Abstract
Background
In the pathogenesis of diabetic retinopathy, damaged retinal mitochondria accelerate apoptosis of retinal capillary cells, and regulation of oxidative stress by manipulating mitochondrial superoxide dismutase (SOD2) protects mitochondrial homeostasis and prevents the development of diabetic retinopathy. Diabetes also activates matrix metalloproteinase-9 (MMP-9), and activated MMP-9 damages retinal mitochondria. Recent studies have shown a dynamic DNA methylation process playing an important role in regulation of retinal MMP-9 transcription in diabetes; the aim of this study is to investigate the role of oxidative stress in MMP-9 transcription.
Methods
The effect of regulation of mitochondrial superoxide on DNA methylation of MMP-9 promoter region was investigated in retinal endothelial cells incubated in the presence or absence of a MnSOD mimetic- MnTBAP, by quantifying the levels of 5 methyl cytosine (5mC) and hydroxyl-methyl cytosine (5hmC). The binding of DNA methylating, and of hydroxymenthylating enzymes (Dnmts and Tets, respectively), at MMP-9 promoter (by chromatin immunoprecipitation) was also evaluated. The in vitro results were confirmed in the retina of diabetic mice overexpressing SOD2.
Results
MnTBAP attenuated glucose-induced decrease in 5mC levels and increase on Dnmt1 binding at the MMP-9 promoter region. MnTBAP also ameliorated alterations in 5hmC levels and Tet binding, regulated MMP-9 transcription, and prevented mitochondrial damage. Similarly, mice overexpressing SOD2 were protected from diabetes-induced alteration in MMP-9 promoter methylation, and its transcription.
Conclusions
Thus, regulation of oxidative stress by pharmacologic/genetic approaches maintains retinal mitochondrial homeostasis by ameliorating epigenetic modifications in the MMP-9 promoter region.
Keywords: Diabetic retinopathy, DNA methylation, Epigenetics, Oxidative stress
Introduction
Oxidative stress remains as one of the major causative factors in the development of diabetic retinopathy; free radical production is increased and their removal is decreased in the retina and mitochondria are damaged, and this accelerates apoptosis of capillary cells [1, 2]. Regulation of oxidative stress by pharmacological means or by manipulating the enzyme responsible for scavenging mitochondrial superoxide, manganese superoxide dismutase (SOD2), protects mitochondrial homeostasis, and prevents the development of diabetic retinopathy in animal models [3, 4]. Diabetes also stimulates secretion of proteinases including matrix metalloproteinase-9 (MMP-9) [5]. The activity and transcription of MMP-9 is increased in diabetes in the retina and its capillary cells, and activated MMP-9 damages the mitochondria, releasing cytochrome c into the cytosol. Regulation of MMP-9 activation by genetic manipulations protects diabetes-induced mitochondrial dysfunction and apoptosis, and also prevents the development of diabetic retinopathy in mice [6, 7]. In addition to role of MMP-9 in increasing oxidative stress by damaging mitochondria, the presence of crucial cysteine residues within its DNA-binding domain also makes MMP-9 sensitive to oxidative stress [8, 9]. Furthermore, activation of redox-sensitive upstream signaling molecules e.g. mitogen-activated protein kinase and nuclear transcriptional factor B (NF-kB), by increased oxidative stress also activates MMP-9 [6].
Gene transcription, in addition to the transcription factors, is also modulated by epigenetic modifications, and diabetes enables many of these epigenetic modifications [10, 11]. Recent work has shown critical role of these epigenetic modifications in many diabetic complications including retinopathy and nephropathy [2, 12]. We have shown that diabetes hypomethylates H3K9 at the MMP-9 promoter, freeing up that lysine 9 (K9) for acetylation, and increased K9 acetylation facilitates the recruitment of transcription factors NF-kB and activator protein-1, AP-1, resulting in MMP-9 transcriptional activation. In addition, increase in oxidative stress inhibits deacetylase sirtuin 1(Sirt1), which hyperacetylates NF-kB and AP-1, increasing their binding at the MMP-9 promoter [13, 14]. MMP-9 transcription is also regulated by DNA methylation [15]. Our recent work has shown a major role of a dynamic DNA methylation process in regulating transcription of retinal MMP-9 in diabetes [16]. While DNA methyl transferases (Dnmts) add a methyl group to the cytosine forming methyl cytosine (5mC), Ten eleven translocases (Tets) hydroxymethylate that cytosine to form 5-hydroxymethyl cytosine, 5hmC [17–19]. In diabetes, the binding of these two redox-sensitive enzymes [20], with opposing functions, is significantly increased at the MMP-9 promoter in diabetes [16]. However, regulation of DNA methylation machinery by oxidative stress plays any role in modulating retinal MMP-9 transcription in diabetes remains unclear.
The goal of this study is to investigate the role of oxidative stress in retinal MMP-9 transcription in diabetes. Using retinal endothelial cells in culture, we have investigated the effect of regulation of mitochondrial superoxide on DNA methylation of MMP-9 promoter region by quantifying the levels of 5mC and 5hmC, and the binding of DNA methylating and hydroxymenthylating enzymes at MMP-9 promoter. The in vitro results are supported by analyzing the same parameters in the retina from diabetic mice genetically manipulated to overexpress mitochondrial superoxide dismutase, a genetic manipulations which protects them from the development of diabetic retinopathy in these mice [2].
