Abstract
Uncoupling protein 1 (UCP1) is an integral membrane protein found in the mitochondrial inner membrane of brown adipose tissue, and facilitates the process of non-shivering thermogenesis in mammals. Its activation by fatty acids, which overcomes its inhibition by purine nucleotides, leads to an increase in the proton conductance of the inner mitochondrial membrane, short-circuiting the mitochondrion to produce heat rather than ATP. Despite 40 years of intense research, the underlying molecular mechanism of UCP1 is still under debate. The protein belongs to the mitochondrial carrier family of transporters, which have recently been shown to utilise a domain-based alternating-access mechanism, cycling between a cytoplasmic and matrix state to transport metabolites across the inner membrane. Here, we review the protein properties of UCP1 and compare them to those of mitochondrial carriers. UCP1 has the same structural fold as other mitochondrial carriers and, in contrast to past claims, is a monomer, binding one purine nucleotide and three cardiolipin molecules tightly. The protein has a single substrate binding site, which is similar to those of the dicarboxylate and oxoglutarate carriers, but also contains a proton binding site and several hydrophobic residues. As found in other mitochondrial carriers, UCP1 has two conserved salt bridge networks on either side of the central cavity, which regulate access to the substrate binding site in an alternating way. The conserved domain structures and mobile inter-domain interfaces are consistent with an alternating access mechanism too. In conclusion, UCP1 has retained all of the key features of mitochondrial carriers, indicating that it operates by a conventional carrier-like mechanism.
Keywords: Thermogenesis, Proton transport, Purine nucleotide inhibition, Alternating access mechanism
1. Introduction
Uncoupling protein 1 (UCP1) is the defining feature of brown fat and is responsible for the unique thermogenic properties of the tissue [1–3]. The protein catalyses the leak of protons across the mitochondrial inner membrane, dissipating the electrochemical proton gradient that would otherwise drive ATP production by ATP synthase. As a result, the energy from the oxidation of respiratory substrates is instead released as heat. Hence, substrate oxidation is ‘uncoupled’ from ADP phosphorylation. UCP1 is the original uncoupling protein that defined the name and, unlike its homologues within the mitochondrial carrier family of metabolite transporters (e.g. UCP2 and 3), is the only carrier that is undisputed in having a proton leak function (see Refs. [4,5]). Brown fat that is rich in UCP1 has long been known to facilitate non-shivering thermogenesis in newborn mammals to help defend body temperature against the cold. However, over recent years it has come to prominence that the tissue is present in adult humans too [6,7]. Humans exhibit both ‘classical’ brown fat, with characteristics similar to the developmental tissue studied in newborns, and ‘beige’ brown fat, a recruitable form in white adipose tissue (see Ref. [8] for an overview); the major depots are found in the supraclavicular region and the neck [6,9]. These tissues utilise UCP1 and, when activated (e.g. by cold exposure), have the potential to contribute significantly to whole-body energy expenditure [7,10–12]. Notably, the occurrence of brown fat in humans correlates with leanness [7,9,13]. In mice, thermogenesis by brown fat has been shown to clear lipids and dispose of glucose from the blood, reducing metabolic disease [14], while the genetic ablation of brown fat [15], or UCP1 specifically [16], induces obesity. Methods to encourage brown adipose tissue and, importantly, activate thermogenesis via UCP1 in the absence of physiological stimuli [11,17,18] could provide therapies to combat obesity and related diseases [19].
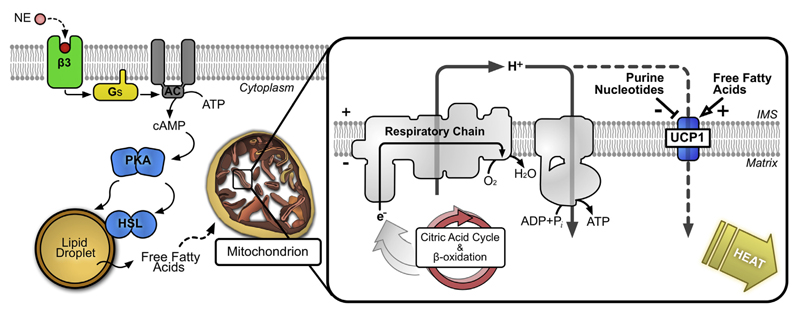
UCP1 activity is well regulated in brown adipocytes, where it is induced in response to cold exposure or over-feeding through the sympathetic nervous system, facilitating adaptive thermogenesis (see Ref. [20] for review). Signals from the brain stimulate the release of catecholamines, such as noradrenaline, in innervated brown adipose tissue, which activate adrenergic receptors (most significantly β3) in the plasma membrane of brown adipocytes to initiate intracellular cAMP-dependent signaling (Fig. 1). Gαs protein, dissociated from stimulated receptors, activates adenylate cyclase, increasing cellular cAMP, which leads to changes in UCP1 activity both acutely and through transcriptional regulation of the UCP1 gene [21–23]. In the acute response, cAMP activates protein kinase A, which, among several targets, is believed to phosphorylate hormone sensitive lipase and the perilipin protein that protects stored lipid droplets from lipase breakdown, activating and deactivating them, respectively [20,24,25]. The resulting lipolysis of triglyceride stores releases free fatty acids into the cytosol, which act as a fuel for mitochondrial oxidation, and, importantly, as the direct activator of UCP1 [26], switching on thermogenesis (Fig. 1). UCP1 binds and is inhibited by cytosolic (Mg2+-free) purine nucleotide (see Ref. [27]), ATP and ADP being the most relevant. The mechanism of how fatty acids activate UCP1 and overcome nucleotide inhibition is debated (see below).
Fig. 1. Signaling and acute activation of UCP1-dependent thermogenesis in brown adipocytes.
Left: norepinephrine (NE) released in response to physiological stimuli (e.g. cold exposure or overfeeding) activates a cAMP-dependent pathway leading to the release of fatty acids from cellular lipid stores. β3, β3-adrenergic receptor; Gs, Gαs protein; AC, adenylate cyclase; PKA, protein kinase A; HSL, hormone sensitive lipase. Right: free fatty acids interact to activate UCP1 in mitochondria, overcoming inhibition of the protein by cytosolic purine nucleotides. Activated UCP1 catalyses the leak of protons across the mitochondrial inner membrane, dissipating the electrochemical proton gradient generated by the respiratory chain that would otherwise drive ATP synthesis. As a result, the energy from the oxidation of respiratory substrates is released as heat.
Up-regulation of the UCP1 gene occurs as part of adaptive thermogenesis following adrenergic stimulation. The signaling process and regulation of UCP1 transcription is multifaceted (see [21,23] for a review), involving the activation of the p38 MAP kinase pathway downstream of protein kinase A stimulation by cAMP. The UCP1 gene has a complex enhancer region upstream of the promoter, where cAMP response elements facilitate transcriptional control [28]. The region contains binding sites for several nuclear receptor transcription factors, including the peroxisome proliferator-activated receptor γ (PPARγ), retinoic X receptor (RXR), 9-cis retinoic acid receptor (RAR) and thyroid hormone receptor (TR), consistent with the transactivation of the UCP1 gene in response to their respective ligands [23]. The active control of expression, tied to physiological cues, means that UCP1 protein concentrations in brown adipose tissue mitochondria can vary significantly. The protein is a relatively abundant mitochondrial carrier in conventional brown fat mitochondria, even in warm adapted animals. In cold adapted animals, however, UCP1 levels can increase to represent as much as ~10% of the mitochondrial inner membrane protein [29].
The features of the UCP1 protein first started to emerge from early studies with isolated mitochondria (see Ref. [30]). The ‘recoupling’ effect of nucleotides on mitochondrial respiration and the demonstration of an externally-exposed GDP-binding site led to the proposal that a ‘nucleotide binding protein’ was responsible for the unusual ion transport properties of brown fat mitochondria [31]. Ricquier and Kader [32] were first to highlight a 32 kDa mitochondrial protein that increases in concentration in association with cold acclimation, which corresponded to the same protein in later 8-azido-ATP labeling studies identified to be responsible for controlling energy dissipation [29]. The purification of the protein, first by using immobilised nucleotide [33] and later by ‘negative chromatography’ methods developed for the ADP/ATP carrier [34,35], led to investigations into the properties of what was then first termed “uncoupling protein”. These studies revealed UCP1 as an integral membrane protein like the ADP/ATP carrier but with distinct nucleotide binding properties. Purine nucleotides (ATP, ADP, GTP and GDP) specifically bind with high affinity at low pH (<6), stabilising the detergent-solubilised protein [35]. The stoichiometry of nucleotide binding and oligomeric state of UCP1 was also addressed, but has since been revised in our recent work (see below and [36]). The identification of the UCP1 gene and amino acid sequence [37,38] confirmed that UCP1 is related to the ADP/ATP carrier, and is a member of the mitochondrial carrier family of metabolite exchangers, which all share the same basic structure and membrane topology [39–41]. They are composed of three ~100-amino acid homologous domains, each comprising two transmembrane α-helices separated by a loop and small α-helix on the matrix side [41]. The first helix of each domain contains the signature motif PX[DE]XX[RK], which is well conserved across the protein family. Following the reconstitution of isolated UCP1 into liposomes, key characteristics associated with the uncoupling activity of isolated brown adipose tissue mitochondria could be demonstrated [42–44]. Namely, proton conductance activated by fatty acids, which can be inhibited by nucleotides. However, despite many studies over the years, the molecular mechanism of UCP1 has not been resolved.
2. Proposed mechanisms of UCP1 activation
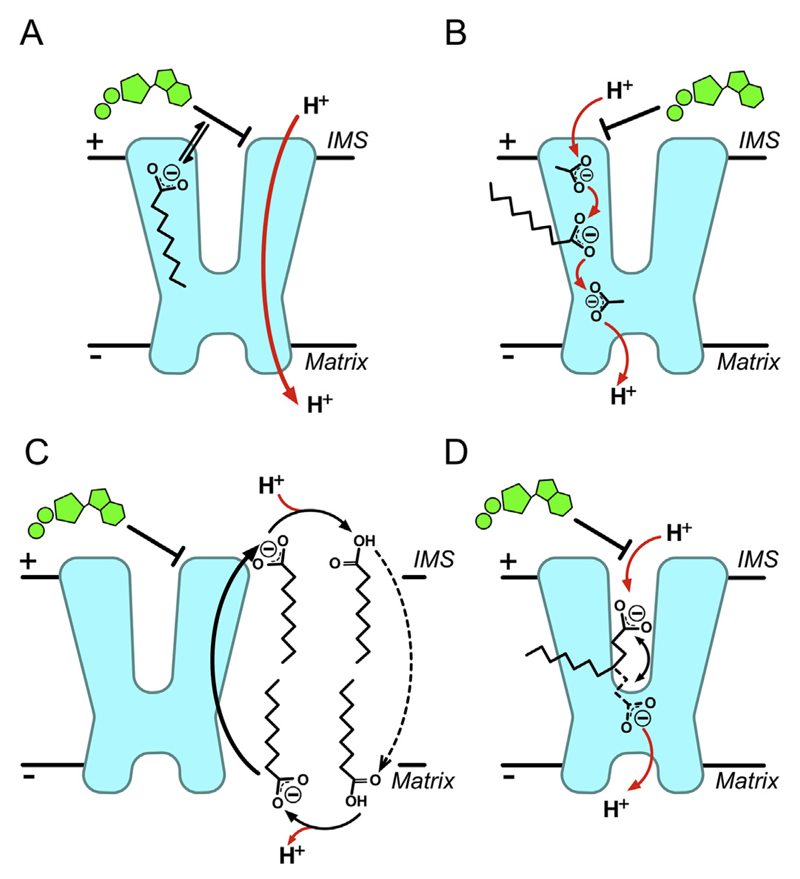
There have been many claims for various cellular metabolites interacting with and regulating UCP1, however, most are controversial and are likely to relate to the many technical difficulties in studying this type of protein (see “Unsubstantiated mechanistic and structural claims” below). What is clear is that proton conductance by UCP1 is activated by free fatty acids and inhibited by purine nucleotides. Several models have been proposed to explain the interplay that occurs with these effectors, each derived from observations largely based on different methodological approaches. Studies with isolated mitochondria have suggested that fatty acids and nucleotides influence uncoupling activity with simple competitive kinetics [45,46], supporting a ‘functional competition’ model (Fig. 2A). In this proposal, fatty acids act on UCP1 purely to remove nucleotide inhibition, either by direct competition at the nucleotide-binding site or allosterically through binding at a separate site [45], to allow net proton transfer by an undefined process. In the absence of either regulator, however, the model predicts UCP1 to be inherently active, which is not consistent with observations with the isolated protein. In liposomes, UCP1 proton conductance does not occur without the addition of fatty acids [47,48], and apparent activator-free activity in isolated mitochondria is likely to relate to endogenous fatty acids from the breakdown of lipids by lipases [48]. Furthermore, in contrast to the situation with isolated mitochondria, fatty acids do not appear to alter the affinity of reconstituted UCP1 for nucleotides [47,48].
