Abstract
Mutations in CCAAT/enhancer binding protein α (CEBPA) occur in 5–10% of cases of acute myeloid leukemia. CEBPA-double-mutated cases usually bear biallelic N- and C-terminal mutations and are associated with a favorable clinical outcome. Identification of CEBPA mutants is challenging because of the variety of mutations, intrinsic characteristics of the gene and technical issues. Several screening methods (fragment-length analysis, gene expression array) have been proposed especially for large-scale clinical use; although efficient, they are limited by specific concerns. We investigated the phenotypic profile of blast and maturing bone marrow cell compartments at diagnosis in 251 cases of acute myeloid leukemia. In this cohort, 16 (6.4%) patients had two CEBPA mutations, whereas ten (4.0%) had a single mutation. First, we highlighted that the CEBPA-double-mutated subset displays recurrent phenotypic abnormalities in all cell compartments. By mutational analysis after cell sorting, we demonstrated that this common phenotypic signature depends on CEBPA-double-mutated multi-lineage involvement. From a multidimensional study of phenotypic data, we developed a classifier including ten core and widely available parameters. The selected markers on blasts (CD34, CD117, CD7, CD15, CD65), neutrophil (SSC, CD64), monocytic (CD14, CD64) and erythroid (CD117) compartments were able to cluster CEBPA-double-mutated cases. In a validation set of 259 AML cases from three independent centers, our classifier showed excellent performance with 100% specificity and 100% sensitivity. We have, therefore, established a reliable screening method, based upon multidimensional analysis of widely available phenotypic parameters. This method provides early results and is suitable for large-scale detection of CEBPA-double-mutated status, allowing gene sequencing to be focused in selected cases.
Introduction
Mutations in the transcription factor CCAAT/enhancer binding protein α (CEBPA) are found in approximately 10% of cases of acute myeloid leukemia (AML).1–3 Most CEBPA-mutant AML exhibit two mutations, which frequently involve a combination of an N-terminal and a C-terminal gene mutation, typically on different alleles. Recent comprehensive data have shown that CEBPA-double-mutated (CEBPA-dm) cases, rather than single mutants, are associated with a common gene expression signature4 and a relatively favorable outcome.4–7 Based on these features, CEBPA-dm AML has been recognized as a separate entity in the revised World Health Organization classification.8 The identification of a CEBPA-dm genotype provides crucial prognostic information, since these patients often lack other main predictors of relapse risk. However, there are several technical issues with CEBPA mutational analysis. First, CEBPA sequencing is known to be difficult because of the high GC content of the gene, which frequently correlates with failure of the polymerase chain reaction, and the presence of background or sequencing artifacts. Sequencing the entire gene enables detection of all mutations but is labor-intensive, especially in a routine context, and requires expertise with unusual variants. Several screening methods have, therefore, been developed. Although efficient and sensitive, polymerase chain reaction-based fragment-length analyses can only detect mutations resulting in a net insertion or deletion and not substitution mutations.9,10 Furthermore, they cannot distinguish a common 6-bp duplication polymorphism from an actual insertion or duplication.11 Next-generation sequencing-based CEBPA studies are able to overcome these difficulties but are not widely available yet. Some reports have proposed gene expression arrays as a screening method for CEBPA-dm status, given the unique profile of these malignancies.4,6,12–14 Although these methods have excellent performance, they require further technology and relative expertise for specific application in this context.
As a screening method for genetic abnormalities, the immunophenotype of AML blasts is often able to predict the main underlying genotypes.15 Generally, the association with phenotype is strong when a few, relevant genetic events are responsible for leukemogenesis [e.g., CBF-related translocations or t(15;17)], whereas it is weaker when genetic heterogeneity is greater (e.g., normal karyotype with several gene mutations). Furthermore, the strength of the correlation with a certain genotype depends on phenotypic aberrations being rare in AML not characterized by that genotype [e.g., cross-lineage CD19 and t(8;21)]. In fact, CEBPA-mutated AML has not been associated with a specific immunophenotype. Rather, it has been described as showing positivity for commonly expressed antigens, such as CD15, CD7, CD34 and HLA-DR on blasts.16 Although associated with a mutant status, this phenotypic profile was not able to screen effectively for CEBPA-mutated cases, since about 25% of them were missed.16 Furthermore, it was based on the strict application of the European Group for Immunological Characterization of Leukemia (EGIL) threshold for positivity (i.e., more than 20% of cells),17 which is probably inadequate for dissecting a shared phenotypic signature, especially for frequently expressed antigens.
In this study, we extensively investigated the immunophenotype of CEBPA-mutated AML by analyzing all bone marrow cell compartments at diagnosis and by comparing each compartment with its corresponding normal counterpart in order to highlight aberrations. Our aim was to develop a screening method for CEBPA-mutated AML based on the phenotypic profile, which would be straightforward, widely available and fast, in order to focus molecular techniques on a narrow subset of AML patients.
Methods
Patients
Patients entering the study had a diagnosis of untreated AML, based on World Health Organization criteria.18 and an available immunophenotypic characterization on bone marrow at diagnosis. When eligible for intensive chemotherapy, patients were treated according to two protocols as specified below. Briefly, from 2006 to March 2007 (protocol 1), patients received standard course induction. High-dose cytarabine was used as first consolidation in patients aged <61 years attaining complete remission. On an intent-to-treat basis, patients aged <55 years with a high-risk karyotype, FLT3-ITD or adverse clinical features were assigned to undergo allogeneic stem cell transplantation. Patients with intermediate cytogenetic risk in the absence of FLT3-ITD and adverse clinical features were allocated to allogeneic stem cell transplantation if a related donor was available. Autologous stem cell transplantation was offered to patients aged <61 years with low-risk cytogenetics, intermediate-risk cytogenetics without sibling donor and high-risk disease not eligible for allogeneic transplantation. From April 2007 to 2013 (protocol 2), patients were enrolled in the Northern Italy Leukemia Group (NILG) AML 02–06 protocol (Eudract code: 2006-003817-42). This protocol included randomization at induction between a standard ICE induction and an experimental, intensified one. Patients randomized to the experimental arm were excluded from the outcome analysis. A more detailed description of treatment protocols is provided in the Online Supplementary Data.
Only intensively treated non-M3 patients were considered for the outcome analysis. The study was approved by the local institutional review board (protocol number: 2013/0024340), and patients were included after giving written informed consent, in accordance with the Declaration of Helsinki.
