Abstract
Inflammatory bowel diseases involve the dynamic interplay of host genetics, microbiome and inflammatory response. Here, we report that NLRP12, a negative regulator of innate immunity, is reduced in human ulcerative colitis by comparing monozygotic twins and other patient cohorts. In parallel, Nlrp12-deficiency in mice caused increased colonic basal inflammation, leading to a less-diverse microbiome, loss of protective gut commensal strains (Lachnospiraceae) and increased colitogenic strains (Erysipelotrichaceae). Dysbiosis and colitis susceptibility associated with Nlrp12-deficency were reversed equally by treatment with antibodies targeting inflammatory cytokines or by administration of beneficial commensal Lachnospiraceae isolates. Fecal transplants from specific pathogen free reared mice into germ-free Nlrp12-deficient mice showed that NLRP12 and the microbiome each contribute to immune signaling that culminates in colon inflammation. These findings reveal a feed-forward loop where NLRP12 promotes specific commensals that can reverse gut inflammation, while cytokine blockade during NLRP12-deficiency can reverse dysbiosis.
The healthy intestine is inhabited by trillions of bacteria and has evolved a fine-tuned balance between pathogen recognition and commensal tolerance1. In contrast, microbial imbalance, referred to as dysbiosis, is highly associated with inflammatory bowel diseases (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), yet IBD treatment is limited to immunotherapies. Hence, understanding the host gene-microbiota interaction that contributes to colitis holds promise in unveiling novel microbiome-based therapeutic options.
A hallmark of IBD is the dysregulated activation of inflammatory cytokines and signaling pathways such as the NF-κB, MAPK or STAT family members2. Several innate immune receptors and sensors known as NLRs (nucleotide-binding domain, leucine-rich repeat proteins or NOD-like receptors) regulate these pathways. NLRs are a family of intracellular innate immune sensors that exert either pro- or anti-inflammatory functions. The NLR family member NOD2 activates NF-κB, and mutations in NOD2 represent the first genetic association with CD susceptibility3, 4. Nod2-deficient mice display gut dysbiosis, which may contribute to exacerbated colitis5, 6, 7. Similarly, loss of the NLRP6 inflammasome is associated with increased pathogenic colitis-associated microbes8. Although the NLRP12 inflammasome has been implicated in the recognition of specific infections9, NLRP12 has non-inflammasome and anti-inflammatory functions by impeding canonical and non-canonical NF-κB10, 11, 12, 13. Although NLRP12 serves as negative regulator of inflammatory signaling in experimental colitis in mice, its impact on the colonic bacterial ecology and translational relevance to humans are completely unknown. Here, we identified an additional function for NLRP12 in restricting intestinal inflammation by promoting beneficial Lachnospiraceae strains.
RESULTS
Exacerbated colitis in Nlrp12−/− mice is microbiota-dependent
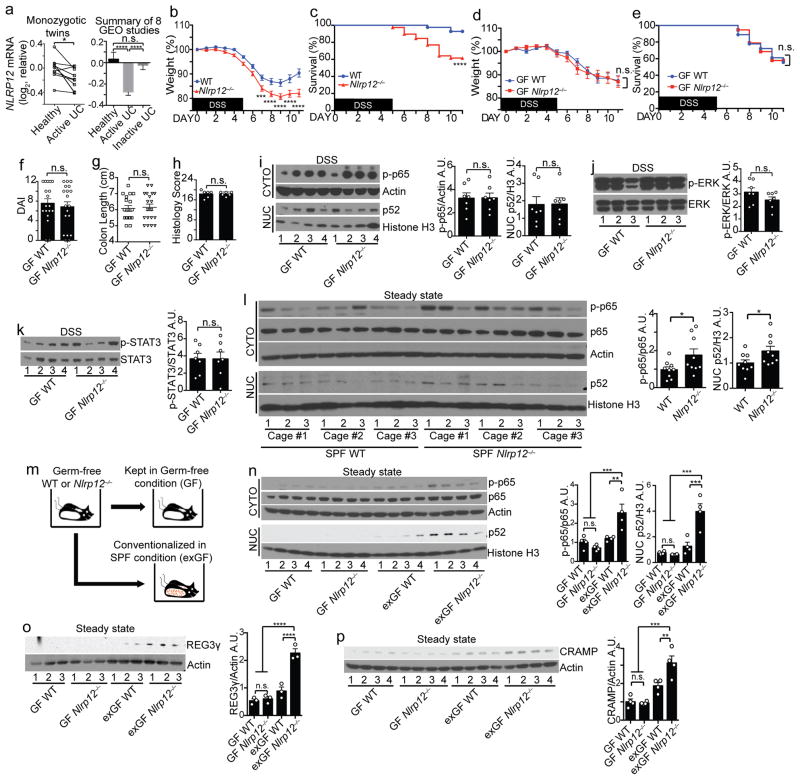
NLRP12 performs an anti-inflammatory role in experimental colitis12, 13. To determine if NLRP12 is clinically relevant to human colitis, we performed a meta-analysis of a paired-comparison between 10 pairs of monozygotic UC and healthy twins and seven additional UC patient gene-profiling studies (Fig. 1a and Supplementary Fig. 1) and found that NLRP12 was significantly down-regulated in active UC cohorts compared to healthy controls or inactive UC cohorts. In animals, conventionally-raised Nlrp12−/− mice displayed more severe colitis compared to wild type mice following the oral administration of 3% dextran sodium sulfate (DSS), which induces experimental colitis. These differences include significantly increased weight loss, mortality, histology score and clinical disease symptoms measured as disease-associated index (DAI), and Nlrp12−/− mice exhibited reduced colon length (indication of increased colonic inflammation) (Fig. 1b–c and Supplementary Fig. 2a–d)12, 13. Additionally, the loss of Nlrp12 resulted in heightened NF-κB, ERK and STAT3 activation (Supplementary Fig. 2e–g). These findings establish a link between reduced NLRP12 expression and human IBD and consolidate NLRP12’s protective role in limiting colon inflammation via suppressing excessive immune signaling.
Figure 1.
Microbiota differentiates colitis severity between WT and Nlrp12−/− mice. (a) NLRP12 gene expression between 10 pairs of monozygotic healthy and UC twins (left panel). Composite of 8 NCBI GEO ulcerative colitis (UC) studies (right panel) (Supplementary Fig. 1 shows the other seven individual studies). (b) Body weight and (c) percent survival of conventionally-raised mice treated with 3% DSS (WT, n=41; Nlrp12−/−, n=42), compiled from 4 independent experiments. (d) Body weight, (e) percent survival, (f) disease-associated index (DAI) and (g) colon length of germ-free (GF) WT (n=18) and Nlrp12−/− (n=19) mice given 1.5% DSS, compiled from 2 independent experiments. (h) Blinded histopathology scoring of colons (n=6/group). (i–k) Representative immunoblots and densitometry of distal colon proteins from DSS-treated GF mice from 2 independent experiments (n=7/group). (l) Immunoblots and densitometry of distal colon cytosolic (CYTO) and nuclear (NUC) protein fractions from untreated specific-pathogen free (SPF) mice (n=9/group). (m) Schematic comparing GF and conventionalized (exGF) mice. (n–p) Immunoblots and densitometry of distal colon proteins from untreated GF and exGF mice (n,p, n=4/group; o, n=3/group). One dot or one lane represents one mouse. Error bars show SEM. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001, and n.s. means no significance by two-tailed paired t test (a, left panel), unpaired t test (a, right panel, b, d, f–l, n–p) or Log-rank (Mantel Cox) test (c and e).
In addition to the association between genetic susceptibility and colon inflammation, mounting evidence supports a role for the microbiota during IBD pathogenesis14, 15, 16. To assess the impact of the microbiota on the increased colitis susceptibility in Nlrp12−/− mice, we subjected germ-free (GF) wild-type and Nlrp12−/− mice to a low-dose 1.5% DSS in drinking water due to the reported increased sensitivity of GF mice to DSS17. In contrast to conventionally-raised mice, GF-wild-type and GF-Nlrp12−/− mice showed indistinguishable weight loss, mortality, DAI, colon length and histology scores following DSS treatment (Fig. 1d–h). Colons from GF-wild-type and GF-Nlrp12−/− mice showed similar levels of activated NF-κB, ERK and STAT3 (Fig. 1i–k). These results implicate a role for the microbiota in severe colitis found in Nlrp12−/− mice.
The presence of NLRP12 curtails the activation of inflammatory cell signaling pathways induced by bacteria-derived ligands11, 12, 13, 18, which suggests Nlrp12−/− mice may present increased basal colonic inflammation in the presence of resident bacteria at the steady state. Indeed, colons from naive specific-pathogen free (SPF) Nlrp12−/− mice displayed elevated NF-κB p65 and p52 activation compared to controls (Fig. 1l). Of note, this basal level of activation was low compared to mice treated with DSS, as these blots required a longer exposure time to visualize the proteins. To determine the role of NLRP12 in regulating basal colon inflammation in the presence of microbiota, we housed GF-wild-type and GF-Nlrp12−/− mice in a SPF vivarium for 4 weeks (Fig. 1m). GF-Nlrp12−/− mice colonized for 4 weeks (exGF-Nlrp12−/−) displayed increased colon NF-κB activation compared to control mice (exGF-wild-type) and animals maintained in sterile isolators (i.e., GF-wild-type and GF-Nlrp12−/− mice) at steady state (Fig. 1n). These results demonstrate that basally-activated NF-κB immune signaling pathways require Nlrp12-deficiency and the microbiota from mice housed in conventional conditions. C-type lectin antimicrobial peptide REG3γ and cathelicidin-related antimicrobial peptide (CRAMP) were increased in the exGF-Nlrp12−/− mice compared to exGF-wild-type mice and control GF animals (Fig. 1o–p), which agrees with previous work that inflammation promotes antimicrobial peptides expression resulting in a dysbiotic intestinal microbiome19. These findings indicate that the intestinal microbiota exacerbated colitis in Nlrp12−/− mice.