Methods
Retinal endothelial cells, isolated from bovine retina, were cultured in Dulbecco’s Modified Eagle medium containing 15% heat inactivated fetal bovine serum, 5% Nu-serum, 50μg/ml heparin and 50μg/ml endothelial cell growth supplement. Cells from 5–7th passage were incubated in 5mM or 20mM glucose media with/without a cell permeable manganese superoxide dismutase mimic, manganese (III) tetrakis (4-benzoic acid)porphyrin chloride (MnTBAP, 250μM) [21]. For osmolarity control, cells were incubated in 20mM mannitol, instead of 20mM glucose.
Mice
Hemizygous SOD2-Tg mice and their wildtype littermates were made diabetic by streptozotocin injection (55mg/kg BW) for 4 consecutive days (Tg-D and WT-D respectively) and their age-matched normal SOD2-Tg and wildtype mice were used as controls (Tg-N and WT-N respectively) [3, 21]. To avoid ketosis, diabetic mice received 01-02IU insulin 4–6 times a week, and 6 months after induction of diabetes, mice were sacrificed by carbon dioxide asphyxiation. The treatment of the animals conformed to the Association for Research in Vision and Ophthalmology Resolution on the Use of Animals in Research, and all procedures performed involving animals were in accordance with the ethical standards of the institution.
Levels of 5hmC and 5hC were quantified in ~250–500ng sonicated gnomic DNA (DNeasy Blood & Tissue Kit, Qiagen, Germantown, MD) using hydroxymethylated/methylated DNA and Immunoprecipitation kits (hMeDIP/MeDIP kits, EPIGENTEK, Farmingdale, NY). 5hMC and 5mC were captured using a high affinity 5-hmC antibody and 5-mC monoclonal antibody respectively, and semi-quantitative PCR was performed on DNA eluted from the plate using primers for MMP-9 promoter region (bovine: Fwd-CTAGCCTGAGAAGGATGAAG and Rev- TTCCTTGGGCTTCTGAGA; mouse Fwd-CAGACGCCACAACACTCCCA and Rev- TCCTCTCCCTGCTCCACCTG). After denaturation at 95°C for 10 minutes and 35 cycles of denaturation at 95°C for 15 seconds, annealing was performed at 55°C for 30 seconds and extension at 72°C for 30 seconds. This was followed by 5 minutes at 72°C. The products were also analyzed on a 2% agarose gel, and visualized by ethidium bromide [16, 22].
Chromatin Immunoprecipitation (ChIP) assay was performed to analyze interactions between Dnmt1/Tet2 and MMP-9 following the methods routinely performed in our laboratory [14, 22]. Sonicated, cross-linked sample (100~120μg) was incubated overnight with Tet2 or Dnmt1 antibody (Tet2- ab135087, Dnmt1- ab13537, Abcam, Cambridge, MA). Immune-precipitated with non-immune IgG (Cell Signaling, Danvers, MA) samples were used as antibody controls. Protein A/G agarose beads (EMD Millipore, Billerica, MA) were employed to pull down the antibody-chromatin complexes. Protein-associated DNA was recovered by incubating the eluent at 65°C, purified by phenol extraction, and precipitated with ethanol, as reported previously [13, 22, 23].
Immunofluorescence technique was employed to confirm binding of Dnmt1 and MMP-9 using the methods employed routinely [13]. Retinal endothelial cells, grown on coverslips in 5mM or 20mM glucose, in the presence or absence of MnTBAP respectively were fixed with 4% paraformaldehyde, and after permeabilization with 0.2% Triton X-100, blocked with 2% BSA for one hour. The cells were the incubated with anti-rabbit MMP-9 antibody and antimouse-Dnmt1 antibody (catalog #s ab38898 and ab13537 respectively, Abcam). After blocking again with 10% goat serum, they were incubated with anti-rabbit-DyLight 488 (green, MMP-9) and anti-mouse-Texas Red (red, Dnmt1) secondary antibodies (catalog #s TI-2000 and DI-1488 respectively, Vector Lab, Burlingame, CA). The cells were then washed, mounted with DAPI containing medium (blue; H-1500; Vector Lab), and imaged at 40X magnification using Zeiss ApoTome fluorescence microscope (Carl Zeiss Inc., Germany).
Activities of Tet and Dnmt were estimated in nuclear fractions using EpiQuik™ Epigenase™ 5mC-Hydroxylase TET Activity/Inhibition and DNA Methyltransferase Activity/Inhibition kits respectively (EPIGENTEK) [16, 24].
Real-time quantitative PCR, SYBR Green-based, was performed to quantify the gene transcripts following the amplification program which included 50°C for 2 minutes and 95°C for 10 minutes-holding satge, 40 cycles of denaturation at 95°C for 15 seconds, annealing and extension at 60°C for 1 minute, and 72°C for 5 minutes. Single peak in the melting curve was used to validate the reaction products. Cycle threshold (Ct) values from β-actin (bovine- Fwd CCTCTATGCCAACACAGTGC and Rev CATCGTACTCCTGCTTGCTG; mouse Fwd- CCTCTATGCCAACACAGTGC and Rev- CATCGTACTCCTGCTTGCTG) were used to normalize the values. Relative fold changes were calculated by considering the values obtained from cells in 5mM glucose or normal mice as 1 [13, 22, 23].