Karyotype
Cytogenetic analysis was performed on bone marrow cells taken at diagnosis and the results are reported according to the International System for Human Cytogenetic Nomenclature.19
Molecular genetics
NPM1, FLT3-ITD and CEBPA mutations were searched for using previously described methods.1,21,22 Further details are reported in the Online Supplementary Data.
Flow cytometry
Technical details about flow cytometry sample handling, reagents, acquisition and analysis are reported in the Online Supplementary File. Data were analyzed with Infinicyt software (Cytognos SL, Salamanca, Spain). Some major bone marrow cell compartments were identified: (i) blasts; (ii) maturing neutrophils; (iii) monocytes; and (iv) mature erythroid cells. A series of 79 phenotypic parameters were defined (24 for blasts, 30 for the neutrophils, 14 for the monocytic compartment and 11 for erythroid cells). Parameters were expressed as percentage of positive cells for an antigen and/or as mean fluorescence intensity (MFI; arbitrary relative linear units, scaled from 0 to 104). Bone marrow samples from 21 healthy donors (male 13, female 8; median age 36 years; range, 20–59) were used to define the normal phenotypic profile (mean value ± two standard deviations for each parameter).
CEBPA mutation analysis on sorted cells
Cell sorting was performed using a FACSAria flow cytometer (BD) on diagnostic fresh bone marrow samples from six patients with CEBPA-dm AML. Some customized tubes were designed based on the phenotypic profile at diagnosis in order to sort specific cell fractions: (i) blasts; (ii) monocytes; (iii) maturing neutrophils; (iv) erythroid lineage cells; and (v) T-lymphocytes. Purity checks were performed to ensure sorting quality. Dead cells were excluded by analyzing forward scatter (FSC) versus side scatter (SSC) dot plots. Doublets were excluded by a FSC-height versus FSC-area dot plot. CEBPA mutational analysis was carried out on sorted cell fractions to reveal clonal multi-lineage involvement.
Statistical analysis
Data were processed using R software (http://cran.rproject.org). Comparisons between groups were performed using the Mann–Whitney U test. P values <0.05 were considered to denote statistically significant differences. Complete remission was defined using established criteria.23 Principal component analysis was used to visualize the similarity of phenotypic profiles, comparing CEBPA-dm cases with other genotypes. We performed Ward hierarchical clustering to reveal recurrent phenotypic aberrations and used Euclidean distance as the distance measure on phenotypic parameters. Consistent with the clustering strategy, we developed a Euclidean distance-based classifier on a selected group of phenotypic parameters to predict CEBPA-dm status. Samples in the validation dataset showing a distance between their normalized phenotypic data and the CEBPA-dm reference vector less than or equal to a classification threshold were considered “highly probable” cases of CEBPA-dm. In order to allow the method to be reproduced, the R script to perform the prediction of CEBPA-dm status is available in the Online Supplementary Data.
Results
Characterization of patients according to CEBPA genotype
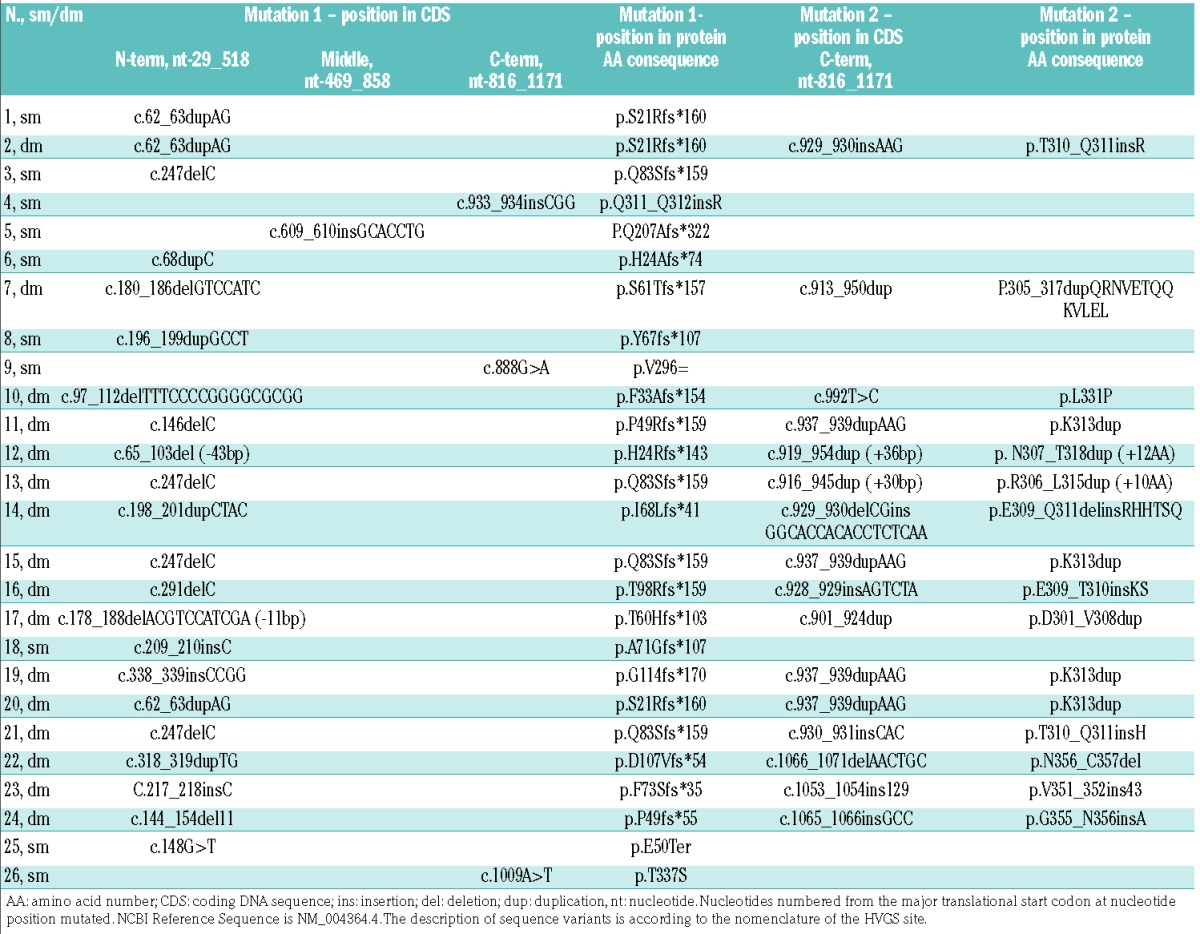
Between 2006 and 2013, 318 consecutive patients were diagnosed with AML at our Institution. Enrollment criteria for the present study were the availability of: (i) a full immunophenotype (i.e., including all required phenotypic parameters) on bone marrow at diagnosis; (ii) karyotype; and (iii) molecular genetics for NPM1, FLT3 and CEBPA. On the basis of these criteria, 67 patients were excluded because of incomplete immunophenotype on bone marrow (n=32), immunophenotype on peripheral blood (n=23), and lack of molecular genetics and unavailability of a diagnostic cryopreserved specimen (n=12). Thus, 251 patients met all criteria and were studied. Their characteristics are summarized in Table 1. In this cohort, 42 CEBPA mutations were identified in 26 patients (10.3%). Sixteen out of the 26 patients (61.5%) had two CEBPA mutations, whereas the remaining ten (38.5%) had a single mutation. The 16 patients with two CEBPA mutations had both an N-terminal truncation mutation resulting in p30 CEBPA and a C-terminal mutation affecting the bZIP domain of CEBPA. A summary of detected mutations is reported in Table 2. According to the number of mutations in the CEBPA gene, we divided our cases into: patients with double N- and C-terminal CEBPA mutations (CEBPA-dm, n=16), patients with a single mutation (CEBPA-sm, n=10), and wild-type patients without any mutation (CEBPA-wt, n=225). As regards clinical and biological features at diagnosis, CEBPA-dm patients were younger, had higher hemoglobin values and reticulocyte percentages and lower platelet counts compared to CEBPA-wt subjects. Consistently with published literature,7 a higher incidence of normal karyotype and no mutations of NPM1 and FLT3 genes were observed in CEBPA-dm cases. CEBPA-sm patients had higher white blood cell counts, as well as higher incidences of normal karyotype and NPM1 mutations with respect to CEBPA-wt cases.