NLRP12-deficiency results in a dysbiotic microbiome
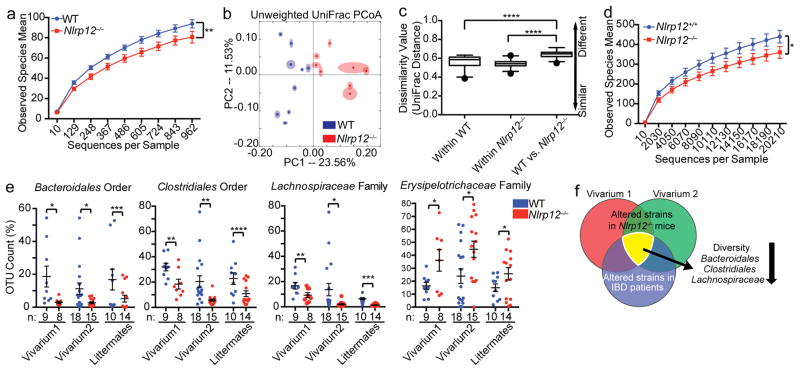
To determine if NLRP12 alters the microbiome, high throughput 16S rRNA gene sequencing analysis was performed on fecal bacterial DNA isolated from untreated wild-type and Nlrp12−/− mice that were originally generated from the same heterozygous Nlrp12+/− parents and raised in our facilities for more than nine generations. Rarefaction analysis was used to compare bacterial diversity within individuals of a group20. Compared to wild-type mice, Nlrp12−/− mice harbored a microbiota with significantly reduced diversity (Fig. 2a) and a different community composition (Fig. 2b). Comparing within and between group dissimilarity indicated that the microbiome difference between the wild-type vs. Nlrp12−/− mice was significantly greater than the differences between animals of each genotype (Fig. 2c, calculated from Fig. 2b). Housing and diet are key factors that can influence the intestinal microbiota21; thus, we repeated the microbiome analysis three years later with mice housed in a second vivarium with different formula chow and observed similar findings (Supplementary Fig. 3a–c). These results demonstrate that Nlrp12−/− mice retained this altered microbiome in different housing conditions.
Figure 2.
Nlrp12-deficiency results in a dysbiotic colonic microbiome. (a) Bacterial diversity and (b) unweighted UniFrac principal coordinate analysis (PCoA) showing microbiota compositional differences between WT (n=9) and Nlrp12−/− (n=8) mice from one experiment in vivarium #1. A second independent experiment was performed in vivarium #2 (Supplementary Fig. 3). Each symbol represents one mouse. (c) Quantification of UniFrac distance from (b) indicated as dissimilarity values. (d) Bacterial diversity of Nlrp12+/+ (n=10) and Nlrp12−/− (n=14) littermates, compiled from 2 independent experiments. (e) Consistently altered intestinal bacterial groups in Nlrp12−/− vs. WT mice from experiments described in (a–d and Supplementary Fig. 3). (f) Venn diagram showing overlapped microbiota changes between Nlrp12−/− vs. WT mice and IBD vs. healthy individuals (Supplementary Table 4 provides detailed information). Error bars denote SEM. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001 determined by two-tailed unpaired t test (a, d, and e) or ANOSIM test (c). The Tukey’s boxplot indicates the first (bottom of the box) and third quartiles (top of the box). The line inside the box represents the median, and the ends of the whiskers indicate the 1.5 interquartile ranges of the upper or lower quartiles (c).
Because the analyzed wild-type and Nlrp12−/− mice had been bred separately for multiple generations, it is possible that familial transmission instead of NLRP12-deficiency was responsible for the altered microbiome22. Therefore, we analyzed feces from wild-type and Nlrp12−/− littermates birthed from the same Nlrp12 heterozygous parents (Nlrp12+/−) and housed separately by genotype after weaning (Supplementary Fig. 3d). Compared to wild-type littermates, Nlrp12−/− mice from this heterozygous mating harbored a microbiota with significantly reduced diversity (Fig. 2d) and increased dissimilarity (Supplementary Fig. 3e) compared to wild-type littermates.
To establish rigor of the microbiota results, we used high throughput 16S rRNA gene sequencing to characterize the microbiome of naive wild-type and Nlrp12−/− mice in three mouse cohorts (vivarium 1, vivarium 2 and littermate studies) (Supplementary Tables 1–3) and observed a consistent decrease in Bacteroidales, Clostridiales and Lachnospiraceae and an increase in Erysipelotrichaceae in the Nlrp12−/− mice compared to wild-type animals (Fig. 2e). This is noteworthy, as IBD patients have been reported to display a similar microbiome profile14, 16, 23 (Fig. 2f and Supplementary Table 4). Thus, we focused on these bacterial groups in further studies.
Fecal transplantation attenuates disease in Nlrp12−/− mice
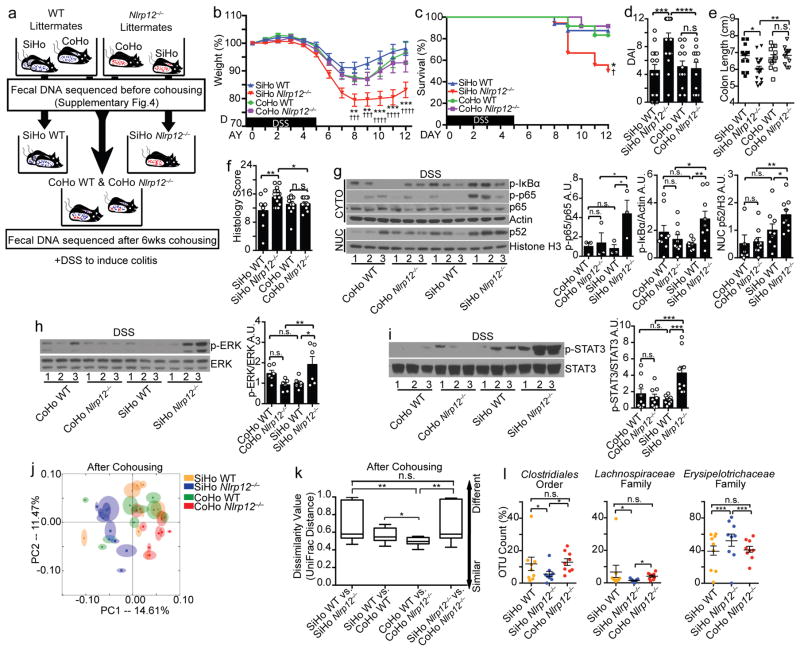
To determine if the altered microbiome observed in Nlrp12−/− mice is responsible for the exacerbated colon inflammation in Nlrp12−/− mice, we conducted microbiota transfer studies by cohousing mice, which leads to the exchange of the microbiota through coprophagia8, 24, 25. Age-and-sex matched wild-type and Nlrp12−/− mice were either single-housed (SiHo) or cohoused (CoHo) for six weeks prior to 3% DSS treatment (Fig. 3a). Control single-housed wild-type and Nlrp12−/− mice showed significant differences in disease severity (Fig. 3b–f). After receiving DSS, Nlrp12−/− mice that were cohoused with wild-type mice (CoHo Nlrp12−/−) displayed reduced disease and immune signaling compared to their SiHo Nlrp12−/− littermates (Fig. 3b–i and Supplementary Fig. 4a). Moreover, CoHo Nlrp12−/− and wild-type cage-mates were similar in all measurements (Fig. 3b–i). As a control, cohousing wild-type animals with inflammasome-deficient Asc−/− mice failed to ameliorate colitis in DSS-treated Asc−/− mice, but instead increased death, DAI and histopathology in wild-type mice as reported (Supplementary Fig. 4b–f)8. This is in contrast to improved survival and lessened colitis in CoHo Nlrp12−/− mice and makes Nlrp12-deficiency the first case where elevated colitis susceptibility resulting from an NLR deficiency can be reversed by transferring normal microbiota.
Figure 3.
Nlrp12−/− mice co-housed with WT mice display attenuated colitis. (a) Schematic of cohousing (CoHo) vs. single-housing (SiHo) strategy, fecal microbiome sequencing and DSS treatment of WT and Nlrp12−/− mice. (b) Body weight and (c) percent survival († indicates statistical significance between SiHo WT vs. SiHo Nlrp12−/−, and * indicates significance between SiHo Nlrp12−/− vs. CoHo Nlrp12−/−), (d) DAI and (e) colon length of littermate mice treated as illustrated in (a) (SiHo WT, n=16; SiHo Nlrp12−/−, n=18; CoHo WT, n=12; CoHo Nlrp12−/−, n=12), compiled from 3 independent experiments. (f) Histopathology scoring of colons (n=10/group). (g–i) Representative immunoblots and densitometry of distal colon proteins from the single- and cohoused mice from 3 independent experiments. (j) PCoA plot showing fecal microbial composition after cohousing (n=9/group). (k) Quantification of UniFrac distance between mice from (j) after cohousing, indicated as dissimilarity values. (l) Significantly altered strains identified by two-way ANOVA test from all sequenced bacteria in SiHo WT, SiHo and CoHo Nlrp12−/− mice (n=9/group). One dot or one lane represents one mouse. Error bars show SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 and n.s. means no significance determined using two-tailed unpaired t test (b), Log-rank (Mantel Cox) test (c), one-way ANOVA Fisher’s LSD test (d–i), ANOSIM test (k) and two-way ANOVA (l).