MMP-9 activity was quantified using an ELISA kit (Amersham, Buckinghamshire, UK), and the final product was quantified spectophotometrically at 405nm using the specific peptide chromogen [7, 14].
Statistical analysis
Each measurement was made in duplicate, and assay was repeated 3 or more times. Data are expressed as mean ± SD. Statistical analysis was performed using the nonparametric Kruskal-Wallis test followed by Mann-Whitney test, and p< 0.05 was considered statistically significant.
Results
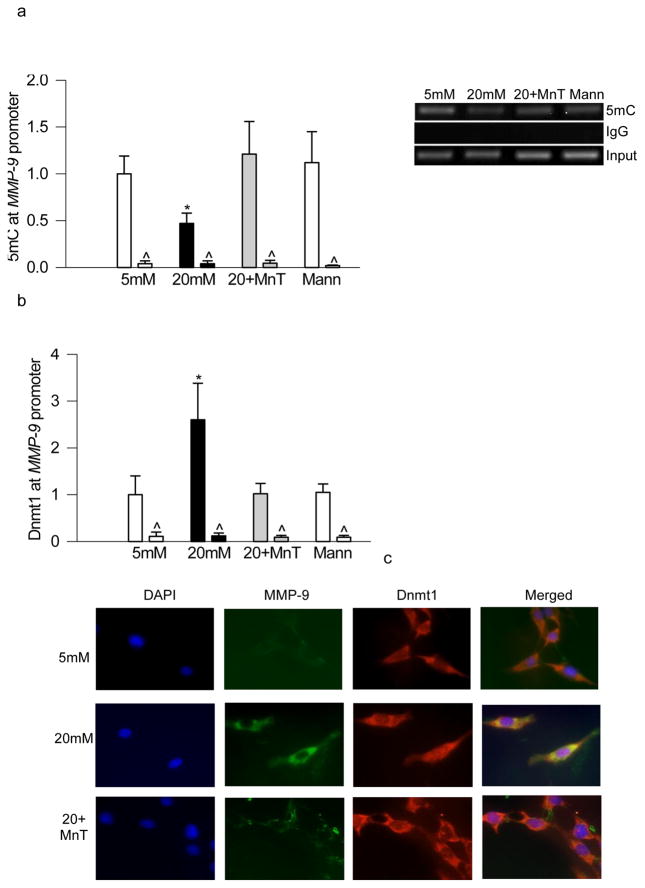
High glucose exposure of retinal endothelial cells, as expected, decreased 5mC levels in the MMP-9 promoter region [16], and regulation of oxidative stress by MnTBAP ameliorated glucose-induced decrease in 5mC levels; the values obtained from cells incubated in high glucose in the presence and absence of MnTBAP were significantly different from each other (Figure 1a). In the same samples, glucose-induced increased Dnmt1 binding at the MMP-9 promoter region was also attenuated by MnTBAP (Figure 1b). However, supplementation of MnTBAP in 5mM glucose medium did not alter either 5mC or Dnmt1 binding. Effect of MnTBAP on DNA methylation was further confirmed by immunoflurorescnce technique; as shown in figure 1c, glucose-induced increased co-localization of Dnmt1 and MMP-9 was significantly decreased by supplementation of MnTBAP in the incubation medium.
Figure 1.
Regulation of oxidative stress prevents glucose-induced decrease in 5mC and increased Dnmt1 binding at the MMP-9 promoter in retinal endothelial cells. Endothelial cells were incubated in 5mM or 20mM glucose for 4 days in the presence or absence of 250μM MnTBAP. (a) 5mC at MMP-9 promoter was quantified using methylated DNA immunoprecipitation kit. (b) Dnmt1 binding was quantified in the crosslinked cells, and IgG was used as a negative antibody control (indicated as ^). Values are represented as mean ± SD from 4–5 samples in each group. (c) Co-localization of Dnmt1 and MMP-9 was performed by immunofluoresence techique using anti-rabbit-DyLight 488 (green) as the secondary antibodies for MMP-9 was anti-mouse-Texas Red (red) for Dnmt1 anti-mouse-Texas Red (red). Cells mounted with DAPI containing medium (blue) w. 5mM and 20mM=cells incubated in 5mM and or 20mM glucose respectively; 20+Mn= cells in 20mM glucose in the presence of MnTBAP; Mann=cells in 20mM mannitol. *p< 0.05 compared to 5mM glucose.