Fig. 2. Proposed mechanisms of proton transport by UCP1.
A) The functional competition model: fatty acids act simply to remove nucleotide inhibition of UCP1, either by direct competition or allosterically [45]. B) The cofactor model: fatty acids associate with UCP1 to provide carboxylate(s) as protonatable groups, which complete a proton transfer pathway within the protein [47]. C) The cycling model: fatty acid anions act as actual transport substrate of UCP1 and are exported, flipping back across the membrane in a protonated state independently of UCP1, to give net proton transfer [49,50]. D) The shuttling model: both protonated or deprotonated fatty acid species can be transported/exchanged by UCP1, however, a restricted ability to release hydrophobic long chain fatty acids from the protein results in net proton transfer only [57]. See main text for details.
Studies predominantly with liposomes have led to two models of UCP1 proton transport, the ‘cofactor’ model and the ‘cycling’ model, both relying on fatty acids as an essential component in the process. In the cofactor model (Fig. 2B), fatty acids associate with UCP1 to provide one or more carboxylate groups to complete a proton transfer pathway within the protein, allowing transport across the membrane [47]. Consistent with a central role of the fatty acid carboxylate, the pH dependence of UCP1 activity correlates well with the pKa of the activating fatty acid used [47]. Alternatively, in the cycling model (Fig. 2C), fatty acid anions act as the actual transport substrate of UCP1 and are exported, flipping back across the membrane in a protonated state independently of UCP1, leading to a net proton transfer [49,50]. This model is supported by the well-established observation that UCP1 transports chloride and other monovalent anions too [51,52]. Of particular relevance, the protein can transport various alkyl-sulfonate anions, low pKa analogues of fatty acids, with rates similar to the proton conductance induced by equivalent fatty acid species [50]. These analogues do not induce proton conductance by UCP1, consistent with their inability to be protonated at physiological pH and flip back across the membrane in accordance with the model [50]. It was claimed that other analogues, such as glucose-O-ω-palmitate, can also activate UCP1 proton conductance but are unable to flip across the membrane due to the hydrophilic moiety at the end of the acyl tail, providing evidence against the cycling model [27,53]. However, the data were not substantiated and later work suggested that the hydrophilic moiety does indeed interfere with the ability of the compound to activate proton conductance by UCP1, contradicting the claim [54]. Other arguments against the model rely on observations that anion (chloride) transport rates are too low to support proton transport by UCP1 [55], which would not be consistent with anion transport underlying the proton transport mechanism. However, as pointed out [56], chloride may be a poor representative anion for transport by UCP1, as fatty acid-like anions are transported at much faster rates.
More recently, patch clamp studies on mitochondrial inner membranes from brown adipose tissue have provided evidence for an alternative model, which has elements of both the cofactor and cycling hypotheses. The ‘shuttling model’ [57] (Fig. 2D) contends that fatty acids are effectively transport substrates, as in the cycling model, but that either protonated or deprotonated fatty acid species are exchanged by UCP1, i.e UCP1 has both fatty acid− uniport and fatty acid−/H+ symport activity. A restricted ability to release long chain fatty acids from the protein on either side of the membrane due to their hydrophobicity results in net proton transfer dependent on the UCP1-associated fatty acid carboxylate, reminiscent of the cofactor model. As such, long chain fatty acids act to ‘shuttle’ protons through UCP1. Long chain fatty acids induced continuous H+ currents via UCP1 in patch clamp recordings, whereas the currents induced by equivalent alkyl sulfonates were carried by alkyl sulfonate movement and found to be transient, consistent with anion transport that is restricted by product build up on one side of the membrane. Accordingly, decreasing the alkyl sulfonate acyl chain length or adding a binding agent to increase the solubility of the species in the aqueous phase could convert transient currents into continuous ones, consistent with full anion transport and the previously proposed cycling model. Of particular significance, however, was the observation that long chain alkyl sulfonates were only potent in inducing currents when introduced to UCP1 from the cytoplasmic side, which is not consistent with the cycling model where fatty acid anion binding at the matrix side for export to the cytosol would be expected to be favoured (cf. Fig. 2C). As such, the shuttling model proposes the binding and transport of the protonated species from the cytosol via UCP1 as well to accommodate these observations. Why the hydrophobic protonated species would favour or rely on UCP1 for this part of the transport cycle, however, rather than flipping directly across the membrane, which they can do rapidly [58], is not clear.
The considerable research efforts outlined here along with the corresponding models have made significant advances to explain the biochemical process of how UCP1 may mediate fatty acid activated proton conductance. However, a robust molecular and structural context for UCP1 is essential to provide the necessary framework and constraints to clarify how the protein is likely to function.
3. Unsubstantiated mechanistic and structural claims
There have been many high profile claims regarding UCP1, which have been refuted or not substantiated by others, particularly on the activation of the protein by various metabolites. Notably, ubiquinone (but not ubiquinol) was reported by Echtay et al. [59,60] to be an obligatory cofactor, necessary for proton conductance by recombinant UCP ‘refolded’ from bacterially produced inclusion bodies. However, the finding could not be reproduced in similar investigations by others [61], and was further disproved through the study of UCP1 expressed in a strain of yeast unable to make ubiquinone [62]. The bacterially produced UCP material may have generated experimental artefacts, particularly with the use of indirect fluorescent methods to assess nucleotide binding and protein integrity in the absence of parallel native protein controls (see discussion below). Nevertheless, at the time, the work was the impetus for further studies into related putative activators of UCPs. Subsequent work (see Echtay et al. [63]) reported that superoxide, instead, activated UCP1, 2 and 3 based on proton leak measurements with isolated mitochondria from various tissues. The claim was further revised to suggest 4-hydroxy-2-nonenal, a lipid breakdown product downstream of superoxide interaction with lipid membranes, was the actual activating species [64]. In either case, the activation of mild uncoupling by UCPs to attenuate the production of reactive oxygen species at high mitochondrial membrane potential was proposed as a feedback mechanism to defend against oxidative stress. Although an attractive hypothesis, other groups were not able to corroborate the claim. In several similar studies with isolated mitochondria, no evidence that either superoxide or 4-hydroxy-2-nonenal activated proton leak by UCPs could be found [65–68]. Furthermore, the study of mice with lower superoxide levels, due to the overexpression of superoxide dismutase, did not show decreased proton leak by UCPs (neither by UCP1 nor UCP3) as would be expected, suggesting that endogenously generated superoxide does not regulate UCP activity [69]. Physiological arguments have also been put forward to suggest that UCP activation in this manner is unlikely [70,71]. More recently, patch clamp investigations on mitochondrial inner membranes from brown adipose tissue have clarified that 4-hydroxy-2-nonenal has no effect on proton transport by UCP1 [57].
The proposal that UCP1 is activated by reactive oxygen species has resurfaced recently in a study by Chouchani et al. [72]. The authors correlated an increase in reactive oxygen species with the onset of thermogenesis in brown adipose tissue of mice kept in the cold, and reported that antioxidants inhibit UCP1-dependent heat production leading to hypothermia. In this study, direct activation of UCP1 by reactive oxygen species was purported to occur through the formation of a sulfenic acid adduct at C253 in UCP1. In addition to the preceding studies that refute a role of reactive oxygen species in activating UCPs [65–71], issues have been raised in response to this work (see Ref. [73]), most notably with regards to the experimental approach used and limited clarity on the link from whole-animal physiology observations to a sulfenylation mechanism of one specific residue of UCP1. Many other mitochondrial carriers that are crucial for the function of the mitochondrion, such as the mitochondrial ADP/ATP carrier, dicarboxylate carrier, oxodicarboxylate carrier, citrate carrier, and aspartate/glutamate carrier, have cysteine residues in similar positions as C253 in the matrix α-helices. They are not essential for function (see supplemental information in Ref. [74]), and substitutions, such as serine, threonine or alanine, are commonly found. These residues are hidden in the cytoplasmic state and exposed in the matrix state [74,75] and their modification by sulfhydryl reagents leads to irreversible inactivation of mitochondrial carriers [74]. In brown adipose mitochondria, UCP1 is more abundant than other mitochondrial carriers and sulfenylation, which is indiscriminate, would need to happen on a large scale for regulation to be effective without affecting the activity of many essential mitochondrial carriers, which also have equivalent cysteine residues. Our measurements of the total mass of native UCP1, purified from brown adipose tissue of newborn lambs, have not revealed any evidence of sulfenylation (unpublished data, Ian Fearnley). The observations of Chouchani et al. [72] could simply relate to false positives generated by the indirect methods used to detect sulfenic acid modifications.
As discussed above, UCP material generated in Escherichia coli and refolded was responsible for the refuted claims that ubiquinone acts as a cofactor [59,60]. Many other studies have also used recombinant material produced in E. coli to report on the behaviour of UCPs [76–82]. For example, UCP1 has been claimed to self-assemble to form tetramers [80] or to have a nucleotide binding site that can be accessed from both sides of the membrane [79]. James Chou and colleagues have also published several high profile NMR studies on mitochondrial carriers produced in a similar manner, including a backbone structure of UCP2 [78,82–84]. In E. coli, recombinant mitochondrial carriers and UCPs are expressed as inclusion bodies, which must be solubilized and refolded in detergent following purification. Typically, the protein is either extracted directly with a zwitterionic detergent, e.g. Fos-Choline-12, also called dodecylphosphocholine (DPC), or solubilized in a known harsh detergent and exchanged into a milder one. Importantly, we, and others, have raised concerns on the structural integrity and functionality of membrane proteins produced in this way. Molecular dynamic simulations with the UCP2 structure and tests with reconstituted UCP1 have shown that the structural integrity of these proteins is compromised in Fos-Choline-12 micelles (DPC) [85]. Through a detailed analysis of thermostability, we have demonstrated that mitochondrial carriers from native sources are relatively unstable, even in mild detergents [86]. Not only did the zwitterionic detergent Fos-Choline-12 (DPC) destabilise native UCP1, but it was also able to solubilize UCP1 from inclusion body material and maintain the protein in a soluble but unfolded state. As such, it is imperative that robust methods are applied to verify the structural integrity and functionality of the material, particularly with recombinantly produced protein. Unfortunately, as outlined in Ref. [86], commonly employed methods, such as fluorescence resonance energy transfer with nucleotide-conjugated fluorophores, monitoring tryptophan fluorescence or analysis of CD spectra, may give results that can be falsely interpreted to suggest the presence of folded carrier protein when native material is not used as a control.
