Abstract
Joubert syndrome and related disorders (JSRD) are a heterogeneous group of ciliopathies defined based on the mid-hindbrain abnormalities that result in the characteristic “molar tooth sign” on brain imaging. The core clinical findings of JSRD are hypotonia, developmental delay, abnormal eye movements and breathing abnormalities. To date, more than 30 JSRD genes that encode proteins important for structure and/or function of cilia have been identified. Here, we present 2 siblings with Joubert syndrome associated with growth hormone deficiency. Whole exome sequencing of the family identified compound heterozygous mutations in KIAA0753, i.e., a missense mutation (p.Arg257Gly) and an intronic mutation (c.2359-1G>C). The intronic mutation alters normal splicing by activating a cryptic acceptor splice site in exon 16. The novel acceptor site skips nine nucleotides, deleting three amino acids from the protein coding frame. KIAA0753 (OFIP) is a centrosome and pericentriolar satellite protein, previously not known to cause Joubert syndrome. We present comprehensive clinical descriptions of the Joubert syndrome patients as well as the cellular phenotype of defective ciliogenesis in the patients’ fibroblasts.
Introduction
Joubert syndrome and related disorders (JSRD) are a clinically and genetically heterogeneous group of ciliopathies defined based on the characteristic constellation of cerebellar abnormalities that result in the diagnostic “molar tooth sign” on axial brain images (Parisi 2009; Romani et al. 2013). Core clinical findings of JSRD are hypotonia, developmental delay, abnormal eye movements and breathing abnormalities. Variable features include retinal dystrophy, ocular colobomas, fibrocystic renal disease and congenital hepatic fibrosis (Gunay-Aygun 2009; Parisi 2009). Although more than 30 JSRD genes have been identified to date (Romani et al. 2013), these genes do not account for all JSRD patients suggesting further genetic heterogeneity (Bachmann-Gagescu et al. 2015).
Ciliogenesis and maintenance of cilia depend on the interaction of the various components of molecular complexes (Bhogaraju et al. 2013; Pedersen and Christensen 2012; Pedersen and Rosenbaum 2008; Wei et al. 2012), including the most recently identified OFIP-OFD1-FOR20 complex (Chevrier et al. 2015). OFIP, also known as KIAA0753, or Moonraker, has been implicated in centriolar duplication (Firat-Karalar et al. 2014; Jakobsen et al. 2011) through a cascade mechanism, whereby KIAA0753 recruits WDR62 to the pericentriolar complex, which in turn recruits CEP63 (Kodani et al. 2015). This ultimately leads to increased intracellular Cyclin Dependent Kinase 2, which promotes centriole duplication in S-phase as required. The centriole–basal body plays an essential role in ciliogenesis. After the centriole turns into a basal body, a new structure called the ‘transition zone” (TZ) forms at its distal end (Czarnecki and Shah 2012). The TZ consists of Y-linkers that densely cross-link the doublet microtubules to the surrounding ciliary membrane; it functions as a “gatekeeper” that regulates entry into the ciliary compartment. Most genes that cause Joubert, Bardet–Biedl or Meckel–Gruber syndromes encode proteins that localize to the basal body or transition zone, underscoring the importance of these structures in ciliary assembly and function (Czarnecki and Shah 2012).
Until the recent report by Chevrier et al., describing mutations in KIAA0753 in a single patient with oral–facial–digital syndrome type VI, no human disease was known to be caused by mutations in this gene (Chevrier et al. 2015). Here, we present 2 siblings with Joubert syndrome associated with growth hormone deficiency but no oral or digital anomalies, in whom we identified biallelic mutations in KIAA0753. We also demonstrate that primary fibroblasts from patients with mutations in KIAA0753 have defects in ciliogenesis.
Materials and methods
Clinical evaluation
The patients and their families were evaluated at the NIH Clinical Center under the clinical protocol “Clinical and Molecular Investigations into Ciliopathies” (www.clinical-trials.gov, trial NCT00068224) approved by the National Human Genome Research Institute (NHGRI) Institutional Review Board. Patients and/or their parents gave written, informed consent for this study. Evaluations at the NIH Clinical center included family history, physical examination, formal neurocognitive evaluation, complete eye examination, magnetic resonance and ultrasonography imaging and comprehensive blood and urine testing including kidney and liver function tests as well as hormone levels.
Whole exome sequencing
Genomic DNA was obtained from leukocytes using standard protocols. For exome sequencing we used the HiSeq 2000 (Illumina, San Diego, CA, USA) (Bentley et al. 2008) that employed 101-bp paired-end read sequencing. Image analysis and base calling were performed using Illumina Genome Analyzer Pipeline software (versions 1.13.48.0) with default parameters. Reads were aligned to a human reference sequence (UCSC assembly hg19, NCBI build 37) using a package called Efficient Large-scale Alignment of Nucleotide Databases (Illumina, San Diego, CA, USA). Genotypes were called at all positions where there were high-quality sequence bases using a Bayesian algorithm called the Most Probable Genotype (Teer and Mullikin 2010) and variants were filtered using the graphical software tool VarSifter v1.5 (Teer et al. 2012). The novel variants identified in this study have been submitted to public database LOVD and now available online (http://databases.lovd.nl/shared/variants/KIAA0753/unique).