To investigate detailed microbiome changes after cohousing wild-type mice with Nlrp12−/− mice, we performed 16S rRNA gene microbiome sequencing analysis on feces from wild-type and Nlrp12−/− mice before and after cohousing (Fig. 3a). Consistent with Fig. 2, we observed significantly different community composition between the wild-type and Nlrp12−/− mice before cohousing (Supplementary Fig. 4g–h). After six weeks of cohousing, the microbiota composition of the cohoused Nlrp12−/− mice was more similar to that of the wild-type mice and distinct from the single-housed Nlrp12−/− littermates (Fig. 3j). The microbiome dissimilarity between cohoused and single-housed Nlrp12−/− mice was similar to that of control single-housed wild-type vs. Nlrp12−/− mice (Fig. 3k, calculated from Fig. 3j). Thus, Nlrp12−/− mice cohoused with wild-type mice developed a microbiota that was closer in community structure to that of wild-type animals. To assess if the transferred microbiota resulted in changes in specific bacteria, we performed a two-way ANOVA test on all sequenced fecal bacteria identified by 16S rRNA gene sequencing. The Clostridiales order and Lachnospiraceae family were significantly higher in frequency, and Erysipelotrichaceae was lower in the cohoused Nlrp12−/− mice compared to the single-housed Nlrp12−/− mice (Fig. 3l). These data demonstrate that transferred microbiota from wild-type mice ameliorated disease susceptibility in Nlrp12−/− mice in a co-housing setting.
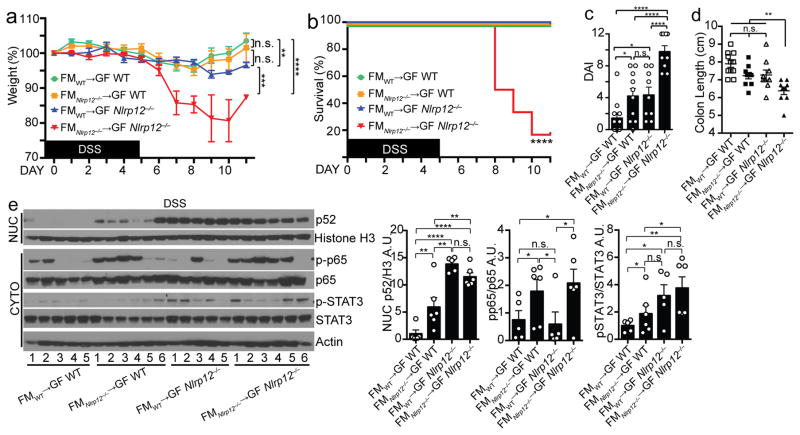
The experiments utilizing Nlrp12+/+ and Nlrp12−/− littermate mice and GF animals suggest NF-κB dysregulation occurs prior to alterations in the microbiota, as Nlrp12−/− mice developed a different microbiome from that of Nlrp12+/+ littermates after weaning (Fig. 2d–e), and GF Nlrp12−/− mice display increased basal colon inflammation when colonized (Fig. 1n). However, compared to single-housed Nlrp12−/− mice, cohoused Nlrp12−/− mice displayed attenuated NF-κB activation during DSS-induced colitis, suggesting the altered microbiota in Nlrp12−/− mice also contributes to heightened NF-κB activation (Fig. 3g). To investigate the causality between NF-κB hyperactivation and altered microbiota in Nlrp12−/− mice, we performed reciprocal fecal transplantation experiments in which GF wild-type mice were reconstituted with the microbiota of SPF Nlrp12−/− mice, and GF Nlrp12−/− mice were reconstituted with the microbiota of SPF wild-type mice via oral gavage once a week for 3 weeks as described24. The animals were given a one-week rest period where no fecal transfer was performed, and the mice were then exposed to DSS in drinking water for 5 days. Transfer of the fecal microbiota (FM) from an SPF wild-type donor into GF wild-type mice (designated as FMwild-type→GF wild-type) resulted in significantly reduced DSS-induced colitis, survival and disease index, but greater colon length compared to the transfer of FM from an SPF Nlrp12−/− donor into GF Nlrp12−/− mice (FMNlrp12−/−→GF Nlrp12−/−) (Fig. 4a–d). However, FMwild-type→GF Nlrp12−/− and FMNlrp12−/−→GF wild-type mice exhibited similar weight loss, survival, disease index and colon length, which were less severe compared to control FMNlrp12−/−→GF Nlrp12−/− mice (Fig. 4a–d). This indicates that Nlrp12−/−-shaped microbiome and genetic deficiency in Nlrp12 are both required to produce the fulminant colitis in Nlrp12−/− mice. Next, we examined the contribution of FM vs. host genetics on immune signaling pathways (Fig. 4e). As expected, colon samples from control FMwild-type→GF wild-type mice exhibited minimal NF-κB and STAT3 activation, whereas FMNlrp12−/−→GF Nlrp12−/− samples showed elevated p52, pp65 and pSTAT3. Colons from FMNlrp12−/−→GF wild-type and FMwild-type→GF Nlrp12−/− mice showed intermediate, but distinct activation patterns. FMNlrp12−/−→GF wild-type colons had elevated pp65 similar to FMNlrp12−/−→GF Nlrp12−/− colons, but reduced p52 nuclear translocation and STAT3 activation. By contrast, FMwild-type→GF Nlrp12−/− colons showed elevated p52 and pSTAT3, but modest pp65 activation. These results indicate that both the microbiome in Nlrp12−/− animals and Nlrp12 gene deficiency are required to cause the full activation of p52, p65 and STAT3 and fulminant colitis.
Figure 4.
Transferring microbiota from Nlrp12−/− mice induces colonic inflammation. (a) Body weight, (b) percent survival, (c) DAI and (d) colon length of GF WT vs. GF Nlrp12−/− mice reconstituted with fecal material (FM) from SPF WT or Nlrp12−/− mice and treated with DSS (FMWT→GF WT, n=11; FMNlrp12−/−→GF WT, n=10; FMWT→GF Nlrp12−/−, n=9; and FMNlrp12−/−→GF Nlrp12−/−, n=9), compiled from 2 independent experiments. (e) Immunoblots and densitometry of distal colon proteins from mice described in (a–d). Error bars show SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 and n.s. indicates no significance determined using two-tailed unpaired t test (a, e), Log-rank (Mantel Cox) test (b) and one-way ANOVA Fisher’s LSD test (c–d).
Lachnospiraceae suppresses colitis in Nlrp12−/− mice
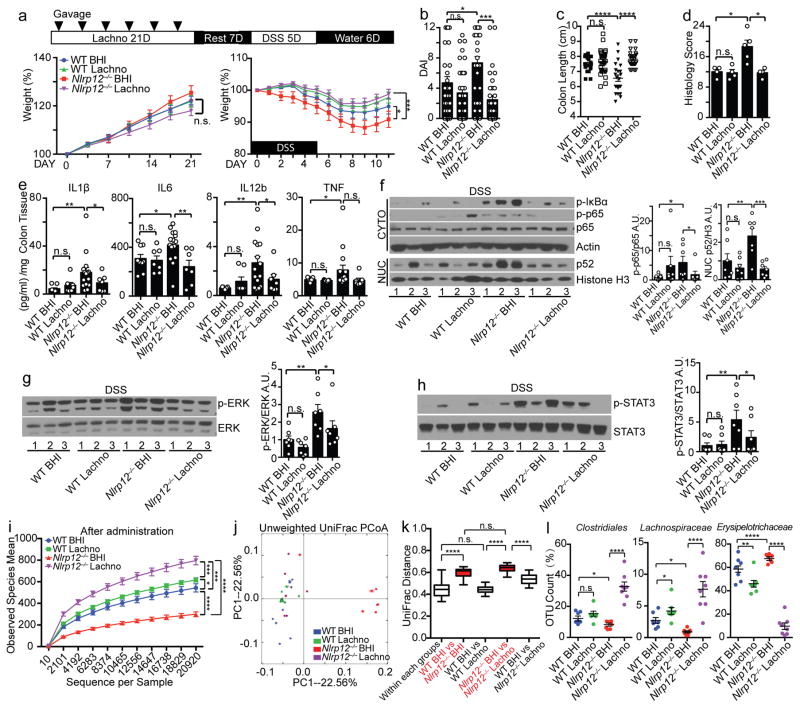
Figure 3 showed that Clostridiales order and Lachnospiraceae family were significantly higher, and Erysipelotrichaceae was lower in Nlrp12−/− mice compared to wild-type mice. Lachnospiraceae strains are decreased in CD14, 16, 23, and Lachnospiraceae limits experimental C. difficile-induced colitis26. To determine if bacterial groups that are lacking in Nlrp12−/− mice can restrict colitis, we inoculated wild-type and Nlrp12−/− mice with 23 strains of Lachnospiraceae via oral gavage for 21 days followed by one week of rest before DSS treatment (Fig. 5a). Mice given Lachnospiraceae and brain-heart infusion (BHI) vehicle control, which was used to grow Lachnospiraceae, had similar weight prior to DSS treatment (Fig. 5a, left panel). Strikingly, DSS-treated Nlrp12−/− mice given Lachnospiraceae displayed reduced colitis including reduced weight loss, DAI, colon histopathology and increased colon lengths compared to Nlrp12−/− mice given vehicle control (Fig. 5a–d and Supplementary Fig. 5). Proinflammatory cytokines (Fig. 5e) and canonical and non-canonical NF-κB, ERK and STAT3 showed similar reductions (Fig. 5f–h).
