Abstract
Optogenetics has revolutionized neuroscience by providing means to control cell signaling with spatiotemporal control in discrete cell types. In this review, we summarize four major classes of optical tools to manipulate neuromodulatory GPCR signaling: opsins (including engineered chimeric receptors); photoactivatable proteins; photopharmacology through caging - photoswitchable molecules; fluorescent protein based reporters and biosensors. Additionally, we highlight technologies to utilize these tools in vitro and in vivo, including Cre dependent viral vector expression and two-photon microscopy. These emerging techniques targeting specific members of the GPCR signaling pathway offer an expansive base for investigating GPCR signaling in behavior and disease states, in addition to paving a path to potential therapeutic developments.
Introduction
G-protein coupled receptors (GPCRs) are critical for neuromodulation. GPCRs modulate neuronal excitability by signaling through the heterotrimeric G-protein families (Gs, Gi/o, Gq G12/13, among others), which can couple to channels or enzymes via direct beta-gamma subunit interactions or amplify intracellular signaling pathways (Figure 1). Gq couples to phospholipase C, cleaving phosphatidylinositol into IP3 and diacylglycerol (DAG), mobilizing the release of calcium from intracellular stores. This is known to facilitate depolarization, in addition to synaptic release in terminals, also dependent on calcium. Gi/o signaling via beta-gamma interactions positively regulates G-protein coupled inward rectifying potassium channels (GIRKs) to hyperpolarize the cell, as well as negatively couples to calcium channels to inhibit synaptic release. Gs signaling utilizes coupling to adenylyl cyclase to amplify the levels of cAMP in a neuron, which can influence excitability through cyclic-gated nucleotide channels (CNGA2), or via enzymatic kinase mediated pathways. GPCRs signal through other intracellular pathways through G-proteins and arrestin to modulate neuron and glial activity independent of ionic changes in the cell.
Figure 1.
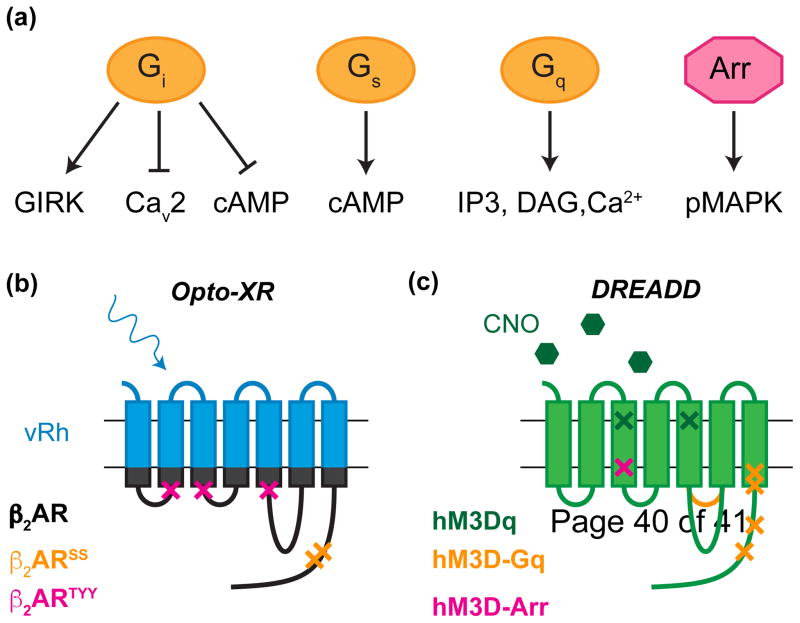
Schematic representations of GPCR signaling in neurons. (a) GPCRs signal through both heterotrimeric G-proteins and arrestins. Gi activates G protein-coupled inwardly-rectifying potassium channels (GIRK) to hyperpolarize the cell and inhibits voltage gated calcium channels (CaV2) to inhibit synaptic release. Gi and Gs respectively inhibit or amplify cyclic AMP (cAMP) production. Gq couples to phospholipase C to generate IP3 and DAG which in turn regulate intracellular calcium stores. Arrestin signaling is predominantly mediated through phosphorylation of MAP kinases. (b) Schematic representation of Opto-β2AR depicting mutations for G-protein (Opto-2AR-SS) or arrestin (Opto-β2AR-LYY) bias (c) Schematic representation of Designer Receptors Exclusively Activated by Designer Drugs depicting mutations for G-protein (hM3D-Arr) or arrestin (hM3D-Gq) bias. (IP3, Inositol triphiosphate; MAPK, mitogen activated protein kinase)
Many neurotransmitters activate GPCRs that initiate intracellular pathways with complex temporal, and spatial effects within a neuron. In the brain, cellular, receptor, and signaling heterogeneity can cloud discrete conclusions. Therefore, GPCR-based discoveries have slowly matured, due to the intrinsic limitations of pharmacological manipulations. These approaches lack both receptor subtype specificity and cellular resolution. Though development of optogenetics provided neuroscientist with the ability to probe neural circuits with discrete spatiotemporal control, most optogenetic applications have focused on using light-sensitive ion channels. To achieve more naturalistic control of neuromodulator signaling, GPCR-based optogenetic and chemogenetic tools were created. These techniques allow for the selective interrogation of specific signaling and cell types in neural circuits with high spatial and temporal resolution in vivo. These tools (used independently or in a multiplexed fashion) offer powerful means for neuromodulation in the brain.
Here we provide a description of recent advances in the development of genetically targeted tools to interrogate neuromodulation, more specifically GPCR signaling in the brain (outlined in Table 1). This review is not meant to be exhaustive; here we focus on recently developed approaches used to characterize GPCR bias, or define either specific endogenous GPCR intracellular pathways, each with potential to be used in the brain to aid in the development of novel therapeutic treatments. For more comprehensive reviews, see Ref. [1–6]. We also compare both optical and chemical techniques. Additionally, we will discuss technological advancements for utilizing these tools in vivo. Finally, we discuss future development and application of optogenetic and chemogenetic tools.
Table 1.
Recently developed optogenetic tools
| Tool | Mechanism | Targeted pathway | Activation | Deactivation/Dissociation |
|---|---|---|---|---|
| Opsins [14*,143] | Conformational change | G-protein and Arrestin | NIRW-UV | Dark or bistable |
| Opto-Chimeras [1,20] | Conformational change | G-protein and Arrestin | Red-Blue | Dark or bistable |
| BLUF photoreceptors [35,37] | Conformational change | Cyclic nucleotides | Blue | Dark |
| LOV domains [42,44,48] | Proximity | P13K | Blue | Dark |
| Cryptochromes [3,51] | Conformational change Proximity |
GTPase, MAPK Kinase, GTPase, PI3K, MAPK, transcription in vivo |
Blue Blue |
Dark Dark |
| Phytochromes [56,59,144] | Conformational change | cAMP | NIRW | NIRW |
| Caged Ligands [145,146] | Proximity Uncaging |
ERK, PI3K, GTPase GPCR |
Red UV |
NIRW Irreversible |
| Photoswitchable molecules [4,67] | Uncaging | GPCR, DAG | UV (Red) | Green (Blue) |
| Photoswitchable tethered ligands [73,74] | Uncaging | GPCR | UV | Green |
| Resonance Energy Transfer (RET) [75] | Proximity, Sensor | kinase activity, 2nd messengers, inter-and intramolecular interactions | Red-Blue | Target protein dependent |
| Genetically encoded calcium indicators (i.e. GCaMP) [5,82] | Sensor | Calcium | Red and Green | Calcium dependent |
| Fluorescent Protein Exchange (FPX) [80] | Sensor | Calcium, PIP2, PKA activation, ERK activation | Red or Green | Target domain dependent |
Optogenetics: control or measurement of cell signaling with light
Optogenetics describes the technology in which cells are modified genetically to express light sensitive proteins allowing for either measuring or controlling cellular signaling with light. Optogenetics debuted in neuroscience as the direct activation of ion channels to regulate neuronal excitability with discrete spatiotemporal control [7]. Current advances in optogenetics target specific members of canonical GPCR signaling pathways with both natural and engineered optically sensitive proteins. Additionally, endogenous receptors are targeted by photoactivatable ligands, a method called photopharmacology [4,8]. Furthermore, other optogenetic approaches utilize genetically encoded reporters of protein-protein interactions or the production of second messengers [9].
Opsins
Naturally occurring Opsins
Opsins are photosensitive GPCRs bound to chromophores. The first GPCR specific optogenetic tools were naturally occurring opsins [10]. Animal opsins are advantageous because they do not require exogenous chromophores (called retinals) to be added to the brain. Naturally occurring opsins are diverse in their spectral and signaling properties, often requiring little mutation or special tuning.
Vertebrate visual opsins: Visual opsins are conopsin and rhodopsin, the latter is classified as a weak candidate for optogenetics due to slow kinetics and bleaching. Conopsins have spectrally diverse excitation spectra. Developing tools from conopsin could in theory allow for the combinatorial activation of two neuronal cell populations. In the eye, conopsins couple to Gα transducin, part of the Gi/o family of G-proteins, and thus optogenetic tools based on conopsin will signal to Gi/o. Thus far, all human conopsins have been characterized in neuronal cultures, with some work in brain tissue [11]. The use of human blue conopsin (vLWO; vertebrate long wavelength opsin) and mouse red conopsin (vSWO; vertebrate short wavelength opsin) for modulating neural circuits has been validated brain slices [12]. Photostimulation of conopsin modulates GIRK currents, a typical GPCR coupling event in neural circuits. Additionally, unlike rhodopsin, conopsins can be repeatedly activated without a noticeable desensitization of the response [12]. Recently, conopsin hyperpolarized DRN neurons in brain slice, and a chimera (see below) expressed in the DRN was able to modulate anxiety behavior [12].
Melanopsin, a non-visual opsin, has also successfully engaged neural circuits [13]. Melanopsins is activated by blue light, but unlike the visual opsins, are bistable and thus can be deactivated by yellow light. Though melanopsin has the potential to couple Gi/o to regulate GIRK channels in heterologous systems, when examined in neurons it is almost exclusively Gq-coupled. Recently two variants of melanopsin have been characterized for transient and sustained Gq activation [14*]. Stimulation of ectopic melanopsin in pyramidal neurons mimics Gq modulation of channels [15].
Gs signaling can be activated by photo-stimulating certain non-mammal opsins. Jellyfish opsin signals through Gs to induce translocation of adenylyl cyclase, increase the production of cAMP, and increase phosphorylation of ERK [16]. A natural guanylyl cyclase fused to an opsin, was recently discovered in fungus [17]. This cyclase, termed Guanylyl Cyclase Rhodopsin (BeCyclOp), does not produce cAMP, unlike previously engineered guanylyl cyclase optogenetic tools. BeCyclOp has a functional spectrum broader than vertebrate opsins, with maximum cGMP production in green (530nm) light and low activity in red and violet light. This cyclase was also functionally expressed in c. elegans muscle cells demonstrating activity comparable to photo-stimulation from channel-based optogenetics [18*].