Missense variant analysis
The effect of missense variations on protein function was evaluated using a variety of pathogenicity prediction programs, including MutationTaster (Schwarz et al. 2014) (http://www.mutationtaster.org/index.html), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2//; Polymorphism Phenotyping v2 (Adzhubei et al. 2010), and SIFT (http://sift.jcvi.org/; Sorting Intolerant From Tolerant (Ng and Henikoff 2001).
RT-PCR, TA cloning and Sanger sequencing
Genomic DNA was extracted from peripheral blood leukocytes using standard protocols. RNA was extracted from control and patient fibroblasts using the Trizol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). RNA was treated with the DNA-free DNase (Applied Biosystems, Austin, TX, USA). RNA concentrations and purity were measured on the Nanodrop ND-1000 instrument (Nanodrop Technologies, Wilmington, DE, USA). First strand cDNA was synthesized using a high-capacity RNA-to-cDNA kit (Applied Biosystems, Austin, TX, USA) according to the manufacturer’s guidelines. For dideoxy sequencing of gDNA, primers were designed to cover specific regions of KIAA0753, and for cDNA sequencing primers were designed to amplify overlapping cDNA fragment of exon 16 (primer sequences available upon request). Direct sequencing of the PCR amplification products was carried out using BigDye 3.1 Terminator chemistry (Applied Biosystems, Austin, TX, USA) and separated on an ABI 3130xl genetic analyzer (Applied Biosystems Austin, TX, USA). Data were evaluated using Sequencher v5.0 software (Gene Codes Corporation, Ann Arbor, MI, USA). For TA cloning, PCR products were cloned in TOPO vector according to manufacturer’s instructions. Random colonies were picked, and the desired region was sequenced as mentioned above.
Fibroblast culture and immunoblotting
Patients’ primary fibroblasts were cultured from a forearm skin biopsy. Control fibroblasts were purchased from ATCC (Manassas, VA, USA). Fibroblasts were cultured in high-glucose (4.5 g/L) DMEM medium supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products, West Sacramento, CA, USA), 2 mM L-glutamine, MEM nonessential amino acid solution and penicillin–streptomycin. Cells were grown to confluency in 75-cm2 flasks, washed twice with ice-cold PBS and scraped into 250 μL of cell lysis buffer containing 50 mM Tris, pH 7; 150 mM NaCl; 0.1% SDS; 0.5% sodium deoxycholate; 1% Triton x-100; and 1 mM EDTA and Complete Mini Protease Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN, USA). Cell lysates were centrifuged (21,000g, for 30 min at 4 °C); and obtained the supernatants for immunoblotting experiments. The concentration of protein was determined by the DCA Protein assay (BioRad, Hercules, CA, USA). Twenty micrograms of total protein samples were loaded onto 4–12% Tris–Glycine gels. Proteins were blotted onto nitrocellulose membranes (Invitrogen, Carlsbad, CA, USA) and blocked for 1 hour in blocking buffer (Li-Cor). After blocking, membranes were probed with rabbit polyclonal against KIAA0753 (Abcam) and loading was controlled by blotting the same membranes with beta actin (mouse anti beta actin, Sigma). IRDye 680RD or 800CW conjugated secondary anti-mouse, anti-rabbit antibodies were used (Li-Cor Biosciences, Lincoln, NE). The antigen–antibody complexes were visualized with a Li-Cor Odyssey Infrared imaging system.
Immunofluorescence microscopy and cilia length measurement
For cilia measurement, cells were grown in chamber slides until 70% confluent, serum-starved for 48 h and fixed using ice-cold methanol for 10 min. After three washes in PBS, cells were blocked with 2% donkey serum and 2% BSA in PBS, and incubated overnight at 4 °C in ARL13B antibody (Proteintech Group, Chicago, IL, USA) and monoclonal anti-gamma tubulin antibody produced in mouse (clone GTU-88, Sigma). After washing three times with PBS, samples were incubated in Donkey anti-mouse IgG (H+L) Alexa flour 555 and donkey anti-rabbit IgG (H+L) Alex flour 488 secondary antibodies (Invitrogen, Walthan, MA, USA), washed and imaged using Zeiss LSM700 confocal laser scanning microscope. Optical sections were collected from the xy plane and a total of ~200 cells (3 replicates) were analyzed per cell line. Nonparametric t test was used to compare control and patient cells.