Table 1.
Characteristics of patients according to CEBPA status.
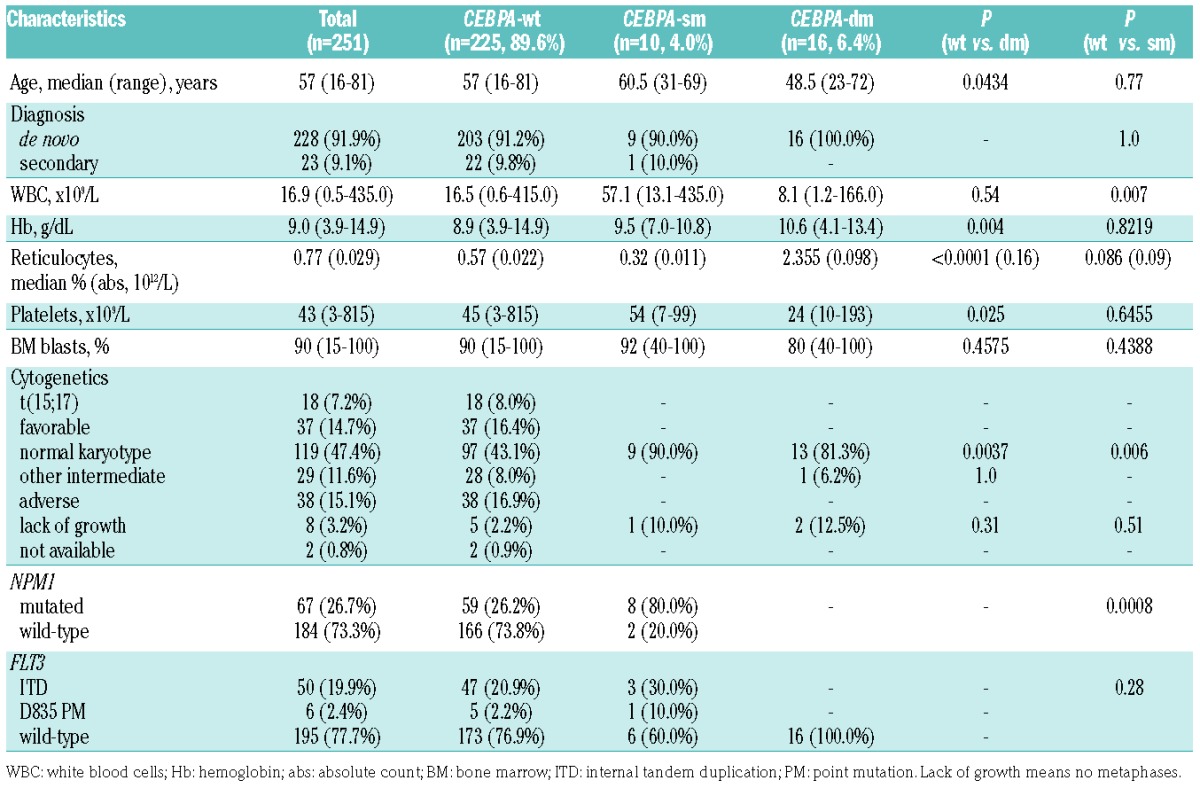
Table 2.
Summary of CEBPA mutations in the primary cohort.
Clinical outcome
Two-hundred and two patients out of 251 had non-M3 AML and were intensively treated. In accordance with previous studies,5–7 CEBPA-dm patients, compared to CEBPA-wt patients, showed a trend toward a higher complete remission rate after the first cycle of treatment (87.5% versus 61.0%, respectively; P=0.0549), longer overall survival (median not reached versus 22.3 months, respectively; P=0.00626; Figure 1A) and longer disease-free survival (median not reached versus 26.8 months, respectively; P=0.0667; Figure 1B). These findings did not change significantly when patients undergoing allogeneic stem cell transplantation were censored at the time of their transplant (Figure 1C,D).
Figure 1.