Figure 5.
Administration of Lachnospiraceae suppresses colitis in Nlrp12−/− mice. (a) Body weight of WT and Nlrp12−/− mice administered Lachnospiraceae (Lachno) or BHI vehicle (left panel) and DSS (right panel), with (b) DAI and (c) colon length after DSS treatment (WT BHI, n=24; WT Lachno, n=33; Nlrp12−/− BHI, n=22; Nlrp12−/− Lachno, n=22), compiled from 3 independent experiments. (d) Histopathology scoring of BHI- or Lachno-treated mice given DSS (WT BHI, n=4; WT Lachno, n=4; Nlrp12−/− BHI, n=5; Nlrp12−/− Lachno, n=4). (e) Colon explant culture cytokines (WT BHI, n=9; WT Lachno, n=9; Nlrp12−/− BHI, n=14; Nlrp12−/− Lachno, n=8), and (f–h) representative immunoblots with densitometry of distal colon proteins from DSS-treated mice given BHI or Lachno from 3 independent experiments (n=7/group). (i) Bacterial diversity and (j) PCoA plot showing microbiome compositional differences quantified by (k) UniFrac distances among the mice administered Lachno or BHI prior to DSS (WT BHI, n=7; WT Lachno, n=8; Nlrp12−/− BHI, n=7; Nlrp12−/− Lachno, n=8). (l) Significantly altered strains among all identified intestinal bacteria by 16s rRNA gene sequencing following Lachno administration. One dot or one lane represents one mouse. Error bars show SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 and n.s. means no significance determined using unpaired t test (a, i), one-way ANOVA Fisher’s LSD test (b–h), ANOSIM test (k) and two-way ANOVA (l).
To examine whether Lachnospiraceae modulates severity of inflammation by altering the composition of the microbiota of Nlrp12−/− mice, we sequenced the microbiome of the mice receiving Lachnospiraceae or control BHI. For both wild-type and Nlrp12−/− mice, the diversity of the microbiota from Lachnospiraceae-receiving mice was significantly increased compared to BHI-treated littermates, although the impact was greater in Nlrp12−/− mice (Fig. 5i). UniFrac distance analysis was used to quantify the dissimilarity between bacterial communities among the groups. The distance between the BHI-treated Nlrp12−/− mice vs. Lachnospiraceae-treated Nlrp12−/− mice was as large as the distance between the control BHI-treated wild-type vs. BHI-treated Nlrp12−/− mice (Fig. 5j–k), indicating that the microbiome in Lachnospiraceae-treated Nlrp12−/− mice was significantly distinct from that of littermate control Nlrp12−/− mice given BHI. Additionally, the dissimilarity distance between the BHI-treated wild-type and Lachnospiraceae-treated Nlrp12−/− mice was significantly decreased compared to the distance between the BHI-treated Nlrp12−/− mice and Lachnospiraceae-treated Nlrp12−/− mice (Fig. 5k), suggesting that Nlrp12−/− mice administered Lachnospiraceae develop a microbiome more reminiscent of wild-type animals. Among all the strains sequenced (Supplementary Table. 5), administration of Lachnospiraceae promoted the growth of Clostridiales in Nlrp12−/− mice, but significantly reduced Erysipelotrichaceae (Fig. 5l). As the abundance of Erysipelotrichaceae was increased in the Nlrp12−/− mice, which positively correlated with disease severity (Fig. 3b–f), administration of Lachnospiraceae may limit colitis in part by suppressing expansion of Erysipelotrichaceae.
Hematopoietic NLRP12 expression prevents dysbiosis
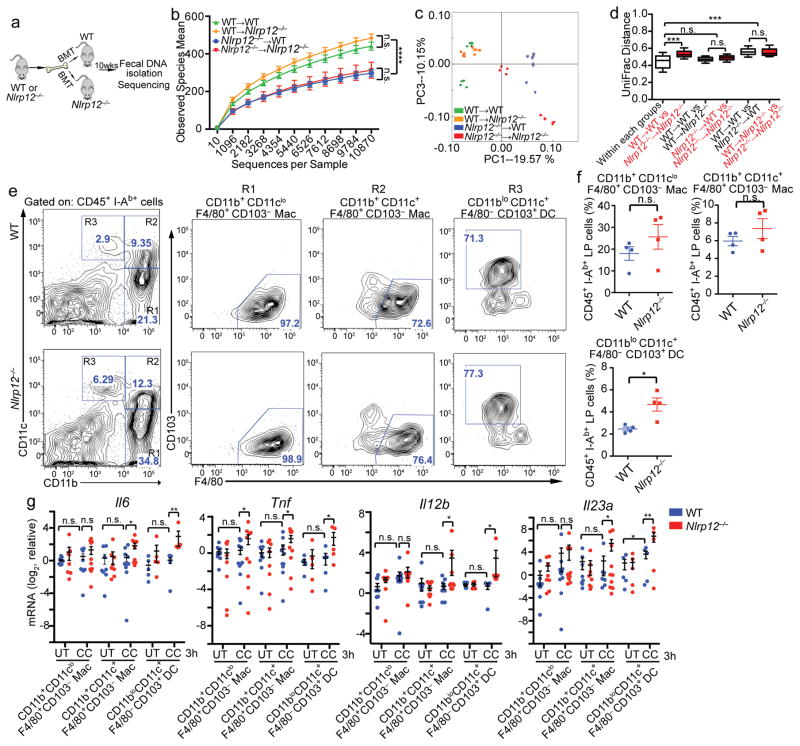
Increased intestinal inflammation in Nlrp12−/− mice is driven by both hematopoietic and non-hematopoietic components12, 13, but the cellular compartment expressing NLRP12 that shapes the intestinal commensals is unknown. To address this, we generated radiation bone marrow chimeric mice by lethally irradiating wild-type and Nlrp12−/− mice and then transferred bone marrow from wild-type and Nlrp12−/− donors to generate wild-type mice containing myeloid cells deficient in Nlrp12 (Nlrp12−/−→wild-type) and Nlrp12−/− mice expressing Nlrp12 only in myeloid cells (wild-type→Nlrp12−/−). We then assessed the microbiome after a 10-week-reconstituition period (Fig. 6a). As expected, control wild-type mice given wild-type bone marrow retained greater microbiota diversity than Nlrp12−/− recipients with Nlrp12−/− bone marrow (Fig. 6b). Wild-type mice with Nlrp12−/− bone marrow (Nlrp12−/−→wild-type) displayed reduced bacterial diversity compared to wild-type→wild-type mice. Additionally, transplantation of wild-type bone marrow into Nlrp12−/− mice (wild-type→Nlrp12−/−) increased bacterial diversity compared to control Nlrp12−/−→Nlrp12−/− mice (Fig. 6b). These results indicate that NLRP12 expression in the hematopoietic compartment dominantly shapes the diversity of the intestinal bacteria. Analysis of bacterial composition confirmed these findings (Fig. 6c–d).
Figure 6.
NLRP12 expression by hematopoietic cells prevents intestinal dysbiosis. (a) Schematic illustrating bone-marrow transplantation (BMT) and fecal DNA collection for microbiome analysis. (b) Bacterial diversity, (c) PCoA plot and (d) UniFrac distance plots showing microbiome compositional differences in WT and Nlrp12−/− mice after BMT (n=8/group), compiled from 2 independent experiments. One dot represents one mouse. (e) Representative concatenated flow cytometry plots, and (f) quantification of colonic lamina propria macrophage and DC subpopulations in SPF mice. Each dot contains pooled cells isolated and sorted from the colons of 2–3 mice. (g) RT-qPCR of proinflammatory cytokines expressed by colon resident macrophages (9 technical replicates/group) and DCs (6 technical replicates/group) stimulated with cecal contents, compiled from 2 independent experiments. Error bars show SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 and n.s. means no significance determined using two-tailed unpaired t test (b, f and g) and ANOSIM test (d).
We next characterized the hematopoietic cells in the colon lamina propria (cLP) that could be contributing to increased basal inflammation in Nlrp12−/− mice. Although NLRP12 suppresses inflammatory pathways in bone marrow-derived macrophages13, 18 and DCs12, there is no evidence to support a role for NLRP12 in colonic macrophages and DCs, which are phenotypically different from their bone marrow-derived counterparts27. Within the cLP, colon resident macrophages are identified as two subsets that express the fractalkine receptor (CX3CR1), F4/80 and CD11b with or without coexpression of CD11c. DCs within the cLP express CD11c and CD103 and are considered CD11b negative or low (Fig. 6e)28. To analyze the impact of NLRP12 on these macrophage and DC populations in the steady state, we isolated cLP cells from naive wild type and Nlrp12−/− mice and gated on CD45+I-Ab+ cells with differential expression of F4/80, CD11b, CD11c and CD103 by flow cytometry. In accordance with increased colonic inflammation in SPF Nlrp12−/− mice, these mice displayed a significantly increased frequency of CD11c+CD11blo cells (designated R3 in Fig. 6e, left panels) and a modestly increased frequency of CD11cloCD11b+ (R1) and CD11c+CD11b+ (R2) populations. These R1–R3 fractions were further stained for F4/80 and CD103. In wild type and Nlrp12−/− mice, the R1 and R2 populations were found to be primarily macrophages, and R3 were DCs (Fig. 6e, right panels). The composite data of multiple animals showed an increased frequency of CD11bloCD11c+F4/80−CD103+ DCs in the lamina propria of Nlrp12−/− mice (Fig. 6f). The frequency of CD11b+CD11cloF4/80+CD103− and CD11b+CD11c+F4/80+CD103− populations, which were characterized as macrophages, were also increased in Nlrp12−/− mice, but this difference was not significant (Fig. 6f). These results suggest that Nlrp12 deficiency increased CD11bloCD11c+F4/80−CD103+ colonic DCs.