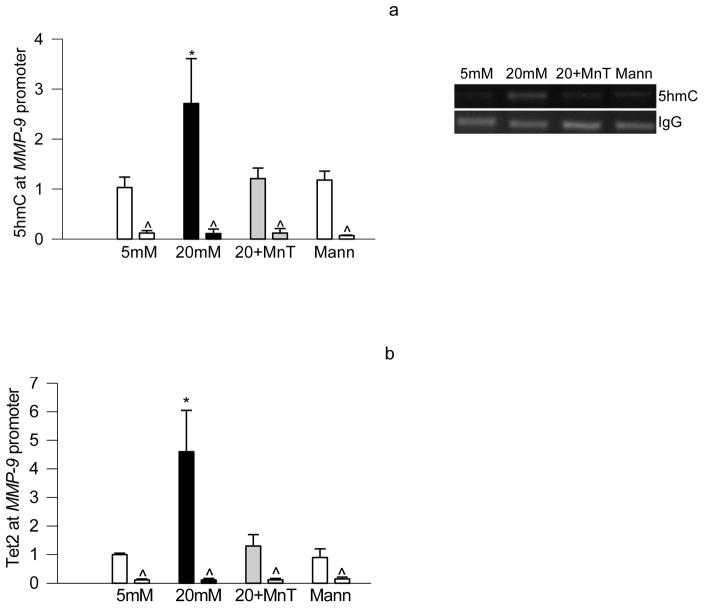
Since DNA methylation is a dynamic process and Tet can oxidize methyl cytosine to hydroxymethyl cytosine, and our recent study has shown that 5mC produced at MMP-9 promoter in hyperglycemic milieu undergoes hydroxymethylation [16]; the effect of MnTBAP on hydroxylmethylation of MMP-9 promoter was determined. To investigate the role of oxidative stress on hydroxymethylation of MMP-9 promoter, 5hmC levels were quantified in the cells exposed to high glucose medium supplemented with MnTBAP. As shown in figure 2, MnTBAP attenuated glucose-induced increase in hydroxymethylation, and this was accompanied by a significant decrease in Tet2 binding at the MMP-9 promoter. The values obtained from cells incubated in the presence of MnTBAP in high glucose medium were similar to those obtained from the cells incubated in 5mM glucose or 20mM mannitol.
Figure 2.
Inhibition of glucose-induced increase in superoxide levels prevents increase in hydroxymethylation in the MMP-9 promoter region. Cells incubated in high glucose in the presence of MnTBAP were analyzed for (a) 5hmC using by hMeDIP immunoprecipitation technique, (b) and Tet2 binding by ChIP technique. Each measurement was made in duplicate in cells from 4–5 different preparations, and each value is presented as mean ± SD from 4–5 different cell preparations, with each measurement made in duplicate. 5mM and 20mM=cells incubated in 5mM and or 20mM glucose respectively; 20+Mn= cells incubated in 20mM glucose in the presence of MnTBAP; Mann=cells in 20mM mannitol. *p< 0.05 compared to 5mM glucose.
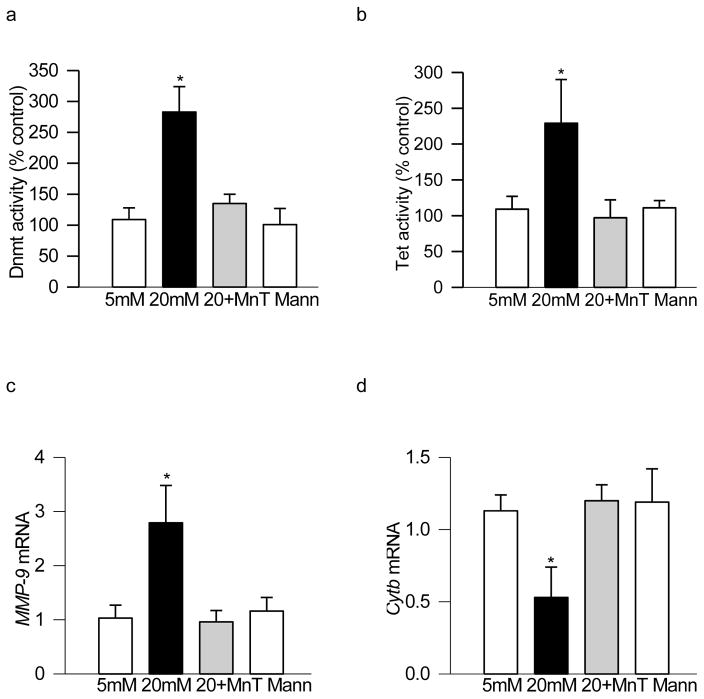
To investigate if oxidative stress is implicated in regulating the enzymes responsible for DNA methylation-hydroxymethylation, the effect of MnTBAP on glucose-induced activation of DNA methylating and hydroxymethylating enzymes was determined. Addition of MnTBAP in the medium prevented glucose-induced increase in both Dnmt and Tet activities (Figure 3a & b). In the same cell preparations, MnTBAP ameliorated increase in MMP-9 transcripts (Figure 3c) experienced by the cells in high glucose conditions, and also prevented mitochondrial DNA damage (Figure 3d); mRNA expression of mitochondrial DNA (mtDNA)-encoded cytochrome b (Cytb) was similar in the cells incubated in high glucose in the presence of MnTBAP and in the cells incubated in normal glucose or in 20mM mannitol (Figure 3d).
Figure 3.
DNA methylation machinery is modulated by oxidative stress: The activities of (a) Dnmt1 and (b) Tet, and mRNA levels of (c) MMP-9 and (d) mtDNA-encoded Cytb were quantified in retinal endothelial cells incubated in 5mM or 20mM glucose for 4 days in the presence or absence of 250μM MnTBAP. The enzyme activities and mRNA values obtained from the cells incubated in 5mM glucose alone are considered as 100% and 1 respectively. Values are represented as mean ± SD from 3–5 samples in each group, with each measurement made in duplicate. 5mM and 20mM=cells incubated in 5mM and or 20mM glucose respectively; 20+Mn= cells in 20mM glucose in the presence of MnTBAP; Mann=cells in 20mM mannitol. *p< 0.05 compared to 5mM glucose.
