Abstract
Nursing scientists have long been interested in complex, context-dependent questions addressing individual- and population-level challenges in health and illness. These critical questions require multilevel data (e.g., genetic, physiologic, biologic, behavioral, affective, and social). Advances in data-gathering methods have resulted in the collection of large sets of complex, multifaceted, and often non-comparable data. Scientific visualization is a powerful methodological tool for facilitating understanding of these multidimensional data sets. Our purpose is to demonstrate the utility of scientific visualization as a method for identifying associations, patterns, and trends in multidimensional data as exemplified in two studies. We describe a brief history of visual analysis, processes involved in scientific visualization, and opportunities and challenges in the use of visualization methods. Scientific visualization can play a crucial role in helping nurse scientists make sense of the structure and underlying patterns in their data to answer vital questions in the field.
Keywords: Big Data, visualization, data analysis, data science, multidimensional data, data integration
“The greatest value of a picture is when it forces us to notice what we never expected to see.”
—Tukey (1977, p. vi)
Scientists in the field of nursing have long been interested in complex, context-dependent questions that address the challenges that individuals and populations face in health and illness situations. These critical questions often require data that cross levels of response, from genetic, physiologic, biologic, behavioral, affective, and social (Lim & Thuemmler, 2015). In the past, methodological and analytic models have favored a more reductionist approach requiring variable-oriented, hypothesis-driven designs, homogeneous samples, and unidimensional and balanced data sets (Sharts-Hopko, 2013). This often results in the design of studies that do not truly answer the question of interest, or a change in the research question to suit the prevailing methodologic and analytic models.
Over the past decade, innovations in health care research processes and technologies that support them have resulted in highly productive methods for gathering large, multidimensional data sets. These complex data sets are particularly powerful for addressing vital questions such as those of interest to scientists in health care (Johnson et al., 2014). These multifaceted data sets can be characterized by complex and unbalanced spatiotemporal elements, multivariate and non-comparable measurement units, and multimodal data streaming or acquisitioned modalities (Kehrer & Hauser, 2013; Vorderstrasse, Melkus, Lewinski, & Johnson, 2015). Due to the increasing complexity of these vital data sets, new sophisticated analytic approaches are being developed that harness the data and support the search for hidden patterns and trends. Scientific visualization, with interactive and dynamic capabilities, is one such methodological tool that can play a crucial role in helping nurse scientists make sense of the structure and underlying patterns that may be held in the data.
Scientific visualization is the use of graphic images to display varying combinations of empirical data, enabling scientists to process, explore, understand, and gain insight from complex data (Fox & Hendler, 2011; Kehrer & Hauser, 2013; Keim et al., 2008). Often used interchangeably with terms such as visual analytics and information visualization, scientific visualization is distinct in bringing together scientific communities from health, social, and behavioral sciences, with technical communities from computer, information visualization, cognitive and perceptual sciences, and interactive and graphic design to allow the human sensory system to systematically process data (Chen, Hou, Hu, & Liu, 2012; Keim et al., 2008). The process of scientific visualization allows the researcher to examine the expected, discover the unexpected, draw insights, generate and test hypotheses, and develop a narrative about their data. The purpose of this article is to give a broad overview of the history of scientific visualization, summarize the cognitive-analytic processes involved, demonstrate its application in nursing science through the presentation of its use in two study exemplars, and describe some of the current methodologic challenges and future opportunities.
The use of graphic visualization of data is not new. Although nursing scientists are familiar with its value for illustrating data in publications and presentations, visualization of graphed data has a long history as a key component of analysis (Lynch, 1985). Its historical roots extend back to the Babylonians of 2500 BC’s use of tables with columns and rows to display transactions (Few, 2009b), tables and graphical depictions in the 10th century of star and celestial body positions (Friendly, 2006), and cartography such as the Map of Tracks of Yu, a map of China dating back before 1100 (Tufte, 1997). By the 18th century, with fundamental statistical theory gaining popularity, William Playfair introduced the core data visualizations of line and bar graphs (Friendly, 2006). Playfair is credited with demonstrating the power of visual data displays for multivariate data in 1821 with a famous time series/bar graph of the prices of wheat, weekly wages, and the ruling monarchs over a 250-year period (Tufte, 1983).