Opto-XRs
GPCR signaling in neural circuits may also be modulated though intracellular signaling that may not be dependent solely on generic Gα subunit coupling dynamics. Therefore, to more closely mimic endogenous signaling, chimeric optogenetic tools have been engineered using opsins with intracellular loops and C-terminal tail of GPCRs endogenously expressed in the central nervous system. The first of this family of “Opto-XRs” were adrenergic receptors Opto-α1AR and Opto-β2AR (Figure 1b). [19,20]. The full profile of Opto-β2AR signaling was recently validated as mimicry of most of the endogenous receptor’s properties. This study was also the first to express Opto-β2AR in vivo to demonstrate real time behavioral responses in endogenous neural circuits (anxiogenesis) [21**,22**]. Additionally, the first prototypes of G-protein and arrestin biased chimeras have been developed and characterized in vitro [21**]. This study reported diverse signaling dynamics were possible with Opto-XRs mutated either in the canonical GPCR DRY motif (Optoβ2-LYY, Addgene), known to be involved in G-protein coupling [23] or the c-terminal serines (Optoβ2-SS, available in Addgene) known to be phosphorylation sites of G-protein coupled receptor kinase (GRK; Figure 1b)[24]. These prototypes differed in canonical ERK signaling, densensitization and internalization patterns. However, future studies and additional prototypes are warranted in this regard, as crystal structures, critical residues for G-protein and arrestin bias, and interactions continue to be identified [25].
Another chimera, Opto-A2AR, from bovine rhodopsin chimera and the Adenosine 2A receptor, was also characterized in vitro and in vivo [26*]. Opto-A2AR mimicked endogenous A2AR by increasing cAMP production and differentially recruiting phosphorylation of only CREB in the hippocampus and only MAPK in the nucleus accumbens (NAc). In the same behavioral paradigm, stimulation of Opto-A2AR in the hippocampus impaired memory consolidation, while stimulation in the Nac did not affect memory but increased locomotor activity. This demonstrates that Opto-A2AR mimics differential A2A signaling in endogenous brain regions which lead to also different behaviors [26*].
OMOR, comprised of is rat rhodopsin with intracellular components of the mu opioid receptor (MOR), mimics endogenous opioid signaling in vitro through Gi/o protein and arrestin mediated signaling, by inhibiting production of cAMP, coupling to GIRK and stimulating ERK phosphorylation. In neuronal slices, saturating concentrations of mu agonist, DAMGO, prevented a subsequent response to photo-stimulation of OMOR-induced GIRK currents suggesting this chimeric receptor accessed the same intracellular pools of downstream effectors. Lastly, OMOR also mimicked MOR’s ability to induce either a preference or aversion depending on the endogenous neural circuit where it was expressed [27**]. This study comprehensively validated the use of OMOR as a proxy for MOR a variety of preparations.
Additional, chimeric GPCRs have been engineered with melanopsin and conopsins and have been utilized in vivo [12,15,28*,29]. Opto-mGluR6, which is human melanopsin with the intracellular components of metabotropic glutamate receptor mGluR6, was used to restore vision through ON bipolar cells in blind mice [28*]. Other non-rhodopsin chimeras are partial chimeras that contain only some of the intracellular domains serotonin receptors. Herlitze and colleagues used the C-terminal tail of serotonin receptors to target opsins to the appropriate signaling domains. This method was also used to create tools from vertebrate conopsins (Gi/o coupled) and human melanopsin (Gq coupled) with the c-tail of 5HT1A and 5-HT2C, respectively. Both tools modulated dorsal raphe circuits involved in anxiety behavior. Recently, another partial melanopsin-serotonin chimera, created only with the intracellular loops of 5-HT2A, was reported to transiently hyperpolarize cells, but unreliable expression prevented quantification of the effects [15].
One exciting possibility with these chimeric Opto-XR approaches is the ability to mimic spatial and temporal properties of neuromodulator signaling at synapses or in vivo. For example, tuning the amount of response via a photo-stimulation event to mirror the release and activation kinetics of a monoamines or neuropeptides will allow us to better understand how these signals are transmitted in time, over small distances, in genetically defined cell types, and neural circuits. In addition, future approaches to define the 2-photon emission spectra of these opsin tools will allow for more advanced imaging studies in vitro in brain sections alongside sensing of signal transduction (i.e GCaMP, or FLIM-based methods; see below). This would allow the investigator to perturb signaling in a native system while capturing neuronal ensemble dynamics, or GPCR signaling in real time. Advances along these avenues are likely to occur as additional Opto-XRs and native GPCR opsins become characterized and validated side-by-side against their native receptor counterparts. Additional work will be needed to validate the expression and localization profiles of these non-native receptor tools, and they may not recapitulate endogenous GPCR trafficking, and may need chaperone sequences added in some cases. These photo-sensitive GPCR constructs could also be used for subcellular optogenetics to investigate bias based on GPCR localization. Localization of signaling is now at the forefront of GPCR research. Gautam et al. demonstrated that Gβγ subunits not only translocate to intracellular membranes upon GPCR activation, but regulate cytoplasmic calcium concentrations [30,31]. Additionally, there is recent and increasing evidence for the sustain propagation of G-protein-mediated signaling within endosomes [32,33]. Photoactivatable proteins are another powerful tool to investigate subcellular GPCR signaling cascades.
Photoactivatable Proteins
In addition to activation of GPCRs, there are other optogenetic tools for the activation/inhibition of 2nd messengers. This subset of tools includes unmodified proteins and engineered/chimeric tools containing naturally derived domains from non-mammalian species. These tools regulate downstream signaling through two mechanisms: allostery and proximity (Figure 2). For a more comprehensive review of these tools, see Ref. [3,34].
Figure 2.
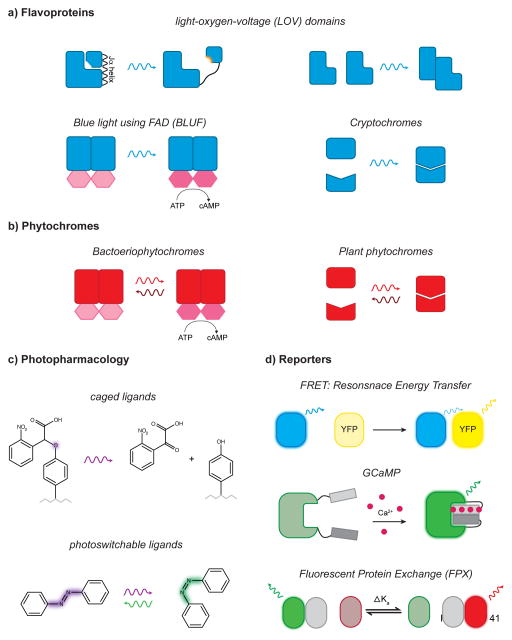
Schematics of Non-GPCR optogenetic tools (a) Blue light sensitive flavoproteins induce conformational changes (LOV and BLUF domains, left panel) or protein interactions (LOV domains and cryptochromes, right panel) (b) Red sensitive phytochromes induce both conformational changes bacteriophytochromes, left panel) or photoswitchable protein interactions (plant phytochromes, right panel). (c) Photopharmacology techniques: peptides may be caged by the addition of a photolabile carboxynitrobenzyl moiety (CNB) to tyrosine (top). Photoswitchable ligands provide reversible caging through photoisomerization of azobenzene (bottom). (d) Ratiometric optogenetic reporters: FRET sensors report protein interactions (top). GCaMP, is a genetically encoded calcium sensor (middle). Fluorescent Protein Exchange sensors rely on the change of affinity of a fluorescence enhancing monomer (bottom). (AsLOV2, Avena sativa phototropin 1 LOV domain; Bphy, hodobacter sphaeroides bacteriophytochrome domain; CIB1, CRY-interacting bHLH1 (helix-loop-helix 1); CRY2, cryptochrome 2; CFP, cyan fluorescent protein; NIRW, near infrared window; PIF, phytochrome interaction factor; PHY, phytochrome domain; YFP, yellow fluorescent protein)
Flavoproteins
Flavoproteins are a diverse set of proteins that contain blue light sensing domains that use either flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) chromophores, both produced in mammalian cells. These include blue light using FAD (BLUF), light-oxygen-voltage (LOV) domains, and cryptochromes (Figure 2a). Flavoprotein domains used for optogenetic tools either initiate enzymatic activity, dimerize, or change in conformation, all in response to light.
Photoreceptors containing BLUF domains modulate cyclic nucleotide production (for a complete review on tools to modulate cAMP signaling see Ref. [35]); Pioneering studies used naturally derived flagellate Photoactivated Adenylyl Cyclase (PACα) to modulate behavior in non-mammalian model organisms [36–39]. Recently, investigators used PAC in rodent neuronal cultures to define the cAMP-PKA pathway that modulates axonal branching, a study capitalizing on the subcellular spatiotemporal control of optogenetics [40]. BLUF domains are the primary way to optogenetically manipulate cAMP aside from GPCRs, though the precise mechanism of activation is unknown. Excitingly, the crystal structure of a smaller version of PAC was characterized from cyanobacteria Oscillatoria acuminate (OaPAC), providing insights on its mechanism of light activation [41**].
LOV domains uniquely offer two mechanisms for tool development, conformational change and dimerization (for review see Ref. [42]). Light induces conformational change in the AsLOV2 domain, excluding the unfolding of the Jα helix, revealing the c-terminus [43]. In the “unmasking” approach, enzymatic or binding domains are fused to the c-terminus [42] (Figure 2a, upper left). Unmasking was used to design a photoactivatable GTPase (PA-Rac1), which was used in brain with some success (NAc cocaine) [44,45]. Recently, AsLOV2 was also used as the foundation for light-induced protein dissociation, an approach called LOV2 trap and release of protein (LOVTRAP). The creators of LOVTRAP generated a small protein (ZDark) with high affinity to the coiled Jα helix, thus protein association only occurs in the dark. In the proof of concept study, LOVTRAP was used to sequester proteins from their signaling domain, but this approach can be extended to diffusible partners [46**]. Dimer association can be achieved with the Vivid (VVD) LOV domains that do not contain the Ja-helix [47,48]. Recently, VVD domains were tuned to create Magnets, a family of photoswitches with dissociation constants ranging on the time scale of seconds to minutes. The authors demonstrated the utility of magnets by creating a photoactivatable phosphatidylinositol 3-kinase with the ability to change the morphology of COS-7 cells [49**].
