Abstract
The mechanistic Target of Rapamycin (mTOR) coordinates eukaryotic cell growth and metabolism with environmental inputs including nutrients and growth factors. Extensive research over the past two decades has established a central role for mTOR in regulating many fundamental cell processes, from protein synthesis to autophagy, and deregulated mTOR signaling is implicated in the progression of cancer and diabetes, as well as the aging process. Here, we review recent advances in our understanding of mTOR function, regulation, and importance in mammalian physiology. We also highlight how the mTOR-signaling network contributes to human disease, and discuss the current and future prospects for therapeutically targeting mTOR in the clinic.
Introduction
In 1964 a Canadian expedition to the isolated South Pacific island of Rapa Nui (also known as Easter Island) collected a set of soil samples with the goal of identifying novel antimicrobial agents. In bacteria isolated from one of these samples, Sehgal and colleagues discovered a compound with remarkable antifungal, immunosuppressive, and antitumor properties (Eng et. al. 1984; Martel et. al. 1977; Vezina et. al 1975). Further analysis of this compound, named rapamycin after its site of discovery (clinically referred to as sirolimus), revealed that it acts in part by forming a gain of function complex with the peptidyl-prolyl-isomerase FKBP12 to inhibit signal transduction pathways required for cell growth and proliferation (Chung et. al., 1992).
Despite these insights, the full mechanism of action of rapamycin remained elusive until 1994 when biochemical studies identified the mechanistic (formerly “mammalian”) Target of Rapamycin (mTOR) as the direct target of the rapamycin-FKBP12 complex in mammals (Brown et. al. 1994; Sabatini et. al. 1994; Sabers et. al 1995), and revealed it to be the homolog of the yeast TOR/DRR genes that had previously been identified in genetic screens for rapamycin resistance (Cafferkey et. al. 1993; Heitman et. al. 1991; Kunz et. al. 1993).
In the more than two decades since these discoveries, studies from dozens of labs across the globe have revealed that the mTOR protein kinase nucleates a major eukaryotic signaling network that coordinates cell growth with environmental conditions and plays a fundamental role in cell and organismal physiology. Many aspects of mTOR function and regulation have only recently been elucidated, and many more questions remain unanswered. In this review, we provide an overview of our current understanding of the mTOR pathway and its role in growth, metabolism, and disease.
mTORC1 and mTORC2
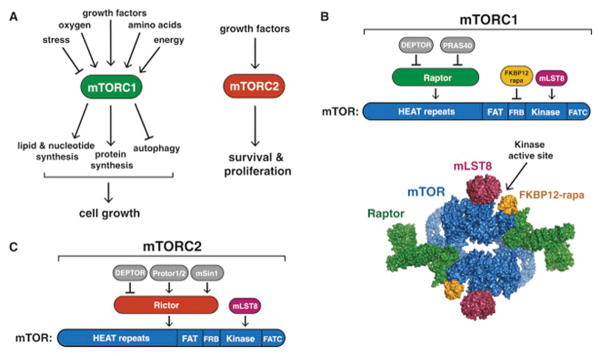
mTOR is a serine/threonine protein kinase in the PI3K-related kinase (PIKK) family that forms the catalytic subunit of two distinct protein complexes, known as mTOR Complex 1 (mTORC1) and 2 (mTORC2) (Fig. 1A). mTORC1 is defined by its three core components: mTOR, Raptor (regulatory protein associated with mTOR), and mLST8 (mammalian lethal with Sec13 protein 8, also known as GβL) (Fig. 1B, Kim et al. 2002; Hara et al. 2002; Kim et al. 2003). Raptor facilitates substrate recruitment to mTORC1 through binding to the TOR signaling (TOS) motif found on several canonical mTORC1 substrates (Nojima et al., 2003; Schalm et al., 2003), and, as described later, is required for the correct subcellular localization of mTORC1. mLST8 by contrast associates with the catalytic domain of mTORC1 and may stabilize the kinase activation loop (Yang et al. 2013), though genetic studies suggest it is dispensible for the essential functions of mTORC1 (Guertin et al., 2006). In addition to these three core components, mTORC1 also contains the two inhibitory subunits PRAS40 (proline-rich Akt substrate of 40 kDa) (Sancak et al. 2007; Vander Haar et al. 2007; Wang et al. 2007) and DEPTOR (DEP domain containing mTOR interacting protein) (Peterson et al. 2009).
Figure 1. mTORC1 and mTORC2.
(A) The mTORC1 and mTORC2 signaling pathways.
(B) mTORC1 subunits and respective binding sites on mTOR. The FKBP12-rapamycin. The 5.9 Å cryo-EM structure of mTORC1 (without DEPTOR and PRAS40, PDB ID: 5FLC) from is depicted as a space filling model and colored by subunit.
(C) mTORC2 subunits and respective binding sites on mTOR.
Structural studies of mTORC1 have yielded significant insights into its assembly, function, and perturbation by rapamycin. Cryo-EM reconstructions of both mTORC1 and yeast TORC1 have revealed that the complex forms a 1 mDa “lozenge”-shaped dimer, with the dimerization interface comprised of contacts between the mTOR HEAT repeats as well as between Raptor and mTOR (Fig. 1B, Aylett et al. 2016; Baretic et al., 2016; Yip et al. 2010). In addition, a crystal structure of the mTOR kinase domain bound to mLST8 showed that the rapamycin-FKBP12 complex binds to the FRB domain of mTOR to narrow the catalytic cleft and partially occlude substrates from the active site (Yang et al., 2013).
While the rapamycin-FKBP12 complex directly inhibits mTORC1, mTORC2 is characterized by its insensitivity to acute rapamycin treatment. Like mTORC1, mTORC2 also contains mTOR and mLST8 (Fig. 1C). Instead of Raptor however, mTORC2 contains Rictor (rapamycin insensitive companion of mTOR), an unrelated protein that likely serves an analogous function (Jacinto et al. 2004; Sarbassov et al. 2004). mTORC2 also contains DEPTOR (Peterson et al. 2009), as well as the regulatory subunits mSin1 (Frias et al. 2006; Jacinto et al. 2006; Yang et al. 2006) and Protor1/2 (Pearce et al. 2007; Thedieck et al. 2007; Woo et al., 2007). Although rapamycin-FKBP12 complexes do not directly bind or inhibit mTORC2, prolonged rapamycin treatment does abrogate mTORC2 signaling, likely due to the inability of rapamycin-bound mTOR to incorporate into new mTORC2 complexes (Lamming et al., 2012).
The mTOR Signaling Network
Downstream of mTORC1
In order to grow and divide, cells must increase production of proteins, lipids, and nucleotides while also suppressing catabolic pathways such as autophagy. mTORC1 plays a central role in regulating all of these processes, and therefore controls the balance between anabolism and catabolism in response to environmental conditions (Fig. 2, A and B). Here we review the critical substrates and cellular processes downstream of mTORC1 and how they contribute to cell growth. Most of the functions discussed here were identified and characterized in the context of mammalian cell lines, while the physiological context in which these processes are important will be discussed in greater detail later.
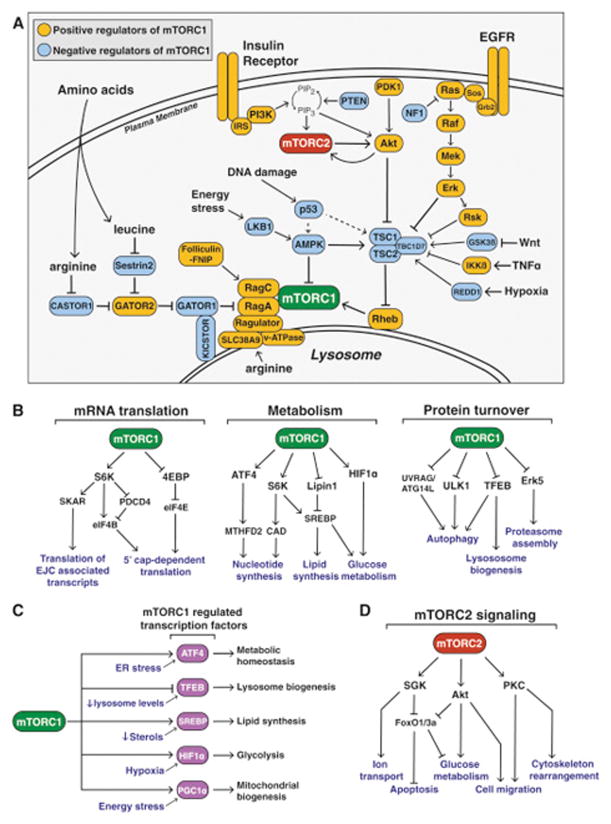
Figure 2. The mTOR Signaling Network.
