Abstract
The importance of microbial root inhabitants for plant growth and health was recognized as early as 100 years ago. Recent insights reveal a close symbiotic relationship between plants and their associated microorganisms, and high structural and functional diversity within plant microbiomes. Plants provide microbial communities with specific habitats, which can be broadly categorized as the rhizosphere, phyllosphere, and endosphere. Plant-associated microbes interact with their host in essential functional contexts. They can stimulate germination and growth, help plants fend off disease, promote stress resistance, and influence plant fitness. Therefore, plants have to be considered as metaorganisms within which the associated microbes usually outnumber the cells belonging to the plant host. The structure of the plant microbiome is determined by biotic and abiotic factors but follows ecological rules. Metaorganisms are co-evolved species assemblages. The metabolism and morphology of plants and their microbiota are intensively connected with each other, and the interplay of both maintains the functioning and fitness of the holobiont. Our study of the current literature shows that analysis of plant microbiome data has brought about a paradigm shift in our understanding of the diverse structure and functioning of the plant microbiome with respect to the following: (i) the high interplay of bacteria, archaea, fungi, and protists; (ii) the high specificity even at cultivar level; (iii) the vertical transmission of core microbiomes; (iv) the extraordinary function of endophytes; and (v) several unexpected functions and metabolic interactions. The plant microbiome should be recognized as an additional factor in experimental botany and breeding strategies.
Keywords: Endosphere, holobiont, microbiome, phyllosphere, plant-microbe interaction, rhizosphere
Introduction
The plant microbiome has been considered one of the key determinants of plant health and productivity for over 100 years, and intensive research on this topic started with Lorenz Hiltner’s work in 1901 (Hartmann et al., 2008). This long research period was influenced by the continuous development of research methods, but it was the application of molecular and omics techniques, as well as novel microscopic techniques combining molecular and analytical tools, that led to the important milestones (Muyzer and Smalla, 1998; Caporaso et al., 2012; Jansson et al., 2012). For example, deeper insights into the structure and function of plant-associated microbial communities of the model plant Arabidopsis were presented by Bulgarelli et al. (2012) and Lundberg et al. (2012), while another study detailed a disease-suppressive rhizosphere microbiome in sugar beet (Mendes et al., 2011). The last century has been characterized by important, diverse, and unexpected discoveries relating to plant-associated microorganisms that were made by applying several research methods, especially combinations thereof. Several selected examples are as follows: (i) the potential of root-associated microbes to suppress soil-borne pathogens, demonstrated by strain selection and field trials (Cook et al., 1995; Weller et al., 2002); (ii) trans-kingdom communication between plants and microbes, analysed by analytical and molecular methods (Hartmann and Schikora, 2012); (iii) plant species-specific rhizosphere microbial communities, obtained by molecular fingerprints and molecular strain analysis (Berg and Smalla, 2009; Hartmann et al., 2009); (iv) the rhizosphere as a reservoir of facultative human pathogens, detected by isolation and characterization of strains (Berg et al., 2005), and deep study of the lettuce metagenome (Berg et al., 2014a); (v) the high diversity and importance of the endophytic (myco) biome visualized especially by fluorescence in situ hybridization and microscopy (Omacini et al., 2001; Rodriguez et al., 2009; Hardoim et al., 2015); and (vi) the detection of abundant endophytic Archaea in trees using molecular markers based on genomics of non-cultivable organisms (Müller et al., 2015).
From protists to humans, all organisms are inhabited by microorganisms. According to the holobiont concept, metaorganisms are co-evolved species assemblages. Moreover, co-evolution has resulted in intimate relationships forming between microbes and their hosts that create specific and stable microbiomes. Therefore, all eukaryotic organisms can be considered to be metaorganisms: an association of the macroscopic hosts and a diverse microbiome consisting of bacteria, archaea, fungi, and protists (even protists can have their own bacterial microbiota, and it has been argued that microbiota play an important role in the evolution of multicellularity; McFall-Ngai et al., 2013). Together, microbiota fulfil all important functions for the holobiont themselves, and also for the ecosystem (Mendes and Raaijmakers, 2015; Vandenkoornhuyse et al., 2015). Interestingly, in addition to the joint fulfilment of tasks, many organisms have ‘outsourced’ some essential functions, including those of their own development, to symbiotic organisms living with them (Gilbert et al., 2012).
The realization that microbial communities colonize virtually every host and have central roles in health and disease throughout the entire life cycle of the hosts has been a revolutionary advance in biological sciences, also directing plant research towards a more holistic view. In addition to the discovery of an immense diversity associated with hosts, research will move from describing the composition of microbial communities to elucidating the principles that govern their assembly, dynamics, and functions (Waldor et al., 2015). Here, we report old and new insights into the plant microbiome with a particular emphasis on the progress in the field, which has been driven by multi-omic technologies, and new computational and microscopic tools. We also discuss the implications for the study of model organisms in experimental botany.