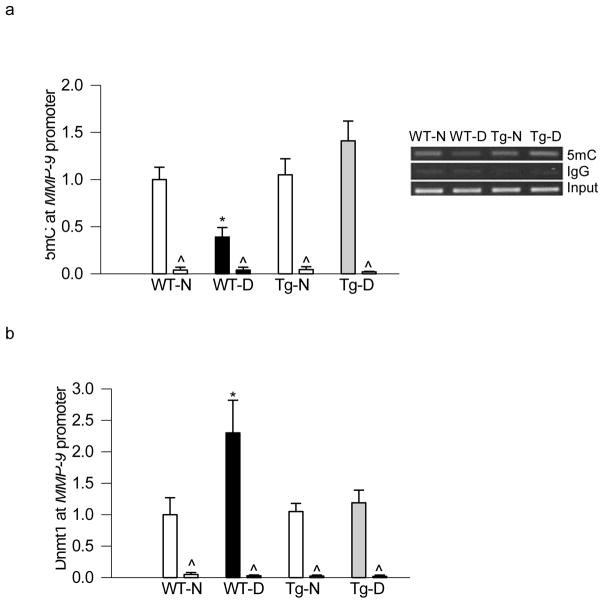
To further confirm our in vitro results in the in vivo model, retina from SOD2-Tg mice, diabetic for 6 months, was analyzed. The levels of 5mC in the MMP-9 promoter region, as expected, were 50% lower in the retina from diabetic mice compared to their-age-matched normal mice, and overexpression of SOD2 prevented diabetes-induced decrease in 5mC (Figure 4a). In the same retina samples, overexpression of SOD2 also prevented increase in Dnmt1 binding in the same MMP-9 promoter region (Figure 4b), and the values obtained from the retina of wildtype diabetic mice were significantly different from those obtained from SOD2-Tg diabetic or normal mice.
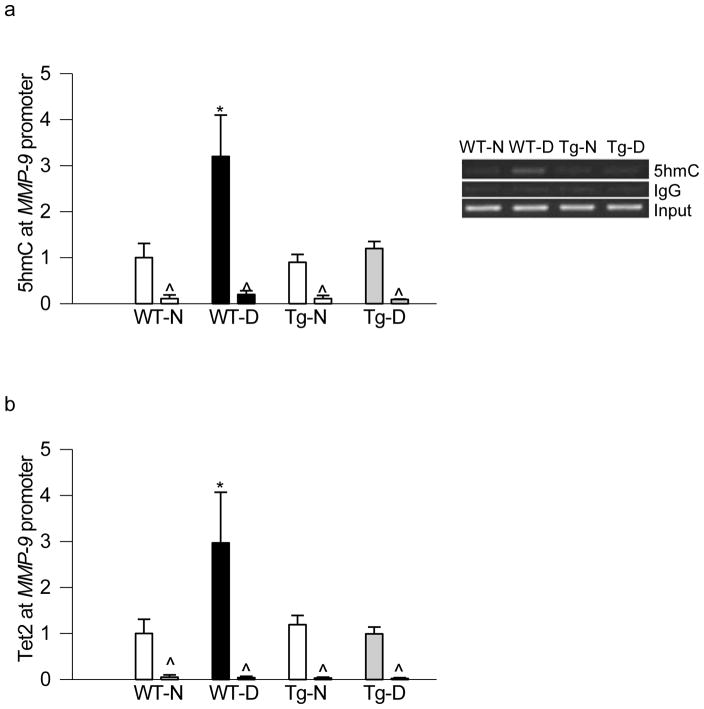
Figure 4.
Overexpression of SOD2 in mice prevents diabetes-induced alteration in retinal MMP-9 promoter methylation. Retina from mice overexpressing SOD2, or their wildtype control mice, diabetic for ~6 months, was analyzed for (a) 5mC levels in the MMP-9 promoter region using methylated DNA immunoprecipitation kit. (b) Dnmt1 binding at MMP-9 promoter region was quantified by ChIP technique. Measurement were made in duplicate in 5–7 animals in each group, and the values are represented as mean ± SD. WT-N and WT-D= wildtype mice, normal and diabetes respectively; Tg-N and Tg-D= SOD2 overexpressing mice, normal and diabetes respectively. *p< 0.05 compared to WT-normal.