To maintain colon homeostasis, macrophages and DCs in the cLP acquire anti-inflammatory phenotypes29 and become anergic to bacterial stimulation27. To test if NLRP12 plays a role in maintaining colonic macrophage and DC quiescence, we exposed cLP macrophages and DCs from naive wild-type and Nlrp12−/− mice to cecal contents from wild-type animals to mimic the cell-resident bacterial interactions in vivo. Consistent with the concept that cLP macrophages and DCs are anergic to bacterial stimulation27, cecal content exposure did not increase inflammatory cytokine gene expression in wild-type macrophages and DCs with the exception of a slight increase in Il23a (encoding p19) expressed by wild-type DCs (Fig. 6g). By contrast, cecal contents significantly increased Il6, Tnf, Il12b (encoding p40) and Il23a expression in Nlrp12−/− CD11b+CD11c+ macrophages and CD11bloCD11c+ DCs compared to wild-type controls (Fig. 6g). Only Tnf was significantly elevated in Nlrp12−/− CD11b+CD11clo macrophages after exposure to cecal contents. These results suggest that NLRP12 maintains homeostasis in colonic macrophages and DCs in the presence of cecal material.
TNF and IL-6 cause microbial dysbiosis in Nlrp12−/− mice
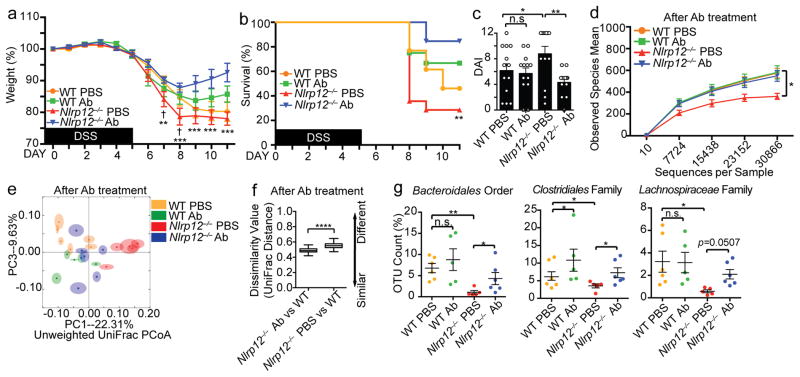
The findings above (Fig. 4) indicate that the Nlrp12 deficiency and Nlrp12−/−-derived microbiome resulted in elevated inflammation. We next directly tested if increased inflammation could drive microbial dysbiosis. Anti-TNF antibody is a common treatment for IBD patients30, while anti-IL-6R antibody has been discussed as a new therapeutic strategy for colitis31, 32. We targeted the inflammatory cytokines TNF and IL-6 in vivo by injecting mice with anti-TNF and anti-IL-6 receptor (IL-6R) antibodies (Ab) prior to and during DSS exposure (Supplementary Fig. 6a–b). Blocking TNF and IL-6 signaling ameliorated DSS-induced colitis in Nlrp12−/− mice indicated by significantly improved weight gain, survival and clinical scores (Fig. 7a–c), while the impact was less pronounced in wild-type controls.
Figure 7.
Anti-IL-6R and anti-TNF suppresses colitis and restores the missing bacterial groups in Nlrp12−/− mice. (a) Body weight, (b) percent survival († indicates statistical significance between WT PBS vs. Nlrp12−/− PBS; * indicates significance between Nlrp12−/− PBS vs. Nlrp12−/− Ab) and (c) DAI of DSS-treated WT and Nlrp12−/− mice injected i.p. with anti-IL-6R and anti-TNF antibodies (Ab) or PBS (n=13/group), compiled from 3 independent experiments. (d) Intestinal microbial diversity, and (e) PCoA showing microbiota compositional differences in Ab-treated WT and Nlrp12−/− mice (n=6/group). (f) Quantification of UniFrac distances from (e). (g) Significantly altered groups from sequenced bacteria in Ab- vs. PBS-treated animals. Error bars show SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 and n.s. indicates no significance determined by two-tailed unpaired t test (a and d), Log-rank (Mantel Cox) test (b), one-way ANOVA Fisher’s LSD test (c), ANOSIM test (f) or two-way ANOVA (g).
To determine if Ab treatment reversed dysbiosis in Nlrp12−/− mice, we performed microbiome analysis before and after Ab treatment. Prior to Ab treatment, naive Nlrp12−/− mice displayed significantly altered microbiome compared to wild-type mice (Supplementary Fig. 6c–e). However, anti-TNF- and anti-IL-6R-treated Nlrp12−/− mice displayed significantly increased intestinal bacterial diversity compared to PBS-treated Nlrp12−/− mice (Fig. 7d) and reduced microbiome compositional differences compared to wild-type mice (Fig. 7e–f), with elevated Bacteroidales and Clostridiales taxa (Fig. 7g). By contrast, PBS-treated Nlrp12−/− mice maintained significantly reduced bacterial diversity, Bacteroidales and Clostridiales compared to wild-type mice. These results indicate that blocking excessive inflammatory cytokines recovered the altered microbiome and reduced colitis in Nlrp12−/− mice.
DISCUSSION
NLRP12 has a protective role in intestinal inflammation by suppressing canonical and noncanonical NF-κB12, 13. In this study, we identified an additional critical role for NLRP12 by regulating gut microbial communities. IBD-profiling studies revealed that NLRP12 expression is negatively correlated with active UC. In addition, an imbalance in the intestinal microbiota, or dysbiosis, has been linked to IBD pathogenesis14, 15, 16. We reveal a link between these two clinical observations by providing direct evidence that NLRP12 promotes microbial symbiosis, which results in reduced colitis susceptibility.
Unlike conventionally-raised Nlrp12−/− mice, which are more susceptible to colitis compared to similarly raised wild-type animals, Nlrp12−/− mice raised under germ-free conditions were indistinguishable from GF-wild-type mice during experimental colitis. This indicates interactions between host cells and resident bacteria are required for the severe disease susceptibility in Nlrp12−/− mice. Intestinal microbial diversity and human disease have a complex reciprocal cause and effect relationship. A reduction of gut microbiome richness is a biomarker for human metabolic and inflammatory disorders33, including IBD24. Loss of commensal diversity due to genetic alterations34 or an unhealthy diet35 correlates with exacerbated colitis. Our results indicate that NLRP12 acts a central component of this relationship by curtailing excessive inflammatory cytokine production to limit intestinal inflammation and maintain commensal diversity and protective microbiota. Neutralization of TNF is a current IBD therapy, and targeting IL-6R is being assessed as a similar therapeutic approach. Our findings indicate that in addition to suppressing inflammatory signaling, targeting these cytokines may also alleviate colitis by reversing dysbiosis.
Many protective intestinal bacterial groups are necessary for maintaining gut homeostasis. Similar to the microbiota found in CD patients14, we observed a decrease in Bacteroidales and Clostridiales (in particular Lachnospiraceae) strains in untreated Nlrp12−/− mice, which correlated with a greater sensitivity to DSS-induced colitis. Nod2- or Nlrp6-deficiency results in a colitogenic microbiota that can exacerbate colitis5, 6, 7, 8. In contrast, this is the first observation where dysbiosis and colitis severity caused by loss of an NLR can be reversed by transferring the microbiota from wild-type animals, further supporting our conclusion that NLRP12 plays a predominate role in maintaining protective commensal bacterial groups and limiting colitogenic strains. In line with this, Nlrp12−/− mice cohoused with wild-type mice exhibited elevated intestinal Clostridiales and Lachnospiraceae, decreased Erysipelotrichaceae and reduced DSS-induced colitis. Although Lachnospiraceae are decreased in IBD patients14, 16, 23, 36, 37 and Clostridium difficile-associated colitis38, little is known about how these protective strains impact the host immune system. Lachnospiraceae are more enriched in the mucosal folds compared to the central lumen39. The spatial location of Lachnospiraceae favors their interaction with lamina propria residual immune cells, which suggests Lachnospiraceae may function as an immune regulator to prevent enteric pathogen adhesion/colonization. Lachnospiraceae are also major producers of the short chain fatty acid (SCFA) propionate in the human gut40, which promotes peripheral regulatory T cell generation41, 42. Loss of Lachnospiraceae species in the Nlrp12−/− mice may result in reduced SCFA production, which could contribute to elevated inflammation in Nlrp12−/− mice. Moreover, there is a negative correlation between intestinal Lachnospiraceae and Erysipelotrichaceae. Erysipelotrichaceae have been linked to elevated TNF levels and chronic intestinal inflammation in SIV-infected animals and HIV-infected patients receiving antiretroviral therapy43, 44, which suggest these organisms appear to be highly inflammatory. In agreement with these findings, we showed that Erysipelotrichaceae-enriched fecal microbiota from Nlrp12−/− mice induced more NF-κB and STAT3 activation. Lachnospiraceae may provide a protective function in part by suppressing over-growth of intestinal Erysipelotrichaceae. Accompanied by the human gene profile outcome where mucosal NLRP12 expression is reduced in active colitis, our results suggest that Lachnospiraceae may be therapeutic when applied to UC subjects with reduced NLRP12 expression.
