Abstract
Resolution and formation of facultative heterochromatin is essential to development, reprogramming, and oncogenesis. The mechanisms underlying these changes are poorly understood due to the inability to study heterochromatin dynamics and structure in vivo. We devised an in vivo approach to investigate these mechanisms and found that topoisomerase II (TOP2), but not TOP1, synergizes with BAF (mSWI/SNF) ATP-dependent chromatin remodeling complexes genome-wide to resolve facultative heterochromatin to accessible chromatin independent of transcription, indicating that changes in DNA topology through (de-)catenation rather than release of torsional stress through swiveling is necessary for heterochromatin resolution. In turn, TOP2 and BAF cooperate to recruit pluripotency factors, explaining some of the instructive roles of BAF complexes. Unexpectedly, we found that TOP2, also plays a role in the reformation of facultative heterochromatin, suggesting that facultative heterochromatin and accessible chromatin exist at different states of catenation or other topologies, which may be critical to their structures.
Facultative heterochromatin refers to regions of the genome that switch from compacted to accessible states, or vice versa, during development, reprogramming, oncogenesis, or in response to stimuli1. Changes in chromatin state alter gene expression, which is important for determining cell identity. Constitutive heterochromatin refers to regions, such as centromeres, that are silent and inaccessible at nearly all times. Facultative heterochromatin is marked with, and produced in part by, the histone modifications H2Aub and H3K27me3, and the Polycomb complexes PRC1 and PRC2, respectively, that place them1,2. Constitutive heterochromatin is typically marked by broad H3K9me3 domains, although it can also be repressed by Polycomb complexes as in X chromosome inactivation1. H3K9me3 domains can also mark facultative heterochromatin, along with absence of histone acetylation, elevated DNA methylation, and the presence of histone H1. The complexity of facultative heterochromatin has contributed to difficulty preparing it in vitro and thus obtaining a clear structure of compacted chromatin3. The mechanisms of resolution and formation of facultative heterochromatin in vivo are also poorly understood. Therefore, novel techniques are necessary to understand the structure and mechanisms by which facultative heterochromatin is resolved and reformed at thousands of loci during development and other cell identity transformations.
To dissect the mechanisms underlying the dynamics of facultative heterochromatin in vivo, we developed a method and mouse called the Chromatin indicator and Assay (CiA) to study the action of chromatin regulators at any haplosufficient locus of any cell type4. In this approach we recruit a chromatin regulator to an endogenous locus with fully native chromatin and thereby avoid the ambiguity associated with recapitulating heterochromatin at exogenous loci or in vitro. We used the Oct4 locus, which undergoes a dramatic transition during development from active and accessible in embryonic stem (ES) cells to highly repressed inaccessible facultative heterochromatin in somatic cells bound by Polycomb complexes5. However, this locus can be reconverted back into an accessible state by induced Pluripotent Stem (iPS) cell reprogramming6. We made use of the previously described Oct4 Chromatin indicator and Assay (CiA:Oct4) locus, in which custom zinc-finger and GAL4 binding sites have been inserted 232 bp upstream of one allele of the endogenous Oct4 promoter for recruitment of chromatin regulators, and eGFP has been inserted into the first exon to monitor expression4 (Fig. 1a). The CiA-modified and unmodified Oct4 alleles have identical expression patterns and histone modifications, including markers of activity in ES cells (H3K4me3 and H3K27ac) and markers of facultative heterochromatin in fibroblasts (H2Aub, H3K27me3, and H3K9me3), indicating that this locus produces native chromatin in its full complexity4,5. We then recruit chromatin regulators by over-expressing the 97 amino-acid FRB domain of mTOR fused to a subunit or domain of a regulatory complex or protein, along with FKBP fused to a zinc-finger (ZnF-FKBP) that binds the custom zinc-finger binding sites. Addition of the Chemical Inducer of Proximity (CIP) rapamycin induces FRB/FKBP dimerization and thus rapidly recruits the FRB-fused chromatin regulator to the locus proportionate to the concentration of rapamycin4.
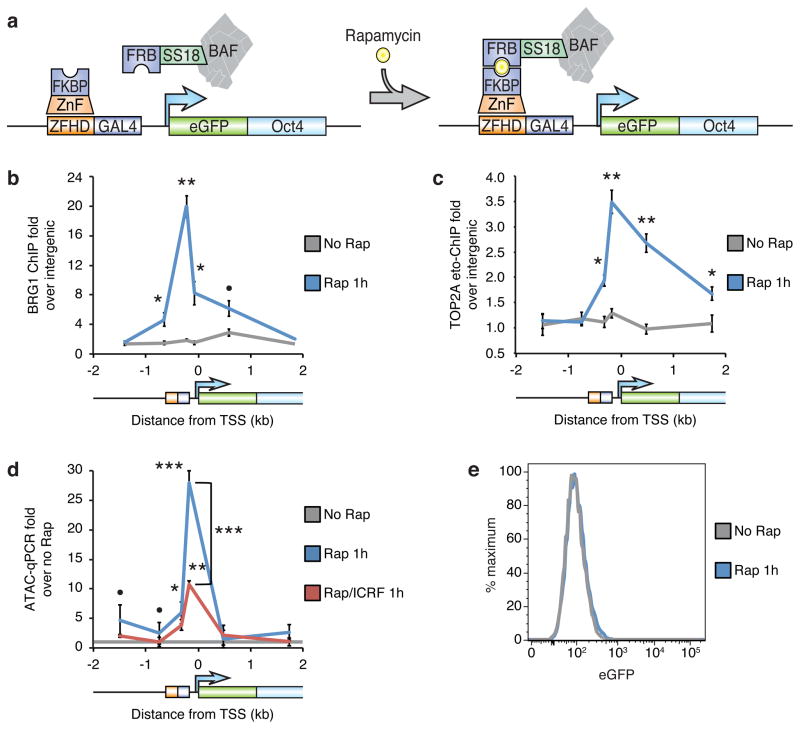
Figure 1. TOP2 is required for transcription-independent BAF-mediated resolution of facultative heterochromatin to accessible chromatin.
(a) Strategy for BAF recruitment to the CiA:Oct4 locus in fibroblasts. The recruitment site is −638 to −232 bp from the TSS. The two primer sets upstream of the recruitment sites target both the CiA:Oct4 and unmodified Oct4 alleles, whereas the other primer sets target only the CiA:Oct4 locus. BRG1 ChIP (b) or TOP2A etoposide-ChIP (c) in fibroblasts with the BAF recruitment system treated with 3 nM rapamycin (Rap) for 1 hour. (d) ATAC-qPCR in cells treated with rapamycin and 1 μM ICRF-193 for 1 hour to inhibit TOP2. (e) eGFP flow cytometry of cells treated with rapamycin for 1 hour. Significance assessed by two-tailed t-tests versus no rapamycin control or as specified: n.s: p ≥ 0.1, ●: p < 0.1, *: p < 0.05, **: p < 0.01, ***: p < 0.001. Lines represent means and error bars represent s.e.m. from 4 (b,d) or 6 (c) cell passages.