4. UCP1 is a monomer binding one purine nucleotide molecule
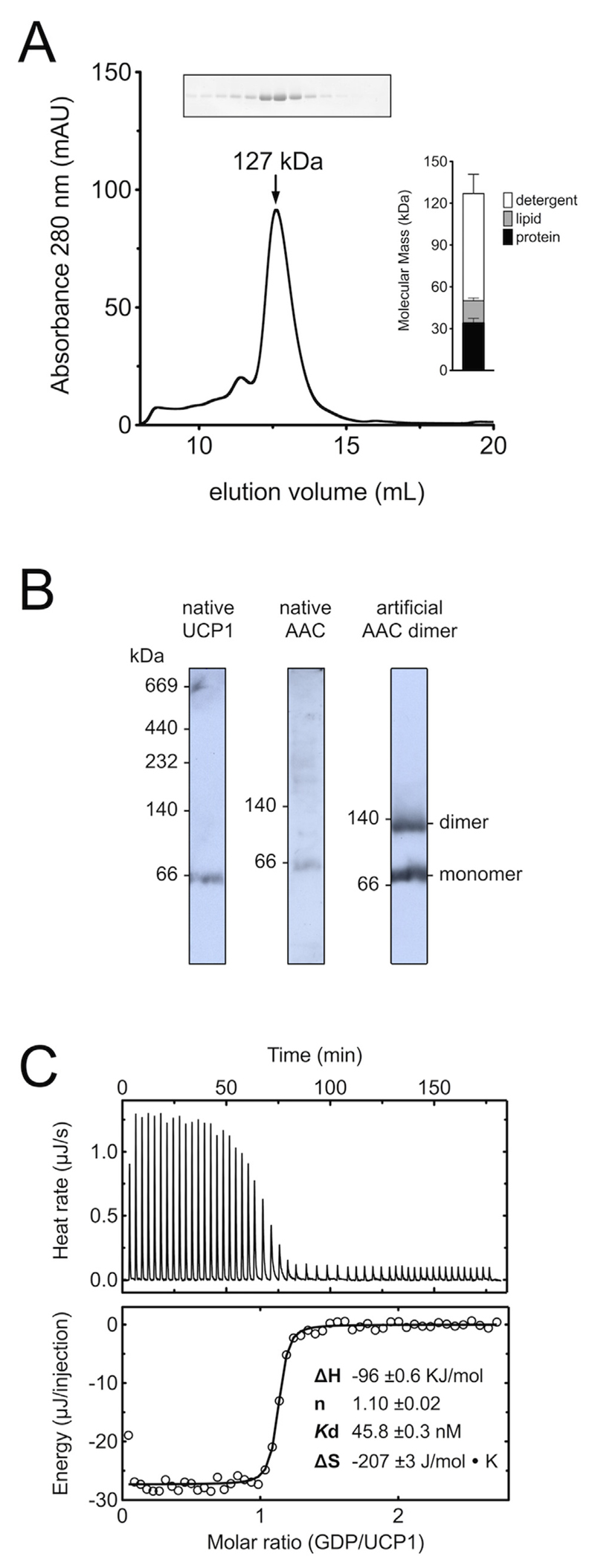
For many years it was thought that UCP1 formed a homo-dimer based on analytical ultracentrifugation [87], nucleotide binding [34,35,88,89] and protein cross-linking studies [35,90]. In fact, it was believed that all mitochondrial carriers were dimers, but subsequent studies have shown that these claims were incorrect (see Ref. [91] for a review). Many of these earlier studies were carried out in Triton X-100, which is difficult to quantify and leads to a large overestimation of the protein concentration in the modified protein assays of Lowry [92]. Moreover, crosslinking of UCP1 via the C-terminal cysteine (C304) does not happen in native membranes unless oxidising conditions are applied [35,90]. We decided to re-examine the oligomeric state of UCP1 by determining the molecular mass of purified protein in decyl maltose neopentyl glycol (10MNG) and cardiolipin micelles by size exclusion chromatography, as we have done for other carriers [93–96]. UCP1 eluted in the major peak corresponding to a total mass of the protein/detergent/lipid micelle of 127 kDa (Fig. 3A) [36]. Since the total mass is the sum of all components [94], the amounts of the associated detergent and lipid were determined too, being 77 kDa and 16 kDa, respectively (Fig. 3A, inset). Therefore, the protein was 34 kDa, which correlates well with the calculated mass of a single UCP1 polypeptide of 33 kDa. The apparent mass of native UCP1 was also analysed by blue native gel electrophoresis [97]. Under optimised conditions the apparent masses of UCP1 and the ADP/ATP carrier were found to be similar, approximately 66 kDa (Fig. 3B), but they were monomeric nonetheless, as bound Coomassie, lipid and detergent cause them to migrate anomalously in blue native gels see Ref. [97] for details).
Fig. 3. UCP1 is a monomer binding GDP with a 1:1 stoichiometry.
(A) Assessment of the molecular mass of UCP1 in 10MNG/cardiolipin by size exclusion chromatography [36]. Inset shows the mass composition of the major peak. (B) The apparent molecular mass of native UCP1 and ADP/ATP carrier in blue native gels [97]. Mitochondrial membrane extracts from lamb brown adipose tissue (UCP1, left), rat liver (ADP/ATP carrier, middle) and yeast mitochondria (monomer and covalently linked dimer of the ADP/ATP carrier AAC3, right) were prepared in 1% dodecylmaltoside and separated in 6–18% (w/v) polyacrylamide gels. (C) Isothermal titration calorimetry of purified UCP1 with GDP [36]. The enthalpy changes associated with the titration of GDP into UCP1 at 4 °C (top), and corresponding isotherms fitted to a one-site binding model with ∆H, Kd, and stoichiometry as fitting parameters (bottom).
Original observations with isolated UCP1 indicated that one nucleotide is bound per two UCP1 [34], but this stoichiometry might have been wrong due to the protein assay used (see above). To obtain an accurate measure of the binding stoichiometry, we used isothermal titration calorimetry. For this purpose, purified UCP1, quantified by amino acid analysis, was titrated with known concentrations of GDP and the resulting isotherms were fitted with an appropriate binding model, which demonstrated that 1.1 GDP molecules bind per UCP1 with a Kd of 46 nM [36] (Fig. 3C). Therefore, our data support a simple model where one UCP1 is capable of binding one nucleotide inhibitor, just as one ADP/ATP carrier is capable of binding one atractyloside or one carboxyatractyloside inhibitor [41,98]. The other mitochondrial carriers that have been examined have also proven to be monomeric [91,93–95] with the exception of the mitochondrial aspartate/glutamate carrier, which forms a dimer through extensive interactions of the N-terminal regulatory domains, but the two carrier domains do not interact [96]. In contrast to earlier claims, therefore, UCP1 functions as a monomer.
5. UCP1 has three tightly bound cardiolipin molecules
Measurements using 31P NMR had previously indicated that purified UCP1 does not bind cardiolipin [99], whereas the ADP/ATP carrier binds three cardiolipin molecules [100]. Furthermore, reconstitution studies had indicated that this lipid is not required for UCP1 activity [101], but may alter the affinity of the protein for purine nucleotides [47,99]. In contrast, this mitochondrial lipid does enhance the activity of mitochondrial carriers [102,103].
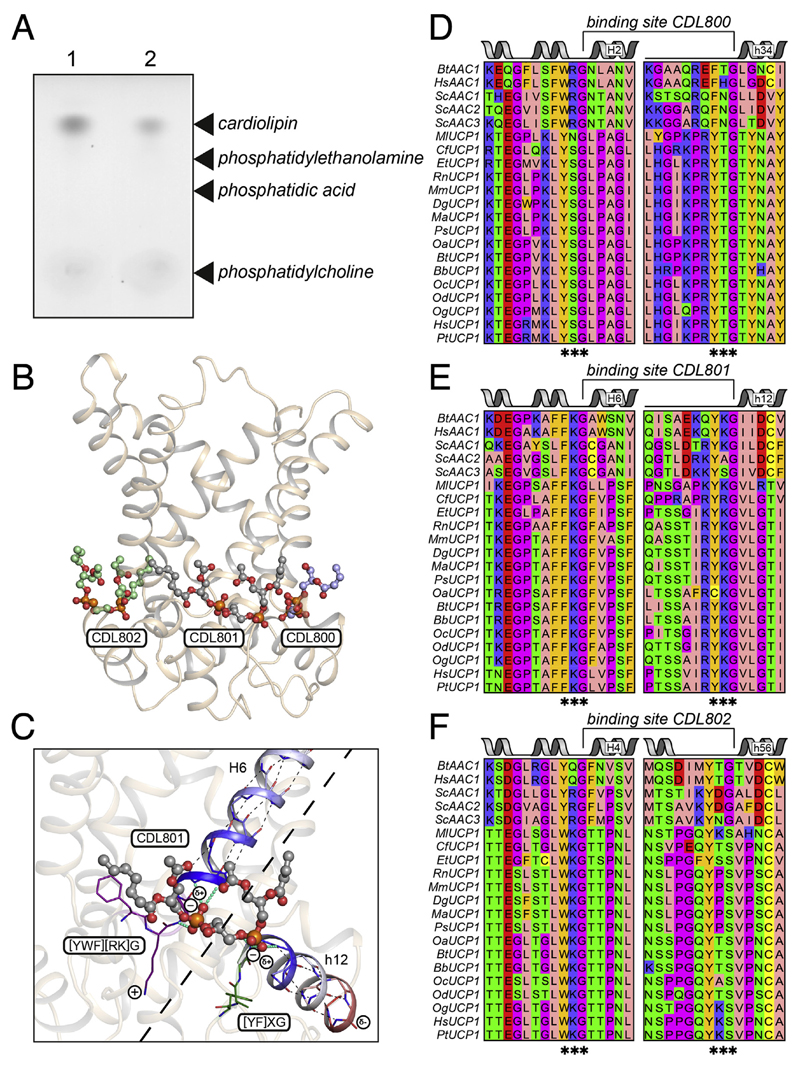
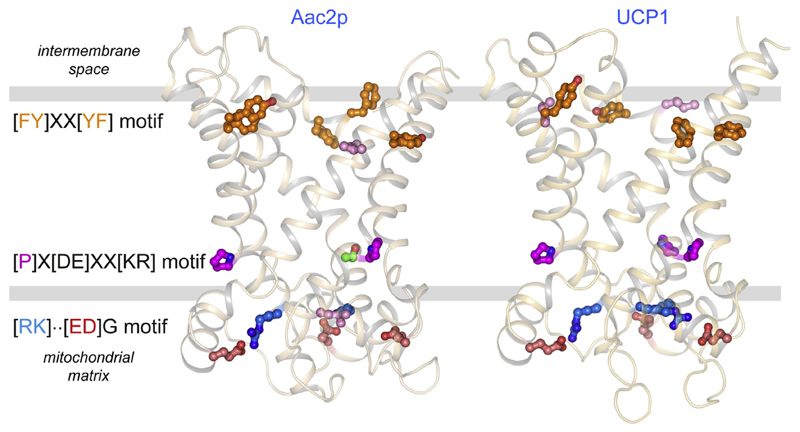
Using a new purification procedure that allows the immobilisation of native UCP1, we discovered that endogenous cardiolipin binds tightly to native UCP1, resisting removal by extensive detergent washes (Fig. 4A) [36]. Quantification showed that approximately six moles of lipid phosphorus were associated with UCP1, consistent with UCP1 binding three molecules of cardiolipin [36]. Three cardiolipin molecules can be observed bound in all available atomic structures of the mitochondrial ADP/ATP carrier [104,105] (Fig. 4B). The key features of cardiolipin binding were deduced by analysing the structures of 27 cardiolipin binding sites [104,105] (Fig. 4C). First, the cardiolipin molecules are bound at the start of the even-numbered and matrix α-helices, bridging the interdomain interface (Fig. 4C). Second, the conserved symmetrical sequence motifs [YWF][RK]G and [YF]XG precede these α-helices. The glycine residues of these motifs are in the loop to helix transitions, functioning as helix breakers. As a consequence, the amide groups at the start of the α-helices are not involved in intra-helical hydrogen bonding and instead use the phosphate groups of cardiolipin as hydrogen bond acceptors. Third, the bound cardiolipin molecules have two formal negative charges [106], which could be compensated by the positive helix dipoles at the N-terminal end of the even-numbered and matrix α-helices. There is no structural evidence that the positively charged residues of the [YWF][RK]G motif interact directly with the phosphate groups [105], as suggested previously [99].
Fig. 4. Three cardiolipin molecules are tightly bound to UCP1.
(A) Endogenous cardiolipin co-purifies with UCP1, as shown by the detection of unsaturated lipid, specifically, following thin layer chromatography of lipid extracts from purified UCP1 after 300 mL washes in decylmaltoside supplemented with GDP plus 14:0 phosphatidylcholine (1) or 14:0 cardiolipin (2) [36]. (B) Lateral view of the yeast ADP/ATP carrier Aac2p showing binding of three cardiolipin molecules CDL800 (light blue), CDL801 (grey) and CDL802 (green) [105]. The protein and cardiolipin molecules are shown in cartoon and ball-and-stick representations, respectively. The cardiolipin acyl chains have only been partially modelled. (C) Detailed view of the binding site for cardiolipin molecule CDL801. Hydrogen bonds within the helices are shown as thin black dotted lines. Residues in the [YWF][KR]G and [YF]XG motifs are shown in violet and green, respectively. Hydrogen bond interactions between protein and cardiolipin are shown as thick cyan dashed lines. The positive and negative helical dipole are shown in blue and red, respectively. (D), (E), and (F) amino acid sequences of the cardiolipin binding sites of CDL800, CDL801, and CDL802, respectively, of the mitochondrial ADP/ATP carrier and uncoupling protein from the following species: Bt, Bos taurus; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Ml, Myotis lucifugus; Cf, Canis familiaris; Et, Echinops telfairi; Rn, Rattus norvegicus; Mm, Mus musculus; Dg, Dicrostonyx groenlandicus; Ma, Mesocricetus auratus; Ps, Phodopus sungorus; Oa, Ovis aries; Bb, Bubalus bubalis; Oc, Oryctolagus cuniculus; Od, Ochotona dauurica; Og, Otolemur garnettii and Pt, Pan troglodytes.
