Abstract
Drug-induced hepatotoxicity (DIH) is a common adverse event that is associated with both antiretroviral (ARV) and anti-tuberculosis drugs (ATD). Moreover, the genetic variations predisposing ARV- and ARV-ATD-induced liver toxicity in African populations are not well investigated, despite the two diseases being the major global health problems in sub-Saharan Africa. We performed a genome-wide association study (GWAS) and replication study to identify the genetic variants linked to the risk of developing DIH due to ARV drugs alone, and ARV-ATD co-treatment in Ethiopian HIV-positive patients. Treatment-naïve newly diagnosed HIV patients (n = 719) with or without tuberculosis (TB) co-infection were enrolled prospectively and received efavirenz-based ARV therapy with or without rifampicin-based short course ATD, respectively. Whole-genome genotyping was performed by using the Illumina Omni Express Exome Bead Chip genotyping array with 951,117 single nucleotide polymorphisms (SNPs) on a total of 41 cases of DIH, and 452 people without DIH (treatment tolerants). The replication study was carried out for 100 SNPs with the lowest p-values (top SNPs) by using an independent cohort consisting of 18 DIH cases and 208 treatment tolerants. We identified a missense SNP rs199650082 (2756G→A, R919Q, p = 1.4 × 10−6, odds ratio [OR] = 18.2, 95% confidence interval [CI] = 7.1–46.9) in an endoplasmic reticulum to the nucleus signaling-1 (ERN1) gene on chromosome 17 to be associated with DIH in the ARV-only cohort. In the ARV-ATD co-treatment groups, rs4842407, a long intergenic noncoding RNAs (lincRNAs) transcript variant on chromosome 12, was associated with DIH (p = 5.3 × 10−7, OR = 5.4, 95% CI = 2.8–10.3). These genetic variants that are putatively associated with DIH due to ARV drugs alone and ARV-ATD co-treatment establish a foundation for future personalized medicine in people with HIV and TB and call for larger studies in independent populations.
Keywords: : antiretroviral drugs, anti-tuberculosis drugs, drug-induced hepatotoxicity, genome-wide association study, GWAS, OMICS, Africa
Introduction
Antiretroviral therapy (ART) has dramatically lowered HIV-/AIDS-related mortality globally, but treatment-associated liver toxicity remains a challenge (Fink and Bloch, 2013; Ugiagbe et al., 2012; Yimer et al., 2011, 2012, 2014). To end the AIDS epidemic, several African countries, including Ethiopia, are currently making important progress in scaling up HIV testing and ART in response to the UNAIDS (2014) 90–90–90 target: 90% of people with HIV diagnosed, 90% of diagnosed people on treatment, and 90% of treated people having fully suppressed viral replication by 2020. On the other hand, antiretroviral (ARV) treatment-associated hepatotoxicity is of increasing concern in the management of patients with HIV/AIDS (Jones and Nunez, 2012; Reisler et al., 2003; Turkova et al., 2009; Ugiagbe et al., 2012; Walker, 2007).
Tuberculosis (TB) is the most common opportunistic infection in people living with HIV/AIDS, and concomitant treatment of the two diseases is challenging because of drug interactions and overlapping toxicities. The risk of developing drug-induced hepatotoxicity (DIH) after treatment initiation is much higher for TB-HIV co-infected patients than for those with HIV or TB mono-infection (Araujo-Mariz et al., 2016; Mugusi et al., 2012; Yimer et al., 2011, 2014). ARV- and anti-tuberculosis drug (ATD)-induced hepatotoxicity have become major clinical challenges because of high morbidity, mortality, and frequent hospitalization as a leading cause of acute liver failure and complications requiring liver transplantation (Elsharkawy et al., 2013; Fink and Bloch, 2013; Nunez, 2010; Turkova et al., 2009).
The type and incidence of DIH varies between populations and geographical locations (Lamar and Nunez, 2011; Mugusi et al., 2012; Yimer et al., 2011). The mechanisms and risk factors in DIH are complex, involving both host genetics and environmental factors (Daly, 2010; Nunez, 2010). Previous candidate gene association studies have reported the potential role of genetic variation in Cytochrome P450 (CYP) 2B6 and ATP-binding cassette subfamily-B member-1 (ABCB1) for ARV-DIH with efavirenz- or nevirapine-based regimens (Mugusi et al., 2012; Ritchie et al., 2006; Yimer et al., 2011, 2012). Studies also reported an association between polymorphisms in drug-metabolizing genes (N-acetyltransferase-2 [NAT2], CYP2E1, glutathione S-transferase-1 [GSTM1], uridine diphosphateglucuronosyltransferase-1A1 [UGT1A1]) and mitochondrial superoxide dismutase (SOD2) with ATD-induced hepatotoxicity (Huang, 2014; Kim et al., 2015; Yimer et al., 2011).
Although candidate gene studies contribute to the discovery of genetic risk variants that are associated with ARV- and ATD-induced hepatotoxicity, the identified variants may account only for a proportion of the genetic variations. Genome-wide association studies (GWAS) have identified variant alleles that are associated with an increased risk of developing DIH after treatment intentions with various classes of drugs (Aithal and Grove, 2015; Daly et al., 2009; Urban et al., 2014). Recently, using GWAS in Ethiopian TB patients, we have reported genetic variations in ATP-/GTP-binding protein-like-4 (AGBL4) and family with sequence similarity 65 member-B (FAM65B) as potential risk factors for developing ATD-induced liver toxicity (Petros et al., 2016).
The genetic variations predisposing ARV- and ARV-ATD-induced liver toxicity in black African populations are not well investigated, despite the two diseases being the major public health problems in sub-Saharan Africa. As the scale of ART is increasing in sub-Saharan African countries, it is imperative to identify the genetic markers that are associated with an increased risk of ARV-induced liver toxicity for subsequent treatment interventions. This study reports on GWAS and replication studies of genetic variants in relation to DIH risk with ARV drugs alone, and ARV-ATD co-treatment, in Ethiopian HIV-positive people with or without TB co-infection.
Materials and Methods
Study participants
The study participants for the present GWAS were included from a recent observational, prospective cohort study where the incidence and patterns of ARV drugs and/or ATD-induced liver toxicities were investigated (Yimer et al., 2014). In brief, newly diagnosed treatment naïve patients enrolled into the following study arms were considered for the present study:
1. HIV-positive patients with CD4 count ≤200 cells/mm3 without TB co-infection receiving efavirenz-based ARV therapy only.
2. TB and HIV co-infected patients with CD4 count ≤200 cells/mm3 receiving both rifampicin-based short-course anti-TB drugs and efavirenz-based ARV therapy at the same time (Fig. 1). Following the World Health Organization (WHO) and national TB-HIV treatment guidelines valid during the study period, all TB-HIV patients initiated rifampicin-based ATD regimen and initiation of ARV was delayed for a maximum of 8 weeks after starting ATD treatment. Patients who developed DIH while on anti-TB drugs alone (before starting ARV therapy) were not included in the present study.
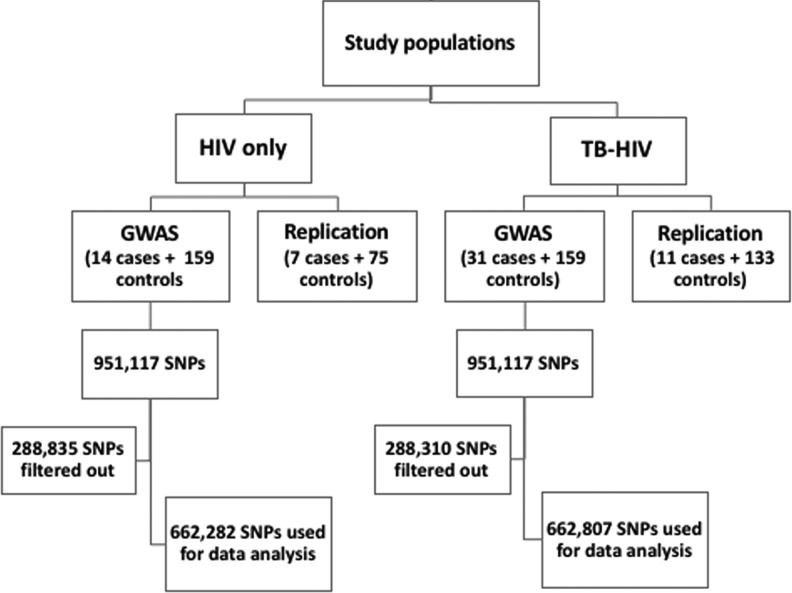
FIG. 1.
