Abstract
Hypoxia-inducible factors (HIFs) belong to a family of transcription factors (TF) responsive to a low O2 availability, which is often a characteristic feature of solid tumors. The alpha subunit of the HIF heterodimer is O2-sensitive, and once stabilized in hypoxia, it functions as a master regulator of various genes involved in hypoxia pathway. Changes in the HIF1A (hypoxia inducible factor 1, alpha subunit) nucleotide sequence or expression has been shown to be associated with development of several diseases. Due to increasing research interest in HIF1A gene a review of association studies was needed. We here reviewed published data on single nucleotide polymorphisms (SNPs) in HIF1A in various diseases; in total, 34 SNPs were tested for an association with 49 phenotypes, and the results were visualized using the Cytoscape software. Among all collected polymorphisms 16 SNPs showed significant associations with 40 different phenotypes, including six SNPs associated with 14 cancer types. Missense SNPs (rs11549465 and rs11549467) within the oxygen-dependent degradation domain were most frequently studied. The study provides a comprehensive tool for researchers working in this area and may contribute to more accurate disease diagnosis and identification of therapeutic targets.
Keywords: HIF1A gene, polymorphism, SNP, human, cancer, association study
Introduction
Mammalian cells rapidly respond and adapt to low oxygen conditions (hypoxia). In hypoxic cells important responses are activated for metabolic, bioenergetics, and redox demands (reviewed in (Majmundar et al., 2010)). A primary transcriptional response to hypoxia is mediated by the hypoxia-inducible factor (HIF), which is known as a pivotal regulator under hypoxia stress. Besides its adaptive response in cellular stress, it carries important roles in physiological and pathological processes (Semenza 2003, Majmundar et al., 2010). The HIF protein complex is a heterodimer consisting of an oxygen sensitive (alpha, A) and an oxygen stable (beta, B) subunit (Wang and Semenza 1993). In mammals three different HIFA isoforms are present, among which HIF1A is expressed ubiquitously while HIF2A and HIF3A expression vary depending on type of tissue cells (Bertout et al., 2008). In normally oxidized conditions (normoxia) A-subunits are hydroxylated at conserved proline residues (p.Pro402 and p.Pro564) (Ivan et al. 2001) by oxygen regulated prolyl hydroxylase domain-containing enzymes (EGLN1, 2, 3 or PHD1, 2, 3) (Wang et al. 1995, Chan et al. 2005, Kaelin and Ratcliffe 2008). Marked HIFA-subunits are recognized by E3 ubiquitin ligase from the von Hippel-Lindau protein complex (pVHL) for proteasomal degradation (Wang et al. 1995, Majmundar et al., 2010). The pVHL can target the N-terminal transactivation domain (N-TAD) within the oxygen-dependent degradation domain (ODD domain), which controls HIF1A degradation by ubiquitin-proteasome pathway and consists of approximately 200 amino acids (Huang et al. 1998) (Supplementary Figure 1). The removal of the ODD domain renders HIF1A stability even under normoxic conditions, consequently resulting in autonomous HIF1A heterodimerisation, DNA binding and transactivation independently from hypoxic signaling (Huang et al. 1998).
Transcription factor HIF1A mediates transcriptional responses to hypoxia for a high number of genes to control cellular oxygen supply and maintain cell viability during periods of low oxygen concentration (Wang et al. 1995, Wenger et al., 2005, Vilela et al., 2008, Keith et al., 2012). Analysis of 98 HIF1A target genes collected from 51 published studies revealed 20 associated pathways (Slemc and Kunej 2016). HIF1A was implicated in: metabolism and redox homeostasis (glucose catabolism, regulation of lipid metabolism), vascular responses in hypoxia (ischemia-induced angiogenesis, endothelial cells), cancer (tumorigenesis, metastasis, tumor angiogenesis, cancer stem cells, regulation by cancer metabolism), inflammation (regulation in inflammatory cells, myeloid cell function, tumor-associated macrophages), and also as part of a systemic response to hypoxia reviewed in (Majmundar et al., 2010). Besides regulating expression of protein-coding genes, it has been shown that HIF1A also regulates noncoding RNA genes (ncRNA) including microRNAs (miRNAs) (Gee et al. 2014) and transcribed-ultraconserved regions (T-UCRs) (Ferdin et al. 2013).
Although HIF1A gene has been a topic of several studies it is not yet included in the Diseasome map, which visualizes known interactions between genes and diseases (Goh et al. 2007). The aim of the present study was to review published reports of associations between HIF1A gene polymorphisms and diseases or phenotypic traits in human and to graphically visualize associations as the gene-disease network.
A literature review of HIF1A polymorphisms, associated diseases and phenotypes was performed using PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and Web of Science (WoS; http://apps.webofknowledge.com/WOS). The literature search was performed on publications published between 2003 and 2015 using keywords, including: HIF1A, polymorphisms, SNP, human, cancer, and association studies. The nomenclature for SNPs was unified to the reference SNP (rs) IDs according to the Human Genome Variation Society (HGVS) guidelines (http://www.hgvs.org/mutnomen/). Versions of dbSNP build 137 from NCBI and Ensembl (release 85) were used to obtain information about variations (biotype of the polymorphism and genomic location). Associations between the HIF1A gene and phenotypes were visualized using Cytoscape 3.2.1 software (Shannon et al. 2003). Associated phenotypes have been sorted into disease categories as in the Diseasome map, which includes 22 categories of diseases and disorders: bone, cancer, cardiovascular, connective tissue, dermatological, developmental, ear, nose and throat, endocrine, gastrointestinal, hematological, immunological, metabolic, muscular, neurological, nutritional, ophthamological, psychiatric, renal, respiratory, skeletal, multiple, and unclassified (Goh et al. 2007). Due to heterogeneity in phenotype nomenclature in publications, the terminology was unified according to the Disease Ontology (DO) database (http://disease-ontology.org/).
Extraction of literature data and editing according to the genomic databases
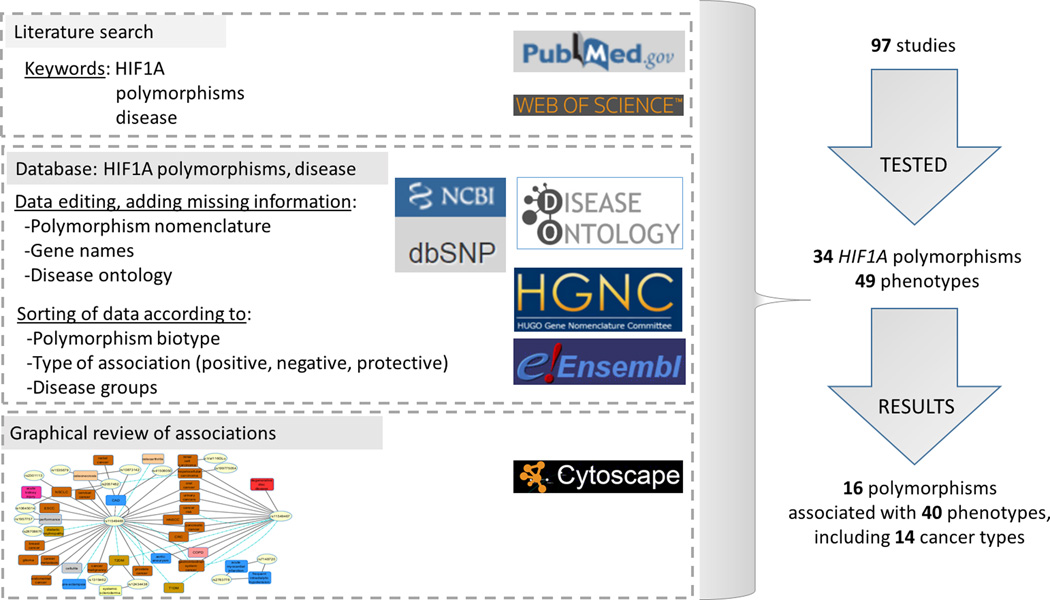
The workflow of the study and main results are presented in Fig. 1. Polymorphisms’ names were edited according the latest genomic browser releases. Usually, articles had polymorphisms defined with rs ID numbers, but in more than 20 cases it was necessary to identify corresponding rs ID number in genomic databases (Prior et al. 2003, Yamada et al. 2005, Hong et al. 2007, Konac et al. 2007, Lee et al. 2008). Two polymorphisms from the published literature (rs62639821 and rs1957577) were not included in the study, since they are now annotated to different genomic locations. Disease and phenotype names used in the studies were edited according to the nomenclature in the DO database, if available. Namely; some studies used different terms for the same disease, or only some aspect(s) of disease development, progression or consequence were recorded. Out of 49 tested phenotypes it was possible to translate 38 phenotypes according to the DO database, however for the rest of the tested diseases and phenotypes an appropriate term was not yet available in the database. Because some of the studied traits are phenotypes and not diseases, it will be necessary in the future studies to combine data from additional ontology databases like Human Phenotype Ontology (HPO) to verify the terminology of all traits.
Figure 1.
Workflow of the study and main results. We performed literature search in PubMed and Web of Science and extracted data related to polymorphisms and phenotype. The collected data were edited and complemented with additional information. Next, we sorted studies according to association type between polymorphism and phenotype (positive, protective, negative). Disease and phenotype terminology was curated according to Disease Ontology database and sorted into classes according to Diseasome map. Collected associations between SNPs and phenotypes were visualized as a network.