Results
KIAA0753 deficient patients exhibit core clinical features of Joubert syndrome
Two siblings, born to non-consanguineous parents of Welsh-Croatian and German background, were prospectively evaluated at the National Institutes of Health (NIH) Clinical Center. Patient 1, a male, was seen at ages 7 and 10.5 years and patient 2, a female, at ages 22 months and 5.5 years. Patient 2 was diagnosed with Joubert syndrome at age 10 months; her older brother (patient 1) was diagnosed at age 6 years after the diagnosis of his sister.
Patient 1
The boy (Fig. 1a–c) had prenatal ultrasonography at 18 weeks of gestation, showing a hypoplastic cerebellar vermis; fetal MRI at 20 weeks confirmed inferior vermis hypoplasia. Birth was full term; weight (3870 g) and length (49.5 cm) were normal. The infant had no breathing abnormalities or feeding difficulty. Hypotonia and global developmental delay were noted at 10 months; he sat unsupported at 9 months, crawled at 18 months, walked independently at 26 months, and spoke his first words at 18 months. He received physical therapy until age 4 years and continued to receive speech therapy at age 10.5 years. At 4 years, an endocrine evaluation for short stature showed low insulin like growth factor-1 (IGF-1) at 22.4 ng/mL (normal 44–117). However, growth hormone stimulation and ACTH stimulation tests were normal. A repeat IGF-1 at age 6 was 36 (normal 33–276).
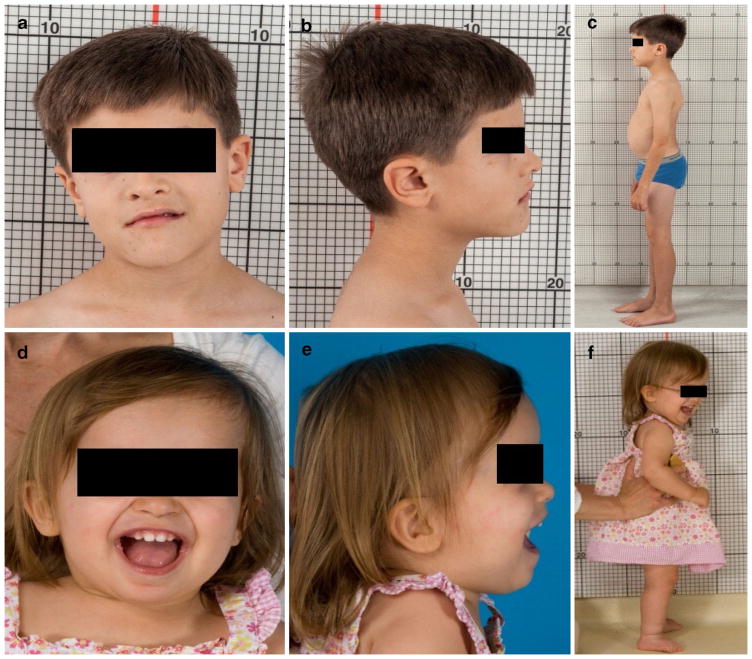
Fig. 1.
Clinical photographs of patients 1 and 2. a–c Patient 1, at age 10.5 years, displayed slight head tilt, bilateral epicanthal folds, downturned corners of the mouth and short stature. d–f Patient 2 at age 22 months had macrocephaly, frontal bossing, epicanthal folds, borderline low-set ears and esotropia
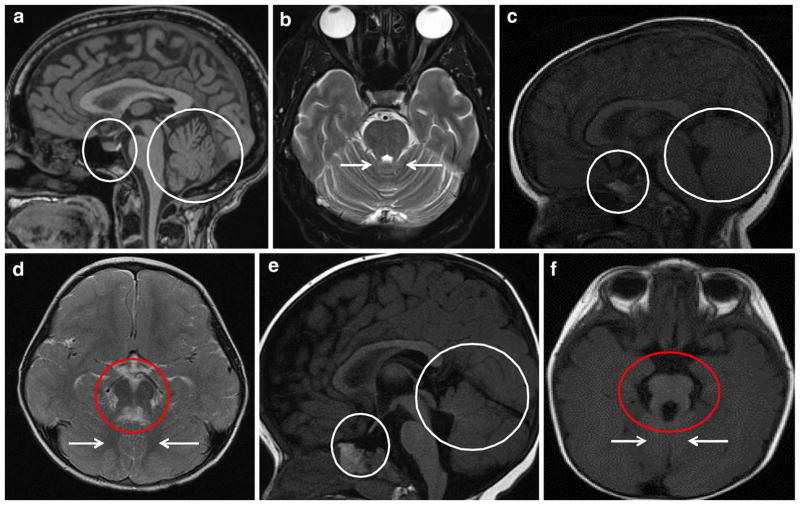
At his first evaluation at the NIH Clinical Center at age 7, height was 108.2 cm (Z score −2.59; <3%; 50th for 5 years of age), weight was 18 kg (3%), and head circumference was 52 cm (50th %). Craniofacial examination was significant for frontal prominence, posteriorly rotated ears, bilateral epicanthal folds and down-turned corners of the mouth (Fig. 1a, b). In terms of vision history, he had no adverse behavior to dim or bright lights and no problems seeing the chalkboard at school. At NIH at age 7, his best-corrected visual acuity was 20/25 in the right eye and 20/25 in the left eye without glasses. Ocular motility assessment showed a very mild bilateral oculomotor apraxia. The remainder of the eye exam was normal; the anterior segment exam was unremarkable and retina and optic nerves were healthy. Formal neurocognitive evaluation was performed at both NIH visits at ages 7 and 10.5 years. On the WISC-IV (Wechsler Intelligence Scale for Children—Fourth Edition), his Verbal Comprehension Index score of 95 and his pro-rated Perceptual Reasoning Index of 92 were in the average range. His Processing Speed Index of 75 was at 5th percentile. In summary, his verbal and perceptual reasoning abilities were in the average range. Repeat neurocognitive evaluation at age 10.5 years yielded similar results. At his NIH visit at age 7 years, serum IGF-1 was low at 29 ng/mL (Tanner I reference range for males: 63–279). Endocrine evaluation confirmed growth hormone deficiency, and he responded well to growth hormone therapy. At the 2nd NIH visit at age 10.5 years, his height was much improved at 138.9 cm (Z score −0.3; 40th %), weight 33.8 kg (60th %) and head circumference 56 cm. Brain MRI showed an ectopic posterior pituitary gland (Fig. 2c) in addition to the molar tooth sign (Fig. 2d).