Cryptochromes are another class of proteins able to create dimers. CRY2-CIB1 heterodimers are widely used in optogenetic tools to modulate protein-protein interaction and regulate protein localization [50,51]. CRY2-CIB1 technology has also been used to inhibit G-protein signaling by recruiting regulator of G-protein signaling 4 (RGS4) to the plasma membrane, providing both the groundwork for tools to optically control G-protein activation with subcellular precision, and new insights into GPCR localized signaling [52**]. Interestingly, CRY2 domains also have the ability to form homodimers. This has been used recently to develop CRY2olig: Light Induced Co-clustering (LINC) which can be used as a mode of activation or inhibition through sequestration. Limitations to LINC are similar to those of co-immunoprecipitation, for the binding partners of the target protein may be sequestered as well. Another consideration is that oligomerization through LINC currently does not have the capability to localize clustering to a specific subcellular domain.
Phytochromes
Tools engineered with phytochromes enhance temporal control of signaling because they are bistable. They are sensitive to both red and neared infrared wavelengths of light (650–750nm). Near red wavelengths are ideal for in vivo models due to the deceased amount of photo toxicity and increase penetrance in tissue [53]. However, red-shifted proteins do have the risk of thermal activation [54]. Phytochromes are derived from plant, fungi and bacteria; and like LOV domains they can regulate signaling through conformational changes and dimerization (Figure 2b).
Bacteriophytochromes are an extremely attractive platform for in vivo applications since not only are they the most sensitive to far red wavelengths, but their chromophore (biliverdin) is present in all mammalian cells [53,55*]. Bacteriophytochromes are enzymatic photoreceptors. Though the exact details of conformational changes that lead to the activation of bacteriophytochromes is unknown, two groups have used crystal structures to engineer optogenetic tools to regulate cyclic nucleotides in eukaryotic cells. Light-activatable phosphodiesterase (LAPD) was based on structure similarities of the regulatory domains of human phosphodiesterase, PDE2, and the light sensing domain of a bacteriophytochrome histidine kinase, PaBPhy. Fusion of the phosphodiesterase and light sensitive domains of these proteins generated a red light-activatable phosphodiesterase that deactivates in far red light [56**]. LAPD was functional in whole zebrafish embryos, but has yet to be used in a behavioral assay. Also guided by structure, Ryu et al. [55*] generated a near infrared window light activated adenylyl cyclase (Ilac) by fusing domains of a bacteriophytochrome diguanylate cyclase and bacterial adenylyl cyclase. The authors used this tool to induce locomotion in wildtype c. elegans through the production of cAMP, an experiment not possible using PAC, due to confounding blue light avoidance in wildtype in c. elegans [38,57].
Optogenetic tools derived from plant phytochromes take advantage of the heterodimerization of the PHY domain with PIF3 or PIF6 [58,59]. These domains can be used to associate known interacting partners, or to translocate a protein of interest to its functional intracellular domain. Phytochrome heterodimers may be preferred over flavoproteins due to their red-shifted sensitivity; however, the chromophore PCB is not endogenous in mammalian cells, limiting its application in neuroscience. Additional genetic engineering would be required to enable cells to synthesize PCB [60]. Phytochromes have been used to generate optogenetic tools for the regulation of phosphoinositides and ERK [61,62]. It is likely that future iterations of phytochrome domains will begin to be used in cultured neurons and in brain tissue as they become refined and more well accepted. Phytochromes offer several advantages for dissecting subdomains of neuronal signaling, and for uncovering the complexity of the intracellular compartments involved in neuromodulation.
Photopharmacology
Studies using classic pharmacological approaches are limited: drugs cannot target a subset of constitutively expressed proteins with spatiotemporal precision. However, spatiotemporally precise drug delivery can be achieved by integrating optical methods, an approach called photopharmacology (Figure 2c). Photopharmacology offers subcellular resolution while potentially preserving properties of the native receptors, including activation and deactivation kinetics, trafficking, and levels of expression [63–65].
For several years, neuroscientists have practiced photopharmacology with caged-ligands [66]. Caged ligands contain a photosensitive moiety that render the ligand inert. These ligands become biologically active only milliseconds after the UV photolysis. Recently, Banghart and colleagues from the Sabatini group have used this technology to target opioid receptors. Endogenous opioid agonists were modified at the terminal tyrosine with a photoactivatable chromophore. These ligands offered the spatiotemporal control needed to study neuropeptide signaling in brain tissue [64] (Figure 2c). In a later study, the same group caged opioid antagonist naloxone to study the deactivation kinetics of opioid signaling in vitro [65].
Though photo-caging is a powerful tool for delivering ligands, it is largely irreversible in most instances. Reversible activation of ligands is achieved through the use of proteins containing photolabile domains, also known as photoswitches. The most common photoswtich used is azobenzene, originally sensitive to UV light; red shifted versions have developed to allow for use in vivo [67,68*] (Figure 2c). A photoswitchable small molecule mu opioid agonist fentanyl has also been developed as well using this approach [69]. Diffusible photoswitches have also been developed to allosterically modulate metabotrophic glutamate receptors [70,71**]. Additional, neuropeptide photo-ligand mimics are in development and offer exciting extensions of this early work. These modified neuropeptides, however, have yet to be applied in vivo. Azobenzene technology can also be extended to second messengers. Recently, photoswitchable DAG was developed and used to control synaptic transmission in hippocampal mouse slices and c. elegans in vivo, illustrating the potential of photoswitches in modulate specific signaling pathways [72*].
Diffusible photosensitive ligands allow for spatial activation, but, like traditional pharmacological methods, lack genetic specificity. Genetic targeting can be achieved with Photoswitchable tethered ligands (PTL). PTLs are photosensitive ligands that will covalently bind to an engineered protein [4]. This approach was developed for metabotropic glutamate receptors (LimGluR), but could be translation to another GPCRs [73]. Although engineering GPCRs for PTL requires minimal mutation, the design of PTLs entails modeling based on crystal structures, possibly limiting their extension to less studied receptors [74]. Nevertheless, the PTL approach is an attractive technology since the minimal mutations may preserve properties of the native receptor and allow for integration of other mutations, including those that generate bias.
Reporters
Not only can optogenetic approaches use light to induce changes in the cell, they can also report localization, intra- or intermolecular interactions, or the state of the cell (voltage, pH, presence of second messengers) (Figure 2d). These are usually fluorescent proteins (FP) containing intrinsic chromophores. Optogenetic reporters are useful for real-time imaging. Optogenetic reporters are advantageous over chemical probes because through mutagenesis their spectral properties can be changed.
Most commonly used are Resonance energy transfer (RET) techniques. The RET approach examine inter and intramolecular interactions by utilizing the overlap of emission and excitation of light-emitting proteins to [75]. In this technique, light emitted from a donor protein excites an acceptor FP within 100 Å [75]. The donor proteins of RET are either bioluminescent (BRET), or fluorescent [in the case of FRET (Fo rster Resonance Energy Transfer)]. Interactions in RET are determined by analyzing intensity. Classically, this is of the relative intensity expressed as a ratiometric output, but can also be determined by the exponential decay of the intensity of the donor, a fluorescent lifetime imaging (FLIM) which is independent of relative protein expression [76]. Recently, a FRET-FLIM expressed in neurons was able to report endogenous adenosine-A2A receptor activity, demonstrating the value of this technology to observe GPCR signaling in the brain [77].
Other optogenetic reports are comprised of single fluorescent proteins. Previously, interacting domains of interest were fused to independent halves of a single FP, creating a fully functional protein based on proximity. However, this approach is irreversible, posing challenges for some applications. Alford and colleagues have developed a method heterodimers. One monomer weakly fluoresces due to a quenched chromophore; the second monomer enhances the fluorescence. Originally, there were two sets of AB heterodimers for either red (RA and RB) or green (GA and GB) fluorescence [78,79]. However, the enhancing partners (monomer B) bound both RA and GA. Through optimization of the monomer B, the authors created a sensor that would fluoresce green or red depending on the preferential affinity of monomer B, a concept coined fluorescent protein exchange (FPX) [80*]. By fusing the monomers to various protein domains, the author created sensors for MAP kinase activity and a variety of second messengers (Ca2+, PIP2, PKA activation via CaMP). Furthermore, FPX sensors can be expressed single polypeptides, reducing confounds of expressing multiple proteins.
Classically, Gq-mediated calcium increases via neuromodulator receptors, was detected by variety of dyes including Fura-based compounds that cannot be targeted to genetically defined neuronal populations. Recently, genetically encoded calcium sensors (GECI) have revolutionized neuroscience with the ability to detect calcium transients as a proxy for action potential firing [5]. GCaMP-type GECIs are the most utilized, and optimized for in vivo applications, although in some cases they have been used in primary cultures [81]. Reporters in the GCaMP family are circularly permutated FPs fused to calmodulin (CaM) and a Ca2+/calmodulin binding domain, M13 [82]. In the absence of Ca2+, the intrinsic chromophore is exposed and therefore quenched by the cytosol. In the presence of CaM and M13 interact, protecting the chromophore from quenching, therefore allowing the FP to fluoresce more effectively (Figure 2d) [82]. The original GCaMP fluoresces green but now they are available in other colors [83]. Though GCaMP has been extensively used to uncover the specific contributions of neuronal ensembles to behavior or evoked circuit activity, its value as a tool to investigate endogenous GPCR signaling through pharmacological approaches is only beginning to be realized [84]. In the next several years, we are likely to see the use of this tool broadened to neuromodulator based questions and in vivo drug screening approaches. Developments in voltage indicators such will also allow for fine temporal precision of measuring neuromodulatory effects on membrane potential [85,86].
Chemogenetics: control of cell signaling with biologically inert molecules
Chemogenetic tools allow for selective modulation of GPCR signaling in specific tissues or cell types. Like optogenetic tools, chemogenetic tools enhance the spatial resolution of GPCR signaling; Chemogenetic tools are also more easily adaptable for behavioral applications (Table 2). For a more extensive review on these tools see Refs [87–89]. The most widely used chemogenetic tools are modified GPCRs (see below). In addition to GPCRs, kinases have also been chemically engineered. By mutating ATP-binding sites allowing for the acceptance of bulky ATP analogues, Shokat and colleagues were able to develop methods to selectively inhibit kinase activity or identify substrates [90–92]. Both of these techniques have been used in neuroscience applications, though not directly related to GPCRs [93]. Mitogen activated protein kinases, commonly downstream of both G-protein and arrestin signaling pathways through GPCRs have also been a subject to this engineering [94]. However, due to the toxicity of thiophosphates, identification of kinase substrates has limited use in vivo.
Table 2.