Visualization of the HIF1A gene - phenotype network
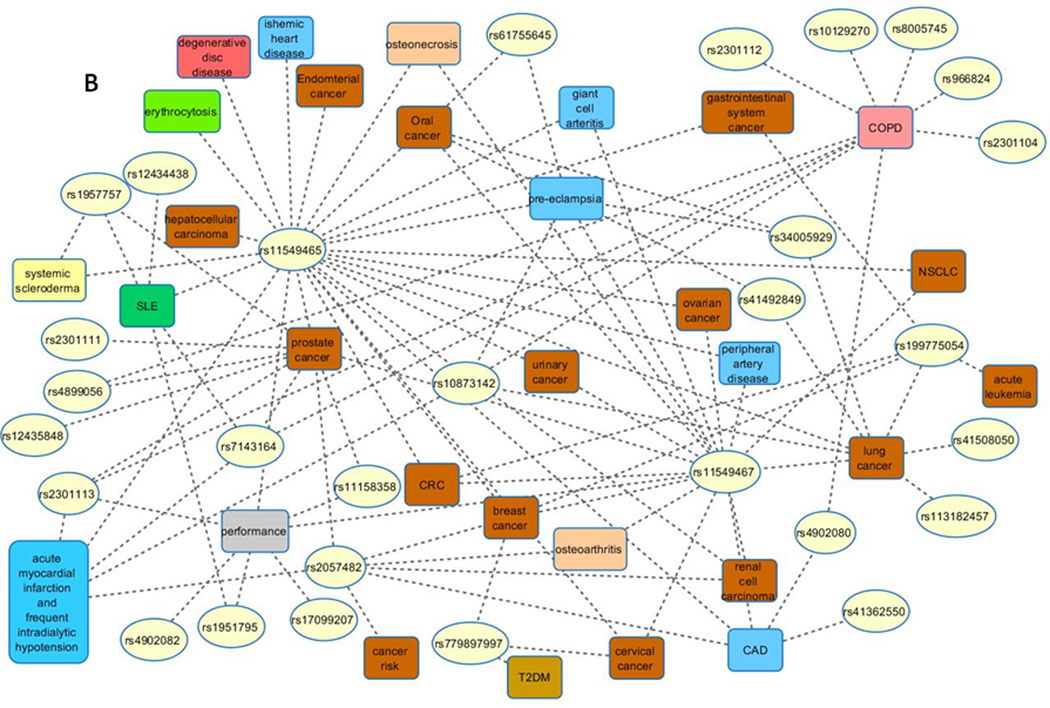
Literature review of 97 association studies in humans revealed that 34 HIF1A SNPs were tested for an association with 49 phenotypes (Figure 2, Supplementary Figure 2). Those 49 phenotypes belong to 11 disease categories: cancer, cardiovascular, bone, renal, hematological, immunological, connective tissue disorder, respiratory, gastrointestinal, muscular and unclassified. Among tested associations 16 SNPs showed positive associations with 40 phenotypes, including 14 cancer types (Figure 2A). Some studies also showed the protective role of polymorphisms; meaning a protective association for the specific disease phenotype (such as loss of articular cartilage, or advanced prostate cancer, and so on); three polymorphisms in nine associations were associated with lower risk for development of different diseases (blue lines in Fig. 2A). The studied HIF1A SNPs are located within various genomic regions: 5’-flanking, exons, introns, 3’-UTR, and 3’-flanking (Supplementary Figure 3). An integrated review revealed that among 34 SNPs, 10 showed variable associations (i.e., either present or absent; Table 1), 6 were always positively associated with the tested phenotypes, and 18 showed no association with the tested phenotypes (Table 2, Fig. 2B). Among all SNPs, missense polymorphisms within the ODD domain (rs11549465 and rs11549467) were most frequently studied and for which variable nomenclature was used in publications. Moreover, several studies that tested associations of these two SNPs with phenotypes showed conflicting results for the same cancer type. For example, opposing results were reported for rs11549465 in prostate cancer susceptibility studies; with association (Chau et al. 2005, Fu et al. 2005, Orr-Urtreger et al. 2007, Jacobs et al. 2008, Foley et al. 2009) and no association (Li et al. 2007, Li et al. 2012).
Figure 2.

Visualization of reported HIF1A genotype-phenotype associations studies in human. (A) Associations between HIF1A SNPs and studied phenotypes. Blue lines denote protective role of polymorphism. (B) Reported negative associations between studied SNPs and phenotypes.
Figure legend: CAD, coronary artery disease; COPD, chronic obstructive pulmonary disease; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; NSCLC, non-small cell lung cancer; OSCC, Oral squamous cell carcinoma; T1DM, diabetes mellitus type 1; T2DM, diabetes mellitus type 2; SLE, systemic lupus erythematosus. The term “performance” is used as a hypernym for: adaptation to living at high altitude, endurance status, maximal oxygen consumption, power-oriented athletes, symptom-limited exercise test duration over time, and response to acute hypoxia.
Table 1.
HIF1A SNPs showing both positive and negative association with disease phenotypes.
| # | SNP ID/substitution |
SNP name as in reference |
Genomic location |
Association with diseases and phenotypes |
|---|---|---|---|---|
| 1 | rs1957757 [T>C] | intron 6 | ||
| (+) | ○ symptom-limited exercise test duration (Sarzynski et al. 2010) | |||
| (−) | • prostate cancer risk (Jacobs et al. 2008) | |||
| ○ systemic sclerosis (Wipff et al. 2009) | ||||
| ○ systemic lupus erythematosus (Feng et al. 2014) | ||||
| 2 | rs12434438 [G>A] | intron 6 | ||
| (+) | ○ systemic sclerosis (Wipff et al. 2009) | |||
| ○ T2DM (together with rs1319462) (Yamada et al. 2005) | ||||
| (−) | ○ Systemic lupus erythematosus (Feng et al. 2014) | |||
| 3 | rs10873142 [C>T] | intron 8 | ||
| (+) | ○ coronary artery disease with stable exertional angina (Hlatky et al. 2007) |
|||
| ○ idiopathic osteonecrosis of the femoral head in men (Hong et al. 2007) | ||||
| (−) | ○ acute myocardial infarction and frequent intradialytic hypotension (Hemodialysis patients) (Zheng et al. 2009) |
|||
| ○ early- onset pre-eclampsia (Andraweera et al. 2014) | ||||
| ○ lung cancer (Konac et al. 2009) | ||||
| ○ COPD (Ding et al. 2015) | ||||
| 4 | rs41508050 [C>T] | exon 10 | ||
| (+) | ○ coronary artery disease with stable exertional angina (Hlatky et al. 2007) |
|||
| Thr418Ile | (−) | • lung cancer (Konac et al. 2009) | ||
| 5 | rs2301113 [C>A] | intron 10 | ||
| (+) | ○ early-stage NSCLC (Liu et al. 2014) | |||
| (−) | ○ prostate cancer risk (Jacobs et al. 2008) | |||
| ○ acute myocardial infarction and frequent intradialytic hypotension (Hemodialysis patients) (Zheng et al. 2009) | ||||
| ○ elite endurance status (Doring et al. 2010) | ||||
| ○ COPD(Ding et al. 2015) | ||||
| 6 | rs11549465 [C>T] | exon 12 | ||
| Pro582Ser | (+) | • cancer risk (He et al. 2013, Yang et al. 2013, Hu et al. 2014, Ye et al. 2014) and cancer metastasis (Zhang et al. 2013) cancer malignancy (Wu et al. 2014) |
||
| • glioma (Xu et al. 2011) | ||||
| • oral cancer risk (together with rs11549467) (Chen et al. 2009) | ||||
| • esophageal squamous cell carcinoma (Ling et al. 2005) | ||||
| • head and neck squamous cell carcinoma (Tanimoto et al. 2003) | ||||
| • renal cell carcinoma (Ollerenshaw et al. 2004) | ||||
| C2028T | • lung cancer (NSCLC) (Koukourakis et al. 2006) | |||
| • breast cancer risk (poor prognosis) (Naidu et al. 2009, Lee et al. 2008, Kim et al. 2008) | ||||
| • CRC risk and metastasis (Kuwai et al. 2004, Kang et al. 2011), ulcerative CRC (Fransen et al. 2006) | ||||
| • prostate cancer risk/susceptibility (Foley et al. 2009, Orr-Urtreger et al. 2007); androgen-independent prostate cancer (Chau et al. 2005, Fu et al. 2005) | ||||
| • pancreatic cancer (Ruiz-Tovar et al. 2012) (Wang et al. 2011) | ||||
| C1772T | • cervical and endometrial cancers (Konac et al. 2007) | |||
| • urinary cancers (Li et al. 2013) | ||||
| • increased female specific cancer risk (Zhao et al. 2009) | ||||
| • gastrointestinal tract cancer risk (Xu et al.) | ||||
| ○ CAD with stable exertional angina (Hlatky et al. 2007) | ||||
| ○ ischemic heart disease (collaterals formation) (Resar et al. 2005); | ||||
| ○ chronic obstructive pulmonary disease susceptibility (with rs11549467) (Putra et al. 2011a) | ||||
| ○ risk of cellulite (Emanuele et al. 2010) | ||||
| ○ abdominal aortic aneurysm (Strauss et al. 2012) | ||||
| ○ acute kidney injury (Kolyada et al. 2009) | ||||
| ○ endurance status (McPhee et al. 2011, Doring et al. 2010) | ||||
| ○ maximal oxygen consumption (Prior et al. 2003) | ||||
| ○ power-oriented athletes and muscle activity (Ahmetov et al. 2008, Cieszczyk et al. 2011, Gabbasov et al. 2012) | ||||
| ✓ knee osteoarthritis (Fernandez-Torres et al. 2015) | ||||
| ✓ diabetic nephropathy (Gu et al. 2012) | ||||
| ✓ prostate cancer (Jacobs et al. 2008) | ||||
| ✓ T2DM (Yamada et al. 2005, Nagy et al. 2009) | ||||
| ✓ T1DM (Nagy et al. 2009) | ||||
| ✓ early-onset pre-eclampsia (Andraweera et al. 2014) | ||||
| (−) | • gastric cancer (Li et al. 2009) | |||
| • breast cancer risk (Zagouri et al. 2012, Vleugel et al. 2005, Meka et al. 2015); sporadic breast cancer (Apaydin et al. 2008) | ||||
| • ovarian cancer (Konac et al. 2007) | ||||
| • digestive cancer (Yang et al. 2014, Sun et al. 2015) | ||||
| • prostate cancer (Li et al. 2007, Li et al. 2012) | ||||
| • cervical cancer (Fu Sl Fau - Miao et al. 2014) | ||||
| • bladder cancer occurrence (Nadaoka et al. 2008) | ||||
| • oral squamous cell carcinoma (Munoz-Guerra et al. 2009, Shieh et al. 2010) | ||||
| • lung cancer (Konac et al. 2009, Kuo et al. 2012) (associated with lung cancer with TP53 LOH) (Putra et al. 2011b), NSCLC (Kim et al. 2010) | ||||
| • renal cell carcinoma (Morris et al. 2009) (Qin et al. 2011) | ||||
| • hepatocellular carcinoma risk (Hsiao et al. 2010) | ||||
| • endometrial cancer risk (potential association with tumorigenesis and increased tumor vasculature) (Horree et al. 2008) | ||||
| • survival of patients with colorectal cancer (Lee et al. 2011) | ||||
| ○ systemic sclerosis (Wipff et al. 2009) | ||||
| ○ acute myocardial infarction and frequent intradialytic hypotension (Zheng et al. 2009) | ||||
| ○ pre-eclampsia (Heino et al. 2008, Kim et al. 2012, Nava-Salazar et al. 2011) | ||||
| ○ erythrocytosis (Percy et al. 2003) | ||||
| ○ giant cell arteritis (Torres et al. 2010) | ||||