Fig. 2.
Brain MRI images of patients 1 and 2 in comparison to normal. a Sagittal T1-weighted and b axial T2-weighted brain MRI images of healthy controls showing normal appearance of pituitary gland (small circle) and cerebellar vermis (large circle), and superior cerebellar peduncles (arrows). c–f Both patients had cerebellar vermis hypoplasia (large circles), dysplastic vermis (arrows) and thickened and horizontally oriented superior cerebellar peduncles resulting in the appearance of “molar tooth sign” on axial MRI images (red circles). c Patient 1 had an ectopic posterior pituitary gland (small circle). c Patient 2’s pituitary stalk appeared to be absent and the pituitary gland itself was very small (small circle). There was a suggestion of a pituitary bright spot immediately posterior to the chiasm, suggesting an ectopic posterior pituitary
Patient 2
The younger sister of patient 1 (Fig. 1d–f) was born at term after an unremarkable pregnancy. Birth weight, length and head circumference were normal at 3520 g, 51.5 and 35.5 cm, respectively. Fast breathing was noted at birth. At 2 months, she did not track faces, smile, or make eye contact. She had hyperopia and esotropia. Brain MRI revealed the “molar tooth sign” (Fig. 2f), and she had global developmental delay. She began sitting at 9 months. At 22 months, she was pulling to stand but not crawling or cruising; first words were at 20 months. At the initial NIH exam at age 22 months, weight was 9.8 kg (<3%; 50% for 13 months), length was 75.5 cm (Z score −2.6; < 3%; 50% for 14 months) and head circumference was 49 cm (90–95%). The anterior fontanel was still open, measuring 4 × 5 cm. There was frontal bossing (Fig. 1d–f), bilateral epicanthal folds, and posteriorly rotated and borderline low-set ears. Serum IGF-1 was low (<25 ng/mL), and growth hormone levels were undetectable with a low peak stimulated growth hormone of 6.3 ng/mL, consistent with growth hormone deficiency. Peak stimulated cortisol level was normal at 56.9 mcg/dL. Thyroid axis testing did not suggest central hypothyroidism but showed mild TSH elevation. Brain MRI, performed at age 7 months and reevaluated at NIH, showed an absent pituitary stalk and a small pituitary gland (Fig. 2e). There was a pituitary bright spot immediately posterior to the chiasm, suggesting an ectopic posterior pituitary. She responded to GH and thyroid hormone treatment with a growth velocity of 13.3 cm in 1 year. At the second NIH visit at age 5 years, after growth hormone therapy, her height was much improved at 110.5 cm (Z score 0; 50th %), weight was 20.7 kg (60th %), and head circumference was 55 cm (>98th %). She had no adverse behavior in dim-lit rooms, and no sensitivity to bright light. At 1 year of age, she was prescribed glasses to help her strabismus. At the NIH at 22 months, she was able to see the 20/80 grading equivalent with each eye. There was mild oculomotor apraxia, and an inferior oblique overaction of each eye. The remainder of the eye exam was unremarkable with normal anterior segment, retina and optic nerves.
