Background: PSII is a protein complex that captures sunlight to drive water oxidation.
Results: Cyanidioschyzon merolae PSII is protected by reversible reaction center-based non-photochemical quenching.
Conclusion: C. merolae PSII employs reaction center non-photochemical quenching as the main photoprotective mechanism.
Significance: We provide the first direct evidence of the PSII reaction center as the primary locus of non-photochemical quenching in the extremophilic red algae.
Keywords: Algae, Fluorescence, Photosynthesis, Photosynthetic Pigments, Photosystem II, Single Particle Analysis, Cyanidoschyzon merolae, Photoprotection
Abstract
Members of the rhodophytan order Cyanidiales are unique among phototrophs in their ability to live in extremely low pH levels and moderately high temperatures. The photosynthetic apparatus of the red alga Cyanidioschyzon merolae represents an intermediate type between cyanobacteria and higher plants, suggesting that this alga may provide the evolutionary link between prokaryotic and eukaryotic phototrophs. Although we now have a detailed structural model of photosystem II (PSII) from cyanobacteria at an atomic resolution, no corresponding structure of the eukaryotic PSII complex has been published to date. Here we report the isolation and characterization of a highly active and robust dimeric PSII complex from C. merolae. We show that this complex is highly stable across a range of extreme light, temperature, and pH conditions. By measuring fluorescence quenching properties of the isolated C. merolae PSII complex, we provide the first direct evidence of pH-dependent non-photochemical quenching in the red algal PSII reaction center. This type of quenching, together with high zeaxanthin content, appears to underlie photoprotection mechanisms that are efficiently employed by this robust natural water-splitting complex under excess irradiance. In order to provide structural details of this eukaryotic form of PSII, we have employed electron microscopy and single particle analyses to obtain a 17 Å map of the C. merolae PSII dimer in which we locate the position of the protein mass corresponding to the additional extrinsic protein stabilizing the oxygen-evolving complex, PsbQ′. We conclude that this lumenal subunit is present in the vicinity of the CP43 protein, close to the membrane plane.
Introduction
Photosystem II (PSII)3 is a multimeric transmembrane complex present in the thylakoid membranes of cyanobacteria, algae, and higher plants that is able to capture solar energy and use it to drive charge separation and water oxidation catalysis. Recently, the crystal structure of PSII isolated from the thermophilic cyanobacterium Thermosynechococcus vulcanus has been reported at a resolution of 1.9 Å (1). This atomic structure has facilitated formation of the working models for understanding the water-splitting reaction based on the previous crystallographic structures of cyanobacterial PSII (2–9). The cyanobacterial complex contains 20 protein subunits with a total molecular mass of 350 kDa. Bound within these subunits are over 1,300 water molecules and 85 cofactors: 35 chlorophyll (Chl) molecules, two pheophytins, 11 β-carotenes, over 20 lipids, two plastoquinones, two heme irons, one non-heme iron, four manganese atoms, 3–4 Ca2+ ions, three Cl− ions, and one bicarbonate (1). The catalytic site is composed of a Mn4Ca cluster surrounded by a number of highly conserved amino acids, mainly derived from the D1 reaction center (RC) subunit and one from the CP43 inner antenna subunit. The model that places three manganese ions and a calcium ion at the corner of a cubane with oxo-bridges and the fourth manganese linked to the cubane via a bridging oxygen (2) has gained support in the latest atomic structure of PSII (1) as well as from quantum mechanical and x-ray spectroscopic considerations (10–13).
Although we now have a detailed structural model of PSII from cyanobacteria at an atomic resolution, no corresponding structure of the eukaryotic PSII complex has been published to date. Hence, there is an urgent need to purify a highly stable eukaryotic PSII complex that would prove suitable for high resolution structural analysis. Equally important, PSII as nature's water-splitting enzyme provides a blueprint in terms of the catalytic rate of water oxidation in ambient conditions for the construction of stable synthetic catalysts operating within solar-to-fuel nanodevices (14–17). A logical approach toward these goals is to obtain a eukaryotic form of PSII using an organism that is likely to provide a robust and highly active form of the complex. To this end, we isolated and characterized the dimeric PSII complex from the extremophilic unicellular red alga Cyanidioschyzon merolae.
C. merolae belongs to the rhodophytan order Cyanidiales, whose members thrive in acidic hot springs (18) and are unique among phototrophs in the ability to live at extremely low pH (pH 0.2–4) and moderately high temperatures (40–56 °C). Despite such an extremely acidic environment, the intracellular pH of these algae is most likely neutral due to the active H+ efflux across the plasma membrane (19). C. merolae is considered to be one of the most primitive eukaryotic phototrophs because it diverged near the root of the red algal lineage that forms a basal group within the photosynthetic eukaryotes (20). The photosynthetic apparatus of this alga is regarded as the closest equivalent of the prokaryotic ancestor of the present day chloroplast (21). As an evolutionary intermediate between the photosynthetic apparatus of prokaryotic cyanobacteria and that of the eukaryotes in the green lineage, it contains a mixture of prokaryotic and eukaryotic structural traits. Whereas red algal photosystem I (PSI) resembles a higher plant complex, with an crescent-shaped Chla-binding light-harvesting antenna system asymmetrically bound on one side of the core complex (22–23), PSII of red algae is structurally similar to the cyanobacterial counterpart in that it contains light-harvesting antenna composed of phycobilisomes, large peripheral membrane complexes formed by phycobiliproteins, instead of Chla/b-binding antenna proteins that constitute the light-harvesting system of green algae and higher plants.
The oxygen-evolving complex (OEC) in C. merolae PSII is stabilized by four extrinsic lumenal subunits: cyanobacteria-like PsbV and PsbU, the evolutionarily conserved PsbO subunit, and an additional 20-kDa subunit PsbQ′ distantly related to the higher plant and green algal PsbQ polypeptides (24). The precise localization of this subunit and its role in stabilization of the OEC are currently unknown, although low resolution single particle analysis and in vitro reconstitution experiments suggested that the red algal PsbQ′ may independently associate with the PSII core complex in the close vicinity of PsbV and the PsbU subunits and is required for effective binding of these extrinsic subunits (22, 25). In contrast, PsbQ in higher plants functionally associates with PSII via its direct interaction with both PsbO and PsbP extrinsic subunits, stabilizing the OEC (reviewed in Ref. 26). Interestingly, PsbQ′ can functionally replace PsbQ in spinach during cross-reconstitution experiments despite the low amino acid sequence homology between both proteins (25). The binding mode for another OEC subunit, PsbV, also differs between cyanobacteria and red alga. In red algae, PsbV binds via other extrinsic subunits, whereas its cyanobacterial counterpart binds directly with the PSII core (reviewed in Ref. 26). All of these lines of evidence point toward significant structural differences on the lumenal side of the PSII complex that was formed at various evolutionary stages.
It is well established that under conditions of excessive irradiation (i.e. when light energy absorption exceeds the capacity and demands of photosynthesis), several photoprotective and optimizing mechanisms are triggered. These mechanisms include regulation of light absorption capacity between PSI and PSII (state transitions; see Ref. 27), fast D1 protein turnover during photoinhibition of PSII, and photoprotective non-photochemical quenching (see Refs. 28 and 29 for recent reviews). Non-photochemical quenching (NPQ) represents a feedback regulatory mechanism that leads to dissipation of excessive light either in the light-harvesting antennae (29) or in the reaction center of PSII (30). The most flexible and dominant component of NPQ, ΔpH-dependent high energy quenching (qE) is a major photoprotective strategy that operates on a time scale of seconds to minutes.
Although mechanistic aspects and molecular components of pH-dependent qE have been widely studied in higher plants and green algae, little is known about molecular mechanisms of PSII fluorescence quenching in phycobilisome-containing red algae. In the red algae ancestor, prokaryotic cyanobacteria, excessive light is dissipated in phycobilisomes, aided by the orange carotenoid protein that is absent in red algae (31). The detailed NPQ mechanisms in red algae are largely unknown, and the NPQ properties are mostly derived from in vivo fluorescence measurements in intact cells. Kirilovsky and colleagues (32, 33) demonstrated the existence of ΔpH-dependent quenching in Rhodella violacea and Porphyridium cruentum strains of mesophylic red algae. These authors have suggested that, in contrast to higher plants and green algae, the dominant part of NPQ in red algae might occur in the RC rather than in the antenna (33). Several photochemical mechanisms have been suggested for the RC type of quenching (reviewed in Ref. 34). Although the real importance and extent of RC quenching in photoprotection is still a matter of debate, especially in comparison with qE in antennae (compare Refs. 29 and 30), the existence of this type of quenching has been proved for PSII isolated from various photothrophs (35, 36). However, the same direct evidence has been missing for red algae.
Here we report a detailed structural and functional analysis of a highly active and robust dimeric PSII complex isolated from C. merolae. We provide the first direct evidence that the red algal PSII complex is rich in zeaxanthin and employs reversible RC-based non-photochemical quenching that is triggered by low pH. These features provide the basis for the remarkable robustness of this complex across a range of extreme light, temperature, and pH conditions. In the first attempt to provide structural details of this eukaryotic form of PSII, we have employed electron microscopy (EM) and single particle analyses to produce a two-dimensional electron density map in which we locate the position of the additional protein mass over and above that found in cyanobacterial PSII. We attribute this mass to the PsbQ′ subunit, which binds to the OEC in the vicinity of the CP43 protein, close to the membrane plane.
MATERIALS AND METHODS
Cell Culturing and Isolation of Thylakoids
C. merolae strain NIES-1332 was obtained from the Microbial Culture Collection of the National Institute for Environmental Studies (Tsukuba, Japan). Liquid cultures of the C. merolae cells were grown in a Versatile Environmental Test Chamber (Sanyo, Japan) in standard 250-ml tissue flasks oriented in an upright position. Cells were suspended in 50 ml of Allen 2 medium, pH 2.5 (37), at 42 °C under continuous 90-μE white light illumination with shaking at 115 rpm. Small scale cultures were grown to OD680 ∼2.5 and then subcultured into 1-liter interim cultures and finally grown as 10-liter cultures in Allen 2 medium until late log phase (until OD682 3.5) in the presence of 5% CO2 administered at a constant flow rate of 3 liters/min under 150-μE continuous white light illumination. For thylakoid preparation, cells were harvested at OD680 ∼2.5 by centrifugation at 5,000 rpm at 4 °C for 10 min. Cell pellets were washed with 50–100 ml of low ionic strength buffer A (40 mm MES-KOH, pH 6.1, 10 mm CaCl2, 5 mm MgCl2, 25% (w/v) glycerol) and then resuspended in 50 ml of buffer A supplemented with 50 μg/ml DNase I and the CompleteTM protease inhibitor mixture (Roche Applied Science). Cells were ruptured by vigorous agitation with 0.1-mm glass beads in a BeadBeater (BioSpec) using 13 cycles of 10 s and interim 4-min cooling off periods. Cell lysate was separated from beads by vacuum filtration. Thylakoids were pelleted by centrifugation at 104,200 × g for 30 min at 4 °C and washed once with buffer A. Final thylakoid pellets were resuspended in buffer A at a Chl concentration of 2–2.5 mg/ml, snap-frozen in liquid N2, and stored at −70 °C prior to use.