To study the resolution of facultative heterochromatin, we recruited BAF (mSWI/SNF) ATP-dependent chromatin remodeling complexes to the heterochromatinized CiA:Oct4 locus in mouse fibroblasts. BAF complexes natively bind the Oct4 locus in ES cells but not in differentiated cells5,7. We believed we could investigate heterochromatin dynamics through BAF recruitment because these complexes create accessible chromatin8–10, oppose Polycomb complexes5,10–13, and play essential roles in both the maintenance of cell identity and transitions that require resolution and formation of heterochromatin14. For example, BAF complexes in ES cells, known as esBAF, are essential for maintaining ES cell identity7,10,15,16 and for reprogramming somatic cells into iPS cells17,18. Alternate BAF assemblies, such as nBAF in post-mitotic neurons, are instructive for neurodevelopment and neuronal reprogramming19,20. Further, exome sequencing studies have revealed that BAF subunit mutations contribute to over ~20% of cancers21–24, largely as tumor suppressors, but also sometimes as oncogenes11. Several human neurologic disorders, including autism, are also likely caused by BAF mutations25,26. It is not fully understood how the chromatin remodeling activities of BAF complexes translate to its instructive roles in cell identity, although there is some evidence showing that it may involve assisting fate-determining transcription factors to bind chromatin8,10,27,28.
BAF complexes also promote decatenation of sister chromatids by recruiting topoisomerase IIα (TOP2A) to chromatin29. This function is essential for cell cycling and genome stability, and likely contributes to the resistance of BAF mutant cancers to topoisomerase II (TOP2) inhibitors30. There are two major classes of metazoan topoisomerases. The type II topoisomerases TOP2A and TOP2B alter the catenation state of DNA (TOP2 can both relieve and introduce catenation31) by creating a double-strand break in one duplex, passing a second duplex through the break, and then re-ligating the break32. In contrast, the type I topoisomerase TOP1 modulates DNA supercoiling by creating a single-strand break, swiveling one side of the double helix, before re-ligating the break. Both classes relax supercoiling in a non-redundant fashion33–36, enhance transcription and maintain accessibility of active long genes by relieving tangles produced by RNA polymerase37–40, and activate poised enhancers solely by breaking DNA41,42. However, it is unknown whether TOP2 or TOP1 are involved in chromatin remodeling or resolution of facultative heterochromatin prior to transcriptional activation, or whether they are relevant to the instructive roles of BAF complexes in cell identity.
Here, we found that recruitment of BAF complexes to facultative heterochromatin causes rapid resolution to accessibility, and that this process requires TOP2 (de-)catenation activity in its initial stages. Further, utilizing conditional knockouts and genome-wide studies, we found that TOP2 synergizes with BAF complexes to resolve facultative heterochromatin at a large swath of regulatory elements, although TOP2 does not appear to be involved in maintenance of accessible chromatin. Synergy between TOP2 and BAF is in turn required for proper targeting of pioneer transcription factors important for fate-determination. To our surprise, by reversing BAF recruitment through rapamycin washout, we found that TOP2 is also required for reformation of facultative heterochromatin. Taken together, these data suggest that facultative heterochromatin and accessible chromatin exist at different catenation or topological states, as TOP2 is not required for the maintenance of either chromatin state but is required for transitions between them in both directions.
RESULTS
TOP2 activity is required for the initial stage of BAF-mediated resolution of facultative heterochromatin
To study the resolution of facultative heterochromatin in vivo, we recruited BAF complexes to the CiA:Oct4 locus in fibroblasts by expressing a SS18-FRB fusion, as SS18 is among the most tightly held BAF subunits21, along with ZnF-FKBP. Addition of 3 nM rapamycin results in robust recruitment of BAF complexes within 1 hour (Fig. 1a,b and Supplementary Fig. 1a,b). This strategy permits fine temporal control over BAF recruitment as well as mimics the transient interactions that chromatin remodelers have with their endogenous chromatin targets due to rapamycin’s high on/off rates5,43–45 (see Discussion for detailed binding kinetics by C.H.).
Consistent with our previous finding that BAF complexes are required to recruit TOP2A to chromatin29,30, recruitment of BAF complexes yielded co-recruitment of active TOP2A (Fig. 1c). Interestingly, TOP2A binding is biased downstream of the recruitment site. Although this is likely in part because the upstream primer sets are not specific to the CiA allele of Oct4 or due to the orientation of the zinc-finger binding sites, this may reflect an intrinsic bias of TOP2A to coding regions. Recruitment of BAF/TOP2A leads to the resolution of inaccessible facultative heterochromatin to accessible chromatin within 1 hour (Fig. 1d). Recruitment of the chromatin remodelers LSH or INO80 do not resolve this locus (Supplementary Fig. 1c,d), indicating this is a specific feature of BAF-mediated chromatin remodeling. However, BAF is not sufficient to re-activate expression of the locus (Fig. 1e), or to co-recruit RNA Polymerase II (Supplementary Fig. 1e), demonstrating that transcription is not required for inducing accessibility. Surprisingly, given the role of TOP2 in relieving tangles produced by transcription37–40, inhibition of TOP2 activity by ICRF-193, a non-covalent pan-TOP2 catalytic inhibitor that blocks both TOP2A and TOP2B46, prevented much of the induction of accessibility (Fig. 1d), despite the lack of transcription and RNA Pol. II (Fig. 1e and Supplementary Fig. 1e), without affecting BAF recruitment (Supplementary Fig. 1f). Further, inhibition of TOP1 with topotecan did not inhibit the development of accessibility (Supplementary Fig. 1g). As the primary role of TOP2 is decatenation and the role of TOP1 is release of supercoiling32, these data indicate that decatenation rather than relief of torsional stress is critical to resolve facultative heterochromatin, although as TOP2 has a role in relieving supercoiling that is non-redundant with TOP133–36, other changes in DNA topology may also be involved.
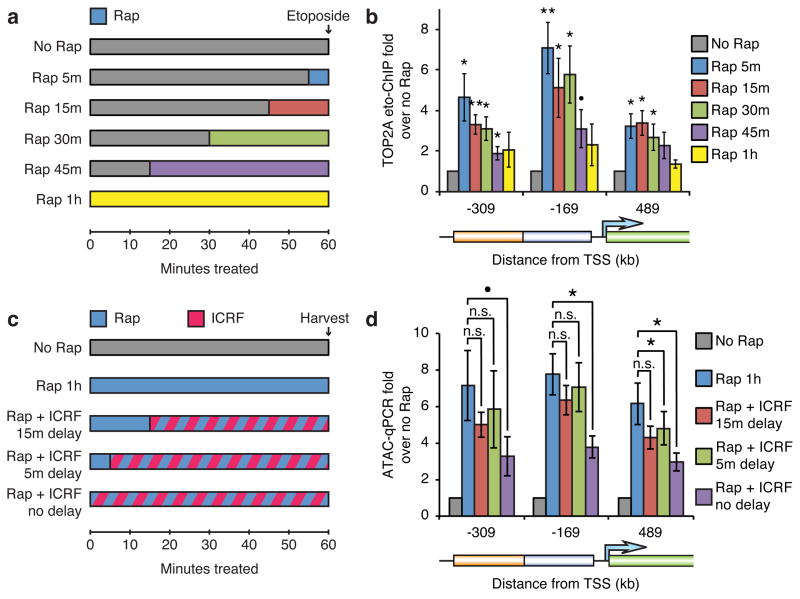
To temporally resolve the role of TOP2 in BAF-mediated resolution of facultative heterochromatin, we probed for decatenation intermediates over a short time-course of accessibility induction (Fig. 2a). We did this by treating cells with etoposide, which crosslinks active TOP2 to chromatin after it creates a double-strand DNA break but before passing the second duplex, and then performing ChIP for TOP2A29,40. Interestingly, maximal TOP2A strand-cleaved intermediates occurred only 5 minutes after initiation of BAF recruitment (Fig. 2b). Fewer intermediates were observed at longer time-points, suggesting that early intermediates quickly resolve as facultative heterochromatin becomes accessible.