On neurocognitive testing at the second NIH visit at age 5 years, the girl smiled often and made eye contact occasionally, but the psychologist was unable to engage her in structured tasks sufficient to administer cognitive testing. She could write her own name, indicate her age using her fingers, and draw recognizable human figures. Based on Vineland Adaptive Behavior Scales-II structured interview with her mother, the patient’s adaptive behaviors were at the 3rd percentile with an Adaptive Behavior Composite of 71 ± 5. She fed herself with a spoon and a fork, was toilet trained for urine during the day, could put on some of her clothing and take her clothing off. Motor skills included walking up and down stairs but not running. Fine motor skills included drawing recognizable figures and coloring and cutting a straight line. In summary, she was functioning in the borderline range across all domains assessed by the Vineland.
Other tests that showed normal results in both patients included echocardiography, complete abdominal ultrasonography, liver and kidney related blood and urine chemistries.
Identification of mutations in KIAA0753
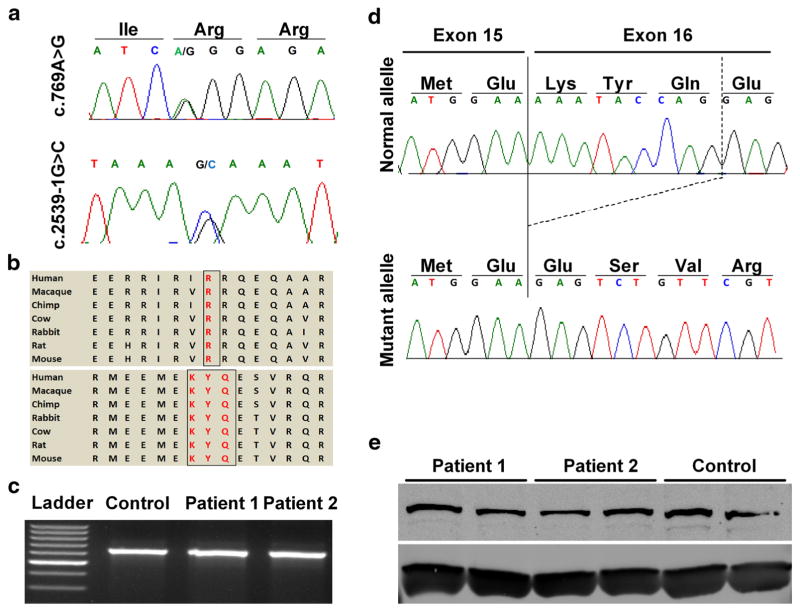
Whole exome sequencing (WES), performed on genomic DNA from both affected individuals and parents, yielded ~48,000 genetic variants in each patient and ~19,000 common to both affected siblings. “Of these 19,000 variants, 652 were either absent or had a minor allele frequency of less than 1% in the online databases including Clinseq (Biesecker et al. 2009) and ExAC (Lek et al. 2016). Using a candidate gene approach, along with filtering using parental variants and a recessive inheritance pattern, we identified biallelic mutations in KIAA0753, segregating with both affected individuals. Dideoxy sequencing confirmed a missense mutation in exon 4 inherited from the mother (NM_014804.2: c.769A>G; p. Arg257Gly), and a splice site mutation preceding exon 16 inherited from the father (NM_014804.2: c.2359-1G>C) (Fig. 3a). Bioinformatic analysis of the Arg257Gly missense predicted the change to be probably damaging, with a PolyPhen-2 (score 0.996), MutationTaster (p value 0.516), and a SIFT (score 0.01). The arginine residue is comparatively bigger than glycine, and its change to glycine alters the charge and proper folding of the protein. The ExAC whole exome database showed no occurrence of either the missense variant or the splice site mutation. Both mutations affect amino acids that are highly conserved across different species (Fig. 3b).
Fig. 3.
Molecular analysis of the patients’ mutations in KIAA0753. a Sequence chromatograms representing the c.769A>G missense mutation and the c.2359-1G>C splice site mutation. b Conservation of protein sequence across different species, encompassing both missense mutation (upper panel) and in-frame deletion (lower panel). c Agarose gel images of PCR products using cDNA specific primers flanking exon 16 showing absence of exon skipping/large deletion in both the patients, compared to wild type. d TA cloning of the PCR products followed by sequencing showing deletion of nine nucleotides encoding 3 amino acids (KYQ), at the beginning of exon 16. e Western blotting, performed in duplicate, for control and both patients showing no significant difference in the protein expression level of KIAA073. Housekeeping protein beta actin (ACTB) was used a loading control
cDNA and protein expression analysis of KIAA0753 in patients’ fibroblasts
In order to further delineate the effects of the canonical splice site mutation (c.2359-1G>C), we PCR amplified cDNAs using primers spanning exon 13 to exon 17. The product (650 bp) showed no discernible size difference between control and patient (Fig. 3c). Cloning of this fragment into the TOPO vector showed colonies containing reference sequence and colonies with 9 base pair deletion, resulting in an in-frame 3 amino acid (p.Lys787_ Gln789del) deletion at the protein level (Fig. 3d). Western blot analysis revealed no significant differences in the expression of full length KIAA0753 (main isoform; NP_055619.2) (Fig. 3e) in either patient, compared to control.