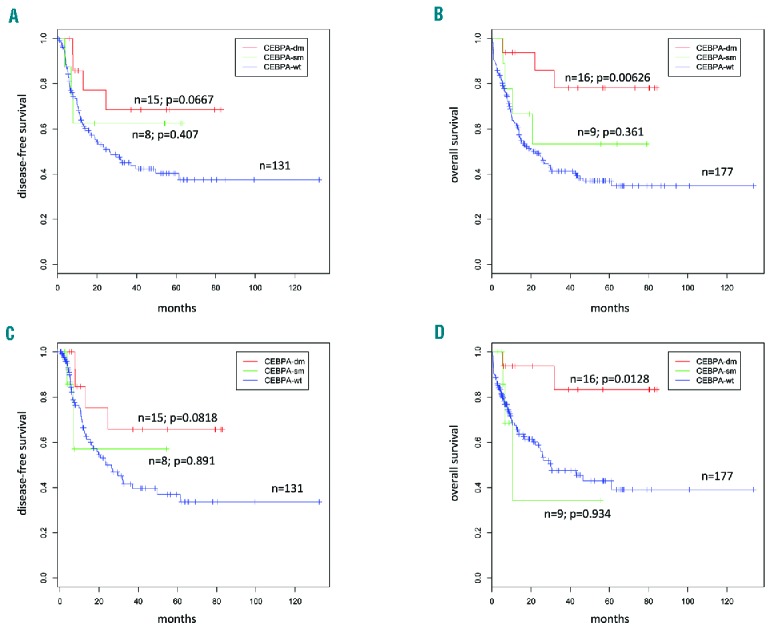
Survival outcomes according to CEBPA gene status. An outcome analysis was carried out for the 202 of 251 patients who were intensively treated. Kaplan Meier curves are stratified on CEBPA status: CEBPA-wild type (blue), single mutants (green), and double mutants (red) with P values representing the comparison versus wild-type patients. (A) Disease-free survival; (B) overall survival; (C) disease-free survival and (D) overall survival after censoring allo-transplanted patients at the date of transplant.
CEBPA status and immunophenotypic findings
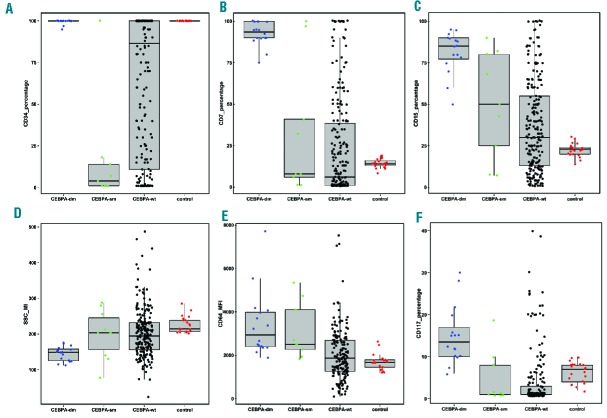
We quantified bone marrow cell compartments at diagnosis and found that their distribution varied widely among patients. The blast compartment represented a median of 45.49% (range, 0.14–97.74) of the global cellularity, the monocytic compartment 5.53% (range, 0.00–90.32) and the neutrophil and erythroid series accounted for 9.29% (range, 0.03–71.76) and 2.32% (range, 0.0–55.96), respectively. Phenotypic parameters were evaluated and compared among CEBPA genotypic groups and also to the cell counterpart in a control group, in order to highlight deviations from the normal phenotypic profile (Online Supplementary Data - Online Supplementary Tables S1–S4). CEBPA-dm cases showed some recurrent abnormalities in blasts and also in major maturing cell compartments in the bone marrow. With respect to control CD34+ cells, blasts from CEBPA-dm patients displayed high and homogeneous expression of immature antigens (CD34, CD117, HLA-DR) with asynchronous maturation (concomitant high expression of CD15, CD65, CD64, cyMPO) and aberrant cross-lineage expression of CD7. Beyond being merely defined as CD7+, CEBPA-dm cases showed a peculiar CD7 expression, since the vast majority of blasts expressed this antigen (Figure 2B). Similar findings were observed for antigens of maturation such as CD15 and CD65 (Figure 2C and Online Supplementary Figure S2). The median level of expression for these antigens was also significantly higher than observed in CEBPA-wt AML (Online Supplementary Table S1). Five out of 16 (31.3%) CEBPA-dm cases displayed cross-lineage CD56 expression on blasts. CEBPA-sm cases showed more heterogeneous phenotypic patterns (Figure 2 and Online Supplementary Figure S2). CEBPA-dm AML displayed several recurrent phenotypic abnormalities in the maturing cell compartment as well. The most frequently observed abnormalities in the neutrophil compartment were low SSC (35.3% of cases; Figure 2D), lower expression of CD65 and higher expression of CD64 compared both to controls and CEBPA-wt and –sm cases (Online Supplementary Figure S3). Monocytic cells, although not quantitatively expanded compared to controls (mean percentage 2.6% versus 4.6%), were recurrently characterized by high expression of CD64 (Figure 2E) and low expression of CD36 (Online Supplementary Figure S4). The erythroid compartment was significantly more represented in CEBPA-dm cases (7.2%) than in CEBPA-wt (1.6%) and CEBPA−sm (0.7%) cases, being similar to control values (8.9%). Furthermore, CEBPA-dm cases shared a significant increase of more immature stages of erythroid series, as revealed by high expression of CD117 (Figure 2F) and CD105, and some antigenic abnormalities (low CD36, low CD71) (Online Supplementary Figure S5).
Figure 2.
Phenotypic profile of blasts according to CEBPA status. Box plots illustrate the distribution of values in CEBPA-dm, -sm, -wt and controls for some core parameters: percentages of (A) CD34, (B) CD15 and (C) CD7 in blasts; (D) SSC signal in neutrophil compartment; (E) CD64 MFI in the monocyte compartment; (F) CD117 in the erythroid compartment. Box plots were generated by R software. Boxes represent the interquartile range containing 50% of the cases; the horizontal line marks the median; dots are single cases.
CEBPA-double-mutated status and multi-lineage involvement
As previously reported, CEBPA-dm AML is often characterized as M1-M2 according to the French-American-British classification.24 Available published data show that about 20–25% of CEBPA-dm cases are associated with multi-lineage dysplasia, as defined by the World Health Organization (i.e. presence of >50% of dysplastic cells in at least 2 cell lineages).18 In our series, five out of 16 (31.3%) CEBPA-dm cases showed multi-lineage dysplasia by morphology. Specifically, erythroid dysplasia was observed in the majority of patients (10 out of 16, 62.5%), which is relatively higher than expected for a de novo, intermediate karyotype category. In this respect, the morphological findings are consistent with phenotypic data: as reported above, maturing cell compartments, and especially the erythroid one, showed aberrant phenotypic patterns that were recurrent in this genotypic subset. We thus investigated whether CEBPA mutations were clonally represented in maturing cell lineages. In order to do this, we performed CEBPA mutational status analysis after separation by fluorescence-activated cell sorting in six out of 16 CEBPA-dm patients from our cohort. Overall, post-sorting acquisition of isolated cell fractions documented a purity of 97±1%. We were able to isolate blast, neutrophil, monocytic and erythroid cell compartments from all six patients; T-lymphocytes were employed as a negative control. In addition to blast cells, all sorted myeloid populations showed a CEBPA-dm status, whereas T lymphocytes were CEBPA-wt. The data from one illustrative case are shown in Figure 3.
Figure 3.
