The zona pellucida (ZP) or vitelline envelope (VE) is a cellular feature unique to animal oocytes that provides a barrier to the external environment. This structure is composed of several glycoproteins that universally contain a ZP domain: a polypeptide of ~260 residues that functions as a polymerization module, adopts a predominantly β-sheet fold, and includes 8–10 cysteine residues that form disulfide bonds (Bork and Sander 1992; Han et al. 2010). Our knowledge of the structural features underlying ZP domains has been enhanced in recent years by the release of multiple crystal structures and molecular evolutionary analyses that, together, reveal a repeated pattern of gene duplication, sequence adaptation, and structural conservation from yeast to humans (Swanson et al. 2011). Within ZP domains are two clusters of conserved cysteine residues, which are often separately described as the ZP-N and ZP-C subdomains or regions. However, biochemical and evolutionary analyses unequivocally support that both ZP-N and ZP-C are bone fide protein domains.
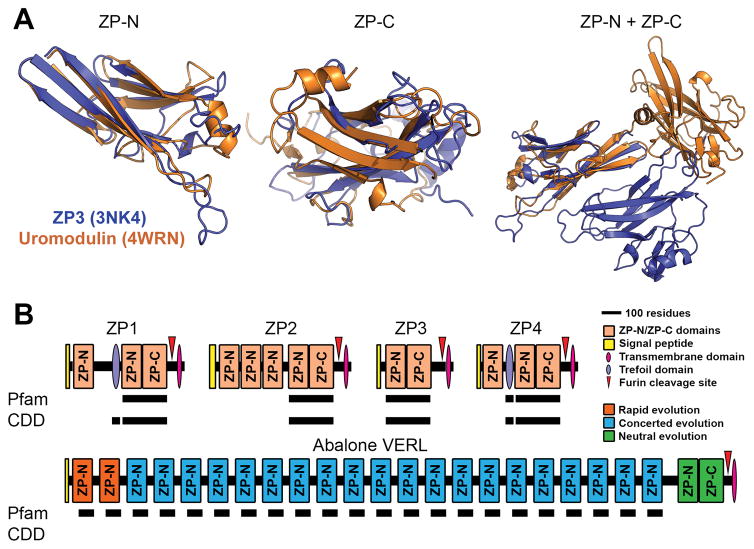
The crystal structures of two “ZP domain”-containing proteins – chicken ZP3 (Protein Data Bank: 3NK4) and human uromodulin (4WRN) – reveal close structural alignment between the ZP-N and ZP-C segments when considered independently, but no global topological conservation between the two motifs (Fig. 1A) (Bokhove et al. 2016; Han et al. 2010). Functionally, biochemical data indicate that only ZP-N is required for protein polymerization (Jovine et al. 2006). Evolutionarily, many ZP/VE proteins contain tandem arrays of ZP-N repeats that evolved independently of one another and from their associated ZP-C motifs (Fig. 1B) (Callebaut et al. 2007; Swanson et al. 2011; Wilburn and Swanson 2016). Therefore, by all common definitions, ZP-N and likely ZP-C are separate protein domains. While ZP-N and ZP-C are often referred to as separate domains in the protein structure literature, the reproduction community commonly still conflates these two motifs as a single domain, such that we contest this is creating a language barrier and challenges to research.
Figure 1. Protein and gene structures of ZP-N/ZP-C domain containing proteins.
(A) Comparison of chicken ZP3 (Protein Data Bank: 3NK4) and human uromodulin (4WRN). Independent alignment of the ZP-N and ZP-C domain reveal close structural similarity of ~3.4 Å and 4.2 Å rmsd, respectively. When viewed together (and aligned based on the ZP-N domain), however, there is no similarity in the positioning of ZP-C relative to ZP-N (adapted from Bokhove et al., 2016). (B) Several classical “ZP domain”-containing proteins often contain N-terminal repeats of the ZP-N domain. Structural databases such as Pfam and the Conserved Domain Database (CDD), however, do not detect these additional repeats in human ZP proteins; Pfam also did not detect the conserved trefoil domain in ZP1. The most extreme case of this ZP-N duplication is in abalone VERL, which has 22 ZP-N repeats with different modes of evolution based on genetic effects and adaptation with coevolving sperm proteins. Pfam includes an additional entry for 78 residues of the VERL repeats (pfam11386), but this annotation does not fully span the repeats or report ZP-N homology. The C-terminal ZP module of VERL is also not detected in either database (adapted from Wilburn and Swanson, 2016).