Thematic cartography was introduced in the early 19th century (Friendly, 2006). For example, John Snow’s celebrated thematic map of the cholera epidemic in 1854 London, with its display of streets, water pump locations, and locations of cholera deaths, supported his hypothesis that cholera was a water-born pathogen (Cameron & Jones, 1983). Charles Minard’s depiction in 1869 of Napoleon’s army march to Russia, their movements, and the temperature they encountered on the return path is noteworthy for its graphical display of multivariate data using space, numbers, events, and times. Tufte (2001) called this geographic flow map the “best statistical graphic ever drawn” (p. 40).
One of the first famous graphic visualizations used by a nurse scientist was Florence Nightingale’s 1858 coxcomb figures contrasting the death toll from disease above the death toll from wounds of men fighting in the Crimean War. She used this graphic to analyze data generated during a particular time and context, and to generate hypotheses about responses in this situation (Cohen, 1984).
Scientists’ fascination with developments in classic statistical theory and hypothesis testing in the early part of the 20th century relegated data visualizations primarily to illustration of statistical analyses in publications and presentations (Friendly, 2006). However, key publications beginning in the latter half of the century moved the field forward. These included mathematician John Tukey’s (1977) Exploratory Data Analysis, highlighting visualization as a critical step in understanding data sets; computer scientist William Cleveland’s (1985), The Elements of Graphing Data; and statistician Edward Tufte’s (1990, 1997, 2001, 2006) set of illuminating books on information design and data visualization describing characteristics of effective data illustrations. These works, as well as the development and rapid progress of computing power, statistical software, and advancements in screen technologies, led the way to resurgence in scientific visualization (Wong & Bergeron, 1994).
Now firmly in the midst of the 21st century’s Big Data era, scientific visualization is being called upon to play a key role in interpreting the vast amount of multidimensional data being generated (Clark, 2013). The ability to harvest large quantities of multivariate and diverse forms (e.g., text, numbers, and photos) of biomedical data places a generation of nurse scientists in a position to advance our understanding of how individuals and populations respond in health and illness. Importantly, Big Data is not only defined by the size of a data set but can also be characterized by traits including velocity, variety, veracity, exhaustivity, relationality, extensionality, and scalability (Kitchin & McArdle, 2016). Data scientists and researchers have described variety as the greatest challenge of the Big Data era (Jin, Wah, Cheng, & Wang, 2015; Leventhal, 2014). Data sets that include diverse data types, incompatible forms, and unbalanced timing of collection require alternative methods of organization, integration, and analysis to gain meaning. Big Data to Knowledge (BD2K), a project of the National Institutes of Health, founded in 2013 in response to the challenges inherent in knowledge extraction from Big Data, is well poised to assist scientists to locate, analyze, and use Big Data (Margolis et al., 2014). Scientific visualization is an emerging, powerful tool for facilitating understanding of large, complex, and multidimensional data sets (Shaw, 2014).
Cognitive-Analytic Processes in Scientific Visualization
An understanding of human perception and cognition is at the base of the science behind translating data into visual representations that can be accurately and meaningfully decoded (Card, Mackinlay, & Shneiderman, 1999). The primary goal of scientific visualization is to encode information in a manner that allows our eyes to discern patterns to engage and amplify cognition. As Few (2009a) explains, seeing (perception), work of the visual cortex, is fast and efficient. Whereas thinking (cognition), work of the cerebral cortex, is slower and less efficient. Scientific visualization is effective because it shifts the balance between perception and cognition. Traditional data analysis is heavily loaded on cognition, whereas visualization analysis makes greater use of perception, allowing us to process information more rapidly and discern patterns more readily (Few, 2009a).
As will be demonstrated in two exemplar studies, scientific visualization can be used with any type of data including numbers, text, images, or a mixture (Abramson & Dohan, 2015; Knigge & Cope, 2006; Onwuegbuzie & Dickinson, 2008). The analytic objective is to transform and integrate data sets into visual representations, using a well-understood, transparent, and reproducible process, such that it allows the researcher to search visually for patterns or trends in varied configurations of the data. An important first step is the organization of the data sets such that they can be queried in as many different ways as necessary, allowing the same data to be seen in varying juxtapositions and enabling a range of comparisons. The use of visual displays for organization of the data by type, timing, and mode can give the researcher a visual inventory and gain a deeper understanding of the possible relationships within the data set.
Once the data sets have been organized, encoding and assimilation strategies are used for reduction, abstraction, transformation, and integration. The use of a graphical interface for navigation and exploration of data sets allows the analyst to search different, correlated views and iteratively select and examine features that appear interesting and illuminating (Shamir & Stolpnik, 2012; Tominski, Abello, & Schumann, 2009; Weber & Hauser, 2014).