Measurement of cilia in patients’ fibroblasts
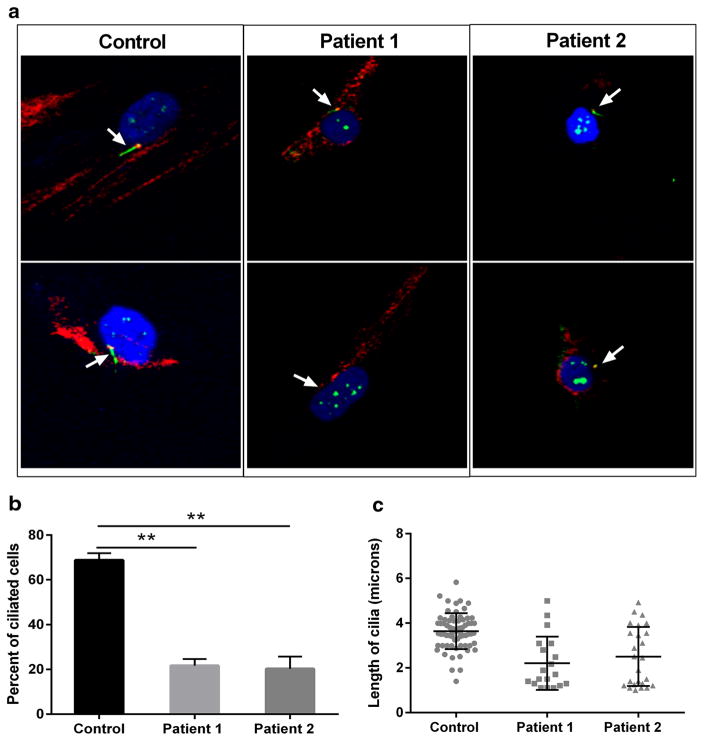
Since KIAA053 is considered to have function in the formation of cilia, we examined cilia formation in primary fibroblasts from both patients and control (Fig. 4a). After 48 h of serum starvation, the percentage of ciliated cells, was significantly lower (~20–30%) in both Patient 1 and Patient 2 when compared to control (Fig. 4b). Control cells had cilia measuring from 2.6 to 5.1 μm, which was not statistically different from the cilia length for Patient 1 (2.17–4.99) or Patient 2 (2.55–4.2) (Fig. 4c).
Fig. 4.
Mutations in KIAA0753 cause defects in ciliogenesis. Fibro-blasts were grown to ~70% confluence, starved for 48 h, methanol fixed. Cilia were stained with ARL13B (green) and centrosome with gamma tubulin (red). Representative cilia images of cells from control, Patient 1, and Patient 2. Z-stacked images were analyzed for percentage of cells with cilia. a Staining identified both reduced length ciliated cells (upper panel) and non-ciliated cells (lower panel) in both patients. b Graph representing the percentage of ciliated cells in patients, comparison to control. c Dot plot demonstrating the length of cilia (in micrometer) in both patients compared to control. A total of ~200 cells (3 replicates) were analyzed for cilia measurement. Error bars represent standard deviation. **p < 0.05 (nonparametric t test)
Discussion
In this study, we present the first report of Joubert syndrome caused by mutations in KIAA0753. The two children presented here have mid-hindbrain abnormalities that result in the “molar tooth” sign on brain MRI characteristic of Joubert syndrome (Parisi 2009; Romani et al. 2013). The clinical findings in our patients including hypotonia, developmental delay, abnormal eye movements, and breathing abnormalities are typical features observed in patients with Joubert syndrome associated with other genes. None of the variable clinical features of Joubert Syndrome, namely retinal dystrophy, ocular colobomas, fibrocystic renal disease, and hepatic fibrosis were detected in our patients. However, in addition to the core Joubert features, both of our patients had growth hormone deficiency in association with structural abnormalities of the pituitary gland on brain MRI. Growth hormone deficiency occurs in a small number of patients with ciliopathies, including a family recently reported with KIAA0556 mutations (Parisi and Glass 1993; Sanders et al. 2015), and an IFT172-related disorder (Lucas-Herald et al. 2015), suggesting the potential importance of ciliary function in the development of the pituitary gland. Whether the pituitary defects in our patients were caused by the KIAA0753 defect remains uncertain; identification of other Joubert syndrome patients with defects in KIAA0753 will help to answer this question. Expression data from online databases (http://www.proteinatlas.org; http://www.biogps.org) show ubiquitous expression of this protein in all human tissues including brain, with higher expression in testis, pituitary and parathyroid glands (Uhlen et al. 2015), suggesting its role in the formation and functioning of pituitary gland. Although there are no convincing reports depicting the direct role of KIAA0753 on neurogenesis, pericentriolar material 1 protein (PCM1) that interacts with KIAA0753, has been shown to play a role in the proliferation and differentiation of neuronal progenitor cells (Zhang et al. 2016). The direct role of KIAA073 in neural and pituitary development is yet to be rigorously examined.