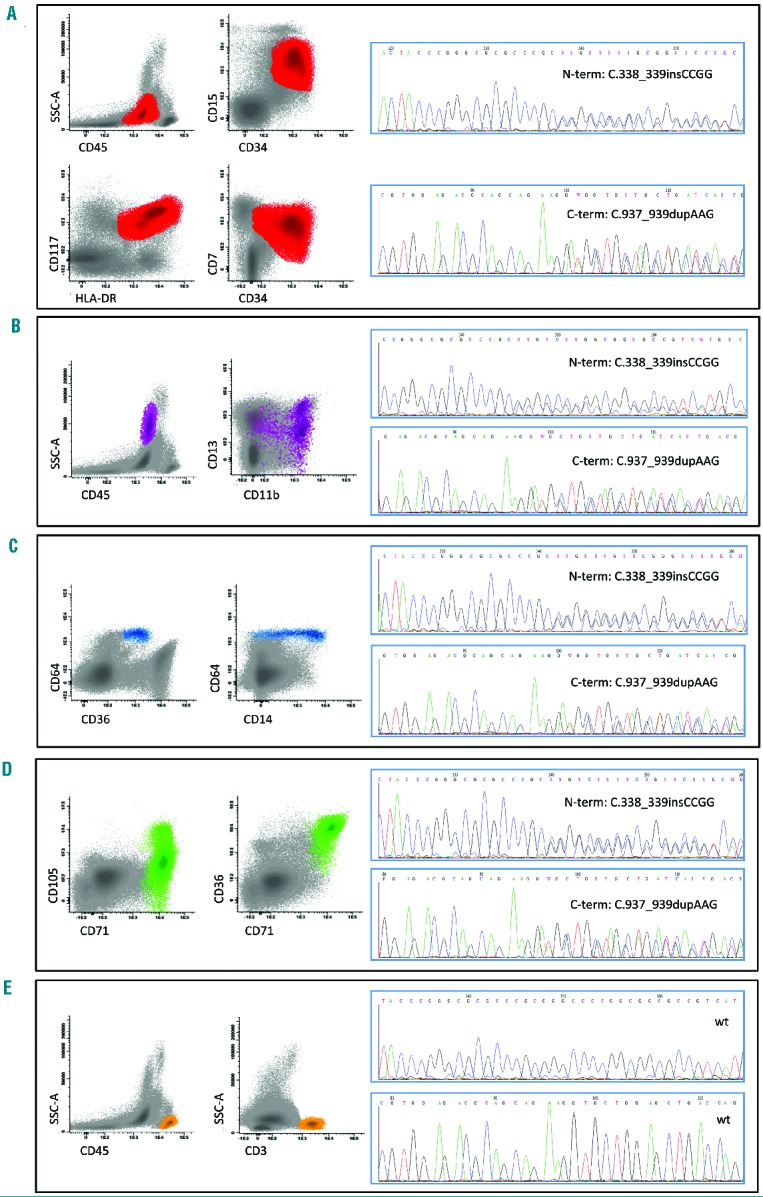
CEBPA mutational analysis on sorted cell fractions in one CEBPA-double-mutated patient. Cell compartments are shown on the left, with core phenotypic parameters for (A) blasts, (B) neutrophils, (C) monocytes, (D) erythroid cells, and (E) T-lymphocytes. In the corresponding plots, ungated cells are in gray whereas the relevant cell population is highlighted by color: red for blasts, purple for neutrophils, blue for monocytes, green for erythroid cells and orange for T-lymphocytes. The relative data from CEBPA mutational analysis are reported on the right, together with mutation type.
Multidimensional analysis and classifier definition
Although recurrent in CEBPA-dm cases, most phenotypic abnormalities showed a variable degree of overlap with the distribution of values observed in CEBPA-wt and CEBPA–sm patients. Consequently, the expression of no single antigen was able to discriminate CEBPA genotype. We, therefore, processed our data by multidimensional analysis in order to verify the capability of the whole phenotypic profile, including blasts and more mature compartments, to separate the genotypic groups. First we used principal component analysis to compare CEBPA-dm cases to some genotypic subsets one by one (Figure 4). With this method we observed a clear distinction of CEBPA-dm cases from cases bearing AML1-ETO, CBFB-MYH11, and NPM1 mutations and a complex karyotype. A partial overlap emerged for CEBPA-sm cases, essentially due to one case (Figure 4E) resembling a CEBPA-dm phenotype, which had a normal karyotype and was NPM1-wt and FLT3-wt; neither homozygosity nor a second CEBPA mutation was identified after gene re-sequencing on sorted blasts (Online Supplementary Figure S6). No more sample was available for additional analyses (e.g., next-generation sequencing).
Figure 4.
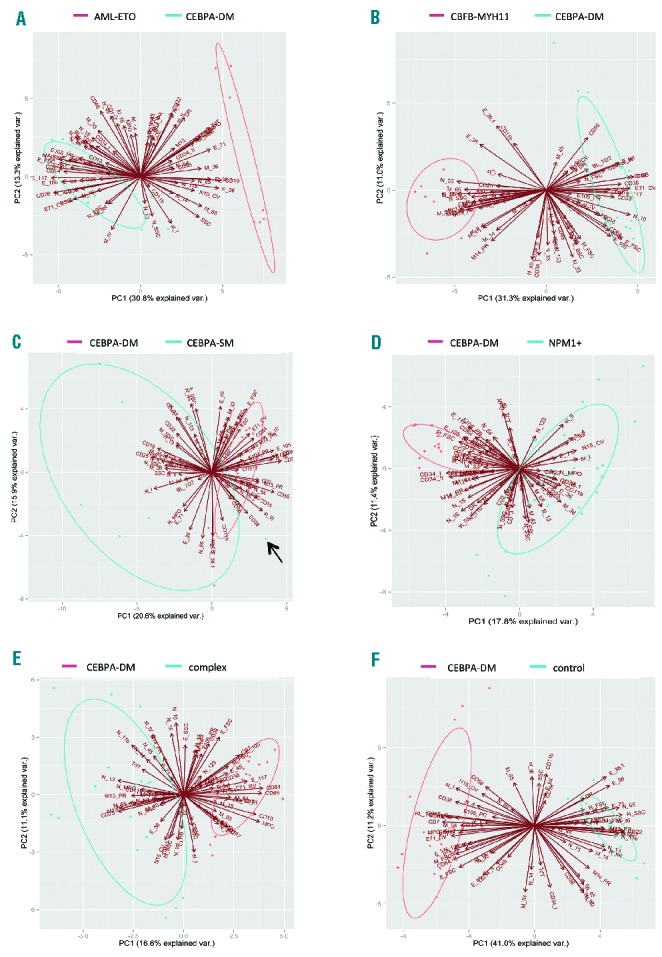
Principal component analysis of CEBPA-dm cases versus other genotypes. The multidimensional analysis of the whole phenotypic profile was able to distinguish CEBPA-dm cases from other genotypic groups: AML bearing (A) AML1-ETO, (B) CBFB-MYH11, (D) NPM1 mutations, (E) complex karyotype. (C) CEBPA-single mutant cases show a wide distribution in the plot area and a partial overlap essentially due to a case (arrow) resembling a CEBPA-dm phenotypic profile. Bi-plots are generated by the combination of the first two principal components (PC), featured by the highest values of variance. Ellipses graphically represent the area of the 95% confidence interval of the distribution for the principal components. Samples outside the ellipse are outliers. Principal component analysis was carried out by R software.