Study design, genotyping, and quality control flow chart. GWAS, genome-wide association study; HIV, human immunodeficiency virus; SNP, single nucleotide polymorphism; TB, tuberculosis.
The study participants were recruited from three health institutions: the Kazanchis and the Beletshachew Health Centers and the Black Lion specialized referral and teaching university hospital in Addis Ababa, Ethiopia. The inclusion criteria were HIV-positive men and non-pregnant women of age ≥18 years. Patients were excluded if they had a history of prior treatment for TB/HIV, known preexisting liver disease, or abnormal liver biochemistry before starting treatment; if they were positive for either hepatitis B virus surface antigen or anti-hepatitis C virus antibody, they were also excluded.
The study protocol and consent procedure were approved by the Institutional Review Board of College of Health Sciences, Addis Ababa University; the National Research Ethics Review Committee of Ethiopia, Ethical Review Board of Karolinska Institutet, Sweden, and Ethical Review Committee of RIKEN, Japan. Written informed consent was obtained from all the study participants before their inclusion in the study.
Drug treatment and patient follow-up
All patients received treatment according to the national and the WHO treatment guidelines for HIV and TB as previously described (Yimer et al., 2014). In brief, all HIV patients received ARV drugs containing efavirenz and lamivudine with stavudine, zidovudine, or tenofovir. TB-HIV coinfected patients also received short-course ATD consisting of rifampicin, isoniazid, pyrazinamide, and ethambutol for the first 2 months followed by rifampicin and isoniazid for the next 4 months. The patients were not on other known hepatotoxic drugs concurrently (except co-trimoxazole, which was given for TB and HIV co-infected patients according to the National Treatment Guideline). Liver function tests were carried out at baseline and on the 1st, 2nd, 4th, 8th, 12th, and 24th weeks after initiation of treatment.
Case definitions, severity grade, and pattern of hepatotoxicity
The criteria set by the International DIH Expert Working Group were used for DIH case definitions and pattern of hepatotoxicity (Aithal et al., 2011). The upper limit of normal (ULN) for liver biochemical parameters used for the study population were alanine aminotransferase (ALT 33 U/L, male; 29 U/L, female), aspartate aminotransferase (AST, 41 U/L), alkaline phosphatase (ALP, 128 U/L), and 1.0 mg/dL for total bilirubin (T Bil) (Yimer et al., 2014). All cases met at least one of the following criteria: (1) ALT ≥5 × ULN, (2) ALP ≥2 × ULN, or (3) ALT ≥3 × ULN along with T Bil ≥2 × ULN. Patients on ARV drugs alone or ARV-ATD co-treatment but who did not fulfill the case definitions and presented no clinical symptoms for DIH during the follow-up period were considered as treatment tolerant controls for the ARV drugs alone and ARV-ATD co-treatment groups, respectively.
The pattern of hepatotoxicity was defined by using an R-value, where R = (ALT/ULN)/(ALP/ULN). Cases were categorized as a hepatocellular (R ≥ 5), cholestatic (R ≤ 2), or mixed (2 < R < 5) pattern of DIH (Aithal et al., 2011). Clinical severity grading was determined by employing the highest measured values for each of the biochemical parameters during the course of DIH. Patients with grades 1 and 2 severities were grouped into a “mild-to-moderate” group, and those with grades 3 and 4 were classified into a “severe” group. Causality assessment for DIH was performed by using the Roussel Uclaf Causality Assessment Method (RUCAM) (Danan and Benichou, 1993).
Genotyping and quality control
Blood samples were collected, and DNA was extracted by using the QIAamp DNA Maxi Kit (QIAGEN GmbH, Hilden, Germany). Genotyping was conducted in RIKEN Center for Integrative Medical Sciences (Yokohama, Japan). For GWAS analysis, genotyping was performed on a total of 41 cases and 452 controls with Illumina Omni Express Exome Bead Chip genotyping array (Illumina, Inc., San Diego, CA, USA) according to the manufacturer's protocol. This array captures 951,117 single nucleotide polymorphisms (SNPs). Replication studies were then carried out for the top 100 SNPs (the lowest p-value) by using an independent cohort of 18 cases and 208 controls. Genotyping for the replication studies was done by using a multiplex polymerase chain reaction (PCR)–based Invader assay (Ohnishi et al., 2001) with the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA).
For data cleaning, as shown in Figure 1, systematic step-wise quality filtering of raw genotyping data was done by using PLINK v1.07 (Purcell et al., 2007). From an initial full set, those SNPs that were not mapped on autosomal chromosomes were filtered out. In addition, SNPs with a call rate <99%, minor allele frequency (MAF) <0.01, or deviated from expected Hardy-Weinberg equilibrium (p < 1.0 × 10−6) were removed from further analysis. A total of 662,282 and 662,807 SNPs passed the quality filter for the ARV drugs alone and ARV-ATD co-treatment groups, respectively. Individuals were checked for gender concordance between recorded clinical data and genotype-determined sex. Samples with a genotyping call rate greater than 99% were included in the analysis. The effect of population structure was assessed through principal component analysis (PCA) implemented in Eigenstrat (Price et al., 2006). Quantile-quantile (QQ) plots comparing the expected and observed p-values were performed in an R-statistical environment. To detect population stratification, genomic control inflation factor (λGC) was also calculated (Devlin and Roeder, 1999).
Statistical analysis
After the quality filter, the tests of associations were done by using PLINK (Purcell et al., 2007). For each SNP, Fisher's exact test using the three genetic inheritance models (dominant, recessive, allele frequency) were carried out to compare allele and genotype frequencies between cases and controls. SNPs were rank ordered according to the minimum p-value in the genetic models. A threshold for genome-wide significance was set as p < 7.5 × 10−8 after Bonferroni correction. SNPs with p-values below 10−5 were considered suggestive of genome-wide significance (Dahlin et al., 2015). Logistic regression analysis adjusted for clinical variables such as sex, body mass index, baseline CD4 count, and HIV viral load as covariates was also performed. These variables were associated with DIH as previously described (Yimer et al., 2008, 2011). A combined analysis of GWAS and replication studies was conducted by using the inverse-variance method (de Bakker et al., 2008). Manhattan plots were generated to visualize the results by using Haploview software (Barrett et al., 2005).
Results
For the GWAS analysis, there were 14 cases and 293 controls in the ARV drug-alone treatment group, and 27 cases and 159 controls in the ARV-ATD co-treatment group that passed the quality filter. The demographics and clinical characteristics of the study participants for ARV-alone and ARV-ATD co-treatment groups are described in Tables 1 and 2, respectively. There were statistically significant differences (p < 0.05) in liver function test values between cases and controls in the GWAS and the replication cohorts of both treatment groups. There were no statistically significant differences in the number of DIH cases and treatment tolerants with regard to the different ARV regimens. In both treatment groups, more than half of the cases had a cholestatic pattern of DIH. Ten percent of the cases developed a severe grade of hepatotoxicity in either a hepatocellular or a mixed pattern. Four participants in each treatment group had developed jaundice and necessitated discontinuation of treatment. There were no cases of liver transplantation. All of the cases had a minimum score of three (“possible”) in the RUCAM scoring system for DIH.