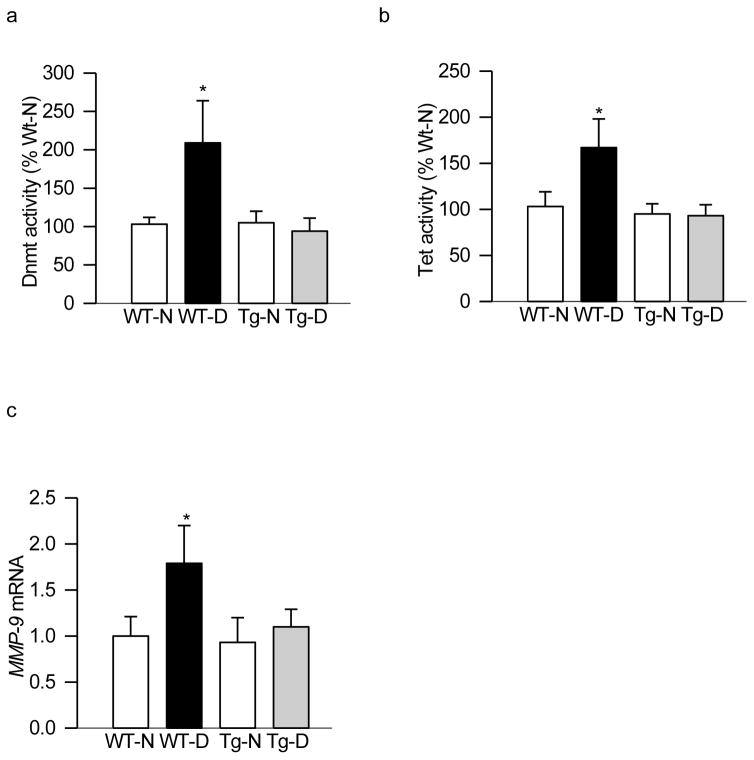
Consistent with our in vitro results, regulation of mitochondrial superoxide levels by overexpression of SOD2 also protected increase in 5hmC levels seen in the MMP-9 promoter in diabetic conditions. In the same animals, the binding of Tet2 at the MMP-9 promoter was also significantly lower in SOD2-Tg diabetic animals compared to age-matched wildtype diabetic animals (Figure 5a & b). Similarly, overexpression of SOD2 also protected diabetes-induced increase in enzymatic activities of Dnmt and Tet, and ameliorated increase in MMP-9 gene expression; values obtained from SOD2-Tg diabetic mice were similar to those obtained from wildtype or SOD2-Tg normal mice (Figure 6a–c).
Figure 5.
Regulation of superoxide radicals in mice ameliorates increase in 5hmC and Tet2 binding at retinal MMP-9 promoter, induced by diabetes. Retina from diabetic mice overexpressing SOD2, or their wildtype control mice, was analyzed for (a) 5hmC levels at MMP-9 promoter (hMeDIP immunoprecipitation method) and (b) Tet2 binding (ChIP technique). Measurement were made in duplicate in 4–6 mice in each group, and mean ± SD are plotted. WT-N and WT-D= wildtype mice, normal and diabetes respectively; Tg-N and Tg-D= SOD2 overexpressing mice, normal and diabetes respectively. *p< 0.05 compared to WT-normal.
Figure 6.
Overexpression of SOD2 in mice prevents diabetes-induced activation of DNA methylation-hydroxymethylation enzymes. Retina from wild type and SOD2 overexpressing normal and diabetic mice was analyzed for (a) Dnmt and (b) Tet activities. (c) In the same retinal samples, gene transcripts of MMP-9 were quantified by SYBR green based qPCR using B-actin as a housekeeping gene. The values are represented as mean ± SD from 4–6 mice in each group, with each measurement made in duplicate. *p< 0.05 compared to WT-normal.
Discussion
Diabetic environment induces many metabolic abnormalities in the retina and its vasculature, e.g., inflammation and oxidative stress are elevated and protein kinase C is activated [1, 2, 25]. Retinal mitochondria become dysfunctional, their membranes leak out cytochrome C into the cytosol, and mtDNA is damaged. Amelioration of oxidative stress by pharmacological means or by genetic manipulation in animal models protect retina and its vasculature from such damage [2, 26]. Many genes implicated in the development of diabetic retinopathy are epigenetically modified, and DNA methylation-hydroxymethylation machinery is activated [2, 16]. We have shown recently that the transcription of MMP-9, an enzyme implicated in retinal mitochondrial damage in the development of diabetic retinopathy, is regulated by a dynamic DNA methylation process [16]. Using both in vitro and in vivo models of diabetic retinopathy, here we provide exciting results demonstrating that this DNA methylation of retinal MMP-9 promoter is under the control of oxidative stress. Amelioration of oxidative stress by chemical/genetic approaches regulates retinal MMP-9 transcription in diabetes via modulating DNA methylation of its promoter region, and this is mediated via protecting the diabetes-induced activation of the machinery responsible for maintaining the DNA methylation process. Thus, regulation of oxidative stress, prevents the activation of retinal MMP-9, which, in turn, protects mitochondrial homeostasis, and, ultimately, the development of diabetic retinopathy.
Methylation of the cytosine residues in CpG dinucleotides influences gene activity without changing the DNA sequences, and aberrant DNA methylation is associated with many pathological conditions, including cancer and diabetes [27–29]. Although there is a balance between reactive oxygen species generation and their utilization/neutralization by enzymatic and non-enzymatic antioxidant systems, in pathological conditions, a redox imbalance is created by increased ROS production and/or reduced antioxidant reserve [2, 30]. ROS damage micro- and macromolecules, and in the pathogenesis of diabetic retinopathy, the levels of oxidatively modified DNA (8-hydroxyguanine, 8-OHdG) are elevated in the retina and its capillary cells [31]. Our results presented here show that the regulation of oxidative stress, despite reducing diabetes-induced increased binding of Dnmt1, prevents decrease in 5mC levels at the MMP-9 promoter, suggesting a role of oxidative stress in maintaining DNA methylation levels. In support, others have shown that although H2O2 treatment can increase the binding of Dnmt1 [32], 8-OHdG negatively affects the methylation of adjacent sites, impeding further DNA methylation [33]. This suggests that attenuation of mitochondrial 8-OHdG accumulation by regulating mitochondrial damage could be assisting in ameliorating Dnmt1 binding.