Comparison of Optogenetic and Chemogenetic Tools
| Tool | Advantage | Disadvantage | Mitigations |
|---|---|---|---|
| Photoactivable Proteins | Microsecond temporal resolution [49**] | Costly equipment | Spectral tuning |
| Subcellular Spatial resolution [52**] Abundant in nature [10] Diversity in wavelength and dynamic range [144] High specificity of signaling proteins/pathway |
Potential thermal activation (long wavelengths) [54] Phototoxicity (short wavelength) Less naturalistic Invasive surgery |
Use of 2-photon to reduce phototoxicity [114] | |
| Opto-XRs | Receptor specific signaling Microsecond temporal resolution Subcellular spatial resolution Fine control of dosage Diversity of tools Greater potential for multiplexing |
Costly Equipment Chimeras Invasive surgery Restrictive actuators |
Newer devices to increase mobility and minimize damage in behavior [117] Use of shorter wavelengths to decrease phototoxicity and increase penetrance |
| DREADDs | Minimally modified Potential for chronic activations over weeks [89] Minimal equipment required Non-restrictive Non-invasive stimulation Many available transgenic lines [88] |
Low temporal resolution [89,97] Lower spatial resolution “Generic” GPCR signaling Lack of diversity Potential for constitutive activity [123] Pharmacological limitations SalB: not water soluable and retains low KOR affinity [112] CNO: can metabolize into clozapine [108], not completely biologically inert [122] |
Spatial Resolution Use to target larger brain structure [118] Target two pathways with one drug [111*] Pharmacology Titration of CNO Alternative agonists for muscarinic DREADDs available [109*] |
The pioneering chemogenetic GPCRs were termed RASSLs, Receptor Activated Solely by Synthetic Ligands. RASSLs tended to have limited use in vivo, due to either low affinity of RASSL ligands, endogenous receptor activation, or constitutive activity [95]. The newest generation of chemogenetic GPCRs, DREADDs (Designer Receptor, Exclusively Activated by Designer Drugs), were designed to only respond to biologically inert synthetic compounds [96]. Through directed molecular evolution of the human muscarinic receptor, the Roth group engineered a family of minimally mutated muscarinic receptors that are only activated by clozapine-N-oxide (CNO), but not endogenous ligands, including acetylecholine. Due to the diversity of muscarinic receptors, DREADDs were originally either Gq (hM3Dq) and Gi/o coupled (hM4Di) (Figure 1c).
Currently, hM3Dq and hM4Di are widely used in vivo to excite and silence neuronal populations, and to modulate gliotransmission [97–99]. Furthermore, the development of DREADDs has been pivotal in selectively modulating GPCR signaling specifically in the brain, notably signaling through pathways that are not directly coupled to excitability through ionic changes. CNO-induced activation of hM3Dq has been validated in its mimicry of acetylecholine-induced M3 receptor activation, aiding in the development of an arrestin-biased and recent Gq-biased DREADD, all of which have been characterized for use in vivo [100–102**] (Figure 1c). Additionally, hM4Di has the ability to inhibit vesicle release at terminals, similar to endogenous Gi-coupled receptors in the brain [99]. Thus, targeting hM4Di to axons offers a tool to silence synapses, independent of the excitability of the cell [103*]. Unlike, hM3Dq and hM4di, Gs-coupled DREADDs (rM3Ds) are chimeras, similar to Opto-β2AR, except the extracellular domain is a Gq DREADD as opposed to rhodopsin [104]. The value of DREADDs is demonstrated by the discovery of novel PKA-dependent GPCR signaling in consummatory behaviors. In an exploratory study, activation of rM3Ds in AgRP neurons increased food intake, through activation of PKA [105*]. Additionally, Gi-coupled postsynaptic receptors, modulated binge alcohol behavior, through inhibition of PKA signaling [106*]. DREADDs have also been used to elucidate sustained Gq-mediated neuronal inactivation through JNK MAP kinase [107]. These experiments highlight the ability of DREADDs to elucidate molecular mechanisms of neuromodulation through G-protein activation.
Though DREADDs demonstrate cell behaviors akin to endogenous GPCR, their translational potential is limited by their ligands. Controversy over the metabolization of CNO to pharmacologically active clozapine in humans and guinea pigs inspired a structure-activity relationship (SAR) analysis of DREADDs aimed to identify alternative agonists [108]. This study yielded “Compound 21”, which is not metabolized in the same pathways as CNO, eliminating the risk for conversion to clozapine [109*]. This study also identified perlapine, a sleep-inducing hypnotic drug, as a highly selective novel agonist for hM3dq [110]. Lack of diversity in DREADDs and their ligands, also limits the potential of multiplexing. However, Alrdin-Kirk and colleagues took advantage of the shared ligand by pairing hM3Dq and rM3Ds to maximally activate dopamine neurons [111*]. In addition to the Gq-coupled excitation by hM3Dq, the inhibitory effects of Gi-coupled dopamine auto-receptors were counteracted by Gs signaling through rM3Ds. With these tools the authors were able to define a cAMP induced mechanism for Graft Induced Dyskinesia in Parkinsonian patients, a phenomenon with no known mechanism and great clinical relevance**. Alternatively, chemogenetic multiplexing can now be achieved with the use of KORD (Kappa Opioid Receptor DREADD) [112]. KORD’s design was driven by the structure of KOR combined with molecular modeling of Salnorvin B (SalB), a low affinity inert ligand of KOR. Because it is pharmacologically distinct, KORD can be used in concert with muscarinic DREADDs to gain bidirectional control of the same neuron [111*,112]. KORD is also kinetically unique from hMD4i DREADDs, exerting shorter lasting effects. KORD has played a pivotal role in expanding the chemogenetic toolbox for modulation of neural circuits.
Applicability of Optogenetic and Chemogenetic Tools
Optogenetic and chemogenetic tools have undoubtedly revolutionized interrogation of specific GPCR signaling, especially in the brain. Both genetic approaches have inherent limitations; thus applicability of each toolset depends on experimental design. It is important to note that both approaches have been used to simulate the endogenous signaling of the other techniques model GPCRs: hMD3q has been used to recapitulate melanopsin signaling in the retina and melanopsin for muscarinic signaling in pyramidal neurons of the cortex [15,113].
Optogenetics provides precise spatial (subcellular) and temporal (microsecond) control of signaling. Temporal control is enhanced with bistable tools (ligand, receptors, effectors) that allow for inactivation by absorption at a second wavelength. Due to the range of spectral properties optogenetic tools from ultraviolet to near infrared wavelength, there is flexibility in the number of available tools for a specific target. Additionally, since photosensitive proteins are abundant in nature, there are many unexplored avenues for tool development.
Currently, there are optogenetic tools available for each step of the GPCR signaling pathway, from ligand binding to downstream signaling events and deactivation. There are some considerations in using optogenetics, although recent advances in technology have addressed some of these concerns. Biophysical considerations include the risk of dark state activity, whether basal enzymatic activity, dimerization or incomplete caging, and limitations of wavelength required for stimulation. Some opotogenetic tools require blue or UV light, short wavelengths with potentially toxic effects and shallow tissue penetration. The use of UV light can be avoided either by delivering two or more photons of lower energy, as in 2-photon microscopy), a method especially useful for commonly UV sensitive photoactivatable ligands. Two-photon microscopy also enhances the spatial resolution, for focusing two beams restricts stimulation to a confined three dimensional space [114]. Furthermore, near IR wavelengths combined with cranial windows allow for the stimulation and imaging of deep brain structures in vivo [115]. Previously, in vivo actuators greatly limited behavioral approaches because were invasive, restrictive, and lacked spatial precision. The latest optogenetic devices minimally damage the brain, liberate the subject’s mobility and precisely deliver light [116,117].
Compared to optogenetics, chemogenetic approaches are generally less technically challenging to use, for they are less invasive, less restrictive and cost-effective. Chemogenetic tools require no specialized equipment for actuation; Due to its high blood brain barrier penetrance, CNO can be administered through local or systemic injection [99,103*,118]. CNO is also water soluble, allowing for chronic exposure through dilution in animal drinking water for extended periods of time (i.e. 2 weeks), without decreases in behavioral effects [89,119–121]. However, some recent studies highlight the need for care and optimization when using DREADD receptors in vivo, including the selection of proper control groups. Previous concerns about the biological activity of CNO were limited to only humans and guinea pigs, however a recent study demonstrated that CNO also exerts behavioral effects on rats that do not express DREADDs [122]. Another group also demonstrated biologically activity of DREADDs in the absence of CNO [123]. These studies highlight the importance of experimental design, including proper controls.
Chemogenetic tools enable the activation of GPCR pathways with spatial resolution, though significantly less so than optogenetic tools. Both tools can be similarly expressed through stereotaxic injections (see below). Optogenetics, however, allows for precise subcellular stimulation of GPCR signaling, not available in chemogenetics [34]. Nevertheless, chemogenetic tools allow for modulation of larger brain areas that cannot be feasibly illuminated through optogenetic approaches [118]. Chemogenetic tools share limitations with classic pharmacological methods: these tools potentially have off-target effects and lack precise temporal control. In addition to the slower kinetics of activation, compared to optogenetic tools, CNO has a slow wash out rate, potentially lasting for at least an two hours [89,97,104]. Therefore, chemogenetics are more suited for investigating slow, long-lasting behavioral effects. In summary, chemogenetic and optogenetic tools differ in their feasibility and spatiotemporal resolution. Each toolset has a set potential strengths and limitations, determined by experimental design. Both tools can be used complimentary to each other to dissect receptor specific from generic G-protein signaling [22**].
Expression Methods
The resolution of optogenetic and chemogenetic tools is defined not only by stimulation but also expression. These tools are expressed either virally or through transgenic animal approaches. Viral delivery is the most widely used route for expressing transgenes [124]. Viral delivery requires less commitment of resources needed to generate and maintain a genetic line. Additionally, viral expression is limited to the site of infection, increasing spatial resolution. Adeno-associated viruses (AAV) and lentiviruses are most commonly used [21**,27**,124]. Preference of viral vector can depend on desired specificity of expression. AAVs spread farther from the injection site, due to their small size, and lentiviruses typically infect smaller regions, but can carry larger cargo when needed. Fine spatial resolution can theoretically be achieved by encoding elements to ensure proper targeting to synapses and other neuronal compartments. Recent efforts have utilized both c-terminal and n-terminal domains for this purpose and further developments will enhance specificity to select neuronal compartments [12,29,103*].