Table 1.
Demographics and Clinical Variables of the Study Participants in Antiretroviral Drug-Alone Treatment Group
| GWAS | Replication | |||||
|---|---|---|---|---|---|---|
| Variables | Cases | Controls | p | Cases | Controls | p |
| No. of patients | 14 | 293 | 7 | 75 | ||
| Sex (M, F) | 5, 9 | 115, 178 | 0.79 | 1, 6 | 8, 67 | 0.77 |
| Age (year), mean (SD) | 35.3 (12.6) | 37.1 (10.6) | 0.31 | 36.1 (8.8) | 33.0 (7.1) | 0.28 |
| BMI (kg/m2), mean (SD) | 20.6 (5.0) | 19.3 (2.8) | 0.11 | 19.4 (2.8) | 19.6 (2.6) | 0.85 |
| CD4 count, mean (SD) | 96.2 (64.2) | 99.2 (53.6) | 0.84 | 93.4 (68.1) | 109.4 (59.2) | 0.51 |
| Viral load, log mean (SD) | 5.0 (0.9) | 4.9 (1.0) | 0.64 | 4.9 (0.9) | 4.7 (1.2) | 0.54 |
| ALT (U/L), mean (SD) | 43.6 (15.4) | 30.1 (13.2) | <0.01 | 51.9 (48.3) | 31.8 (16.0) | <0.01 |
| AST (U/L), mean (SD) | 48.4 (13.0) | 36.1 (16.7) | <0.01 | 66.4 (23.5) | 34.2 (14.3) | <0.01 |
| ALP (U/L), mean (SD) | 199.8 (75.1) | 120.3 (63.1) | <0.01 | 203.5 (89.3) | 118.0 (36.2) | <0.01 |
| T Bil (mg/dL), mean (SD) | 0.7 (0.9) | 0.5 (0.3) | 0.70 | 0.6 (0.3) | 0.6 (1.1) | 0.97 |
| ARV drugs, N (%) | ||||||
| D4T/3TC/EFV | 7 (50.0) | 148 (50.5) | 0.97 | 3 (42.9) | 33 (44.0) | 0.96 |
| ZDV/3TC/EFV | 4 (28.6) | 91 (31.1) | 0.84 | 4 (57.1) | 42 (56.0) | 0.96 |
| TDF/3TC/EFV | 3 (21.4) | 54 (18.4) | 0.78 | |||
| DIH type, N (%) | ||||||
| Cholestatic | 10 (71.4) | 5 (71.4) | ||||
| Hepatocellular | 2 (14.3) | 1 (14.3) | ||||
| Mixed | 2 (14.3) | 1 (14.3) | ||||
| Severity grade, N (%) | ||||||
| Mild to moderate | 13 (92.6) | 6 (85.7) | ||||
| Severe | 1 (7.4) | 1 (14.3) | ||||
| RUCAM scale, N (%) | ||||||
| Definite (score >8) | 9 (64.3) | 4 (57.1) | ||||
| Probable (score 6–8) | 3 (21.4) | 2 (28.6) | ||||
| Possible (score 3–5) | 2 (14.3) | 1 (14.3) | ||||
3TC, lamivudine; ALP, alkaline phosphatase; ALT, alanine aminotransferase; ARV, antiretroviral; AST, aspartate aminotransferase; BMI, body mass index; D4T, stavudine; DIH, drug induced hepatotoxicity; EFV, efavirenz; GWAS, genome-wide association study; RUCAM, Roussel Uclaf Causality Assessment Method; SD, standard deviation; TDF, tenofovir; T Bil, total bilirubin; ZDV, zidovudine.
Table 2.
Demographics and Clinical Variables of the Study Participants in Antiretroviral and Anti-Tuberculosis Drugs Co-Treatment Group
| GWAS | Replication | |||||
|---|---|---|---|---|---|---|
| Variables | Cases | Controls | p | Cases | Controls | p |
| No. of patients | 27 | 159 | 11 | 133 | ||
| Sex (M, F) | 15, 12 | 73, 86 | 0.35 | 5, 6 | 69, 64 | 0.68 |