(A) The signaling pathways upstream of mTORC1 and mTORC2. Positive regulators of mTORC1 signaling are shown in yellow, while negative regulators are shown in blue. mTORC1 and mTORC2 are shown in green and red, respectively.
(B) The major signaling pathways downstream of mTORC1 signaling in mRNA translation, metabolism, and protein turnover.
(C) mTORC1 controls the activity of several transcription factors that can also be independently regulated by cell stress.
(D) The major signaling pathways downstream of mTORC2 signaling.
Protein synthesis
mTORC1 promotes protein synthesis largely through the phosphorylation of two key effectors, p70S6 Kinase 1 (S6K1) and eIF4E Binding Protein (4EBP) (Fig 2B). mTORC1 directly phosphorylates S6K1 on its hydrophobic motif site, Thr389, enabling its subsequent phosphorylation and activation by PDK1. S6K1 phosphorylates and activates several substrates that promote mRNA translation initiation including eIF4B, a positive regulator of the 5′cap binding eIF4F complex (Holz et al., 2005). S6K1 also phosphorylates and promotes the degradation of PDCD4, an inhibitor of eIF4B (Dorello et al. 2006), and enhances the translation efficiency of spliced mRNAs via its interaction with SKAR, a component of exon-junction complexes (Ma et al., 2008).
The mTORC1 substrate 4EBP is unrelated to S6K1 and inhibits translation by binding and sequestering eIF4E to prevent assembly of the eIF4F complex. mTORC1 phosphorylates 4EBP at multiple sites to trigger its dissociation from eIF4E (Brunn et al., 1997; Gingras et al. 1999), allowing 5′cap-dependent mRNA translation to occur. Although it has long been appreciated that mTORC1 signaling regulates mRNA translation, whether and how it affects specific classes of mRNA transcripts has been debated. Global ribosome foot printing analyses however revealed that while acute mTOR inhibition moderately suppresses general mRNA translation, it most profoundly affects mRNAs containing pyrimidine-rich 5′ TOP or “TOP-like” motifs, which includes most genes involved in protein synthesis (Hsieh et al. 2012; Thoreen et al. 2012).
Lipid, nucleotide, and glucose metabolism
Growing cells require sufficient lipids for new membrane formation and expansion. mTORC1 promotes de novo lipid synthesis through the sterol responsive element binding protein (SREBP) transcription factors, which control the expression of metabolic genes involved in fatty acid and cholesterol biosynthesis (Porstmann et al., 2008). While SREBP is canonically activated in response to low sterol levels, mTORC1 signaling can also activate SREBP independently through both an S6K1-dependent mechanism (Duvel et al., 2010) as well as through the phosphorylation of an additional substrate, Lipin1, which inhibits SREBP in the absence of mTORC1 signaling (Peterson et al., 2011).
Recent studies established that mTORC1 also promotes the synthesis of nucleotides required for DNA replication and ribosome biogenesis in growing and proliferating cells. mTORC1 increases the ATF4-dependent expression of MTHFD2, a key component of the mitochondrial tetrahydrofolate cycle that provides one-carbon units for purine synthesis (Ben-Sahra et al., 2016). Additionally, S6K1 phosphorylates and activates carbamoyl-phosphate synthetase (CAD), a critical component of the de novo pyrimidine synthesis pathway (Ben-Sahra et al., 2013; Robataille et al., 2013).
mTORC1 also facilitates growth by promoting a shift in glucose metabolism from oxidative phosphorylation to glycolysis, which likely facilitates the incorporation of nutrients into new biomass. mTORC1 increases the translation of the transcription factor HIF1α (Fig. 2C), which drives the expression of several glycolytic enzymes such as phospho-fructo kinase (PFK) (Duvel et al., 2010). Furthermore, mTORC1-dependent activation of SREBP leads to increased flux through the oxidative pentose phosphate pathway (PPP), which utilizes carbons from glucose to generate NADPH and other intermediary metabolites needed for proliferation and growth.
Regulation of protein turnover
In addition to the various anabolic processes outlined above, mTORC1 also promotes cell growth by suppressing protein catabolism (Fig. 1B), most notably autophagy. An important early step in autophagy is the activation of ULK1, a kinase that forms a complex with ATG13, FIP2000, and ATG101 and drives autophagosome formation. Under nutrient replete conditions, mTORC1 phosphorylates ULK1, thereby preventing its activation by AMPK, a key activator or autophagy (Kim et al., 2011). Thus, the relative activity of mTORC1 and AMPK in different cellular contexts largely determines the extent of autophagy induction. mTORC1 also regulates autophagy in part by phosphorylating and inhibiting the nuclear translocation of the transcription factor TFEB, which drives the expression of genes for lysosomal biogenesis and the autophagy machinery (Martina et al., 2012; Roçzniak-Ferguson et al., 2012; Settembre et al., 2012).
The second major pathway responsible for protein turnover is the ubiquitin-proteasome system (UPS), through which proteins are selectively targeted for degradation by the 20S proteasome following covalent modification with ubiquitin. Two recent studies found that acute mTORC1 inhibition rapidly increases proteasome-dependent proteolysis through either a general increase in protein ubiquitylation, or an increased abundance of proteasomal chaperones via inhibition of Erk5 (Fig. 2B, Rousseau et al., 2016, Zhao et al., 2015). However, another study found that genetic hyper-activation of mTORC1 signaling also increases proteasome activity, through elevated expression of proteasome subunits downstream of Nrf1 (Zhang et al., 2014). One possible explanation for this discrepancy is that while acute mTORC1 inhibition promotes proteolysis to restore free amino acid pools, prolonged mTORC1 activation also triggers a compensatory increase in protein turnover to balance the increased rate of protein synthesis. Given that the UPS is responsible for the majority of protein degradation in human cells, precisely how mTORC1 regulates this process is an important question going forward.
Downstream of mTORC2
While mTORC1 regulates cell growth and metabolism, mTORC2 instead controls proliferation and survival primarily by phosphorylating several members of the AGC (PKA/PKG/PKC) family of protein kinases (Fig. 2D). The first mTORC2 substrate to be identified was PKCα, a regulator of the actin cytoskeleton (Jacinto et al., 2004, Sarbassov et al., 2004). More recently, mTORC2 has also been shown to phosphorylate several other members of the PKC family, including PKCδ (Gan et al., 2012), PKCζ (Li and Gao, 2014), as well as PKCγ and PKCε (Thomanetz et al., 2013), all of which regulate various aspects of cytoskeletal remodeling and cell migration.
The most important role of mTORC2 however is likely the phosphorylation and activation of Akt, a key effector of insulin/PI3K signaling (Sarbassov et al., 2005). Once active, Akt promotes cell survival, proliferation, and growth through the phosphorylation and inhibition of several key substrates including the FoxO1/3a transcription factors, the metabolic regulator GSK3β, and the mTORC1 inhibitor TSC2. However while mTORC2-dependent phosphorylation is required for Akt to phosphorylate some substrates in vivo, such as FoxO1/3a, it is dispensable for the phosphorylation of others including TSC2 (Guertin et al., 2006; Jacinto et. al., 2006). Finally, mTORC2 also phosphorylates and activates SGK1, another AGC-kinase that regulates ion transport as well as cell survival (Garcia-Martinez and Alessi, 2008).
Upstream of mTORC1
The mTORC1-dependent shift towards increased anabolism should only occur in the presence of pro-growth endocrine signals as well as sufficient energy and chemical building blocks for macromolecular synthesis. In mammals, these inputs are largely dependent on diet, such that mTORC1 is activated following feeding to promote growth and energy storage in tissues such as the liver and muscle, but inhibited during fasting conserve limited resources. Here we discuss the cellular pathways upstream of mTORC1 and the mechanisms through which they control mTORC1 activation.
Growth Factors
Studies of rapamycin in the early 1990s revealed that mTORC1 is a downstream mediator of several growth factor and mitogen-dependent signaling pathways, all of which inhibit a key negative regulator of mTORC1 signaling known as the Tuberous Sclerosis Complex (TSC) complex. TSC is a heterotrimeric complex comprised of TSC1, TSC2, and TBC1D7 (Dibble et al., 2012), and functions as a GTPase activating protein (GAP) for the small GTPase Rheb (Inoki et al., 2003; Tee et al., 2003), which directly binds and activates mTORC1 (Long et al., 2005; Sancak et al., 2007). Although Rheb is an essential activator of mTORC1, exactly how it stimulates mTORC1 kinase activity remains unknown.