Purification of PSII
Purification of the dimeric PSII complex was performed according to a modified protocol of Adachi et al. (38). Thylakoids (1 mg/ml Chl) were solubilized with 1% (w/v) dodecyl-β-d-maltoside (DDM; Biomol) at 4 °C for 40 min in the dark. Solubilized membranes were centrifuged at 104,200 × g for 30 min at 4 °C, and the filtered supernatants were applied onto a DEAE TOYOPEARL 650 M column equilibrated with buffer A (40 mm MES-KOH, pH 6.1, 10 mm CaCl2, 5 mm MgCl2, 25% (w/v) glycerol) supplemented with 0.03% DDM. The loaded column was washed with 2 column volumes of the medium ionic strength buffer (0.09 m NaCl, 40 mm MES-KOH, pH 6.1, 3 mm CaCl2, 25% (w/v) glycerol, 0.03% (w/v) DDM), and crude PSII was eluted with the high ionic strength buffer (0.23 m NaCl, 40 mm MES-KOH, pH 6.1, 3 mm CaCl2, 25% (w/v) glycerol, 0.03% (w/v) DDM). The PSII-containing fractions were pooled and dialyzed overnight in 5 liters of buffer A containing 0.03% DDM at 4 °C in the dark. Crude PSII was loaded onto a DEAE TOYOPEARL 650 S column equilibrated with buffer A supplemented with 0.03% DDM. After washing the column with 3 column volumes of the wash buffer (0.05 m NaCl, 40 mm MES-KOH, pH 6.1, 3 mm CaCl2, 25% (w/v) glycerol, 0.03% DDM), pure PSII (devoid of residual free carotenoids and phycobilisomes) was eluted with a continuous NaCl gradient (0.05–0.15 M NaCl, 40 mm MES-KOH, pH 6.1, 3 mm CaCl2, 25% (w/v) glycerol, 0.03% DDM) to separate PSII monomers and dimers. Fractions containing PSII dimers were identified by size exclusion chromatography (SEC), using a Biosep SEC-4000 column (Phenomenex, Torrance, CA) equilibrated with a carrier buffer (20 mm MES-KOH, pH 6.5, 10 mm MgCl2, 3 mm CaCl2, 0.5 m mannitol, 0.05% DDM) at 2 ml/min. PSII dimer fractions were pooled and then concentrated using VivaSpin-20 (100,000 molecular weight cut-off) concentrators (Sartorius Stedim) to at least 2 mg/ml Chl and stored upon snap freezing at −70 °C until further use.
PSII Activity Measurement
Functional activity of the isolated PSII (5 μg of Chl) was measured using an oxygen Clark-type electrode (Hansatech). Measurements were performed at 30 °C in buffer A in the presence of 0.125 mm 2,6-dichloro-p-benzoquinone (Sigma) and 2.5 mm potassium ferricyanide (POCH, Gliwice, Poland) as the exogenous electron acceptors. Samples (1 μg/ml Chl) were illuminated with a standard white light intensity of 6,000 μE/m2/s or as described, using a KL 2500 LCD white light source (Schott, Mayence, Germany). Activities were calculated from initial rates of oxygen evolution curves. The values of light intensity (600–25,000 μE/m2/s), temperature (15–55 °C), and buffer pH (3–7.5) were permutated, depending on the experimental settings. The herbicide sensitivity of the QB site was monitored following preincubation of PSII for 1 min in the dark with 0.001–20 μm 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU; Sigma) prior to oxygen evolution measurement under standard conditions. Each measurement was repeated three times for each DCMU concentration.
Carotenoid Content Analysis
Analytical HPLC was performed according to a modified method described before (39), using a maximum flow rate of 1 ml/min and a Nucleosil 100 C18 column (Teknokroma, Barcelona, Spain). Pigments were extracted from cells grown at 90 μE/m2/s and the corresponding PSII samples (0.5 mg Chl) with a 1-ml acetone/methanol (7:2, v/v) mixture. The extract was concentrated to 1 mg/ml Chl, and samples (20 μg of Chl) were analyzed on a C18 column. The content of each carotenoid species was expressed as a ratio of the area under the pigment-corresponding peak to the area under the Chl peak. Pigment molar ratios were calculated using extinction coefficients of 83.2, 91.7, and 125.3 mm−1 cm−1 for zeaxanthin, Chla, and β-carotene, respectively (40).
LC-MS/MS Identification of PSII Proteins
PSII proteins after acetone precipitation were dissolved in 0.1% (w/v) RapiGestTM surfactant (Waters) in 50 mm ammonium bicarbonate. After reduction and subsequent alkylation of cysteine residues, proteins were digested in sequencing grade trypsin (Sigma-Aldrich) at 30 °C overnight. Reaction was stopped by the addition of trifluoroacetic acid to 1% (v/v) final concentration. For small PSII subunits that do not contain trypsin cleavage sites, additional digestion in solution as well as in-gel digestions with chymotrypsin (Sigma-Aldrich) were carried out. Digested peptides were separated using a NanoAcquity Ultra Performance LC system (Waters) connected with a mass spectrometer. Peptides were loaded onto a Symmetry® C18 (5 μm; 180 μm × 20 mm) trap column (Waters) at a flow rate of 10 μl/min in 99% buffer A (0.1% formic acid in water) and 1% buffer B (0.1% formic acid in acetonitrile) for 3 min. Trapped peptides were separated on a BEH 130 C18 (1.7 μm; 75 μm × 200 mm) analytical column (Waters) equilibrated in 97% buffer A and 3% buffer B. The column was eluted at a constant flow rate of 300 nl/min at 35 °C with a linear gradient of buffer B distributed as follows: 3–40% B in the first 145 min; 40–85% B in 145–151 min; and 85% B in 151–165 min. In the next 5 min, concentration of buffer B was decreased from 85 to 3%, and the column was equilibrated for an additional 10 min before the next injection. Online MSE analyses were performed in a positive ionization mode on the Synapt G2 HDMS mass spectrometer (Waters). Fragmentation spectra were recorded in the range of 50–2,000 Da (1.0 s/single scan), and the transfer collision energy was ramped in the range of 15–35 V. The mass accuracy of the raw data was corrected using leucine enkephalin (2 ng/μl, 1 μl/min flow rate, 556.2771 Da/e [M + H]+) that was infused into the mass spectrometer as a lock mass during sample analysis. Each sample was analyzed at least three times and was mixed with bovine serum albumin tryptic digest (60 fmol) as an internal standard. For protein identification, peak lists were created from the raw data sets and used to search for proteins in a randomized C. merolae protein data bank using ProteinLynx Global Server version 2.4 software (Waters).
77 K Fluorescence Measurement
Steady-state fluorescence spectra were collected using a modified Shimadzu RF-5301PC spectrofluorometer (41) at 77 K and excitation wavelengths of 440 and 580 nm. PSII samples (10 μg/ml Chl) in a buffer composed of 40 mm MES, pH 6.1, 10 mm CaCl2, 5 mm MgCl2, 25% (w/v) glycerol, and 0.03% (w/v) DDM were loaded into prechilled cuvettes and then frozen in liquid nitrogen for 5 min in the dark prior to collecting emission spectra.
Room Temperature Absorbance and Redox Spectroscopy
All measurements were carried out at room temperature using a Shimadzu UV 1800 spectrophotometer. To estimate the content of cytochrome c550 and cytochrome b559, redox difference spectra (oxidized − reduced) were calculated. For redox spectroscopy, the PSII dimer was diluted to 0.01 mg/ml Chl. 1 ml of the sample was used to prepare the base line in the wavelength range between 600 and 500 nm. Subsequently, a grain of potassium ferricyanide (K3[Fe(CN)6]; POCH) was mixed with the sample for 30 s to oxidize the total cytochrome pool prior to collection of the oxidized spectra. For the reduction of cytochromes, a 1-ml sample from the same PSII suspension was mixed with a grain of sodium dithionite (Na2S2O4; POCH) for 30 s. prior to collection of the reduced absorbance spectra. To ensure complete oxidation or reduction of cytochromes, an additional grain of ferricyanide or dithionite was added before reacquiring the oxidized and reduced spectra, respectively. The cytochrome c550 concentration was calculated from the maximal absorption at 550 nm and the extinction coefficient of 27.0 mm−1 cm−1 (42). For calculation of the cytochrome b559 content, maximum absorbance at 557 nm and an extinction coefficient of 25.1 mm−1 cm−1 were used (42). Each spectrum was recorded three times.
In Vivo Chlorophyll a Fluorescence and Non-photochemical Quenching
An FL-100 fluorometer (Photon Systems Instruments, Brno, Czech Republic) has been used for measurements of NPQ of maximal fluorescence in vivo. Fluorescence signal was detected in the 690–750 nm range. Maximal fluorescence of dark-adapted cells (FM) was measured at a saturating flash (466 nm; Δt = 500 ms; ∼2,000 μE/m2/s) applied before and after a short period of low intensity blue light (466 nm; duration 60 s; intensity 7 μE/m2/s). The same setup has been used to exclude the effect of state transitions on the FM value during phycobilisome binding in cyanobacteria (31). NPQ was induced by blue actinic light (466 nm; Δλ, 20 nm; 750 μE/m2/s, duration 150 s). NPQ value was calculated from quenching of maximal fluorescence in light (FM′) at a saturation flash (466 nm; Δt = 500 ms; ∼2,000 μE/m2/s), according to the Stern-Volmer formula, NPQ = (FM − FM′)/FM′. Recovery from the NPQ state was measured in the dark following a given light period. Cells used for in vivo measurements were cultivated in the thermostated bioreactor (38 °C) bubbled with air, at a continuous irradiation of 80 μE/m2/s.
Variable Fluorescence and Effective Antenna Size Measurement in Vitro
Variable fluorescence (FV) and the effective antennae size of pure PSII (σPSII) were measured with a custom designed fluorometer FL3500 (Photon Systems Instruments) equipped with a fast repetition rate fluorescence protocol. The single-turnover flash was induced by application of a series of short subsaturating flashes (1.5 μs long, λ = 463 nm) to dark-adapted samples (e.g. see Ref. 43). The measured fluorescence rise during the single turnover flash was fitted according to the model described before (44), giving the effective PSII cross-section parameter σPSII (43). Variable fluorescence (FV) was calculated as a difference between maximum (FM) and minimal (F0) fluorescence measured before (F0) and after (FM) application of saturating flashes. For measurements at various pH values, purified dimeric PSII samples (0.15–0.2 μg Chl/ml) were resuspended in the thermostated (22 °C) buffer containing 40 mm MES, 10 mm CaCl2, 5 mm MgCl2, 25% (w/v) glycerol, and 0.03% (w/v) DDM, in a pH range of 3–7.5. Samples were kept in the dark for 2 min in a buffer of given pH prior to the fluorescence measurement. The reversibility of pH-induced changes was tested by incubating the concentrated sample for 2 min in a buffer of particular pH, followed by transfer to pH 7 by the addition of an appropriate amount of KOH. After a 4-min recovery at pH 7, the sample was used for fluorescence measurements. In all measurements, 1,4-benzoquinone (22.6 μm) and potassium ferricyanide (48.8 μm) were used as the exogenous electron acceptors of PSII.
SDS-Polyacrylamide Gel Electrophoresis
SDS-PAGE was carried out using the Tris-Tricine system (45, 46). Protein bands (5 μg of Chl/lane) were resolved overnight on an 18% polyacrylamide gel in the presence of 6 m urea at a constant voltage of 25 V. Proteins were visualized with Coomassie Brilliant Blue R-250 using standard procedures. Prior to SDS-PAGE, samples were treated with 4 volumes of ether/methanol (1:1, v/v) overnight at −20 °C to remove pigments. Total protein was collected by centrifugation at 13,500 rpm at 4 °C for 10 min, air-dried at room temperature, and then resuspended in 200 μl of 1× SDS-PAGE sample buffer (12% (w/v) SDS, 30% (w/v) glycerol, 0.05% (w/v) Coomassie Blue G-250, 150 mm Tris-HCl, pH 7.0).