The aforementioned motifs are conserved in the equivalent sites in UCP1 of different species (Fig. 4D, E, and F). In the human UCP1 the [YWF][RK]G and [YF]XG motifs are YSG and YTG for the CDL800 site, FKG and YKG for the CDL801 site, and WKG and YKS for the CDL801 site, respectively. All of the sequences have glycine residues at the start of the even-numbered and matrix α-helices except for the last motif where the glycine is replaced by a serine residue. However, serine is also a common helix breaker as its side chain can form a hydrogen bond with the backbone of the α-helix, consistent with this notion. Therefore, in contrast to earlier claims, UCP1 does have three tightly bound cardiolipin molecules that bridge the inter-domain interfaces. Although the role of the cardiolipin molecules has not been resolved, they could be important for stability of the carrier proteins by cross-linking the three domains on the matrix side [36,41,86] and/or they could protect the dynamic interdomain interfaces (see also below) [105].
6. The substrate binding site of UCP1 and mitochondrial carriers
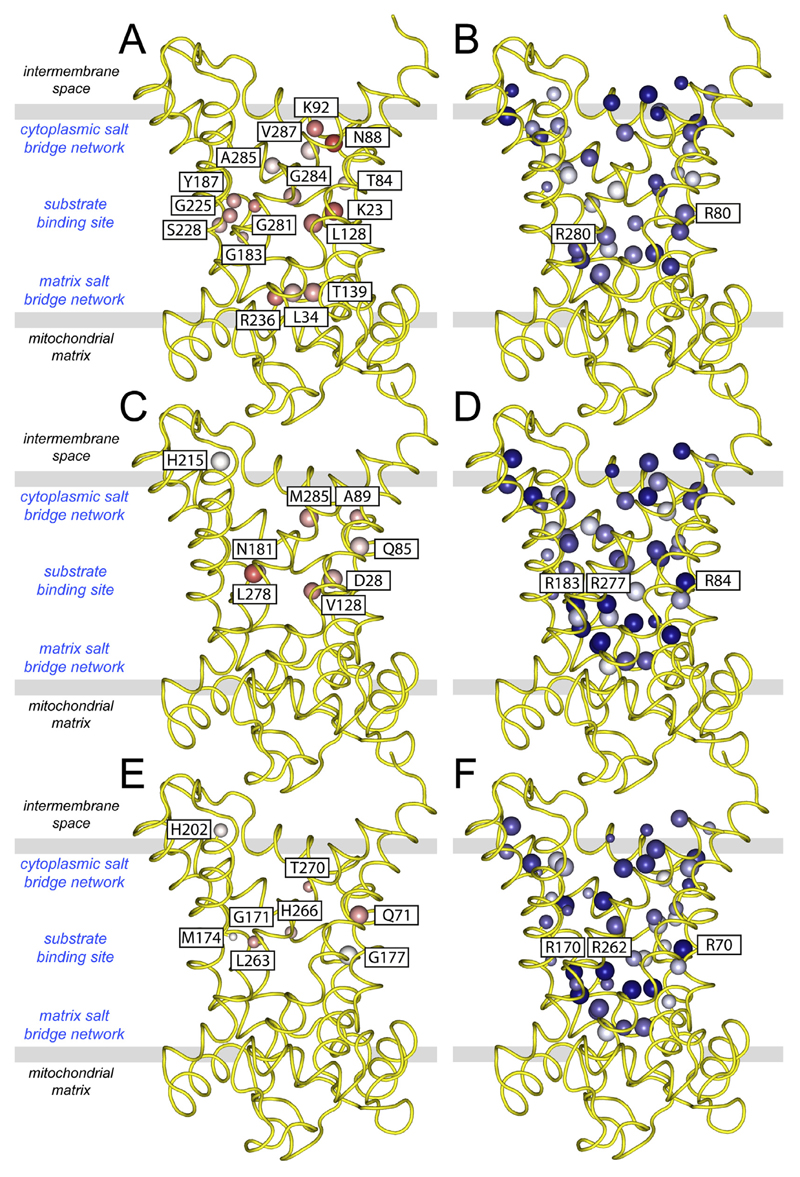
Mitochondrial carriers consist of three homologous sequence repeats [39,40] and they have a high degree of three-fold pseudosymmetry in their structures [41,98,105]. So it is likely that a common ancestor of all mitochondrial carriers consisted of three identical domains. Over time, different functions evolved through mutations of key amino acid residues, introducing asymmetry [107]. Since the substrates are not symmetrical, they require a site with asymmetric residues for binding. In contrast, for the maintenance of the three-fold symmetrical structure and mechanism, symmetrical residues need to be conserved. Based on sequence information only, a score was devised to assess the degree of symmetry and conservation of all residues in mitochondrial carriers, including UCP1 [107]. For all known subfamilies conserved asymmetric residues with the appropriate chemistry for substrate binding were found to cluster in the central cavity, corresponding roughly to the middle of the membrane [107]. For example, this approach successfully identified the residues of the binding site in the central cavity of the human mitochondrial ADP/ATP carrier isoform 1, comprising the asymmetric residues G183, Y187, S228, R236 and K23 (Fig. 5A) and the symmetric residues R80 and R280 (Fig. 5B), which couple substrate binding to a symmetric transport mechanism. These residues have also been flagged up by other computational methods [103,108–110]. On either side of the central cavity one can find symmetric negatively and positively charged residues, which form three-fold salt bridge networks (see below) (Fig. 5B) [107].
Fig. 5. Asymmetric and symmetric residues in the central cavity of mitochondrial carriers and uncoupling protein.
Symmetry analysis was carried out on symmetry-related residues of the human (A and B) mitochondrial ADP/ATP carrier, (C and D) uncoupling protein, (E and F) dicarboxylate carrier. Lateral views from the membrane are shown. Conservation of the amino acid residues in the subfamily of orthologues is represented by the size of the sphere, whereas the average symmetry scores is represented by a colour scale from highly asymmetric (red) to neutral (white) to symmetrical (blue). The asymmetric residues are all labelled (left panels) and the symmetrical contact points of the substrate binding site are also labelled (right panels). The symmetrical residues belong to highly conserved motifs, such as the GXXXG and [FY]XX[YF] motifs and the charged residues of the cytoplasmic and matrix salt bridge network.
UCP1 is one of the most symmetrical of all members of the mitochondrial carrier family and it has very few highly conserved asymmetric residues in the cavity (Fig. 5C). In human UCP1, the most noteworthy residues are the asymmetric Q85 and D28 (Fig. 5C) and the symmetric R84, R183 and R277 (Fig. 5D) at the contact points of the common substrate binding site [111,112]. The asymmetric D28 is in the vicinity of the substrate binding site and could be involved in proton transport [113]. The same symmetry-related arginine triplet is also found in the dicarboxylate (Fig. 5E) and oxoglutarate carriers [107]. Sequence and phylogenetic analyses have shown that UCP1 likely evolved from dicarboxylate carriers and acquired a relatively small number of mutations in adaptation to its new role in thermogenesis [107,114], which was a relatively late development [115]. Based on this analysis it is not clear whether UCP1 has lost its ability to transport dicarboxylates all together, a function retained in UCP2 [116].
7. The binding of purine nucleotides to UCP1
Purine nucleotides inhibit UCP1 with high affinity by binding from the cytosolic side in a 1:1 inhibitor:protein stoichiometry [36]. However, in contrast to the ADP/ATP carrier (Fig. 5A) or GTP/GDP transporters [107], symmetry analysis has not revealed large-scale adaptations to accommodate nucleotide binding in UCP1. Nucleotides most likely adopt a different orientation in this carrier protein. Past proposals did not have the benefit of a carrier structure as a framework to guide interpretation of mutagenesis data (e.g. Refs. [53,55,117]). With the benefit of structural information [41,105], we have described how nucleotides may interact with UCP1 [36]. Nucleotides most likely occupy the central cavity of UCP1 in a similar manner to carboxyatractyloside inhibition of the ADP/ATP carrier [41,105]. GDP increases UCP1 stability [86] and resistance to trypsinolysis [118], much like the effects of carboxyatractyloside on the ADP/ATP carrier [119], consistent with inhibition in the cytoplasmic state of the protein. In the carrier substrate-binding site identified for UCP1 [112], the protein has a triplet of arginine residues, related by three-fold pseudo-symmetry, which would allow the binding of the phosphate moieties of the nucleotide by electrostatic attractions. In support of this notion, mutagenesis of these arginine residues in UCP1 abolishes nucleotide binding [55,117]. Furthermore, other carriers that conserve these arginine residues can also be inhibited by purine nucleotides, albeit with much lower affinity, such as the mitochondrial dicarboxylate carrier [120,121] or the oxoglutarate carrier (Dr. Magnus Monne', unpublished results). Interaction of nucleotides with UCP1 in this manner would be consistent with the loss of binding following the covalent modification of E190 by Woodward's Reagent K [122]. This residue contributes to the pH regulation of nucleotide inhibition and is predicted to face the central cavity of UCP1, toward the cytosolic side, where adduct formation would interfere with binding. Compared to the mitochondrial nucleotide carriers, there are no specific adaptations in UCP1 for binding of the bases [107], which could explain why there is little discrimination between adenine and guanine moieties. However, if the phosphate moieties bind to the arginine triplet, the purine base will be oriented towards the cytoplasmic side of the central cavity where it may have further interactions with other conserved residues (e.g. E190 [123]).
8. The binding of the fatty acid activator
There has been much debate on how fatty acids interact with UCP1 to bring about proton conductance (see proposed mechanisms, above). In describing the various models, the fatty acid acyl chain is often depicted going through the side of UCP1 leaving the carboxylic group in the central cavity (e.g. Refs. [55,57]). This kind of arrangement is unlikely, however, as a channel would mean that there is discontinuous electron density in the core of the membrane. It would also impede any conformational changes. An alternative proposal is that fatty acid anions bind to the outside surface of UCP1 for transport across the membrane at the lipid-protein interface as part of a cycling model, as suggested [124]. However, the outside of UCP1 is highly hydrophobic, as it is imbedded in the membrane, and there are no obvious asymmetric adaptations on the surface of the protein that could facilitate either ionic or specific hydrophobic interactions with a transport substrate.
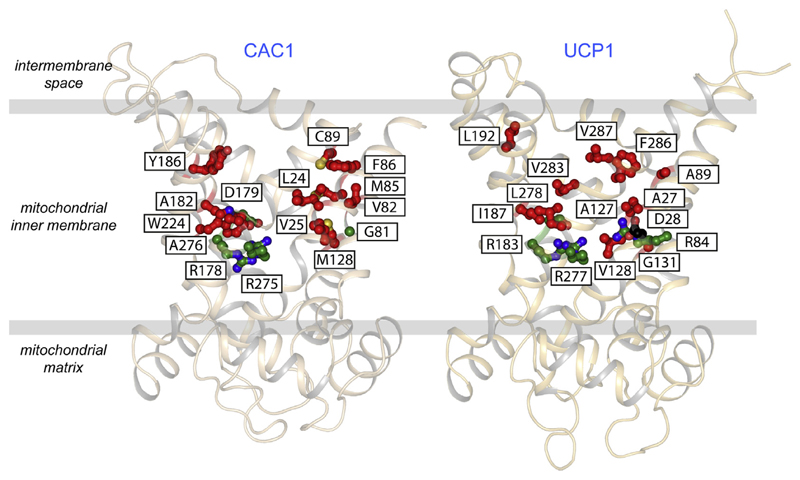
The third possibility is that the fatty acid binds in the central cavity as part of the mechanism. The triplet of arginine residues could be the binding site for the carboxylate group, due to electrostatic attractions. In the water phase of the cavity the hydrophobic acyl chain could have a minimised surface area, so the volume could be quite small. The residues that interact with the tail would have to be hydrophobic in nature. The mitochondrial carnitine/acylcarnitine carrier (CAC1) transports fatty acids covalently linked to carnitine and thus provides a good comparison for the same logistical issues. This carrier has a fairly large number of hydrophobic residues in the water-filled cavity (see Fig. 6), in particular on transmembrane α-helix H2, which have been shown to be important for substrate specificity [125]. On the whole the total number of hydrophobic residues in the water-filled cavity of UCP1 is quite similar (Fig. 6), but here they are mostly on transmembrane α-helix H6. On this basis it is certainly possible, but not proven, that fatty acid molecules are bound in the central cavity in a similar position as the substrates in other mitochondrial carriers. If true this scenario would imply that fatty acids would compete with nucleotides to bind to UCP1, as has been suggested [45] (see functional competition model), if fatty acids bind from the cytosol [57]. Yet, paradoxically, fatty acids have also been shown to increase the binding of a nucleotide derivative (mant-GDP) in pull down experiments with isolated brown adipose tissue mitochondria [118]. We have shown that ADP, as a substrate of the ADP/ATP carrier, can increase inhibitor binding by shifting the isolated protein to the appropriate state through transport cycling [75] (i.e. by increasing the rate of an otherwise inherently slow transition between cytoplasmic and matrix states). By analogy, shifting an otherwise static mixed population of UCP1 between states by fatty acid-induced cycling, would allow a cytoplasmic state conformation to be continuously sampled and only then the full nucleotide binding site population realised. This scenario would explain the fatty-acid induced increase in mant-GDP binding sites observed previously where no change in nucleotide affinity was apparent [118], but only if fatty acids are accepted to be transport substrates of UCP1. In our thermostability measurements with isolated carrier protein, we have found that fatty acids induce similar destabilising shifts in UCP1 as ADP does to the ADP/ATP carrier, which is also sensitive to cardiolipin, suggesting fatty acids do indeed behave like transport substrates [86].