Numerous growth factor pathways converge on TSC (Fig. 2A, reviewed in Huang and Manning, 2008), including the insulin/insulin-like growth factor-1 (IGF-1) pathway, which triggers the Akt-dependent multisite phosphorylation of TSC2. This phosphorylation inhibits TSC by dissociating it from the lysosomal membrane, where at least some fraction of cellular Rheb localizes (Menon et al., 2014). Similarly, receptor tyrosine kinase-dependent Ras signaling activates mTORC1 via the MAP Kinase Erk and its effector p90RSK, both of which also phosphorylate and inhibit TSC2. It is unclear however whether these inputs also control the localization of TSC, or rather inhibit its GAP activity through a distinct mechanism. Additional growth factor pathways upstream of TSC include Wnt and the inflammatory cytokine TNFα, both of which activate mTORC1 through the inhibition of TSC1. Precisely how the TSC complex integrates these numerous signals and their relative impact on mTORC1 activity in various contexts however remains an open question.
Energy, oxygen, and DNA damage
mTORC1 also responds to intracellular and environmental stresses that are incompatible with growth such as low ATP levels, hypoxia, or DNA damage. A reduction in cellular energy charge, for example during glucose deprivation, activates the stress responsive metabolic regulator AMPK, which inhibits mTORC1 both indirectly, through phosphorylation and activation of TSC2, as well as directly through the phosphorylation of Raptor (Gwinn et al., 2008; Inoki et al., 2003b; Shaw et al., 2004). Interestingly, glucose deprivation also inhibits mTORC1 in cells lacking AMPK, through inhibition of the Rag GTPases, suggesting that mTORC1 senses glucose through more than one mechanism (Efeyan et al., 2013; Kalender et al., 2010). Similarly, hypoxia inhibits mTORC1 in part through AMPK activation, but also through the induction of REDD1 (Regulated in DNA damage and development 1), which activates TSC (Brugarolas et al., 2004). Finally, the DNA damage-response pathway inhibits mTORC1 through the induction of p53 target genes including the AMPK regulatory subunit (AMPKβ), PTEN, and TSC2 itself, all of which increase TSC activity (Feng et al., 2007).
Amino Acids
In addition to glucose-dependent insulin release, feeding also leads to an increase in serum amino acid levels due to the digestion of dietary proteins. As amino acids are not only essential building blocks of proteins but also sources of energy and carbon for many other metabolic pathways, mTORC1 activation is tightly coupled to diet-induced changes in amino acid concentrations.
A breakthrough in the understanding of amino acid sensing by mTORC1 came with the discovery of the heterodimeric Rag GTPases as components of the mTORC1 pathway (Kim et al., 2008; Sancak et al., 2008). The Rags are obligate heterodimers of RagA or RagB with RagC or RagD, and are tethered to the lysosomal membrane through their association with the pentameric Ragulator complex comprised of MP1, p14, p18, HBXIP and c7ORF59 (Sancak et al., 2010; Bar-Peled et al., 2012). Amino acid stimulation converts the Rags to their active nucleotide-bound state, allowing them to bind Raptor and recruit mTORC1 to the lysosomal surface, where Rheb is also located. This pathway architecture therefore forms an “AND-gate”, whereby mTORC1 signaling is only on when both the Rags and Rheb are activated, explaining why both growth factors and amino acids are required for mTORC1 activation.
Despite these insights, the identities of the direct amino acid sensors upstream of mTORC1 have been elusive until very recently. It is now clear that mTORC1 senses both intra-lysosomal and cytosolic amino acids through distinct mechanisms. Amino acids inside the lysosomal lumen alter the Rag nucleotide state through a mechanism dependent on the lysosomal v-ATPase, which interacts the Ragulator-Rag complex to promote the guanine-nucleotide exchange factor (GEF) activity of Ragulator towards RagA/B (Zoncu et al., 2011; Bar-Peled et al., 2012). The lysosomal amino acid transporter SLC38A9 interacts with the Rag-Ragulator-v-ATPase complex and is required for arginine to activate mTORC1, making it a promising candidate to be a lysosomal amino acid sensor (Jung et al., 2015; Rebsamen et al., 2015; Wang et al., 2015).
Cytosolic leucine and arginine signal to mTORC1 through a distinct pathway comprised of the GATOR1 and GATOR2 complexes (Bar-Peled et al., 2013). GATOR1 consists of DEPDC5, Nprl2 and Nprl3, and inhibits mTORC1 signaling by acting as a GAP for RagA/B. The recently identified KICSTOR complex (consisting of Kaptin, ITFG2, c12orf66, and SZT2) tethers GATOR1 to the lysosomal surface and is necessary for the appropriate control of the mTORC1 pathway by nutrients (Wolfson et al., 2017). GATOR2 by contrast is a pentameric complex comprised of Mios, WDR24, WDR59, Seh1L and Sec13, and is a positive regulator of mTORC1 signaling that interacts with GATOR1 at the lysosomal membrane (Bar-Peled et al., 2013).
An important insight into the mechanism of cytosolic amino acid sensing came with the identification of Sestrin2 as a GATOR2 interacting protein that inhibits mTORC1 signaling under amino acid deprivation (Chantranupong et al., 2014; Parmigiani et al., 2014). Subsequent biochemical and structural analyses established that Sestrin2 is a direct leucine sensor upstream of mTORC1 that binds and inhibits GATOR2 function in the absence of leucine, and dissociates from it upon leucine binding (Saxton et al., 2016a; Wolfson et al., 2016). Furthermore, the affinity of Sestrin2 for leucine determines the sensitivity of mTORC1 signaling to leucine in cultured cells, demonstrating that Sestrin2 is the primary leucine sensor for mTORC1 in this context. It remains to be seen whether and in what tissues leucine concentrations fluctuate within the relevant range to be sensed by Sestrin2 in vivo, as the levels of interstitial or cytosolic leucine are unknown. Interestingly, another recent study found that Sestrin2 is transcriptionally induced upon prolonged amino acid starvation via the stress-responsive transcription factor ATF4 (Ye et al., 2015), suggesting that Sestrin2 functions as both an acute leucine sensor as well as an indirect mediator of prolonged amino acid starvation.
Cytosolic arginine also activates mTORC1 through the GATOR1/2-Rag pathway by directly binding the recently identified arginine sensor CASTOR1 (Cellular Arginine Sensor for mTORC1). Much like Sestrin2, CASTOR1 binds and inhibits GATOR2 in the absence of arginine, and dissociates upon arginine binding to enable the activation of mTORC1 (Chantranupong et al., 2016; Saxton et al., 2016b). Thus, both leucine and arginine stimulate mTORC1 activity at least in part by releasing inhibitors from GATOR2, establishing GATOR2 as a central node in the signaling of amino acids to mTORC1. Importantly however, the molecular function of GATOR2 and the mechanisms through which Sestrin2 and CASTOR1 regulate it are unknown.
Several additional mechanisms through which amino acids regulate mTORC1 signaling have also recently been reported, including the identification of the Folliculin-FNIP2 complex as a GAP for RagC/D that activates mTORC1 in the presence of amino acids (Petit et al., 2013; Tsun et al., 2013). Another study found that the amino acid glutamine, which is utilized as a nitrogen and energy source by proliferating cells, activates mTORC1 independently of the Rag GTPases through the related Arf family GTPases (Jewell et al., 2015). Finally, a recent report found that the small polypeptide SPAR associates with the v-ATPase-Ragulator complex to suppress mTORC1 recruitment to lysosomes, though how this occurs is unclear (Matsumoto et al., 2016).
Upstream of mTORC2
In contrast to mTORC1, mTORC2 primarily functions as an effector of insulin/PI3K signaling (Fig. 2A). Like most PI3K regulated proteins, the mTORC2 subunit mSin1 contains a phosphoinositide-binding PH domain that is critical for the insulin-dependent regulation of mTORC2 activity. The mSin1 PH domain inhibits mTORC2 catalytic activity in the absence of insulin, and this autoinhibition is relieved upon binding to PI3K-generated PIP3 at the plasma membrane (Liu et al., 2015). mSin1 can also be phosphorylated by Akt, suggesting the existence of a positive-feedback loop whereby partial activation of Akt promotes the activation of mTORC2, which in turn phosphorylates and fully activates Akt (Yang et al., 2015). Another study found that PI3K promotes the association of mTORC2 with ribosomes to activate its kinase activity, although the mechanistic basis for this is unclear (Zinzalla et al., 2011).