Electron Microscopy
Protein samples were negatively stained with 2% uranyl acetate on glow-discharged carbon-coated copper grids. Images were recorded on a Philips CM120 electron microscope with a LaB6 filament, operating at 120 kV, with a Gatan 4000 SP 4K slow scan CCD camera. The magnification used was ×133,000, compatible with a pixel size (after binning the images) of 2.25 Å at the specimen level. GRACE software was used for semiautomated data acquisition (47). From about 2,000 recorded EM images, over 20,000 particle projections were selected. Processing of single particles was performed with the Groningen image-processing (Grip) software package, including multireference alignments, multivariate statistical analysis, and classification. The resolution of the final EM maps was determined with Fourier ring correlation and the 3σ criterion (48) and was calculated at 17 Å.
RESULTS
Purification and Compositional Analysis of the C. merolae PSII Dimer
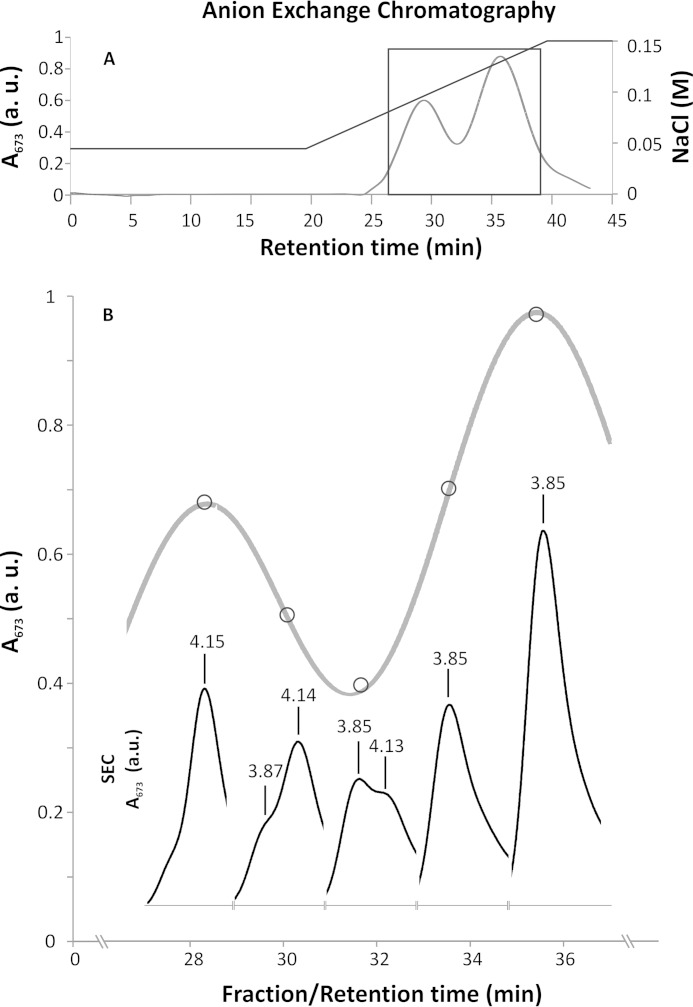
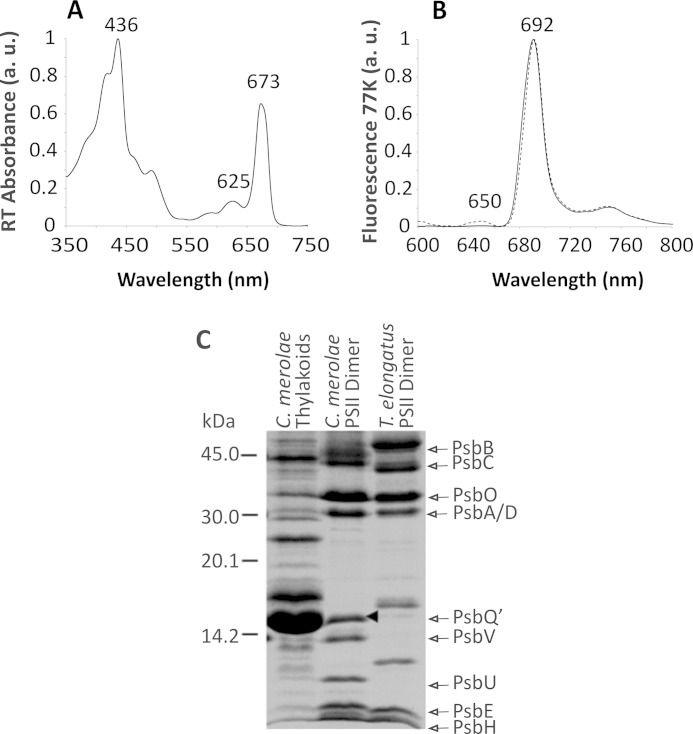
We set out to purify a highly active, intact, and homogenous preparation of the C. merolae dimeric PSII with the aim of examining its capacity to adapt to various extreme conditions as well as obtaining the insight into the eukaryotic PSII molecular structure. Following solubilization of thylakoids with DDM, we separated PSII monomers and dimers using a two-step anion exchange chromatography (AEC) approach (see Fig. 1A). In the second step, we purified a robust and highly active PSII dimer (yield 3.5%) that was stable for up to 120 h of dark incubation at 17 °C (see Fig. 3B). The homogeneity of the sample was verified by SEC, which showed a single elution peak with a retention time corresponding to the PSII dimer (Fig. 1B). The purity of the PSII complex was additionally confirmed spectroscopically by identification of characteristic peaks at 673 nm in the room temperature absorbance spectrum and 692 nm in the 77 K emission spectrum (see Fig. 2, A and B), similar to other red algal PSII preparations from Cyanidium caldarium and P. cruentum (22, 49). The PSII dimer purity was monitored for each AEC fraction by measuring a ratio of A673/A625. For the pure dimeric PSII preparation free of phycobilisomes, the A673/A625 ratio was estimated at ∼5, yielding 99% homogeneous sample.
FIGURE 1.
Anion exchange chromatography purification of the C. merolae PSII dimer. A, AEC chromatogram from the second step of PSII purification on a DEAE ToyoPearl 650 S column. B, selected section of the AEC chromatogram (boxed in A). Elution fractions were analyzed by SEC (inset). The SEC chromatogram of every fraction analyzed is positioned directly below a circle that marks the position of the corresponding fraction in the AEC chromatogram. Consecutive SEC chromatograms show a transition between the PSII monomer and dimer (retention times of 4.15 and 3.85 min, respectively). a.u., arbitrary units.
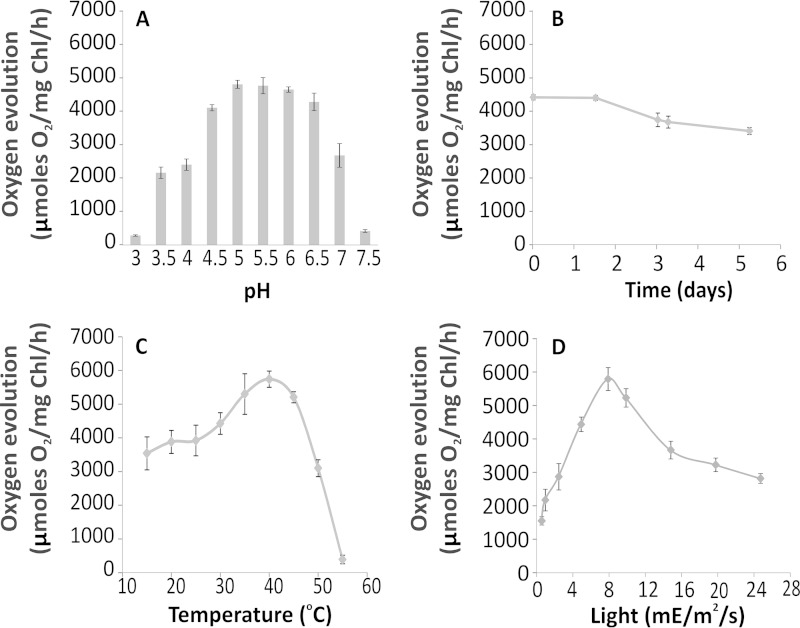
FIGURE 3.
PSII activity is sustained in various extreme conditions. A, PSII maintains nearly full activity of 4,500 μmol of O2/mg of Chl/h in a relatively broad range of pH (between 5 and 6.5) with standard 5,000 μE/m2/s light intensity and 30 °C. B, when stored at 17 °C for a period of 5 days, PSII retains nearly 80% of its activity measured in standard conditions. C, PSII has a distinctive temperature optimum, with a maximum activity at 40 °C, measured in standard conditions. D, upon increasing light intensity, PSII reaches a maximum activity of ∼6,000 μmol O2/mg Chl/h at 8,000 μE/m2/s, which lowers to 3,000 μmol of O2/mg of Chl/h at 25,000 μE/m2/s. Each data point represents an average value from three independent measurements. Error bars, S.D.
FIGURE 2.
Spectroscopic and compositional analyses of the C. merolae PSII dimer. Room temperature absorbance spectrum of PSII dimer (A) shows the red peak at 673 nm, characteristic of PSII. The PSII complex purity was expressed as a ratio of A673/A625 and was estimated at ∼5, confirming a complete removal of residual phycobilisomes. The 77 K steady-state fluorescence emission spectra (B) were taken at excitation wavelengths of 440 nm (solid line) and 580 nm (dashed line). The excitation wavelength of 580 nm was used to detect any residual contamination with phycobilisomes that emit fluorescence at 625 nm. The 440-nm wavelength excited Chla to produce a symmetric emission peak at 692 nm, characteristic of PSII. C, SDS-PAGE protein profile of the C. merolae PSII dimer. Samples (5 μg of Chl/lane) were resolved on a 18% Tris-Tricine gel. The positions of PsbA/C and PsbB/C as well as PsbO, PsbQ′, PsbV, and PsbU were identified by Western blotting and MS/MS analyses. The position of PsbQ′ is marked with an arrowhead in C. a.u., arbitrary units.
We then analyzed the subunit composition of the purified C. merolae PSII dimer by biochemical and mass spectrometry approaches. Fig. 2C and supplemental Table S1 show that the reaction center subunits (D1 and D2), inner antenna subunits (CP43 and CP47), four extrinsic subunits stabilizing the OEC (PsbO, PsbU, PsbV, and PsbQ′), and the majority of small intrinsic subunits were all present in our preparation. In addition, we detected an auxiliary subunit Psb27 and low molecular weight subunit PsbW in our dimer preparations (see supplemental Table S1).
We estimated the stoichiometry of PSII/cytochrome c550 (PsbV) and PSII/cytochrome b559 by measuring redox difference spectra in the presence of ferricyanide (oxidized) or dithionite (reduced). The redox difference spectra showed two characteristic peaks at 550 and 557 nm, corresponding to the fully reduced forms of cytochrome c550 and cytochrome b559, respectively (data not shown). The molar ratios of PSII/cytochrome c550 and PSII/cytochrome b559 were estimated at 0.954 ± 0.014 and 0.953 ± 0.038, respectively, indicating 1:1 stoichiometry of both cytochromes and PSII (data not shown).
Biochemical Activity and Robustness of the C. merolae PSII
First, we tested the herbicide sensitivity of the QB site to DCMU. We observed rapid inactivation of oxygen-evolving activity upon the DCMU treatment, with inhibition of more than 95% of PSII activity at a 10 μm concentration of the herbicide (data not shown). We estimated the value of the inhibition constant Ki at 0.46 ± 0.012 μm DCMU, using the Dixon analysis (50). This value agrees well with the previously reported Ki value of 0.33 μm for DCMU inhibition in spinach thylakoids (51) or in isolated Thermosynechococcus elongatus PSII (52).