| Age (year), mean (SD) | 37.9 (10.7) | 35.7 (9.8) | 0.30 | 37.6 (9.5) | 37.3 (10.0) | 0.92 |
| BMI (kg/m2), mean (SD) | 18.9 (2.4) | 19.0 (2.6) | 0.92 | 19.8 (3.9) | 18.7 (3.2) | 0.29 |
| CD4 count, mean (SD) | 74.7 (55.1) | 90.6 (49.9) | 0.13 | 109.2 (61.6) | 89.8 (52.8) | 0.23 |
| Viral load, log mean (SD) | 5.3 (0.6) | 4.9 (0.9) | 0.04 | 5.0 (0.9) | 4.9 (0.9) | 0.71 |
| ALT (U/L), mean (SD) | 49.3 (19.8) | 30.5 (13.1) | <0.01 | 63.3 (28.2) | 31.1 (14.7) | <0.01 |
| AST (U/L), mean (SD) | 69.8 (40.2) | 36.2 (15.3) | <0.01 | 91.5 (54.8) | 37.6 (16.4) | <0.01 |
| ALP (U/L), mean (SD) | 220.7 (94.4) | 121.3 (68.8) | <0.01 | 206.5 (95.8) | 118.0 (64.6) | <0.01 |
| T Bil (mg/dL), mean (SD) | 0.9 (0.5) | 0.6 (0.3) | <0.01 | 0.8 (0.5) | 0.5 (0.4) | 0.06 |
| ARV drugs, N (%) | ||||||
| D4T/3TC/EFV | 11 (40.7) | 65 (40.8) | 0.99 | 2 (18.2) | 29 (21.8) | 0.78 |
| ZDV/3TC/EFV | 7 (25.9) | 47 (29.6) | 0.70 | 3 (27.3) | 32 (24.1) | 0.81 |
| TDF/3TC/EFV | 9 (33.3) | 47 (29.6) | 0.69 | 6 (54.5) | 72 (54.1) | 0.98 |
| DIH type, N (%) | ||||||
| Cholestatic | 21 (77.8) | 6 (54.5) | ||||
| Hepatocellular | − | 3 (27.3) | ||||
| Mixed | 6 (22.2) | 2 (18.2) | ||||
| Severity grade, N (%) | ||||||
| Mild to moderate | 24 (88.9) | 10 (90.9) | ||||
| Severe | 3 (11.1) | 1 (9.1) | ||||
| RUCAM scale, N (%) | ||||||
| Definite (score >8) | 17 (63.0) | 7 (63.6) | ||||
| Probable (score 6–8) | 7 (25.9) | 3 (27.3) | ||||
| Possible (score 3–5) | 3 (11.1) | 1 (9.1) | ||||
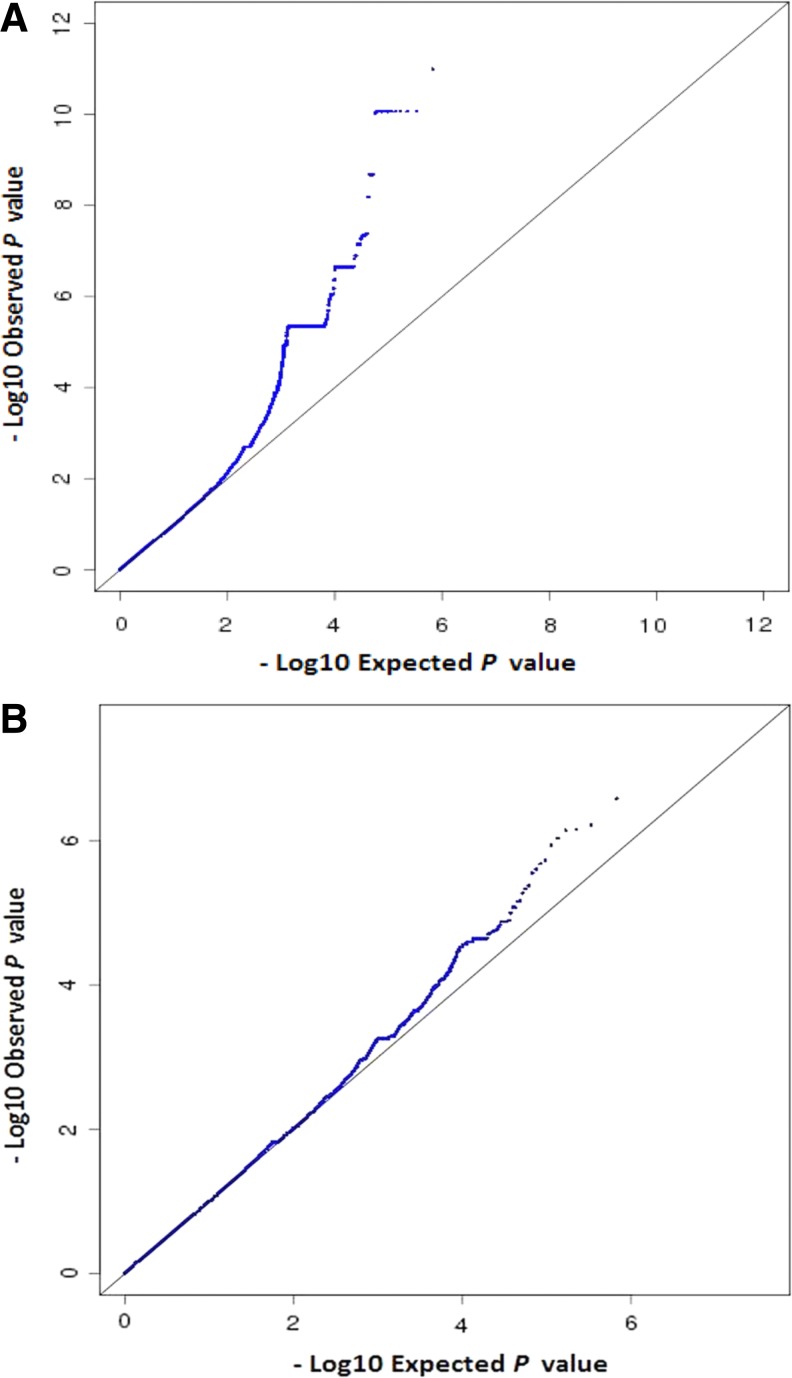
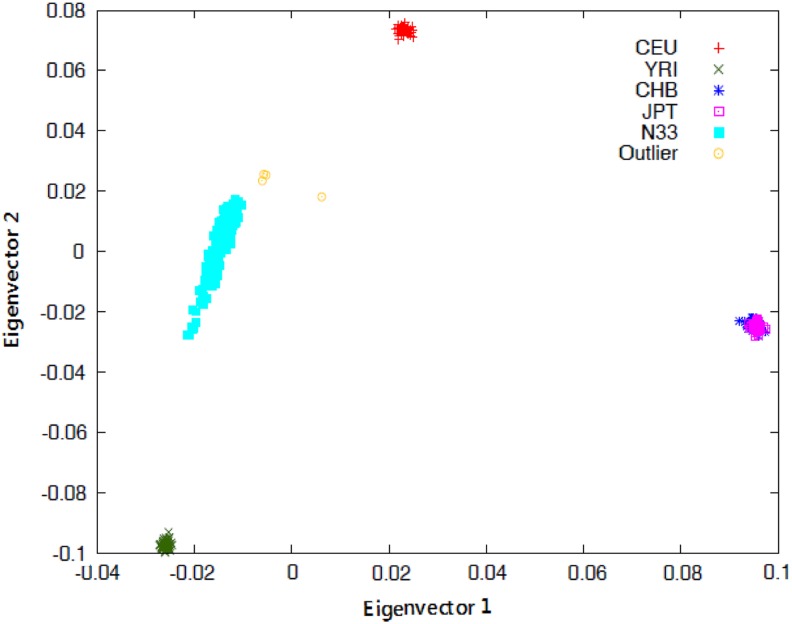
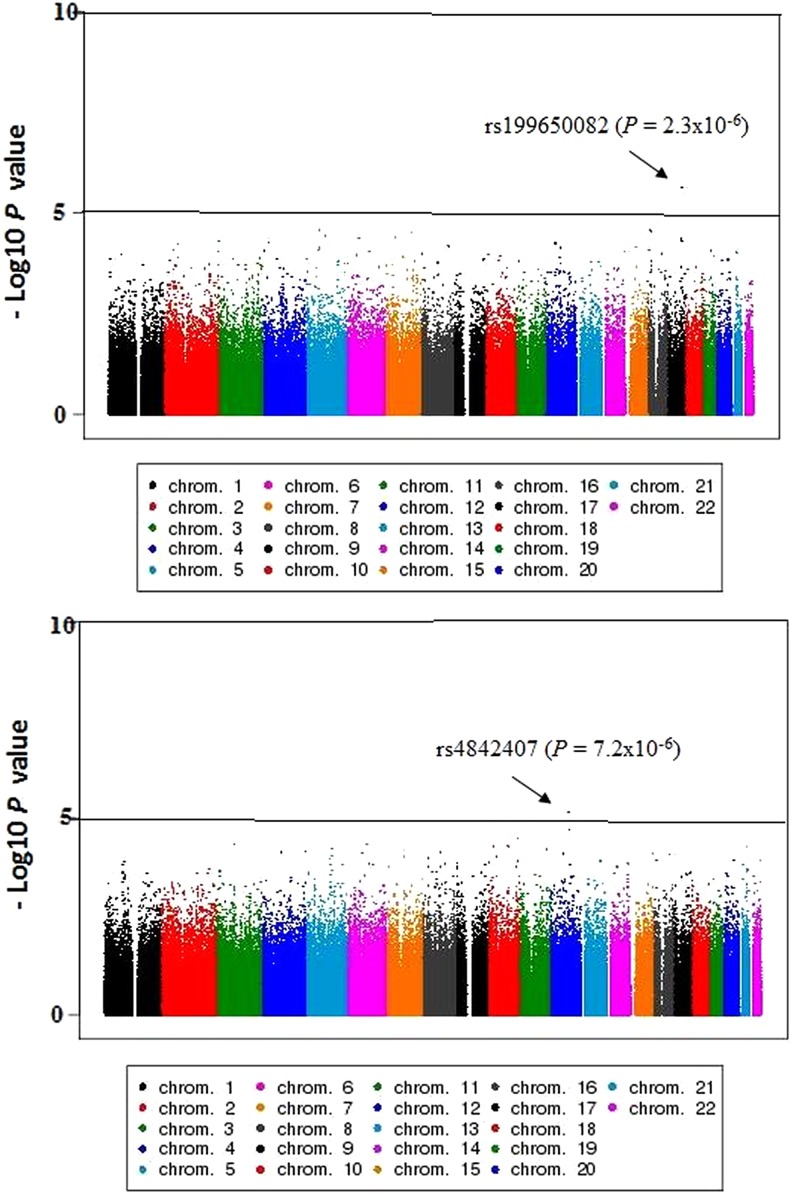
The QQ plots for the observed versus expected p-values are shown in Figure 2. The genomic inflation values were closer to one (λGC 0.98 and 1.00), indicating no systemic test statistic inflation, and population stratification was reasonably controlled. The PCA plot presented in Figure 3 indicated that our study population is distinct from the HapMap reference samples, even from the West African cluster. The Manhattan plots for the GWAS analyses are shown in Figure 4. The SNPs with the lowest p-value (top SNPs) in the GWAS after adjustment for sex, body mass index, CD4 count, and HIV viral load were nonsynonymous SNP (rs199650082, p = 2.3 × 10−6, odds ratio [OR] = 21.4, 95% confidence interval [CI] = 7.5–60.6) in an endoplasmic reticulum to nucleus signaling-1 (ERN1) gene on chromosome 17, and rs4842407 (p = 7.2 × 10−6, OR = 5.9, 95% CI = 2.7–12.8) within the intron of non-coding RNA on chromosome 12 for ARV drug-alone and ARV-ATD co-treatment groups, respectively (Supplementary Table S1).