Structural databases such as Pfam and the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) only allow for searching of motifs that encompass the combined ZP-N and ZP-C, such that independent gene duplication and co-option events are missed without rigorous manual annotation (Swanson et al. 2011). As an example, when human ZP2 is queried against either aforementioned database, only the C-terminal “ZP domain” is reported whereas the preceding three ZP-N repeats are ignored (1B). A prime example of how this nomenclature may be problematic for the community is in an excellent report using transgenic mice by Avella et al. (2014), in which the authors carefully demonstrate that replacement of mouse ZP2 residues 51–149 with the orthologous human segment was sufficient to allow ZP recognition by human sperm; what the authors do not discuss is that these residues almost exclusively comprise the first ZP-N domain (indeed, they do not acknowledge any domains in ZP2 beyond a single “ZP domain”). In the marine mollusk abalone, the first and second ZP-N repeats of the egg coat protein VERL are rapidly evolving and likely also confer species-specific gamete recognition (Fig. 1B), suggesting recurrent evolutionary and functional patterns in animal fertilization proteins that were previously missed (Wilburn and Swanson 2016). Currently, Pfam includes a profile for a partial segment of each VERL repeat, which successfully identifies both the rapidly evolving and homogenized repeats (Fig. 1B); if properly annotated, all ZP-N repeats of ZP2 might have been similarly identified. More recently, Bokhove et al. (2016) referred to the combined ZP-N/ZP-C unit as a “ZP module,” which is likely a better term to refer to the often (but not always) co-occurring domains. Hence, we propose that a concerted effort should be made to replace “ZP domain” with “ZP module,” as well as to treat both ZP-N and ZP-C as independent protein domains. Such a change in nomenclature will facilitate their identification and the biological interpretation of their function.
Acknowledgments
This work was supported by National Institutes of Health grants R01-HD076862 to WJS and F32-GM116298 to DBW. The authors have no conflicts of interest.
References
- Avella MA, Baibakov B, Dean J. A single domain of the ZP2 zona pellucida protein mediates gamete recognition in mice and humans. The Journal of cell biology. 2014;205(6):801–809. 55. doi: 10.1083/jcb.201404025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhove M, Nishimura K, Brunati M, Han L, de Sanctis D, Rampoldi L, Jovine L. A structured interdomain linker directs self-polymerization of human uromodulin. Proceedings of the National Academy of Sciences. 2016;113(6):1552–1557. doi: 10.1073/pnas.1519803113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C. A large domain common to sperm receptors (Zp2 and Zp3) and TGF-β type III receptor. FEBS Letters. 1992;300(3):237–240. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP, Monget P. Isolated ZP-N domains constitute the N-terminal extensions of Zona Pellucida proteins. Bioinformatics. 2007;23(15):1871–1874. doi: 10.1093/bioinformatics/btm265. [DOI] [PubMed] [Google Scholar]
- Han L, Monné M, Okumura H, Schwend T, Cherry AL, Flot D, Matsuda T, Jovine L. Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3. Cell. 2010;143(3):404–415. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Jovine L, Janssen WG, Litscher ES, Wassarman PM. The PLACI-homology region of the ZP domain is sufficient for protein polymerisation. BMC Biochemistry. 2006;7:11. doi: 10.1186/1471-2091-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Aagaard JE, Vacquier VD, Monné M, Sadat Al Hosseini H, Jovine L. The molecular basis of sex: linking yeast to human. Molecular Biology and Evolution. 2011;28(7):1963–1966. doi: 10.1093/molbev/msr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilburn DB, Swanson WJ. From molecules to mating: Rapid evolution and biochemical studies of reproductive proteins. Journal of Proteomics. 2016;135:12–25. doi: 10.1016/j.jprot.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]