| ○ peripheral artery disease (Bahadori et al. 2010) | ||||
| ○ osteonecrosis (Chachami et al. 2013) | ||||
| ○ premature coronary artery disease (Lopez-Reyes et al. 2014) | ||||
| ○ ischemic heart disease (coronary artery collaterals) (Alidoosti et al. 2011) | ||||
| ○ lumbar disc degeneration (Lin et al. 2013) | ||||
| ○ systemic lupus erythematosus (Feng et al. 2014) | ||||
| ○ power-oriented athletes (Eynon et al. 2010) | ||||
| ○ response to acute hypoxia (Hennis et al. 2010) | ||||
| ○ idiopathic osteonecrosis of the femoral head in men (Hong et al. 2007) | ||||
| ○ acute mountain sickness (Droma et al. 2008) | ||||
| 7 | rs11549467 [G>A] | exon 12 | ||
| Ala588Thr | (+) | • cancer risk (Zhou et al. 2014, Yang et al. 2013, Liu and Zhang 2013, Hu et al. 2014) | ||
| • urinary cancers (Li et al. 2013) | ||||
| • digestive cancers (Yang et al. 2014, Sun et al. 2015) | ||||
| • oral squamous cell carcinoma (Chen et al. 2009, Munoz-Guerra et al. 2009) | ||||
| • head and neck squamous cell carcinoma (Tanimoto et al. 2003) | ||||
| • gastric cancer (Li et al. 2009) | ||||
| • ulcerative CRC (together with rs11549467) (Fransen et al. 2006) | ||||
| • hepatocellular carcinoma risk (Hsiao et al. 2010) | ||||
| • renal cell carcinoma (Ollerenshaw et al. 2004) | ||||
| • pancreatic cancer (Ruiz-Tovar et al. 2012, Wang et al. 2011) | ||||
| • prostate cancer (Li et al. 2012) | ||||
| ○ chronic obstructive pulmonary disease susceptibility (with rs11549465) (Putra et al. 2011a) | ||||
| ○ abdominal aortic aneurysm (Strauss et al. 2012) | ||||
| ○ lumbar disc degeneration (Lin et al. 2013) | ||||
| ✓ breast cancer risk (Zhao et al. 2009) | ||||
| ✓ T1DM (Nagy et al. 2009) | ||||
|
G1790A; G2046A |
(−) | • oral squamous cell carcinoma (Shieh et al. 2010) | ||
| • breast cancer risk (Naidu et al. 2009, Apaydin et al. 2008, Kim et al. 2008) | ||||
| • lung cancer (Konac et al. 2009), (associated with adenocarcinoma with 1p34 LOH) (Putra et al. 2011b)) and lung carcinoma (NSCLC) (Koukourakis et al. 2006, Kuo et al. 2012, Kim et al. 2010) | ||||
| • CRC risk, progression and metastasis (Knechtel et al. 2010, Szkandera et al. 2010) | ||||
| • prostate cancer (Li et al. 2007, Orr-Urtreger et al. 2007, Chau et al. 2005) | ||||
| • ovarian, cervical and endometrial cancers (Konac et al. 2007, Fu Sl Fau - Miao et al. 2014) | ||||
| • renal cell carcinoma (Qin et al. 2011, Morris et al. 2009) | ||||
| • bladder cancer (Nadaoka et al. 2008) | ||||
| • survival of patients with colorectal cancer(Lee et al. 2011) | ||||
| ○ pre-eclampsia (Heino et al. 2008, Kim et al. 2012, Nava-Salazar et al. 2011) | ||||
| ○ peripheral artery disease (Bahadori et al. 2010) | ||||
| ○ giant cell arteritis (Torres et al. 2010) | ||||
| ○ coronary artery disease with stable exertional angina (Hlatky et al. 2007) | ||||
| ○ osteonecrosis (Chachami et al. 2013) | ||||
| ○ premature coronary artery disease (Lopez-Reyes et al. 2014) | ||||
| ○ endurance status (Doring et al. 2010) | ||||
| ○ knee osteoarthritis (Fernandez-Torres et al. 2015) | ||||
| 8 | rs199775054 [G>C] |
exon 12 | ||
| Ala593Pro | (+) | • hepatocellular carcinomas (Park et al. 2009) | ||
| Ala593Pro | (−) | • colon, gastric, breast, lung cancer and acute leukemia (Park et al. 2009) |
||
| 9 | rs113182457 rs60361955 [insGT] |
intron 13 | ||
| GT dinucleotide repeat |
(+) | ○ lung carcinoma (NSCLC) (Koukourakis et al. 2006) | ||
| GT14 allele | possible (+) |
○ adaptation to living at high altitude (Suzuki et al. 2003) | ||
| rs10645014 | (−) | • lung cancer (Konac et al. 2009) | ||
| 10 | rs2057482 [T>C] | 3’-UTR | ||
| (+) | • cervical cancer (Fu Sl Fau - Miao et al. 2014) | |||
| • rectal cancer risk (Frank et al. 2010) | ||||
| ○ idiopathic osteonecrosis of the femoral head in men (Hong et al. 2007) | ||||
| ○ early-stage NSCLC (Liu et al. 2014) | ||||
| ○ premature coronary artery disease (Lopez-Reyes et al. 2014) | ||||
| ✓ hepatocellular carcinoma (Guo et al. 2015) | ||||
| (−) | • cancer risk (Hu et al. 2014) | |||
| Ex15+197C>T | • breast cancer risk (Lee et al. 2008) | |||
| • renal cell carcinoma (Qin et al. 2011) | ||||
| • prostate cancer (Li et al. 2012) | ||||
| ○ acute myocardial infarction and frequent intradialytic hypotension (Zheng et al. 2009) | ||||
| ○ knee osteoarthritis (Fernandez-Torres et al. 2015) | ||||
| C191T | ○ coronary artery disease with stable exertional angina (Hlatky et al. 2007) |
Legend:
(+) observed association, (−) no association.
• Cancer
○ Other diseases and phenotypes
✓ Protective role of polymorphism
Table 2.
HIF1A SNPs showing both positive and negative association with disease phenotypes.
| # | SNP ID [substitution] |
Synonym, as named in the reference |
Genomic location |
Association with diseases and phenotypes |
|---|---|---|---|---|
| ASSOCIATION | ||||
| 1 | rs2783778 [C>T] | - | 5’-flanking | ○ acute myocardial infarction and frequent intradialytic hypotension (Hemodialysis patients) (Zheng et al. 2009) |
| 2 | rs7148720 [T>C] | - | 5’-flanking | ○ acute myocardial infarction and frequent intradialytic hypotension (Hemodialysis patients)(Zheng et al. 2009) |
| 3 | rs1535679 [A>C] | −2755C>A | 5’-flanking | ○ idiopathic osteonecrosis of the femoral head in men (Hong et al. 2007) |
| 4 | rs28708675 [A>T] | A-2500T | 5’-flanking | ○ maximal oxygen consumption (Prior et al. 2003) |
| 5 | rs1319462 [G>A] | - | 3’-flanking | ○ T2DM (together with rs12434438) (Yamada et al. 2005) |
| 6 | rs1957755 [G>A] | - | intron 4 | ○ symptom-limited exercise test duration (Sarzynski et al. 2010) |
| NO ASSOCIATION | ||||
| 1 | rs41362550 [T>C] | - | 5’-flanking | ○ CAD with stable exertional angina (Hlatky et al. 2007) |
| 2 | rs7143164 [G>C] | - | intron 1 | ○ acute myocardial infarction and frequent intradialytic hypotension (Hemodialysis patients) (Zheng et al. 2009) |
| ○ systemic lupus erythematosus (Feng et al. 2014) | ||||
| ○ COPD (Ding et al. 2015) | ||||
| • prostate cancer (Jacobs et al. 2008) | ||||
| 3 | rs1951795 [A>C] | - | intron 1 | ○ elite endurance status (Doring et al. 2010) |
| ○ systemic lupus erythematosus (Feng et al. 2014) | ||||
| 4 | rs12435848 [A>G] | - | intron 1 | • prostate cancer risk (Jacobs et al. 2008) |
| 5 | rs2301104 [G>C] | - | intron 1 | ○ COPD (Ding et al. 2015) |
| 6 | rs10129270 [G>A] | - | intron 1 | ○ COPD (Ding et al. 2015) |
| 7 | rs8005745 [T>A] | - | intron 1 | ○ COPD (Ding et al. 2015) |
| 8 | rs779897997[C>A] | S28Y | exon 2 | ○ T2DM (Yamada et al. 2005) |
| C111A | • ovarian, cervical and endometrial cancers (Konac et al. 2007) |
|||
| • breast cancer (Apaydin et al. 2008, Naidu et al. 2009) | ||||
| 9 | rs4899056 [T>C] | - | intron 4 | • prostate cancer (Jacobs et al. 2008) |
| ○ COPD (Ding et al. 2015) | ||||
| 10 | rs11158358 [G>C] | - | intron 6 | • prostate cancer (Jacobs et al. 2008) |
| ○ elite endurance status (elite endurance athletes (EEA)) (Doring et al. 2010) | ||||
| 11 | rs2301111 [G>C] | - | intron 7 | • prostate cancer (Jacobs et al. 2008) |
| 12 | rs966824 [T>C] | - | intron 7 | ○ COPD (Ding et al. 2015) |
| 13 | rs41492849 [C>T] | C1720T | exon 12 | • OSCC (Shieh et al. 2010) |
| • lung cancer (Konac et al. 2009) | ||||
| 14 | rs34005929 [G>A] | 1740G>A | exon 12 | ○ pre-eclampsia (Heino et al. 2008) |
| • OSCC (Shieh et al. 2010) | ||||
| • lung cancer (Konac et al. 2009) | ||||
| 15 | rs61755645 [A>T] | A1828T | exon 12 | ○ pre-eclampsia (Heino et al. 2008) |
| • OSCC (Shieh et al. 2010) | ||||
| 16 | rs4902080 [T>C] | - | intron 12 | ○ CAD with stable exertional angina (Hlatky et al. 2007) |
| ○ COPD (Ding et al. 2015) | ||||
| 17 | rs4902082 [C>T] | T+140 C | intron 14 | ○ maximal oxygen consumption (Prior et al. 2003) |
| 18 | rs17099207 [G>A] | - | 3’-flanking HIF1A and |
○ elite endurance status (elite endurance athletes (EEA)) (Doring et al. 2010) |
Legend:
• Cancer
○ Other diseases and phenotypes
Abbreviations: OSCC, oral squamous cell carcinoma; CAD, coronary artery disease; RCC, renal cell carcinoma; CRC, colorectal cancer; T2DM, diabetes mellitus (type 2); T1DM, diabetes mellitus (type 1); NSCLC, non-small-cell lung cancer.