FIG. 2.
QQ plots for the observed versus expected p-values. (A) ARV drug-alone treatment group (λGC = 0.98). (B) ARV-ATD drug co-treatment group (λGC = 1.00). ARV, antiretroviral; ARV-ATD, antiretroviral and anti-tuberculosis; QQ, quantile-quantile.
FIG. 3.
PCA plot of the study population. Ethiopians coded as N33 with the HapMap reference population. CEU, Utah residents of Northern and Western European ancestry; CHB, Han Chinese individuals from Beijing, China; JPT, Japanese individuals from Tokyo, Japan; PCA, principal component analysis; YRI, Yoruba trios from Ibadan, Nigeria.
FIG. 4.
−Log10 p values of logistic regression across chromosomes. (A) ARV drug-alone treatment group. (B) ARV-ATD drugs co-treatment group.
The top SNPs for the replication studies are shown in Supplementary Table S2. The top SNPs in the combined analysis after adjustment for covariates were rs199650082 (p = 1.4 × 10−6, OR = 18.2, 95% CI = 7.1–46.9), and rs4842407 (p = 5.3 × 10−7, OR = 5.4, 95% CI = 2.8–10.3) for ARV drug-alone and ARV-ATD co-treatment groups, respectively (Table 3). Top SNPs for DILI in pharmacokinetic genes and human leukocyte antigen (HLA) region for ARV drug-alone and ARV-ATD co-treatment groups are indicated in Supplementary Table S3.
Table 3.
Top Single Nucleotide Polymorphisms Associated with Drug-Induced Hepatotoxicity in the Combined Analysis for Antiretroviral Drug-Alone and Antiretroviral and Anti-Tuberculosis Drugs Co-Treatment Groups
| Group | SNP | Chr (loci) | Alleles (RA) | Study | Cases/controls | MAF | p_min | p_adj | OR (95% CI) | Gene/loci |
|---|---|---|---|---|---|---|---|---|---|---|
| ARV | rs199650082 | 17 (62121526) | T/C (T) | GWAS | 14/293 | 0.03 | 4.3 × 10−6 | 2.3 × 10−6 | 21.4 (7.5 − 60.6) | ERN1 |
| Rep | 7/66 | 0.02 | 2.6 × 10−1 | 2.3 × 10−1 | 6.8 (0.3–15.8) | |||||
| Comb | 21/359 | 0.03 | 1.8 × 10−6 | 1.4 × 10−6 | 18.2 (7.1–46.9) | |||||
| rs7804397 | 7 (116857547) | A/C (A) | GWAS | 14/293 | 0.09 | 7.3 × 10−6 | 3.0 × 10−5 | 7.1 (3.1–16.2) | ST7 | |
| Rep | 7/66 | 0.08 | 5.9 × 10−1 | 1.0 × 100 | NA | |||||
| Comb | 21/359 | 0.09 | 1.1 × 10−5 | 3.0 × 10−5 | 7.1 (3.4–14.9) | |||||
| rs16947045 | 17 (61770954) | T/C (T) | GWAS | 14/293 | 0.05 | 1.5 × 10−5 | 4.7 × 10−5 | 7.4 (2.9–18.7) | MAP3K3 | |
| Rep | 7/66 | 0.05 | 1.0 × 100 | 1.0 × 100 | NA | |||||
| Comb | 21/359 | 0.05 | 3.4 × 10−5 | 4.7 × 10−5 | 7.4 (3.1–17.5) | |||||
| ARV-ATD | rs4842407 | 12 (79201073) | T/C (C) | GWAS | 27/159 | 0.44 | 2.3 × 10−7 | 7.2 × 10−6 | 5.9 (2.7–12.8) | LOC642550 |
| Rep | 8/108 | 0.44 | 1.6 × 10−2 | 2.2 × 10−2 | 4.2 (1.2–14.4) | |||||
| Comb | 35/267 | 0.44 | 3.0 × 10−7 | 5.3 × 10−7 | 5.4 (2.8–10.3) | |||||
| rs11012476 | 10 (21292923) | A/G (A) | GWAS | 27/159 | 0.03 | 5.7 × 10−5 | 5.1 × 10−5 | 13.7 (4.3–43.5) | NEBL | |
| Rep | 8/107 | 0.02 | 2.4 × 10−2 | 1.9 × 10−2 | 16.9 (1.6–178) | |||||
| Comb | 35/266 | 0.03 | 9.6 × 10−7 | 2.8 × 10−6 | 14.3 (5.3–39.9) | |||||
| rs251891 | 5 (115050362) | A/C (A) | GWAS | 27/159 | 0.10 | 1.2 × 10−4 | 6.0 × 10−5 | 5.8 (2.7–12.2) | TMED7 | |
| Rep | 8/104 | 0.13 | 8.9 × 10−3 | 1.3 × 10−2 | 4.5 (1.4–13.9) | |||||
| Comb | 35/263 | 0.11 | 3.5 × 10−6 | 2.5 × 10−6 | 5.2 (2.9–9.5) |
ARV-ATD, antiretroviral and anti-tuberculosis drugs co-treatment; Chr (loci), chromosome, and chromosomal loci based on NCBI built 37; CI, confidence interval; Comb, combined analysis; ERN1, endoplasmic reticulum to nucleus signaling-1; MAF, minor allele frequency; NA, not applicable; OR, odds ratio; p_adj, logistic p-value after adjustment for covariates; p_min, minimum p-value among allelic, dominant, and recessive models of Fisher's exact test, and p-value of inverse variance combined analysis; RA, risk allele; Rep, replication study; SNP, single nucleotide polymorphism.
Discussion
Genetic determinants of HIV treatment response are currently firmly on the global health research agenda as efforts to improve access to treatment scale up (Soko et al., 2016). In this broader and global context, using a candidate gene approach, we have investigated pharmacogenetic markers for ARV alone (n = 285) or with ATD co-treatment-induced liver toxicity (n = 353) (Yimer et al., 2011, 2012). As a continuation, we evaluated the patterns of ARV and/or ATD drug-induced liver toxicities in a large prospective cohort study (n = 1060) (Yimer et al., 2014). In the present study, we carried out GWAS and a replication study in a total of 719 patients treated with either ARV drugs alone or ATD to identify genetic variants associated with ARV-induced liver toxicity. Our preliminary finding indicates the potential role of a missense SNP rs199650082 (2756G→A, R919Q) in an ERN1 gene on chromosome 17, and rs4842407 located in the long intergenic noncoding RNA (lincRNA) on chromosome 12 for ARV-alone- and ARV-ATD-induced liver toxicity, respectively. To our knowledge, this is the first GWAS for ART-associated DIH.
The participants for the current study were selected from a recent prospective cohort study (Yimer et al., 2014) by considering DIH cases developed during ART treatment only (in HIV only patients) or TB-HIV patients who developed DIH after initiation of ART therapy. In the latter cohort, anti-TB therapy was initiated 4–8 weeks before ART initiation. The median time for DIH onset among Ethiopian HIV patients who initiated ART alone was 4 weeks; whereas for TB-HIV patients receiving anti-TB therapy alone, the median DIH onset was 2 weeks after treatment initiation (Yimer et al., 2014). TB-HIV patients who developed DIH during prior anti-TB therapy (before starting ART) were excluded from this study.