DNA methylation is a dynamic process, and hydroxylation of 5mC can significantly reduce methylation of the target cytosine causing DNA hypomethylation [34]. In diabetes, despite increased binding of Dnmt1 and decreased 5mC levels, retinal MMP-9 transcription is increased and 5hmC levels are elevated [16]. Here we show that regulation of oxidative stress, in addition to affecting DNA methylation of the MMP-9 promoter, also maintains its hydroxymethylation; addition of MnTBAP to the cells in high glucose condition, or overexpression of SOD2 in mice, prevents hyperglycemia-induced increase in 5hmC levels and recruitment of Tet2.
Methylation status of DNA is maintained by a group of enzymes with opposing functions, and the addition of a methyl group from S-adenosyl-methionine to cytosine is catalyzed by Dnmts resulting in formation of 5mC. Although there are three major Dnmts that are enzymatically active in mammals, Dnmt1 and Dnmt3a are the most active isoforms in neurons [35]. In the pathogenesis of diabetic retinopathy, Dnmt1 is the only isoform overexpressed in the retina and its capillary cells [16, 22]. Here we show that retinal Dnmt1 is also under the control of oxidative stress; regulation of mitochondrial superoxide levels by chemical or genetic approaches regulates activation of Dnmt1 seen in hyperglycemic milieu. Consistent with our results, others have shown that oxidative stress affects Dnmts and alters DNA methylation [33]. Hydroxymethyl marks on DNA are actively removed by Tets, and methyl group on the cytosine isoxidized to form, 5hmC [34]. Our data clearly show that, in addition to regulation of DNA methylation of MMP-9 promoter by oxidative stress, MnTBAP supplementation during high glucose incubation of retinal endothelial cells, or diabetic mice overexpressing SOD2, prevents increase in the formation of 5hmC at the promoter. This is accompanied by decrease in the binding of Tet2, suggesting a major role of oxidative stress in hydroxymethylation process. To further strengthen this, we show that the same in vitro/in vivo manipulations of MnSOD also ameliorate diabetes-induced increase in retinal Tet activity. Consistent with this, Tets are activated under high oxygen conditions by alpha ketoglutarate, a cofactor produced in the citric acid cycle [36], and N-acetyl-l-cysteine, a known antioxidant, diminishes global DNA hypomethylation [37].
Thus, regulation of oxidative stress by chemical/genetic approaches, which prevents hyperglycemia-induced accelerated capillary cell apoptosis, and the development of diabetic retinopathy [3, 4], also regulates the DNA methylating- hydroxymenthylating machinery. This prevents epigenetic modifications of the MMP-9 promoter, and helps further in maintaining retinal mitochondrial homeostasis.
Acknowledgments
We thank Mangayarkarasi ThandampallayamAjjeya for her help with the maintenance of animal colony and Dr. Arul J. Duraisamy for immunohistochemical analysis. This study was supported in part by grants from the National Institutes of Health (EY014370, EY017313 and EY022230) and from the Thomas Foundation to RAK, and an unrestricted grant to the Ophthalmology Department from Research to Prevent Blindness. The sponsor had no role in the design or conduct of this research.
Footnotes
Conflict of interest: RAK and YS have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Ethical approval: All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
References
- 1.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 2.Kowluru RA, Kowluru A, Mishra M, Kumar B. Oxidative stress and epigenetic modifications in the pathogenesis of diabetic retinopathy. Prog Ret Eye Res. 2015;48:40–61. doi: 10.1016/j.preteyeres.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanwar M, Chan PS, Kern TS, Kowluru RA. Oxidative damage in the retinal mitochondria of diabetic mice: possible protection by superoxide dismutase. Invest Ophthalmol Vis Sci. 2007;48:3805–3811. doi: 10.1167/iovs.06-1280. [DOI] [PubMed] [Google Scholar]
- 4.Kowluru RA, Odenbach S. Effect of long-term administration of alpha lipoic acid on retinal capillary cell death and the development of retinopathy in diabetic rats. Diabetes. 2004;53:3233–3238. doi: 10.2337/diabetes.53.12.3233. [DOI] [PubMed] [Google Scholar]
- 5.Das A, McLamore A, Song W, McGuire PG. Retinal neovascularization is suppressed with a matrix metalloproteinase inhibitor. Arch Ophthalmol. 1999;117:498–503. doi: 10.1001/archopht.117.4.498. [DOI] [PubMed] [Google Scholar]
- 6.Mohammad G, Kowluru RA. Diabetic retinopathy and signaling mechanism for activation of matrix metalloproteinase-9. J Cell Physiol. 2012;227:1052–1061. doi: 10.1002/jcp.22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowluru RA, Mohammad G, dos Santos JM, Zhong Q. Abrogation of MMP-9 gene protects against the development of retinopathy in diabetic mice by preventing mitochondrial damage. Diabetes. 2011;60:3023–3033. doi: 10.2337/db11-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto T, Akuta T, Tamura F, van Der Vliet A, Akaike T. Molecular mechanism for activation and regulation of matrix metalloproteinases during bacterial infections and respiratory inflammation. Biol Chem. 2004;385:997–1006. doi: 10.1515/BC.2004.130. [DOI] [PubMed] [Google Scholar]