As their name implies, genetically targeted technologies gain resolution through genetic identity. Lentiviruses have larger packaging capabilities, permitting the use of transcriptional promoters to target expression neuronal types, an approach not available with AAVs [125]. Cell-type specific promoters however may not a drive sufficient amount of expression [126]. To overcome this, viruses dependent on DNA recombination, especially through Cre recombinase, have been developed [127]. Recombinases usually are driven by cell-type specific promoters in the genome of a transgenic line or in an independent virus [128]*. More discrete populations of neurons can be targeted using intersectional approaches, utilizing other recombinases such as FLP and DRE [129,130]. Recombinase technology can also target neurons that project to the injection site through the use of Canine-adenovirus (CAV) [131]. CAV viruses have the ability to infect neurons through retrograde axonal transport [132]. DREADDs have been successfully expressed in vivo through this method [133,134]. Another, very promising retro-AAV (rAAV2) was recently developed, that will open even more possibilities for retrograde expression [135]. Finally, recombinases are also subject to optogenetic stimulation, photoactivatable Cre-recombinase is continually being optimized for the spatial control gene regulation in vivo [136–138]. These approaches of expression allow for further control of discrete cell type manipulations within neural circuits offering even greater specificity and reductionism of neuromodulator pathways in vitro and in vivo.
Development
There are many unexplored avenues of optogenetic and chemogenetic tool development. These tools can be developed to model other GPCR signaling components, such as G11/12 signaling family. Additionally, the spatial resolution of DREADDs would significantly increase with photoactivatable ligands. Tuning and optimization through mutation can also to allow for the development of new tools with different signaling dynamics, including signaling bias, kinetics, spectral properties and subcellular localization [21**,139]. Many optogenetic tools are still in the conceptual stages and have yet to be optimized for expression in neurons or in vivo. In a recent proof of concept report, light induced secretion was developed using UV8R, a plant photoreceptor [140*]. Impressively, the authors used this method to optically induce secretion of a reporter protein at specific dendritic branch points [140*]. Validating this approach with functional signaling proteins will significantly expand the neuroscience optogenetic toolbox for GPCRs. Full characterization of existing tools and their ability to simulate endogenous in vivo signaling in varying neuronal cell types will be crucial since subcellular localization cell-type specific expression can define GPCR bias in vivo. Finally, analysis and comparison of the biophysical properties of currently available optogenetic tools in vitro and in vivo will aid the community in selecting the most appropriate tools for specific biological inquiries [141].
Developing advances in many front of biological technology will foster the creation of many genetically targeted approaches. Genome sequencing has dramatically increased the diversification of the available library of genomic sequences from biological organisms. Currently, tens of thousands of sequences for opsins and photoactivatable proteins have been reported [10]. Natural proteins hold potential as resources for opsin based tools with unique spectral properties with minimal mutagenesis [10]. Advances in technology, such as 2-photon stimulation and imaging have revitalized the development of optogenetic tools sensitive to short wavelengths, such as the recently discovered bistable vertebrate Gi/o coupled neuropsin [142]. Structural biology, through high resolution crystal structures and developing modelling software, will continue to inspire the development of optogenetic and chemogenetic tools far into the foreseeable future.
Concluding remarks
Optogenetics and chemogenetics have not only revolutionized the study of neuronal circuitry and but have great potential in dissecting the role of specific GPCR signaling pathways. Multiplexing within or between tools enables bidirectional neuromodulatory control of signaling pathways in a single neuronal population. Also, multiplexing of optogenetic actuators and reporters of different spectral occupancy can further define pre and post synaptic signaling involved in behavior and disease states. These tools also have translational potential, providing reductionist approaches for understanding drugable neural circuits that may lead to novel chemical entities for drug discovery.
Highlights.
Optogenetic tools provide precise spatial and temporal control of GPCR signaling.
Light-sensitive proteins can be engineered for GPCR-specific applications.
Advances in imaging technology are paving new in vitro and in vivo GPCR research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Readings
- 1.Kleinlogel S. Optogenetic user’s guide to Opto-GPCRs. Front Biosci Landmark Ed. 2016;21:794–805. doi: 10.2741/4421. [DOI] [PubMed] [Google Scholar]
- 2.Zhou XX, Pan M, Lin MZ. Investigating neuronal function with optically controllable proteins. Front Mol Neurosci. 2015 doi: 10.3389/fnmol.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K, Cui B. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 2015;33:92–100. doi: 10.1016/j.tibtech.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broichhagen J, Trauner D. The in vivo chemistry of photoswitched tethered ligands. Curr Opin Chem Biol. 2014;21:121–127. doi: 10.1016/j.cbpa.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Enterina JR, Wu L, Campbell RE. Emerging fluorescent protein technologies. Curr Opin Chem Biol. 2015;27:10–17. doi: 10.1016/j.cbpa.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Gautier A, Gauron C, Volovitch M, Bensimon D, Jullien L, Vriz S. How to control proteins with light in living systems. Nat Chem Biol. 2014;10:533–541. doi: 10.1038/nchembio.1534. [DOI] [PubMed] [Google Scholar]
- 7.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 8.Lerch MM, Hansen MJ, van Dam GM, Szymanski W, Feringa BL. Emerging Targets in Photopharmacology. Angew Chem Int Ed. 2016 doi: 10.1002/anie.201601931. [DOI] [PubMed] [Google Scholar]
- 9.Alford SC, Wu J, Zhao Y, Campbell RE, Knöpfel T. Optogenetic reporters. Biol Cell. 2013;105:14–29. doi: 10.1111/boc.201200054. [DOI] [PubMed] [Google Scholar]
- 10.Mathes T. Natural Resources for Optogenetic Tools. In: Kianianmomeni A, editor. Optogenetics. Springer; New York: 2016. pp. 19–36. [DOI] [PubMed] [Google Scholar]
- 11.Karunarathne WKA, Giri L, Kalyanaraman V, Gautam N. Optically triggering spatiotemporally confined GPCR activity in a cell and programming neurite initiation and extension. Proc Natl Acad Sci. 2013;110:E1565–E1574. doi: 10.1073/pnas.1220697110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masseck OA, Spoida K, Dalkara D, Maejima T, Rubelowski JM, Wallhorn L, Deneris ES, Herlitze S. Vertebrate Cone Opsins Enable Sustained and Highly Sensitive Rapid Control of Gi/o Signaling in Anxiety Circuitry. Neuron. 2014;81:1263–1273. doi: 10.1016/j.neuron.2014.01.041. [DOI] [PubMed] [Google Scholar]
- 13.Koizumi A, Tanaka KF, Yamanaka A. The manipulation of neural and cellular activities by ectopic expression of melanopsin. Neurosci Res. 2013;75:3–5. doi: 10.1016/j.neures.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 14*.Spoida K, Eickelbeck D, Karapinar R, Eckhardt T, Mark MD, Jancke D, Ehinger BV, König P, Dalkara D, Herlitze S, et al. Melanopsin Variants as Intrinsic Optogenetic On and Off Switches for Transient versus Sustained Activation of G Protein Pathways. Curr Biol. 2016;26:1206–1212. doi: 10.1016/j.cub.2016.03.007. This report introduces human and mouse melanopsin variants as tools to control Gq signaling transiently or sustainably, respectively. [DOI] [PubMed] [Google Scholar]
- 15.McGregor KM, Bécamel C, Marin P, Andrade R. Using melanopsin to study G protein signaling in cortical neurons. J Neurophysiol. 2016;116:1082–1092. doi: 10.1152/jn.00406.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailes HJ, Zhuang L-Y, Lucas RJ. Reproducible and Sustained Regulation of Gαs Signalling Using a Metazoan Opsin as an Optogenetic Tool. PLOS ONE. 2012;7:e30774. doi: 10.1371/journal.pone.0030774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avelar GM, Schumacher RI, Zaini PA, Leonard G, Richards TA, Gomes SL. A Rhodopsin-Guanylyl Cyclase Gene Fusion Functions in Visual Perception in a Fungus. Curr Biol. 2014;24:1234–1240. doi: 10.1016/j.cub.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Gao S, Nagpal J, Schneider MW, Kozjak-Pavlovic V, Nagel G, Gottschalk A. Optogenetic manipulation of cGMP in cells and animals by the tightly light-regulated guanylyl-cyclase opsin CyclOp. Nat Commun. 2015;6:8046. doi: 10.1038/ncomms9046. This report characterizes natural specific guanylyl cyclase as a tool for optogenetic manipulation of cGMP signaling in sensory neurons to modulate c. elegans behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J-M, Hwa J, Garriga P, Reeves PJ, RajBhandary UL, Khorana HG. Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry (Mosc) 2005;44:2284–2292. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- 20.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 21**.Siuda ER, McCall JG, Al-Hasani R, Shin G, Il Park S, Schmidt MJ, Anderson SL, Planer WJ, Rogers JA, Bruchas MR. Optodynamic simulation of β-adrenergic receptor signalling. Nat Commun. 2015;6:8480. doi: 10.1038/ncomms9480. The authors in this reported validated opto-β2AR mimicry of kinetics in vitro and behavioral responses modulated endogenous neural circuits. They also developed novel G protein and arrestin biased opto-β2ARs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Siuda ER, Al-Hasani R, McCall JG, Bhatti DL, Bruchas MR. Chemogenetic and Optogenetic Activation of Gαs Signaling in the Basolateral Amygdala Induces Acute and Social Anxiety-Like States. Neuropsychopharmacology. 2016;41:2011–2023. doi: 10.1038/npp.2015.371. The authors employed a complimentary chemogenetic and optogenetic approach to define endogenous Gs-mediated signaling pathways in behavioral models of anxiety. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shenoy SK, Drake MT, Nelson CD, Houtz DA, Xiao K, Madabushi S, Reiter E, Premont RT, Lichtarge O, Lefkowitz RJ. beta-Arrestin-dependent, G Protein-independent ERK1/2 Activation by the beta2 Adrenergic Receptor. J Biol Chem. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 24.Vaughan DJ, Millman EE, Godines V, Friedman J, Tran TM, Dai W, Knoll BJ, Clark RB, Moore RH. Role of the G Protein-coupled Receptor Kinase Site Serine Cluster in β2-Adrenergic Receptor Internalization, Desensitization, and β-Arrestin Translocation. J Biol Chem. 2006;281:7684–7692. doi: 10.1074/jbc.M500328200. [DOI] [PubMed] [Google Scholar]