All the TB-HIV patients who developed liver toxicity (cases) in the present study tolerated the prior anti-TB therapy and developed DIH after initiating ART. Therefore, it is plausible to assume that the DIH event is more likely due to ART but not due to the concomitantly administered anti-TB drugs, although synergistic effects between ARV and anti-TB drugs to elicit the event cannot be ruled out. If this assumption holds true, findings from the present study may reflect possible genetic markers for ARV-induced liver toxicity.
In the present study, we identified SNPs that are suggestive of importance to predict DIH. The top SNP (rs199650082) in the combined analysis of ARV drugs treatment group is a missense SNP (R919Q [2756G→A]) in the ERN1 gene. The protein encoded by this gene is the endoplasmic reticulum (ER) to nucleus signaling 1 protein, a human homologue of the yeast serine/threonine-protein kinase/endoribonuclease inositol-requiring enzyme 1 (IRE1) gene product (Tirasophon et al., 1998). This protein possesses intrinsic kinase and endoribonuclease activities, and it is important in altering gene expression as a response to ER-based stress signals (Chen et al., 2014a; Itzhak et al., 2014). Emerging evidence indicates that stress in ER makes a substantial contribution to the pathogenesis of DIH (Chen et al., 2014b). Perhaps the ARV drugs have contributed to stress in ER.
Another top SNP (rs16947045) identified in the combined analysis of the ARV drug treatment group was located in mitogen-activated protein kinase-3 (MAP3K3/MEKK3). This gene directly regulates the stress-activated protein kinase pathway that participates in the regulation of cellular responses to various extracellular signals (Ellinger-Ziegelbauer et al., 1997). Although further analysis is required to clarify the functional importance of ERN1 and MAP3K3 genes, variation in the expression of proteins that are encoded by these genes might contribute to individual differences for ARV-DIH susceptibility.
In the ARV-ATD co-treatment group, the top SNP (rs4842407) had a consistently strong signal in the GWAS, the replication study, and the combined analyses, both before and after adjustment for covariates. The SNP rs4842407 is a lincRNAs transcript variant located between LOC642550 and SYT1 on chromosome 12. Long noncoding RNAs (lncRNAs) are transcribed RNA molecules (>200 nucleotides in length) that structurally resemble mRNAs but do not encode proteins. Thousands of lincRNAs are now known; however, many of their functions are still unknown.
Recently, lincRNAs are emerging as important regulators in a wide range of biological processes, including proliferation, apoptosis, and differentiation (Cai et al., 2016; Shi et al., 2013). Liver-expressed lincRNA promoters show greater enrichment for proximal binding of liver transcription factors than protein-coding gene promoters, which may reflect the higher conservation of liver lincRNA promoters (Melia et al., 2016). In fact, recently, hepatic lncRNAs are reported to be involved in the progression of liver fibrosis (He et al., 2014; Yu et al., 2016; Zheng et al., 2015). Previous GWAS studies revealed some SNPs within the lincRNAs as disease associated (Hirano et al., 2015; Radtke et al., 2009).
The significant association of rs4842407 with ARV-ATD-induced hepatotoxicity in the present study may indicate its relevance in determining predisposition to DIH in TB-HIV coinfected patients receiving dual treatment for the two diseases. Alternatively, this SNP might be in linkage disequilibrium (LD) with other SNPs that are implicated in the development of DIH. If the results are replicated in a larger sample size, rs4842407 may serve as a marker for DIH susceptibility during ARV-ATD co-treatment.
Candidate gene studies conducted on genetic risk factors contributing to ARV drugs and/or ATD-induced hepatotoxicity identified genetic variants in genes involved in drug metabolism (CYP2B6, NAT2), drug transporter proteins (ABCB1), and HLA region (Daly, 2016; Mugusi et al., 2012; Yimer et al., 2011, 2012). In addition, variants in AGBL4 and FAM65B were identified as potential risk factors for ATD-induced liver toxicity through GWAS (Petros et al., 2016). In the current study, we did not find genetic variants that passed genome-wide significance level in both the pharmacokinetic-related genes and HLA region. But we found a possible association between SNPs rs11642957 (p = 9.5 × 10−5) in ATP-binding cassette subfamily-C member-1 (ABCC1, membrane-bound drug transporter) and rs9276370 (p = 2.8 × 10−4) in the vicinity of HLA-DQA2 in the ARV drug-alone and ARV-ATD co-treatment groups, respectively (Supplementary Table S3). Due to these drugs, DIH may be related to the combined effect of the newly identified variants, the pharmacokinetic and immune-related gene variants.
The incidence of ARV- and ATD-induced liver toxicity displays wide variability among populations. While receiving the same type of treatment regimen, HIV (16% vs. 6%) and TB-HIV co-infected (30% vs. 10%) patients from Ethiopia presented a higher incidence of DIH compared with patients from Tanzania (Mugusi et al., 2012; Yimer et al., 2011 2012, 2014). Ethiopians display a distinct pharmacogenetic profile and CYP enzyme activities compared with other populations, including Caucasian, Asians, and other Black African populations. Higher CYP3A (Gebeyehu et al., 2011), CYP2A6 (Aklillu et al., 2014), CYP2B6 (Ngaimisi et al., 2013), SLCO1B1 (Aklillu et al., 2016), and CYP2D6 (Aklillu et al., 1996) enzyme activities and unique distribution of the respective variant alleles in Ethiopians compared to other populations were reported previously.
Indeed, the PCA plot of the whole-genome genotyping data in the present study indicated distinct clusters for Ethiopians compared with HapMap data for Caucasians, Chinese, Japanese, and Nigerians (Fig. 3). This further corroborates the need for more population-specific pharmacogenetic studies in Africa to identify genetic markers for drug-induced adverse events such as liver toxicity. Black Africans are characterized by greater levels of genetic diversity, more within-group genetic heterogeneity, and low LD between loci compared with non-African populations (Campbell and Tishkoff, 2008; Teo et al., 2010). HIV and TB infection remain major problems in sub-Saharan Africa, and treatment is scaled up. Hence, the identification of genetic biomarkers that predict the safety and efficacy of ARV and ATD treatment in different black African populations is crucial.
There are several limitations of this study. First, as DIH is a rare event, identifying a large number of cases for GWAS can be a constraint for achieving large study samples. For the present study, notably 4 years of research and work were required to collect the 59 cases, which resulted in a relatively limited number of case samples for sub-group analyses. Second, populations of African ancestry display greater genetic diversity and lower levels of LD among chromosomal loci (Campbell and Tishkoff, 2008; Teo et al., 2010). The low levels of LD are disadvantageous when screening the genome for associations by using the current SNP-genotyping approaches that essentially rely on the principle of LD mapping.
Although GWAS in populations of African ancestry is challenging due to a less degree of LD, the high level of genetic diversity and weak LD with neighboring SNPs in Africans ancestry is considered a powerful tool for fine mapping causal disease or phenotype-associated variants globally (Campbell and Tishkoff, 2008; Peprah et al., 2015). Therefore, additional studies with a larger sample size using higher-density SNP array or next-generation sequencing may be required to discover susceptibility variants. Our study may represent an important first step in applying GWAS to identify genetic variants of ARV drug-alone- or ARV-ATD co-treatment-induced hepatotoxicity in black African populations.
The third limitation is that as drug combinations are the current treatment protocols for TB/HIV infections, we cannot affirm that the risk variants identified correspond only to a single drug or multiple drugs. As treatment of TB/HIV consists of combination therapy, it is not possible to study individual ARV or ATD drugs in TB-/HIV-infected patients for ethical reasons. Thus, the identification of genetic risk factors for ARV-ATD co-treatment-induced liver toxicity is both important and relevant for future clinical applications, particularly in the current era when ARV drugs are being rolled for extensive use in Africa that bears a large burden of HIV and TB.