HIF1A and cancer risk
Among 16 HIF1A SNPs associated with 40 different phenotypes six SNPs have been associated with increased risk for 14 cancer types: rs113182457 (Koukourakis et al. 2006), rs11549465 (Tanimoto et al. 2003, Ollerenshaw et al. 2004, Chau et al. 2005, Fu et al. 2005, Ling et al. 2005, Fransen et al. 2006, Koukourakis et al. 2006, Hong et al. 2007, Konac et al. 2007, Orr-Urtreger et al. 2007, Jacobs et al. 2008, Kim et al. 2008, Lee et al. 2008, Chen et al. 2009, Foley et al. 2009, Naidu et al. 2009, Zhao et al. 2009, Kang et al. 2011, Wang et al. 2011, Xu et al. 2011, Ruiz-Tovar et al. 2012, He et al. 2013, Li et al. 2013, Xu et al. 2013,Yang et al. 2013, Zhang et al. 2013, Hu et al. 2014, Wu et al. 2014, Ye et al. 2014, Fernandez-Torres et al. 2015), rs11549467 (Tanimoto et al. 2003, Ollerenshaw et al. 2004, Fransen et al. 2006, Chen et al. 2009, Li et al. 2009, Munoz-Guerra et al. 2009, Zhao et al. 2009, Hsiao et al. 2010, Wang et al. 2011, Li et al. 2012, Ruiz-Tovar et al. 2012, Li et al. 2013, Liu and Zhang 2013, Yang et al. 2013, Hu et al. 2014, Yang et al. 2014, Zhou et al. 2014, Sun et al. 2015), rs199775054 (Park et al. 2009), rs2057482 (Frank et al. 2010, Fu Sl Fau - Miao et al. 2014, Liu et al. 2014, Guo et al. 2015) and rs2301113 (Liu et al. 2014). These six SNPs were most frequently associated with breast, lung, colorectal (CRC), gastric, prostate, oral cancer and renal cell carcinoma (RCC). It is known that solid tumors frequently have low levels of O2, which can be a result of cancer cells growing more rapidly than their supporting vascular network. Hypoxic stress might also be caused by a perfusion defect as a result of abnormal tumor blood vessel structure and function (Majmundar et al., 2010). Consequently, these events cause HIF1A levels to increase in solid tumors (Bertout et al., 2008), but its levels can additionally be increased by HIF-independent pathways (Majmundar et al., 2010). In addition, HIF1A has a capability to directly reprogram the metabolic state in cells, which is important in hypoxic settings such as vascular disease and cancer (Majmundar et al., 2010). Thus, HIF1A in hypoxic cells regulates the transcription of many genes involved in key aspects of cancer biology, including immortalization, maintenance of stem cell pools, cellular differentiation, genetic instability, vascularization, metabolic reprogramming, autocrine growth factor signaling, invasion/metastasis, and treatment failure (Semenza 2007). Increased expression of HIF1A also often associates with poor clinical prognosis of many cancer types (Semenza 2007).
HIF1A and association with other phenotypes and diseases
Genetic variability of HIF1A was also found to be associated with cardiovascular system diseases like: ischemic heart disease, coronary artery disease (CAD) with stable exertional angina, premature coronary artery disease, pre-eclampsia, acute myocardial infarction and frequent intradialytic hypotension. Polymorphisms associated with cardiovascular system diseases were: rs10873142 (Hlatky et al. 2007), rs11549465 (Resar et al. 2005, Hlatky et al. 2007, Strauss et al. 2012, Andraweera et al. 2014), rs11549467 (Strauss et al. 2012), rs2057482 (Lopez-Reyes et al. 2014), rs2783778 (Zheng et al. 2009), rs41508050 (Hlatky et al. 2007) and rs7148720 (Zheng et al. 2009). Cardiovascular diseases like atherosclerosis in the heart, brain, and limb muscle, are known to be susceptible to ischemic injury (Beckman et al., 2002, Kett-White et al. 2002). Besides, myocardial ischemia is the most common cause of cardiac hypoxia in clinical medicine and it occurs when O2 delivery cannot meet myocardial metabolic requirements in the heart (Shohet and Garcia 2007). Expression of HIF1A is essential and sufficient for promoting reperfusion in ischemic skeletal muscle (Majmundar et al., 2010). Moreover, reduced HIF1A activation was also found in hypoxic skin wounds of aged diabetic mice (Liu et al. 2008), emphasizing the role of age in ischemic response. This is important since peripheral arterial disease is associated with age (Beckman et al., 2002). Pro-angiogenic roles of HIF1A were also associated with hypertrophic cardiomyopathy, myocardial infarction, skin wound healing, and retinal neovascularization (reviewed in (Majmundar et al., 2010)). Many studies suggested that the occurrence of local hypoxia in the muscle causes a drop in O2 pressure within the myocyte during exercise (Richardson et al. 1995, Richardson et al., 2001).
Because the cardiovascular system subsequently influences human body performance, HIF1A variations have been associated with the following performance related phenotypes: maximal oxygen consumption, adaptation to living at high altitude, lumbar disc degeneration, idiopathic osteonecrosis of the femoral head, power-oriented athlete performance and muscle activity, endurance status, and symptom-limited exercise test duration over time (rs113182457 (Suzuki et al. 2003), rs10873142 (Hong et al. 2007), rs11549465 (Prior et al. 2003, Hong et al. 2007, Ahmetov et al. 2008, Doring et al. 2010, Cieszczyk et al. 2011, McPhee et al. 2011, Gabbasov et al. 2012), rs11549467 (Lin et al. 2013), rs1535679 (Hong et al. 2007), rs1957757 and rs1957755 (Sarzynski et al. 2010), rs2057482 (Hong et al. 2007), and rs28708675 (Prior et al. 2003)). Some polymorphisms were also associated with other phenotypes such as systemic sclerosis (rs12434438 (Wipff et al. 2009)), acute kidney injury (rs11549465 (Kolyada et al. 2009)), cellulite (rs11549465 (Emanuele et al., 2010)) and chronic obstructive pulmonary disease (rs11549465 (Putra et al. 2011a, Putra et al., 2011b)). Moreover, HIF1A SNPs were shown to be involved in metabolic disorders such as: diabetic nephropathy (rs11549465 (Gu et al. 2013)), which is a result of longstanding type 1 diabetes mellitus (T1DM) (rs11549465 (Nagy et al. 2009)) and type 2 diabetes mellitus (T2DM) (rs11549465 (Yamada et al. 2005, Nagy et al. 2009), rs12434438 (Yamada et al. 2005), and rs1319462 (Yamada et al. 2005)).
Most frequently studied missense SNPs within ODD domain
The review of genotype-phenotype studies revealed that HIF1A association studies were most often focused on two missense SNPs located within the ODD domain: rs11549465 (p.Pro582Ser; C1772T), and rs11549467 (p.Ala588Thr; A1790G). For these two SNPs opposing associations with phenotype were found. In addition, both rs11549465 and rs11549467 are germline SNPs within the ODD/pVHL interaction domain (Clifford et al. 2001). However, rs11549465 is also known to have an ability to enhance transactivation (Huang et al. 1998, Tanimoto et al. 2003), but was not identified as a site for HIF1A hydroxylation and is not mediating pVHL binding (Yamada et al. 2005). Moreover, these results are in agreement with a previous study by Percy et al. (Percy et al. 2003), who showed that the substitution p.Pro582Ser in vitro does not prevent VHL binding to a fragment of HIF1A after hydroxylation at p.Pro564. In the present study we have also collected published negative associations between polymorphisms and phenotypes, since they could be tested also in other populations or in higher number of subjects of the same populations to re-evaluate significance.
As in most association studies, also HIF1A association studies focused on missense SNPs and much less attention was given to synonymous (sSNP) and other non-coding SNPs. This could reflect a long-term assumption that sSNPs are inconsequential, since the primary amino-acid sequence of the protein stays unchanged. Although HIF1A association studies mostly focused on missense SNPs within the ODD domain affecting oxygen-dependent proteolysis, polymorphisms within other HIF1A domains have also been shown to affect HIF1A activity. Possible scenarios that may disrupt the role of HIF1A functional domains and therefore affect HIF1A stability and its role as the main TF in hypoxia are: (1) variations within the bHLH domain may prevent binding of HIF1A to HRE recognition sites within the promoter region of target genes, and likewise variations within HRE may create or destruct HRE binding sites for HIF1A to influence downstream targets; (2) variations within the PAS domain could affect dimerization with ARNT (HIF1B), which would result in HIF1A inability to function as a transcriptional regulator; (3) variations within the ODD domain could affect stability of a protein in normoxia, since conserved proline residues (p.Pro402 and p.Pro564) are usually targeted for VHL/proteasome degradation; (4) variations within the nuclear localization signal (NLS) may have an effect on translocation of HIF1A into the cell nucleus by nuclear transport; and (5) variations within N-TAD and C-TAD could influence transcriptional activation of HIF1A and interaction with its co-activators. Our literature review showed that HIF1A variations outside the ODD domain also have functional effects and were associated with diseases and phenotypes.
HIF1A and clinical trials
There are several clinical studies recorded in the database ClinicalTrials (https://clinicaltrials.gov/) that collates all publicly and privately supported clinical studies of human participants conducted around the world. Although 31 ongoing or already completed clinical trials included HIF1A in various cancers we found only one that specifically explores the effect of a HIF1A polymorphism in breast cancer (clinical trial identifier: NCT01935102; (Allegrini et al. 2014)). A missense SNP rs11549465 within the oxygen-dependent degradation domain that has also been extensively reviewed in our study, was tested in the aforementioned clinical trial. Specifically, the trial was aimed to identify interactions between the vascular endothelial growth factor A (VEGFA) SNPs with SNPs in several other genes: KDR (VEGFR2), CXCL8 (IL-8), HIF1A, HIF2A, and THBS1 (TSP-1) for possible differential bevacizumab response in a population of metastatic breast cancer. The multifactor dimensionality reduction (MDR) analyses found no interactions involving HIF1A genotypes. However, we need to add, that the sample size, especially for interaction analyses, was quite small, possibly leading to low power of statistical detection. For example, for HIF1A SNP rs11549465 the study involved only 8 individuals with the CC genotype, 100 heterozygotes and only 4 with the TT genotype. Nevertheless, the study was able to detect a significant interaction between KDR (VEGFR2) rs11133360 and CXCL8 (IL-8) rs4073 genotypes that identified a favorable genetic profile predicting a better therapeutic outcome. We can expect more such studies exploring HIF1A SNPs in clinical trials especially in terms of predicting their effect on favorable or unfavorable therapy outcomes in several other cancers mentioned also in our study. Apart from such pharmacogenetics clinical trials, we can anticipate also gene therapy trials to correct the most damaging germ line mutations in HIF1A as was recently shown for editing the sickle cell anemia mutation by the CRISPR/Cas9 approach (DeWitt et al. 2016).