It is beyond the scope of this article to give a description of the technical computing, interactive, and graphic design attributes needed for successful data visualizations, nor catalogue the range of visualization techniques. It is critical that nurse researchers planning to use visual analysis form partnerships with scientists in computing, engineering, and graphic design such as those in centers such as Duke University’s Renaissance Computing Institute (RENCI; at http://www.renci.org/) and Stanford’s Hana Immersive Visualization Environment (HIVE; at https://icme.stanford.edu/computer-resources/hive). Importantly, evaluation of the visualization system or tool is a critical step in the scientific visualization process. The conduct of a systematic assessment of the tool’s merit, ability to support the analysis task, decision making, and reasoning is central to ensuring that a rigorous and verifiable process has been used to draw inferences (Keim, Mansmann, Schneidewind, & Ziegler, 2006). The focus of the evaluation may be on different aspects such as functional testing, performance benchmarks, effectiveness of the display, user studies, and assessment of its impact (Keim et al., 2006). As North (2006) explained, given that insight is the main purpose of visualization, investigators must evaluate the degree to which the visualization system or tool generates insight. A range of evaluation designs are available including Visual Data Analysis and Reasoning (VDAR), Collaborative Data Analysis (CDA), and Heuristic Evaluation (HE). VDAR focuses on the tool’s ability to support visual analysis and reasoning with outputs that are both quantifiable (e.g., number of insights obtained) and subjective (e.g., perspectives on quality of data analysis experience). CDA evaluations focus on how an information visualization tool supports collaborative analysis and/or collaborative decision-making processes (Lam, Bertini, Isenberg, Plaisant, & Carpendale, 2012). Collaborative systems should support task work, the actions required to complete the task, and teamwork, the actions required to complete the task as a group. HE is an evaluation method that can be used as part of a CDA evaluation (Lam et al., 2012). HE is a technique well known in human–computer interaction but has had limited application in the field of information visualization.
To summarize, the use of visual analysis requires a multidisciplinary team with members who have some level of knowledge and understanding of (a) how differing types of data are transformed and assimilated for subsequent manipulation, (b) the cognitive benefits that visualization can provide and how the researcher’s goals align with these benefits, (c) issues surrounding data set and visual interaction, and (d) visualization techniques best suited for representing specific types of data (e.g., time series, hierarchical, textual; Kerren, Stasko, & Dykes, 2008). For example, interactive visualizations are suitable for analyzing high-dimensional data that have a large number of time points, and for researchers searching for correlations among attributes, or generating and testing hypotheses (Weber & Hauser, 2014). Interesting applications of interactive visualizations abound in the literature. For example, sociologists Abramson and Dohan (2015) illustrate the use of an ethnoarray, loosely adapted from the microarray or graphical heatmap approach, for analyzing, representing, and sharing ethnographic data.
Visualization Analysis: Two Study Exemplars
In this section, we present two ongoing research studies that illustrate differing applications of scientific visualization. We draw from these two longitudinal studies to (a) describe the aims, (b) detail the multidimensional nature of the data types and forms, (c) give examples of visual displays rendered through the integration of one or more data types, (d) describe the analytic lines resulting from the visual display examples, and (e) describe the initial stages of evaluation processes of the tools and analytic procedures being used to assess the merit, utility, and ability to derive insight of the visualization tools used. Our aim here is not toward reporting study results, but to demonstrate our early-stage use of visualization analysis for organizing, analyzing, and integrating the multidimensional data sets produced in our studies.
1. Decision Making for Infants With Complex Life-Threatening Conditions.
Docherty and Brandon (National Institutes of Health (NIH): National Institute of Nursing Research (NINR); R01NR010548; Principal Investigator [PI]: Docherty) studied parental decision making about life-sustaining treatments over time for infants with life-threatening conditions. The aim of this study was to explore and describe the trajectory of health-related decision making for infants with complex life-threatening conditions. The study focused on understanding how, when, and what types of decisions parents made and how health care providers influenced decision making. Additional factors with the potential to influence parents’ decision making were evaluated across the infant’s illness trajectory including treatment events, perceptions of infant discomfort and hopefulness for the infant, parents’ psychological distress, and the demographic characteristics of the case.
Using a multiple case study design, data were collected on decision-making events from multiple perspectives (parents, physicians, nurses, social workers) using a range of data types. Data included narrative interviews, rating scales, medical records, historical events, infant illness acuity, and standardized measures of parent psychological distress and well-being. Infant medical record data extraction included medical treatments and procedures, illness events, and provider narrative notes. The frequency of data collection varied based on data type. For example, all medical record data were collected 3 times a week, while parent interviews were timed to coincide with life-threatening events or at least monthly. The length of data collection across the 33 cases ranged from 10 days to 18 months depending upon “when” and “if” the infant died during the study.