Identifying individuals at risk for DIH before drug treatment would improve drug safety. Although genetic associations with ATD- and ARV drug-induced hepatotoxicity susceptibility have been reported, none have yet been strong enough to be useful for managing DIH in the clinical practice. Identifying non-genetic factors, such as measurement of biomarkers in body fluids to predict individuals at risk for DIH before drug treatment, could have a potential for prevention. Large-scale “-omics” technologies are powerful tools for molecular profiling of complex disorders such as idiosyncratic DIH. The application of the new technologies, such as transcriptomics (study alterations in gene expression as a result of toxic compounds), proteomics (characterizes patterns of altered protein expression), and metabonomics (characterizes patterns of altered metabolites in blood or urine) besides pharmacogenomics, offer the potential to reveal biomarkers, identify individuals at risk, and clarify the pathogenesis of idiosyncratic hepatotoxicity (Fontana, 2014; Thulin, 2014) (No. 3458).
Proteomics investigations revealed elevated levels of cadherin 5 (CDH5) and fatty acid-binding protein (FABP) as protein biomarkers for DIH (Mikus et al., 2016). However, the detection of idiosyncratic hepatotoxicants with the currently available in vitro methods remains challenging, as idiosyncratic drug reactions are unpredictable and mostly immune mediated (Van Summeren et al., 2012). The integration of data from different research platforms (proteomics, transcriptomics, metabolomics, genomics) by using a systems biology approach may provide an opportunity to identify biomarkers for the prevention of idiosyncratic DIH in clinical practice.
Conclusions
Using genome-wide genotyping, we identified potential genetic variants associated with DIH due to ARV drugs alone and ARV-ATD co-treatment in Ethiopian HIV-infected patients with or without TB co-infection. The results provide evidence that in addition to genetic variants previously identified by candidate gene association studies, other variants also influence the risk of developing DIH by ARV or ARV-ATD drugs. These genetic variants that are putatively associated with DIH due to ARV drugs alone and ARV-ATD co-treatment establish a foundation for future personalized medicine in people with HIV and TB in Africa and call for larger discovery and replication studies in independent populations.
Supplementary Material
Abbreviations Used
- ABCB1
ATP-binding cassette subfamily-B member-1
- AGBL4
ATP-/GTP-binding protein-like-4
- ALP
alkaline phosphatase
- ALT
alanine aminotransferase
- ART
antiretroviral therapy
- ARV
antiretroviral
- AST
aspartate aminotransferase
- ATD
anti-tuberculosis drugs
- CI
confidence interval
- CYP
cytochrome P450
- DIH
drug-induced hepatotoxicity
- ER
endoplasmic reticulum
- ERN1
endoplasmic reticulum to nucleus signaling-1
- FAM65B
family with sequence similarity 65 member-B
- GWAS
genome-wide association studies
- HIV/AIDS
human immunodeficiency virus/acquired immune deficiency syndrome
- HLA
human leukocyte antigen
- LD
linkage disequilibrium
- lincRNAs
long intergenic noncoding RNAs
- lncRNAs
long noncoding RNAs
- MAP3K3
mitogen-activated protein kinase-3
- NAT2
N-acetyltransferase-2
- OR
odds ratio
- PCA
principal component analysis
- PLINK
PuTTY Link
quantile-quantile
- RUCAM
Roussel Uclaf Causality Assessment Method
- SNP
single nucleotide polymorphism
- TB
tuberculosis
- T Bil
total bilirubin
- ULN
upper limit of normal
- WHO
World Health Organization
Acknowledgments
The authors would like to express their heartfelt gratitude to all study participants. They convey their sincere appreciation to members of the Laboratory for International Alliance on Genomic Research for their kind support and technical assistance. All authors met the ICMJE criteria for authorship. The study was funded by Biobank Japan Project, grants from European and Developing Countries Clinical Trials Partnership (grant No. CG_TA.05.40204_005), and Swedish Research Council (grant No. VR 2015-03295). This work was supported in part by the NIH/Fogarty International Center Global Infectious Diseases grant D43TW009127. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of this article.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Aithal GP, and Grove JI. (2015). Genome-wide association studies in drug-induced liver injury: Step change in understanding the pathogenesis. Semin Liver Dis 35, 421–431 [DOI] [PubMed] [Google Scholar]
- Aithal GP, Watkins PB, Andrade RJ, et al. (2011). Case definition and phenotype standardization in drug-induced liver injury. Clin Pharmacol Ther 89, 806–815 [DOI] [PubMed] [Google Scholar]
- Aklillu E, Djordjevic N, Carrillo JA, Makonnen E, Bertilsson L, and Ingelman-Sundberg M. (2014). High CYP2A6 enzyme activity as measured by a caffeine test and unique distribution of CYP2A6 variant alleles in Ethiopian population. OMICS 18, 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aklillu E, Habtewold A, Ngaimisi E, et al. (2016). SLCO1B1 gene variations among Tanzanians, Ethiopians, and Europeans: Relevance for African and worldwide precision medicine. OMICS 20, 538–545 [DOI] [PubMed] [Google Scholar]
- Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, and Ingelman-Sundberg M. (1996). Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther 278, 441–446 [PubMed] [Google Scholar]
- Araujo-Mariz C, Lopes EP, Acioli-Santos B, et al. (2016). Hepatotoxicity during treatment for tuberculosis in people living with HIV/AIDS. PLoS One 11, e0157725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, and Daly MJ. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 [DOI] [PubMed] [Google Scholar]
- Cai L, Chang H, Fang Y, and Li G. (2016). A comprehensive characterization of the function of lincRNAs in transcriptional regulation through long-range chromatin interactions. Sci Rep 6, 36572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, and Tishkoff SA. (2008). African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet 9, 403–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xu S, Liu L, et al. (2014a). Cab45S inhibits the ER stress-induced IRE1-JNK pathway and apoptosis via GRP78/BiP. Cell Death Dis 5, e1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Melchior WB, Jr, and Guo L. (2014b). Endoplasmic reticulum stress in drug- and environmental toxicant-induced liver toxicity. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 32, 83–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin A, Litonjua A, Lima JJ, et al. (2015). Genome-wide association study identifies novel pharmacogenomic loci for therapeutic response to montelukast in asthma. PLoS One 10, e0129385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AK. (2010). Drug-induced liver injury: Past, present and future. Pharmacogenomics 11, 607–611 [DOI] [PubMed] [Google Scholar]
- Daly AK. (2016). Are polymorphisms in genes relevant to drug disposition predictors of susceptibility to drug-induced liver injury? Pharm Res. DOI: 10.1007/s11095-016-2091-1. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AK, Donaldson PT, Bhatnagar P, et al. (2009). HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet 41, 816–819 [DOI] [PubMed] [Google Scholar]
- Danan G, and Benichou C. (1993). Causality assessment of adverse reactions to drugs—I. A novel method based on the conclusions of international consensus meetings: Application to drug-induced liver injuries. J Clin Epidemiol 46, 1323–1330 [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, and Voight BF. (2008). Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet 17, R122–R128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, and Roeder K. (1999). Genomic control for association studies. Biometrics 55, 997–1004 [DOI] [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H, Brown K, Kelly K, and Siebenlist U. (1997). Direct activation of the stress-activated protein kinase (SAPK) and extracellular signal-regulated protein kinase (ERK) pathways by an inducible mitogen-activated protein Kinase/ERK kinase kinase 3 (MEKK) derivative. J Biol Chem 272, 2668–2674 [DOI] [PubMed] [Google Scholar]
- Elsharkawy AM, Schwab U, McCarron B, et al. (2013). Efavirenz induced acute liver failure requiring liver transplantation in a slow drug metaboliser. J Clin Virol 58, 331–333 [DOI] [PubMed] [Google Scholar]