Figure 2. Active TOP2 is recruited immediately upon BAF recruitment and is required for the initial stage of accessibility induction.
(a) Strategy for timecourse of active TOP2/strand-cleaved decatenation intermediate detection during BAF recruitment. (b) TOP2A etoposide-ChIP in cells treated with 3 nM rapamycin for various times before fixation with 100 μM etoposide. (c) Strategy for timecourse of TOP2 inhibition during BAF recruitment. (d) ATAC-qPCR in cells treated with rapamycin and 1 μM ICRF-193 after various delays. Significance assessed by t-tests as before. Bars represent means and error bars represent s.e.m. from 6 (b) or 3 (d) cell passages.
To further define the time-point when TOP2 functions, we recruited BAF and then inhibited TOP2 after various delays (Fig. 2c). Consistent with the detection of strand-cleaved intermediates within 5 minutes, a 5 minute delay in ICRF-193 treatment after BAF recruitment largely eliminated the effect of TOP2 inhibition in suppressing maximal accessibility induction, indicating that TOP2 is important for resolving heterochromatin but not maintaining accessibility (Fig. 2d). As TOP2 executes its function very early in the process of heterochromatin resolution, it is unlikely that its role is to relive tangles created by BAF-mediated remodeling. As BAF recruitment leads to neither RNA Pol. II binding (Supplementary Fig. 1e) nor detectable expression (Fig. 1e), it is also unlikely that the role of TOP2 is relief of tangles generated by transcription. Rather, these data indicate that the initial attack on heterochromatin by BAF and TOP2 involves nearly immediate strand-cleavage to relieve tangled DNA (or alter DNA topology in other ways), which is likely intrinsic to the structure of facultative heterochromatin.
TOP2 synergizes with BAF complexes to resolve facultative heterochromatin at regulatory elements genome-wide
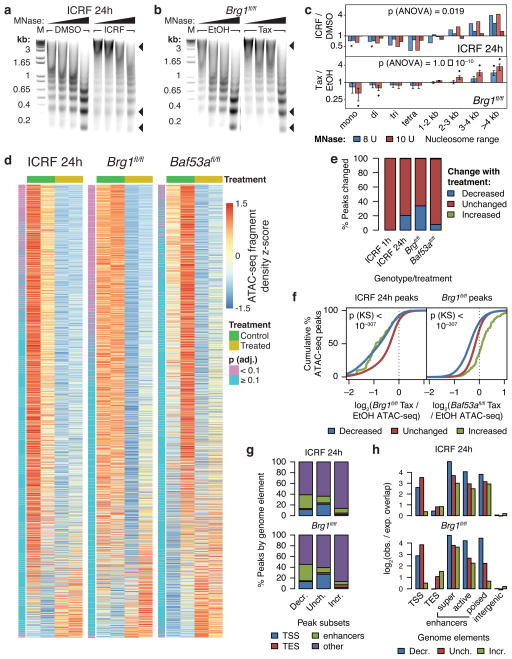
To assess the prevalence of the synergy between TOP2 and BAF complexes in resolving facultative heterochromatin, we performed Micrococcal Nuclease (MNase) digests on native chromatin from ES cells treated with ICRF-193 for 24 hours, as well as Brg1 conditional knockout (Brg1fl/fl; actin-CreER)10 and Baf53a conditional knockout (Baf53afl/−; actin-CreER) ES cells (Supplementary Fig. 2a–c). MNase preferentially releases nucleosomes from accessible chromatin and thus can be used to assay global changes in chromatin accessibility47–49. BRG1 is the primary catalytic subunit in esBAF complexes, whereas BAF53a is a non-catalytic subunit dispensable for in vitro remodeling15. TOP2 inhibition, as well as BAF subunit deletion, increased the amount of chromatin refractory to MNase digestion, especially of larger fragments, which likely reflect heterochromatin (Fig. 3a–c and Supplementary Fig. 2d,e). This indicates that TOP2 and BAF complexes antagonize heterochromatin.
Figure 3. TOP2 and BAF promote accessibility of enhancers and promoters genome-wide.
DNA gel electrophoresis of MNase digests of ES cells treated with 1 μM ICRF-193 for 24 hours (a) or Brg1fl/fl; actin-CreER (b) ES cells treated with tamoxifen (Tax) to knockout Brg1 using 6, 8, 10, or 12 units of MNase. Arrowheads point to different nucleosome species. M: DNA marker. Uncropped gel images are shown in Supplementary Data Set 1. (c) Fold-change densitometry of MNase digests in log-scale. Significance of treatment assessed by t-tests as before. Overall significance of the interaction effect of treatment and size range assessed by three-way ANOVA. Bars represent actual values from 2 cell passages (ICRF) or means (Brg1fl/fl) and error bars represent s.e.m. from 4 cell passages. (d) Row-wise z-score heatmaps of ATAC-seq fragment density at ATAC-seq peaks sorted by edgeR-adjusted fold-change. (e) Percentage of ATAC-seq peaks altered upon ICRF-193 or tamoxifen treatment. (f) Cumulative probability distributions of specified log2 fold-change ATAC-seq fragment density over specified ATAC-seq peaks. Significance between “Decreased” and “Unchanged” distributions assessed by Kolmogorov-Smirnov tests. (g) Classification of ATAC-seq peaks by genome element. (h) log2 observed/expected overlap of ATAC-seq peaks by genome elements.
To pinpoint the specific loci affected by TOP2 inhibition or BAF subunit deletion, we performed ATAC-seq on two independent cell passages. Accessible sites were classified as exhibiting “Decreased” or “Increased” accessibility upon ICRF-193 treatment or subunit knockout if their FDR was less than 10%. Consistent with our MNase results, 24 hours of TOP2 inhibition, as well as Brg1 deletion, reproducibly reduced accessibility at 20.3% to 33.7% of sites over the genome, respectively (Fig. 3d,e, Supplementary Fig. 2f, and Supplementary Table 1). Deletion of Baf53a was more modest, resulting in decreased accessibility at 7.8% of sites. However, 1 hour of ICRF-193 treatment did not reproducibly affect chromatin accessibility in three cell passages (Fig. 3e), suggesting that TOP2 is involved in establishment of accessibility, presumably after mitosis or DNA synthesis, rather than maintenance of chromatin state. This is consistent with our result that maximal decatenation intermediates occur only 5 minutes after BAF recruitment (Fig. 2b), and that blocking full accessibility induction requires TOP2 inhibition within the first 5 minutes (Fig. 2d). Decreased (but not increased) accessibility upon 24 hours of ICRF-193 treatment is predictive of decreased accessibility upon Brg1 deletion, which is in turn predictive of changes upon Baf53a deletion (Fig. 3f). Therefore, these perturbations largely affect the same sites (Supplementary Fig. 2g). Accessibility is decreased at all classes of enhancers, as well as promoters to a lesser degree (Fig. 3g,h and Supplementary Fig. 2h), consistent with the binding profiles of BAF complexes7,50. These studies confirm that TOP2 and BAF cooperate to resolve facultative heterochromatin and promote accessibility of regulatory regions.