Recent insights into the plant microbiome
Plants harbour different microbial communities specific for each plant organ, for example the phyllosphere (Vorholt, 2012), rhizosphere (Berendsen et al., 2012; Philippot et al., 2013), and endosphere (Hardoim et al., 2015). The rhizosphere is the most studied habitat owing to its enormous potential for plant nutrition and health (Berendsen et al., 2012; Hirsch and Mauchline, 2012; Bakker et al., 2013; Mendes et al., 2013). It has been known for many years that the rhizosphere enriches specific microbial species/genotypes in comparison to soil and inner tissues, but modern technologies provide much deeper insights and expand our understanding of plant–microbe interactions (Bais et al., 2006; Doornbos et al., 2012). A current model shows the occurrence of seed-borne microorganisms (Christin Zachow and Gabriele Berg, personal communication) and the attraction of microbes to nutrients such as carbohydrates and amino acids (Moe, 2013) in combination with plant-specific secondary metabolites (Weston and Mathesius, 2013). Plant root exudates play important roles as both chemo-attractants and repellents (Badri and Vivanco, 2009). Additionally, plant defence signalling plays a role in this process (Doornbos et al., 2012). The importance of the rhizosphere microbiome can be underlined by the number of species: in the metagenomes studied in our group, we found up to 1200 prokaryotic species (extracted 16S rRNA genes annotated using the Greengenes reference database). Moreover, a higher number of species was found in medicinal and wild plants than on crops grown in intensive agriculture (Martina Köberl and Gabriele Berg, personal communication). For comparative analyses, all metagenomes were rarefied at a sequencing depth of 1.7 × 107 sequences; the actual species diversity is even much higher. The abundances measured sum up to 109–1011 bacterial cells colonizing each gram of the root, which often not only outnumbers the cells of the host plants but also represent more microbes than people existing on Earth. While the well-studied rhizosphere represents the soil–plant interface, the phyllosphere forms the air–plant interface. This microhabitat is also of special interest owing to its large and exposed surface area and its connection to the air microbiome, especially airborne pathogens (Vorholt, 2012). In our metagenomes, we found a lower microbial diversity in the phyllosphere than in the rhizosphere, but the overall diversity was quite large and comprised up to 900 species (Armin Erlacher and Gabriele Berg, personal communication). In general, leaves have different strategies to trigger microbial colonization, for example (antimicrobial) wax layers, (antimicrobial) secondary metabolites, trichomes, and hairs, and the microbial composition seems to be highly individual but also plant-dependent. However, an overview of a broader range of plant phyla is still missing. Recently, the majority of the research has been focused on the endosphere of plants. Although endophytes were defined by De Bary in 1866 as ‘any organisms occurring within plant tissues’, their existence was ignored until the end of the last century, and very often these organisms were considered contaminants. Now, the organisms inhabiting the endosphere are well-accepted and, moreover, their intimate interaction with the plant makes them the focus of (biotechnological) interest. Seeds also harbour a surprisingly diverse microbiome in their endosphere (Johnston-Monje and Raizada, 2011). There are many more micro-environments described, for example the endorhiza (root), the anthosphere (flower), the spermosphere (seeds), and the carposphere (fruit), but their specific microbiome is less studied.
All plant organs are colonized by microorganisms (Fig. 1). The composition of the plant microbiome is influenced by different factors, including plant age or developmental stage, plant species or cultivar, and plant health. In addition, a multitude of abiotic factors modulate the structural and functional diversity of the plant-associated microbiome, including soil properties, nutrient status, and climatic conditions (reviewed in Berg and Smalla, 2009). The colonization of plants by microorganisms is not random; it is a targeted process that is underlined by the existence of specific co-occurrence patterns and microbial networks (Cardinale et al., 2015). These networks are related to a colonization resistance pattern, which determines the potential for allochthonous microorganisms (pathogens but also biological control agents) to invade the autochthonous community. In addition, structural diversity is paramount to the preventive avoidance of pathogen invasion/outbreaks (van Elsas et al., 2012).
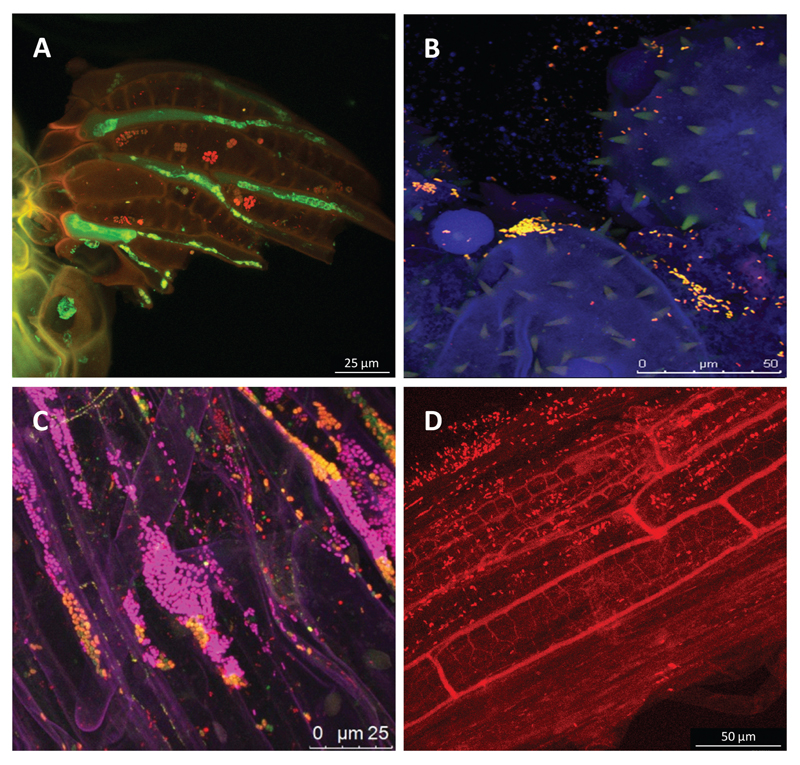
Fig. 1.