Conclusions
Hypoxia is a characteristic feature for many pathological settings; hence, a stable HIF1A protein is critical for cells to adapt, thrive and survive in a hypoxic environment. The present HIF1A genotype-phenotype integrated review and manually curated graphical presentation could help researchers in better planning of future experiments as well as identifying traits and pathologies likely to be affected by HIF1A variability. Potential novel discoveries about functional HIF1A variations might be important to improve our understanding of HIF1A regulation. In addition, altered HIF1A regulation and its direct influence on various cellular pathways could better explain the role of HIF1A on a cellular level and its contribution to human complex diseases like cancer, diabetes, and obesity, or its contribution to the function of immune response and cardiovascular systems.
Supplementary Material
Acknowledgments
Supported by
This work was supported by the Slovenian Research Agency (ARRS) through the Research program Comparative genomics and genome biodiversity [grant number P4-0220] and PhD project to JF. Dr Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. This work was supported by National Institutes of Health (NIH/NCATS) grant UH3TR00943-01 through the NIH Common Fund, Office of Strategic Coordination (OSC). Work in Dr. Calin’s laboratory is supported in part by the grant NIH/NCI 1 R01 CA182905-01, the UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), Aim at Melanoma Foundation and the Miriam and Jim Mulva research funds, the UT MD Anderson Cancer Center Brain SPORE (2P50CA127001), a Developmental Research award from Leukemia SPORE, a CLL Moonshot Flagship project, a 2015 Knowledge GAP MDACC grant, an Owens Foundation grant, and the Estate of C. G. Johnson, Jr,.
References
- Ahmetov II, Hakimullina AM, Lyubaeva EV, Vinogradova OL, Rogozkin VA. Effect of HIF1A gene polymorphism on human muscle performance. Bull Exp Biol Med. 2008;146(3):351–353. doi: 10.1007/s10517-008-0291-3. [DOI] [PubMed] [Google Scholar]
- Alidoosti M, Ghaedi M, Soleimani A, Bakhtiyari S, Rezvanfard M, Golkhu S, Mohammadtaghvaei N. Study on the role of environmental parameters and HIF-1A gene polymorphism in coronary collateral formation among patients with ischemic heart disease. Clinical Biochemistry. 2011;44(17–18):1421–1424. doi: 10.1016/j.clinbiochem.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Allegrini G, Coltelli L, Orlandi P, Fontana A, Camerini A, Ferro A, Cazzaniga M, Casadei V, Lucchesi S, Bona E, Di Lieto M, Pazzagli I, Villa F, Amoroso D, Scalese M, Arrighi G, Molinaro S, Fioravanti A, Finale C, Triolo R, Di Desidero T, Donati S, Marcucci L, Goletti O, Del Re M, Salvadori B, Ferrarini I, Danesi R, Falcone A, Bocci G. Pharmacogenetic interaction analysis of VEGFR-2 and IL-8 polymorphisms in advanced breast cancer patients treated with paclitaxel and bevacizumab. Pharmacogenomics. 2014;15(16):1985–1999. doi: 10.2217/pgs.14.140. [DOI] [PubMed] [Google Scholar]
- Andraweera PH, Dekker GA, Thompson SD, Dissanayake VHW, Jayasekara RW, Roberts CT. Hypoxia-inducible factor-1a gene polymorphisms in early and lateonset preeclampsia in Sinhalese women. Placenta. 2014;35:491–495. doi: 10.1016/j.placenta.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Apaydin I, Konac E, Onen HI, Akbaba M, Tekin E, Ekmekci A. Single nucleotide polymorphisms in the hypoxia-inducible factor-1alpha (HIF-1alpha) gene in human sporadic breast cancer. Arch Med Res. 2008;39(3):338–345. doi: 10.1016/j.arcmed.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Bahadori B, Uitz E, Mayer A, Harauer J, Dam K, Truschnig-Wilders M, Pilger E, Renner W. Polymorphisms of the hypoxia-inducible factor 1 gene and peripheral artery disease. Vascular Medicine. 2010;15(5):371–374. doi: 10.1177/1358863X10379674. [DOI] [PubMed] [Google Scholar]
- Beckman JA, Creager MA, Libby P. Diabetes and atherosclerosis: epidemiology, pathophysiology, and management. JAMA. 2002;287(19):2570–2581. doi: 10.1001/jama.287.19.2570. [DOI] [PubMed] [Google Scholar]
- Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8(12):967–975. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachami Georgia, Kalousi Alkmini, Papatheodorou Loukia, Lyberopoulou Aggeliki, Nasikas Vasileios, Tanimoto Keiji, Simos George, Malizos N Konstantinos, Georgatsou Eleni. An Association Study between Hypoxia Inducible Factor-1alpha (HIF-1a) Polymorphisms and Osteonecrosis. Plos One. 2013;8(11) doi: 10.1371/journal.pone.0079647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Yen SE, Giaccia AJ. Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1 alpha. Mol Cell Biol. 2005;25(15):6415–6426. doi: 10.1128/MCB.25.15.6415-6426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau CH, Permenter MG, Steinberg SM, Retter AS, Dahut WL, Price DK, Figg WD. Polymorphism in the hypoxia-inducible factor 1alpha gene may confer susceptibility to androgen-independent prostate cancer. Cancer Biol Ther. 2005;4(11):1222–1225. doi: 10.4161/cbt.4.11.2091. [DOI] [PubMed] [Google Scholar]
- Chen MK, Chiou HL, Su SC, Chung TT, Tseng HC, Tsai HT, Yang SF. The association between hypoxia inducible factor-1alpha gene polymorphisms and increased susceptibility to oral cancer. Oral Oncol. 2009;45(12):e222–e226. doi: 10.1016/j.oraloncology.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Cieszczyk P, Eider J, Arczewska A, Ostanek M, Leonska-Duniec A, Sawczyn S, Ficek K, Jascaniene N, Kotarska K, Sygit K. The Hif1a Gene Pro582ser Polymorphism in Polish Power-Orientated Athletes. Biology of Sport. 2011;28(2):111–114. [Google Scholar]
- Clifford SC, Astuti D, Hooper L, Maxwell PH, Ratcliffe PJ, Maher ER. The pVHL-associated SCF ubiquitin ligase complex: molecular genetic analysis of elongin B and C, Rbx1 and HIF-1alpha in renal cell carcinoma. Oncogene. 2001;20(36):5067–5074. doi: 10.1038/sj.onc.1204602. [DOI] [PubMed] [Google Scholar]
- DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, Heo SJ, Mitros T, Munoz DP, Boffelli D, Kohn DB, Walters MC, Carroll D, Martin DI, Corn JE. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8(360):360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Yang D, Xun X, Wang Z, Sun P, Xu D, He P, Niu H, Jin T. Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Hainan population: a case-control study. Int J Chron Obstruct Pulmon Dis. 2015;10:7–13. doi: 10.2147/COPD.S73042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring F, Onur S, Fischer A, Boulay MR, Perusse L, Rankinen T, Rauramaa R, Wolfarth B, Bouchard C. A common haplotype and the Pro582Ser polymorphism of the hypoxia-inducible factor-1alpha (HIF1A) gene in elite endurance athletes. J Appl Physiol. 2010;108(6):1497–1500. doi: 10.1152/japplphysiol.01165.2009. [DOI] [PubMed] [Google Scholar]
- Droma Y, Ota M, Hanaoka M, Katsuyama Y, Basnyat B, Neupane P, Arjyal A, Pandit A, Sharma D, Ito M, Kubo K. Two Hypoxia Sensor Genes and Their Association with Symptoms of Acute Mountain Sickness in Sherpas. Aviation Space and Environmental Medicine. 2008;79(11):1056–1060. doi: 10.3357/asem.2361.2008. [DOI] [PubMed] [Google Scholar]
- Emanuele E, Bertona M, Geroldi D. A multilocus candidate approach identifies ACE and HIF1A as susceptibility genes for cellulite. J Eur Acad Dermatol Venereol. 2010;24(8):930–935. doi: 10.1111/j.1468-3083.2009.03556.x. [DOI] [PubMed] [Google Scholar]
- Eynon N, Alves AJ, Meckel Y, Yamin C, Ayalon M, Sagiv M. Is the interaction between HIF1A P582S and ACTN3 R577X determinant for power/sprint performance? Metabolism-Clinical and Experimental. 2010;59(6):861–865. doi: 10.1016/j.metabol.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Feng CC, Ye QL, Zhu Y, Leng RX, Chen GM, Yang J, Cen H, Yang XK, Li R, Xu WD, Pan HF, Ye DQ. Lack of association between the polymorphisms of hypoxia-inducible factor 1A (HIF1A) gene and SLE susceptibility in a Chinese population. Immunogenetics. 2014;66(1):9–13. doi: 10.1007/s00251-013-0743-4. [DOI] [PubMed] [Google Scholar]
- Ferdin J, Nishida N, Wu X, Nicoloso MS, Shah MY, Devlin C, Ling H, Shimizu M, Kumar K, Cortez MA, Ferracin M, Bi Y, Yang D, Czerniak B, Zhang W, Schmittgen TD, Voorhoeve MP, Reginato MJ, Negrini M, Davuluri RV, Kunej T, Ivan M, Calin GA. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell Death and Differentiation. 2013;20(12):1675–1687. doi: 10.1038/cdd.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Torres J, Hernandez-Diaz C, Espinosa-Morales R, Camacho-Galindo J, Galindo-Sevilla Ndel C, Lopez-Macay A, Zamudio-Cuevas Y, Martinez-Flores K, Santamaria-Olmedo MG, Pineda C, Granados J, Martinez-Nava GA, Gutierrez M, Lopez-Reyes AG. Polymorphic variation of hypoxia inducible factor-1 A (HIF1A) gene might contribute to the development of knee osteoarthritis: a pilot study. BMC Musculoskelet Disord. 2015;16:218. doi: 10.1186/s12891-015-0678-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley R, Marignol L, Thomas AZ, Cullen IM, Perry AS, Tewari P, O’Grady A, Kay E, Dunne B, Loftus B, Watson WR, Fitzpatrick JM, Woodson K, Lehman T, Hollywood D, Lynch TH, Lawler M. The HIF-1alpha C1772T polymorphism may be associated with susceptibility to clinically localised prostate cancer but not with elevated expression of hypoxic biomarkers. Cancer Biol Ther. 2009;8(2):118–124. doi: 10.4161/cbt.8.2.7086. [DOI] [PubMed] [Google Scholar]