TOP2 synergizes with BAF complexes to promote binding of pioneer pluripotency factors to chromatin
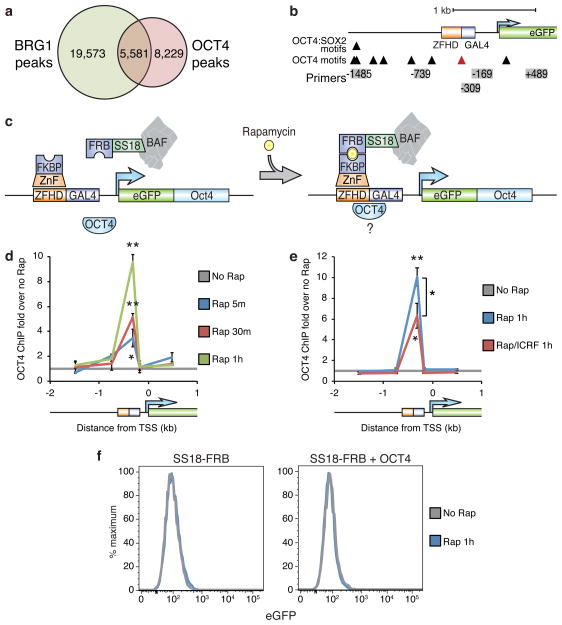
Chromatin accessibility is essential for DNA binding by transcription factors51. However, some fate-determining transcription factors, known as pioneer factors, are able to penetrate chromatin barriers and alter cell identity47,52. It is unclear whether these factors are self-sufficient for this capability or rely on other preexisting factors for creating an accessible environment, so we tested whether TOP2 or BAF complexes regulate pioneer factor recruitment. OCT4 (also known as POU5F1) is a key pioneer, pluripotency, and reprogramming factor6 that co-localizes with BAF complexes at 5,581 of 13,810 sites in ES cells (p < 10−307, bootstrapping) (Fig. 4a)7,10. To determine if TOP2 and/or BAF complexes can assist OCT4 binding, we made use of an OCT4 motif within the CiA:Oct4 DNA binding array (Fig. 4b) and expressed OCT4 in CiA:Oct4 fibroblasts along with our BAF recruitment system (Fig. 4c and Supplementary Fig. 3a). Strikingly, BAF recruitment lead to OCT4 concomitantly binding its motif in the array (Fig. 4d and Supplementary Fig. 3b). TOP2 activity is required for maximal OCT4 binding (Fig. 4e) despite the failure of both BAF and OCT4 recruitment to re-activate expression (Fig. 4f). In contrast, direct recruitment of OCT4 by expression of OCT4-GAL4 in the absence of BAF recruitment (Supplementary Fig. 3c) neither induced accessibility (Supplementary Fig. 3d) nor BAF occupancy (Supplementary Fig. 3e). These studies indicate that TOP2 and BAF complexes facilitate OCT4’s pioneering capability, which is consistent with the requirement of BAF complexes for efficient iPS cell reprogramming17,18.
Figure 4. TOP2 is required for optimal BAF-mediated recruitment of OCT4.
(a) Venn diagram of overlapping BRG1 and OCT4 ChIP-seq peaks in ES cells. (b) Diagram of OCT4 motifs at the CiA:Oct4 locus. Red arrowhead indicates the OCT4 motif in the DNA binding 19 bp downstream from the zinc-finger recruitment site. (c) Strategy for testing BAF/TOP2 pioneering for OCT4 using BAF recruitment to the CiA:Oct4 locus in fibroblasts while expressing exogenous OCT4. OCT4 ChIP in fibroblasts with the BAF recruitment system over-expressing OCT4 treated with 3 nM rapamycin (d) in the presence of 1 μM ICRF-193 (e). (f) eGFP flow cytometry of fibroblasts over-expressing OCT4 treated with rapamycin for 1 hour. Significance assessed by t-tests as before. Lines represent means and error bars represent s.e.m. from 3 cell passages (d,e).
To determine if changes in accessibility upon TOP2 inhibition or Brg1 deletion are predictive of effects on transcription factor recruitment genome-wide, we compared our ATAC-seq data to STAT3 ChIP-seq in Brg1fl/fl ES cells. STAT3 is critical to pluripotency in mice and most STAT3 sites are dependent on BAF complexes10. Indeed, loss of accessibility upon Brg1 deletion is strongly predictive of the magnitude of loss of STAT3 binding (Fig. 5a). Moderate loss of STAT3 binding also occurs at sites of “Unchanged” accessibility, which may be due to changes that do not pass the threshold for “Decreased” accessibility. Alternatively, BAF complexes may physically recruit STAT3 to chromatin in addition to making its binding sites accessible.
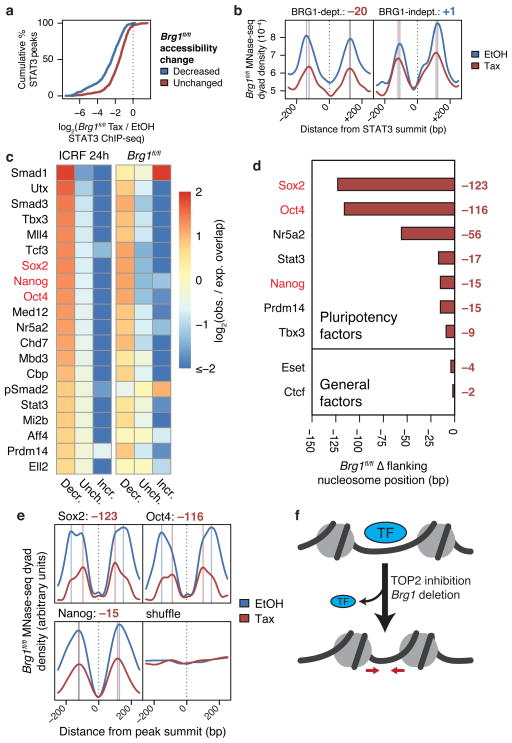
Figure 5. TOP2 and BAF are required for recruitment of pluripotency factors genome-wide.
(a) Cumulative probability distributions of log2 fold-change STAT3 ChIP-seq fragment density in Brg1fl/fl ES cells at STAT3 peaks overlapping specified ATAC-seq peaks. (b) MNase-seq nucleosome dyad density across specified subsets of linker-bound STAT3 sites in Brg1fl/fl ES cells. Vertical lines indicate nucleosome positions and values are changes in nucleosome positioning (bp, tamoxifen – EtOH). (c) Heatmap of log2 observed/expected change in accessibility of accessible ChIP-seq peaks from various datasets, sorted by enrichment for decreased accessibility upon ICRF-193 treatment. Top 20 ChIP-seq datasets are shown. Changes in Brg1fl/fl MNase-seq mean flanking nucleosome positions (d) and nucleosome dyad density profiles (e) around specified linker-bound ChIP-seq peaks or 38,864 randomly shuffled sites that exclude TSSs, TESs, or enhancers. (f) Model for loss accessibility, nucleosome spacing, and transcription factor binding.
We also performed MNase-seq on Brg1fl/fl ES cells, using constant reaction times and enzyme units according to methods used previously to study chromatin remodelers28,53–55, to assess changes in nucleosome structure. We sequenced mono-nucleosomes from light digests of native chromatin to preserve the sensitivity of MNase to chromatin accessibility, preserve fragile nucleosomes, which often flank regulatory regions28,47–49,56, and prevent over-digestion of accessible nucleosomes. We can then assess changes in nucleosome positioning, which are consistent across different levels of digestion48, as well as changes in nucleosome “density”, which is convolved with nucleosome occupancy and accessibility47–49. Indeed, nucleosome density decreased around linker-bound STAT3 sites upon Brg1 deletion, which likely reflects decreased accessibility10,47–49 (Fig. 5b). Additionally, spacing between the nucleosomes flanking BRG1-dependent STAT3 sites decreased substantially more than at BRG1-independent STAT3 sites. Therefore, the reduction in spacing likely reflects reduction in STAT3 binding, which is predominantly dependent on BAF function10.