Fig. 6. Hydrophobic amino acid residues in the cavity of the carnitine/acylcarnitine carrier and uncoupling protein.
Lateral view of the human carnitine/acylcarnitine carrier CAC1 (left) and human uncoupling protein UCP1 (right), modelled on yeast Aac3p [105] and bovine AAC1 [41], both shown in cartoon representation. Substrate binding site, proton binding site and hydrophobic residues (Gly, Ala, Val, Leu, Ile, Phe, Tyr, Trp, Met, and Cys) are shown in green, black and red ball-and-stick representations, respectively. Hydrophobic residues in the cavity that are common to all mitochondrial carriers, such as those belonging to the GXXXG and [FY]XX[YF] motifs, were excluded in the analysis.
9. UCP1 has a cytoplasmic and matrix salt bridge network
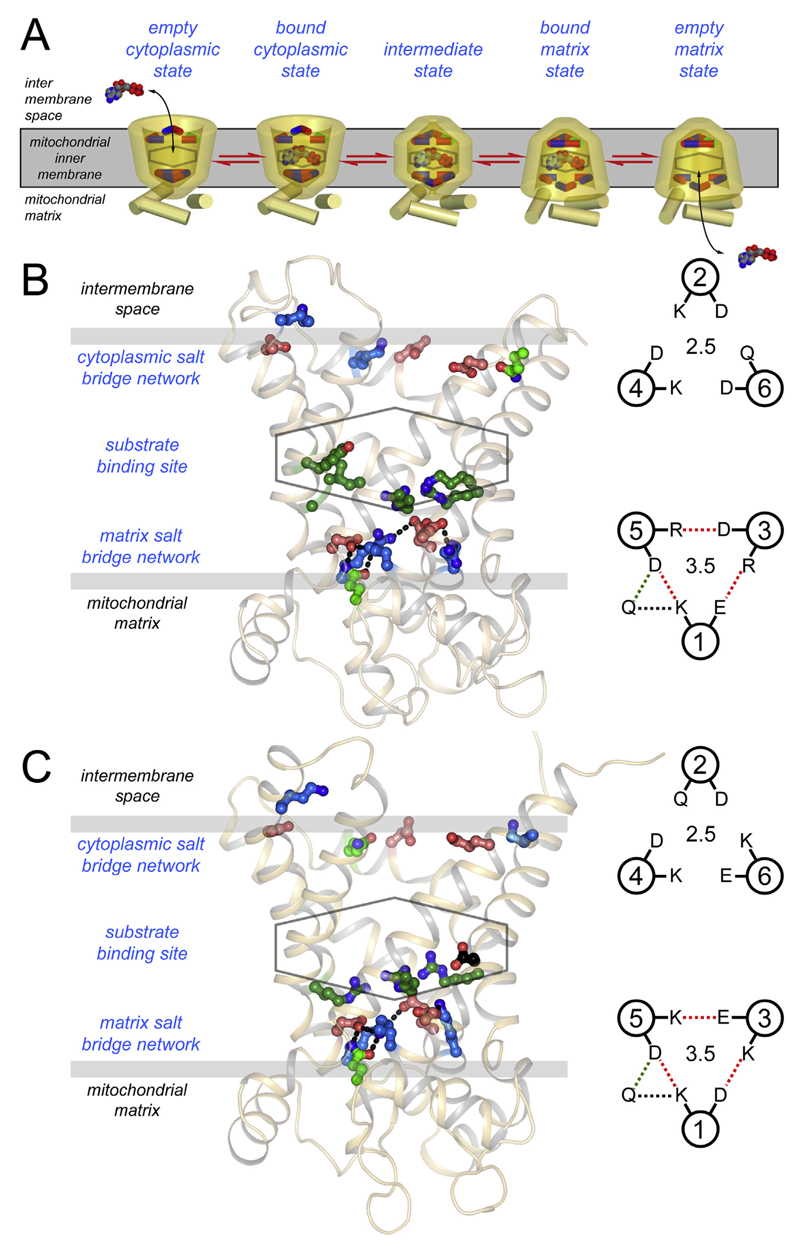
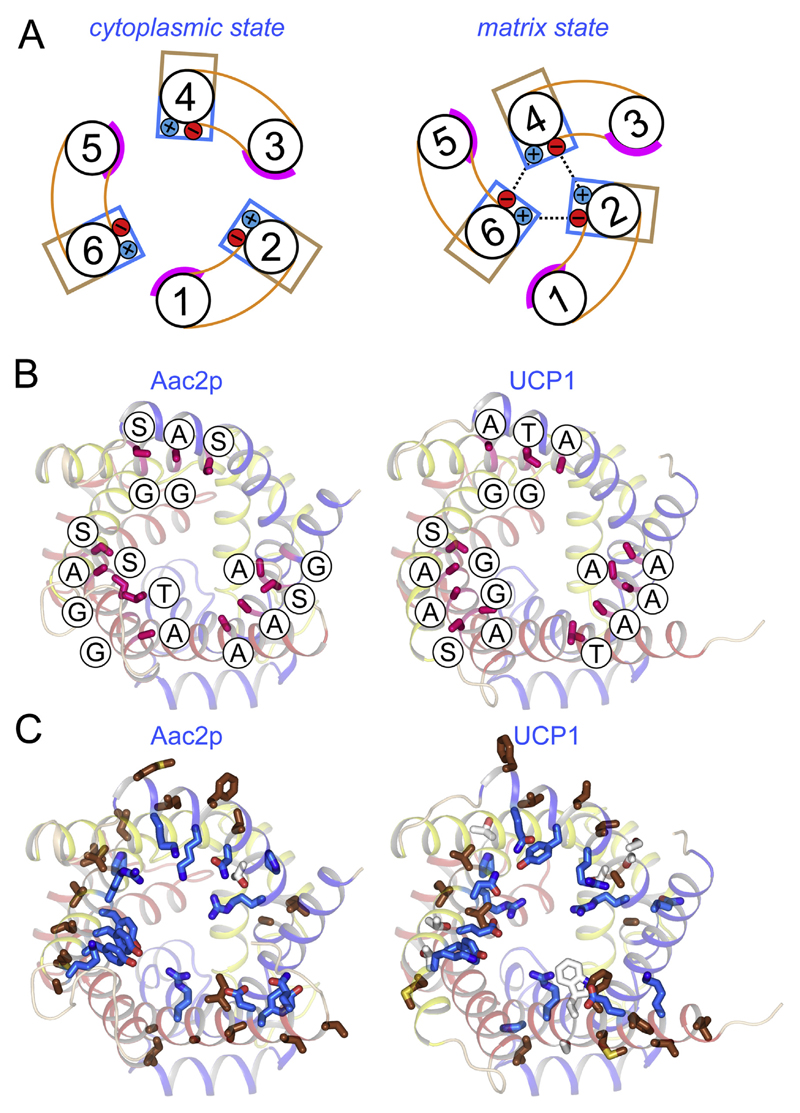
An alternating access transport mechanism for mitochondrial carriers has been proposed on the basis of inhibitor and substrate binding [126–128] as well as sequence and structural features [105–107]. Mitochondrial carriers cycle between a cytoplasmic state and matrix state in which the central substrate binding site is alternately accessible to each of these compartments. Key elements of this mechanism are two salt bridge networks on either side of the membrane that regulate access to this site in an alternating way (Fig. 7A) [107].
Fig. 7. Main functional elements of the mitochondrial ADP/ATP carrier and uncoupling protein.
A) The reversible transport states of the mitochondrial carrier named in blue. Disruption and formation of the cytoplasmic and matrix salt bridge networks, top and bottom respectively, have been proposed to be involved in the opening and closing of the carrier to either side of the membrane in an alternating way (107). B) Lateral view of the yeast ADP/ATP carrier Aac2p [105] and C) uncoupling protein 1, modelled on the yeast ADP/ATP carrier Aac3p [105] and bovine AAC1 [41], both shown in cartoon representation. The proposed central substrate binding site (stick), the cytoplasmic salt bridge network (top, ball-and-stick), and the matrix salt bridge network (bottom, ball-and-stick) are also shown. Positively charged, negatively charged and polar residues are shown in blue, red, and green, respectively. Also shown on the right are the matrix (bottom) and cytoplasmic network (top) in schematic representations.
Charged residues of the signature motif PX[DE]XX[RK] form a three-fold pseudo-symmetrical salt bridge network on the matrix side when the carrier is in the cytoplasmic state [41,129]. Beneath the network, bracing both residues of the salt bridges by hydrogen bonds, are conserved glutamine residues [105] (Fig. 7B). Mitochondrial carriers have different numbers of glutamine braces, thereby modulating the overall interaction energy of the matrix network. The matrix network of UCP1 has three salt bridges and one glutamine brace (Fig. 7C), just like the ADP/ATP carrier (cf. Fig. 7B).
Analysis of the pseudo-symmetry of mitochondrial carriers and UCP1 revealed a highly conserved and symmetrical [FY][DE]XX[RK] motif on the cytoplasmic side of the carrier (Fig. 7), which also has the propensity to form a salt bridge network [107]. In the cytoplasmic state the charged residues are not engaged in interactions [41,105], but they form a cytoplasmic salt bridge network when the carriers are in the matrix state [75,105]. The residues of the cytoplasmic network are located on the even-numbered α-helices, whereas those of the matrix network are on the odd-numbered α-helices. Both networks are at the water-membrane interface on either side of the carrier, where solute access to the central substrate binding site may be controlled. In the mechanism, substrate binding in one conformation allows the conversion to the other conformation by the disruption and formation of these networks, causing the alternating opening and closing of the carrier to either side of the membrane (Fig. 7A). UCP1 has a cytoplasmic network similar to that of the ADP/ATP carrier of yeast, consisting of two salt bridges and one hydrogen bond (Fig. 7B and C). The complete conservation of both salt bridge networks in UCP1 suggests that it also has an alternating access mechanism. In agreement with this notion it has been shown that stability and proteolysis sensitivity changes in the presence of fatty acids, suggesting that UCP1 cycles between states [86,118].
10. UCP1 has other conserved symmetrical features compatible with a dynamic carrier-like mechanism
Symmetry analysis has highlighted other highly conserved symmetrical features of mitochondrial carriers and UCP1 [107], which are likely to be important for the three-fold pseudo-symmetrical structure and mechanism.
One of the symmetrical features are the proline residues of the aforementioned motif PX[DE]XX[RK], which are found at the kinks of the L-shaped odd-numbered α-helices [41] (Fig. 8). The proline residues break the hydrogen bonding arrangement of the helices, allowing the kink to occur [41,105]. In agreement, a serine substitution in the second domain of the yeast ADP/ATP carriers forms a hydrogen bond with the backbone, mimicking proline (Fig. 8) [105]. UCP1 has three proline residues, indicating that it also has L-shaped α-helices. It has been proposed that the L- shape allows formation and disruption of the matrix salt bridge network to be coupled to the disruption and formation of the cytoplasmic salt bridge network in an alternating way [105].
Fig. 8. Conserved symmetrical elements of the mitochondrial ADP/ATP carrier and uncoupling protein.
Lateral view of the yeast ADP/ATP carrier Aac2p [105] (left) and uncoupling protein 1 (right), a comparative model using the yeast Aac3p [105] and bovine AAC1 [41] structures, in cartoon representation. Aromatic and hydrophobic residues of the [FY]XX[YF] motif are shown in orange and pink ball-and-stick representations, respectively. Proline residues of the PX[DE]XX[KR] motif are shown in a magenta ball-and-stick representations, whereas the serine residue in Aac2p, which mimics a proline residue, is shown in a green ball-and-stick representation. Positively charged residues, found two positions after the PX[DE]XX[KR] motif, and negatively charged residues of the [ED]G motif are shown in blue and red ball-and-stick representations, respectively, whereas a hydrophobic substitution of a positively charged residue in Aac2p is shown in pink.
A second set of highly conserved symmetrical features are residues of the [FY]XX[YF] motif, which precede those of the cytoplasmic network. Although residues of this motif are largely aromatic, hydrophobic substitutions are also found. Due to their position, it has been proposed that formation of the cytoplasmic salt bridge network in the matrix state would bring these residues together to form a hydrophobic layer as part of the gate that closes the carrier to the cytoplasmic side [130,131]. The residues of UCP1 in these positions are either hydrophobic or aromatic, and so they could form an equivalent hydrophobic layer, consistent with a carrier mechanism.