In sum, reciprocal fecal transplantation experiments, cohousing and bacteria transfer experiments collectively indicate that the interaction between aberrant immune signaling and dysbiotic microbiota in Nlrp12−/− animals does not proceed in a one-way linear trajectory, but instead in a vicious feed-forward cycle in which increased inflammation driven by loss of NLRP12 expression produces a proinflammatory microbiota that further escalates inflammation (Supplementary Fig. 7).
METHODS
Mice
All animal procedures were approved by the University of North Carolina Chapel Hill (UNC) Institutional Animal Care and Use Committee (IACUC) according to US National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animal numbers were empirically determined to optimize numbers necessary for statistical significance based on our previous reports utilizing these disease models (minimum of 4–7 animals/group). Animals were excluded if exhibiting signs of disease not associated with the conditions of the colitis (e.g., fight wounds and malocclusion). No randomization method was used due to the single-housing and co-housing strategy required for the microbiome studies, ad libitum administration of DSS in drinking water and use of littermate controls.
Wild type (WT) C57BL/6J mice were obtained from Jackson laboratory and house-raised at UNC for at least 9 generations. Nlrp12−/− and Asc−/− mice are on C57BL/6J background after at least 9 generations of backcross. Conventionally raised mice were bred and housed in specific-pathogen-free (SPF) conditions, and germ-free (GF) animals were generated and housed in the National Gnotobiotic Rodent Resource Center of North Carolina at Chapel Hill or in the Center for Gastrointestinal Biology and Disease, Gnotobiotic Core at North Carolina State University. For cohousing experiments, 4-week old mice originating from the same breeders were divided for either single-housed (SiHo) or cohoused (CoHo) with age- and sex-matched mice for 6 weeks. CoHo mice were compared to their SiHo littermates as controls. For conventionalization study, GF animals were transferred into SPF condition and housed for 4 weeks. Conventionalized (exGF) animals were compared to mice kept in the GF condition as controls. For the reciprocal fecal transplantation study, GF wild-type or Nlrp12−/− mice were orally gavaged once a week for three weeks with a PBS suspension of feces derived from SPF wild-type or Nlrp12−/− mice following the previously described protocol24. The mice were given DSS 1 week after the final gavage. For adoptive bone marrow transplantation study, wild-type and Nlrp12−/− chimeric mice were generated as previously described12. No antibiotics were used during the reconstitution phase, and fecal DNA was collected 10 weeks after the transplantation for microbiome analysis.
Experimental Colitis
Experimental colitis was initiated by treating mice with 1.5%–3% dextran sulfate sodium (DSS) (36,000–50,000 M.Wt, MP Biomedicals) in autoclaved drinking water for 5 days as indicated. DSS was then replaced by normal autoclaved water for 6–7 days. Body weight was monitored daily, and disease-associated index (DAI) was determined by an investigator blinded to experimental conditions on day 8 post-DSS treatment according to the following parameters: 1) Weight loss (0 points=0% weight loss from baseline, 1 point=1–5% weight loss, 2 points=5–10% weight loss, 3 points=10–20% weight loss and 4 points=more than 20% weight loss); 2) Rectal bleeding (0 points=negative, 2 points=positive hemoccult test and 4 points=gross bleeding); and 3) Stool consistency (0 points=normal, 2 points=semiformed stool and 4 points=liquid that adhere to the anus). The sum of these 3 parameters results in the total DAI ranging from 0 (healthy) to 12 (maximal activity of colitis)12.
Histopathology
Colons were Swiss-rolled, fixed in 10% neutral-buffered formalin and paraffin embedded and processed for histological analysis. Five-micron-thick colon sections were stained with hematoxylin and eosin (H&E) and semi-quantitatively scored for histopathology by a board-certified veterinary pathologist in a blind manner. Histology score represented the sum of each histological alterations outlined below. This system assessed inflammation, epithelial defects, area of inflammation, area of epithelial defect, crypt atrophy and dysplasia/neoplasia by giving each parameter a separate score (0–4) for severity and extent as previously described12.
Immunoblotting
Colons were excised, opened longitudinally and washed with cold PBS. The distal-most-3 cm section of each colon was collected and mechanically homogenized in NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo Scientific) containing Complete Protease Inhibitor and PhosphoStop (Roche) per manufacturer’s protocol. Cytosolic and nuclear lysates were subjected to SDS-PAGE and immunoblotting. The following primary antibodies were use for immunoblot analysis: anti-pIκBα (Ser32) (14D4) (cat. no 2859); anti-pIKKα/β (Ser176/180) (16A6) (cat. no. 2697); anti-pp65 (Ser536) (93H1) (cat. no. 3033); anti-NIK (cat. no. 4994); anti-pERK1/2 (Thr202/Tyr204) (D13.14.4E) (cat. no. 4370); anti-p65 (D14E12) (cat. no. 8242); anti-pSTAT3 (Tyr705) (cat. no. 9131); and anti-STAT3 (79D7) (cat. no. 4904) (Cell Signaling Technology); anti-p52 (C-5) (cat. no. sc-7386); anti-ERK1 (C-16) (cat. no. sc-93); anti-ERK2 (C-14) (cat. no. sc-154); anti-CRAMP (G-1) (cat. no. sc-166055) and anti-Actin-HRP (C-11) (cat. no. sc-1615) (Santa Cruz Biotechnology); anti-REG3γ (cat. no. ab198216) (Abcam); and anti-Histone H3 (cat. no. 07-690) (Millipore). Goat anti-rabbit-HRP (cat. no. 111-035-144) and goat anti-mouse-HRP (cat. no. 115-035-146) (Jackson Laboratories) were used as secondary antibodies. Protein densitometry were quantified by Image J software.
Colon Explant Cultures
Colons were excised, washed several times in cold PBS containing 2x penicillin/streptomycin (Sigma Aldrich) and cultured for 15 hours in RPMI media (Gibco) containing 2x penicillin/streptomycin at 37°C. Supernatants were collected, centrifuged to clear debris and assessed for cytokines by Luminex Bio-Plex System per manufacturers instructions.
Fecal DNA Extraction
Fecal samples were collected from live mice, snap-frozen and stored at −80°C. DNA was isolated by incubating fecal material at 65°C for 30 min in Lysing Matrix E tubes (MP Biomedicals) containing 200 mM NaCl, 100 mM Tris, 20 mM EDTA (pH 8.0), SDS and proteinase K (Qiagen). Phenol:Choloroform:Isoamyl alcohol (Invitrogen) was added, and the samples were homogenized at 4°C for 3 min using a bead beater homogenizer. The samples were centrifuged at 8000 rpms for 3 min at 4°C, and the supernatant was incubated with Phenol:Chloroform (Invitrogen) for 10 min at room temperature. The samples were centrifuged at 13,000 rpms for 5 min at 4°C, and the aqueous phase was incubated with isopropanol and 3M sodium acetate, pH 5.2, at −20°C for 15 hrs to precipitate DNA. The precipitated DNA was collected by centrifugation at 13000 rpm at 4°C for 20 min, washed twice with 100% cold ethanol and resuspended in TE buffer. The DNA was further purified using a DNeasy Blood and Tissue Kit (Qiagen) according to the manufactures protocol.
16S rRNA Gene Sequencing and Data Analysis
Fecal DNA samples were amplified by PCR using barcoded primer pairs targeting the V1–V2 region of the 16S gene. PCR amplicons were sequenced at the V1–V2 region using Roche 454 for the vivarium #1 study37, and the same V1–V2 region was sequenced using an Mi-Seq Illumina sequencer38 for all other described experiments. The resulting bacterial sequence fragments were clustered into Operational Taxonomic Units (OTUs) and aligned to microbial genes with 97% sequence similarity from Greengenes Database using UCLUST method in QIIME. Bacterial taxa summarization and rarefaction analyses of microbial diversity or compositional differences (dissimilarity value indicated by Unweighted UniFrac Distance) were calculated in QIIME (1.8.0) as previously described20 using QIIME scripts (including pick_open_reference_otus.py, summarize_taxa.py, alpha_rarefaction.py, jackknifed_beta_diversity.py and make_distance_boxplots.py). PCoA plots indicating compositional difference were generated by QIIME script, make_2d_plots.py. Each point represents one mouse, and the ellipses represent the interquartile range (IQR) during the rarefaction analyses45 (see http://www.wernerlab.org/teaching/qiime/overview for scripts details). All 16S rRNA microbiome sequences have been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena) and QIITA (https://qiita.ucsd.edu). The accession codes are listed in the Data Availability section.