The plant as a natural metaorganism visualized by fluorescence in situ hybridization (A-C) and confocal laser scanning microscopy. (A) Phyllosphere of a Sphagnum leave, (B) bacteria on pumpkin pollen, (C) bacteria in the rhizosphere of lettuce, and (D) root of an oilseed rape inoculated with the DsRed-labelled biocontrol agent Pseudomonas trivialis 3Re2-7.
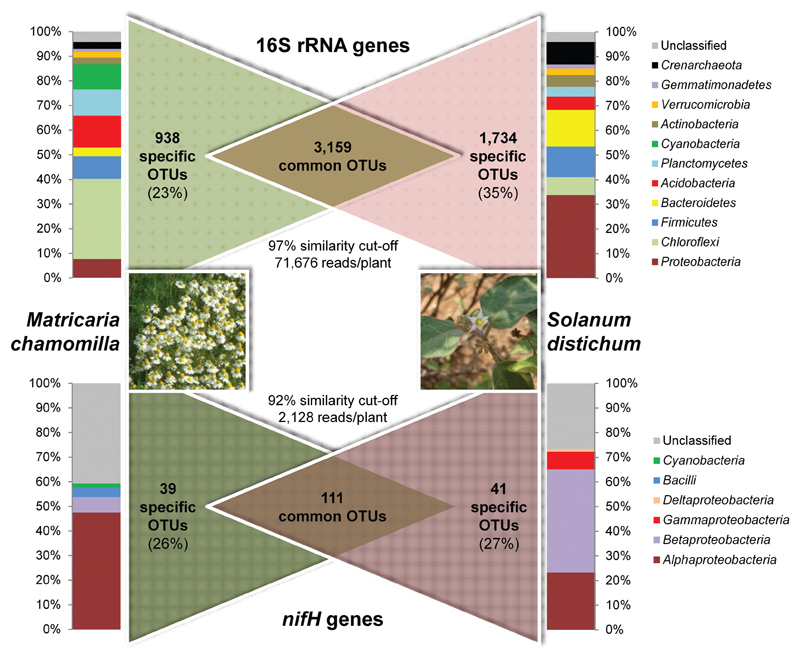
As an example of outstandingly high microbial diversity, we will have a closer look at the associated microbiomes of medicinal plants. Figure 2 visualizes and compares the rhizosphere colonization of the German chamomile, Matricaria chamomilla L., and the African nightshade, Solanum distichum Schumach. and Thonn. The calculated Shannon indices (H’) for their prokaryotic rhizosphere diversity were in the range of 9.4–9.7 (16S rRNA genes at a similarity of 97%). Both medicinal plants were cultivated under desert farming conditions and organic management in Egypt (Köberl et al., 2011). However, despite their being grown in direct proximity to one another, their structural (16S rRNA genes) as well as functional (diazotrophic community, nifH genes encoding the nitrogenase reductase subunit) colonization profiles revealed a high degree of plant specificity, with around 30% of specific operational taxonomic units for each investigated community. In spite of the clearly plant-specific selection of their associated bacterial microbiomes, indigenous Bacillus and Paenibacillus strains of native desert soil with promising antagonistic properties against a wide range of soil-borne phytopathogens were enriched in all investigated plant roots (Köberl et al., 2011, 2013a).
Fig. 2.
Taxonomic composition and Venn diagrams of the 16S rRNA and nifH gene communities inhabiting the rhizosphere of medicinal plants (German chamomile [Matricaria chamomilla L.] and African nightshade [Solanum distichum Schumach. and Thonn.]). Both plants were cultivated in direct proximity to each other under field conditions (loamy sand soil) and were investigated in four independent replicate samples by amplicon sequencing. Singletons, operational taxonomic units defined by only a single observation, were removed and not considered in either dataset.