Unexpectedly, mTORC2 signaling is also regulated by mTORC1, due to the presence of a negative feedback loop between mTORC1 and insulin/PI3K signaling. mTORC1 phosphorylates and activates Grb10, a negative regulator of insulin/IGF-1 receptor signaling upstream of Akt and mTORC2, (Hsu et al., 2011; Yu et al., 2011), while S6K1 also suppresses mTORC2 activation through the phosphorylation-dependent degradation of insulin receptor substrate 1 (IRS1) (Harrington et al., 2004; Shah et al., 2004). This negative feedback regulation of PI3K and mTORC2 signaling by mTORC1 has numerous implications for the pharmacological targeting of mTOR in disease, discussed below.
Evolutionary conservation of the TOR pathway
One remarkable feature of the TOR pathway is its conservation as a major growth regulator in virtually all eukaryotes. Like mammals, S. cerevisiae also have two distinct TOR containing complexes, TORC1 and TORC2 (reviewed in Loewith and Hall, 2011) as well as homologs of Raptor (Kog1), mLST8 (Lst8), Rictor (Avo3), and mSin1 (Avo1), although several additional components are yeast or mammal specific. Furthermore yeast TORC1 also primarily controls cell growth and anabolic metabolism, including the activation of protein synthesis and inhibition of autophagy, while yeast TORC2 primarily functions to activate AGC family kinases such as YPK1, the homologue of mammalian SGK1.
As with mTORC1, yeast TORC1 also senses and responds to a diverse array of environmental stimuli, although the specific inputs and upstream signaling components differ in several respects, as one would expect given the vastly different environmental conditions that are relevant for these organisms (Fig. 3A). For example, hormone and growth factor receptor signaling are developments specific to multicellular organisms, and the mTORC1 regulator TSC is not found in S. cerevisiae. Instead, yeast TORC1 appears to be primarily sensitive to direct biosynthetic inputs such as carbon, nitrogen and phosphate sources.
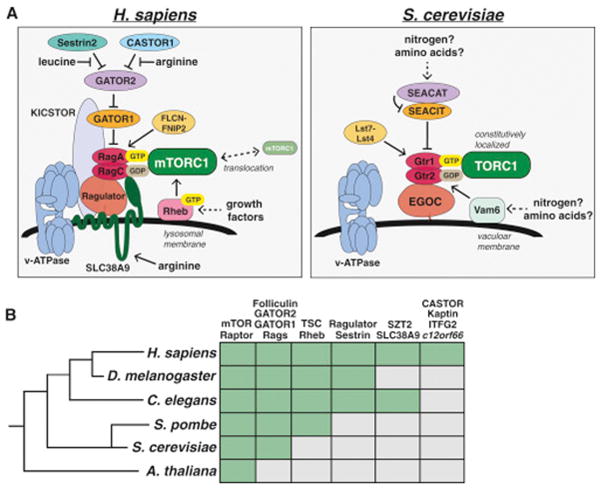
Figure 3. Evolutionary Conservation of the mTOR Pathway.
(A) The nutrient sensing pathway upstream of mammalian mTORC1 (left) and yeast TORC1 (right).
(B) Phylogenetic tree depicting the presence (green box) of key mTORC1 regulators in various model organisms.
Unlike mammals, yeast are able to synthesize all 20 amino acids, and starvation of individual amino acids like leucine or arginine does not inhibit TORC1 signaling in wild-type strains. Leucine deprivation does inhibit TORC1 in leucine-auxotrophs however (Binda et al., 2009), suggesting there may be a mechanism for signaling amino acid levels to TORC1, although this could also be due to the sensing of nitrogen sources that are perturbed in this context. Both the Rag GTPases and the GATOR1/2 complexes are present in S. cerevisiae in the form of Gtr1/2 and the SEACIT/SEACAT complexes, respectively (Fig. 3A, Panchaud et al., 2009), while the yeast EGO Complex is a structural homolog of Ragulator that interacts with Gtr1/2 and likely serves an analogous function (Powis et al., 2015; Zhang et al., 2012). In contrast to mammals however, amino acids do not affect the localization of yeast TORC1, which is constitutively bound to the Gtr-Ego complex at the vacuolar membrane (Binda et al., 2009), suggesting an alternative sensing mechanism exists. Consistent with this, the mammalian amino acid sensors SLC38A9, Sestrin2, and CASTOR1 all lack clear homologs in yeast.
While most of the TORC1 pathway components are also well conserved in other multicellular model organisms, the direct amino acid sensors appear to have diverged (Fig. 3B). For example, although both SLC38A9 and CASTOR1 are conserved throughout many metazoan lineages, they are absent in D. melanogaster, suggesting that this organism either does not sense arginine or does so through a distinct mechanism. Both D. melanogaster and C. elegans do have a Sestrin homolog however, and dmSestrin also binds leucine (Wolfson et al., 2016). Interestingly, both ceSestrin and dmSestrin contain subtle differences in their leucine binding pockets predicted to reduce their affinity for leucine relative to human Sestrin2, likely enabling the sensing of physiologically relevant leucine levels in these organisms (Saxton et al., 2016a).
Physiological roles of mTOR
Changes in available energy sources following fasting or feeding require alterations in whole body metabolism to maintain homeostasis. Under starvation, levels of nutrients and growth factors drop, inducing a catabolic state in which energy stores are mobilized to maintain essential functions (Fig. 4A). Alternatively, high levels of nutrients in the fed-state trigger a switch towards anabolic growth and energy storage. Consistent with its role in coordinating anabolic and catabolic metabolism at the cellular level, physiological studies in mice have revealed that mTOR signaling is essential for proper metabolic regulation at the organismal level as well. Importantly, however, constitutive mTOR activation is also associated with negative physiological outcomes, indicating that the proper modulation of mTOR signaling in response to changing environmental conditions is crucial (Fig. 4B).
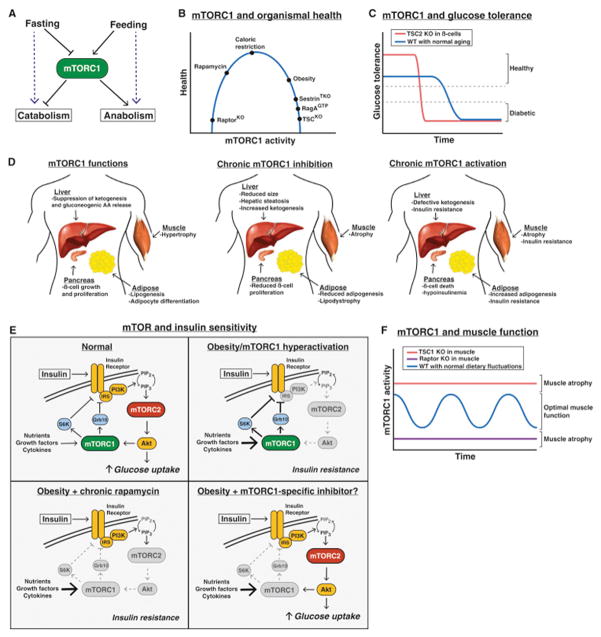
Figure 4. Physiological Roles of mTOR.
(A) mTORC1 controls the balance between anabolism and catabolism in response to fasting and feeding.
(B) The effect of cumulative mTORC1 activity on overall health.
(C) The effect of mTORC1 hyperactivation in pancreatic β-cells on glucose tolerance over time.
(D) The normal functions of mTORC1 in the liver, muscle, pancreas, and adipose tissue (left), and the consequences of chronic mTORC1 inhibition (middle) or activation (right).
(E) Deregulation of mTORC1 signaling in insulin resistance/diabetes, and the effect of rapamycin or a theoretical mTORC1 specific inhibitor.
Glucose homeostasis
When blood glucose levels drop, the liver activates a compensatory response involving the induction of autophagy, gluconeogenesis, and the release of alternative energy sources in the form of ketone bodies. Several lines of evidence suggest that regulation of mTORC1 signaling is crucial for the response of the liver to diet. For example, mice with liver specific deletion of TSC1, which have constitutively activated mTORC1 signaling, fail to generate ketone bodies during fasting due to sustained mTORC1-dependent suppression of PPARa, a transcriptional activator of ketogenic genes (Sengupta et al., 2010). The importance of inhibiting mTORC1 in the liver during fasting has also been observed through the generation mice with a while-body knock-in of a constitutively active allele of RagA (RagAGTP). Although they develop normally, these mice die rapidly after birth due to an inability to maintain blood glucose levels during the perinatal fasting period (Efeyan et al., 2013). Further analysis of these mice revealed that sustained mTORC1 activity during this fasting period prevents the induction of autophagy in the liver, which is critical for supplying free amino acids for gluconeogenesis. As a result, RagAGTP mice display fatal hypoglycemia in response to fasting, consistent with a similar phenotype in autophagy deficient mice (Kuma et al., 2004).
mTORC1 signaling also plays an important role in glucose homeostasis by regulating pancreatic β-cell function. Studies using β-cell specific TSC2 knock out (β-TSC2KO) mice revealed that hyperactivation of mTORC1 has a biphasic effect on β-cell function, with young β-TSC2KO mice exhibiting increased β-cell mass, higher insulin levels, and improved glucose tolerance (Mori et al., 2009; Shigeyama et al., 2008). This effect is reversed in older β-TSC2KO mice, which more rapidly develop reduced β-cell mass, lower insulin levels, and hyperglycemia. Thus, high mTORC1 activity in the pancreas is initially beneficial for glucose tolerance, but also leads to a faster decline in β-cell function over time (Fig. 4C).