- 9.Tyagi SC, Rodriguez WPA, Roberts AM, Falcone JC, Passmore JC, Fleming JT, Joshua IG. Hyperhomocysteinemic diabetic cardiomyopathy: oxidative stress, remodeling, and endothelial-myocyte uncoupling. J Card Pharmacol Ther. 2005;10:1–10. doi: 10.1177/107424840501000101. [DOI] [PubMed] [Google Scholar]
- 10.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.VaissiPre T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 2010;299:F14–25. doi: 10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra M, Flaga J, Kowluru RA. Molecular mechanism of transcriptional regulation of matrix metalloproteinase-9 in diabetic retinopathy. J Cell Physiol. 2016;231:1709–1718. doi: 10.1002/jcp.25268. [DOI] [PubMed] [Google Scholar]
- 14.Zhong Q, Kowluru RA. Regulation of matrix metalloproteinase-9 by epigenetic modifications and the development of diabetic retinopathy. Diabetes. 2013;62:2559–2568. doi: 10.2337/db12-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chicoine E, Esteve PO, Robledo O, Van Themsche C, Potworowski EF, St-Pierre Y. Evidence for the role of promoter methylation in the regulation of MMP-9 gene expression. Biochem Biophys Res Comm. 2002;297:765–772. doi: 10.1016/s0006-291x(02)02283-0. [DOI] [PubMed] [Google Scholar]
- 16.Kowluru RA, Shan Y, Mishra M. Dynamic DNA methylation of matrix metalloproteinase-9 in the development of diabetic retinopathy. Lab Invest. 2016 doi: 10.1038/labinvest.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majumdar S, Buckles E, Estrada J, Koochekpour S. Aberrant DNA methylation and prostate cancer. Curr Genomics. 2011;12:486–505. doi: 10.2174/138920211797904061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang KA, Zhang R, Kim GY, Bae SC, Hyun JW. Epigenetic changes induced by oxidative stress in colorectal cancer cells: methylation of tumor suppressor RUNX3. Tumor Biol. 2012;33:403–412. doi: 10.1007/s13277-012-0322-6. [DOI] [PubMed] [Google Scholar]
- 21.Santos JM, Tewari S, Goldberg AFX, Kowluru RA. Mitochondria biogenesis and the development of diabetic retinopathy. Free Rad Biol Med. 2011;51:1849–1860. doi: 10.1016/j.freeradbiomed.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra M, Kowluru RA. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2015;56:5133–5142. doi: 10.1167/iovs.15-16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra M, Zhong Q, Kowluru RA. Epigenetic modifications of Keap1 regulate its interaction with the protective factor Nrf2 in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55:7256–7265. doi: 10.1167/iovs.14-15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tewari S, Zhong Q, Santos JM, Kowluru RA. Mitochondria DNA replication and DNA methylation in the metabolic memory associated with continued progression of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2012;53:4881–4888. doi: 10.1167/iovs.12-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank RN. Diabetic Retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 26.Kowluru RA, Santos JM, Mishra M. Epigenetic modifications and diabetic retinopathy. BioMed Res Intl. 2013;2013:635284. doi: 10.1155/2013/635284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H, Liu X, Deng Y, Qing H. DNA methylation, a hand behind neurodegenerative diseases. Front Aging Neurosci. 2013;5:1–16. doi: 10.3389/fnagi.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maghbooli Z, Larijani B, Emamgholipour S, Amini M, Keshtkar A, Pasalar P. Aberrant DNA methylation patterns in diabetic nephropathy. J Diabetes Metab Disord. 2014;13:1–8. doi: 10.1186/2251-6581-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manev H, Dzitoyeva S, Chen H. Mitochondrial DNA: A Blind spot in neuroepigenetics. Biomol Concepts. 2012;3:107–115. doi: 10.1515/bmc-2011-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowluru RA. Mitochondria damage in the pathogenesis of diabetic retinopathy and in the metabolic memory associated with its continued progression. Curr Med Chem. 2013;20:3226–233. doi: 10.2174/09298673113209990029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madsen-Bouterse SA, Mohammad G, Kanwar M, Kowluru RA. Role of mitochondrial DNA damage in the development of diabetic retinopathy, and the metabolic memory phenomenon associated with its progression. Antiox Redox Signal. 2010;13:797–805. doi: 10.1089/ars.2009.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu L, Khor TO, Lee JH, Boyanapalli SS, Huang Y, Wu TY, et al. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011;13:606–614. doi: 10.1208/s12248-011-9300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Q, Ni X. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr Drug Targets. 2015;16:13–19. doi: 10.2174/1389450116666150113121054. [DOI] [PubMed] [Google Scholar]
- 34.Ponnaluri VK, Maciejewski JP, Mukherji M. A mechanistic overview of TET-mediated 5-methylcytosine oxidation. Biochem Biophys Res Comm. 2013;436:115–120. doi: 10.1016/j.bbrc.2013.05.077. [DOI] [PubMed] [Google Scholar]
- 35.Yu NK, Baek SH, Kaang BK. DNA methylation-mediated control of learning and memory. Mol Brain. 2011;4:1–9. doi: 10.1186/1756-6606-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chia N, Wang L, Lu X, Senut MC, Brenner C, Ruden DM. Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics. 2011;6:853–856. doi: 10.4161/epi.6.7.16461. [DOI] [PubMed] [Google Scholar]
- 37.Han ZJ, Song G, Cui Y, Xia HF, Ma X. Oxidative stress is implicated in arsenic-induced neural tube defects in chick embryos. Int J Dev Neurosci. 2011;29:673–680. doi: 10.1016/j.ijdevneu.2011.06.006. [DOI] [PubMed] [Google Scholar]