We then proceeded with a detailed examination of the catalytical activity of the C. merolae PSII complex across a broad range of extreme conditions (see Fig. 3). The oxygen-evolving activity of PSII varied between 4,000 and 6,000 μmol of O2/mg of Chl/h for different preparations, with an average activity of 4,500 μmol of O2/mg of Chl/h under the standard experimental conditions (30 °C, pH 6.3, white light of 5,000–8,000 μE/m2/s). It was one of the highest recorded oxygen-evolving activities obtained under our standard experimental conditions and exceeded that of PSII isolated from the thermophilic cyanobacteria used to obtain the x-ray structures of the complex (1–3, 53). Moreover, the C. merolae dimeric PSII complex maintained its full activity in a relatively broad range of pH (between 5 and 6.5) when illuminated with white light of 5,000 μE/m2/s at 30 °C (see Fig. 3A). It also exhibited a distinctive temperature optimum, with a maximum activity at 40 °C, when measured in standard conditions (see Fig. 3C). Moreover, the C. merolae PSII dimer retained its average oxygen-evolving activity when illuminated with high white light intensities (up to 20,000 μE/m2/s; see Fig. 3D), indicating that the oxygen-evolving complex remained intact when subjected to light intensities well in excess of the saturation level.
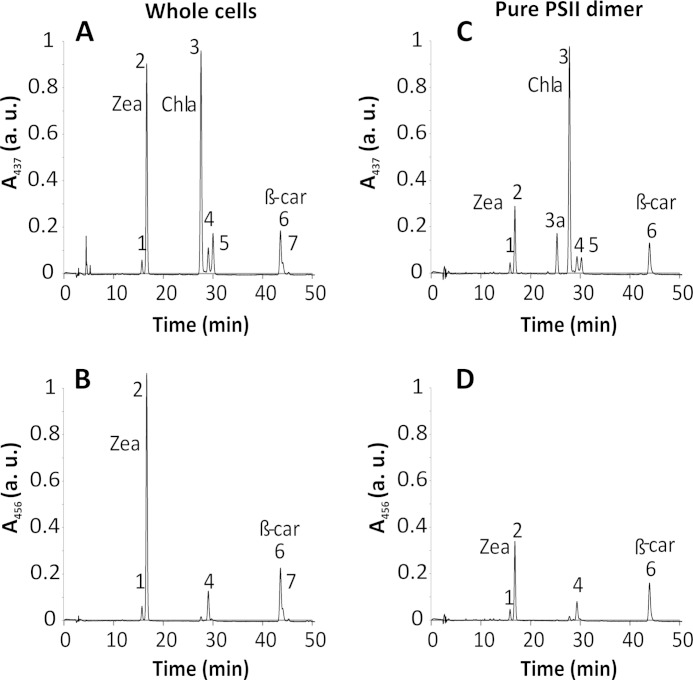
We determined the total pigment composition of C. merolae cells and of PSII dimers, using HPLC analysis (see Fig. 4 and Table 1). Both carotenoids and Chla were detected at 437 nm, whereas exclusive carotenoid peaks were traced at 456 nm, as shown in Fig. 4. The whole cell extract yielded a distinctive peak corresponding to zeaxanthin (Zea) (see Fig. 4, A and B), which nearly equalled the main Chla peak (see Fig. 4A). In contrast, the amount of β-carotene was 3-fold lower compared with Chla (Fig. 4A). The level of Zea in the pure PSII dimer was 3-fold lower compared with the whole cells (Fig. 4, compare A and C), whereas the relative ratio of Zea to β-carotene was 2-fold lower in the purified PSII dimer compared with the whole cells (Fig. 4, compare B and D; see Table 1). We did not detect any peaks corresponding to violaxanthin or antheraxanthin, either in the whole cells or in the purified PSII.
FIGURE 4.
HPLC pigment analysis of C. merolae cells and PSII dimer. Total pigments were analyzed by HPLC, and their absorbance was measured at 437 nm (for carotenoids and Chla) and at 456 nm (for carotenoids only). Peak identities are as follows. 1, Zea (cis isomer); 2, Zea; 3a, oxidized Chla; 3, Chla; 4, β-cryptoxanthin; 5, Chla′; 6, β-carotene; 7, β-carotene (cis isomer). Peaks were assigned to the corresponding pigments by LC-MS according to Ref. 39. Whole cell extract yielded a distinctive peak corresponding to Zea (A and B) nearly identical to the peak of Chla (A), whereas the β-carotene peak was 3-fold lower than that of Chla. The Zea abundance in the pure PSII dimer was significantly lower than in whole cells (C); however, the relative contribution of Zea and β-carotene was similar (D) when PSII was compared with the whole cell extract (see Table 1). a.u., arbitrary units.
TABLE 1.
Relative abundance of zeaxanthin and β-carotene in whole cells and purified PSII
Molar ratios of pigments were calculated by integration of an area underneath the relevant peak and using extinction coefficients, as described under “Materials and Methods.” S.D. values were calculated from two independent measurements (n = 2).
| Source | Zea/Chla | Zea/β-carotene | β-Carotene/Chla |
|---|---|---|---|
| PSII | 0.275 ± 0.033 | 2.140 ± 0.270 | 0.131 ± 0.015 |
| Intact cells | 0.880 ± 0.033 | 4.230 ± 0.180 | 0.210 ± 0.007 |
Investigation into the in Vivo and in Vitro Photoprotection Mechanisms in C. merolae
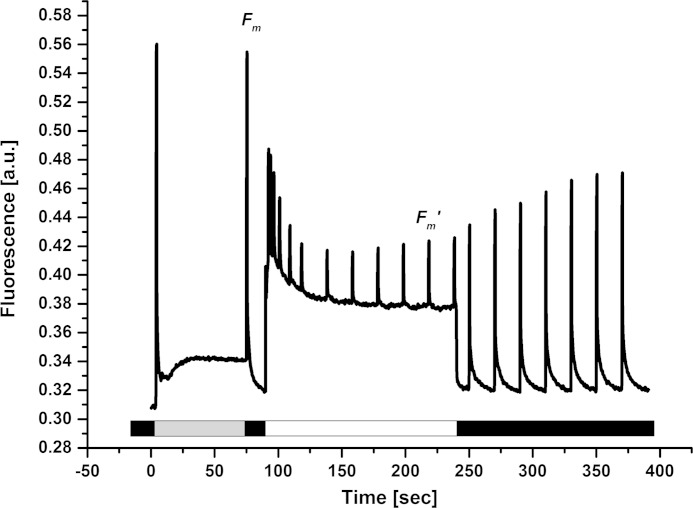
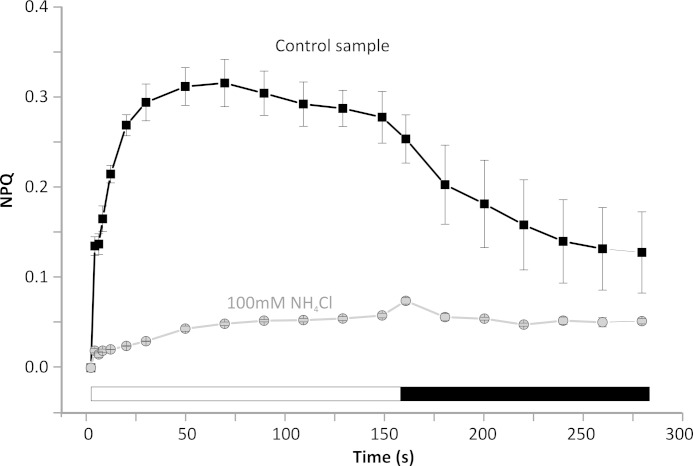
In order to gain an insight into the photoprotective mechanisms in C. merolae, we set out to dissect the locus of NPQ both in vivo (intact cells) and in vitro (isolated PSII dimers). We investigated the presence and extent of photoprotective NPQ in vivo by measuring the variable Chla fluorescence (FV) in intact C. merolae cells (Fig. 5). A typical measuring protocol has been applied to exclude the effect of state transitions on the maximal fluorescence in dark (FM) that is known to be affected by the phycobilisome binding in cyanobacteria (54). There were no significant changes in the FM value after a short period of illumination with low intensity blue light, excluding the putative influence of state transitions on the measured FM values. The FM value obtained in the dark (see Fig. 5) was then used for calculation of the NPQ kinetics upon transition of the cells to light (Fig. 6). The exposure of dark-adapted C. merolae cells to high intensity blue light resulted in pronounced quenching of maximal fluorescence (FM′), in a process that was reversible in the dark (see Fig. 5). These results confirmed the presence of NPQ in C. merolae cells in vivo. Maximal NPQ value was around 0.4 (Fig. 6), and resembled the value determined for cyanobacteria (54), indicating a similar amount of energy that is dissipated during NPQ despite different quenchers employed by both phototrophs. The NPQ was inhibited in vivo after the addition of an uncoupler NH4Cl (Fig. 6), demonstrating the role of acidification of the lumen in triggering NPQ in C. merolae cells.
FIGURE 5.
Fluorescence quenching in C. merolae cells exposed to blue light. Cells were adapted for 20 min in the dark and then used for in vivo measurements of NPQ. Low intensity blue light was used (∼7 μE/m2/s; dark gray bar) to reach maximal fluorescence in the dark (FM). Quenching of maximal fluorescence upon high intensity blue light exposure (750 μE/m2/s; white bar) and its recovery in the dark (black bar) were observed. The NPQ value was calculated based on the Stern-Volmer formula, (FM − FM′)/FM′. a.u., arbitrary units.
FIGURE 6.
Non-photochemical quenching kinetics in C. merolae cells. The induction of NPQ was measured in dark-adapted cells exposed to blue light (750 μE/m2/s; white bar) followed by recovery in the dark (black bar). Curves were measured either without any inhibitors (control sample) or after the addition of an uncoupler (100 mm NH4Cl) that disrupts lumen acidification. Data represent averages and S.D. (error bars) for n = 3.
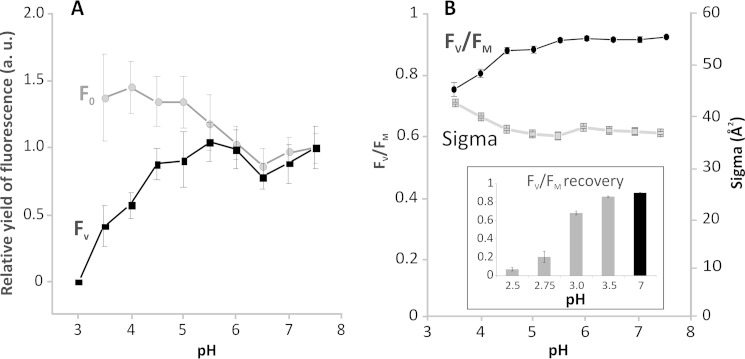
In order to gain the first glimpse into the mechanism of photoprotection in isolated C. merolae dimeric PSII complexes, we measured the effect of various pH levels (pH range of 3–7.5) on variable Chla fluorescence, FV. Photoprotective NPQ that is triggered by low pH can be demonstrated by detection of a decrease of FV (see Ref. 29 for a recent review). As shown in Fig. 7A, variable fluorescence of PSII was progressively reduced at pH below 4.5 and completely disappeared at pH 3. This FV decrease was correlated with the reduction of maximal PSII efficiency, FV/FM (see Fig. 7B), and with a decrease of the oxygen evolution rate at this pH range (compare Figs. 3A and 7). These data indicate that the reduction of photochemical PSII efficiency at low pH is due to stimulation of non-photochemical pathways of de-excitation. In contrast to the pH-dependent FV decrease, there was no quenching of minimal fluorescence (F0) at low pH (see Fig. 7A). Because we have not found any external LHC-type antennae attached to the isolated PSII dimers (see supplemental Table S1), this selective quenching of variable fluorescence (FV) suggests the presence of the NPQ locus in the C. merolae PSII reaction center. A similar mechanism has been proposed for PSII isolated from higher plants (35, 36).
FIGURE 7.