Functions and ecology of the plant microbiome
Many functions of the plant microbiome are essential for the host. We would like to start with the germination procedure, the first step of a plants’ life cycle. Interestingly, many plants cannot start their life without microorganisms, such as mosses, which belong to the oldest land plants on Earth (Hornschuh et al., 2006), and orchids, whose very small seeds need the help of specific fungi, often Rhizoctonia, to germinate (Jacquemyn et al., 2015). The germination-promoting fungus Rhizoctonia comprises beneficial organisms as well as pathogens. To avoid any pathogenic interaction after germination, the host plant digests their helping fungus completely. In these cases of germination support, microorganisms are essential, and this may be one reason that these keystone microorganisms are vertically transmitted as shown for Sphagnum mosses (Bragina et al., 2012). A positive impact on germination has also been found for cosmopolitan plant-associated bacteria like Stenotrophomonas (Alavi et al., 2013).
The fact that plant-associated microorganisms stimulate plant growth and nurture plants is well known, as in the examples of rhizobia and mycorrhizal fungi. The mechanisms by which these microorganisms support plant growth include the production of phytohormones, the fixation of nitrogen, and the mobilization of phosphorus and minerals such as iron (Tkacz and Poole, 2015). Plant-specific nitrogen-fixing communities from the rhizosphere of medicinal plants are visualized in Fig. 2.
The plant microbiome is also involved in pathogen suppression, and it is especially the root microbiome that acts as a protective shield against soil-borne pathogens (Weller et al., 2002). The mechanisms are well studied and include several direct interactions with plant pathogens as well as indirect interactions via the plant by stimulation of the plant immune system (Lugtenberg and Kamilova, 2009). The most recent research has shown that the microbiome is not only involved in coping with biotic stress, it is also involved in protection against abiotic stress (Bragina et al., 2013). For example, the plant microbiome has been shown to be involved in protection against high salinities and drought (Yang et al., 2009; Rolli et al., 2015). Recently, we found that the plant microbiome is also involved in cold acclimation, a primary factor limiting the geographical distribution of plants as well as the growth and yield of crops in certain areas (Mohammad Etemadi and Gabriele Berg, personal communication). Under the challenge of climate change, this function is an important aspect.
The plant microbiota also influences the composition of plant secondary metabolites and the resulting development of different metabotypes. This has been shown for the taste of strawberries (Zabetakis et al., 1999; Verginer et al., 2010) and the production of bioactive compounds in medicinal plants (Köberl et al., 2013b; Schmidt et al., 2014). In a study on Arabidopsis thaliana, the rhizosphere microbiome could be linked to insect feeding behaviour, which was most probably a result of microbiome-driven changes in the leaf metabolome (Badri et al., 2013). Peñuelas et al. (2014b) showed that the removal of the floral microbiome of Sambucus nigra resulted in a reduced floral terpene emission, which plays a key role in pollination and consequently in fruit and seed production.
Recent studies also revealed the direct impact of the root microbiome on plant phenology. Wagner et al. (2014) demonstrated, for instance, that natural soil microorganisms have an impact on the flowering time of a wild Arabidopsis relative, Boechera stricta. Similarly, Panke-Buisse et al. (2015) demonstrated that successful transplantation of rhizosphere microbiomes from Arabidopsis thaliana to Brassica rapa had an impact on their flowering times, resulting in similar shifts in flowering phenology. Additional essential roles of the plant microbiome for phenotypic and epigenetic plasticity as well as the evolution of plants were suggested by Partida-Martínez and Heil (2011).
Co-evolution of plants and associated microbial communities has already been hypothesized based on culture-dependent results obtained for the rhizosphere of ancient and modern wheat cultivars (Germida and Siciliano, 2001). Co-evolution was recently shown to be prevalent amongst other plants, such as maize, sugar beet, and lettuce, by the application of deep sequencing techniques (Berg et al., 2014b; Cardinale et al., 2015). Crop breeding has been identified as a strong driver of natural evolution (Berg et al., 2013). In some cases, the breeding strategy was targeted against pathogens, but historically it was mainly a random selection process for plant phenotypes.
Implications for experimental botany
Experimental botany involves the study of plant behaviour and physiology under varying conditions. In gnotobiotic systems, plants are investigated under sterile conditions. Under such conditions it is important to use highly sensitive molecular detection methods to check sterility, because some microbes can reach a non-culturable but viable state as endophytes. According to our experience, it is sometimes very difficult to work under axenic conditions. This has been extensively discussed in a review by Partida-Martínez and Heil (2011). In in vitro systems (climate chamber, greenhouse), standard soil or artificial substrates are very often used. If they are not sterilized prior to usage, they can provide the plants with a microbiome. However, this is often completely different from those of natural soils and has a strong impact on the rhizosphere and endosphere microbiome (Zachow et al., 2014). Field studies present the most natural conditions, but owing to the strong impact of environmental factors, they show the highest variability. It is also important to consider that agricultural systems and systems intensively used by humans are often characterized by a shift (often a reduction) in microbial diversity. This may also be extended to plants raised in pot experiments, where we expect a reduced diversity or altered structure of the microbiomes. This could explain situations where pot experiments cannot clearly explain the responses of plants in natural environments.