- Frank B, Hoffmeister M, Klopp N, Illig T, Chang-Claude J, Brenner H. Single nucleotide polymorphisms in Wnt signaling and cell death pathway genes and susceptibility to colorectal cancer. Carcinogenesis. 2010;31(8):1381–1386. doi: 10.1093/carcin/bgq082. [DOI] [PubMed] [Google Scholar]
- Fransen K, Fenech M, Fredrikson M, Dabrosin C, Soderkvist P. Association between ulcerative growth and hypoxia inducible factor-1alpha polymorphisms in colorectal cancer patients. Mol Carcinog. 2006;45(11):833–840. doi: 10.1002/mc.20209. [DOI] [PubMed] [Google Scholar]
- Fu Sl Fau - Miao J, Miao J Fau - Ding B, Ding B Fau - Wang XL, Wang Xl Fau - Cheng WJ, Cheng Wj Fau - Dai HH, Dai Hh Fau - Han SP, Han SP. A polymorphism in the 3’ untranslated region of Hypoxia-Inducible Factor-1 alpha confers an increased risk of cervical cancer in a Chinese population. Neoplasma. 2014;61(1):63–69. [PubMed] [Google Scholar]
- Fu XS, Choi E, Bubley GJ, Balk SP. Identification of hypoxia-inducible factor-1alpha (HIF-1alpha) polymorphism as a mutation in prostate cancer that prevents normoxia-induced degradation. Prostate. 2005;63(3):215–221. doi: 10.1002/pros.20190. [DOI] [PubMed] [Google Scholar]
- Gabbasov RT, Arkhipova AA, Borisova AV, Hakimullina AM, Kuznetsova AV, Williams AG, Day SH, Ahmetov II. The HIF1A gene Pro582Ser polymorphism in Russian strength athletes. J Strength Cond Re. 2012;27(8):2055–2058. doi: 10.1519/JSC.0b013e31827f06ae. [DOI] [PubMed] [Google Scholar]
- Gee Harriet E, Cristina Ivan George, Calin A, Mircea Ivan. HypoxamiRs and Cancer: From Biology to Targeted Therapy. Antioxidants & Redox Signaling. 2014;21(8):1220–1238. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu HF, Zheng X, Abu Seman N, Gu T, Botusan IR, Sunkari VG, Lokman EF, Brismar K, Catrina SB. Impact of the Hypoxia-Inducible Factor-1 alpha (HIF-1alpha) Pro582Ser Polymorphism on Diabetes Nephropathy. Diabetes Care. 2013;36(2):415–421. doi: 10.2337/dc12-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Li D, Chen Y, An J, Wang K, Xu Z, Chen Z, Xing J. SNP rs2057482 in HIF1A gene predicts clinical outcome of aggressive hepatocellular carcinoma patients after surgery. Sci Rep. 2015;5:11846. doi: 10.1038/srep11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Pengfei, Han Qi, Liu Jiajia, Liu Dongjuan, Zhao Xin, Hu Ting, Jiang Lu, Dan Hongxia, Zeng Xin, Li Jing, Wang Jiayi, Chen Qianming. The Association between Hypoxia-Inducible Factor-1 α Gene C1772T Polymorphism and Cancer Risk: A Meta-Analysis of 37 Case-Control Studies. PLoS ONE. 2013;8(12):e83441. doi: 10.1371/journal.pone.0083441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino S, Kaare M, Andersson S, Laivuori H. Non-synonymous sequence variants within the oxygen-dependent degradation domain of the HIF1A gene are not associated with pre-eclampsia in the Finnish population. BMC Med Genet. 2008;9:96. doi: 10.1186/1471-2350-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennis PJ, Bussell C, Darlison MG. The Lack of Associations Between Alleles at the Hypoxia-Inducible Factor 1A C1772T Loci and Responses to Acute Hypoxia. Wilderness & Environmental Medicine. 2010;21(3):219–228. doi: 10.1016/j.wem.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Hlatky MA, Quertermous T, Boothroyd DB, Priest JR, Glassford AJ, Myers RM, Fortmann SP, Iribarren C, Tabor HK, Assimes TL, Tibshirani RJ, Go AS. Polymorphisms in hypoxia inducible factor 1 and the initial clinical presentation of coronary disease. Am Heart J. 2007;154(6):1035–1042. doi: 10.1016/j.ahj.2007.07.042. [DOI] [PubMed] [Google Scholar]
- Hong JM, Kim TH, Chae SC, Koo KH, Lee YJ, Park EK, Choi JY, Ryoo HM, Kim SY. Association study of hypoxia inducible factor 1alpha (HIF1alpha) with osteonecrosis of femoral head in a Korean population. Osteoarthritis Cartilage. 2007;15(6):688–694. doi: 10.1016/j.joca.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Horree N, Groot AJ, van Hattem WA, Heintz APM, Vooijs M, van Diest PJ. HIF-1A gene mutations are associated with higher microvessel density in endometrial carcinomas. Histopathology. 2008;52(5):637–639. doi: 10.1111/j.1365-2559.2008.02991.x. [DOI] [PubMed] [Google Scholar]
- Hsiao PC, Chen MK, Su SC, Ueng KC, Chen YC, Hsieh YH, Liu YF, Tsai HT, Yang SF. Hypoxia inducible factor-1alpha gene polymorphism G1790A and its interaction with tobacco and alcohol consumptions increase susceptibility to hepatocellular carcinoma. J Surg Oncol. 2010;102(2):163–169. doi: 10.1002/jso.21539. [DOI] [PubMed] [Google Scholar]
- Hu X, Fang Y, Zheng J, He YZ, Zan X, Lin S, Li X, Li H, You C. The association between HIF-1 alpha polymorphism and cancer risk: a systematic review and meta-analysis. Tumor Biology. 2014;35(2):903–916. doi: 10.1007/s13277-013-1160-x. [DOI] [PubMed] [Google Scholar]
- Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998;95(14):7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jacobs EJ, Hsing AW, Bain EB, Stevens VL, Wang Y, Chen J, Chanock SJ, Zheng SL, Xu J, Thun MJ, Calle EE, Rodriguez C. Polymorphisms in angiogenesis-related genes and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(4):972–977. doi: 10.1158/1055-9965.EPI-07-2787. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Kang MJ, Jung SA, Jung JM, Kim SE, Jung HK, Kim TH, Shim KN, Yi SY, Yoo K, Moon IH. Associations between single nucleotide polymorphisms of MMP2, VEGF, and HIF1A genes and the risk of developing colorectal cancer. Anticancer Res. 2011;31(2):575–584. [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kett-White R, Hutchinson PJ, Al-Rawi PG, Czosnyka M, Gupta AK, Pickard JD, Kirkpatrick PJ. Cerebral oxygen and microdialysis monitoring during aneurysm surgery: effects of blood pressure, cerebrospinal fluid drainage, and temporary clipping on infarction. J Neurosurg. 2002;96(6):1013–1019. doi: 10.3171/jns.2002.96.6.1013. [DOI] [PubMed] [Google Scholar]
- Kim HO, Jo YH, Lee J, Lee SS, Yoon KS. The C1772T genetic polymorphism in human HIF-1alpha gene associates with expression of HIF-1alpha protein in breast cancer. Oncol Rep. 2008;20(5):1181–1187. [PubMed] [Google Scholar]
- Kim SJ, Hwang SH, Kim IJ, Lee MK, Lee CH, Lee SY, Lee EY. The association of F-18-deoxyglucose (FDG) uptake of PET with polymorphisms in the glucose transporter gene (SLC2A1) and hypoxia-related genes (HIF1A, VEGFA, APEX1) in non-small cell lung cancer. SLC2A1 polymorphisms and FDG-PET in NSCLC patients. Journal of Experimental & Clinical Cancer Research. 2010;29:8. doi: 10.1186/1756-9966-29-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park Y, Lim SY, Lee JHBY, Yang JH, Ryu HM. Hypoxia inducible factor-1alpha gene polymorphisms in Korean patients with pre-eclampsia. Journal of Endocrinological Investigation. 2012;35(7):6. doi: 10.3275/8009. [DOI] [PubMed] [Google Scholar]
- Knechtel G, Szkandera J, Stotz M, Hofmann G, Langsenlehner U, Krippl P, Samonigg H, Renner W, Dehchamani D, Gerger A. Single nucleotide polymorphisms in the hypoxia-inducible factor 1 gene and colorectal cancer risk. Onkologie. 2010;33:225–226. doi: 10.1002/mc.20655. [DOI] [PubMed] [Google Scholar]
- Kolyada AY, Tighiouart H, Perianayagam MC, Liangos O, Madias NE, Jaber BL. A genetic variant of hypoxia-inducible factor-1alpha is associated with adverse outcomes in acute kidney injury. Kidney Int. 2009;75(12):1322–1329. doi: 10.1038/ki.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konac E, Dogan I, Onen HI, Yurdakul AS, Ozturk C, Varol A, Ekmecki A. Genetic Variations in the Hypoxia-Inducible Factor-1 alpha Gene and Lung Cancer. Experimental Biology and Medicine. 2009;234(9):1109–1116. doi: 10.3181/0902-RM-49. [DOI] [PubMed] [Google Scholar]
- Konac E, Onen HI, Metindir J, Alp E, Biri AA, Ekmekci A. An investigation of relationships between hypoxia-inducible factor-1 alpha gene polymorphisms and ovarian, cervical and endometrial cancers. Cancer Detect Prev. 2007;31(2):102–109. doi: 10.1016/j.cdp.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Papazoglou D, Giatromanolaki A, Panagopoulos I, Maltezos E, Harris AL, Gatter KC, Sivridis E. C2028T polymorphism in exon 12 and dinucleotide repeat polymorphism in intron 13 of the HIF-1alpha gene define HIF-1alpha protein expression in non-small cell lung cancer. Lung Cancer. 2006;53(3):257–262. doi: 10.1016/j.lungcan.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Kuo WH, Shih CM, Lin CW, Cheng WE, Chen SC, Chen W, Lee YL. Association of hypoxia inducible factor-1 alpha polymorphisms with susceptibility to non-small-cell lung cancer. Translational Research. 2012;159(1):42–50. doi: 10.1016/j.trsl.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Kuwai T, Kitadai Y, Tanaka S, Kuroda T, Ochiumi T, Matsumura S, Oue N, Yasui W, Kaneyasu M, Tanimoto K, Nishiyama M, Chayama K. Single nucleotide polymorphism in the hypoxia-inducible factor-1alpha gene in colorectal carcinoma. Oncol Rep. 2004;12(5):1033–1037. [PubMed] [Google Scholar]