- Fink DL, and Bloch E. (2013). Liver transplantation for acute liver failure due to efavirenz hepatotoxicity: The importance of routine monitoring. Int J STD AIDS 24, 831–833 [DOI] [PubMed] [Google Scholar]
- Fontana RJ. (2014). Pathogenesis of idiosyncratic drug-induced liver injury and clinical perspectives. Gastroenterology 146, 914–928.e911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebeyehu E, Engidawork E, Bijnsdorp A, Aminy A, Diczfalusy U, and Aklillu E. (2011). Sex and CYP3A5 genotype influence total CYP3A activity: High CYP3A activity and a unique distribution of CYP3A5 variant alleles in Ethiopians. Pharmacogenomics J 11, 130–137 [DOI] [PubMed] [Google Scholar]
- He Y, Wu YT, Huang C, et al. (2014). Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim Biophys Acta 1842, 2204–2215 [DOI] [PubMed] [Google Scholar]
- Hirano T, Yoshikawa R, Harada H, Harada Y, Ishida A, and Yamazaki T. (2015). Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Mol Cancer 14, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS. (2014). Recent progress in genetic variation and risk of antituberculosis drug-induced liver injury. J Chin Med Assoc 77, 169–173 [DOI] [PubMed] [Google Scholar]
- Itzhak D, Bright M, McAndrew P, et al. (2014). Multiple autophosphorylations significantly enhance the endoribonuclease activity of human inositol requiring enzyme 1alpha. BMC Biochem 15, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, and Nunez M. (2012). Liver toxicity of antiretroviral drugs. Semin Liver Dis 32, 167–176 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim SH, Lee JH, et al. (2015). Superoxide dismutase gene (SOD1, SOD2, and SOD3) polymorphisms and antituberculosis drug-induced hepatitis. Allergy Asthma Immunol Res 7, 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar ZS, and Nunez M. (2011). Higher risk of severe drug-induced liver injury among Hispanic HIV-infected patients after initiation of highly active antiretroviral therapy. J Int Assoc Physicians AIDS Care (Chic) 10, 183–186 [DOI] [PubMed] [Google Scholar]
- Melia T, Hao P, Yilmaz F, and Waxman DJ. (2016). Hepatic long intergenic noncoding RNAs: High promoter conservation and dynamic, sex-dependent transcriptional regulation by growth hormone. Mol Cell Biol 36, 50–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikus M, Drobin K, Gry M, et al. (2016). Elevated levels of circulating CDH5 and FABP1 in association with human drug-induced liver injury. Liver Int 37, 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugusi S, Ngaimisi E, Janabi M, et al. (2012). Liver enzyme abnormalities and associated risk factors in HIV patients on efavirenz-based HAART with or without tuberculosis co-infection in Tanzania. PLoS One 7, e40180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngaimisi E, Habtewold A, Minzi O, et al. (2013). Importance of ethnicity, CYP2B6 and ABCB1 genotype for efavirenz pharmacokinetics and treatment outcomes: A parallel-group prospective cohort study in two sub-Saharan Africa populations. PLoS One 8, e67946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez M. (2010). Clinical syndromes and consequences of antiretroviral-related hepatotoxicity. Hepatology 52, 1143–1155 [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki H, and Nakamura Y. (2001). A high-throughput SNP typing system for genome-wide association studies. J Hum Genet 46, 471–477 [DOI] [PubMed] [Google Scholar]
- Peprah E, Xu H, Tekola-Ayele F, and Royal CD. (2015). Genome-wide association studies in Africans and African Americans: Expanding the framework of the genomics of human traits and disease. Public Health Genomics 18, 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros Z, Lee MM, Takahashi A, et al. (2016). Genome-wide association and replication study of anti-tuberculosis drugs-induced liver toxicity. BMC Genomics 17, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, and Reich D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke I, Mullighan CG, Ishii M, et al. (2009). Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci U S A 106, 12944–12949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisler RB, Han C, Burman WJ, Tedaldi EM, and Neaton JD. (2003). Grade 4 events are as important as AIDS events in the era of HAART. J Acquir Immune Defic Syndr 34, 379–386 [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Haas DW, Motsinger AA, et al. (2006). Drug transporter and metabolizing enzyme gene variants and nonnucleoside reverse-transcriptase inhibitor hepatotoxicity. Clin Infect Dis 43, 779–782 [DOI] [PubMed] [Google Scholar]
- Shi X, Sun M, Liu H, Yao Y, and Song Y. (2013). Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett 339, 159–166 [DOI] [PubMed] [Google Scholar]
- Soko ND, Masimirembwa C, and Dandara C. (2016). Pharmacogenomics of Rosuvastatin: A glocal (global+local) African perspective and expert review on a statin drug. OMICS 20, 498–509 [DOI] [PubMed] [Google Scholar]
- Teo Y, Small K, and Kwiatkowski D. (2010). Methodological challenges of genome-wide association analysis in Africa. Nat Rev Genet 11, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulin P, Nordahl G, Gry M, et al. (2014). Keratin-18 and microRNA-122 complement alanine aminotransferase as novel safety biomarkers for drug-induced liver injury in two human cohorts. Liver Int 34, 367–378 [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, and Kaufman RJ. (1998). A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12, 1812–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkova A, Ball C, Gilmour-White S, Rela M, and Mieli-Vergani G. (2009). A paediatric case of acute liver failure associated with efavirenz-based highly active antiretroviral therapy and effective use of raltegravir in combination antiretroviral treatment after liver transplantation. J Antimicrob Chemother 63, 623–625 [DOI] [PubMed] [Google Scholar]
- Ugiagbe RA, Malu AO, Bojuwoye BJ, and Onunu AN. (2012). Incidence of hepatotoxicity of highly active antiretroviral therapy in a tertiary health centre in Nigeria. Niger Postgrad Med J 19, 127–132 [PubMed] [Google Scholar]
- UNAIDS. (2014). 90-90-90 An ambitious treatment target to help end the AIDS epidemic. Geneva: www.unaids.org/en/resources/documents/2014/90-90-90 Accessed February19, 2017 [Google Scholar]
- Urban TJ, Daly AK, and Aithal GP. (2014). Genetic basis of drug-induced liver injury: Present and future. Semin Liver Dis 34, 123–133 [DOI] [PubMed] [Google Scholar]
- Van Summeren A, Renes J, van Delft JH, Kleinjans JC, and Mariman EC. (2012). Proteomics in the search for mechanisms and biomarkers of drug-induced hepatotoxicity. Toxicol In Vitro 26, 373–385 [DOI] [PubMed] [Google Scholar]
- Walker UA. (2007). Antiretroviral therapy-induced liver alterations. Curr Opin HIV AIDS 2, 293–298 [DOI] [PubMed] [Google Scholar]
- Yimer G, Aderaye G, Amogne W, et al. (2008). Anti-tuberculosis therapy-induced hepatotoxicity among Ethiopian HIV-positive and negative patients. PLoS One 3, e1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimer G, Amogne W, Habtewold A, et al. (2012). High plasma efavirenz level and CYP2B6*6 are associated with efavirenz-based HAART-induced liver injury in the treatment of naive HIV patients from Ethiopia: A prospective cohort study. Pharmacogenomics J 12, 499–506 [DOI] [PubMed] [Google Scholar]
- Yimer G, Gry M, Amogne W, et al. (2014). Evaluation of patterns of liver toxicity in patients on antiretroviral and anti-tuberculosis drugs: A prospective four arm observational study in Ethiopian patients. PLoS One 9, e94271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimer G, Ueda N, Habtewold A, et al. (2011). Pharmacogenetic & pharmacokinetic biomarker for efavirenz based ARV and rifampicin based anti-TB drug induced liver injury in TB-HIV infected patients. PLoS One 6, e27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Lu Z, Chen B, Dong P, and Zheng J. (2016). Identification of a novel lincRNA-p21-miR-181b-PTEN signaling cascade in liver fibrosis. Mediators Inflamm 2016, 9856538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Dong P, Mao Y, et al. (2015). lincRNA-p21 inhibits hepatic stellate cell activation and liver fibrogenesis via p21. FEBS J 282, 4810–4821 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.