Fudim L, Ornoy A, Szyf M. Gestational Diabetes Alters Offspring DNA Methylation Profiles in Human and Rat: Identification of Key Pathways Involved in Endocrine System Disorders, Insulin Signaling, Diabetes Signaling, and ILK Signaling. Endocrinology. 2015;156:2222–2238. doi: 10.1210/en.2014-1643. doi:2210.1210/en.2014-1643. Epub 2014 Dec 2216. [DOI] [PubMed] [Google Scholar]
- 82.Finer S, Mathews C, Lowe R, Smart M, Hillman S, Foo L, Sinha A, Williams D, Rakyan VK, Hitman GA. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum Mol Genet. 2015;24:3021–3029. doi: 10.1093/hmg/ddv013. doi:3010.1093/hmg/ddv3013. Epub 2015 Jan 3029. [DOI] [PubMed] [Google Scholar]
- 83.El Hajj N, Pliushch G, Schneider E, Dittrich M, Muller T, Korenkov M, Aretz M, Zechner U, Lehnen H, Haaf T. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes. 2013;62:1320–1328. doi: 10.2337/db12-0289. doi:1310.2337/db1312-0289. Epub 2012 Dec 1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang BT, Dayeh TA, Volkov PA, Kirkpatrick CL, Malmgren S, Jing X, Renstrom E, Wollheim CB, Nitert MD, Ling C. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol Endocrinol. 2012;26:1203–1212. doi: 10.1210/me.2012-1004. doi:1210.1210/me.2012-1004. Epub 2012 May 1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ling C, Del Guerra S, Lupi R, Ronn T, Granhall C, Luthman H, Masiello P, Marchetti P, Groop L, Del Prato S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–622. doi: 10.1007/s00125-007-0916-5. doi:610.1007/s00125-00007-00916-00125. Epub 02008 Feb 00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang BT, Dayeh TA, Kirkpatrick CL, Taneera J, Kumar R, Groop L, Wollheim CB, Nitert MD, Ling C. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia. 2011;54:360–367. doi: 10.1007/s00125-010-1967-6. doi:310.1007/s00125-00010-01967-00126. Epub 02010 Nov 00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dayeh T, Volkov P, Salo S, Hall E, Nilsson E, Olsson AH, Kirkpatrick CL, Wollheim CB, Eliasson L, Ronn T, Bacos K, Ling C. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. doi:1004110.1001371/journal.pgen.1004160. eCollection 1002014 Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, Calonne E, Volkmar U, Igoillo-Esteve M, Naamane N, Del Guerra S, Masini M, Bugliani M, Marchetti P, Cnop M, Eizirik DL, Fuks F. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hall E, Dayeh T, Kirkpatrick CL, Wollheim CB, Dekker Nitert M, Ling C. DNA methylation of the glucagon-like peptide 1 receptor (GLP1R) in human pancreatic islets. BMC Med Genet. 2013;14:76. doi: 10.1186/1471-2350-1114-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gillberg L, Ling C. The potential use of DNA methylation biomarkers to identify risk and progression of type 2 diabetes. Front Endocrinol (Lausanne) 2015;6:43. doi: 10.3389/fendo.2015.00043. eCollection 02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ronn T, Ling C. DNA methylation as a diagnostic and therapeutic target in the battle against Type 2 diabetes. Epigenomics. 2015;7:451–460. doi: 10.2217/epi.15.7. doi:410.2217/epi.2215.2217. [DOI] [PubMed] [Google Scholar]
- 92.Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, Nilsson E, Tornberg A, Dekker Nitert M, Eriksson KF, Jones HA, Groop L, Ling C. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. doi:1003510.1001371/journal.pgen.1003572. Epub 1002013 Jun 1003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lindholm ME, Marabita F, Gomez-Cabrero D, Rundqvist H, Ekstrom TJ, Tegner J, Sundberg CJ. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics. 2014;9:1557–1569. doi: 10.4161/15592294.2014.982445. doi:1510.4161/15592294.15592014.15982445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Redondo MJ, Yu L, Hawa M, Mackenzie T, Pyke DA, Eisenbarth GS, Leslie RD. Heterogeneity of type I diabetes: analysis of monozygotic twins in Great Britain and the United States. Diabetologia. 2001;44:354–362. doi: 10.1007/s001250051626. [DOI] [PubMed] [Google Scholar]