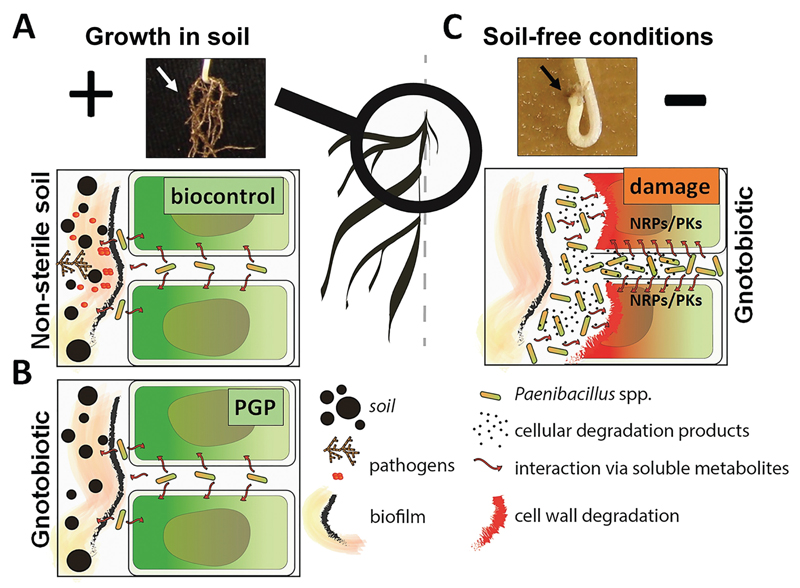
When conducting experiments, we should also consider the ecological/symbiotic continuum of associated microbiota, i.e. when outcomes of interactions may depend on general experimental conditions. Such effects may create secondary functional regulation that influences the outcome of experiments initially designed for plants only. This was confirmed in a study by Rybakova et al. (2015b), which showed that experimental design strongly influenced the outcome of the experiment. The authors found that the positive impact of Paenibacillus treatments on plant growth in soil was completely reversed under soil-free gnotobiotic conditions, where bacteria had a destructive effect on the host plants (Fig. 3). This negative impact could be associated with the microbiome shift induced by Paenibacillus spp. applied to the seeds of B. napus and B. oleracea and/or with the toxic secondary metabolites produced by these bacteria under artificial plant growth conditions (Rybakova et al., 2015a; Timmusk et al., 2015).
Fig. 3.
Controversial Paenibacillus–plant interaction depends on the plant growth conditions. The images to the left show how the interaction with Paenibacillus spp. may improve plant health when plants are grown in soil, while the images on the right side illustrate the destructive behaviour of Paenibacillus spp. in soil-free conditions. (A) The Paenibacillus–plant interaction in non-sterile soil where the growth of pathogens is reduced by the secondary metabolites produced by the Paenibacillus spp.; the access of pathogens to the plant root cells is blocked by the biofilm produced by Paenibacillus spp. (B) Illustration of plant growth promotion (PGP) by secondary metabolites produced by Paenibacillus spp. in the absence of other bacteria when plants are grown in sterile soil. (C) Possible scenarios of how Paenibacillus spp. can damage plant cells by local overproduction of toxic secondary metabolites (e.g. nonribosomal peptides [NRPs] and polyketides [PKs]). Stunting of the root system and inhibition of plant growth may also result from degradation of the plant root cells by Paenibacillus spp. in the absence of other competing microorganisms and a low nutrient environment.
Associated microbiota may additionally contribute to microbial loops, which could have an effect on the host plants. Microbes usually respond to much smaller differences in general conditions, especially in the water content. In contrast to the less sensitive reaction of the host organisms, even small water pulses lead to activation of microbial physiology.
The plant hormone indole-3-acetic acid (IAA) is another example of intense plant–microbe interplay. Phylogenetic evidence suggests that IAA biosynthesis evolved independently in bacteria, microalgae, fungi, and plants, which leads to the hypothesis that natural selection might have favoured IAA as a widespread physiological code in these microorganisms and their interactions (Fu et al., 2015). Recent research is more and more targeted on communication via volatile organic compounds (VOCs). Microbes use a diverse spectrum of VOCs to communicate with each other as well as with their host (Bitas et al., 2013; Peñuelas et al., 2014a). VOCs of certain rhizobacteria are not only able to directly promote plant growth and suppress the growth of pathogens, but can also induce the plant’s systemic resistance, enabling the plant to better defend itself (Ryu et al., 2003; Lee et al., 2012). Besides VOCs, several other bacterial components like flagella, lipopolysaccharides, siderophores, or the quorum-sensing molecules of Gram-negative bacteria are capable of induced systemic resistance signalling pathway activation (Lugtenberg and Kamilova, 2009; Hartmann and Schikora, 2012; Schenk and Schikora, 2015).
Altogether, the (non)existence of microorganisms should be considered in the interpretation of all plant experiments. Moreover, cultivar-specific microbiomes should be considered in experimental as well as breeding strategies. So far, we do not know anything about the plant-associated microbiome at the beginning and end of physiological experiments. We suggest that such characterization may complete our understanding of how the microbiome could affect the results. In particular, the contribution of plant microbiomes to hormone-triggered responses could be of importance. We should therefore be able to consider the physiological responses of plants in a wider context, in the sense of a ‘metaphysiology’.