This biphasic effect of mTORC1 signaling is reminiscent of diet-induced (type 2) diabetes progression, in which pancreatic β-cells initially expand and produce more insulin to compensate for an increased glycemic load, but eventually undergo exhaustion. Indeed, obese or high fat diet (HFD) treated mice have high mTORC1 signaling in many tissues, including the pancreas, likely due to increased levels of circulating insulin, amino acids, and pro-inflammatory cytokines (Khamzina et al., 2005). Increased mTORC1 signaling in these tissues also contributes to peripheral insulin resistance due to enhanced feedback inhibition of insulin/PI3K/Akt signaling, which is prevented in mice lacking S6K1/2 (Fig. 4D, Um et al., 2004).
That mTORC1 hyperactivation from genetic or dietary manipulation results in insulin resistance has led many to speculate that mTORC1 inhibitors could improve glucose tolerance and protect against type 2 diabetes. Paradoxically, however, chronic pharmacological inhibition of mTORC1 using rapamycin has the opposite effect, causing insulin resistance and impaired glucose homeostasis (Fig. 4D, Cunningham et al., 2007). This result is explained at least in part by the fact that prolonged rapamycin treatment also inhibits mTORC2 signaling in vivo (Lamming et al., 2012). As mTORC2 directly activates Akt downstream of insulin/PI3K signaling, it is not surprising that mTORC2 inhibition disrupts the physiological response to insulin. Consistent with this, liver specific Rictor knockout mice have severe insulin resistance and glucose intolerance (Hagiwara et al., 2012; Yuan et al., 2012), as do mice lacking Rictor in the muscle or fat (Kumar et al., 2008 & 2010).
Muscle mass and function
Although the importance of mTOR signaling in promoting muscle growth is well appreciated by basic scientists and bodybuilders alike, the mechanisms underlying this process are still poorly understood, in part due to the difficulty of genetically manipulating multinucleate myocytes in vivo. Nonetheless, early studies of mTOR signaling in the muscle revealed that mTORC1 activation is associated with muscle hypertrophy (Bodine et al., 2001) and that both IGF-1 and leucine promote hypertrophy through the activation of mTORC1 signaling in cultured cells and in mice (Anthony et al., 2000; Rommel et al., 2001). Moreover, muscle specific Raptor knockout mice display severe muscle atrophy and reduced body weight leading to early death (Bentzinger et al., 2008). This dramatic phenotype is also observed in muscle specific mTOR knockout mice, but not Rictor deficient mice, suggesting that the critical functions of mTOR in skeletal muscle are through mTORC1 (Bentzinger et al., 2008; Risson et al., 2009).
While acute activation of mTORC1 signaling in vivo does promote muscle hypertrophy in the short-term (Bodine et al., 2001) chronic mTORC1 activation in the muscle through loss of TSC1 also results in severe muscle atrophy, low body mass, and early death, primarily due to a lack the inability to induce autophagy in this tissue (Fig. 4D, Castets et al, 2013). Considering that turnover of old or damaged tissue plays a critical role in muscle growth, these results suggest that alternating periods of high and low mTORC1 activity, as occurs with normal feeding and fasting cycles, is essential for maintaining optimal muscle health and function (Fig. 4F).
An accumulating body of evidence suggests that muscle contraction also activates mTORC1 in the muscle; potentially explaining at least in part how increased muscle use promotes anabolism (Baar and Esser, 1999). A recent study found that mechanical stimuli activate mTORC1 signaling by inducing the phosphorylation of Raptor (Frey et al., 2014), although how this occurs in not clear. Understanding how mTORC1 can integrate the distinct signals from insulin, amino acids, and mechanical force in the muscle will be an important goal going forward and may inform approaches for treating muscle wasting disorders such as those associated with disuse and aging.
Adipogenesis and lipid homeostasis
Many studies over the last two decades also reveal a role for mTOR in promoting adipocyte formation and lipid synthesis in response to feeding and insulin (Fig. 4D, reviewed in Lamming and Sabatini, 2013). mTORC1 promotes adipogenesis and enhanced lipogenesis in cell culture and in vivo, consistent with adipocyte-specific raptor knock out (Ad-RapKO) mice displaying lipodystrophy and hepatic steatosis (Lee et al., 2016). However, the role of mTORC1 in adipose is complicated by the fact that Ad-RapKO mice are also resistant to diet induced obesity due to reduced adipogenesis, suggesting mTORC1 inhibition in this tissue can have both positive and negative effects (Polak et al., 2008).
Similarly, the loss of mTORC2 activity in adipocytes primarily results in insulin resistance due to reduced Akt activity (Kumar et al., 2010), but also in less lipid synthesis in part due to reduced expression of ChREBPβ, a master transcription factor for lipogenic genes (Tang et al., 2016). mTORC2 has also been shown to promote lipogenesis in the liver as well, suggesting a general role for mTORC2 in lipid synthesis (Hagiwara et al., 2012; Yuan et al., 2012) Thus, both mTORC1 and mTORC2 play important roles in multiple aspects of adipocyte function and lipid metabolism.
Immune function
Early studies into the biological properties of rapamycin revealed a role in blocking lymphocyte proliferation, leading to its eventual clinical approval as an immunosuppressant for kidney transplants in 1999. The immunosuppressive action of rapamycin is largely attributed to its ability to block T-cell activation, a key aspect of the adaptive immune response (reviewed in Powell et al., 2012). Mechanistically, mTORC1 facilitates the switch towards anabolic metabolism that is required for T-cell activation and expansion, and lies downstream of several activating signals present in the immune microenvironment including IL-2, the co-stimulatory receptor CD28, as well as amino acids. Interestingly, mTORC1 inhibition during antigen presentation results in T-cell anergy, whereby cells fail to activate upon subsequent antigen exposure (Zheng et al., 2007). As the induction of T-cell anergy via nutrient depletion or other inhibitory signals is a mechanism utilized by tumors in immune evasion, these data suggest that promoting mTORC1 activation in immune cells may actually be beneficial in some contexts, such as cancer immunotherapy.
Recent studies have also found a role for mTORC1 in influencing T-cell maturation, as rapamycin promotes the differentiation and expansion of CD4+FoxP3+ Regulatory T-cells and CD8+ memory T-cells while suppressing CD8+ and CD4+ effector T-cell populations (Araki et al., 2009; Haxhanisto et al., 2008), consistent with the metabolic profiles of these cell types. Indeed, a recent report found that during the asymmetric division of activated CD8+ T-cells, high mTORC1 activity is high in the “effector-like” daughter cell, but low in the “memory-like” daughter cell, due to the asymmetric partitioning of amino acid transporters (Polizzi et al., 2016; Verbist et al., 2016). Thus, the role of mTOR signaling in the immune system is clearly more complex than previously thought. Given the current clinical use of mTOR inhibitors in both immunosuppression and cancer, a more comprehensive understanding of how mTOR signaling influences overall immune responses in vivo will be a critical goal going forward.
Brain function
mTOR has also emerged as an important regulator of numerous neurological processes including neural development, circuit formation, and the neural control of feeding (reviewed in Lipton and Sahin, 2014). The deletion of either Raptor or Rictor in neurons causes reduced neuron size, and early death, suggesting that signaling by both mTORC1 and mTORC2 is important for proper brain development. Conversely, the impact of hyperactive mTORC1 signaling in the brain is best observed in human patients with Tuberous Sclerosis Complex (TSC), who exhibit a range of debilitating neurological disorders including epilepsy, autism, and the presence of benign brain tumors.