To assess the extent that TOP2 and BAF regulate transcription factor recruitment, we compared our ATAC-seq and MNase-seq data to 121 publically available ChIP-seq datasets in ES cells (Supplementary Table 2). TOP2 and BAF specifically promote accessibility of pluripotency factor binding sites, such as SOX2, OCT4, and NANOG6 (Fig. 5c and Supplementary Fig. 4). Interestingly, despite loss of accessibility at pluripotency factors as well as widespread loss of accessibility assessed by MNase titrations (Fig. 3b,c), we did not observe global differences in nucleosome phasing (Supplementary Fig. 5a). However, Brg1 deletion resulted in flanking nucleosomes moving into sites normally occupied by pluripotency factors, but did not result in substantial shifts around sites of general factors like ESET or CTCF, randomly shuffled sites, or regions beyond ~0.75 kb from pluripotency factor sites (Fig. 5d–f and Supplementary Fig. 5b,c). Also, although BAF complexes maintain accessibility of motifs of both OCT4 and ASCL1, a fate-determining pioneer factor for neural progenitors that is not expressed in ES cells57 (Supplementary Fig. 5d), BAF promotes spacing of nucleosomes around OCT4 but not ASCL1 motifs in ES cells (Supplementary Fig. 5e). These results indicate that, as with STAT3, BAF is critical for specifically allowing pluripotency factors to access their binding sites. Indeed, loss of accessibility and collapse of flanking nucleosome spacing predicts loss of both OCT4 and SOX2 binding to their specific sites upon TOP2 inhibition or Brg1 deletion (Supplementary Fig. 5f). This is similar to the dependence of TAL1 on BRG1 for targeting and for spacing flanking nucleosomes during erythrocyte differentiation28. Our finding that TOP2 and BAF complexes promote targeting of pluripotency factors helps explain the instructive role that BAF complexes play in ES cells7,10,14–16 and iPS cell formation17,18.
TOP2 activity is also crucial for the reformation of facultative heterochromatin
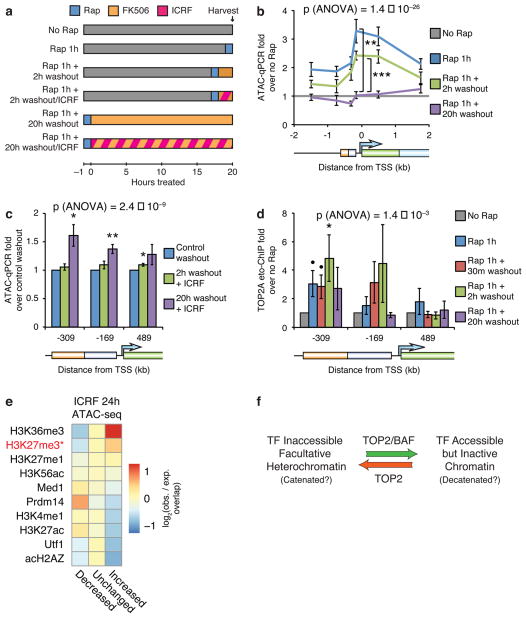
Given our discovery that TOP2 synergizes with BAF complexes to resolve facultative heterochromatin by altering catenation or other topological states of DNA, and that TOP2 catalyzes both decatenation and catenation in vivo31, we hypothesized that heterochromatin and accessible chromatin exist at different catenation or topological states. Therefore, TOP2 may also have a role in the reformation of facultative heterochromatin. To test this, we reversed BAF recruitment after 1 hour by washing out rapamycin while adding 100 nM FK506, which disrupts residual rapamycin-mediated FRB/FKBP dimerization4 (Fig. 6a). This completely reforms facultative heterochromatin within 20 hours and partially reforms it within 2 hours as assessed by accessibility loss (Fig. 6b and Supplementary Fig. 6a), giving us the first assay for facultative heterochromatin reformation. The rate of facultative heterochromatin reformation is slower than accessibility induction, suggesting that assembly of facultative heterochromatin is more complex than its resolution. Interestingly, inhibiting TOP2 by adding ICRF-193 simultaneously with rapamycin washout and FK506 addition (Fig. 6a) significantly impairs the loss of accessibility (Fig. 6c), indicating that TOP2 also has a role in heterochromatin reformation.
Figure 6. TOP2 is required for reformation of facultative heterochromatin.
(a) Strategy for BAF recruitment and rapamycin washout using 100 nM FK506 with TOP2 inhibition using 1 μM ICRF-193. ATAC-qPCR in fibroblasts treated with 3 nM rapamycin for 1 hour and subsequently washed out with FK506 (b) in the presence of ICRF-193 (c). (d) TOP2A etoposide-ChIP in cells treated with rapamycin and subsequently washed out with FK506. (e) Heatmap of log2 observed/expected change in accessibility of various ChIP-seq peaks, sorted by enrichment for increased accessibility upon ICRF-193 treatment. Top 10 ChIP-seq datasets are shown. *: does not include bivalent peaks. (f) Model for the role of TOP2 and BAF in resolution and reformation of heterochromatin. Significance of individual primer sets assessed by t-tests as specified (b), versus control washout (c), or versus no rapamycin control (d). Overall significance of the effect of washout (b,d) or ICRF-193 (c) assessed by three-way ANOVA. Lines and bars represent means and error bars represent s.e.m. from 5 (b,c) or 9 (d) cell passages.
To further determine if TOP2 produces inaccessibility, we assayed TOP2 strand-cleaved intermediates. Remarkably, TOP2 stand-cleaved intermediates increased two hours after BAF washout and then subsequently resolved (Fig. 6d), indicating that TOP2 functions during the process of reforming inaccessibility. Importantly, reformation of heterochromatin occurs in the absence of transcription, therefore this function of TOP2 is also distinct from the role of TOP2 in transcription. Consistently, non-bivalent H3K27me3 sites in ES cells, which mark facultative heterochromatin1, are enriched for increased accessibility upon TOP2 inhibition (Fig. 6e and Supplementary Fig. 6b), suggesting that TOP2 has a role in making facultative heterochromatin inaccessible genome-wide. The enrichment for increased accessibility upon TOP2 inhibition at H3K36me3 sites, which mark gene bodies and transcription end sites of active genes, is likely a consequence of the role of TOP2 in facilitating transcriptional elongation37–40, and thus distinct from the role of TOP2 in modulating facultative heterochromatin outside the context of transcription.
DISCUSSION
The mechanisms of the resolution and formation of facultative heterochromatin have largely been a mystery. For the first time, our system of rapamycin-mediated BAF recruitment to a heterochromatinized locus has given us the ability to study in vivo chromatin state transitions in real-time. This system is especially advantageous because we predict that it mimics physiologic chromatin remodeler binding43,44. Given the Kd for FKBP-rapamycin binding to FRB of ~12 nM45, and a typical rate constant kon for diffusion-limited biomolecular binding of ~106 M−1 s−1, we estimated the mean residence time (ton) of recruited BAF complexes:
Given a concentration of 3 nM rapamycin, we estimated the equilibrium fraction (f) of bound FKBP-rapamycin-FRB ternary complexes:
Therefore only ~20% of FKBP-rapamycin-FRB ternary complexes are bound at any given time, resulting in binding lifetimes on the order of tens of seconds, which is similar to dwell times of chromatin remodelers binding to endogenous sites43,44. Empirical evidence shows that BAF recruitment to the CiA:Oct4 locus is dynamic as we have recently shown (and replicated independently here) that rapamycin washout leads to loss of BAF binding in less than an hour5 (Supplementary Fig. 6a). It is likely that recruited BAF complexes remain on chromatin longer than predicted from rapamycin’s on/off rates because of the complexes’ numerous DNA and histone binding domains14. This system may in fact reveal a more accurate view of chromatin remodeler binding as photobleaching measures dynamics of freely diffusible complexes that may not reflect that of active complexes stably bound to chromatin43,44.