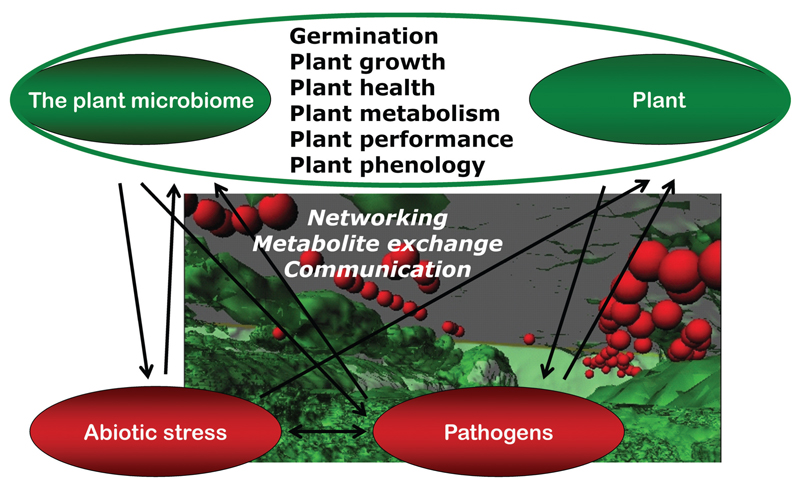
The overall conclusion is that each plant has to be considered as a metaorganism containing many more microbial cells than plant cells. The structure of the plant microbiome is determined by biotic and abiotic factors but follows clear ecological rules. The metabolism and morphology of plants and their microbiota are innately connected with each other, and an intense interplay of both maintains the functioning of the holobiont (Fig. 4).
Fig. 4.
Model visualizing the interplay within the plant holobiont.
Acknowledgement
We would like to thank Timothy Mark (Graz) for English revision. This study was partly supported by the EU-Egypt Innovation Fund (RDI ENPI/2014/342–707) and the Austrian Science Fund FWF (J 3638 to MK, co-funded by the European Commission, and I 882 to GB and MG) and by the European Union in frame of FP7-KBBE-2013-7-single-stage (BIOCOMES; No. 612713). The cooperation of GB was funded by a project in the Austrian Centre of Industrial Biotechnology, which has been supported by the Austrian BMWFW, BMVIT, SFG, Standortagentur Tirol, and ZIT through the Austrian FFG-COMET-Funding Program.
References
- Alavi P, Starcher MR, Zachow C, Müller H, Berg G. Root-microbe systems: the effect and mode of interaction of Stress Protecting Agent (SPA) Stenotrophomonas rhizophila DSM14405T. Frontiers in Plant Science. 2013;4:141. doi: 10.3389/fpls.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Vivanco JM. Regulation and function of root exudates. Plant, Cell & Environment. 2009;32:666–681. doi: 10.1111/j.1365-3040.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- Badri DV, Zolla G, Bakker MG, Manter DK, Vivanco JM. Potential impact of soil microbiomes on the leaf metabolome and on herbivore feeding behavior. The New Phytologist. 2013;198:264–273. doi: 10.1111/nph.12124. [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annual Review of Plant Biology. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- Bakker PA, Berendsen RL, Doornbos RF, Wintermans PC, Pieterse CM. The rhizosphere revisited: root microbiomics. Frontiers in Plant Science. 2013;4:165. doi: 10.3389/fpls.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends in Plant Science. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environmental Microbiology. 2005;7:1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- Berg G, Erlacher A, Smalla K, Krause R. Vegetable microbiomes: is there a connection among opportunistic infections, human health and our “gut feeling”? Microbial Biotechnology. 2014a;7:487–495. doi: 10.1111/1751-7915.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Grube M, Schloter M, Smalla K. The plant microbiome and its importance for plant and human health. Frontiers in Microbiology. 2014b;5:491. doi: 10.3389/fmicb.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology. 2009;68:1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- Berg G, Zachow C, Müller H, Philipps J, Tilcher R. Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy. 2013;3:648–656. [Google Scholar]
- Bitas V, Kim HS, Bennett JW, Kang S. Sniffing on microbes: diverse roles of microbial volatile organic compounds in plant health. Molecular Plant-Microbe Interactions. 2013;26:835–843. doi: 10.1094/MPMI-10-12-0249-CR. [DOI] [PubMed] [Google Scholar]
- Bragina A, Berg C, Cardinale M, Shcherbakov A, Chebotar V, Berg G. Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. The ISME Journal. 2012;6:802–813. doi: 10.1038/ismej.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina A, Cardinale M, Berg C, Berg G. Vertical transmission explains the specific Burkholderia pattern in Sphagnum mosses at multi-geographic scale. Frontiers in Microbiology. 2013;4:394. doi: 10.3389/fmicb.2013.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale M, Erlacher A, Grube M, Berg G. Bacterial networks and co-occurrence relationships in the lettuce root microbiome. Environmental Microbiology. 2015;17:239–252. doi: 10.1111/1462-2920.12686. [DOI] [PubMed] [Google Scholar]
- Cook RJ, Thomashow LS, Weller DM, Fujimoto D, Mazzola M, Bangera G, Kim DS. Molecular mechanisms of defense by rhizobacteria against root disease. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4197–4201. doi: 10.1073/pnas.92.10.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doornbos RF, Van Loon LC, Bakker PA. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. Agronomy for Sustainable Development. 2012;32:227–243. [Google Scholar]
- Fu SF, Wei JY, Chen HW, Liu YY, Lu HY, Chou JY. Indole-3-acetic acid: a widespread physiological code in interactions of fungi with other organisms. Plant Signaling & Behavior. 2015;10:e1048052. doi: 10.1080/15592324.2015.1048052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germida J, Siciliano S. Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biology and Fertility of Soils. 2001;33:410–415. [Google Scholar]