Mouse colonic lamina propria macrophage and dendritic cells isolation and stimulation
Mouse colons were opened longitudinally and washed with cold PBS to remove the fecal contents. Pooled colons from 2 mice were further cut into 1.5 cm pieces and washed twice with 30 ml of HBSS containing 10% FBS and 2mM EDTA on an orbital shaker at 250 rpms for 30 min at 37°C. After washing, the colons were finely minced and digested with 20 ml of HBSS containing 10% FBS, 1.5 mg/ml Type VIII Collagenase (C2139; Sigma-Aldrich) and 40 μg/ml DNase I (4527; Sigma-Aldrich) at 200 rpms for 20 min at 37°C. After the digestion, the digested colonic lamina propria (cLP) cells were filtered through a 100 μm strainer, centrifuged at 1,500 rpm for 5 min at 4 °C and resuspended in 2 ml MACS buffer for flow cytometric analysis or fluorescence-activated cell sorting (FACS)46.
For flow cytometric analysis, cLP cells were incubated with mouse Fcγ II/III receptor blocker (553141; BD Pharmingen) for 15 min on ice and were then stained with the following labeled antibodies for 30 min on ice: Brilliant Violet 421-conjugated anti-CD45 (103133; Biolegend), PE-conjugated anti-IAb (553552; BD Pharmingen), APC-Cy7-conjugated anti-CD11c (117324; Biolegend), PE-Cy7-conjugated anti-CD11b (101216; Biolegend), APC-conjugated anti-CD103 (17-1031-80; eBioscience), and Alexa Fluor 488-conjugated anti-F4/80 (123120; Biolegend). Samples were washed twice in MACS buffer and were analyzed on a BD LSR II flow cytometry at the UNC Flow Cytometry Core Facility. Dead cells and doublets were excluded from the cLP mononuclear based on appropriate forward and side scatter plots. Antigen-presenting cells (APC) were defined as CD45+I-Ab+. Macrophage and dendritic cell (DC) populations were firstly categorized by the expression of CD11b and CD11c and then further characterized by CD103 and F4/80 expression as previously described46.
For cLP macrophage and DC FACS separation and ex vivo stimulation, CD11b+ and CD11c+ microbeads (Miltenyi Biotec) were used to enrich CD11b+ or CD11c+ positive cells. The enriched cells were stained with labeled antibodies aforementioned. Stained cells were sorted to purify the indicated population at the UNC Flow Cytometry Core Facility. Sorted macrophages and DCs were seeded in a 96-well plates (0.5~1X105 cells per well) with 200 μl RPMI medium containing 10% FBS. For cecal content stimulation, cecal contents were generated from wild-type mice as described elsewhere47 and were added to the cell culture medium at a 1:200 dilution for 3 hours. Total RNA was then isolated by TRIzol according to the manufacturer’s instructions (Thermo Fisher Scientific) for qRT-PCR analysis.
Design of Lachnospiraceae specific primers
Lachnospiraceae strains were identified utilizing an adapted version of the plate wash PCR technique48. This allowed for rapid screening by PCR using phylotype specific primers of the 16S rRNA gene to screen selective media plates in order to identify conditions that enrich for specific bacteria. To generate Lachnospiraceae specific primers the near full length 16S rRNA gene sequence from 34 bacterial strains were used to generate CLUSTALW multiple-sequence alignments. The 16S rRNA gene sequences used were the following: 9 of the most represented bacterial sequences from the murine large intestine found in Berry, et al.49 (Akkermansia muciniphila, Alistipes finegoldii, Bacteroides acidifaciens, Barnesiella intestinihominis, Clostridium propionicum, Enterococcus faecalis, Eubacterium siraeum, Oscillibacter valericigenes, Ruminococcus flavefaciens), 4 Firmicutes (Clostridium difficile, Enterococcus faecalis, Lactobacillus murinus, and Staphylococcus xylosus), 6 Lachnospiraceae (Clostridium aldenense, Clostridium bolteae, Clostridium citroniae, Clostridium indolis, Clostridium propionicum, Clostridium xylosus), 3 Lachnospiraceae strains isolated in Reeves et al.26 (D4, G11, and E7), and 12 of the most represented Lachnospiraceae-classified OTUs from Reeves, et al.26. Areas of homology between Lachnospiraceae sequences that were not homologous to other Firmicutes and Bacteroidetes were used to create 4 Lachnospiraceae specific 16S rRNA forward gene primers: Lachno419F (5′ –GAC GCC GCG TGA GTG AAG AAG TAT- 3′), Lachno428F (5′ –GTA AAG CTC TAT CAG CAG GGA AGA- 3′), Lachno481F (5′ –GAC GGT ACC TGA CTA AGA AGC CC- 3′), and Lachno462F (5′ –GTC CAC AGG ACT TTG GAC GG- 3′). The primer LachF (5′-CC GCA TAA GCG CAC AGC- 3′) from Reeves, et al.26 was also utilized in this study. These forward primers were used with the 16S rRNA reverse gene primer 1492R (5′-GGT TAC CTT GTT ACG ACT T- 3′) for PCR amplification. One Lachnospiraceae specific reverse primer was also created: Lachno1261R (5′-TCG CTT CCC TTT GTT TAC GC- 3′), which was used with the 16S rRNA forward gene primer 8F (5′-AGA GTT TGA TCC TGG CTC AG- 3′) for PCR amplification. The specificity and coverage of the Lachnospiraceae primers (Supplementary Table 6) was obtained using the Ribosomal Database Project Probe Match tool (https://rdp.cme.msu.edu/probematch/search.jsp accessed on May 19, 2012). Additionally, primers were tested with Gram-negative bacteria Bacteroides fragilis Escherichia coli; Gram-positive bacteria Lactobacillus murinus, Staphylococcus aureus, Staphylococcus pneumonia; Clostridium difficile strain VPI 10463 to ensure specificity to Lachnospiraceae.
PCR was performed with 1 μl of template DNA (approximately 100 ng), 20 pmol of each primer, 8mM dNTP master mix (Promega-U1511), 1 unit GoTaq DNA polymerase (Promega- M3005), PCR buffer (Promega- M3005) and water in a total of 25 μl per reaction. PCR reaction was performed under the following cycling conditions: 95°C for 2 min, 30 cycles of 95°C for 30 sec, annealing at 57°C for 45 sec, and extension at 72°C for 90 sec, 72°C for 10 min.
Bacterial isolation and selective growth conditions
Three of the Lachnospiraceae isolates (Lachnospiraceae D4, G11, and E7) used in this study were reported previously26. The remaining 20 strains were isolated from murine cecal contents, cecal tissue and stool. Mouse stool was collected into a sterile tube directly from a restrained mouse and immediately transferred into an anaerobic chamber (Coy Industries, Grass Lake, MI). Ceca from C57BL/6 mice were collected in a sterile manner and immediately transferred into an anaerobic chamber. A sterile scalpel was used to open the cecum and separate cecal content and cecal tissue. Cecal content was added to a sterile tube and diluted into anaerobic 1X phosphate-buffered saline (PBS). Using a sterile syringe 2 ml of PBS was gently injected into the cecum to remove any additional cecal content. The cecal tissue was then added to 1 ml of anaerobic PBS and placed into a sterile container for tissue homogenization (50 μm pore size) using a Medimachine tissue homogenizer (BD Biosciences). The tissue was grinded for 15 seconds and homogenized tissue was used for plating. The cecal contents, cecal tissue homogenization and stool was serial diluted in anaerobic PBS and plated in duplicate onto brain heart infusion agar (BD Biosciences) with 0.01% cysteine (BHI).
A variety of media conditions were used to enrich for Lachnospiraceae isolates. Bacterial growth from the first agar plate was collected and genomic bacterial DNA was isolated using an Easy-DNA (Invitrogen) kit. Using the Lachnospiraceae specific primers and PCR conditions described above, we identified several media conditions that were enriched for Lachnospiraceae strains. Once a media condition was identified that enriched for Lachnospiraceae strains, single colonies from the duplicate plate were used to inoculate 1 ml of BHI + 5% fetal bovine serum (FBS) into a sterile 96-well plate. These cultures were grown anaerobically for 3 days at 37°C, and then 1 μl of the liquid culture was used as a template for the PCR reaction described above. The Lachnospiraceae-specific primers from (Supplementary Table 6) were used to identify potential Lachnospiraceae isolates. If any of the primers used yielded a successful PCR reaction, 50 μl of the corresponding culture was plated for single colonies anaerobically on BHI + 5% FBS for one to three days at 37°C. A single colony was used to inoculate a 5 ml BHI + 5% FBS culture that grew anaerobically for one to three days at 37°C. This culture was used to create 20% final concentration glycerol stocks of all the isolates that were stored at −80°C.
The following media conditions were used to isolate Lachnospiraceae strains in this study: BHI + 5% FBS; BHI + 1 μg/mL aztreonam + 10 μg/mL colistin + 2 μg/mL gentamicin; and BHI + 0.5 μg/mL ampicillin + 2 μg/mL erythromycin + 0.25 μg/mL vancomycin. Single colonies from the duplicate plate were used to inoculate 1 ml of BHI + 5% FBS. Specific media conditions used to isolate each Lachnospiraceae strains are defined in (Supplementary Table 7).
Taxonomic classification of bacterial isolates
Genomic DNA was isolated using an Easy-DNA (Invitrogen) kit. PCR reaction conditions were described above and PCR product cleanup was performed using ExoSAP-IT (Affymetrix) per the manufactures protocol. Near full-length 16S rRNA amplicons were sequenced at the University of Michigan DNA Sequencing Core using primers 8F (5′-AGA GTT TGA TCC TGG CTC AG- 3′), 515F (5′-GTG CCA GCM GCC GCG GTA- 3′), E939R (5′-CTT GTG CGG GCC CCC GTC AAT TC- 3′), and 1492R (5′-GGT TAC CTT GTT ACG ACT T- 3′). CLUSTALW multiple-sequence alignments were generated for each isolate and a near-full length 16S rRNA gene consensus sequence was obtained. The consensus sequence was taxonomically classified using the RDP classifier (https://rdp.cme.msu.edu/classifier/classifier.jsp accessed between 6-29-2012 and 3-7-2013)50. Bacterial strains that classified to the family Lachnospiraceae were used in this study ((Supplementary Table 7)).