- 25.DeVree BT, Mahoney JP, Vélez-Ruiz GA, Rasmussen SGF, Kuszak AJ, Edwald E, Fung J-J, Manglik A, Masureel M, Du Y, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 2016;535:182–186. doi: 10.1038/nature18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Li P, Rial D, Canas PM, Yoo J-H, Li W, Zhou X, Wang Y, van Westen GJP, Payen M-P, Augusto E, et al. Optogenetic activation of intracellular adenosine A2A receptor signaling in the hippocampus is sufficient to trigger CREB phosphorylation and impair memory. Mol Psychiatry. 2015;20:1339–1349. doi: 10.1038/mp.2014.182. This report describes opto-A2AR a chimera that mimics endogenous signaling pathways of Adenosine A2A receptor Gs signaling, including bias in different neural circuits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Siuda ER, Copits BA, Schmidt MJ, Baird MA, Al-Hasani R, Planer WJ, Funderburk SC, McCall JG, Gereau RW, Bruchas MR. Spatiotemporal Control of Opioid Signaling and Behavior. Neuron. 2015;86:923–935. doi: 10.1016/j.neuron.2015.03.066. This report introduced and characterized an optogenetic chimeric GPCR that mimics both endogenous Gi/o opioid signaling in vitro and endogenous opioid mediated behavioral effects based on expression within endogenous neural circuits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28**.van Wyk M, Pielecka-Fortuna J, Löwel S, Kleinlogel S. Restoring the ON Switch in Blind Retinas: Opto-mGluR6, a Next-Generation, Cell-Tailored Optogenetic Tool. PLOS Biol. 2015;13:e1002143. doi: 10.1371/journal.pbio.1002143. The authors developed the first optogenetic chimera of a class C GPCR. The translational potential of this tool was demonstrated through the restoration of vision in mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spoida K, Masseck OA, Deneris ES, Herlitze S. Gq/5-HT2c receptor signals activate a local GABAergic inhibitory feedback circuit to modulate serotonergic firing and anxiety in mice. Proc Natl Acad Sci. 2014;111:6479–6484. doi: 10.1073/pnas.1321576111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giri L, Patel AK, Karunarathne WKA, Kalyanaraman V, Venkatesh KV, Gautam N. A G-Protein Subunit Translocation Embedded Network Motif Underlies GPCR Regulation of Calcium Oscillations. Biophys J. 2014;107:242–254. doi: 10.1016/j.bpj.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karunarathne AWK, O’Neill PR, Martinez-Espinosa PL, Kalyanaraman V, Gautam N. All G protein βγ complexes are capable of translocation on receptor activation. Biochem Biophys Res Commun. 2012;421:605–611. doi: 10.1016/j.bbrc.2012.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsvetanova NG, Irannejad R, von Zastrow M. G Protein-coupled Receptor (GPCR) Signaling via Heterotrimeric G Proteins from Endosomes. J Biol Chem. 2015;290:6689–6696. doi: 10.1074/jbc.R114.617951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomsen ARB, Plouffe B, Cahill TJ, III, Shukla AK, Tarrasch JT, Dosey AM, Kahsai AW, Strachan RT, Pani B, Mahoney JP, et al. GPCR-G Protein-β-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell. 2016;166:907–919. doi: 10.1016/j.cell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karunarathne WKA, O’Neill PR, Gautam N. Subcellular optogenetics – controlling signaling and single-cell behavior. J Cell Sci. 2015;128:15–25. doi: 10.1242/jcs.154435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paramonov VM, Mamaeva V, Sahlgren C, Rivero-Muller A. Genetically-encoded tools for cAMP probing and modulation in living systems. Exp Pharmacol Drug Discov. 2015 doi: 10.3389/fphar.2015.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- 37.Schröder-Lang S, Schwärzel M, Seifert R, Strünker T, Kateriya S, Looser J, Watanabe M, Kaupp UB, Hegemann P, Nagel G. Fast manipulation of cellular cAMP level by light in vivo. Nat Methods. 2007;4:39–42. doi: 10.1038/nmeth975. [DOI] [PubMed] [Google Scholar]
- 38.Weissenberger S, Schultheis C, Liewald JF, Erbguth K, Nagel G, Gottschalk A. PACα– an optogenetic tool for in vivo manipulation of cellular cAMP levels, neurotransmitter release, and behavior in Caenorhabditis elegans. J Neurochem. 2011;116:616–625. doi: 10.1111/j.1471-4159.2010.07148.x. [DOI] [PubMed] [Google Scholar]
- 39.Nagahama T, Suzuki T, Yoshikawa S, Iseki M. Functional transplant of photoactivated adenylyl cyclase (PAC) into Aplysia sensory neurons. Neurosci Res. 2007;59:81–88. doi: 10.1016/j.neures.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Z, Tanaka KF, Matsunaga S, Iseki M, Watanabe M, Matsuki N, Ikegaya Y, Koyama R. Photoactivated adenylyl cyclase (PAC) reveals novel mechanisms underlying cAMP-dependent axonal morphogenesis. Sci Rep. 2016;5:19679. doi: 10.1038/srep19679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Ohki M, Sugiyama K, Kawai F, Tanaka H, Nihei Y, Unzai S, Takebe M, Matsunaga S, Adachi S, Shibayama N, et al. Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium. Proc Natl Acad Sci. 2016;113:6659–6664. doi: 10.1073/pnas.1517520113. This study reports the crystal structure of a previously uncharacterized photoactivated adenylyl cyclase (oaPAC), providing insight on the mechanism of light activation of BLUF PACs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pudasaini A, El-Arab KK, Zoltowski BD. LOV-based optogenetic devices: light-driven modules to impart photoregulated control of cellular signaling. Biophysics. 2015;2:18. doi: 10.3389/fmolb.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konold PE, Mathes T, Weiβenborn J, Groot ML, Hegemann P, Kennis JTM. Unfolding of the C-Terminal Jα Helix in the LOV2 Photoreceptor Domain Observed by Time-Resolved Vibrational Spectroscopy. J Phys Chem Lett. 2016;7:3472–3476. doi: 10.1021/acs.jpclett.6b01484. [DOI] [PubMed] [Google Scholar]
- 44.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dietz DM, Sun H, Lobo MK, Cahill ME, Chadwick B, Gao V, Koo JW, Mazei-Robison MS, Dias C, Maze I, et al. Rac1 is essential in cocaine-induced structural plasticity of nucleus accumbens neurons. Nat Neurosci. 2012;15:891–896. doi: 10.1038/nn.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46**.Wang H, Vilela M, Winkler A, Tarnawski M, Schlichting I, Yumerefendi H, Kuhlman B, Liu R, Danuser G, Hahn KM. LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nat Methods. 2016;13:755–758. doi: 10.1038/nmeth.3926. This report describes the generation of LOVTRAP, an optogenetic technique for reversible protein dissociation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoltowski BD, Crane BR. Light activation of the LOV protein vivid generates a rapidly exchanging dimer. Biochemistry (Mosc) 2008;47:7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational Switching in the Fungal Light Sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Kawano F, Suzuki H, Furuya A, Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun. 2015;6:6256. doi: 10.1038/ncomms7256. The authors of this study diversified the optogenetic tool box by tuning LOV domain dimers, thus creating a family of dimers (MAGNETs) with a broad range of dissociation kinetics. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 51.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52**.O’Neill PR, Gautam N. Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol Biol Cell. 2014;25:2305–2314. doi: 10.1091/mbc.E14-04-0870. The authors of this study used CRY2-CIB1 technology to inhibit G protein signaling with subcellular resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem Soc Rev. 2013;42:3441–3452. doi: 10.1039/c3cs35458j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo D-G, Yue WWS, Ala-Laurila P, Yau K-W. Activation of Visual Pigments by Light and Heat. Science. 2011;332:1307–1312. doi: 10.1126/science.1200172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55*.Ryu M-H, Kang I-H, Nelson MD, Jensen TM, Lyuksyutova AI, Siltberg-Liberles J, Raizen DM, Gomelsky M. Engineering adenylate cyclases regulated by near-infrared window light. Proc Natl Acad Sci. 2014;111:10167–10172. doi: 10.1073/pnas.1324301111. This report describes the development of an far red sensitive adenylyl cyclase. The authors demonstrated the utility of this tool by modulating wildtype c elegans behavior without affecting the blue light avoidance response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Gasser C, Taiber S, Yeh C-M, Wittig CH, Hegemann P, Ryu S, Wunder F, Möglich A. Engineering of a red-light–activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci. 2014;111:8803–8808. doi: 10.1073/pnas.1321600111. The authors in this study engineered the first light-activated phosphodiesterase based on structural similarities between bacteriophytochrome receptors and human phosphodiesterase 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwards SL, Charlie NK, Milfort MC, Brown BS, Gravlin CN, Knecht JE, Miller KG. A Novel Molecular Solution for Ultraviolet Light Detection in Caenorhabditis elegans. PLOS Biol. 2008;6:e198. doi: 10.1371/journal.pbio.0060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 59.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Müller K, Zurbriggen MD, Weber W. Control of gene expression using a red- and far-red light–responsive bi-stable toggle switch. Nat Protoc. 2014;9:622–632. doi: 10.1038/nprot.2014.038. [DOI] [PubMed] [Google Scholar]