Dissection of the NPQ locus in C. merolae PSII. A, relative yields of minimal (F0) and variable fluorescence (FV) of isolated PSII as a function of pH. Values represent averages and S.D. values from six measurements (n = 6) done with two independent PSII isolations. Data are normalized to fluorescence parameters at pH 7.5. For pH 3 and lower, there was no detectable variable fluorescence; therefore, other physiological parameters indicating closed (FM) or opened (F0) RC were calculated only in the pH range 3.5–7.5. B, maximal quantum yield (FV/FM) and effective antennae size (σPSII) of isolated PSII as a function of pH. Values represent averages and S.D. values from six measurements (n = 6) done with two independent PSII preparations. Inset, reversibility of PSII efficiency (FV/FM) after a treatment with low pH. All values were measured at pH 7, either after 2-min treatment at low pH (2.5–3.5) followed by 4 min of recovery at pH 7 (gray bars) or directly at pH 7 without prior low pH treatment (black bar). Data represent averages and S.D. (error bars) for n = 3. a.u., arbitrary units.
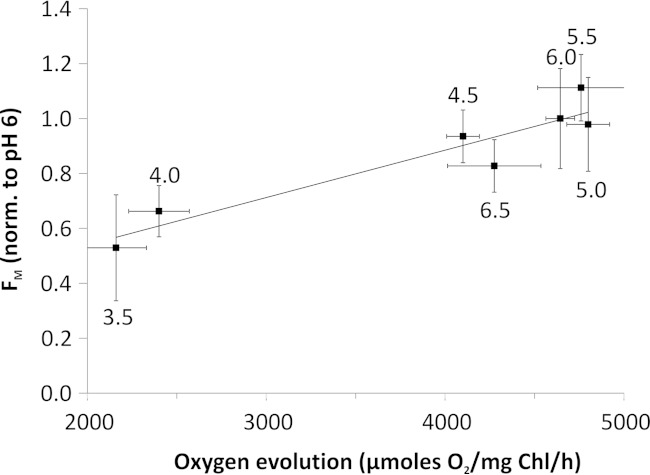
We observed a clear reversibility of low pH-induced changes in maximal PSII efficiency FV/FM, upon transfer of PSII from low to neutral pH. The maximal efficiency of PSII almost fully recovered upon transfer from pH 3.5, with almost 70% recovery of the FV/FM ratio following incubation of PSII at pH 3 (see Fig. 7B, inset), at which no variable fluorescence FV was observed (Fig. 7A). These data clearly confirm the presence of reversible photoprotective NPQ in the dimeric C. merolae PSII exposed to extreme low pH. No NPQ was observed when PSII was exposed to pH below 3 (see Fig. 7B, inset), most likely due to the irreversible damage to the OEC on the donor side and denaturation of the PSII complex. We have tested correlation between low pH-induced changes in FM and inhibition of oxygen evolution rates. We observed linear correlation between FM quenching and reduction in oxygen evolution rates at low pH (pH 3.5–6.5) (see Fig. 8). Conversely, no such correlation was observed at higher pH (pH 7 and 7.5), when the O2 evolution rate was reduced, whereas FM remained non-quenched (see Figs. 3A and 7A). This is in line with the fact that only low pH can trigger NPQ in vivo.
FIGURE 8.
Correlation between low pH induced maximal fluorescence quenching and inhibition of oxygen evolution in the isolated C. merolae PSII. Values of FM were normalized at pH 6 and plotted with values of PSII activity at a given pH (numerical values by the data points). A linear correlation with the correlation factor R2 = 0.89 (p = 0.0014) of both data sets was observed, indicating that within the pH range of 3.5–6.5, the loss of activity is related to quenching of fluorescence and not to irreversible damage to OEC. No such correlation was observed at pH 7 and 7.5. Error bars, S.D.
We further explored the mechanism of NPQ in C. merolae PSII by measuring the effective antennae size of PSII (σPSII) corresponding to the inner antenna Chls (see Table 2). In intact C. merolae cells, the antennae size of PSII was ∼30% higher compared with isolated thylakoid membranes or dimeric PSII complexes, indicating a partial loss in delivering excitation energy to PSII upon solubilization of this complex from the membranes. Because the effective antennae size of the C. merolae PSII was similar to that observed for the higher plant complex (see Ref. 36), our PSII dimer was isolated in a fully native and functional form. There was no significant difference in σPSII for isolated PSII and thylakoids that contain both PSI and PSII (see Table 2). In isolated PSII, we did not observe any decrease of σPSII at pH 3.5, a pH value at which FV was already significantly reduced (Fig. 7, compare A and B). The constant (or slightly increasing) values of σPSII at low pH preclude the possibility of de-excitation of excess light occurring within the internal PSII-core antennae CP43 and CP47 because such a mechanism would imply a decrease in both σPSII and FV.
TABLE 2.
Effective PSII antennae size (σPSII ) in intact C. merolae cells and isolated PSII
Values represent averages and S.D. from six measurements (n = 6) done with two independent PSII isolations.
| Source | Intact cells | Thylakoids | PSII (pH 6) | PSII (pH 3.5) |
|---|---|---|---|---|
| σPSII | 52.8 ± 1.6 | 38.1 ± 0.7 | 37.1 ± 0.9 | 41.8 ± 1.3 |
Structural Analysis of the C. merolae PSII Dimer
In order to get an impression of subunit organization within the C. merolae PSII dimer, we visualized the complex by negative stain electron microscopy combined with single particle analyses. Single particle averaging of a large set of dimers showed that the particles were mostly present in top lumenal or side view positions. Within the top views, there was little variation, allowing for the final projection map to be obtained at a resolution of 17 Å, as depicted in Fig. 9A. In the class of side view projections, we detected some variation at the site of the extrinsic subunits, which resulted in a partition of the data set into three classes (Fig. 9, B–D). As a main difference between the classes, there is a stronger or fainter presence of the 12-kDa subunit (PsbU) at the top of the other extrinsic subunits (orange arrows in Fig. 9, C and D). In addition, the visibility of the cytochrome c550 (PsbV) subunit is variable (blue arrows in Fig. 9, B and D), despite the apparent 1:1 stoichiometry of PsbV/PSII (data not shown). The size of a side view C. merolae PSII dimer particle (Fig. 9, B–D) was estimated at 21.2 × 11 nm, which is in a good agreement with the size of the PSII dimers isolated from another red alga, P. cruentum (49), as well as dimensions of the cyanobacterial PSII dimer calculated from the atomic crystal structure (19.9 × 11.6 nm; see Ref. 1).
FIGURE 9.
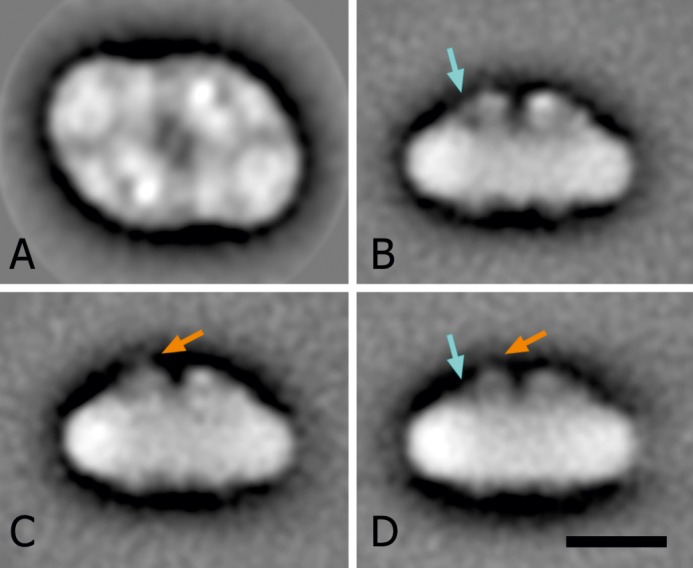
Projection maps of the top and side views of C. merolae. A, final projection map of the top view, as seen from the lumenal side of the membrane; the sum is composed of the best 2,048 particles, according to their correlation coefficient in the last alignment step. Two-fold rotational symmetry was imposed after analysis. B–D, averaged images of three classes of side views. The sums are composed of 493, 588, and 353 projections, respectively. As a main difference between the classes, the orange arrows indicate a stronger (C) or fainter (D) presence of the 12-kDa subunit on top of the other extrinsic subunits. Blue arrows show a stronger (B) and weaker (D) visibility of the cytochrome c550 subunit. Scale bar (A–D), 10 nm.
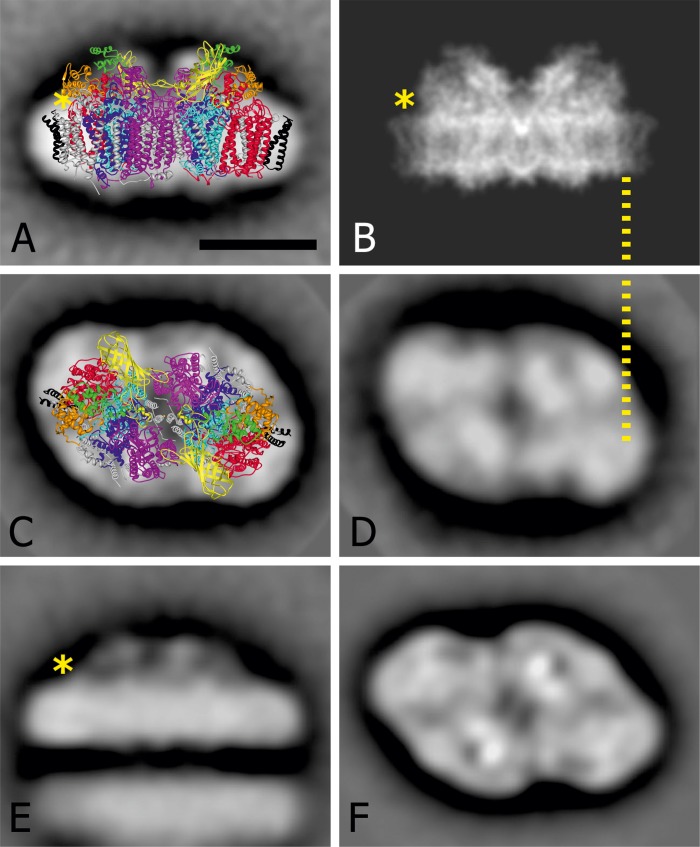
Comparison with the structural work on cyanobacterial PSII dimers could give us a hint of where PsbQ′, an extra extrinsic subunit of C. merolae over and above the cyanobacterial homologues (PsbO, PsbU, and PsbV), is located. First, the side views were analyzed because in this position we observed less overlap between the PSII subunits, allowing for the distinctive differences between the maps to be dissected. Indeed, we identified an additional protein mass visible in the map of C. merolae (Fig. 10A) compared with the high resolution structure of PSII from T. vulcanus (Fig. 10B) digitally truncated to 8 Å for comparison of protein densities. The yellow asterisk indicates the position where the C. merolae PSII dimer seems to have protein mass that is absent in the cyanobacterial structure (Fig. 10, compare A and B). The most obvious candidate for the location in this position is the additional lumenal extrinsic subunit PsbQ′.
FIGURE 10.
Comparison of dimeric PSII projection maps to visualize differences between C. merolae and T. elongatus at the subunit level. A and C, side and lumenal top view projection maps of the C. merolae PSII dimer particles, respectively. In A, the 1.9 Å structure of PSII from T. vulcanus (1) (Protein Data Bank coordinates 3ARC) was overlaid onto the top view EM projection map of the C. merolae complex. The D1 (PsbA) and D2 (PsbD) subunits are colored in light blue and dark blue, respectively. The CP43 (PsbC) and CP47 (PsbB) subunits are depicted in red and magenta, respectively. The PsbZ subunit is highlighted in black. The extrinsic subunits PsbV, PsbU, and PsbO are shown in orange, green, and yellow, respectively. For clarity, all remaining subunits are shown in gray. The overall size of PSII dimer particles in A is 21.2 × 11 nm. B, a high resolution structure of T. vulcanus PSII seen from aside and truncated at 8 Å resolution (1). D and E, top and side views of T. elongatus PsbZ-less PSII dimers (82); F, top view of T. elongatus PSII containing PsbZ (83). The dotted line correlates the position of an additional density with the CP43 subunit in the side view map (B) and in the top view (D). *, location where PsbQ′ is located in the C. merolae PSII complex and is absent in the T. vulcanus and the T. elongatus complex. Scale bar, 10 nm.