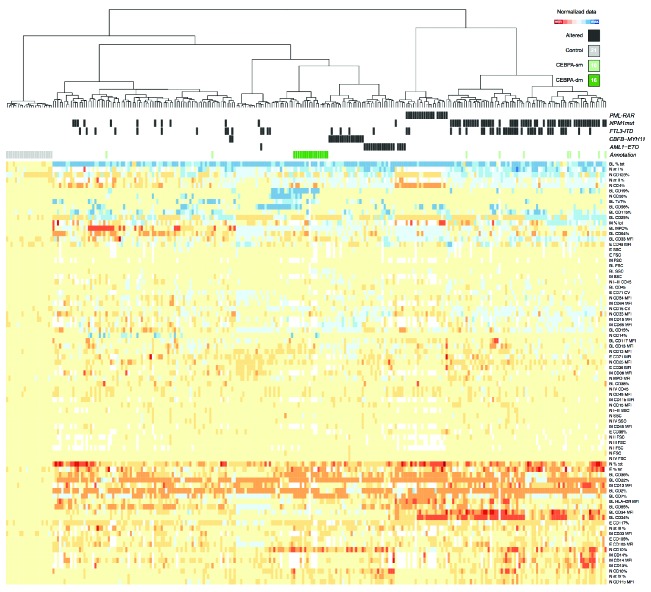
We then carried out an unsupervised clustering analysis (Figure 5). This approach was able to collect CEBPA-dm cases into a well-separated cluster. CEBPA-sm cases did not group separately, probably due to the influence on phenotype of other relevant gene mutations (e.g., NPM1). We also carried out hierarchical clustering within selected subsets, such as the intermediate-risk karyotype category (Online Supplementary Figure S7). Since CEBPA-mutated AML has been associated with EGIL-based positivity for CD7 on blasts,17 we repeated our analysis within CD7+ cases in our cohort (Online Supplementary Figure S8). Our systematic approach provided clustering of CEBPA-dm patients even in these subgroup analyses. Given the average poor prognostic significance of CD7 expression in AML, we studied outcome in CD7+ cases (Online Supplementary Figure S9): CEBPA-dm was confirmed to have a favorable impact in this phenotypic context.
Figure 5.
Unsupervised hierarchical clustering according to genotypic groups. Cluster analysis of controls (n=21) and AML cases (n=251) based on the phenotypic parameters of all bone marrow cell compartments at diagnosis. The CEBPA-double-mutated subset clearly grouped in a separate cluster (dark green in the upper bar). CEBPA-single mutated cases displayed a heterogeneous distribution (light green in the upper bar). Columns represent individual bone marrow samples; rows represent the normalized log2 ratios of each parameter analyzed in a given cell compartment divided by the mean value obtained for that parameter in all control samples. The value of each parameter is represented in a color code according to control values: blue represents expression greater than the mean, red represents expression lower than the mean, white when not available; color intensity represents the magnitude of the deviation from the mean. Cluster analysis was carried out using R software.
To gain insight into potential influences of additional genetic changes on phenotype, we studied 12 (out of 16) CEBPA-dm cases for mutations of TET2 and GATA2 genes, which are known to be enriched in this subset (Online Supplementary Table S5). The presence of a mutated status did not influence clustering in the whole cohort nor within the CEBPA-dm group (data not shown).
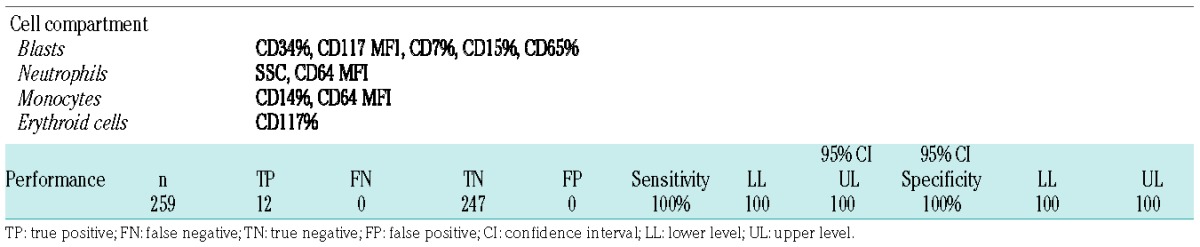
In order to define a suitable classifier we carried out a selection of parameters from the initial group of 79. Selection criteria were first based on coupled comparisons of single phenotypic parameters among CEBPA-dm versus CEBPA-wt, CEBPA-sm or controls. We then selected and tested several restricted groups of parameters in principal component analysis and hierarchical clustering. Finally, we chose one set of ten parameters (Table 3) that preserved the ability to separate CEBPA-dm cases in principal component analysis (Online Supplementary Figure S10) and clustering analysis (Online Supplementary Figure S11). The selected markers were: CD34, CD117, CD7, CD15, CD65 on blasts; SSC, CD64 on cells of the neutrophil compartment; CD14, CD64 on the monocytic compartment and CD117 on erythroid cells. Furthermore, we studied the efficacy of the parameter set at clustering in a group of AML samples (n=94), with data also acquired by a FACSCanto II flow cytometer (Online Supplementary Figure S12) in order to prove that the method was not affected by the instrument type. This classifier was thus tested as a potential screening method for CEBPA-dm genotype in AML.
Table 3.
Parameters of the classifier according to cell compartment and performance in the validation cohort as far as concerns prediction of a CEBPA-double-mutated status.