- 61.Idevall-Hagren O, De Camilli P. Detection and manipulation of phosphoinositides. Biochim Biophys Acta BBA - Mol Cell Biol Lipids. 2015;1851:736–745. doi: 10.1016/j.bbalip.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toettcher JE, Weiner OD, Lim WA. Using Optogenetics to Interrogate the Dynamic Control of Signal Transmission by the Ras/Erk Module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tadevosyan A, Villeneuve LR, Fournier A, Chatenet D, Nattel S, Allen BG. Caged ligands to study the role of intracellular GPCRs. Methods. 2016;92:72–77. doi: 10.1016/j.ymeth.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 64.Banghart MR, Sabatini BL. Photoactivatable Neuropeptides for Spatiotemporally Precise Delivery of Opioids in Neural Tissue. Neuron. 2012;73:249–259. doi: 10.1016/j.neuron.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banghart MR, Williams JT, Shah RC, Lavis LD, Sabatini BL. Caged Naloxone Reveals Opioid Signaling Deactivation Kinetics. Mol Pharmacol. 2013;84:687–695. doi: 10.1124/mol.113.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kramer RH, Mourot A, Adesnik H. Optogenetic pharmacology for control of native neuronal signaling proteins. Nat Neurosci. 2013;16:816–823. doi: 10.1038/nn.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong M, Babalhavaeji A, Samanta S, Beharry AA, Woolley GA. Red-Shifting Azobenzene Photoswitches for in Vivo Use. Acc Chem Res. 2015;48:2662–2670. doi: 10.1021/acs.accounts.5b00270. [DOI] [PubMed] [Google Scholar]
- 68.Konrad DB, Frank JA, Trauner D. Synthesis of Redshifted Azobenzene Photoswitches by Late-Stage Functionalization. Chem Weinh Bergstr Ger. 2016;22:4364–4368. doi: 10.1002/chem.201505061. [DOI] [PubMed] [Google Scholar]
- 69*.Schönberger M, Trauner D. A Photochromic Agonist for μ-Opioid Receptors. Angew Chem Int Ed. 2014;53:3264–3267. doi: 10.1002/anie.201309633. This report describes the development of a photoswitchable opioid agonist. [DOI] [PubMed] [Google Scholar]
- 70.Broichhagen J, Johnston NR, von Ohlen Y, Meyer-Berg H, Jones BJ, Bloom SR, Rutter GA, Trauner D, Hodson DJ. Allosteric Optical Control of a Class B G-Protein-Coupled Receptor. Angew Chem Int Ed. 2016;55:5865–5868. doi: 10.1002/anie.201600957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71**.Rovira X, Trapero A, Pittolo S, Zussy C, Faucherre A, Jopling C, Giraldo J, Pin J-P, Gorostiza P, Goudet C, et al. OptoGluNAM4.1, a Photoswitchable Allosteric Antagonist for Real-Time Control of mGlu4 Receptor Activity. Cell Chem Biol. 2016;23:929–934. doi: 10.1016/j.chembiol.2016.06.013. This study describes the development of the first photoswitchable negative allosteric modulator of a GPCR and demonstrated its ability to modulate behavior. [DOI] [PubMed] [Google Scholar]
- 72*.Frank JA, Yushchenko DA, Hodson DJ, Lipstein N, Nagpal J, Rutter GA, Rhee J-S, Gottschalk A, Brose N, Schultz C, et al. Photoswitchable diacylglycerols enable optical control of protein kinase C. Nat Chem Biol. 2016;12:755–762. doi: 10.1038/nchembio.2141. The authors extended the use of photoswitches beyond pharmacology to modulate a DAG, an intracellular signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Levitz J, Pantoja C, Gaub B, Janovjak H, Reiner A, Hoagland A, Schoppik D, Kane B, Stawski P, Schier AF, et al. Optical control of metabotropic glutamate receptors. Nat Neurosci. 2013;16:507–516. doi: 10.1038/nn.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reiner A, Levitz J, Isacoff EY. Controlling ionotropic and metabotropic glutamate receptors with light: principles and potential. Curr Opin Pharmacol. 2015;20:135–143. doi: 10.1016/j.coph.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Unen J, Woolard J, Rinken A, Hoffmann C, Hill SJ, Goedhart J, Bruchas MR, Bouvier M, Adjobo-Hermans MJW. A Perspective on Studying G-Protein-Coupled Receptor Signaling with Resonance Energy Transfer Biosensors in Living Organisms. Mol Pharmacol. 2015;88:589–595. doi: 10.1124/mol.115.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goedhart J, Hink MA, Jalink K. An introduction to fluorescence imaging techniques geared towards biosensor applications. Methods Mol Biol Clifton NJ. 2014;1071:17–28. doi: 10.1007/978-1-62703-622-1_2. [DOI] [PubMed] [Google Scholar]
- 77.Chen Y, Saulnier JL, Yellen G, Sabatini BL. A PKA activity sensor for quantitative analysis of endogenous GPCR signaling via 2-photon FRET-FLIM imaging. Front Pharmacol. 2014;5:56. doi: 10.3389/fphar.2014.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alford SC, Ding Y, Simmen T, Campbell RE. Dimerization-dependent green and yellow fluorescent proteins. ACS Synth Biol. 2012;1:569–575. doi: 10.1021/sb300050j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alford SC, Abdelfattah AS, Ding Y, Campbell RE. A Fluorogenic Red Fluorescent Protein Heterodimer. Chem Biol. 2012;19:353–360. doi: 10.1016/j.chembiol.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80*.Ding Y, Li J, Enterina JR, Shen Y, Zhang I, Tewson PH, Mo GCH, Zhang J, Quinn AM, Hughes TE, et al. Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange. Nat Methods. 2015;12:195–198. doi: 10.1038/nmeth.3261. The authors described a new approach to create biosensors based on the reversible exchange in monomers of dimerization dependent fluorescent proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 83.Inoue M, Takeuchi A, Horigane S, Ohkura M, Gengyo-Ando K, Fujii H, Kamijo S, Takemoto-Kimura S, Kano M, Nakai J, et al. Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat Methods. 2015;12:64–70. doi: 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- 84.Partridge JG. Utilizing GCaMP transgenic mice to monitor endogenous Gq/11-coupled receptors. Neuropharmacology. 2015;6:42. doi: 10.3389/fphar.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark-Margulies N, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lou S, Adam Y, Weinstein EN, Williams E, Williams K, Parot V, Kavokine N, Liberles S, Madisen L, Zeng H, et al. Genetically Targeted All-Optical Electrophysiology with a Transgenic Cre-Dependent Optopatch Mouse. J Neurosci. 2016;36:11059–11073. doi: 10.1523/JNEUROSCI.1582-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burnett CJ, Krashes MJ. Resolving Behavioral Output via Chemogenetic Designer Receptors Exclusively Activated by Designer Drugs. J Neurosci. 2016;36:9268–9282. doi: 10.1523/JNEUROSCI.1333-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roth BL. DREADDs for Neuroscientists. Neuron. 2016;89:683–694. doi: 10.1016/j.neuron.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith KS, Bucci DJ, Luikart BW, Mahler SV. DREADDS: Use and application in behavioral neuroscience. Behav Neurosci. 2016;130:137–155. doi: 10.1037/bne0000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 91.Liu Y, Shah K, Yang F, Witucki L, Shokat KM. Engineering Src family protein kinases with unnatural nucleotide specificity. Chem Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- 92.Islam K. Allele-Specific Chemical Genetics: Concept, Strategies, and Applications. ACS Chem Biol. 2015;10:343–363. doi: 10.1021/cb500651d. [DOI] [PubMed] [Google Scholar]
- 93.Ultanir SK, Hertz NT, Li G, Ge W-P, Burlingame AL, Pleasure SJ, Shokat KM, Jan LY, Jan Y-N. Chemical Genetic Identification of NDR1/2 Kinase Substrates AAK1 and Rabin8 Uncovers Their Roles in Dendrite Arborization and Spine Development. Neuron. 2012;73:1127–1142. doi: 10.1016/j.neuron.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Habelhah H, Shah K, Huang L, Burlingame AL, Shokat KM, Ronai Z‘ev. Identification of New JNK Substrate Using ATP Pocket Mutant JNK and a Corresponding ATP Analogue. J Biol Chem. 2001;276:18090–18095. doi: 10.1074/jbc.M011396200. [DOI] [PubMed] [Google Scholar]
- 95.Conklin BR, Hsiao EC, Claeysen S, Dumuis A, Srinivasan S, Forsayeth JR, Guettier J-M, Chang WC, Pei Y, McCarthy KD, et al. Engineering GPCR signaling pathways with RASSLs. Nat Methods. 2008;5:673–678. doi: 10.1038/nmeth.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alexander GM, Rogan SC, Abbas AI, Armbruster BN, Pei Y, Allen JA, Nonneman RJ, Hartmann J, Moy SS, Nicolelis MA, et al. Remote Control of Neuronal Activity in Transgenic Mice Expressing Evolved G Protein-Coupled Receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scofield MD, Boger HA, Smith RJ, Li H, Haydon PG, Kalivas PW. Gq-DREADD Selectively Initiates Glial Glutamate Release and Inhibits Cue-induced Cocaine Seeking. Biol Psychiatry. 2015;78:441–451. doi: 10.1016/j.biopsych.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mahler SV, Vazey EM, Beckley JT, Keistler CR, McGlinchey EM, Kaufling J, Wilson SP, Deisseroth K, Woodward JJ, Aston-Jones G. Designer receptors show role for ventral pallidum input to ventral tegmental area in cocaine seeking. Nat Neurosci. 2014;17:577–585. doi: 10.1038/nn.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alvarez-Curto E, Prihandoko R, Tautermann CS, Zwier JM, Pediani JD, Lohse MJ, Hoffmann C, Tobin AB, Milligan G. Developing Chemical Genetic Approaches to Explore G Protein-Coupled Receptor Function: Validation of the Use of a Receptor Activated Solely by Synthetic Ligand (RASSL) Mol Pharmacol. 2011;80:1033–1046. doi: 10.1124/mol.111.074674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nakajima K, Wess J. Design and functional characterization of a novel, arrestin-biased designer G protein-coupled receptor. Mol Pharmacol. 2012;82:575–582. doi: 10.1124/mol.112.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102*.Hu J, Stern M, Gimenez LE, Wanka L, Zhu L, Rossi M, Meister J, Inoue A, Beck-Sickinger AG, Gurevich VV, et al. A G Protein-biased Designer G Protein-coupled Receptor Useful for Studying the Physiological Relevance of Gq/11-dependent Signaling Pathways. J Biol Chem. 2016;291:7809–7820. doi: 10.1074/jbc.M115.702282. This study describes the development of an Gq-biased DREADD receptor, demonstrating its use in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103*.Stachniak TJ, Ghosh A, Sternson SM. Chemogenetic Synaptic Silencing of Neural Circuits Localizes a Hypothalamus→Midbrain Pathway for Feeding Behavior. Neuron. 2014;82:797–808. doi: 10.1016/j.neuron.2014.04.008. The authors of this study targeted DREADDs to axonal terminals, creating a method to chemogenetically silence synapses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guettier J-M, Gautam D, Scarselli M, Azua IR, de Li JH, Rosemond E, Ma X, Gonzalez FJ, Armbruster BN, Lu H, et al. A chemical-genetic approach to study G protein regulation of β cell function in vivo. Proc Natl Acad Sci. 2009;106:19197–19202. doi: 10.1073/pnas.0906593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105**.Nakajima K, Cui Z, Li C, Meister J, Cui Y, Fu O, Smith AS, Jain S, Lowell BB, Krashes MJ, et al. Gs-coupled GPCR signalling in AgRP neurons triggers sustained increase in food intake. Nat Commun. 2016;7:10268. doi: 10.1038/ncomms10268. This study uses DREADDs in vivo to define a novel Gs-mediated intracellular pathway involved in feeding behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106*.Pleil KE, Rinker JA, Lowery-Gionta EG, Mazzone CM, McCall NM, Kendra AM, Olson DP, Lowell BB, Grant KA, Thiele TE, et al. NPY signaling inhibits extended amygdala CRF neurons to suppress binge alcohol drinking. Nat Neurosci. 2015;18:545–552. doi: 10.1038/nn.3972. The authors of this study used DREADDs in vivo to elucidate a novel postsynaptic intracellular mechanism involved in binge drinking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bellocchio L, Ruiz-Calvo A, Chiarlone A, Cabanas M, Resel E, Cazalets J-R, Blázquez C, Cho YH, Galve-Roperh I, Guzmán M. Sustained Gq-Protein Signaling Disrupts Striatal Circuits via JNK. J Neurosci. 2016;36:10611–10624. doi: 10.1523/JNEUROSCI.1192-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Löffler S, Körber J, Nubbemeyer U, Fehsel K. Comment on “Impaired Respiratory and Body Temperature Control Upon Acute Serotonergic Neuron Inhibition. Science. 2012;337:646–646. doi: 10.1126/science.1222519. [DOI] [PubMed] [Google Scholar]