Mouse Colonization with Lachnospiraceae Bacteria
Mice were orally gavaged with a mixture containing the 23 Lachnospiraceae stains (~1X108 bacteria) described above in brain-heart infusion broth (BHI) twice a week for 3 weeks. BHI broth was used as vehicle control. The mice were given DSS 1 week after the final gavage.
In vivo anti-IL-6R and anti-TNF Treatment
Mice were given 2 mg/kg body weight of anti-IL-6R (Tocilizumab, Genentech) and anti-TNF (Infliximab, Janssen Biotech) via intraperitoneal injection twice a week for four weeks prior to DSS exposure and during the 11 day DSS treatment period. Fecal DNA was collected from wild-type littermates and Nlrp12−/− littermates before and after the 4-week antibody treatment period, but prior to DSS treatment.
Metadata Study of Human NLRP12 profiling and microbiome changes in UC Patients
Raw data from 8 NCBI GEO human UC studies were renormalized and analyzed by Genespring GX (Agilent Tech). The following studies were used: GSE22619, GSE42911, GSE14580, GSE16879, GSE13367, GSE65114, GSE21231 and GSE57945. In the summary panels, the healthy group includes the samples from healthy participants and normal un-inflamed tissues of the patients, the active group includes the samples from the patients’ inflamed tissues and the inactive group includes un-inflamed tissue samples of patients in remission due to the treatment (anti-TNF, steroids and others).
For microbiome changes in IBD patients, we downloaded the raw 16S rRNA gene sequencing file published by Dirk Gevers et al.14 from the open-source microbiome deposition site: QIITA (https://qiita.ucsd.edu/) under study ID: 1939. This file includes 16S microbiome sequencing results from 28 healthy controls, 63 colonic Crohn’s disease patients (cCD), 156 ileal Crohn’s disease patients (iCD) and 24 UC patients. Raw sequencing data was reanalyzed as aforementioned. Significantly altered bacterial strains were identified by comparing IBD (cCD, iCD and UC) patients against healthy participants.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 6 software. Significance between two groups was determined by unpaired, two-tailed Student’s t test, and significance between multiple groups was determined using one-way analysis of variance (ANOVA) with Fisher’s LSD test. A paired, two-tailed Student’s t test was used to compare NLRP12 mRNA between monozygotic twins in (Fig. 1a). A two-way ANOVA test was used to identify the significantly changed microbial groups among all strains identified by 16s microbiome sequencing between different experimental conditions. Statistical significance for survival studies was determined by Log-rank (Mantel Cox) test. For immunoblots and dot plots, one dot or lane represents one mouse. Microbiome compositional dissimilarity was displayed by PCoA plots and quantified by UniFrac Distance values, and significant separation of the microbiome composition was determined by ANOSIM test using Qiime 1.8.0. The distribution of the UniFrac Distance values were displayed in Tukey’s boxplots, which display the first (bottom of the box) and third quartiles (top of the box), the median (the line inside the box) and 1.5 interquartile range of the upper or lower quartile (whiskers). For all statistical comparisons, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 and n.s. means no significance. The error bar represents mean ± standard error of the mean (SEM).
Supplementary Material
Acknowledgments
We would like to thank the UNC-NCSU Center for Gastrointestinal Biology and Disease (CGIBD), the UNC Microbiome Core Facility, Susan L. Tonkonogy and the Gnotobiotic Core at North Carolina State University, Maureen A. Bower and Jeremy W. Herzog at the National Gnotobiotic Rodent Resource Center and Crohn's and Colitis Foundation of America Gnotobiotic Animal Facility at UNC, the UNC Lineberger Comprehensive Cancer Center (LCCC) Animal Histopathology Core Facility, the UNC LCCC Animal Studies Core Facility and the UNC Flow Cytometry Core Facility for core and technical support. This work was supported by National Institute of Health (NIH) grants RO1 CA156330 (National Cancer Institute), P01 DK094779 (National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)), U19 AI067798 (National Institute of Allergy and Infectious Disease (NIAID)) and R37 AI029564 (NIAID) to J.P.-Y.T., P40 OD010995 (Office of the Director (OD), P30 DK034987 (NIDDK) and the Crohn's and Colitis Foundation of America to R.B.S and U19 AI090871 (NIAID) to V.B.Y. J.E.W. was supported in part by the NIH postdoctoral fellowship F32-DK088417-01 (NIDDK) and in part by a postdoctoral fellowship from the American Cancer Society, PF-13-401-01-TBE. W-C.C was supported by a postdoctoral fellowship from the National Multiple Sclerosis Society, FG 1968-A-1. A.D.T. was supported by the postdoctoral fellowship F32-DK098916 (NIDDK).
Footnotes
Author Contributions
L.C. and J.E.W. contributed equally to this manuscript. L.C., J.E.W and J.P-Y.T designed the experiments and wrote the manuscript, with critical input from R.B.S and V.B.Y. S.A.M performed the histopathological scoring. C.D.P, N.M., S.E.P. and R.B.S. contributed to the fecal DNA isolation and 16S rRNA gene sequencing experiments. W.C.C contributed to the immunoblots, cytokine measurements and flow cytometric analysis. M.J.K and V.B.Y generated the purified Lachnospiraceae strains. R.B.S generated the germ-free mice. A.D.T, W.J.B and G.K.M. generated the radiation bone marrow chimeric mice.
Competing Financial Interests: The authors declare no competing financial interests
Data Availability
All 16S rRNA microbiome sequences raw files that support the findings of this study have been deposited in The European Nucleotide Archive (http://www.ebi.ac.uk/ena) with primary access codes: “PRJEB18700”, “PRJEB18699” and “PRJEB18679”; and in the open-source microbiome sharing database QIITA (https://qiita.ucsd.edu) with the Study IDs: “10427”, “10428”, “10429”, “10818” and “10820”. The other data are available from the corresponding author upon reasonable request.
REFERECES
- 1.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karrasch T, Jobin C. NF-kappaB and the intestine: friend or foe? Inflamm Bowel Dis. 2008;14:114–124. doi: 10.1002/ibd.20243. [DOI] [PubMed] [Google Scholar]
- 3.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 4.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 5.Couturier-Maillard A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramanan D, Tang MS, Bowcutt R, Loke P, Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014;41:311–324. doi: 10.1016/j.immuni.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W, et al. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J Exp Med. 2013;210:2465–2476. doi: 10.1084/jem.20122490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vladimer GI, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37:96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams KL, et al. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem. 2005;280:39914–39924. doi: 10.1074/jbc.M502820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lich JD, et al. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol. 2007;178:1256–1260. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- 12.Allen IC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaki MH, et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20:649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2016 doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitajima S, Morimoto M, Sagara E, Shimizu C, Ikeda Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp Anim. 2001;50:387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- 18.Zaki MH, Man SM, Vogel P, Lamkanfi M, Kanneganti TD. Salmonella exploits NLRP12-dependent innate immune signaling to suppress host defenses during infection. Proc Natl Acad Sci U S A. 2014;111:385–390. doi: 10.1073/pnas.1317643111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuczynski J, et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol. 2012;Chapter 1(Unit 1E):5. doi: 10.1002/9780471729259.mc01e05s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmody RN, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumgart M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 24.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun. 2012;80:3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 28.Cerovic V, Bain CC, Mowat AM, Milling SW. Intestinal macrophages and dendritic cells: what’s the difference? Trends Immunol. 2014;35:270–277. doi: 10.1016/j.it.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peyrin-Biroulet L. Anti-TNF therapy in inflammatory bowel diseases: a huge review. Minerva Gastroenterol Dietol. 2010;56:233–243. [PubMed] [Google Scholar]
- 31.Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest. 2011;121:3375–3383. doi: 10.1172/JCI57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka T, Narazaki M, Kishimoto T. Anti-interleukin-6 receptor antibody, tocilizumab, for the treatment of autoimmune diseases. FEBS Lett. 2011;585:3699–3709. doi: 10.1016/j.febslet.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 33.Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 34.Kawamoto S, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 35.Sonnenburg ED, et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lepage P, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Haberman Y, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubert AM, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014;5:e01021–01014. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reichardt N, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handley SA, et al. SIV Infection-Mediated Changes in Gastrointestinal Bacterial Microbiome and Virome Are Associated with Immunodeficiency and Prevented by Vaccination. Cell Host Microbe. 2016;19:323–335. doi: 10.1016/j.chom.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinh DM, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt R, et al. Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front Microbiol. 2014;5:64. doi: 10.3389/fmicb.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geem D, Medina-Contreras O, Kim W, Huang CS, Denning TL. Isolation and characterization of dendritic cells and macrophages from the mouse intestine. J Vis Exp. 2012:e4040. doi: 10.3791/4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo SU, et al. Distinct Commensals Induce Interleukin-1beta via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity. 2015;42:744–755. doi: 10.1016/j.immuni.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol. 2004;70:4748–4755. doi: 10.1128/AEM.70.8.4748-4755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berry D, et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 2012;6:2091–2106. doi: 10.1038/ismej.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.