DISCUSSION
In the present work, we isolated and purified the dimeric PSII complex from the red alga C. merolae and demonstrated its remarkable robustness over a range of extreme conditions, including high light illumination, high temperatures, and extreme pH range conditions that usually lead to destablization of the OEC and inhibition of PSII function. The oxygen-evolving activity of the isolated complex is one of the highest reported to date and exceeds the activity of dimeric PSII complexes isolated from, for example, the thermophilic cyanobacteria T. elongatus and T. vulcanus used to obtain medium to high resolution structures of PSII (1, 2, 53). Although the pH optimum for the activity of the C. merolae PSII complex is at pH 5–5.5, the complex sustains over 40–60% of its oxygen-evolving activity at pH 3.5 and pH 7, which is unprecendented for other PSII preparations isolated from thermophilic cyanobacteria (55–57), mesophylic and other thermoacidophilic red algae (38, 49), green algae (58), and higher plants (59, 60). Moreover, the C. merolae complex increases its activity at relatively high temperatures up to 45 °C as well as retaining its intactness over a prolonged period of incubation at room temperatures (see Fig. 3), which may make this complex amenable to application in biometic solar-to-fuel devices often intended to operate at elevated temperatures over extended periods of time (61, 62).
One important question arises: what are the molecular mechanisms responsible for the remarkable robustness of C. merolae PSII, especially under extreme illumination conditions? Under the conditions of imbalance between energy capture and utilization (e.g. in high light), accumulation of excitons in external and intrinsic antennae of both photosystems increases the occurrence of singlet Chl-excited states (1Chl*). Under excess light, 1Chl* species often undergo intersystem crossing to form the Chl triplet state (3Chl*) (63). This species reacts with molecular oxygen to form highly reactive oxygen species, including singlet oxygen (1O2), when excitations are not rapidly quenched. This can occur when the capacity for productive photochemical quenching is exceeded and non-photochemical quenching is insufficient or not yet sufficiently turned on (64–66).
Our carotenoid and chlorophyll fluorescence quenching analyses point to the involvement of both zeaxanthin and the primary oxidant, P680+, being responsible for the photoprotection of thylakoids and PSII under high light irradiance. It is well established that carotenoids are essential for photoprotection in oxygenic photosynthesis (reviewed in Refs. 28, 63, 67, and 68). Moreover, they affect assembly and stability of the PSII reaction center and light-harvesting antenna subunits (69, 70).
Of all of the carotenoids, Zea plays a particularly important role in photoprotection. High light-induced synthesis of Zea and its binding to the specific proteins enhance photoprotection by decreasing the yield of potentially dangerous Chl-excited states. A recent study using time-resolved differencial spectroscopy demonstrated increased efficiency of this molecule in controlling 3Chl* species formation (71). Zea is known to be involved in several types of photoprotection of the PSII reaction centers that occur at various time scales: (i) rapid feedback de-excitation quenching (qE) of 1Chl*, (ii) a slowly inducible quenching (qZ) caused by Zea binding to LHC upon exchange with violaxanthin, and (iii) a long term irreversible quenching associated with PSII photoinhibition (71).
In this work, we have identified a considerable amount of Zea both in intact C. merolae cells and in the isolated PSII complex (see Fig. 4 and Table 1), indicating that this xanthophyll may play an important photoprotective role in reactive oxygen species scavenging in the lipid phase of C. merolae thylakoids. Because the light-harvesting antenna of red algal PSII is formed by phycobilisomes, it is unlikely that Zea would be involved in slowly inducible quenching qZ. Importantly, our pigment quantification suggests that in the C. merolae PSII complex, Zea may also be present inside the PSII core complex, where it may take a direct role in quenching of 3Chl* species. Taking into account the pigment molar ratios presented in Table 1 and normalizing these values to 35 Chla molecules/PSII, as determined in the latest atomic structure (1), we estimate 4–5 β-carotenes and 8–10 Zea molecules in our PSII complex. Because we would expect 11 β-carotenes/PSII (1), it is possible that Zea may replace some β-carotenes in the C. merolae PSII complex, perhaps at the boundary of CP43 and associated low molecular weight subunits. This intriguing possibility needs to be supported by a more detailed study.
Our present data showing insensitivity of the effective PSII antennae size to low pH-induced quenching (Fig. 7B) seems to exclude a direct role of C. merolae PSII inner antennae in quenching. The pH insensitivity of the effective antennae size during NPQ is characteristic of the NPQ locus present within the closed reaction centers upon charge recombination (34, 72). It is therefore possible that some Zea molecules putatively bound in the vicinity of the charge separation pathway in C. merolae PSII could be involved in the RC-based quenching observed in the present study. Similarly, Fleming and colleagues (73) observed that the mechanism of non-radiative deactivation of 1Chl* during excess light occurs by excitation transfer to a Chl-Zea heterodimer, followed by ultrafast Car+· formation. Such a process was observed for Zea bound within the peripheral light-harvesting antenna of PSII. Whether a similar mechanism exists for Zea putatively bound within the RC of red algal PSII remains to be elucidated.
Alternatively, Zea could play a direct antioxidant role in scavenging free radicals and singlet oxygen molecules produced in high light in the lipid phase of the thylakoid membrane in the vicinity of PSII macrodomains (74). In light of the high lipid content of the cyanobacterial PSII dimer shown in its latest atomic structure (1), it is tempting to speculate that free Zea molecules bound within the C. merolae PSII complex-lipid interface may protect these intrinsic lipid molecules from 1O2-mediated peroxidation. A similar NPQ-independent protective role of Zea in scavenging singlet oxygen has been observed for the ch1 mutant of Arabidopsis thaliana devoid of the LHCII antenna (75). In addition, Zea identified in this study in intact cells of C. merolae might play a role as a thylakoid membrane-rigidifying molecule (76, 77), allowing this extremophilic alga to thrive at high temperatures.
Similarly, Gantt and colleagues (78) showed that in another unicellular red alga, P. cruentum, the cellular content of Zea increased with growth irradiance, confirming a role for this carotenoid in photoprotection. In the latter work, the ratio of cellular Zea/β-carotene incrementally increased upon high light exposure of Porphyridium cells and was similar to the ratio obtained in this study for similar conditions of illumination (2.6 and 2.8, respectively). The relative content and composition of carotenoids and Chl are also similar in C. merolae (see Table 1 in this study and Ref. 39) and P. cruentum cells (78). Importantly, we did not detect violaxanthin either in whole cells or purified PSII (see Fig. 4) and PSI (not shown), confirming the previous data of Gantt and colleagues (39), who showed that this dieepoxide xanthophyll is absent in C. merolae cells. Instead, in this organism, Zea most likely forms by conversion of β-carotene via intermediate β-cryptoxanthin (79), a process catalyzed by β-carotene 3-hydroxylase enzymes, such as the CrtR cyanobacterial type β-carotene hydroxylase or cytochrome P450 (39).
In addition to the Zea-mediated photoprotection, our fluorescence measurements clearly point to the pH-dependent reaction center quenching as the main photoprotective mechanism in the C. merolae PSII. This type of quenching seems to sustain a high oxygen-evolving activity of C. merolae PSII even at extremely high irradiation (see Fig. 3). Kirilovsky and colleagues (32, 33) have shown that non-photochemical quenching in mesophilic red algae is triggered by low pH and may occur in the PSII reaction center rather than the inner antenna. However, a direct proof for this type of photoprotective mechanism and dissection of its exact locus were missing because their work was performed exclusively in intact cells. Due to this limited experimental approach, the authors could only suggest the reaction center-based mechanism of NPQ in red algae based on indirect measurements of F0 pH insensitivity during stimulation of non-photochemical quenching (32, 33).
Here, we provide the first direct evidence of the PSII reaction center as the primary locus of pH-dependent NPQ in the extremophilic red algae. This type of quenching can occur through (i) energy dissipation via alternative electron transport after primary radical pair production, (ii) charge recombination between Pheo− and P680+ (80), (iii) rapid charge recombination between QA− and P680+, and (iiii) accumulation of P680+ that can act as a direct quencher (30, 36). All four types of RC-based quenching are consistent with pH insensitivity of effective antenna size during quenching. Indeed, our measurements of the absorbance cross-section of isolated C. merolae PSII revealed no decrease in the functional antenna size at low pH (see Fig. 7B), which supports a reaction center-based quenching mechanism in this extremophile when exposed to saturating light levels. Importantly, we detected significantly decreased FV values at low pH, a phenomenon that was rapidly reversible upon exposure of PSII to physiological pH (see Fig. 7B). We also observed a significant correlation between FM (and FV) quenching and inhibition of oxygen evolution in isolated PSII at low pH (see Fig. 8) and a lack thereof at higher pH. These data exclude the possibility of the OEC inactivation as the primary cause for fluorescence quenching in the isolated C. merolae PSII and confirm the presence of reversible low pH-induced NPQ in this complex.
We previously obtained three-dimensional crystals of the C. merolae PSII dimer (81) with the aim of solving the structure of this complex. Because we were unable to significantly improve the resolution of x-ray diffraction patterns obtained from these crystals, we applied electron microscopy coupled with single particle analyses of the isolated PSII dimer to gain an insight into the structure of this eukaryotic complex, in particular its lumenal side. Comparison with structural work on cyanobacterial PSII dimers allowed us to suggest an assignment of the position for the PsbQ′, an extra extrinsic subunit over and above the cyanobacterial-like PsbO, PsbU, and PsbV subunits stabilizing the OEC in the C. merolae complex (24–26). In our difference map in which the atomic structure of cyanobacterial PSII was modeled onto the averaged side and lumenal top view projections of the C. merolae PSII dimer particles, the additional density attributed to PsbQ′ is present close to the lumenal side of the membrane, in the vicinity of the CP43 inner antenna subunit (see Fig. 10). Our assignment of the PsbQ′ position is supported by previous EM studies of cyanobacterial PSII (82, 83), whereby the side view dimer projection lacks the additional mass identified in our C. merolae PSII dimer particles (Fig. 10E). Another position would be in the center, because the EM and x-ray data also differ here, such that in the EM map, the extrinsic protein densities for both monomers appear to be more compact. However, this location is unlikely, because the cyanobacterial map of T. elongatus is essentially the same in this position (Fig. 10E).
Our assignment of PsbQ′ differs somewhat from that presented for the dimeric PSII particles from a closely related red alga, C. caldarium (22), in that it places this additional OEC subunit closer to the membrane in the vicinity of the CP43 intrinsic antenna subunit. Vacha and colleagues proposed positioning of PsbQ′ more toward the lumenal space, between the PsbU and PsbV subunits (22). However, in contrast to the present work, they were unable to reveal any additional protein densities in the projection maps of C. caldarium compared with the equivalent maps of P. cruentum (49) and T. elongatus (82) PSII dimer particles.