- 109*.Chen X, Choo H, Huang X-P, Yang X, Stone O, Roth BL, Jin J. The First Structure–Activity Relationship Studies for Designer Receptors Exclusively Activated by Designer Drugs. ACS Chem Neurosci. 2015;6:476–484. doi: 10.1021/cn500325v. This structure activity relationship study identified DREADDs alternative ligands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stille G, Sayers A, Lauener H, Eichenberger E. 6-(4-Methyl-1-piperazinyl)morphanthridine (Perlapine), a new tricyclic compound with sedative and sleep-promoting properties. A pharmacological study. Psychopharmacologia. 1973;28:325–337. doi: 10.1007/BF00422753. [DOI] [PubMed] [Google Scholar]
- 111**.Aldrin-Kirk P, Heuer A, Wang G, Mattsson B, Lundblad M, Parmar M, Björklund T. DREADD Modulation of Transplanted DA Neurons Reveals a Novel Parkinsonian Dyskinesia Mechanism Mediated by the Serotonin 5-HT6 Receptor. Neuron. 2016;90:955–968. doi: 10.1016/j.neuron.2016.04.017. This work exemplifies the use of combinations of DREADD multiplexing (rM3Ds, hM3Dq, KORD) to define novel GPCR mechanisms in CNS disease models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vardy E, Robinson JE, Li C, Olsen RHJ, DiBerto JF, Giguere PM, Sassano FM, Huang X-P, Zhu H, Urban DJ, et al. A New DREADD Facilitates the Multiplexed Chemogenetic Interrogation of Behavior. Neuron. 2015;86:936–946. doi: 10.1016/j.neuron.2015.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Milosavljevic N, Cehajic-Kapetanovic J, Procyk CA, Lucas RJ. Chemogenetic Activation of Melanopsin Retinal Ganglion Cells Induces Signatures of Arousal and/or Anxiety in Mice. Curr Biol. 2016;26:2358–2363. doi: 10.1016/j.cub.2016.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mostany R, Miquelajauregui A, Shtrahman M, Portera-Cailliau C. Two-Photon Excitation Microscopy and Its Applications in Neuroscience. In: Verveer PJ, editor. Advanced Fluorescence Microscopy. Springer; New York: 2015. pp. 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hübener M, Keck T, Knott G, Lee W-CA, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim T-i, McCall JG, Jung YH, Huang X, Siuda ER, Li Y, Song J, Song YM, Pao HA, Kim R-H, et al. Injectable, Cellular-Scale Optoelectronics with Applications for Wireless Optogenetics. Science. 2013;340:211–216. doi: 10.1126/science.1232437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jeong J-W, McCall JG, Shin G, Zhang Y, Al-Hasani R, Kim M, Li S, Sim JY, Jang K-I, Shi Y, et al. Wireless Optofluidic Systems for Programmable In Vivo Pharmacology and Optogenetics. Cell. 2015;162:662–674. doi: 10.1016/j.cell.2015.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Robinson S, Todd TP, Pasternak AR, Luikart BW, Skelton PD, Urban DJ, Bucci DJ. Chemogenetic Silencing of Neurons in Retrosplenial Cortex Disrupts Sensory Preconditioning. J Neurosci. 2014;34:10982–10988. doi: 10.1523/JNEUROSCI.1349-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chang SE, Todd TP, Bucci DJ, Smith KS. Chemogenetic manipulation of ventral pallidal neurons impairs acquisition of sign-tracking in rats. Eur J Neurosci. 2015;42:3105–3116. doi: 10.1111/ejn.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Poyraz FC, Holzner E, Bailey MR, Meszaros J, Kenney L, Kheirbek MA, Balsam PD, Kellendonk C. Decreasing Striatopallidal Pathway Function Enhances Motivation by Energizing the Initiation of Goal-Directed Action. J Neurosci. 2016;36:5988–6001. doi: 10.1523/JNEUROSCI.0444-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Urban DJ, Zhu H, Marcinkiewcz CA, Michaelides M, Oshibuchi H, Rhea D, Aryal DK, Farrell MS, Lowery-Gionta E, Olsen RHJ, et al. Elucidation of The Behavioral Program and Neuronal Network Encoded by Dorsal Raphe Serotonergic Neurons. Neuropsychopharmacology. 2016;41:1404–1415. doi: 10.1038/npp.2015.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.MacLaren DA, Browne RW, Shaw JK, Radhakrishnan SK, Khare P, España RA, Clark SD. Clozapine-n-oxide administration produces behavioral effects in Long-Evans rats - implications for designing DREADD experiments. eneuro. 2016 doi: 10.1523/ENEURO.0219-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saloman JL, Scheff NN, Snyder LM, Ross SE, Davis BM, Gold MS. Gi-DREADD Expression in Peripheral Nerves Produces Ligand-Dependent Analgesia, as well as Ligand-Independent Functional Changes in Sensory Neurons. J Neurosci. 2016;36:10769–10781. doi: 10.1523/JNEUROSCI.3480-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sizemore RJ, Seeger-Armbruster S, Hughes SM, Parr-Brownlie LC. Viral vector-based tools advance knowledge of basal ganglia anatomy and physiology. J Neurophysiol. 2016;115:2124–2146. doi: 10.1152/jn.01131.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Parr-Brownlie LC, Bosch-Bouju C, Schoderboeck L, Sizemore R, Abraham W, Hughes SM. Lentiviral vectors as tools to understand central nervous system biology in mammalian model organisms. Front Mol Neurosci. 2015 doi: 10.3389/fnmol.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kügler S. Tissue-Specific Promoters in the CNS. In: Manfredsson FP, editor. Gene Therapy for Neurological Disorders. Springer; New York: 2016. pp. 81–91. [DOI] [PubMed] [Google Scholar]
- 127.Schnütgen F, Doerflinger N, Calléja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 128.Gompf HS, Budygin EA, Fuller PM, Bass CE. Targeted genetic manipulations of neuronal subtypes using promoter-specific combinatorial AAVs in wild-type animals. Front Behav Neurosci. 2015 doi: 10.3389/fnbeh.2015.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brust RD, Corcoran AE, Richerson GB, Nattie E, Dymecki SM. Functional and Developmental Identification of a Molecular Subtype of Brain Serotonergic Neuron Specialized to Regulate Breathing Dynamics. Cell Rep. 2014;9:2152–2165. doi: 10.1016/j.celrep.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hnasko TS, Perez FA, Scouras AD, Stoll EA, Gale SD, Luquet S, Phillips PEM, Kremer EJ, Palmiter RD. Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc Natl Acad Sci. 2006;103:8858–8863. doi: 10.1073/pnas.0603081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Junyent F, Kremer EJ. CAV-2 — why a canine virus is a neurobiologist’s best friend. Curr Opin Pharmacol. 2015;24:86–93. doi: 10.1016/j.coph.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 133.Marchant NJ, Whitaker LR, Bossert JM, Harvey BK, Hope BT, Kaganovsky K, Adhikary S, Prisinzano TE, Vardy E, Roth BL, et al. Behavioral and Physiological Effects of a Novel Kappa-Opioid Receptor-Based DREADD in Rats. Neuropsychopharmacology. 2016;41:402–409. doi: 10.1038/npp.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Augur IF, Wyckoff AR, Aston-Jones G, Kalivas PW, Peters J. Chemogenetic Activation of an Extinction Neural Circuit Reduces Cue-Induced Reinstatement of Cocaine Seeking. J Neurosci. 2016;36:10174–10180. doi: 10.1523/JNEUROSCI.0773-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tervo DGR, Hwang B-Y, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons [Internet] Neuron. doi: 10.1016/j.neuron.2016.09.021. [date unknown], 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schindler SE, McCall JG, Yan P, Hyrc KL, Li M, Tucker CL, Lee J-M, Bruchas MR, Diamond MI. Photo-activatable Cre recombinase regulates gene expression in vivo. Sci Rep. 2015;5:13627. doi: 10.1038/srep13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol. 2016;12:425–430. doi: 10.1038/nchembio.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kawano F, Okazaki R, Yazawa M, Sato M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering [Internet] Nat Chem Biol. 2016 doi: 10.1038/nchembio.2205. advance online publication. [DOI] [PubMed] [Google Scholar]
- 139.O’Neill PR, Gautam N. Optimizing optogenetic constructs for control over signaling and cell behaviours. Photochem Photobiol Sci. 2015;14:1578–1585. doi: 10.1039/c5pp00171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140*.Chen D, Gibson ES, Kennedy MJ. A light-triggered protein secretion system. J Cell Biol. 2013;201:631–640. doi: 10.1083/jcb.201210119. This proof of concept study demonstrated optical control over the secretion of proteins from [stores] at individual dendritric branches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hallett RA, Zimmerman SP, Yumerefendi H, Bear JE, Kuhlman B. Correlating in Vitro and in Vivo Activities of Light-Inducible Dimers: A Cellular Optogenetics Guide. ACS Synth Biol. 2016;5:53–64. doi: 10.1021/acssynbio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yamashita T, Ohuchi H, Tomonari S, Ikeda K, Sakai K, Shichida Y. Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc Natl Acad Sci. 2010;107:22084–22089. doi: 10.1073/pnas.1012498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gutierrez DV, Mark MD, Masseck O, Maejima T, Kuckelsberg D, Hyde RA, Krause M, Kruse W, Herlitze S. Optogenetic Control of Motor Coordination by Gi/o Protein-coupled Vertebrate Rhodopsin in Cerebellar Purkinje Cells. J Biol Chem. 2011;286:25848–25858. doi: 10.1074/jbc.M111.253674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural Photoreceptors as a Source of Fluorescent Proteins, Biosensors, and Optogenetic Tools. Annu Rev Biochem. 2015;84:519–550. doi: 10.1146/annurev-biochem-060614-034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fehrentz T, Schönberger M, Trauner D. Optochemical Genetics. Angew Chem Int Ed. 2011;50:12156–12182. doi: 10.1002/anie.201103236. [DOI] [PubMed] [Google Scholar]
- 146.Sreekumar R, Ikebe M, Fay FS, Walker JW. Biologically active peptides caged on tyrosine. In: Marriot G, editor. Caged Compounds. Academic Press; 1998. pp. 78–94. [DOI] [PubMed] [Google Scholar]