Highly conserved and symmetrical positively charged residues, found two residue positions after the PX[DE]XX[KR] motif, interact with the negatively charged residues of the symmetrical [ED]G motif to link the C-terminal end of the odd-numbered α-helices with the matrix α-helices. These interactions could stabilise the structure of the domains, allowing its highly tilted shape. The mitochondrial ADP/ATP carrier has one, possibly two, of these interactions [41,105], as one is prevented by a hydrophobic substitution, whereas UCP1 has the propensity to form three of these interactions, highlighting again that UCP1 is more symmetrical. The peculiar shape of the domains, which is maintained by this interaction, allows the opening and closing of the carrier to either side of the membrane in an alternating way.
Finally, analyses of the inter-domain interfaces of mitochondrial carriers have indicated that the even-numbered α-helices may rotate across the surface of the odd-numbered α-helices as part of the transport cycle (Fig. 9A) [105]. The helical surface of the odd-numbered α-helices is smooth, as it has residues with short side chains or glycine residues of the symmetrical GXXXG motif, which is conserved in the mitochondrial ADP/ATP carrier and uncoupling protein (Fig. 9B). On the even-numbered α-helices in the interface, hydrophobic residues face the membrane whereas hydrophilic residues face the cavity, neither pointing towards the interface, which is also conserved in both proteins (Fig. 9C). The conservation of these features in the inter-domain interface indicates that UCP1 has retained the ability to execute these dynamic changes as part of its mechanism. The three tightly bound cardiolipin molecules will span this interface and hold the domains together at the matrix side during these rotations.
Fig. 9. Uncoupling protein 1 has retained residues consistent with a dynamic inter-domain interface.
A) Schematic representation of the α-helical arrangement of mitochondrial carriers in the cytoplasmic (left) and matrix state (right), caused by rotation of the three domains [105]. The smooth surfaces of the odd numbered helices are shown in magenta, whereas the hydrophilic and hydrophobic residues of the even-numbered α-helices are represented by blue and brown boxes, respectively. The formation of the cytoplasmic salt bridge network is also shown. B) and C) The structures of the ADP/ATP carrier (left) and uncoupling protein 1 (right), viewed from the cytoplasmic side. Domain 1, 2 and 3 are coloured blue, yellow, and red, respectively. B) Residues of the odd-numbered α-helices in the inter-domain interface have no or small side chains, including the residues of the conserved GXXXG motif (shown in magenta and labelled). (C) Residues of the even-numbered α-helices in the inter-domain interface are hydrophobic (brown), facing the membrane, or hydrophilic (blue), facing the cavity.
11. Conclusions
In this review we have investigated the sequence and protein properties of UCP1 and compared them to those of other members of the mitochondrial carrier family. UCP1 functions as a monomer, binds a single purine nucleotide as part of its regulation, and has three tightly bound cardiolipin molecules that are important for the stability and mechanism of this membrane protein [36,86]. Sequence and phylogenetic analyses have shown that UCP1 likely evolved from dicarboxylate carriers and acquired a relatively small number of mutations in adaptation to its new role in thermogenesis [107], which was a relatively late development [114]. Remnants of its function as a dicarboxylate carrier are the symmetric arginine triplet and Q85 in its substrate binding site [107]. Among the most striking adaptations, away from that function, are D28 and E190, which could be key residues for binding of protons and purine nucleotides, respectively. Another possible adaptation is a number of hydrophobic residues in the central water-filled cavity, which could facilitate the binding of the acyl chain moiety of fatty acids.
We have also examined whether key features of an alternating access mechanism have been retained in UCP1. UCP1 has two salt bridge networks on either side of the central substrate binding site [107], indicating that formation and disruption of these networks is important for UCP1 function. They also have conserved prolines to preserve the L-shaped odd-numbered α-helices, [RK] … [ED]G interactions to preserve the shape of the domain, a hydrophobic layer as part of the cytoplasmic gate, and dynamic inter-domain interfaces, all pointing towards a dynamic mechanism in which the three domains rotate to link the disruption and formation of these two networks.
After reviewing all of the highly conserved protein features of UCP1, it is clear that all of the key functional properties of mitochondrial carriers are conserved. Taken together they may indicate that fatty acid activated proton conductance by UCP1 may have a carrier-like mechanism. Alternatively, these conserved features could reflect an additional more conventional transport function of the protein.
Abbreviations
- UCP
uncoupling protein
- AAC
mitochondrial ADP/ADP carrier
- DPC
dodecylphosphocholine (Fos-Choline-12)
- 10MNG
decyl maltose neopentyl glycol
References
- [1].Matthias A, Ohlson KB, Fredriksson JM, Jacobsson A, Nedergaard J, Cannon B. Thermogenic responses in brown fat cells are fully UCP1-dependent. UCP2 or UCP3 do not substitute for UCP1 in adrenergically or fatty acid-induced thermogenesis. J Biol Chem. 2000;275:25073–25081. doi: 10.1074/jbc.M000547200. [DOI] [PubMed] [Google Scholar]
- [2].Golozoubova V, Hohtola E, Matthias A, Jacobsson A, Cannon B, Nedergaard J. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. FASEB J Off Publ Fed Am Soc Exp Biol. 2001;15:2048–2050. doi: 10.1096/fj.00-0536fje. [DOI] [PubMed] [Google Scholar]
- [3].Nedergaard J, Golozoubova V, Matthias A, Asadi A, Jacobsson A, Cannon B. UCP1: the only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochim Biophys Acta. 2001;1504:82–106. doi: 10.1016/s0005-2728(00)00247-4. [DOI] [PubMed] [Google Scholar]
- [4].Divakaruni AS, Brand MD. The regulation and physiology of mitochondrial proton leak. Physiol (Bethesda, Md.) 2011;26:192–205. doi: 10.1152/physiol.00046.2010. [DOI] [PubMed] [Google Scholar]
- [5].Nedergaard J, Cannon B. The 'novel' 'uncoupling' proteins UCP2 and UCP3: what do they really do? Pros and cons for suggested functions. Exp Physiol. 2003;88:65–84. doi: 10.1113/eph8802502. [DOI] [PubMed] [Google Scholar]
- [6].Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- [7].Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lidell ME, Betz MJ, Enerback S. Two types of brown adipose tissue in humans. Adipocyte. 2014;3:63–66. doi: 10.4161/adip.26896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- [10].Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, Nuutila P. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- [11].Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- [12].Yoneshiro T, Aita S, Matsushita M, Kameya T, Nakada K, Kawai Y, Saito M. Brown adipose tissue, whole-body energy expenditure, and thermogenesis in healthy adult men. Obesity (Silver Spring, Md.) 2011;19:13–16. doi: 10.1038/oby.2010.105. [DOI] [PubMed] [Google Scholar]
- [13].Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, Kawai Y, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, Eychmuller A, et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- [15].Lowell BB, SS V, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- [16].Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- [17].Sell H, Berger JP, Samson P, Castriota G, Lalonde J, Deshaies Y, Richard D. Peroxisome proliferator-activated receptor gamma agonism increases the capacity for sympathetically mediated thermogenesis in lean and ob/ob mice. Endocrinology. 2004;145:3925–3934. doi: 10.1210/en.2004-0321. [DOI] [PubMed] [Google Scholar]
- [18].Nedergaard J, Cannon B. The browning of white adipose tissue: some burning issues. Cell Metab. 2014;20:396–407. doi: 10.1016/j.cmet.2014.07.005. [DOI] [PubMed] [Google Scholar]
- [19].Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- [20].Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- [21].Collins S, Yehuda-Shnaidman E, Wang H. Positive and negative control of Ucp1 gene transcription and the role of beta-adrenergic signaling networks. Int J Obes (2005) 2010;34(Suppl 1):S28–S33. doi: 10.1038/ijo.2010.180. [DOI] [PubMed] [Google Scholar]
- [22].Collins S, Cao W, Robidoux J. Learning new tricks from old dogs: beta-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol Endocrinol (Baltimore, Md.) 2004;18:2123–2131. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- [23].Klingenspor M. Cold-induced recruitment of brown adipose tissue thermogenesis. Exp Physiol. 2003;88:141–148. doi: 10.1113/eph8802508. [DOI] [PubMed] [Google Scholar]
- [24].Chaudhry A, Granneman JG. Differential regulation of functional responses by beta-adrenergic receptor subtypes in brown adipocytes. Am J Physiol. 1999;277:R147–R153. doi: 10.1152/ajpregu.1999.277.1.R147. [DOI] [PubMed] [Google Scholar]
- [25].Shih MF, Taberner PV. Selective activation of brown adipocyte hormone-sensitive lipase and cAMP production in the mouse by beta 3-adrenoceptor agonists. Biochem Pharmacol. 1995;50:601–608. doi: 10.1016/0006-2952(95)00185-3. [DOI] [PubMed] [Google Scholar]
- [26].Locke RM, Rial E, Scott ID, Nicholls DG. Fatty acids as acute regulators of the proton conductance of hamster brown-fat mitochondria. Eur J Biochem. 1982;129:373–380. doi: 10.1111/j.1432-1033.1982.tb07060.x. [DOI] [PubMed] [Google Scholar]
- [27].Klingenberg M, Echtay KS, Bienengraeber M, Winkler E, Huang SG. Structure-function relationship in UCP1. Int J Obes Relat Metab Disord J Int Assoc Study Obes. 1999;23(Suppl 6):S24–S29. doi: 10.1038/sj.ijo.0800939. [DOI] [PubMed] [Google Scholar]
- [28].Kozak UC, Kopecky J, Teisinger J, Enerback S, Boyer B, Kozak LP. An up-stream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol Cell Biol. 1994;14:59–67. doi: 10.1128/mcb.14.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Heaton GM, Wagenvoord RJ, Kemp A, Jr, Nicholls DG. Brown-adipose-tissue mitochondria: photoaffinity labelling of the regulatory site of energy dissipation. Eur J Biochem. 1978;82:515–521. doi: 10.1111/j.1432-1033.1978.tb12045.x. [DOI] [PubMed] [Google Scholar]
- [30].Nicholls DG, Rial E. A history of the first uncoupling protein, UCP1. J Bioenerg Biomembr. 1999;31:399–406. doi: 10.1023/a:1005436121005. [DOI] [PubMed] [Google Scholar]
- [31].Rafael J, Heldt HW. Binding of guanine nucleotides to the outer surface of the inner membrane of guinea pig brown fat mitochondria in correlation with the thermogenic activity of the tissue. FEBS Lett. 1976;63:304–308. doi: 10.1016/0014-5793(76)80117-2. [DOI] [PubMed] [Google Scholar]
- [32].Ricquier D, Kader JC. Mitochondrial protein alteration in active brown fat: a soidum dodecyl sulfate-polyacrylamide gel electrophoretic study. Biochem Biophys Res Commun. 1976;73:577–583. doi: 10.1016/0006-291x(76)90849-4. [DOI] [PubMed] [Google Scholar]
- [33].Ricquier D, Gervais C, Kader JC, Hemon P. Partial purification by guanosine-5'-diphosphate–agarose affinity chromatography of the 32,000 molecular weight polypeptide from mitochondria of brown adipose tissue. FEBS Lett. 1979;101:35–38. [PubMed] [Google Scholar]
- [34].Lin CS, Klingenberg M. Isolation of the uncoupling protein from brown adipose tissue mitochondria. FEBS Lett. 1980;113:299–303. doi: 10.1016/0014-5793(80)80613-2. [DOI] [PubMed] [Google Scholar]
- [35].Lin CS, Klingenberg M. Characteristics of the isolated purine nucleotide binding protein from brown fat mitochondria. Biochemistry. 1982;21:2950–2956. doi: 10.1021/bi00541a023. [DOI] [PubMed] [Google Scholar]