- Gilbert SF, Sapp J, Tauber AI. A symbiotic view of life: we have never been individuals. The Quarterly Review of Biology. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiology and Molecular Biology Reviews. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Rothballer M, Schmid M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant and Soil. 2008;312:7–14. [Google Scholar]
- Hartmann A, Schikora A. Quorum sensing of bacteria and trans-kingdom interactions of N-acyl homoserine lactose with eucaryotes. Journal of Chemical Ecology. 2012;38:704–713. doi: 10.1007/s10886-012-0141-7. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Schmid M, van Tuinen D, Berg G. Plant driven selection of microbes. Plant and Soil. 2009;321:235–257. [Google Scholar]
- Hirsch PR, Mauchline TH. Who’s who in the plant root microbiome? Nature Biotechnology. 2012;30:961–962. doi: 10.1038/nbt.2387. [DOI] [PubMed] [Google Scholar]
- Hornschuh M, Grotha R, Kutschera U. Moss-associated methylobacteria as phytosymbionts: an experimental study. Naturwissenschaften. 2006;93:480–486. doi: 10.1007/s00114-006-0137-7. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Waud M, Merckx VS, Lievens B, Brys R. Mycorrhizal diversity, seed germination and long-term changes in population size across nine populations of the terrestrial orchid Neottia ovata. Molecular Ecology. 2015;24:3269–3280. doi: 10.1111/mec.13236. [DOI] [PubMed] [Google Scholar]
- Jansson JK, Neufeld JD, Moran MA, Gilbert JA. Omics for understanding microbial functional dynamics. Environmental Microbiology. 2012;14:1–3. doi: 10.1111/j.1462-2920.2011.02518.x. [DOI] [PubMed] [Google Scholar]
- Johnston-Monje D, Raizada MN. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One. 2011;6:e20396. doi: 10.1371/journal.pone.0020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köberl M, Müller H, Ramadan EM, Berg G. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS One. 2011;6:e24452. doi: 10.1371/journal.pone.0024452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köberl M, Ramadan EM, Adam M, Cardinale M, Hallmann J, Heuer H, Smalla K, Berg G. Bacillus and Streptomyces were selected as broad-spectrum antagonists against soilborne pathogens from arid areas in Egypt. FEMS Microbiology Letters. 2013a;342:168–178. doi: 10.1111/1574-6968.12089. [DOI] [PubMed] [Google Scholar]
- Köberl M, Schmidt R, Ramadan EM, Bauer R, Berg G. The microbiome of medicinal plants: diversity and importance for plant growth, quality and health. Frontiers in Microbiology. 2013b;4:400. doi: 10.3389/fmicb.2013.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Farag MA, Park HB, Kloepper JW, Lee SH, Ryu CM. Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS One. 2012;7:e48744. doi: 10.1371/journal.pone.0048744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. Plant-growth promoting rhizobacteria. Annual Review of Microbiology. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, et al. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiology Reviews. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, De Bruijn I, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Mendes R, Raaijmakers JM. Cross-kingdom similarities in microbiome functions. The ISME Journal. 2015;9:1905–1907. doi: 10.1038/ismej.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moe LA. Amino acids in the rhizosphere: from plants to microbes. American Journal of Botany. 2013;100:1692–1705. doi: 10.3732/ajb.1300033. [DOI] [PubMed] [Google Scholar]
- Müller H, Berg C, Landa BB, Auerbach A, Moissl-Eichinger C, Berg G. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Frontiers in Microbiology. 2015;6:138. doi: 10.3389/fmicb.2015.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- Omacini M, Chaneton EJ, Ghersa CM, Müller CB. Symbiotic fungal endophytes control insect host-parasite interaction webs. Nature. 2001;409:78–81. doi: 10.1038/35051070. [DOI] [PubMed] [Google Scholar]
- Panke-Buisse K, Poole AC, Goodrich JK, Ley RE, Kao-Kniffin J. Selection on soil microbiomes reveals reproducible impacts on plant function. The ISME Journal. 2015;9:980–989. doi: 10.1038/ismej.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Martínez LP, Heil M. The microbe-free plant: fact or artifact? Frontiers in Plant Science. 2011;2:100. doi: 10.3389/fpls.2011.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñuelas J, Asensio D, Tholl D, Wenke K, Rosenkranz M, Piechulla B, Schnitzler JP. Biogenic volatile emissions from the soil. Plant, Cell & Environment. 2014a;37:1866–1891. doi: 10.1111/pce.12340. [DOI] [PubMed] [Google Scholar]