- Lee JY, Choi JY, Lee KM, Park SK, Han SH, Noh DY, Ahn SH, Kim DH, Hong YC, Ha E, Yoo KY, Ambrosone CB, Kang D. Rare variant of hypoxia-inducible factor-1alpha (HIF-1A) and breast cancer risk in Korean women. Clin Chim Acta. 2008;389(1–2):167–170. doi: 10.1016/j.cca.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim J, Sohn JG, Chae SK, Moon YS, Kang JH, Park BW, Park JSJY, Choi GS. No association of the hypoxia-inducible factor-1alpha gene polymorphisms with survival in patients with colorectal cancer. Medical Oncology. 2011;28(4):6. doi: 10.1007/s12032-010-9618-9. [DOI] [PubMed] [Google Scholar]
- Li DW, Liu JK, Zhang WH, Ren JC, Yan L, Liu HN, Xu ZH. Association between HIF1A P582S and A588T Polymorphisms and the Risk of Urinary Cancers: A Meta-Analysis. Plos One. 2013;8(5):9. doi: 10.1371/journal.pone.0063445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bubley GJ, Balk SP, Gaziano JM, Pollak M, Stampfer MJ, Ma J. Hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms, circulating insulin-like growth factor binding protein (IGFBP)-3 levels and prostate cancer. Prostate. 2007;67(12):1354–1361. doi: 10.1002/pros.20589. [DOI] [PubMed] [Google Scholar]
- Li K, Zhang Y, Dan Z, Wang Y, Ren ZC. Association of the hypoxia inducible factor-1alpha gene polymorphisms with gastric cancer in Tibetans. Biochem Genet. 2009;47(9–10):625–634. doi: 10.1007/s10528-009-9254-2. [DOI] [PubMed] [Google Scholar]
- Li P, Cao Q, Shao PF, Cai HZ, Zhou H, Chen JW, Qin C, Zhang ZD, Ju XB, Yin CJ. Genetic polymorphisms in HIF1A are associated with prostate cancer risk in a Chinese population. Asian J Androl. 2012;14(6):864–869. doi: 10.1038/aja.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Wen-Ping, Wang Xue-Jin, Wang Cong-Ren, Zhang Li-Qun, Wang Fa-Sheng, Lin Jian-Hua, Li Neng. Polymorphism in the Hypoxia-Inducible Factor 1alpha Gene May Confer Susceptibility to LDD in Chinese Cohort. Plos One. 2013;8(8):e73158. doi: 10.1371/journal.pone.0073158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling TS, Shi RH, Zhang GX, Zhu H, Yu LZ, Ding XF. Common single nucleotide polymorphism of hypoxia-inducible factor-1alpha and its impact on the clinicopathological features of esophageal squamous cell carcinoma. Chin J Dig Dis. 2005;6(4):155–158. doi: 10.1111/j.1443-9573.2005.00223.x. [DOI] [PubMed] [Google Scholar]
- Liu BY, Liu QC, Song Y, Li XF, Wang YJ, Wan SG, Zhang ZP, Su HC. Polymorphisms of HIF1A gene are associated with prognosis of early stage non-small-cell lung cancer patients after surgery. Medical Oncology. 2014;31(4):9. doi: 10.1007/s12032-014-0877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Jie, Zhang Hong-xin. 1790 G/A polymorphism, but not 1772 C/T polymorphism,is significantly associated with Cancers: An update study. Gene. 2013;523:58–63. doi: 10.1016/j.gene.2013.03.129. [DOI] [PubMed] [Google Scholar]
- Liu L, Marti GP, Wei X, Zhang X, Zhang H, Liu YV, Nastai M, Semenza GL, Harmon JW. Age-dependent impairment of HIF-1alpha expression in diabetic mice: Correction with electroporation-facilitated gene therapy increases wound healing, angiogenesis, and circulating angiogenic cells. J Cell Physiol. 2008;217(2):319–327. doi: 10.1002/jcp.21503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Reyes A, Rodriguez-Perez JM, Fernandez-Torres J, Martinez-Rodriguez N, Perez-Hernandez N, Fuentes-Gomez AJ, Aguilar-Gonzalez CA, Alvarez-Leon E, Posadas-Romero C, Villarreal-Molina T, Pineda C, Vargas-Alarcon G. The HIF1A rs2057482 polymorphism is associated with risk of developing premature coronary artery disease and with some metabolic and cardiovascular risk factors. The Genetics of Atherosclerotic Disease (GEA) Mexican Study. Experimental and Molecular Pathology. 2014;96(3):405–410. doi: 10.1016/j.yexmp.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JS, Perez-Schindler J, Degens H, Tomlinson D, Hennis P, Baar K, Williams AG. HIF1A P582S gene association with endurance training responses in young women. Eur J Appl Physiol. 2011;111(9):2339–2347. doi: 10.1007/s00421-011-1869-4. [DOI] [PubMed] [Google Scholar]
- Meka PB, Cingeetham A, Nanchari SR, Damineni S, Tipirisetti N, Gorre M, Jarjapu S, Annamaneni S, Digumarthi R, Satti V. HIF-1alpha (1772C>T) polymorphism as marker for breast cancer development. Tumour Biol. 2015;36(5):3215–3220. doi: 10.1007/s13277-014-2949-y. [DOI] [PubMed] [Google Scholar]
- Morris MR, Hughes DJ, Tian YM, Ricketts CJ, Lau KW, Gentle D, Shuib S, Serrano-Fernandez P, Lubinski J, Wiesener MS, Pugh CW, Latif F, Ratcliffe PJ, Maher ER. Mutation analysis of hypoxia-inducible factors HIF1A and HIF2A in renal cell carcinoma. Anticancer Res. 2009;29(11):4337–4343. [PubMed] [Google Scholar]
- Munoz-Guerra MF, Fernandez-Contreras ME, Moreno AL, Martin ID, Herraez B, Gamallo C. Polymorphisms in the hypoxia inducible factor 1-alpha and the impact on the prognosis of early stages of oral cancer. Ann Surg Oncol. 2009;16(8):2351–2358. doi: 10.1245/s10434-009-0503-8. [DOI] [PubMed] [Google Scholar]
- Nadaoka J, Horikawa Y, Saito M, Kumazawa T, Inoue T, Narita S, Yuasa T, Satoh S, Nishiyama H, Ogawa O, Tsuchiya N, Habuchi T. Prognostic significance of HIF-1alpha polymorphisms in transitional cell carcinoma of the bladder. Journal of Urology. 2008;179(4):320–320. doi: 10.1002/ijc.23256. [DOI] [PubMed] [Google Scholar]
- Nagy G, Kovacs-Nagy R, Kereszturi E, Somogyi A, Szekely A, Nemeth N, Hosszufalusi N, Panczel P, Ronai Z, Sasvari-Szekely M. Association of hypoxia inducible factor-1 alpha gene polymorphism with both type 1 and type 2 diabetes in a Caucasian (Hungarian) sample. BMC Med Genet. 2009;10:79. doi: 10.1186/1471-2350-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu R, Har YC, Taib NA. Associations between hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms and risk of developing breast cancer. Neoplasma. 2009;56(5):441–447. doi: 10.4149/neo_2009_05_441. [DOI] [PubMed] [Google Scholar]
- Nava-Salazar Sonia, Sanchez-Rodriguez Elly N, Mendoza-Rodriguez Adriana C, Moran Carlos, Romero-Arauz Juan F, Cerbon Marco A. Polymorphisms in the hypoxia-inducible factor 1 alpha gene in Mexican patients with preeclampsia: A case-control study. BMC Research Notes. 2011;4:6. doi: 10.1186/1756-0500-4-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollerenshaw M, Page T, Hammonds J, Demaine A. Polymorphisms in the hypoxia inducible factor-1alpha gene (HIF1A) are associated with the renal cell carcinoma phenotype. Cancer Genet Cytogenet. 2004;153(2):122–126. doi: 10.1016/j.cancergencyto.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Bar-Shira A, Matzkin H, Mabjeesh NJ. The homozygous P582S mutation in the oxygen-dependent degradation domain of HIF-1 alpha is associated with increased risk for prostate cancer. Prostate. 2007;67(1):8–13. doi: 10.1002/pros.20433. [DOI] [PubMed] [Google Scholar]
- Park SW, Chung NG, Hur SY, Kim HS, Yoo NJ, Lee SH. Mutational analysis of hypoxia-related genes HIF1alpha and CUL2 in common human cancers. APMIS. 2009;117(12):880–885. doi: 10.1111/j.1600-0463.2009.02550.x. [DOI] [PubMed] [Google Scholar]
- Percy MJ, Mooney SM, McMullin MF, Flores A, Lappin TR, Lee FS. A common polymorphism in the oxygen-dependent degradation (ODD) domain of hypoxia inducible factor-1alpha (HIF-1alpha) does not impair Pro-564 hydroxylation. Mol Cancer. 2003;2:31. doi: 10.1186/1476-4598-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior SJ, Hagberg JM, Phares DA, Brown MD, Fairfull L, Ferrell RE, Roth SM. Sequence variation in hypoxia-inducible factor 1alpha (HIF1A): association with maximal oxygen consumption. Physiol Genomics. 2003;15(1):20–26. doi: 10.1152/physiolgenomics.00061.2003. [DOI] [PubMed] [Google Scholar]
- Putra AC, Tanimoto K, Arifin M, Antariksa B, Hiyama K. Genetic variations in detoxification enzymes and HIF-1alpha in Japanese patients with COPD. Clin Respir J. 2011a;7(1):7–15. doi: 10.1111/j.1752-699X.2011.00255.x. [DOI] [PubMed] [Google Scholar]
- Putra AC, Tanimoto K, Arifin M, Hiyama K. Hypoxia-inducible factor-1alpha polymorphisms are associated with genetic aberrations in lung cancer. Respirology. 2011b;16(5):796–802. doi: 10.1111/j.1440-1843.2011.01972.x. [DOI] [PubMed] [Google Scholar]
- Qin C, Cao Q, Ju X, Wang M, Meng X, Zhu J, Yan F, Li P, Ding Q, Chen J, Gu M, Zhang W, Yin C, Zhang Z. The polymorphisms in the VHL and HIF1A genes are associated with the prognosis but not the development of renal cell carcinoma. Annals of Oncology. 2012;23(4):981–989. doi: 10.1093/annonc/mdr325. [DOI] [PubMed] [Google Scholar]
- Resar JR, Roguin A, Voner J, Nasir K, Hennebry TA, Miller JM, Ingersoll R, Kasch LM, Semenza GL. Hypoxia-inducible factor 1alpha polymorphism and coronary collaterals in patients with ischemic heart disease. Chest. 2005;128(2):787–791. doi: 10.1378/chest.128.2.787. [DOI] [PubMed] [Google Scholar]
- Richardson RS, Newcomer SC, Noyszewski EA. Skeletal muscle intracellular PO(2) assessed by myoglobin desaturation: response to graded exercise. J Appl Physiol (1985) 2001;91(6):2679–2685. doi: 10.1152/jappl.2001.91.6.2679. [DOI] [PubMed] [Google Scholar]