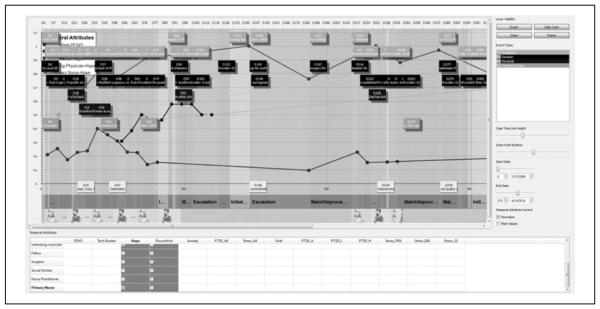
Scientific visualization was used to organize, integrate, and analyze this multidimensional data set and search for patterns over time and typologies in decision making. An Informatics Case Browser for Visual Analysis of Trajectories (NIH-NINR: 3R01NR010548-02S1: PI: Docherty) was developed to display, integrate, and allow for manipulation of the data overlays across multiple trajectories embedded in each case. As seen in Figure 1, each care day is categorized by one of four foci of care goal (initiation of curative or experimental treatments, maintenance of treatments, escalation, and with-drawal of care), and displayed as columns of color. This encoding was the result of the integration and assimilation of illness, caregiving, and treatment data extracted from the infant medical record. These numerical and textual data (e.g., ventilator settings, nutrition support, and health care provider progress notes) were assimilated to create four mutually exclusive categories. The patterns in color allowed the search for relationships between how the focus of care changed in relation to other data such as decision-making events, parent well-being or distress, or provider hopefulness.
Figure 1.
Case browser overview. (The color version of this figure is available in the online version at http://wjn.sagepub.com/supplemental.)
In Figure 1, trajectory events are displayed as one of four colored rectangles: decisions events, treatment events, personal events, and context of care events. This categorization resulted from the integration of narrative interview data and infant medical record data. Other time-series data are displayed along the infant’s illness trajectory and, despite varied scales of measurement, are transformed into a comparable scale through a normalization function. Figure 1 displays patterns in infant illness acuity, and ratings of hopefulness by mother, father, attending physician, and primary nurse.
The ability to play with varying configurations and change the ordering of data across the infant illness trajectory allowed for investigating and hypothesizing on case factors associated with decision making. For example, in Figure 2, two critical epochs of decision making became visible along the infant’s illness trajectory (Day 70 and Day 217) because of their association with daily care goals focused on initiating new curative treatments and escalating care. In addition, shortly following Day 70, an intersecting pattern emerges in the nurse practitioner’s ratings of the infant’s level of discomfort (increasing) and her hopefulness for the infant (decreasing).
Figure 2.
Co-occurrence of key events and provider perceptions of hope and discomfort.
Given that the analysis work on the Decision Making Study is ongoing, and the Informatics Case Browser for Visual Analysis of Trajectories is in the prototype stage of visualization development as defined by Lam et al. (2012), a systematized evaluation process is currently under construction. Aspects of a VDAR evaluation are being implemented to assess the Case Browser’s ability to exploit the visual perception skills of the research team to overlay a range of data types and explore/analyze data relationships. Although this informatics tool is highly effective at integrating and assimilating different data types (as seen in Figures 1 and 2), it can also preserve the complexity by presenting multiple types of views. In doing so, it enables the investigative team to process, explore, understand, and gain insight from the data. The evaluation metrics, thus far, include detailed description of the number and types of insights derived during analysis. They also include detailed field notes taken during weekly analysis team meetings on the quality of the data analysis experience. Field notes are also taken during live presentations of findings derived from the visualization analytic lines. For example, a presentation was recently given during the medical center pediatric grand rounds. Following the presentation that included a display of several graphic visualizations of the relationship between provider hopefulness and their perceptions of infant discomfort, two neonatologists approached the research team to describe a deeper understanding of the patterns that had been described during the presentation. Detailed field notes were taken of their impressions of these displays and the meaningfulness of the knowledge being derived.