- Peñuelas J, Farré-Armengol G, Llusia J, Gargallo-Garriga A, Rico L, Sardans J, Terradas J, Filella I. Removal of floral microbiota reduces floral terpene emissions. Scientific Reports. 2014b;4:6727. doi: 10.1038/srep06727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nature Reviews Microbiology. 2013;11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- Rodriguez RJ, White JF, Jr, Arnold AE, Redman RS. Fungal endophytes: diversity and functional roles. The New Phytologist. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- Rolli E, Marasco R, Vigani G, et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environmental Microbiology. 2015;17:316–331. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- Rybakova D, Cernava T, Köberl M, Liebminger S, Etemadi M, Berg G. Endophytes-assisted biocontrol: novel insights in ecology and the mode of action of Paenibacillus. Plant and Soil. 2015a doi: 10.1007/s11104-015-2526-1. [DOI] [Google Scholar]
- Rybakova D, Schmuck M, Wetzlinger U, Varo-Suarez A, Murgu O, Müller H, Berg G. Kill or cure? The interaction between endophytic Paenibacillus and Serratia strains and the host plant is shaped by plant growth conditions. Plant and Soil. 2015b doi: 10.1007/s11104-015-2572-8. [DOI] [Google Scholar]
- Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Paré PW, Kloepper JW. Bacterial volatiles promote growth in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk ST, Schikora A. AHL-priming functions via oxylipin and salicylic acid. Frontiers in Plant Science. 2015;5:784. doi: 10.3389/fpls.2014.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Köberl M, Mostafa A, Ramadan EM, Monschein M, Jensen KB, Bauer R, Berg G. Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Frontiers in Microbiology. 2014;5:64. doi: 10.3389/fmicb.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S, Kim SB, Nevo E, Abd El Daim I, Ek B, Bergquist J, Behers L. Sfp-type PPTase inactivation promotes bacterial biofilm formation and ability to enhance wheat drought tolerance. Frontiers in Microbiology. 2015;6:387. doi: 10.3389/fmicb.2015.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacz A, Poole P. Role of root microbiota in plant productivity. Journal of Experimental Botany. 2015;66:2167–2175. doi: 10.1093/jxb/erv157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. The importance of the microbiome of the plant holobiont. The New Phytologist. 2015;206:1196–1206. doi: 10.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- van Elsas JD, Chiurazzi M, Mallon CA, Elhottova D, Kristufek V, Salles JF. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1159–1164. doi: 10.1073/pnas.1109326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verginer M, Siegmund B, Cardinale M, Müller H, Choi Y, Míguez CB, Leitner E, Berg G. Monitoring the plant epiphyte Methylobacterium extorquens DSM 21961 by real-time PCR and its influence on the strawberry flavor. FEMS Microbiology Ecology. 2010;74:136–145. doi: 10.1111/j.1574-6941.2010.00942.x. [DOI] [PubMed] [Google Scholar]
- Vorholt JA. Microbial life in the phyllosphere. Nature Reviews Microbiology. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- Wagner MR, Lundberg DS, Coleman-Derr D, Tringe SG, Dangl JL, Mitchell-Olds T. Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecology Letters. 2014;17:717–726. doi: 10.1111/ele.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor MK, Tyson G, Borenstein E, et al. Where next for microbiome research? PLoS Biology. 2015;13:e1002050. doi: 10.1371/journal.pbio.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller DM, Raaijmakers JM, Gardener BB, Thomashow LS. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annual Review of Phytopathology. 2002;40:309–348. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- Weston LA, Mathesius U. Flavonoids: their structure, biosynthesis and role in the rhizosphere, including allelopathy. Journal of Chemical Ecology. 2013;39:283–297. doi: 10.1007/s10886-013-0248-5. [DOI] [PubMed] [Google Scholar]
- Yang J, Kloepper JW, Ryu CM. Rhizosphere bacteria help plants tolerate abiotic stress. Trends in Plant Science. 2009;14:1–4. doi: 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zabetakis I, Moutevelis-Minakakis P, Gramshaw JW. The role of 2-hydroxypropanal in the biosynthesis of 2,5-dimethyl-4-hydroxy-2H-furan-3-one in strawberry (Fragaria x ananassa, cv. Elsanta) callus cultures. Food Chemistry. 1999;64:311–314. [Google Scholar]
- Zachow C, Müller H, Tilcher R, Berg G. Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima – ancestor of all beet crops – and modern sugar beets. Frontiers in Microbiology. 2014;5:415. doi: 10.3389/fmicb.2014.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]