The comparison with the cyanobacterial data further shows a surprising similarity between the top lumenal views of both C. merolae (Fig. 10C) and T. elongatus (Fig. 10D). There are no significant differences in surface densities, indicating that the extra subunit of C. merolae is not visible, most likely due to an overlap with the membrane-integrated moiety of the PSII dimer, possibly the CP43 intrinsic antenna subunit. The close similarity in the surface and shape points to a highly similar composition of the membrane-bound subunits. The map, as depicted in Fig. 10D, was obtained from a PSII particle lacking the PsbZ protein. This protein was, however, present in the map shown in Fig. 10F (83). Thus, it seems that our C. merolae PSII particles do not contain a subunit homologous to the cyanobacterial PsbZ subunit. Interestingly, in the side view projection map of the C. merolae PSII dimer, the position of the PsbO subunit seems to be slightly shifted toward the pseudosymmetry axis of the dimer, compared with the crystal structure of the cyanobacterial counterpart (see Fig. 10A). A similar phenomenon was observed for subpopulations of the dimeric PSII from a green alga Chlamydomonas reinhardtii with altered organization of the LHCII antenna,4 suggesting a certain level of structural flexibility within the OEC region.
Interestingly, our mass spectrometry analysis showed the presence of the PSII auxiliary subunit Psb27 and a low molecular weight subunit PsbW implied in the regulation of the PSII repair cycle (84) and stabilization of the PSII dimers in higher plants (85). Both proteins were detected in all rounds of MS/MS analysis from three independent preparations, suggesting their stable association with the population of C. merolae PSII (probably in its inactive form), which most likely undergoes a repair cycle. However, the stoichiometry of their binding to PSII as well as the turnover rate for the C. merolae PSII are presently unknown and will be a subject of future studies.
Supplementary Material
Acknowledgment
We thank A. K. Jagielski (Department of Metabolic Regulation, Faculty of Biology, University of Warsaw) for assistance in mass spectrometry data collection.
This work was supported in part by the Polish Ministry of Science and Higher Education and by European Science Foundation Grant 844/N-ESFEuroSolarFuels/10/2011/0 (to J. K.) and was partially performed with the use of CePT infrastructure financed by the European Union (European Regional Development Fund within the Operational Program “Innovative Economy” for 2007–2013) (to R. M. and M. G.).

This article contains supplemental Table S1 and an additional reference.
M. Webber-Birungi, B. Drop, S. Yadav, R. Croce, and E. J. Boekema, unpublished data.
- PSII
- photosystem II
- PSI
- photosystem I
- AEC
- anion exchange chromatography
- Chl
- chlorophyll
- DCMU
- 3-(3,4-dichlorophenyl)-1,1-dimethylurea
- DDM
- dodecyl-β-d-maltoside
- F0
- minimal fluorescence
- FV
- variable fluorescence
- FM
- maximal fluorescence of dark-adapted samples
- FM′
- maximal fluorescence in light
- FV/FM
- maximal quantum yield of PSII
- NPQ
- non-photochemical quenching
- OEC
- oxygen evolving complex
- RC
- reaction center
- SEC
- size exclusion chromatography
- σPSII
- effective antennae size of PSII
- Zea
- zeaxanthin
- μE
- microeinstein(s)
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- qE
- high energy quenching
- qZ
- slowly inducible quenching.
REFERENCES
- 1. Umena Y., Kawakami K., Shen J. R., Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 [DOI] [PubMed] [Google Scholar]
- 2. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 3. Kargul J., Maghlaoui K., Murray J. W., Deak Z., Boussac A., Rutherford A. W., Vass I., Barber J. (2007) Purification, crystallization and X-ray diffraction analyses of the T. elongatus PSII core dimer with strontium replacing calcium in the oxygen-evolving complex. Biochim. Biophys. Acta 1767, 404–413 [DOI] [PubMed] [Google Scholar]
- 4. Murray J. W., Maghlaoui K., Kargul J., Ishida N., Lai T.-L., Rutherford A. W., Sugiura M., Boussac A., Barber J. (2008) X-ray crystallography identifies two chloride binding sites in the oxygen evolving centre of Photosystem II. Energy Environ. Sci. 1, 161–166 [Google Scholar]
- 5. Murray J. W., Maghlaoui K., Kargul J., Sugiura M., Barber J. (2008) Analysis of xenon binding to photosystem II by x-ray crystallography. Photosynth. Res. 98, 523–527 [DOI] [PubMed] [Google Scholar]
- 6. Kamiya N., Shen J. R. (2003) Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-Å resolution. Proc. Natl. Acad. Sci. 100, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 8. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 9. Yano J., Kern J., Sauer K., Latimer M. J., Pushkar Y., Biesiadka J., Loll B., Saenger W., Messinger J., Zouni A., Yachandra V. K. (2006) Where water is oxidized to dioxygen. Structure of the photosynthetic Mn4Ca cluster. Science 314, 821–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sproviero E. M., Shinopoulos K., Gascón J. A., McEvoy J. P., Brudvig G. W., Batista V. S. (2008) QM/MM computational studies of substrate water binding to the oxygen-evolving centre of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1149–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dau H., Grundmeier A., Loja P., Haumann M. (2008) On the structure of the manganese complex of photosystem II. Extended-range EXAFS data and specific atomic-resolution models for four S-states. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1237–1243; discussion 1243–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cox N., Messinger J. (2013) Reflections on substrate water and dioxygen formation. Biochim. Biophys. Acta, in press [DOI] [PubMed] [Google Scholar]
- 13. Siegbahn P. E. (2013) Water oxidation mechanism in photosystem II, including oxidations, proton release pathways, O—O bond formation and O2 release. Biochim. Biophys. Acta, in press [DOI] [PubMed] [Google Scholar]
- 14. Badura A., Esper B., Ataka K., Grunwald C., Wöll C., Kuhlmann J., Heberle J., Rögner M. (2006) Light-driven water splitting for (bio-)hydrogen production. Photosystem II as the central part of a bioelectrochemical device. Photochem. Photobiol. 82, 1385–1390 [DOI] [PubMed] [Google Scholar]
- 15. Vittadello M., Gorbunov M. Y., Mastrogiovanni D. T., Wielunski L. S., Garfunkel E. L., Guerrero F., Kirilovsky D., Sugiura M., Rutherford A. W., Safari A., Falkowski P. G. (2010) Photoelectron generation by photosystem II core complexes tethered to gold surfaces. ChemSusChem 3, 471–475 [DOI] [PubMed] [Google Scholar]
- 16. Kato M., Cardona T., Rutherford A. W., Reisner E. (2012) Photoelectrochemical water oxidation with photosystem II integrated in a mesoporous indium-tin oxide electrode. J. Am. Chem. Soc. 134, 8332–8335 [DOI] [PubMed] [Google Scholar]
- 17. Barber J., Tran P. D. (2013) From natural to artificial photosynthesis. J. R. Soc. Interface 10, 20120984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ciniglia C., Yoon H. S., Pollio A., Pinto G., Bhattacharya D. (2004) Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol. Ecol. 13, 1827–1838 [DOI] [PubMed] [Google Scholar]
- 19. Enami I., Adachi H., Shen J.-R. (2010) Mechanisms of acido-tolerance and characteristics of photosystems in an acidophilic and thermophilic red alga, Cyanidium caldarium. in Red Algae in the Genomic Age (Seckbach J., Chapman D. J., eds) pp. 375–389, Springer, Dordrecht, The Netherlands [Google Scholar]
- 20. Nozaki H., Matsuzaki M., Takahara M., Misumi O., Kuroiwa H., Hasegawa M., Shin-i T., Kohara Y., Ogasawara N., Kuroiwa T. (2003) The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids. J. Mol. Evol. 56, 485–497 [DOI] [PubMed] [Google Scholar]
- 21. Wolfe G. R., Cunningham F. X., Durnford D., Green B. R., Gantt E. (1994) Evidence for a common origin of chloroplasts with light-harvesting complexes of different pigmentation. Nature 367, 566–568 [Google Scholar]
- 22. Gardian Z., Bumba L., Schrofel A., Herbstova M., Nebesarova J., Vacha F. (2007) Organisation of photosystem I and photosystem II in red alga Cyanidium caldarium. Encounter of cyanobacterial and higher plant concepts. Biochim. Biophys. Acta 1767, 725–731 [DOI] [PubMed] [Google Scholar]
- 23. Busch A., Nield J., Hippler M. (2010) The composition and structure of photosystem I-associated antenna from Cyanidioschyzon merolae. Plant J. 62, 886–897 [DOI] [PubMed] [Google Scholar]
- 24. Ohta H., Suzuki T., Ueno M., Okumura A., Yoshihara S., Shen J.-R., Enami I. (2003) Extrinsic proteins of photosystem II. An intermediate member of PsbQ protein family in red algal PSII. Eur. J. Biochem. 270, 4156–4163 [DOI] [PubMed] [Google Scholar]
- 25. Enami I., Kikuchi S., Fukuda T., Ohta H., Shen J. R. (1998) Binding and functional properties of four extrinsic proteins of photosystem II from a red alga, Cyanidium caldarium, as studied by release-reconstitution experiments. Biochemistry 37, 2787–2793 [DOI] [PubMed] [Google Scholar]
- 26. Bricker T. M., Roose J. L., Fagerlund R. D., Frankel L. K., Eaton-Rye J. J. (2012) The extrinsic proteins of photosystem II. Biochim. Biophys. Acta 1817, 121–142 [DOI] [PubMed] [Google Scholar]
- 27. Kargul J., Barber J. (2008) Photosynthetic acclimation. Structural reorganisation of light harvesting antenna. Role of redox-dependent phosphorylation of major and minor chlorophyll a/b binding proteins. FEBS J. 275, 1056–1068 [DOI] [PubMed] [Google Scholar]
- 28. Horton P., Johnson M. P., Perez-Bueno M. L., Kiss A. Z., Ruban A. V. (2008) Photosynthetic acclimation. Does the dynamic structure and macro-organisation of photosystem II in higher plant grana membranes regulate light harvesting states? FEBS J. 275, 1069–1079 [DOI] [PubMed] [Google Scholar]
- 29. Ruban A. V., Johnson M. P., Duffy C. D. (2012) The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta 1817, 167–181 [DOI] [PubMed] [Google Scholar]
- 30. Ivanov A. G., Sane P. V., Hurry V., Oquist G., Huner N. P. (2008) Photosystem II reaction centre quenching. Mechanisms and physiological role. Photosynth. Res. 98, 565–574 [DOI] [PubMed] [Google Scholar]
- 31. Kirilovsky D., Kaňa R., Prášil O. (2013) Mechanisms modulating energy arriving at reaction centers in cyanobacteria. in Non-photochemical Quenching and Thermal Energy Dissipation in Plants, Algae and Cyanobacteria (Demmig-Adams B., Adams W., Garab G., Govindjee, eds) Springer, Netherlands, Dordrecht, in press [Google Scholar]
- 32. Delphin E., Duval J. C., Etienne A. L., Kirilovsky D. (1996) State transitions or ΔpH-dependent quenching of photosystem II fluorescence in red algae. Biochemistry 35, 9435–9445 [DOI] [PubMed] [Google Scholar]
- 33. Delphin E., Duval J. C., Etienne A. L., Kirilovsky D. (1998) ΔpH-dependent photosystem II fluorescence quenching induced by saturating, multiturnover pulses in red algae. Plant Physiol. 118, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vass I., Cser K. (2009) Janus-faced charge recombinations in photosystem II photoinhibition. Trends Plant Sci. 14, 200–205 [DOI] [PubMed] [Google Scholar]
- 35. Krieger A., Moya I., Weis E. (1992) Energy-dependent quenching of chlorophyll-a fluorescence. Effect of pH on stationary fluorescence and picosecond-relaxation kinetics in thylakoid membranes and photosystem-II preparations. Biochim. Biophys. Acta 1102, 167–176 [Google Scholar]