Our studies indicate that changes in catenation are vital to chromatin state transitions mediated by BAF chromatin remodeling (Fig. 6f). In contrast to earlier studies showing that TOP2 is required for transcription37–42, the resolution of inaccessible heterochromatin by BAF/TOP2 shown here occurs in the absence of transcription, DNA replication, or RNA Pol. II. Given that TOP2 acts to facilitate resolution of facultative heterochromatin early in the process, it is also unlikely that TOP2 acts to undo tangles introduced by BAF chromatin remodeling. Rather, our data suggest that facultative heterochromatin and accessible chromatin exist at different catenation or topological states, which may affect chromatin compaction and might explain the difficulties in obtaining a structure for heterochromatin3. Interestingly, TOP2 can sense the level of chromatin compaction in vitro, which may determine whether it promotes resolution or formation of facultative heterochromatin58. Our discovery that TOP2 can assist both resolution and formation of repressed chromatin will allow us to re-evaluate other intriguing observations, such as the prolonged therapeutic activation of the dormant allele of Ube3a by transient topoisomerase inhibition59.
Our results also suggest a new interpretation of the function of pioneer transcription factors. These proteins are thought to be able to invade heterochromatic regions and thereby impart instructive functions for cell identity47,52. For example, during the first 48 hours of reprogramming, ectopically expressed pluripotency factors can bind non-H3K9me3-modified inaccessible chromatin60. However, our results indicate that many of these factors, such as OCT4 and SOX26, are largely dependent on TOP2 and BAF complexes for binding to their recognition sites. This is evident from our observation of TOP2/BAF-dependent binding of ectopic OCT4 to an inaccessible locus and also from the genome-wide collapse of nucleosomes toward sites of pioneer factor binding upon Brg1 deletion. Interestingly, directed OCT4 recruitment is not sufficient for induction of accessibility or BAF binding. These findings likely explain the instructive functions of BAF complexes in reprogramming and development7,8,10,15–20. Further study is needed to determine whether the role of BAF complexes in facilitating iPS cell reprogramming17,18, along with TOP2, is to assist initial binding of pluripotency factors to H3K9me3-negative inaccessible chromatin60, promote subsequent binding to sites within H3K9me3-positive facultative heterochromatin, or operate through another mechanism.
In recent other studies, we have found that BAF complexes can directly evict PRC15,13. It is currently unclear how displacement of PRC1 is related to recruitment of TOP2 activity or if these functions are independent. However, it is likely that opposition between BAF and PRC1 affects chromatin compaction. Interestingly, consistent with our previous study showing that BAF activity does not necessarily affect nucleosome occupancy5, we found here that Brg1 deletion does not affect global nucleosome phasing, suggesting that the primary role of BAF complexes is antagonizing higher-order heterochromatic structures. As mutations in BAF subunits cause a number of human neurodevelopmental diseases25,26 and contribute to more than 20% of all human cancers21–24, understanding the mechanisms of BAF functions is critical to therapeutic development. The cooperation between BAF and TOP2 to modulate the accessible genome and pioneer transcription factor binding that we describe in this study likely contributes to the mechanisms of these diseases.
METHODS
Cell culture
SV40 large T antigen-transformed CiA:Oct4 adult mouse fibroblasts4 and Brg1+/+, Brg1fl/fl; actin-CreER, Baf53afl/fl; actin-CreER, and Baf53afl/−; actin-CreER ES cells10 were cultured as described previously. To delete conditional alleles, cells were treated with 0.8 μM of tamoxifen for 24 hours, passaged, and then grown for another 48 hours in tamoxifen-free media before harvesting for a total of 72 hours of growth after initiation of tamoxifen treatment.
Lentiviral preparation and infection
Lentiviruses were produced in Lenti-X 293T cells (Clontech) using polyethylenimine transfection. Media were changed 24 hours after transfection and after another 48 hours media were collected and centrifuged for 2 hours at 20,000 RPM. Viral pellets were re-suspended in PBS and used to infect cells in the presence of 10 μg/ml Polybrene (Santa Cruz). Cells were selected with 2 μg/ml puromycin, 10 μg/ml blastocidin, and/or 100 μg/ml hygromycin as appropriate beginning 48 hours after infection.
Assay of Transposase Accessible Chromatin (ATAC)
ATAC libraries were prepared as described previously with some modifications61. For sequencing, cells were stained with Annexin V-FITC (eBioscience) and 7-AAD (BD Biosciences) and 50,000 viable cells were FACS sorted. For qPCR, cells were counted with a ViCELL Cell Counter (Beckman Coulter) and 50,000 viable cells were used. Those cells were washed with PBS and then with RSB Buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2). Nuclei were obtained by re-suspending cells in 0.5 ml Lysis Buffer (0.1% Tween-20/RSB Buffer) and incubated for 10 minutes on ice. Nuclei were pelleted by centrifuging for 10 minutes at 500xg, re-suspended in 50 μl of Transposition Mix (1X Tagmentation DNA Buffer, 2.5 μl Tagment DNA Enzyme (Illumina)), and incubated for 30 minutes at 37 °C. DNA was purified with a MinElute PCR Purification Kit (QIAGEN) and libraries were amplified by PCR with barcoded Nextera primers (Illumina). For sequencing, libraries were size-selected using Agencourt AMPure XP (Beckman Coulter) for fragments between ~50 – 1000 bp according to the manufacturer’s instructions. Libraries were paired-end sequenced by the Stanford Functional Genomics Facility on a NextSeq Desktop Sequencer (Illumina). For qPCR, libraries were diluted 1:100. Primers used for ATAC-qPCR are available upon request.
Micrococcal Nuclease (MNase) digestion
MNase digests were performed as described previously62. 20 million cells were washed in PBS and nuclei were isolated by re-suspending cells in Lysis Buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 0.15 mM spermine, 0.5 M spermidine) and incubating for 5 minutes on ice. Nuclei were washed with MNase Digestion Buffer (10 mM Tris-HCl pH 7.5, 15 mM NaCl, 60 mM KCl, 0.15 mM spermine, 0.5 mM spermidine) and re-suspended with 800 μl of MNase Digestion Buffer with 1 mM CaCl. Nuclei were split into 8 tubes containing 6, 8, 10, 12, 14, 16, 18, or 20 U of MNase (New England BioLabs). Reactions were incubated for precisely 5 minutes at 37 °C and stopped by adding 150 μl of Stop Buffer (20 mM EDTA pH 8.0, 20 mM EGTA pH 8.0, 0.4% SDS). After Proteinase K (New England BioLabs) digestion, DNA was purified by phenol/chloroform extraction and ethanol precipitation. DNA was run on a 2% agarose/ethidium bromide electrophoresis gel. For sequencing, mononucleosome bands from 12 U digests were isolated using a QIAquick Gel Extraction Kit (QIAGEN), and libraries were constructed as described previously63. ImageJ (version 1.50i) was used for gel densitometry.