Our patients harbor compound heterozygous mutations, a missense variant (c.769A>G; p.Arg257Gly) and a 3 amino acid in-frame deletion (p.Lys787_Gln789del), in highly conserved regions of KIAA0753; these changes are predicted to be pathogenic according to various prediction tools. Our patients’ fibroblasts produced KIAA0753 at normal levels. Neither p.Arg257Gly nor p.Lys787_Gln789del are truncating so significant reduction in mRNA expression is unlikely. The fact that the KIAA0753 protein levels are normal is not surprising; it is likely that these mutations impair the function of the protein, since they are located in conserved domains and are predicted to be deleterious.
KIAA0753 (OFIP) is a centrosome and pericentriolar satellite protein that interacts with FOR20, a centrosomal protein essential for S-phase progression during cell cycle (Shen et al. 2013). KIAA0753 regulates centriole duplication by recruiting WDR62, CEP63 and finally CDK2 to the centrosome. Accumulation of CDK2 at the centrosome promotes duplication of centrioles (Firat-Karalar et al. 2014). The cells lacking functional KIAA0753 is expected to have decreased proliferation rate. However, the fibroblasts derived from our patients, showed fairly normal rate of cell division on tissue culture, compared to control cells which might be due to the mild mutations in our patients. In addition to its role in centriole duplication, KIAA0753 interacts with OFD1 with the help of FOR20, and this ternary complex is required for microtubule stability and formation of cilia. The C-terminal region of OFIP (300–967 and 667–967 residues) interacts with OFD1 and their interaction is strongly favored in the presence of FOR20 (Chevrier et al. 2015). FOR20 helps to stabilize the interaction of OFIP with OFD1. This triple protein complex (OFIP, OFD1 and FOR20) is later recruited to pericentriolar satellites through binding of the OFIP N-terminus (1–299 amino acids) to the pericentriolar material 1 protein (PCM1) that resides in centrosome (Chevrier et al. 2015). Our missense mutation (p.Arg257Gly) lies in this N terminal region, which is essential for the interaction of OFIP with PCM1. The second mutation (p.Lys787_Gln789del) lies in the C-terminal region of OFIP the part of OFIP that interacts with OFD1 (Chevrier et al. 2015). The location of our mutations in these two conserved functional domains suggests they are pathogenic. In addition, siRNA mediated depletion of the OFIP, OFD1 and FOR20 complex resulted in a decrease in the length of primary cilia, but not by a significant amount (15–20% decrease) (Chevrier et al. 2015). We found similar decrease in cilia length, although not significant, along with significantly fewer ciliated cells.
Chevrier et al. (Chevrier et al. 2015) recently reported a patient with oral-facial-digital syndrome type VI who was compound heterozygous for a nonsense (c.1891A>T; p.Lys631*) and a splice site variant (c.1546-3C>A) in KIA00753. They showed that the protein product of this gene (which they referred to as “OFIP”), associates with centrosome/centriole and pericentriolar satellites in human cells and forms a complex with FOR20 and OFD1. Our patients had a relatively mild ciliopathy phenotype, consistent with our relatively mild KIAA0753 mutations in comparison to the null mutations of the OFD type VI patient. Therefore, KIAA0753 represents another example of the same ciliopathy gene causing different clinical syndromes depending on the severity of the mutation. Genetic overlap among ciliopathies is reported in the literature, especially the Joubert, Meckel-Gruber, Bardet-Biedl and oral-facial-digital syndromes (Czarnecki and Shah 2012). For example, mutations in TMEM67 cause Meckel-Gruber syndrome, as well as Joubert syndrome (Parisi and Glass 1993). Therefore, it is not surprising that this is also the case for KIAA0753.
In conclusion, certain mutations in KIAA0753 cause impaired ciliogenesis and result in Joubert syndrome. Further studies on animal models are needed to understand the cellular differences between the KIAA0753-related oral-facial-digital and Joubert syndromes.
Acknowledgments
The authors thank the Joubert Syndrome and Related Disorders Foundation for their extensive support and the patients and their families who generously participated in this investigation. This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, USA.
Funding This work was supported by the Intramural Research Program at the National Human Genome Research Institute, National Institutes of Health.