- [36].Lee Y, Willers C, Kunji ER, Crichton PG. Uncoupling protein 1 binds one nucleotide per monomer and is stabilized by tightly bound cardiolipin. Proc Natl Acad Sci U S A. 2015;112:6973–6978. doi: 10.1073/pnas.1503833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Aquila H, Link TA, Klingenberg M. The uncoupling protein from brown fat mitochondria is related to the mitochondrial ADP/ATP carrier. Analysis of sequence homologies and of folding of the protein in the membrane. EMBO J. 1985;4:2369–2376. doi: 10.1002/j.1460-2075.1985.tb03941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bouillaud F, Weissenbach J, Ricquier D. Complete cDNA-derived amino acid sequence of rat brown fat uncoupling protein. J Biol Chem. 1986;261:1487–1490. [PubMed] [Google Scholar]
- [39].Saraste M, Walker JE. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982;144:250–254. doi: 10.1016/0014-5793(82)80648-0. [DOI] [PubMed] [Google Scholar]
- [40].Aquila H, Link TA, Klingenberg M. Solute carriers involved in energy transfer of mitochondria form a homologous protein family. FEBS Lett. 1987;212:1–9. doi: 10.1016/0014-5793(87)81546-6. [DOI] [PubMed] [Google Scholar]
- [41].Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- [42].Bouillaud F, Ricquier D, Gulik-Krzywicki T, Gary-Bobo CM. The possible proton translocating activity of the mitochondrial uncoupling protein of brown adipose tissue. Reconstitution studies in liposomes. FEBS Lett. 1983;164:272–276. doi: 10.1016/0014-5793(83)80300-7. [DOI] [PubMed] [Google Scholar]
- [43].Klingenberg M, Winkler E. The reconstituted isolated uncoupling protein is a membrane potential driven Hþ translocator. EMBO J. 1985;4:3087–3092. doi: 10.1002/j.1460-2075.1985.tb04049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jezek P, Drahota Z, Ring K. The activating effect of fatty acid on the mitochondrial uncoupling protein reconstituted in liposomes. J Lipid Mediat. 1990;2:85–94. [PubMed] [Google Scholar]
- [45].Shabalina IG, Jacobsson A, Cannon B, Nedergaard J. Native UCP1 displays simple competitive kinetics between the regulators purine nucleotides and fatty acids. J Biol Chem. 2004;279:38236–38248. doi: 10.1074/jbc.M402375200. [DOI] [PubMed] [Google Scholar]
- [46].Shabalina IG, Ost M, Petrovic N, Vrbacky M, Nedergaard J, Cannon B. Uncoupling protein-1 is not leaky. Biochim Biophys Acta. 2010;1797:773–784. doi: 10.1016/j.bbabio.2010.04.007. [DOI] [PubMed] [Google Scholar]
- [47].Winkler E, Klingenberg M. Effect of fatty acids on H+ transport activity of the reconstituted uncoupling protein. J Biol Chem. 1994;269:2508–2515. [PubMed] [Google Scholar]
- [48].Jezek P, Orosz DE, Modriansky M, Garlid KD. Transport of anions and protons by the mitochondrial uncoupling protein and its regulation by nucleotides and fatty acids. A new look at old hypotheses. J Biol Chem. 1994;269:26184–26190. [PubMed] [Google Scholar]
- [49].Skulachev VP. Fatty acid circuit as a physiological mechanism of uncoupling of oxidative phosphorylation. FEBS Lett. 1991;294:158–162. doi: 10.1016/0014-5793(91)80658-p. [DOI] [PubMed] [Google Scholar]
- [50].Garlid KD, Orosz DE, Modriansky M, Vassanelli S, Jezek P. On the mechanism of fatty acid-induced proton transport by mitochondrial uncoupling protein. J Biol Chem. 1996;271:2615–2620. doi: 10.1074/jbc.271.5.2615. [DOI] [PubMed] [Google Scholar]
- [51].Nicholls DG, Lindberg O. Brown-adipose-tissue mitochondria. The influence of albumin and nucleotides on passive ion permeabilities. Eur J Biochem. 1973;37:523–530. doi: 10.1111/j.1432-1033.1973.tb03014.x. [DOI] [PubMed] [Google Scholar]
- [52].Jezek P, Orosz DE, Garlid KD. Reconstitution of the uncoupling protein of brown adipose tissue mitochondria. Demonstration of GDP-sensitive halide anion uniport. J Biol Chem. 1990;265:19296–19302. [PubMed] [Google Scholar]
- [53].Klingenberg M, Huang SG. Structure and function of the uncoupling protein from brown adipose tissue. Biochim Biophys Acta. 1999;1415:271–296. doi: 10.1016/s0005-2736(98)00232-6. [DOI] [PubMed] [Google Scholar]
- [54].Breen EP, Gouin SG, Murphy AF, Haines LR, Jackson AM, Pearson TW, Murphy PV, Porter RK. On the mechanism of mitochondrial uncoupling protein 1 function. J Biol Chem. 2006;281:2114–2119. doi: 10.1074/jbc.M511575200. [DOI] [PubMed] [Google Scholar]
- [55].Klingenberg M, Echtay KS. Uncoupling proteins: the issues from a biochemist point of view. Biochim Biophys Acta. 2001;1504:128–143. doi: 10.1016/s0005-2728(00)00242-5. [DOI] [PubMed] [Google Scholar]
- [56].Jaburek M, Varecha M, Jezek P, Garlid KD. Alkylsulfonates as probes of uncoupling protein transport mechanism. Ion pair transport demonstrates that direct H(þ) translocation by UCP1 is not necessary for uncoupling. J Biol Chem. 2001;276:31897–31905. doi: 10.1074/jbc.M103507200. [DOI] [PubMed] [Google Scholar]
- [57].Fedorenko A, Lishko PV, Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2012;151:400–413. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kamp F, Hamilton JA. pH gradients across phospholipid membranes caused by fast flip-flop of un-ionized fatty acids. Proc Natl Acad Sci U S A. 1992;89:11367–11370. doi: 10.1073/pnas.89.23.11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Echtay KS, Winkler E, Klingenberg M. Coenzyme Q is an obligatory cofactor for uncoupling protein function. Nature. 2000;408:609–613. doi: 10.1038/35046114. [DOI] [PubMed] [Google Scholar]
- [60].Echtay KS, Winkler E, Frischmuth K, Klingenberg M. Uncoupling proteins 2 and 3 are highly active H(þ) transporters and highly nucleotide sensitive when activated by coenzyme Q (ubiquinone) Proc Natl Acad Sci U S A. 2001;98:1416–1421. doi: 10.1073/pnas.98.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jaburek M, Garlid KD. Reconstitution of recombinant uncoupling proteins: UCP1, -2, and -3 have similar affinities for ATP and are unaffected by coenzyme Q10. J Biol Chem. 2003;278:25825–25831. doi: 10.1074/jbc.M302126200. [DOI] [PubMed] [Google Scholar]
- [62].Esteves TC, Echtay KS, Jonassen T, Clarke CF, Brand MD. Ubiquinone is not required for proton conductance by uncoupling protein 1 in yeast mitochondria. Biochem J. 2004;379:309–315. doi: 10.1042/BJ20031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, Harper JA, Roebuck SJ, Morrison A, Pickering S, Clapham JC, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- [64].Echtay KS, Esteves TC, Pakay JL, Jekabsons MB, Lambert AJ, Portero-Otin M, Pamplona R, Vidal-Puig AJ, Wang S, Roebuck SJ, Brand MD. A signalling role for 4-hydroxy-2-nonenal in regulation of mitochondrial uncoupling. EMBO J. 2003;22:4103–4110. doi: 10.1093/emboj/cdg412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Couplan E, del Mar Gonzalez-Barroso M, Alves-Guerra MC, Ricquier D, Goubern M, Bouillaud F. No evidence for a basal, retinoic, or superoxide-induced uncoupling activity of the uncoupling protein 2 present in spleen or lung mitochondria. J Biol Chem. 2002;277:26268–26275. doi: 10.1074/jbc.M202535200. [DOI] [PubMed] [Google Scholar]
- [66].Shabalina IG, Petrovic N, Kramarova TV, Hoeks J, Cannon B, Nedergaard J. UCP1 and defense against oxidative stress. 4-Hydroxy-2-nonenal effects on brown fat mitochondria are uncoupling protein 1-independent. J Biol Chem. 2006;281:13882–13893. doi: 10.1074/jbc.M601387200. [DOI] [PubMed] [Google Scholar]
- [67].Shabalina IG, Hoeks J, Kramarova TV, Schrauwen P, Cannon B, Nedergaard J. Cold tolerance of UCP1-ablated mice: a skeletal muscle mitochondria switch toward lipid oxidation with marked UCP3 up-regulation not associated with increased basal, fatty acid- or ROS-induced uncoupling or enhanced GDP effects. Biochim Biophys Acta. 2010;1797:968–980. doi: 10.1016/j.bbabio.2010.02.033. [DOI] [PubMed] [Google Scholar]
- [68].Nabben M, Shabalina IG, Moonen-Kornips E, van Beurden D, Cannon B, Schrauwen P, Nedergaard J, Hoeks J. Uncoupled respiration, ROS production, acute lipotoxicity and oxidative damage in isolated skeletal muscle mitochondria from UCP3-ablated mice. Biochim Biophys Acta. 2011;1807:1095–1105. doi: 10.1016/j.bbabio.2011.04.003. [DOI] [PubMed] [Google Scholar]
- [69].Silva JP, Shabalina IG, Dufour E, Petrovic N, Backlund EC, Hultenby K, Wibom R, Nedergaard J, Cannon B, Larsson NG. SOD2 overexpression: enhanced mitochondrial tolerance but absence of effect on UCP activity. EMBO J. 2005;24:4061–4070. doi: 10.1038/sj.emboj.7600866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Cannon B, Shabalina IG, Kramarova TV, Petrovic N, Nedergaard J. Uncoupling proteins: a role in protection against reactive oxygen specieseor not? Biochim Biophys Acta. 2006;1757:449–458. doi: 10.1016/j.bbabio.2006.05.016. [DOI] [PubMed] [Google Scholar]
- [71].Nicholls DG. The physiological regulation of uncoupling proteins. Biochim Biophys Acta. 2006;1757:459–466. doi: 10.1016/j.bbabio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- [72].Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, Robinson AJ, et al. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature. 2016;532:112–116. doi: 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nicholls DG, Rial E. A novel regulatory mechanism for the brown-fat uncoupling protein? Nat Struct Mol Biol. 2016;23:364–365. doi: 10.1038/nsmb.3221. [DOI] [PubMed] [Google Scholar]
- [74].Bamber L, Harding M, Monne M, Slotboom DJ, Kunji ER. The yeast mitochondrial ADP/ATP carrier functions as a monomer in mitochondrial membranes. Proc Natl Acad Sci U S A. 2007;104:10830–10834. doi: 10.1073/pnas.0703969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].King MS, Kerr M, Crichton PG, Springett R, Kunji ER. Formation of a cytoplasmic salt bridge network in the matrix state is a fundamental step in the transport mechanism of the mitochondrial ADP/ATP carrier. Biochim Biophys Acta. 2016;1857:14–22. doi: 10.1016/j.bbabio.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Jelokhani-Niaraki M, Ivanova MV, McIntyre BL, Newman CL, McSorley FR, Young EK, Smith MD. A CD study of uncoupling protein-1 and its transmembrane and matrix-loop domains. Biochem J. 2008;411:593–603. doi: 10.1042/bj20071326. [DOI] [PubMed] [Google Scholar]
- [77].Ivanova MV, Hoang T, McSorley FR, Krnac G, Smith MD, Jelokhani-Niaraki M. A comparative study on conformation and ligand binding of the neuronal uncoupling proteins. Biochemistry. 2010;49:512–521. doi: 10.1021/bi901742g. [DOI] [PubMed] [Google Scholar]
- [78].Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011;476:109–113. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhu R, Rupprecht A, Ebner A, Haselgrubler T, Gruber HJ, Hinterdorfer P, Pohl EE. Mapping the nucleotide binding site of uncoupling protein 1 using atomic force microscopy. J Am Chem Soc. 2013;135:3640–3646. doi: 10.1021/ja312550k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hoang T, Smith MD, Jelokhani-Niaraki M. Expression, folding, and proton transport activity of human uncoupling protein-1 (UCP1) in lipid membranes: evidence for associated functional forms. J Biol Chem. 2013;288:36244–36258. doi: 10.1074/jbc.M113.509935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hoang T, Smith MD, Jelokhani-Niaraki M. Toward understanding the mechanism of ion transport activity of neuronal uncoupling proteins UCP2, UCP4, and UCP5. Biochemistry. 2012;51:4004–4014. doi: 10.1021/bi3003378. [DOI] [PubMed] [Google Scholar]