Chromatin Immuno-Precipitation (ChIP)
10 – 30 million cells were fixed in suspension for 15 minutes in 1% formaldehyde. Excess formaldehyde was quenched by addition of glycine to 100 mM. Fixed cells were washed, pelleted, and flash-frozen. Pellets were thawed, re-suspended in NP Rinse 1 buffer (50 mM HEPES-KOH pH 8.0, 140 mM NaCl, 1 mM EDTA pH 8.0, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100), and incubated for 10 minutes on ice to isolate nuclei. Nuclei were washed once with NP Rinse 2 buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 200 mM NaCl) and then washed twice with Shearing Buffer (0.1% SDS, 1 mM EDTA pH 8.0, 10 mM Tris-HCl pH 8.0). Pellets were re-suspended in 900 μl of Shearing Buffer with protease inhibitors and sonicated in a Covaris E220 sonicator for 10 – 12 minutes to obtain DNA fragments between approximately 200 – 1000 bp. Insoluble material was pelleted by centrifuging for 15 minutes at 16,100xg. Chromatin was then immunoprecipitated overnight at 4 °C with antibodies bound to Protein G Dynabeads (Life Technologies) in ChIP Buffer (50 mM HEPES-KOH pH 7.5, 300 mM NaCl, 1 mM EDTA pH 8.0, 1% Triton X-100, 0.1% DOC, 0.1% SDS). The chromatin-beads slurry was washed with a magnetic rack four times with ChIP Buffer, once with DOC Buffer (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% DOC, 1 mM EDTA pH 8.0), and once with TE Buffer. Chromatin was eluted with Elution Solution (0.1 M NaHCO3, 1% SDS). After RNase A (Life Technologies) and Proteinase K (New England BioLabs) digestion and de-crosslinking at 65 °C overnight, DNA was extracted with phenol/chloroform and precipitated with ethanol. Primers used for ChIP-qPCR are available upon request. Etoposide-ChIP was performed as described previously by fixing cells for 10 minutes by adding etoposide to 100 μM directly to media instead of formaldehyde fixation and using anti-TOP2A (Santa Cruz F-12) antibodies29.
Sequencing analysis
Raw reads were mapped to the mm9/NCBI37 Mus musculus genome using Bowtie (version 0.12.9)64. Nuclear ATAC-seq fragments smaller than 235 bp were used to call accessible sites. Accessible sites and ChIP-seq peaks were called by Model-based Analysis for ChIP-seq 2 (MACS2) (version 2.1.0)65. Peaks overlapping ENCODE “blacklist” regions were discarded66. Public raw ultra-high-throughput sequencing was processed using the SRA Toolkit (version 2.3.5)67. Downstream analysis was aided by Bedtools (version 2.17.0)68. Genome annotations were acquired from the UCSC Genome Browser (http://genome.ucsc.edu/)69,70. Putative enhancers were defined as H3K4me1 sites overlapping p300 that are at least 500 bp away from the nearest annotated TSS71,72. These sites were further partitioned as active or poised by presence or absence, respectively, of H3K27ac71. Super-enhancers were defined as enhancers with high Med1 density as described previously73.
To determine which peaks are significantly altered in knockout/treated cells versus wild-type/control cells, we merged peaks from both cell types and all replicates. Non-redundant read/fragment counts was calculated over the set of merged peaks that are common to all replicates either by extending reads by the spline-smoothed mode of the distance between opposing reads (for single-end) or by counting overlapping fragments (for paired-end). Non-redundant fragment depth was used as normalization factors for edgeR (version 3.12.1) analysis of differential peak density74. Peaks were deemed altered between samples if the False Discovery Rate (FDR) < 10%.
Where biological replicates are not available, non-redundant read/fragment density was calculated over the set of merged peaks either by extending reads by the spline-smoothed mode of the distance between opposing reads (for single-end) or by counting overlapping fragments (for paired-end) and normalizing by unique sequencing depth. Peaks were deemed altered between samples if read/fragment density is changed by more than 1.5-fold in either direction and if the change has a FDR < 5% as determined by SICER (version 1.1)75.
Overlap enrichment scores between two sets of genomic intervals were calculated as observed/expected number of intervals from the first set overlapped (at least one bp shared) by intervals from the second. The expected overlap is calculated by performing 1000 trials randomizing the locations of the first set of peaks and then counting the number of randomized peaks overlapped by the second set. P-values are estimated by fitting a Poisson distribution.
For MNase-seq analysis, nucleosome positions were determined using NPS (version 1.3.2)76. Dyad centers of fragments ranging from 130 – 180 bp were used to determine average nucleosome densities around ChIP-seq peak summits. Peaks were filtered for summits that occupy linkers as defined by summits that are > 75 bp away from NPS-called nucleosome dyads. Shuffled sites were determined by randomly positioning 1 kb intervals around transcription start sites to non-overlapping blacklist-subtracted genomic coordinates. For phasogram analysis, the distances between any given read and any other nearby read on the same strand were tabulated as described previously77. For this analysis, only high-confidence nucleosome positions (at least three reads at the same position) were considered.
To locate known motifs in the genome, we used Homer (version 4.7.2)78. Motifs were filtered for occupying linkers as with peak summits.
Western blots
Cells were lysed for at least 30 minutes at 4 °C in RIPA buffer (50mM Tris-HCl pH 8.0, 150mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40). Lysates were centrifuged for 30 minutes at 14,000xg and the supernatant was flash-frozen. Total protein concentration was measured by Bradford assay (BioRad). 10 μg of protein was boiled in gel loading buffer (Life Technologies) with 50 mM of dithiothreitol and loaded onto 4–10% BisTris NuPage gels (Life Technologies). Bands were transferred to PVDF membranes (BioRad) and then membranes were blocked in 5% BSA/TBST. Blots were then incubated with primary antibodies for 1 hour at room temperature. Proteins were detected using the LI-COR detection system.
Antibodies
For ChIP, anti-BAF155, anti-BRG1 (J1B), anti-RNA Polymerase II (Santa Cruz N-20), anti-V5 (Life Technologies R960-25), anti-OCT4 (Santa Cruz N-19), and anti-SOX2 (Santa Cruz Y-17) antibodies were used. For etoposide-ChIP, anti-TOP2A (Santa Cruz F-12) antibodies were used. For western blots, anti-BRG1 (J1B), anti-HSP90 (BD Biosciences 68), anti-BAF53a (Novus Biologicals NB100-61628), anti-SS18 (Santa Cruz H-80), anti-V5 (Life Technologies R960-25), and anti-GAPDH (Santa Cruz 6C5) antibodies were used.
Public ChIP-seq datasets
Raw reads from publically available sequencing datasets were acquired from the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) (Supplementary Table 2)67. Datasets were re-analyzed according to the procedure described here.
Statistics
Statistical significance of differences in experiments using the BAF recruitment system, MNase densitometry, and ChIP-qPCR was determined using two-tailed Student’s t-tests (heteroskedastic). Overall significance for specified effects in these experiments were determined by ANOVAs. FDR for changes in ATAC-seq density was assessed using edgeR74. FDR for changes in STAT3 ChIP-seq density was assessed using SICER75. Significance between different cumulative distributions was determined by Kolmogorov-Smirnov tests. Individual p-values for overlap contingency heatmaps/tables were calculated by hypergeometric tests (right-tailed). Overall p-values for such tables were calculated by chi-squared tests. Significance for overlap between sets of ChIP-seq peaks was determined by bootstrapping Poisson distributions.