- 36. Bruce D., Samson G., Carpenter C. (1997) The origins of nonphotochemical quenching of chlorophyll fluorescence in photosynthesis. Direct quenching by P680(+) in photosystem II enriched membranes at low pH. Biochemistry 36, 749–755 [DOI] [PubMed] [Google Scholar]
- 37. Minoda A., Sakagami R., Yagisawa F., Kuroiwa T., Tanaka K. (2004) Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 45, 667–671 [DOI] [PubMed] [Google Scholar]
- 38. Adachi H., Umena Y., Enami I., Henmi T., Kamiya N., Shen J. R. (2009) Towards structural elucidation of eukaryotic photosystem II. Purification, crystallization and preliminary x-ray diffraction analysis of photosystem II from a red alga. Biochim. Biophys. Acta 1787, 121–128 [DOI] [PubMed] [Google Scholar]
- 39. Cunningham F. X., Jr., Lee H., Gantt E. (2007) Carotenoid biosynthesis in the primitive red alga Cyanidioschyzon merolae. Eukaryot. Cell 6, 533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oren A., Kühl M., Karsten U. (1995) An endoevaporitic microbial mat within a gypsum crust. Zonation of phototrophs, photopigments, and light penetration. Mar. Ecol. Prog. Ser. 128, 151–159 [Google Scholar]
- 41. Rudowska Ł., Gieczewska K., Mazur R., Garstka M., Mostowska A. (2012) Chloroplast biogenesis. Correlation between structure and function. Biochim. Biophys. Acta. 1817, 1380–1387 [DOI] [PubMed] [Google Scholar]
- 42. Kaminskaya O., Kern J., Shuvalov V. A., Renger G. (2005) Extinction coefficients of cytochromes b559 and c550 of Thermosynechococcus elongatus and cyt b559/PS II stoichiometry of higher plants. Biochim. Biophys. Acta 1708, 333–341 [DOI] [PubMed] [Google Scholar]
- 43. Quigg A., Kotabová E., Jarešová J., Kaňa R., Setlík J., Sedivá B., Komárek O., Prášil O. (2012) Photosynthesis in Chromera velia represents a simple system with high efficiency. PLoS One 7, e47036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kolber Z. S., Prasil O., Falkowski P. G. (1998) Measurements of variable chlorophyll fluorescence using fast repetition rate techniques. Defining methodology and experimental protocols. Biochim. Biophys. Acta 1367, 88–106 [DOI] [PubMed] [Google Scholar]
- 45. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 46. Schägger H. (2006) Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 47. Oostergetel G. T., Keegstra W., Brisson A. (1998) Automation of specimen selection and data acquisition for protein electron crystallography. Ultramicroscopy 74, 47–59 [Google Scholar]
- 48. van Heel M. (1987) Similarity measures between images. Ultramicroscopy 21, 95–100 [Google Scholar]
- 49. Bumba L., Havelková-Dousová H., Husák M., Vácha F., (2004) Structural characterization of photosystem II complex from red alga Porphyridium cruentum retaining extrinsic subunits of the oxygen-evolving complex. Eur. J. Biochem. 271, 2967–2975 [DOI] [PubMed] [Google Scholar]
- 50. Dixon M. (1953) The determination of enzyme inhibitor constants. Biochem. J. 55, 170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lavergne J. (1982) Mode of action of 3-(3,4-dichlorophenyl)-1,1-dimethylurea. Evidence that the inhibitor competes with plastoquinone for binding to a common site on the acceptor side of photosystem II. Biochim. Biophys. Acta 682, 345–353 [Google Scholar]
- 52. Boussac A., Sugiura M., Rappaport F. (2011) Probing the quinone binding site of photosystem II from Thermosynechococcus elongatus containing either PsbA1 or PsbA3 as the D1 protein through the binding characteristics of herbicides. Biochim. Biophys. Acta 1807, 119–129 [DOI] [PubMed] [Google Scholar]
- 53. Koua F. H., Umena Y., Kawakami K., Shen J. R. (2013) Structure of Sr-substituted photosystem II at 2.1 A resolution and its implications in the mechanism of water oxidation. Proc. Natl. Acad. Sci. U.S.A. 110, 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaňa R., Kotabová E., Komárek O., Šedivá B., Papageorgiou G. C., Govindjee, Prášil O. (2012) The slow S to M fluorescence rise in cyanobacteria is due to a state 2 to state 1 transition. Biochim. Biophys. Acta 1817, 1237–1247 [DOI] [PubMed] [Google Scholar]
- 55. Shen J. R., Kamyía N. (2000) Crystallization and the crystal properties of the oxygen-evolving photosystem II from Synechococcus vulcanus. Biochemistry 39, 14739–14744 [DOI] [PubMed] [Google Scholar]
- 56. Boussac A., Rappaport F., Carrier P., Verbavatz J. M., Gobin R., Kirilovsky D., Rutherford A. W., Sugiura M. (2004) Biosynthetic Ca2+/Sr2+ exchange in the photosystem II oxygen-evolving enzyme of Thermosynechococcus elongatus. J. Biol. Chem. 279, 22809–22819 [DOI] [PubMed] [Google Scholar]
- 57. Roose J. L., Kashino Y., Pakrasi H. B. (2007) The PsbQ protein defines cyanobacterial photosystem II complexes with highest activity and stability. Proc. Natl. Acad. Sci. U.S.A. 104, 2548–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cullen M., Ray N., Husain S., Nugent J., Nield J., Purton S. (2007) A highly active histidine-tagged Chlamydomonas reinhardtii photosystem II preparation for structural and biophysical analysis. Photochem. Photobiol. Sci. 6, 1177–1183 [DOI] [PubMed] [Google Scholar]
- 59. Wang Z. G., Xu T. H., Liu C., Yang C. H. (2010) Fast isolation of highly active photosystem II core complexes from spinach. J. Integr. Plant Biol. 52, 793–800 [DOI] [PubMed] [Google Scholar]
- 60. Pagliano C., Chimirri F., Saracco G., Marsano F., Barber J. (2011) One-step isolation and biochemical characterization of a highly active plant PSII monomeric core. Photosynth. Res. 108, 33–46 [DOI] [PubMed] [Google Scholar]
- 61. de Groot H. J. (2010) Integration of catalysis with storage for the design of multi-electron photochemistry devices for solar fuel. Appl. Magn. Reson. 37, 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nocera D. G. (2012) The artificial leaf. Acc. Chem. Res. 45, 767–776 [DOI] [PubMed] [Google Scholar]
- 63. Telfer A. (2005) Too much light? How β-carotene protects the photosystem II reaction centre. Photochem. Photobiol. Sci. 4, 950–956 [DOI] [PubMed] [Google Scholar]
- 64. Kok B. (1956) On the inhibition of photosynthesis by intense light. Biochim. Biophys. Acta 21, 234–244 [DOI] [PubMed] [Google Scholar]
- 65. Aro E. M., Virgin I., Andersson B. (1993) Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143, 113–134 [DOI] [PubMed] [Google Scholar]
- 66. Long S. P., Humphries S., Falkowski P. G. (1994) Photoinhibition of photosynthesis in nature. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 633–662 [Google Scholar]
- 67. Frank H. A., Cogdell R. J. (1993) Photochemistry of carotenoids. in Carotenoids in Photosynthesis (Young A., Britton G., eds) pp. 252–326, Chapman and Hall, London [Google Scholar]
- 68. Szabó I., Bergantino E., Giacometti G. M. (2005) Light and oxygenic photosynthesis. Energy dissipation as a protection mechanism against photo-oxidation. EMBO Rep. 6, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Santabarbara S., Casazza A. P., Ali K., Economou C. K., Wannathong T., Zito F., Redding K. E, Rappaport F., Purton S. (2013) The requirement for carotenoids in the assembly and function of the photosynthetic complexes in Chlamydomonas reinhardtii. Plant Physiol. 161, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sozer O., Komenda J., Ughy B., Domonkos I., Laczkó-Dobos H., Malec P., Gombos Z., Kis M. (2010) Involvement of carotenoids in the synthesis and assembly of protein subunits of photosynthetic reaction centers of Synechocystis sp. PCC 6803. Plant Cell Physiol. 51, 823–835 [DOI] [PubMed] [Google Scholar]
- 71. Dall'Osto L., Holt N. E., Kaligotla S., Fuciman M., Cazzaniga S., Carbonera D., Frank H. A., Alric J., Bassi R. (2012) Zeaxanthin protects plant photosynthesis by modulating chlorophyll triplet yield in specific light-harvesting antenna subunits. J. Biol. Chem. 287, 41820–41834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Koblizek M., Ciscato M., Komenda J., Kopecky J., Siffel P., Masojidek J. (1999) Photoadaptation in the green alga Spongiochloris sp. A three-fluorometer study. Photosynthetica 37, 307–323 [Google Scholar]
- 73. Holt N. E., Zigmantas D., Valkunas L., Li X. P., Niyogi K. K., Fleming G. R. (2005) Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307, 433–436 [DOI] [PubMed] [Google Scholar]
- 74. Johnson M. P., Havaux M., Triantaphylidès C., Ksas B., Pascal A. A., Robert B., Davison P. A., Ruban A. V., Horton P. (2007) Elevated zeaxanthin bound to oligomeric LHCII enhances the resistance of Arabidopsis to photooxidative stress by a lipid-protective, antioxidant mechanism. J. Biol. Chem. 282, 22605–22618 [DOI] [PubMed] [Google Scholar]
- 75. Havaux M., Dall'osto L., Bassi R. (2007) Zeaxanthin has enhanced antioxidant capacity with respect to all other xantophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145, 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Havaux M. (1998) Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci. 3, 147–151 [Google Scholar]
- 77. Gruszecki W. I., Strzałka K. (2005) Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta 1740, 108–115 [DOI] [PubMed] [Google Scholar]
- 78. Cunningham F. X., Dennenberg R. J., Mustardy L., Jursinic P. A., Gantt E. (1989) Stoichiometry of photosystem I, photosystem II, and phycobilisomes in the red alga Porphyridium cruentum as a function of growth irradiance. Plant Physiol. 91, 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cunningham F. X., Jr., Gantt E. (2005) A study in scarlet. Enzymes of ketocarotenoid biosynthesis in the flowers of Adonis aestivalis. Plant J. 41, 478–492 [DOI] [PubMed] [Google Scholar]
- 80. Cser K., Vass I. (2007) Radiative and non-radiative charge recombination pathways in photosystem II studied by thermoluminescence and chlorophyll fluorescence in the cyanobacterium Synechocystis 6803. Biochim. Biophys. Acta 1767, 233–243 [DOI] [PubMed] [Google Scholar]
- 81. Kargul J., Boehm M., Morgner N., Robinson C. V., Nixon P. J., Barber J. (2012) Compositional and structural analyses of the photosystem II from the red alga Cyanidioschyzon merolae. in Photosynthesis: Research for Food, Fuel and Future (Lu C., Zhang L., Kuang T., eds) pp. 59–63, Zhejiang University Press/Springer-Verlag, Hangzhao, Beijing, China [Google Scholar]
- 82. Kuhl H., Rögner M., Van Breemen J. F., Boekema E. J. (1999) Localization of cyanobacterial PS II donor-side subunits by electron microscopy and the supramolecular organization of PS II in the thylakoid membrane. Eur. J. Biochem. 266, 453–459 [DOI] [PubMed] [Google Scholar]
- 83. Arteni A. A., Nowaczyk M., Lax J., Kouril R., Rögner M., Boekema E. J. (2005) Single particle electron microscopy in combination with mass spectrometry to investigate novel complexes of membrane proteins. J. Struct. Biol. 149, 325–331 [DOI] [PubMed] [Google Scholar]
- 84. Nowaczyk M. M., Hebeler R., Schlodder E., Meyer H. E., Warscheid B., Rögner M. (2006) Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18, 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shi L. X., Lorković Z. J., Oelmuller R., Schroder W. P. (2000) The low molecular mass PsbW protein is involved in the stabilization of the dimeric photosystem II complex in Arabidopsis thaliana. J. Biol. Chem. 275, 37945–37950 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.