Validation of the classifier on an independent cohort
In order to validate the classifier prospectively, we used a large independent cohort (n=259) of unselected AML cases from three centers (Bergamo, Brescia and Venice). FCS files, blinded as regards clinical and biological features, were sent electronically to the coordinating center. The files were then analyzed and parameters tabulated. A group of controls (n=21) from both centers was analyzed in parallel to provide a homogeneous reference frame. The SSC signal of neutrophils was normalized on lymphocyte SSC. Applying our Euclidean distance-based classifier, a score was attributed to each case of the validation cohort (Online Supplementary Table S6). Below a defined threshold, 12 AML cases were considered as “highly probable” CEBPA-dm. Twelve out of the 12 turned out to bear double CEBPA mutations. Of note, no CEBPA-dm cases were missed by the classifier (i.e., there were no false negatives). Ten out of the 12 CEBPA-dm cases had a combination of N- and C-terminal mutations. The remaining two cases showed different mutation patterns: one had one N-terminal mutation and a nonsense mutation (c.569C>A) in the middle of the coding sequence; the other had two bi-allelic C-terminal mutations confirmed by next-generation sequencing (Online Supplementary File – Online Supplementary Table S7). The validation set included six CEBPA-sm cases, which were not highlighted by the classifier. Considering CEBPA-dm genotype as the target, the sensitivity and specificity of the classifier were both 100%, as were the positive and negative predictive values (Table 3). Our classifier was thus validated as a reliable screening method for CEBPA-dm status on an independent cohort of AML cases.
Discussion
The identification of CEBPA-dm status in AML has major clinical importance, allowing relapse risk to be stratified properly for post-remission treatment. However, most molecular screening methods for its detection have a number of technical problems. In our study, we developed an immunophenotype-based screening approach. Through an extensive phenotypic analysis of a cohort of 251 AML cases, we found that several phenotypic aberrations occurred recurrently on blasts and on maturing cell compartments in the subset of CEBPA-dm cases. Blasts showed features of maturation asynchrony with expression of CD34 and CD117 concomitant with high-intensity CD15, CD65 and MPO. Further, there was cross-lineage expression of CD7 by the whole blast cell population (Figure 1B). This finding is consistent with previous reports correlating the expression of CD7 in AML to loss of wild-type CEBPA due to mutations4,16 or silencing by epigenetic mechanisms.26–28 The neutrophil compartment showed reduced SSC signals and overexpression of CD64, with the latter also being seen in monocytes. The erythroid series was quantitatively expanded in CEBPA-dm cases in comparison to both CEBPA-wt and CEBPA-sm cases, especially at its more immature stages. In fact, the lack of normal CEBPA function has been associated with an imbalance of the transcriptional program of hematopoietic cells, highlighted by the gene expression profile (upregulation of genes involved in erythroid differentiation, downregulation of HOX gene members),29–31 by microRNA (over-expression of the miR-181 family)31 and long non-coding RNA (induction of UCA1 lncRNA)32 signatures. The functional consequences of CEBPA disruption would thus lead to a block in granulocytic differentiation and a preferential redirection toward the erythroid lineage.31 This is consistent with the frequent observation of erythroid dysplasia in CEBPA-dm patients in a previous study25 and in our cohort. To get insight into these data, we documented a CEBPA-dm status in all sorted myeloid cell compartments in six CEBPA-dm AML cases (Figure 3). Our findings are a proof-of-principle of the correlation between phenotypic abnormalities and CEBPA-dm status, indicating the multi-lineage involvement and thus common clonal origin of different lineages. Moreover these data account for the observed phenotypic homogeneity, due to “CEBPA-mutated dependent” pathways of maturation.
The multidimensional analysis of the entire phenotypic profile was able to separate CEBPA-dm cases efficiently from all the other genotypes. These results are coherent with reported gene expression profile data4,6,13 and the common phenotypic signature further confirms that CEBPA-dm represents a distinct AML subset. From the initial list of 79 parameters, we built a classifier from a core group of ten parameters (Table 3), strictly required by basic AML diagnostic recommendations.20 We then applied this classifier to an independent validation set of AML cases (n=259) from three other centers. Our classifier performed extremely well (Table 3) in terms of sensitivity and specificity (100%), and no CEBPA-dm cases were missed. This is probably the most important feature such a screening technique should have in order to avoid overtreatment (i.e. allogeneic transplantation) of patients with a favorable outcome with chemotherapy. The concomitant presence of an FLT3-ITD mutation in one patient in the validation dataset did not affect its correct classification as CEBPA-dm. The profile of one CEBPA-sm case in the primary cohort overlapped that of the CEBPA-dm group in principal component analysis. Interestingly, this case had a normal karyotype and no NPM1 or FLT3 mutations, suggesting that in this genetic context, a single mutation might affect the immunophenotype similarly to CEBPA-dm status. It is worth noting that the application of the classifier was not impaired by intrinsic interlaboratory variability or by the use of different instruments, suggesting high reproducibility besides stringent standardization of the method.
Beyond being technically challenging, interpretation of the CEBPA mutation pattern can sometimes be debatable and still crucial in individual cases in terms of prognosis. The study of functional consequences of CEBPA mutations suggests that the key point of convergence is the exclusive formation of p30/p30 homodimers.33 This scenario is supposed to be shared by bi-allelic N-terminal and C-terminal mutations, as well as by the rarer combinations of two N-terminal mutations or an N-terminal mutation with a frameshift/nonsense mutation in the central part of CEBPA.33 One case from the validation set displayed the latter pattern and one showed an even rarer7 combination of two C-terminal bi-allelic mutations. Of note, both of these cases clustered together with the other CEBPA-dm cases (Online Supplementary File – Online Supplementary Table S7). The phenotypic profile might be useful to suspect bi-allelic mutations occurring on the same gene region, because of the difficult interpretation of Sanger sequencing in such a context. In contrast, it has been reported that about 10% of non-homozygous CEBPA-dm cases carry gene mutations in two different subclones, an event of uncertain significance for leukemogenesis and prognosis.12 Our data suggest that the phenotype-based classifier might pick up a shared phenotypic signature downstream to several mutation patterns, all leading to a peculiar functional CEBPA disruption, independently of mutation type. It could, therefore, enable this “classical” mutation pattern to be distinguished from alternative combinations of gene lesions. We have thus drawn a workflow embedding the classifier in the diagnostic work-up of AML (Online Supplementary Figure S13). This would provide insight into CEBPA-related leukemogenesis and obviously translate into quickly available prognostic information.
Being based on phenotypic data, our approach provides very early results and this goes beyond the mere speeding up of a focused molecular study. Although it is well-recognized that main genetic prognostic factors drive only the post-complete remission phase, knowledge about them since the outset is often meaningful for the clinical management of patients with AML.