Data availability
Sequencing data have been deposited in the Gene Expression Omnibus under accession code GSE94041. Other data is available upon request.
Supplementary Material
Acknowledgments
This work was supported by funds from the Howard Hughes Medical Institute, the Simons Foundation Autism Research Initiative, and by NIH grants NS046789 and CA163915 to G.R.C. E.L.M. was supported by the Lucille P. Markey Stanford Graduate Fellowship in Biomedical Research and the Stanford University Genetics & Developmental Biology Training Program NIH-NIGMS T32 GM007790. D.C.H. was supported by NCI career transition award K99CA184043. C.H. is supported by NCI career transition award K99CA187565. ATAC-seq libraries were prepared with advice from Beijing Wu. We used the BioX3 cluster, which is supported by NIH S10 Shared Instrumentation Grant 1S10RR02664701, for sequencing analysis. Much thanks to Andrew Koh and Christopher Weber for technical advice and support.
Footnotes
ACCESSION CODES
ATAC-seq and MNase-seq reads have been deposited into the Gene Expression Omnibus (GEO) under the accession code GSE94041.
DATA AVAILABILITY
Sequencing data have been deposited in the Gene Expression Omnibus under accession code GSE94041. Other data is available upon request.
AUTHOR CONTRIBUTIONS
The studies of TOP2/BAF function were designed and conducted by E.L.M., G.R.C., and D.C.H. The CiA system was conceived by G.R.C., adapted for analysis of BAF mechanisms by C.K., and adapted for analysis of LSH and INO80 mechanisms by J.P.C. ATAC-seq was conceived by W.J.G. and J.D.B., performed by E.L.M. and C.Y.C, and analyzed by E.L.M. MNase assays and sequencing was performed by E.L.M., K.Z., and K.C. and analyzed by E.L.M. Binding kinetics were calculated by C.H. E.L.M. and G.R.C. wrote the manuscript with input from the co-authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boettiger AN, et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–22. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghirlando R, Felsenfeld G. Chromatin structure outside and inside the nucleus. Biopolymers. 2013;99:225–32. doi: 10.1002/bip.22157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–60. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadoch C, et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet. 2016 doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5187–91. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao X, et al. A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015;16:284. doi: 10.1186/s13059-015-0840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris SA, et al. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho L, et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nature cell biology. 2011;13:903–13. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson BG, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–28. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanton BZ, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet. 2016 doi: 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5181–6. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem cells. 2009;27:317–28. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 17.Kleger A, et al. Increased reprogramming capacity of mouse liver progenitor cells, compared with differentiated liver cells, requires the BAF complex. Gastroenterology. 2012;142:907–17. doi: 10.1053/j.gastro.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Singhal N, et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–55. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 19.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staahl BT, Crabtree GR. Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Curr Opin Neurobiol. 2013;23:903–13. doi: 10.1016/j.conb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodges C, Kirkland JG, Crabtree GR. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nature reviews Genetics. 2013;14:347–59. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santen GW, Kriek M, van Attikum H. SWI/SNF complex in disorder: SWItching from malignancies to intellectual disability. Epigenetics : official journal of the DNA Methylation Society. 2012;7 doi: 10.4161/epi.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hainer SJ, Fazzio TG. Regulation of Nucleosome Architecture and Factor Binding Revealed by Nuclease Footprinting of the ESC Genome. Cell Rep. 2015;13:61–9. doi: 10.1016/j.celrep.2015.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu G, et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–8. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dykhuizen EC, et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature. 2013;497:624–7. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijdeven RH, et al. Genome-Wide Identification and Characterization of Novel Factors Conferring Resistance to Topoisomerase II Poisons in Cancer. Cancer Res. 2015;75:4176–87. doi: 10.1158/0008-5472.CAN-15-0380. [DOI] [PubMed] [Google Scholar]
- 31.Mertz JE, Miller TJ. In vivo catenation and decatenation of DNA. Mol Cell Biol. 1983;3:126–31. doi: 10.1128/mcb.3.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–41. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez X, Diaz-Ingelmo O, Martinez-Garcia B, Roca J. Chromatin regulates DNA torsional energy via topoisomerase II-mediated relaxation of positive supercoils. EMBO J. 2014;33:1492–501. doi: 10.15252/embj.201488091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–83. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naughton C, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–95. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kouzine F, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King IF, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thakurela S, et al. Gene regulation and priming by topoisomerase IIalpha in embryonic stem cells. Nat Commun. 2013;4:2478. doi: 10.1038/ncomms3478. [DOI] [PubMed] [Google Scholar]
- 39.Tiwari VK, et al. Target genes of Topoisomerase IIbeta regulate neuronal survival and are defined by their chromatin state. Proc Natl Acad Sci U S A. 2012;109:E934–43. doi: 10.1073/pnas.1119798109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sano K, Miyaji-Yamaguchi M, Tsutsui KM, Tsutsui K. Topoisomerase IIbeta activates a subset of neuronal genes that are repressed in AT-rich genomic environment. PLoS One. 2008;3:e4103. doi: 10.1371/journal.pone.0004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madabhushi R, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puc J, et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015;160:367–80. doi: 10.1016/j.cell.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erdel F, Schubert T, Marth C, Langst G, Rippe K. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc Natl Acad Sci U S A. 2010;107:19873–8. doi: 10.1073/pnas.1003438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol Biol Cell. 2008;19:3308–22. doi: 10.1091/mbc.E08-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–21. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 46.Tanabe K, Ikegami Y, Ishida R, Andoh T. Inhibition of topoisomerase II by antitumor agents bis(2,6-dioxopiperazine) derivatives. Cancer Res. 1991;51:4903–8. [PubMed] [Google Scholar]
- 47.Iwafuchi-Doi M, et al. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol Cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mieczkowski J, et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat Commun. 2016;7:11485. doi: 10.1038/ncomms11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez J, Tsukiyama T. ATR-like kinase Mec1 facilitates both chromatin accessibility at DNA replication forks and replication fork progression during replication stress. Genes Dev. 2013;27:74–86. doi: 10.1101/gad.202978.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–83. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaret KS, Lerner J, Iwafuchi-Doi M. Chromatin Scanning by Dynamic Binding of Pioneer Factors. Mol Cell. 2016;62:665–7. doi: 10.1016/j.molcel.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Dieuleveult M, et al. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016;530:113–6. doi: 10.1038/nature16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hainer SJ, et al. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 2015;29:362–78. doi: 10.1101/gad.253534.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orvis T, et al. BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res. 2014;74:6486–98. doi: 10.1158/0008-5472.CAN-14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu G, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–92. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wapinski OL, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–35. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krasnow MA, Cozzarelli NR. Catenation of DNA rings by topoisomerases. Mechanism of control by spermidine. J Biol Chem. 1982;257:2687–93. [PubMed] [Google Scholar]
- 59.Huang HS, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–9. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui K, Zhao K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol Biol. 2012;833:413–9. doi: 10.1007/978-1-61779-477-3_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leinonen R, Sugawara H, Shumway M International Nucleotide Sequence Database C. The sequence read archive. Nucleic Acids Res. 2011;39:D19–21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kent WJ, et al. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic acids research. 2012 doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 73.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–8. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valouev A, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–20. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the Gene Expression Omnibus under accession code GSE94041. Other data is available upon request.