Footnotes
Compliance with ethical standards
Conflict of interest All authors declare no conflict of interest.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann-Gagescu R, Dempsey JC, Phelps IG, O’Roak BJ, Knutzen DM, Rue TC, Ishak GE, Isabella CR, Gorden N, Adkins J, Boyle EA, de Lacy N, O’Day D, Alswaid A, Ramadevi AR, Lingappa L, Lourenco C, Martorell L, Garcia-Cazorla A, Ozyurek H, Haliloglu G, Tuysuz B, Topcu M, Chance P, Parisi MA, Glass IA, Shendure J, Doherty D. Joubert syndrome: a model for untangling recessive disorders with extreme genetic heterogeneity. J Med Genet. 2015;52:514–522. doi: 10.1136/jmedgenet-2015-103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, Lorentzen E. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, Bouffard GG, Chines PS, Cruz P, Hansen NF, Teer JK, Maskeri B, Young AC, Program NCS, Manolio TA, Wilson AF, Finkel T, Hwang P, Arai A, Remaley AT, Sachdev V, Shamburek R, Cannon RO, Green ED. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier V, Bruel AL, van Dam TJ, Franco B, Scalzo ML, Lembo F, Audebert S, Baudelet E, Isnardon D, Bole A, Borg JP, Kuentz P, Thevenon J, Burglen L, Faivre L, Riviere JB, Huynen MA, Birnbaum D, Rosnet O, Thauvin-Robinet C. OFIP/KIAA0753 forms a complex with OFD1 and FOR20 at pericentriolar satellites and centrosomes and is mutated in one individual with Oral-Facial-Digital Syndrome. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv488. [DOI] [PubMed] [Google Scholar]
- Czarnecki PG, Shah JV. The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 2012;22:201–210. doi: 10.1016/j.tcb.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates JR, 3rd, Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol. 2014;24:664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunay-Aygun M. Liver and kidney disease in ciliopathies. Am J Med Genet C Semin Med Genet. 2009;151C:296–306. doi: 10.1002/ajmg.c.30225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, Uhlen M, Hyman AA, Andersen JS. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Yu TW, Johnson JR, Jayaraman D, Johnson TL, Al-Gazali L, Sztriha L, Partlow JN, Kim H, Krup AL, Dammermann A, Krogan NJ, Walsh CA, Reiter JF. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. Elife. 2015 doi: 10.7554/eLife07519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Herald AK, Kinning E, Iida A, Wang Z, Miyake N, Ikegawa S, McNeilly J, Ahmed SF. A case of functional growth hormone deficiency and early growth retardation in a child with IFT172 mutations. J Clin Endocrinol Metab. 2015;100:1221–1224. doi: 10.1210/jc.2014-3852. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi MA. Clinical and molecular features of Joubert syndrome and related disorders. Am J Med Genet C Semin Med Genet. 2009;151C:326–340. doi: 10.1002/ajmg.c.30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Glass I. Joubert syndrome and related disorders. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews®. University of Washington; Seattle, WA: 1993. [PubMed] [Google Scholar]
- Pedersen LB, Christensen ST. Regulating intraflagellar transport. Nat Cell Biol. 2012;14:904–906. doi: 10.1038/ncb2569. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (Ift): role in ciliary assembly, resorption and signalling. Ciliary Funct Mamm Dev. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Romani M, Micalizzi A, Valente EM. Joubert syndrome: congenital cerebellar ataxia with the molar tooth. Lancet Neurol. 2013;12:894–905. doi: 10.1016/S1474-4422(13)70136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AA, de Vrieze E, Alazami AM, Alzahrani F, Malarkey EB, Sorusch N, Tebbe L, Kuhns S, van Dam TJ, Alhashem A, Tabarki B, Lu Q, Lambacher NJ, Kennedy JE, Bowie RV, Hetterschijt L, van Beersum S, van Reeuwijk J, Boldt K, Kremer H, Kesterson RA, Monies D, Abouelhoda M, Roepman R, Huynen MH, Ueffing M, Russell RB, Wolfrum U, Yoder BK, van Wijk E, Alkuraya FS, Blacque OE. KIAA0556 is a novel ciliary basal body component mutated in Joubert syndrome. Genome Biol. 2015;16:293. doi: 10.1186/s13059-015-0858-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. Mutation-Taster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Shen M, Cai Y, Yang Y, Yan X, Liu X, Zhou T. Centrosomal protein FOR20 is essential for S-phase progression by recruiting Plk1 to centrosomes. Cell Res. 2013;23:1284–1295. doi: 10.1038/cr.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet. 2010;19:R145–R151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exomescale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Wei Q, Zhang YX, Li YJ, Zhang Q, Ling K, Hu JH. The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol. 2012;14:950. doi: 10.1038/ncb2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kim PJ, Chen Z, Lokman H, Qiu L, Zhang K, Rozen SG, Tan EK, Je HS, Zeng L. MiRNA-128 regulates the proliferation and neurogenesis of neural precursors by targeting PCM1 in the developing cortex. Elife. 2016 doi: 10.7554/eLife11324. [DOI] [PMC free article] [PubMed] [Google Scholar]