In conclusion, we established a reliable and straightforward screening method, based simply on the multidimensional analysis of widely available phenotypic parameters, suitable for large-scale detection of CEBPA-dm status and potentially able to overcome technical issues related to molecular methods. Our approach provides very early results, allowing entire CEBPA sequencing to be performed in only selected cases. The method has high specificity and sensitivity, as demonstrated in an independent AML cohort. This is of major clinical significance, since CEBPA-dm patients show a favorable prognosis, and knowledge about the CEBPA genotype status permits the use of proportional treatment modalities.
Supplementary Material
Acknowledgments
The authors would like to thank the Istituto Toscano Tumori, Ente Cassa di Risparmio di Firenze (2009-15520) and Regione Toscana (Bando Salute 2009 – research n 46) for funding this study.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/3/529
References
- 1.Pabst T, Mueller B, Zhang P, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-(C/EBP), in acute myeloid leukemia. Nat Genet. 2001;27(3):263–270. [DOI] [PubMed] [Google Scholar]
- 2.Gombart A, Hofmann WK, Kawano S, et al. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99(4):1332–1340. [DOI] [PubMed] [Google Scholar]
- 3.Preudhomme C, Sagot C, Boissel N, et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood. 2002;100(8):2717–2723. [DOI] [PubMed] [Google Scholar]
- 4.Wouters B, Löwenberg B, Erpelinck-Verschueren C, van Putten W, Valk P, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113(13):3088–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918. [DOI] [PubMed] [Google Scholar]
- 6.Dufour A, Schneider F, Metzeler K, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol. 2010;28(4):570–577. [DOI] [PubMed] [Google Scholar]
- 7.Green C, Koo K, Hills R, Burnett A, Linch D, Gale R. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol. 2010;28(16):2739–2747. [DOI] [PubMed] [Google Scholar]
- 8.Arber D, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20): 2391–2405. [DOI] [PubMed] [Google Scholar]
- 9.Benthaus T, Schneider F, Mellert G, et al. Rapid and sensitive screening for CEBPA mutations in acute myeloid leukaemia. Br J Haematol. 2008;143(2):230–239. [DOI] [PubMed] [Google Scholar]
- 10.Fuster O, Barragán E, Bolufer P, et al. Fragment length analysis screening for detection of CEBPA mutations in intermediate-risk karyotype acute myeloid leukemia. Ann Hematol. 2011;91(1):1–7. [DOI] [PubMed] [Google Scholar]
- 11.Wouters BJ, Louwers I, Valk PJ, Löwenberg B, Delwel R. A recurrent in-frame insertion in a CEBPA transactivation domain is a polymorphism rather than a mutation that does not affect gene expression profiling-based clustering of AML. Blood. 2007;109(1): 389–390. [DOI] [PubMed] [Google Scholar]
- 12.Behdad A, Weigelin H, Elenitoba-Johnson K, Betz B. A clinical grade sequencing-based assay for CEBPA mutation testing report of a large series of myeloid neoplasms. J Mol Diagn. 2015;17(1):76–84. [DOI] [PubMed] [Google Scholar]
- 13.Taskesen E, Bullinger L, Corbacioglu A, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117(8):2469–2475. [DOI] [PubMed] [Google Scholar]
- 14.Van Vliet MH, Burgmer P, de Quartel L, et al. Detection of CEBPA double mutants in acute myeloid leukemia using a custom gene expression array. Gen Test Mol Biomakers. 2013;17(5):395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrusák O, Porwit-MacDonald A. Antigen expression patterns reflecting genotype of acute leukemias. Leukemia. 2002;16(7): 1233–1258. [DOI] [PubMed] [Google Scholar]
- 16.Lin LI, Chen CY, Lin DT, et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11(4):1372–1379. [DOI] [PubMed] [Google Scholar]
- 17.Bene MC, Castoldi G, Knapp W, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia. 1995;9(10): 1783–1786. [PubMed] [Google Scholar]
- 18.Vardiman J, Harris N, Brunning R. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–2302. [DOI] [PubMed] [Google Scholar]
- 19.Mitelman FP. An International System for Human Cytogenetic Nomenclature. 1995. S. Karger, Basel. [Google Scholar]
- 20.Döhner H, Estey E, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. [DOI] [PubMed] [Google Scholar]
- 21.Noguera NI, Ammatuna E, Zangrilli D, et al. Simultaneous detection of NPM1 and FLT3-ITD mutations by capillary electrophoresis in acute myeloid leukemia. Leukemia. 2005;19(8):1479–1482. [DOI] [PubMed] [Google Scholar]
- 22.Falini B, Martelli M, Bolli N, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108(6): 1999–2005. [DOI] [PubMed] [Google Scholar]
- 23.Cheson B, Bennett J, Kopecky K, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21(24):4642–4649. [DOI] [PubMed] [Google Scholar]
- 24.Fröhling S, Schlenk R, Stolze I, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22(4):624–633. [DOI] [PubMed] [Google Scholar]
- 25.Bacher U, Schnittger S, Macijewski K, et al. Multilineage dysplasia does not influence prognosis in CEBPA-mutated AML, supporting the WHO proposal to classify these patients as a unique entity. Blood. 2012;119(20):4719–4722. [DOI] [PubMed] [Google Scholar]
- 26.Röhrs S, Scherr M, Romani J, Zaborski M, Drexler H, Quentmeier H. CD7 in acute myeloid leukemia: correlation with loss of wild-type CEBPA, consequence of epigenetic regulation. J Hematol Oncol. 2010; 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wouters BJ, Jordà MA, Keeshan K, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110(10):3706–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasan A, Alpermann T, Haferlach C, et al. Frequency and prognostic impact of CEBPA proximal, distal and core promoter methylation in normal karyotype AML: a study on 623 cases. PLoS One. 2013;8(2):e54365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heath V, Suh HC, Holman M, et al. C/EBPalpha deficiency results in hyperproliferation of hematopoietic progenitor cells and disrupts macrophage development in vitro and in vivo. Blood. 2004;104(6): 1639–1647. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P, Iwasaki-Arai J, Iwasaki H, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21(6): 853–863. [DOI] [PubMed] [Google Scholar]
- 31.Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signature associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26(31):5078–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes JM, Legnini I, Salvatori B, et al. C/EBPalpha-p30 protein induces expression of the oncogenic long non-coding RNA UCA1 in acute myeloid leukemia. Oncotarget. 2015;6(21):18534–18544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohlsson E, Schuster MB, Hasemann M, Porse BT. The multifaceted functions of C/EBP in normal and malignant hematopoiesis. Leukemia. 2015;30(4):767–775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.