- 95.Stefan M, Zhang W, Concepcion E, Yi Z, Tomer Y. DNA methylation profiles in type 1 diabetes twins point to strong epigenetic effects on etiology. J Autoimmun. 2014;50:33–7. doi: 10.1016/j.jaut.2013.1010.1001. Epub 2013 Nov 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, Daunay A, Busato F, Mein CA, Manfras B, Dias KR, Bell CG, Tost J, Boehm BO, Beck S, Leslie RD. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7:e1002300. doi: 10.1371/journal.pgen.1002300. doi:1002310.1001371/journal.pgen.1002300. Epub 1002011 Sep 1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Elboudwarej E, Cole M, Briggs FB, Fouts A, Fain PR, Quach H, Quach D, Sinclair E, Criswell LA, Lane JA, Steck AK, Barcellos LF, Noble JA. Hypomethylation within gene promoter regions and type 1 diabetes in discordant monozygotic twins. J Autoimmun. 2016;68:23–9. doi: 10.1016/j.jaut.2015.1012.1003. Epub 2016 Jan 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rui J, Deng S, Lebastchi J, Clark PL, Usmani-Brown S, Herold KC. Methylation of insulin DNA in response to proinflammatory cytokines during the progression of autoimmune diabetes in NOD mice. Diabetologia. 2016;59:1021–1029. doi: 10.1007/s00125-016-3897-4. doi:1010.1007/s00125-00016-03897-00124. Epub 02016 Feb 00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Husseiny MI, Kuroda A, Kaye AN, Nair I, Kandeel F, Ferreri K. Development of a quantitative methylation-specific polymerase chain reaction method for monitoring beta cell death in type 1 diabetes. PLoS One. 2012;7:e47942. doi: 10.1371/journal.pone.0047942. doi:47910.41371/journal.pone.0047942. Epub 0042012 Oct 0047929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fisher MM, Watkins RA, Blum J, Evans-Molina C, Chalasani N, DiMeglio LA, Mather KJ, Tersey SA, Mirmira RG. Elevations in Circulating Methylated and Unmethylated Preproinsulin DNA in New-Onset Type 1 Diabetes. Diabetes. 2015;64:3867–3872. doi: 10.2337/db15-0430. doi:3810.2337/db3815-0430. Epub 2015 Jul 3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Akirav EM, Lebastchi J, Galvan EM, Henegariu O, Akirav M, Ablamunits V, Lizardi PM, Herold KC. Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci U S A. 2011;108:19018–19023. doi: 10.1073/pnas.1111008108. doi:19010.11073/pnas.1111008108. Epub 1111002011 Nov 1111008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Intine RV, Sarras MP., Jr Metabolic memory and chronic diabetes complications: potential role for epigenetic mechanisms. Curr Diab Rep. 2012;12:551–559. doi: 10.1007/s11892-012-0302-7. doi:510.1007/s11892-11012-10302-11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brennan EP, Ehrich M, O’Donovan H, Brazil DP, Crean JK, Murphy M, Sadlier DM, Martin F, Godson C, van den Boom D, Maxwell AP, Savage DA. DNA methylation profiling in cell models of diabetic nephropathy. Epigenetics. 2010;5:396–401. doi: 10.4161/epi.5.5.12077. Epub 2010 Jul 2011. [DOI] [PubMed] [Google Scholar]
- 104.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33. doi: 10.1186/1755-8794-1183-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, Siraj ES, Madaio M. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6:20–28. doi: 10.4161/epi.6.1.13362. Epub 2011 Jan 2011. [DOI] [PubMed] [Google Scholar]
- 106.Smyth LJ, McKay GJ, Maxwell AP, McKnight AJ. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9:366–376. doi: 10.4161/epi.27161. doi:310.4161/epi.27161. Epub 22013 Nov 27119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maghbooli Z, Larijani B, Emamgholipour S, Amini M, Keshtkar A, Pasalar P. Aberrant DNA methylation patterns in diabetic nephropathy. J Diabetes Metab Disord. 2014;13:69. doi: 10.1186/2251-6581-1113-1169. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Phang JM, Liu W, Hancock C. Bridging epigenetics and metabolism: role of non-essential amino acids. Epigenetics. 2013;8:231–236. doi: 10.4161/epi.24042. doi:210.4161/epi.24042. Epub 22013 Feb 24019. [DOI] [PMC free article] [PubMed] [Google Scholar]