The fact that mTORC1 hyperactivation in TSC patients correlates with a high occurrence of epileptic seizures (90% of TSC patients) and autistic traits (50%) suggests that deregulated mTORC1 signaling may also be involved in epilepsy and autism more generally. Indeed, mTORC1 hyperactivation in mice through neural loss of Tsc1 or Tsc2 leads to severe epileptic seizures that are prevented by rapamycin treatment (Zeng et al., 2008), and mutations in components of the GATOR1 and KICSTOR complexes have been linked to epilepsy in humans (Basel-Vanagaite et al., 2013; Ricos et al., 2016).
The importance of mTORC1 in this tissue stems in part from its role in promoting activity-dependent mRNA translation near synapses, a critical step in neuronal circuit formation. Consistent with this, the NMDA receptor antagonist ketamine acutely activates mTORC1 signaling in mouse neurons, which coincides with an increased translation of synaptic proteins (Li et al., 2010). The role of mTORC1 in regulating autophagy is likely also important, as autophagy dysfunction is strongly implicated in the pathogenesis of neurodegenerative disorders including Parkinson’s disease and Alzheimer’s disease (AD). Inhibiting mTOR signaling has beneficial effects on mouse models of AD (Spilman et al., 2010), and it remains to be seen whether similar results will be seen in humans.
mTOR in Cancer
As discussed above, mTORC1 functions as a downstream effector for many frequently mutated oncogenic pathways including the PI3K/Akt pathway as well as the Ras/Raf/Mek/Erk (MAPK) pathway, resulting in mTORC1 hyperactivation in a high percentage of human cancers (Fig. 5A). Furthermore, the common tumor suppressors TP53 and LKB1 are negative regulators of mTORC1 upstream of TSC1 and TSC2, which are also tumor suppressors originally identified through genetic analysis of the familial cancer syndrome TSC. Several components of the nutrient sensing input to mTORC1 have also been implicated in cancer progression, including all three subunits of the GATOR1 complex, which are mutated with low frequency in glioblastoma (Bar-Peled et al., 2013), as well as RagC, which was recently found to be mutated at high frequency (~18%) in follicular lymphoma (Okosun et al., 2015). Additionally, mutations in the gene encoding folliculin (FLCN) are the causative lesion in the Birt-Hogg-Dube hereditary cancer syndrome (Nickerson et al., 2002), which manifests similarly to TSC. Finally, mutations in MTOR itself are also found in a variety of cancer subtypes, consistent with a role for mTOR in tumorigenesis (Grabiner et al., 2014; Sato et al., 2010).
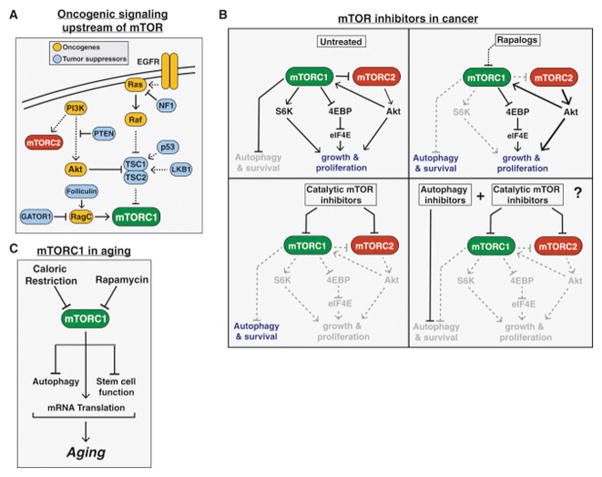
Figure 5. mTOR in Cancer and Aging.
(A) The common tumor suppressors and oncogenes upstream of mTORC1 leading to increased mTORC1 signaling in a wide variety of cancers.
(B) The varying effects of rapalogs, catalytic mTOR inhibitors, a combination of an mTOR inhibitor and autophagy inhibitor on cancer proliferation and survival.
(C) The role of mTORC1 signaling in aging.
mTORC2 signaling is also implicated in cancer largely due to its role in activating Akt, which drives pro-proliferative processes such as glucose uptake and glycolysis while also inhibiting apoptosis. Indeed, at least some PI3K/Akt driven tumors appear to rely on mTORC2 activity, as Rictor is essential in mouse models of prostate cancer driven by PTEN loss, as well as in human prostate cancer cell lines that lack PTEN (Guertin et al., 2009; Hietakangas et al., 2008).
While many mTORC1-driven processes likely contribute to tumorigenesis, the translational program initiated by the phosphorylation of 4EBP is likely the most critical, at least in mouse models of Akt-driven prostate cancer and T-cell lymphoma (Hsieh et al., 2010 & 2012). Consistent with this, a variety of Akt and Erk driven cancer cell lines are dependent on 4EBP phosphorylation, and the ratio of 4EBP to eIF4E expression correlates well with their sensitivity to mTOR inhibitors (Alain et al., 2012; She et al., 2010).
The first mTOR inhibitors approved for use in cancer were a class of rapamycin derivatives known as “rapalogs”. The rapalog temsirolimus (Pfizer) was first approved for treatment of advanced renal cell carcinoma in 2007, followed by everolimus (Novartis) in 2009. Although a small number of “extraordinary responders” have been reported, these rapalogs have been less successful in the clinic than anticipated from pre-clinical cancer models.
Several explanations for this lack of efficacy have been suggested. The first came with the realization that, as allosteric inhibitors, the rapalogs block the phosphorylation of some but not all mTORC1 substrates (Fig. 5B, Choo et al., 2008; Feldman et al., 2009; Kang et al., 2013; Thoreen et al., 2009). In particular, the phosphorylation of 4EBP is largely insensitive to rapamycin. Second, inhibiting mTORC1 releases the negative feedback on insulin/PI3K/Akt signaling, and therefore may paradoxically promote cell survival and prevent apoptosis in some contexts. Indeed, increased Akt signaling has been observed in biopsies of cancer patients following everolimus treatment, and may help explain why rapalogs have largely cytostatic, but not cytotoxic, effects on tumors (Tabernero et al., 2008). Finally, mTORC1 inhibition also induces autophagy, which can help maintain cancer cell survival poorly vascularized, nutrient poor microenvironments such as in pancreatic tumors. Indeed, mTORC1 inhibition also promotes macropinocytosis, whereby extracellular proteins are internalized and degraded to provide amino acids for nutrient-starved tumors (Palm et al., 2015). These data suggest that combining rapalogs with autophagy inhibitors may improve efficacy, consistent with a recent phase 1 clinical trial using temsirolomus and the autophagy inhibitor hydrochloroquine in melanoma patients, which showed an improvement over temsirolomus alone (Rangwala et al., 2014).
In order to address some of the drawbacks of the rapalogs, “second generation,” ATP-competitive catalytic inhibitors against mTOR have also been developed and are now in clinical trials. Unlike rapamycin, these compounds directly inhibit the catalytic activity of mTOR and therefore fully suppress both mTORC1 and mTORC2 (Fig. 5B), making them more effective than rapalogs in a variety of preclinical cancer models. Although these second-generation mTOR inhibitors initially suppress Akt signaling due to inhibition of mTORC2, the release of negative feedback on Insulin/PI3K signaling eventually overcomes this blockade and Akt is reactivated following long-term treatment (Rodrik-Outmezguine et al., 2011). One possible solution to this problem may be the utilization of dual PI3K/mTOR inhibitors, which inhibit the closely related catalytic domains of both PI3K and mTOR, thereby more fully blocking phosphorylation of both Akt and 4EBP (Fig. 5B). These inhibitors have also shown some promise in preclinical and early clinical trial data, but have raised concerns over dose-limiting toxicities.
The cases where rapalogs have had the most success to date have generally involved mutations in the mTOR pathway itself, such as in TSC1 or MTOR (Iyer et al., 2012; Wagle et al., 2014a). Even in these cases, however, an exquisite initial response has been followed by additional mutations in the kinase or FRB domains of MTOR, leading to acquired resistance (Wagle et al., 2014b). One creative way to overcome these resistance mutations may be with the recently described “third-generation” mTOR inhibitor called “RapaLink,” in which the ATP-competitive mTOR inhibitor is chemically linked to rapamycin, enabling inhibition of mTOR mutants that are resistant to either MLN0128 or rapamycin alone (Rodrik-Outmezguine et al., 2016).
mTOR in Aging
mTOR signaling is strongly implicated in the aging process of diverse organisms including yeast, worms, flies, and mammals. This was first observed through studies in the nematode C. elegans, which found that reduced expression of the homologs of mTOR (ceTOR, formerly let-363) or Raptor (daf-15) extend life span (Vellai et al., 2003; Jia et al., 2004). Subsequent genetic studies found that reduced TOR signaling also promotes longevity in Drosophila (Kapahi et al., 2004), budding yeast, (Kaeberlin et al., 2005) as well as mice (Lamming et al., 2012; Wu et al., 2013). Consistent with this, the mTOR inhibitor rapamycin is currently the only pharmacological treatment proven to extend life span in all of these model organisms (Bjedov et al., 2010; Harrison et al., 2009; Powers et al., 2006; Robida-Stubbs et al., 2012).