- [82].Berardi MJ, Chou JJ. Fatty acid flippase activity of UCP2 is essential for its proton transport in mitochondria. Cell Metab. 2014;20:541–552. doi: 10.1016/j.cmet.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bruschweiler S, Yang Q, Run C, Chou JJ. Substrate-modulated ADP/ATP-transporter dynamics revealed by NMR relaxation dispersion. Nat Struct Mol Biol. 2015;22:636–641. doi: 10.1038/nsmb.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sounier R, Bellot G, Chou JJ. Mapping conformational heterogeneity of mitochondrial nucleotide transporter in uninhibited states. Angew Chem Int ed Engl. 2015;54:2436–2441. doi: 10.1002/anie.201408417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zoonens M, Comer J, Masscheleyn S, Pebay-Peyroula E, Chipot C, Miroux B, Dehez F. Dangerous liaisons between detergents and membrane proteins. The case of mitochondrial uncoupling protein 2. J Am Chem Soc. 2013;135:15174–15182. doi: 10.1021/ja407424v. [DOI] [PubMed] [Google Scholar]
- [86].Crichton PG, Lee Y, Ruprecht JJ, Cerson E, Thangaratnarajah C, King MS, Kunji ER. Trends in thermostability provide information on the nature of substrate, inhibitor, and lipid interactions with mitochondrial carriers. J Biol Chem. 2015;290:8206–8217. doi: 10.1074/jbc.M114.616607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lin CS, Hackenberg H, Klingenberg EM. The uncoupling protein from brown adipose tissue mitochondria is a dimer. A hydrodynamic study. FEBS Lett. 1980;113:304–306. doi: 10.1016/0014-5793(80)80614-4. [DOI] [PubMed] [Google Scholar]
- [88].Nedergaard J, Cannon B. [3H]GDP binding and thermogenin amount in brown adipose tissue mitochondria from cold-exposed rats. Am J Physiol. 1985;248:C365–C371. doi: 10.1152/ajpcell.1985.248.3.C365. [DOI] [PubMed] [Google Scholar]
- [89].Jakobs P, Braun A, Jezek P, Trommer WE. Binding of ATP to uncoupling protein of brown fat mitochondria as studied by means of spin-labeled ATP derivatives. FEBS Lett. 1991;284:195–198. doi: 10.1016/0014-5793(91)80683-t. [DOI] [PubMed] [Google Scholar]
- [90].Winkler E, Klingenberg M. Photoaffinity labeling of the nucleotide-binding site of the uncoupling protein from hamster brown adipose tissue. Eur J Biochem. 1992;203:295–304. doi: 10.1111/j.1432-1033.1992.tb19859.x. [DOI] [PubMed] [Google Scholar]
- [91].Kunji ER, Crichton PG. Mitochondrial carriers function as monomers. Biochim Biophys Acta. 2010;1797:817–831. doi: 10.1016/j.bbabio.2010.03.023. [DOI] [PubMed] [Google Scholar]
- [92].Rodriguez-Vico F, Martinez-Cayuela M, Zafra MF, Garcia-Peregrin E, Ramirez H. A procedure for the simultaneous determination of lipid and protein in biomembranes and other biological samples. Lipids. 1991;26:77–80. doi: 10.1007/BF02544029. [DOI] [PubMed] [Google Scholar]
- [93].Bamber L, Harding M, Butler PJ, Kunji ER. Yeast mitochondrial ADP/ATP carriers are monomeric in detergents. Proc Natl Acad Sci U S A. 2006;103:16224–16229. doi: 10.1073/pnas.0607640103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Kunji ER, Harding M, Butler PJ, Akamine P. Determination of the molecular mass and dimensions of membrane proteins by size exclusion chromatography. Methods. 2008;46:62–72. doi: 10.1016/j.ymeth.2008.10.020. [DOI] [PubMed] [Google Scholar]
- [95].Harborne SP, Ruprecht JJ, Kunji ER. Calcium-induced conformational changes in the regulatory domain of the human mitochondrial ATP-Mg/Pi carrier. Biochim Biophys Acta. 2015;1847:1245–1253. doi: 10.1016/j.bbabio.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Thangaratnarajah C, Ruprecht JJ, Kunji ER. Calcium-induced conformational changes of the regulatory domain of human mitochondrial aspartate/glutamate carriers. Nat Commun. 2014;5:5491. doi: 10.1038/ncomms6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Crichton PG, Harding M, Ruprecht JJ, Lee Y, Kunji ER. Lipid, detergent, and Coomassie Blue G-250 affect the migration of small membrane proteins in blue native gels: mitochondrial carriers migrate as monomers not dimers. J Biol Chem. 2013;288:22163–22173. doi: 10.1074/jbc.M113.484329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kunji ER, Harding M. Projection structure of the atractyloside-inhibited mitochondrial ADP/ATP carrier of Saccharomyces cerevisiae. J Biol Chem. 2003;278:36985–36988. doi: 10.1074/jbc.C300304200. [DOI] [PubMed] [Google Scholar]
- [99].Klingenberg M. Cardiolipin and mitochondrial carriers. Biochim Biophys Acta. 2009;1788:2048–2058. doi: 10.1016/j.bbamem.2009.06.007. [DOI] [PubMed] [Google Scholar]
- [100].Beyer K, Klingenberg M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry. 1985;24:3821–3826. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
- [101].Winkler E, Klingenberg M. An improved procedure for reconstitution of the uncoupling protein and in-depth analysis of H+/OH- transport. Eur J Biochem. 1992;207:135–145. doi: 10.1111/j.1432-1033.1992.tb17030.x. [DOI] [PubMed] [Google Scholar]
- [102].Kramer R, Palmieri F. Molecular aspects of isolated and reconstituted carrier proteins from animal mitochondria. Biochim Biophys Acta. 1989;974:1–23. doi: 10.1016/s0005-2728(89)80160-4. [DOI] [PubMed] [Google Scholar]
- [103].Mifsud J, Ravaud S, Krammer EM, Chipot C, Kunji ER, Pebay-Peyroula E, Dehez F. The substrate specificity of the human ADP/ATP carrier AAC1. Mol Membr Biol. 2013;30:160–168. doi: 10.3109/09687688.2012.745175. [DOI] [PubMed] [Google Scholar]
- [104].Nury H, Dahout-Gonzalez C, Trezeguet V, Lauquin G, Brandolin G, Pebay-Peyroula E. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 2005;579:6031–6036. doi: 10.1016/j.febslet.2005.09.061. [DOI] [PubMed] [Google Scholar]
- [105].Ruprecht JJ, Hellawell AM, Harding M, Crichton PG, McCoy AJ, Kunji ER. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc Natl Acad Sci U S A. 2014;111:E426–E434. doi: 10.1073/pnas.1320692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Olofsson G, Sparr E. Ionization constants pKa of cardiolipin. PloS One. 2013;8:e73040. doi: 10.1371/journal.pone.0073040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Robinson AJ, Overy C, Kunji ER. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc Natl Acad Sci U S A. 2008;105:17766–17771. doi: 10.1073/pnas.0809580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang Y, Tajkhorshid E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc Natl Acad Sci U S A. 2008;105:9598–9603. doi: 10.1073/pnas.0801786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Dehez F, Pebay-Peyroula E, Chipot C. Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J Am Chem Soc. 2008;130:12725–12733. doi: 10.1021/ja8033087. [DOI] [PubMed] [Google Scholar]
- [110].Krammer EM, Ravaud S, Dehez F, Frelet-Barrand A, Pebay-Peyroula E, Chipot C. High-chloride concentrations abolish the binding of adenine nucleotides in the mitochondrial ADP/ATP carrier family. Biophys J. 2009;97:L25–L27. doi: 10.1016/j.bpj.2009.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kunji ER, Robinson AJ. The conserved substrate binding site of mitochondrial carriers. Biochim Biophys Acta. 2006;1757:1237–1248. doi: 10.1016/j.bbabio.2006.03.021. [DOI] [PubMed] [Google Scholar]
- [112].Robinson AJ, Kunji ER. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci U S A. 2006;103:2617–2622. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Echtay KS, Winkler E, Bienengraeber M, Klingenberg M. Site-directed mutagenesis identifies residues in uncoupling protein (UCP1) involved in three different functions. Biochemistry. 2000;39:3311–3317. doi: 10.1021/bi992448m. [DOI] [PubMed] [Google Scholar]
- [114].Hughes DA, Jastroch M, Stoneking M, Klingenspor M. Molecular evolution of UCP1 and the evolutionary history of mammalian non-shivering thermogenesis. BMC Evol Biol. 2009;9:4. doi: 10.1186/1471-2148-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Oelkrug R, Goetze N, Exner C, Lee Y, Ganjam GK, Kutschke M, Muller S, Stohr S, Tschop MH, Crichton PG, Heldmaier G, et al. Brown fat in a protoendothermic mammal fuels eutherian evolution. Nat Commun. 2013;4:2140. doi: 10.1038/ncomms3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Vozza A, Parisi G, De Leonardis F, Lasorsa FM, Castegna A, Amorese D, Marmo R, Calcagnile VM, Palmieri L, Ricquier D, Paradies E, et al. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc Natl Acad Sci U S A. 2014;111:960–965. doi: 10.1073/pnas.1317400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Modriansky M, Murdza-Inglis DL, Patel HV, Freeman KB, Garlid KD. Identification by site-directed mutagenesis of three arginines in uncoupling protein that are essential for nucleotide binding and inhibition. J Biol Chem. 1997;272:24759–24762. doi: 10.1074/jbc.272.40.24759. [DOI] [PubMed] [Google Scholar]
- [118].Divakaruni AS, Humphrey DM, Brand MD. Fatty acids change the conformation of uncoupling protein 1 (UCP1) J Biol Chem. 2012;287:36845–36853. doi: 10.1074/jbc.M112.381780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Azzu V, Parker N, Brand MD. High membrane potential promotes alkenal-induced mitochondrial uncoupling and influences adenine nucleotide translocase conformation. Biochem J. 2008;413:323–332. doi: 10.1042/BJ20080321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Borecky J, Nogueira FT, de Oliveira KA, Maia IG, Vercesi AE, Arruda P. The plant energy-dissipating mitochondrial systems: depicting the genomic structure and the expression profiles of the gene families of uncoupling protein and alternative oxidase in monocots and dicots. J Exp Bot. 2006;57:849–864. doi: 10.1093/jxb/erj070. [DOI] [PubMed] [Google Scholar]
- [121].Palmieri L, Picault N, Arrigoni R, Besin E, Palmieri F, Hodges M. Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem J. 2008;410:621–629. doi: 10.1042/BJ20070867. [DOI] [PubMed] [Google Scholar]
- [122].Winkler E, Wachter E, Klingenberg M. Identification of the pH sensor for nucleotide binding in the uncoupling protein from brown adipose tissue. Biochemistry. 1997;36:148–155. doi: 10.1021/bi962178x. [DOI] [PubMed] [Google Scholar]
- [123].Echtay KS, Bienengraeber M, Klingenberg M. Mutagenesis of the uncoupling protein of brown adipose tissue. Neutralization of E190 largely abolishes pH control of nucleotide binding. Biochemistry. 1997;36:8253–8260. doi: 10.1021/bi970513r. [DOI] [PubMed] [Google Scholar]
- [124].Jezek P, Jaburek M, Garlid KD. Channel character of uncoupling protein-mediated transport. FEBS Lett. 2010;584:2135–2141. doi: 10.1016/j.febslet.2010.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Tonazzi A, Console L, Giangregorio N, Indiveri C, Palmieri F. Identification by site-directed mutagenesis of a hydrophobic binding site of the mitochondrial carnitine/acylcarnitine carrier involved in the interaction with acyl groups. Biochim Biophys Acta. 2012;1817:697–704. doi: 10.1016/j.bbabio.2012.02.007. [DOI] [PubMed] [Google Scholar]
- [126].Erdelt H, Weidemann MJ, Buchholz M, Klingenberg M. Some principle effects of bongkrekic acid on the binding of adenine nucleotides to mitochondrial membranes. Eur J Biochem. 1972;30:107–122. doi: 10.1111/j.1432-1033.1972.tb02077.x. [DOI] [PubMed] [Google Scholar]
- [127].Klingenberg M, Buchholz M. On the mechanism of bongkrekate effect on the mitochondrial adenine-nucleotide carrier as studied through the binding of ADP. Eur J Biochem. 1973;38:346–358. doi: 10.1111/j.1432-1033.1973.tb03067.x. [DOI] [PubMed] [Google Scholar]
- [128].Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta. 2008;1778:1978–2021. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- [129].Nelson DR, Felix CM, Swanson JM. Highly conserved charge-pair networks in the mitochondrial carrier family. J Mol Biol. 1998;277:285–308. doi: 10.1006/jmbi.1997.1594. [DOI] [PubMed] [Google Scholar]
- [130].Cappello AR, Curcio R, Valeria Miniero D, Stipani I, Robinson AJ, Kunji ER, Palmieri F. Functional and structural role of amino acid residues in the even-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J Mol Biol. 2006;363:51–62. doi: 10.1016/j.jmb.2006.08.041. [DOI] [PubMed] [Google Scholar]
- [131].Kunji ERS. Structural and mechanistic aspects of mitochondrial transport proteins, in: Comprehensive Biophysics. Elsevier. 2012:174–205. [Google Scholar]