- Richardson RS, Noyszewski EA, Kendrick KF, Leigh JS, Wagner PD. Myoglobin O2 desaturation during exercise. Evidence of limited O2 transport. J Clin Invest. 1995;96(4):1916–1926. doi: 10.1172/JCI118237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Tovar J, Fernandez-Contreras ME, Martin-Perez E, Gamallo C. Association of thymidylate synthase and hypoxia inducible factor-1alpha DNA polymorphisms with pancreatic cancer. Tumori. 2012;98(3):364–369. doi: 10.1177/030089161209800314. [DOI] [PubMed] [Google Scholar]
- Sarzynski MA, Rankinen T, Sternfeld B, Grove ML, Fornage M, Jacobs DR, Sidney S, Bouchard C. Association of Single-Nucleotide Polymorphisms From 17 Candidate Genes With Baseline Symptom-Limited Exercise Test Duration and Decrease in Duration Over 20 Years The Coronary Artery Risk Development in Young Adults (CARDIA) Fitness Study. Circulation-Cardiovascular Genetics. 2010;3(6):531–538. doi: 10.1161/CIRCGENETICS.110.957183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Evaluation of HIF-1 inhibitors as anticancer agents. Drug Discov Today. 2007;12(19–20):853–859. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh TM, Chang KW, Tu HF, Shih YH, Ko SY, Chen YC, Liu CJ. Association between the polymorphisms in exon 12 of hypoxia-inducible factor-1alpha and the clinicopathological features of oral squamous cell carcinoma. Oral Oncol. 2010;46(9):e47–e53. doi: 10.1016/j.oraloncology.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Shohet RV, Garcia JA. Keeping the engine primed: HIF factors as key regulators of cardiac metabolism and angiogenesis during ischemia. J Mol Med (Berl) 2007;85(12):1309–1315. doi: 10.1007/s00109-007-0279-x. [DOI] [PubMed] [Google Scholar]
- Slemc Lucija, Kunej Tanja. Transcription factor HIF1A: downstream targets, associated pathways, polymorphic hypoxia response element (HRE) sites, and initiative for standardization of reporting in scientific literature. Tumor Biology. 2016:1–11. doi: 10.1007/s13277-016-5331-4. [DOI] [PubMed] [Google Scholar]
- Strauss E, Waliszewski K, Oszkinis G, Staniszewski R. Gene-environment interaction for the HIF1-A 1772C>T polymorphisms and cigarette smoking increase susceptibility to abdominal aortic aneurysm. Przegl Lek. 2012;69(10):744–749. [PubMed] [Google Scholar]
- Sun X, Liu YD, Gao W, Shen SH, Li M. HIF-1alpha −1790G>A polymorphism significantly increases the risk of digestive tract cancer: a meta-analysis. World J Gastroenterol. 2015;21(5):1641–1649. doi: 10.3748/wjg.v21.i5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kizaki T, Hitomi Y, Nukita M, Kimoto K, Miyazawa N, Kobayashi K, Ohnuki Y, Ohno H. Genetic variation in hypoxia-inducible factor 1alpha and its possible association with high altitude adaptation in Sherpas. Med Hypotheses. 2003;61(3):385–389. doi: 10.1016/s0306-9877(03)00178-6. [DOI] [PubMed] [Google Scholar]
- Szkandera J, Knechtel G, Stotz M, Hofmann G, Langsenlehner U, Krippl P, Langsenlehner T, Dehchamani D, Samonigg H, Renner W, Gerger A. Association of hypoxia inducible factor 1-alpha gene polymorphisms and colorectal cancer prognosis. Onkologie. 2010;33:226–226. [PubMed] [Google Scholar]
- Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K, Kumazaki T, Oue N, Yasui W, Imai K, Nakachi K, Poellinger L, Nishiyama M. Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis. 2003;24(11):1779–1783. doi: 10.1093/carcin/bgg132. [DOI] [PubMed] [Google Scholar]
- Torres O, Palomino-Morales R, Vazquez-Rodriguez TR, Gamallo C, Morado IC, Miranda-Filloy JA, Amigo-Diaz E, Callejas-Rubio JL, Fernandez-Gutierrez B, Castaneda S, Martin J, Gonzalez-Gay MA. Lack of association between hypoxia inducible factor-1 alpha gene polymorphisms and biopsy-proven giant cell arteritis. Clinical and Experimental Rheumatology. 2010;28(1):S40–S45. [PubMed] [Google Scholar]
- Vilela BJ, Carvalho LC, Amancio S. Integration of stress produced reactive oxygen species in the stomatal regulation of micropropagated Vitis vinifera L. plantlets impaired in ABA signaling. Plant Signal Behav. 2008;3(8):558–559. doi: 10.4161/psb.3.8.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleugel MM, Greijer AE, van der Wall E, van Diest PJ. Mutation analysis of the HIF-1 alpha oxygen-dependent degradation domain in invasive breast cancer. Cancer Genetics and Cytogenetics. 2005;163(2):168–172. doi: 10.1016/j.cancergencyto.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A. 1993;90(9):4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Xiuchao, Liu Yingwei, Ren He, Yuan Zhanna, Li Shasha, Sheng Jun, Zhao Tiansuo, Chen Yong, Liu Fenghua, Wang Feng, Huang He, Hao Jihui. Polymorphisms in the hypoxia-inducible factor-1α gene confer susceptibility to pancreatic cancer. Cancer Biology & Therapy. 2011;12(5):383–387. doi: 10.4161/cbt.12.5.15982. [DOI] [PubMed] [Google Scholar]
- Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005(306):re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- Wipff J, Dieude P, Avouac J, Tiev K, Hachulla E, Granel B, Diot E, Sibilia J, Mouthon L, Meyer O, Kahan A, Boileau C, Allanore Y. Association of hypoxia-inducible factor 1A (HIF1A) gene polymorphisms with systemic sclerosis in a French European Caucasian population. Scand J Rheumatol. 2009;38(4):291–294. doi: 10.1080/03009740802629432. [DOI] [PubMed] [Google Scholar]
- Wu G, Yan WF, Zhu YZ, Sun PC. Hypoxia-inducible factor-1alpha (HIF-1alpha) C1772T polymorphism significantly contributes to the risk of malignancy from a meta-analysis. Tumour BIology. 2014;35(5):9. doi: 10.1007/s13277-013-1538-9. [DOI] [PubMed] [Google Scholar]
- Xu G, Wang M, Xie W, Bai X. Hypoxia-inducible factor-1 alpha C1772T gene polymorphism and glioma risk: a hospital-based case-control study from China. Genet Test Mol Biomarkers. 2011;15(6):461–464. doi: 10.1089/gtmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- Xu J, Xu L, Li L, You Q, Cha L. HIF-1alpha C1772T polymorphism and gastrointestinal tract cancer risk: a meta-analysis and meta-regression analysis. Genet Test Mol Biomarkers. 2013;17(12):918–925. doi: 10.1089/gtmb.2013.0325. [DOI] [PubMed] [Google Scholar]
- Yamada N, Horikawa Y, Oda N, Iizuka K, Shihara N, Kishi S, Takeda J. Genetic variation in the hypoxia-inducible factor-1alpha gene is associated with type 2 diabetes in Japanese. J Clin Endocrinol Metab. 2005;90(10):5841–5847. doi: 10.1210/jc.2005-0991. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang C, Zhu HC, Qin Q, Zhao LJ, Liu J, Xu LP, Zhang Q, Cai J, Ma JX, Cheng HY, Sun XC. HIF-1alpha P582S and A588T polymorphisms and digestive system cancer risk-a meta-analysis. Tumour Biology. 2014;35(3):6. doi: 10.1007/s13277-013-1375-x. [DOI] [PubMed] [Google Scholar]
- Yang Xi, Zhu Hong-Cheng, Zhang Chi, Qin Qin, Liu Jia, Xu Li-Ping, Zhao Lian-Jun, Zhang Qu, Cai Jing, Ma Jian-Xin, Cheng Hong-Yan, Sun Xin-Chen. HIF-1α 1772 C/T and 1790 G/A Polymorphisms Are Significantly Associated with Higher Cancer Risk: An Updated Meta-Analysis from 34 Case-Control Studies. PLoS ONE. 2013;8(11):e80396. doi: 10.1371/journal.pone.0080396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Wang M Fau - Hu Shuang, Hu S Fau - Shi Yingqiang, Shi Y Fau - Zhang Xiefu, Zhang X Fau - Zhou Ye, Zhou Y Fau - Zhao Chunlin, Zhao C Fau - Wang Guojun, Wang G Fau - Wen Jianguo, Wen J Fau - Zong Hong, Zong H. Hypoxia-inducible factor-1alpha C1772T polymorphism and cancer risk: a meta-analysis including 18,334 subjects. Cancer Invest. 2014;32(4):126–135. doi: 10.3109/07357907.2014.883527. [DOI] [PubMed] [Google Scholar]
- Zagouri F, Sergentanis TN, Gazouli M, Tsigginou A, Dimitrakakis C, Papaspyrou I, Eleutherakis-Papaiakovou E, Chrysikos D, Theodoropoulos G, Zografos GC, Antsaklis A, Dimopoulos AM, Papadimitriou CA. HSP90, HSPA8, HIF-1 alpha and HSP70-2 polymorphisms in breast cancer: a case-control study. Molecular Biology Reports. 2012;39(12):10873–10879. doi: 10.1007/s11033-012-1984-2. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen Y, Bin Z, Shi B, Weng WJ, Chen ZP, Guo NN, Hua YB, Zhu LJ. Hypoxia-Inducible Factor-1 alpha Polymorphisms and Risk of Cancer Metastasis: A Meta-Analysis. Plos One. 2013;8(8):6. doi: 10.1371/journal.pone.0070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Lv J, Zhao J, Nzekebaloudou M. Hypoxia-inducible factor-1alpha gene polymorphisms and cancer risk: a meta-analysis. J Exp Clin Cancer Res. 2009;28:159. doi: 10.1186/1756-9966-28-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Hwang YH, Kim SK, Kim S, Son MJ, Ro H, Sung SA, Lee HH, Chung WK, Joo KW, Yang J. Genetic polymorphisms of hypoxia-inducible factor-1 alpha and cardiovascular disease in hemodialysis patients. Nephron Clin Pract. 2009;113(2):c104–C111. doi: 10.1159/000228542. [DOI] [PubMed] [Google Scholar]
- Zhou Yuqiao, Lin Lin, Wang Yun, Jin Xin, Zhao Xin, Liu Dongjuan, Hu Ting, Jiang Lu, Dan Hongxia, Zeng Xin, Li Jing, Wang Jiayi, Chen Qianming. The association between hypoxia-inducible factor-1α gene G1790A polymorphism and cancer risk: ameta-analysis of 28 case-control studies. BMC Cancer cell international. 2014;14(37):11. doi: 10.1186/1475-2867-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.