The only other intervention shown to extend lifespan in such a wide range of organisms is caloric restriction (CR), defined as a reduction in nutrient intake without malnutrition. Given the critical role of mTORC1 in sensing nutrients and insulin, this has led many to speculate that the beneficial effects of CR on life span are also due to reduced mTORC1 signaling. Indeed, CR-like regimens do not further extend life span in yeast, worms, or flies with reduced mTOR signaling, suggesting an overlapping mechanism (Kaeberlin et al., 2005; Hansen et al., 2007; Kapahi et al., 2004).
While there is now a general consensus that mTOR signaling plays a key role in mammalian aging, the mechanism through which this occurs is still unclear. Several lines of evidence suggest that the general reduction in mRNA translation during mTORC1 inhibition slows aging by reducing the accumulation of proteotoxic and oxidative stress, consistent with the observation that loss of the mTORC1 substrate S6K1 also extends life span in mammals (Selman et al., 2009). A related possibility is that inhibition of mTORC1 slows aging by increasing autophagy, which helps clear damaged proteins and organelles such as mitochondria, the accumulation of which are also associated with aging and aging-related diseases. Finally, another model suggests that the attenuation of adult stem cells in various tissues plays a central role in organismal aging, and mTOR inhibition boosts the self-renewal capacity of both hematopoietic and intestinal stem cells in mice (Chen et al., 2009; Yilmaz et al., 2012). Ultimately, the importance of mTORC1 signaling in aging likely reflects its unique capacity to regulate such a wide variety of key cellular functions (Fig 5C).
The observation that mTOR inhibition extends life span and delays the onset of age-associated diseases in mammals has led many to speculate that mTOR inhibitors could be used to enhance longevity in humans. The major drawback of prolonged rapamycin treatment in humans however is the potential for side effects such as immunosuppression and glucose intolerance. There is reason for optimism however, as a trial in healthy elderly humans using everolimus showed safety and even improved immune function (Mannick et al., 2014), and alternative dosing regimens have been proposed that can promote longevity with reduced side effects (Arriola Apelo et al., 2016). Given that many of the negative metabolic side effects associated with mTOR inhibitors are due to inhibition of mTORC2, while the anti-aging effects are due to inhibition of mTORC1, the development of mTORC1 specific inhibitors would be particularly beneficial.
Perspectives
It is now clear that the mTOR pathway plays a central role in sensing environmental conditions and regulating nearly all aspects metabolism at both the cellular and organismal level. In just the last several years, many new insights into both mTOR function and regulation have been elucidated, and extensive genetic and pharmacological studies in mice have enhanced our understanding of how mTOR dysfunction contributes to disease.
While many inputs to mTORC1 have now been identified and their mechanisms of sensing characterized, an integrated understanding of the relative importance of these signals and the contexts in which they are important remains largely unclear. For example, it remains mysterious which inputs to the TSC complex are dominant and how this depends on the physiological context. Similar questions exist for nutrient sensing by the Rag GTPases, specifically regarding the purpose of sensing of both lysosomal and cytosolic amino acids, as well as the tissues in which the nutrient sensors such as Sestrin2, CASTOR1, and SLC38A9 are most important. Such insights will likely require both deeper biochemical studies of these complexes in vitro as well as improved mouse models that enable more nuanced perturbation and monitoring of these sensors in vivo.
The major focus of mTOR research going forward however will be to address whether these molecular insights can improve the therapeutic targeting of mTOR in the clinic. Although rapalogs and catalytic mTOR inhibitors have been successful in the context of immunosuppression and a small subset of cancer types, clear limitations have arisen which limit their utility. Specifically, given the critical functions of mTOR in most human tissues, complete catalytic inhibition causes severe dose-limiting toxicities, while rapalogs also suffer from the drawbacks associated with lack of tissue specificity and unwanted disruption of mTORC2. Future work should focus on the development of mTOR-targeting therapeutics outside of these two modalities, such as truly mTORC1-specific inhibitors for use in diabetes, neurodegeneration, and life-span extension, or tissue-specific mTORC1 agonists for use in muscle wasting diseases and immunotherapy. Such approaches will likely require going beyond targeting mTOR directly to instead developing compounds that modulate tissue-specific receptors or signaling molecules upstream of mTOR, such as the recently characterized amino acid sensors Sestrin2 and CASTOR1, which contain small-molecule binding pockets and specifically regulate mTORC1. Ultimately, such insights may enable the rational targeting of mTOR signaling to unlock the full therapeutic potential of this remarkable pathway.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, et al. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer research. 2012;72:6468–6476. doi: 10.1158/0008-5472.CAN-12-2395. [DOI] [PubMed] [Google Scholar]
- Anthony JC, Yoshizawa F, Anthony TG, Vary TC, Jefferson LS, Kimball SR. Leucine stimulates translation initiation in skeletal muscle of postabsorptive rats via a rapamycin-sensitive pathway. The Journal of nutrition. 2000;130:2413–2419. doi: 10.1093/jn/130.10.2413. [DOI] [PubMed] [Google Scholar]
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriola Apelo SI, Neuman JC, Baar EL, Syed FA, Cummings NE, Brar HK, Pumper CP, Kimple ME, Lamming DW. Alternative rapamycin treatment regimens mitigate the impact of rapamycin on glucose homeostasis and the immune system. Aging cell. 2016;15:28–38. doi: 10.1111/acel.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. Architecture of human mTOR complex 1. Science. 2016;351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- Baar K, Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. The American journal of physiology. 1999;276:C120–127. doi: 10.1152/ajpcell.1999.276.1.C120. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baretic D, Berndt A, Ohashi Y, Johnson CM, Williams RL. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nature communications. 2016;7:11016. doi: 10.1038/ncomms11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Hershkovitz T, Heyman E, Raspall-Chaure M, Kakar N, Smirin-Yosef P, Vila-Pueyo M, Kornreich L, Thiele H, Bode H, et al. Biallelic SZT2 mutations cause infantile encephalopathy with epilepsy and dysmorphic corpus callosum. American journal of human genetics. 2013;93:524–529. doi: 10.1016/j.ajhg.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJ, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell metabolism. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Molecular cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell metabolism. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nature cell biology. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes & development. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Cafferkey R, Young PR, McLaughlin MM, Bergsma DJ, Koltin Y, Sathe GM, Faucette L, Eng WK, Johnson RK, Livi GP. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Molecular and cellular biology. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M, Frank S, Tintignac LA, Sinnreich M, Ruegg MA. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell metabolism. 2013;17:731–744. doi: 10.1016/j.cmet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165:153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell reports. 2014;9:1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Science signaling. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Molecular cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CP, Sehgal SN, Vezina C. Activity of rapamycin (AY-22,989) against transplanted tumors. The Journal of antibiotics. 1984;37:1231–1237. doi: 10.7164/antibiotics.37.1231. [DOI] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS biology. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer research. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- Frey JW, Jacobs BL, Goodman CA, Hornberger TA. A role for Raptor phosphorylation in the mechanical activation of mTOR signaling. Cellular signalling. 2014;26:313–322. doi: 10.1016/j.cellsig.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Current biology: CB. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Gan X, Wang J, Wang C, Sommer E, Kozasa T, Srinivasula S, Alessi D, Offermanns S, Simon MI, Wu D. PRR5L degradation promotes mTORC2-mediated PKC-delta phosphorylation and cell migration downstream of Galpha12. Nature cell biology. 2012;14:686–696. doi: 10.1038/ncb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) The Biochemical journal. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer discovery. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, Mullholland DJ, Magnuson MA, Wu H, Sabatini DM. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Developmental cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell metabolism. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. The Journal of cell biology. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. The Journal of experimental medicine. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva N, Hall M. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM. TOR complex 2 is needed for cell cycle progression and anchorage-independent growth of MCF7 and PC3 tumor cells. BMC cancer. 2008;8:282. doi: 10.1186/1471-2407-8-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. The Biochemical journal. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes & development. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, Pirun M, Sander C, Socci ND, Ostrovnaya I, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nature cell biology. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, Tagliabracci VS, Guan KL. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347:194–198. doi: 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development (Cambridge, England) 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Jung J, Genau HM, Behrends C. Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Molecular and cellular biology. 2015;35:2479–2494. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B, Kemp BE, Bardeesy N, Dennis P, Schlager JJ, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell metabolism. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Current biology: CB. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamzina L, Veilleux A, Bergeron S, Marette A. Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology. 2005;146:1473–1481. doi: 10.1210/en.2004-0921. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Molecular cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nature cell biology. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kumar A, Harris TE, Keller SR, Choi KM, Magnuson MA, Lawrence JC., Jr Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances Basal glycogen synthase activity. Molecular and cellular biology. 2008;28:61–70. doi: 10.1128/MCB.01405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Lawrence JC, Jr, Jung DY, Ko HJ, Keller SR, Kim JK, Magnuson MA, Harris TE. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes. 2010;59:1397–1406. doi: 10.2337/db09-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Sabatini DM. A Central role for mTOR in lipid homeostasis. Cell metabolism. 2013;18:465–469. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PL, Tang Y, Li H, Guertin DA. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Molecular metabolism. 2016;5:422–432. doi: 10.1016/j.molmet.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, Li XY, Aghajanian G, Duman RS. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao T. mTORC2 phosphorylates protein kinase Czeta to regulate its stability and activity. EMBO reports. 2014;15:191–198. doi: 10.1002/embr.201338119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Gan W, Chin YR, Ogura K, Guo J, Zhang J, Wang B, Blenis J, Cantley LC, Toker A, et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer discovery. 2015;5:1194–1209. doi: 10.1158/2159-8290.CD-15-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Current biology: CB. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Mannick JB, Del Giudice G, Lattanzi M, Valiante NM, Praestgaard J, Huang B, Lonetto MA, Maecker HT, Kovarik J, Carson S, et al. mTOR inhibition improves immune function in the elderly. Science translational medicine. 2014;6:268ra179. doi: 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- Martel RR, Klicius J, Galet S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Canadian journal of physiology and pharmacology. 1977;55:48–51. doi: 10.1139/y77-007. [DOI] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Inoki K, Opland D, Munzberg H, Villanueva EC, Faouzi M, Ikenoue T, Kwiatkowski DJ, Macdougald OA, Myers MG, Jr, et al. Critical roles for the TSC-mTOR pathway in beta-cell function. American journal of physiology Endocrinology and metabolism. 2009;297:E1013–1022. doi: 10.1152/ajpendo.00262.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer cell. 2002;2:157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. The Journal of biological chemistry. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- Okosun J, Wolfson RL, Wang J, Araf S, Wilkins L, Castellano BM, Escudero-Ibarz L, Al Seraihi AF, Richter J, Bernhart SH, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nature genetics. 2016;48:183–188. doi: 10.1038/ng.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell. 2015;162:259–270. doi: 10.1016/j.cell.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud N, Peli-Gulli MP, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Science signaling. 2013;6:ra42. doi: 10.1126/scisignal.2004112. [DOI] [PubMed] [Google Scholar]
- Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan KL, Karin M, Budanov AV. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell reports. 2014;9:1281–1291. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. The Biochemical journal. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. The Journal of cell biology. 2013;202:1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Cybulski N, Feige JN, Auwerx J, Ruegg MA, Hall MN. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell metabolism. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Pollizzi KN, Sun IH, Patel CH, Lo YC, Oh MH, Waickman AT, Tam AJ, Blosser RL, Wen J, Delgoffe GM, et al. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8(+) T cell differentiation. Nature immunology. 2016;17:704–711. doi: 10.1038/ni.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell metabolism. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annual review of immunology. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes & development. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricos MG, Hodgson BL, Pippucci T, Saidin A, Ong YS, Heron SE, Licchetta L, Bisulli F, Bayly MA, Hughes J, et al. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Annals of neurology. 2016;79:120–131. doi: 10.1002/ana.24547. [DOI] [PubMed] [Google Scholar]
- Risson V, Mazelin L, Roceri M, Sanchez H, Moncollin V, Corneloup C, Richard-Bulteau H, Vignaud A, Baas D, Defour A, et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. The Journal of cell biology. 2009;187:859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell metabolism. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Science signaling. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer discovery. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de Stanchina E, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534:272–276. doi: 10.1038/nature17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nature cell biology. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- Rousseau A, Bertolotti A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536:184–189. doi: 10.1038/nature18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. The Journal of biological chemistry. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Molecular cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current biology: CB. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29:2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, Schwartz TU, Sabatini DM. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016a;351:53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016b;536:229–233. doi: 10.1038/nature19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Current biology: CB. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. The EMBO journal. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Current biology: CB. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, Solit DB, Rosen N. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeyama Y, Kobayashi T, Kido Y, Hashimoto N, Asahara S, Matsuda T, Takeda A, Inoue T, Shibutani Y, Koyanagi M, et al. Biphasic response of pancreatic beta-cell mass to ablation of tuberous sclerosis complex 2 in mice. Molecular and cellular biology. 2008;28:2971–2979. doi: 10.1128/MCB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, Richardson A, Strong R, Galvan V. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PloS one. 2010;5:e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, Martinelli E, Ramon y Cajal S, Jones S, Vidal L, et al. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:1603–1610. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- Tang Y, Wallace M, Sanchez-Gurmaches J, Hsiao WY, Li H, Lee PL, Vernia S, Metallo CM, Guertin DA. Adipose tissue mTORC2 regulates ChREBP-driven de novo lipogenesis and hepatic glucose metabolism. Nature communications. 2016;7:11365. doi: 10.1038/ncomms11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Current biology: CB. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jeno P, Arrieumerlou C, Hall MN. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PloS one. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomanetz V, Angliker N, Cloetta D, Lustenberger RM, Schweighauser M, Oliveri F, Suzuki N, Ruegg MA. Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. The Journal of cell biology. 2013;201:293–308. doi: 10.1083/jcb.201205030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. The Journal of biological chemistry. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Molecular cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nature cell biology. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Verbist KC, Guy CS, Milasta S, Liedmann S, Kaminski MM, Wang R, Green DR. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature. 2016;532:389–393. doi: 10.1038/nature17442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. The Journal of antibiotics. 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- Wagle N, Grabiner BC, Van Allen EM, Hodis E, Jacobus S, Supko JG, Stewart M, Choueiri TK, Gandhi L, Cleary JM, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer discovery. 2014a;4:546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Grabiner BC, Van Allen EM, Amin-Mansour A, Taylor-Weiner A, Rosenberg M, Gray N, Barletta JA, Guo Y, Swanson SJ, et al. Response and acquired resistance to everolimus in anaplastic thyroid cancer. The New England journal of medicine. 2014b;371:1426–1433. doi: 10.1056/NEJMoa1403352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Harris TE, Roth RA, Lawrence JC., Jr PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. The Journal of biological chemistry. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Wyant GA, Gu X, Orozoco JM, Condon KJ, Petri S, Kedir J, Scaria SM, Abu-Remaileh M, et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017 doi: 10.1038/nature21423. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SY, Kim DH, Jun CB, Kim YM, Haar EV, Lee SI, Hegg JW, Bandhakavi S, Griffin TJ, Kim DH. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. The Journal of biological chemistry. 2007;282:25604–25612. doi: 10.1074/jbc.M704343200. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell reports. 2013;4:913–920. doi: 10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Murashige DS, Humphrey SJ, James DE. A Positive Feedback Loop between Akt and mTORC2 via SIN1 Phosphorylation. Cell reports. 2015;12:937–943. doi: 10.1016/j.celrep.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes & development. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Palm W, Peng M, King B, Lindsten T, Li MO, Koumenis C, Thompson CB. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes & development. 2015;29:2331–2336. doi: 10.1101/gad.269324.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Molecular cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Pino E, Wu L, Kacergis M, Soukas AA. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. The Journal of biological chemistry. 2012;287:29579–29588. doi: 10.1074/jbc.M112.386854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LH, Xu L, Gutmann DH, Wong M. Rapamycin prevents epilepsy in a mouse model of tuberous sclerosis complex. Annals of neurology. 2008;63:444–453. doi: 10.1002/ana.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Peli-Gulli MP, Yang H, De Virgilio C, Ding J. Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1. Structure (London, England: 1993) 2012;20:2151–2160. doi: 10.1016/j.str.2012.09.019. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:15790–15797. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Collins SL, Lutz MA, Allen AN, Kole TP, Zarek PE, Powell JD. A role for mammalian target of rapamycin in regulating T cell activation versus anergy. Journal of immunology (Baltimore, Md: 1950) 2007;178:2163–2170. doi: 10.4049/jimmunol.178.4.2163. [DOI] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]