2. Visualization of Multidimensional Data Obtained From a Virtual Environment Self-Management Intervention
Johnson and Vorderstrasse (NIH:National Library of Medicine (NLM); R21 LM 010727; PI: Johnson) conducted a pilot study with the primary aim of evaluating the feasibility and preliminary effects of a theoretically grounded, technologically based, bio-behavioral intervention using a virtual environment (VE) to facilitate Type 2 diabetes self-management and control (Johnson et al., 2014). Using Social Cognitive Theory as the theoretical basis for this pilot study, a large multidimensional data set was generated from 20 participants over 6 months study participation. This data set included numbers, text, videos, and still images of participant avatars in the VE. The quantitative data included number of participant log-ins and overall time spent in the VE, avatar movements in the VE on X, Y, Z axes, frequency of participant avatar interactions with objects in the site (e.g., clicking on informational items in the grocery store or restaurant), proxemics data (direction participant avatar is facing and proximity to other avatars when interacting with other participant avatars), survey data, and biomarker data. The qualitative data included voice and text conversations, observational data of participant avatar interactions, email, and forum discussions. The video and still images included videos of the synchronous classes and images of the avatar interactions within the VE.
The purpose of this visualization pilot study was to dynamically map and spatially illustrate the possible complex relationships, patterns, and trends across this multidimensional data set. The development of visual images allowed for the study of patterns of action and make connections between these patterns more salient (funded by the Renaissance Computing Institute, Duke University, North Carolina). The goal was to develop a design and analytic model with this smaller data set that could be applied to our currently funded multisite randomized controlled trial (NIH-NHLBI-1R01-HL118189-01) in which similar data are being collected with a significantly larger sample size (300) and over a longer period of time (18 months per participant; Vorderstrasse et al., 2015). Visualization of this multidimensional data provided multiple views of these data beyond the traditional 2D perspective. Using animation, volume, shapes, and color allowed a deeper exploration of the data through visual images. Using animation allowed the combining of multiple types of data and highlighted aspects of the temporal data.
Visualizations of these data were created using the R environment, R-Shiny libraries, which included a large inventory of visualization approaches and supported web deployment, and D3.js JavaScript libraries. These libraries not only added power and flexibility but also allowed animation of the visualizations.
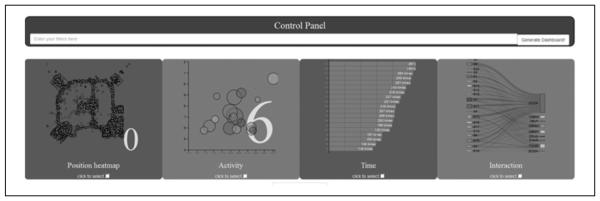
A wide range of visualizations have been created out of the preliminary data. One example that shows the use of color, volume, shape, utility, and animation is the dashboard. The dashboard allows the visualization of different types of data within one screen at the same study time points (see Figure 3), for example, simultaneous viewing on the heat map of the geographic places visited by participants (Figure 3, Panel 1), compared with activity and biomarker data (Panel 2), or compared with number of log-ins in the site (Panel 3) or interactions with various types of objects in the site (Panel 4). These dynamic portraits assist in uncovering evolutionary patterns of change over time. Using animation to visually represent these data allows salient patterns to be more readily apparent and the development of a deeper understanding of how the data are affected by time and group dynamics. Thus, the use of animation of these different types of related data shows representations that are not possible to perceive with traditional graphs.
Figure 3.
Dashboard of multiple visualizations.
Significant challenges remain in integrating the variety of data being gathered in the multisite randomized controlled trial. For example, although the quantitative data have been readily transformed, integrated, and assimilated to render visualizations, the research team is working to determine how to visually combine the results from the qualitative data analysis (conversations, forums, email, and text), videos, and images from the VE to make other possible relationships visible across these multidimensional data. Given that the dynamic mapping and spatial illustrations being conducted in this pilot study remain in the design phase of visualization development (Lam et al., 2012), a systematic evaluation is currently being constructed.
Opportunities and Challenges in Using Scientific Visualization
Scientific visualization is an important methodology that can be used by nurse scientists to understand the features and trends represented in their multidimensional data sets. The tremendous growth in computing power, advances in screen technologies, and development of the discipline of visualization scientists have created a range of opportunities for nurse scientists to put visual analysis to work on their complex heterogeneous data sets. Ongoing development of user friendly visualization software systems is allowing scientists with little experience in graphic processes to venture into this field (Wu & Hsu, 2013). Visualization is rapidly being considered critical to the ability to process complex data and generate and test hypotheses about the world around us (Fox & Hendler, 2011).
However, a number of challenges remain in the use of visual analytics to aid in the understanding of complex data. Some of these challenges center on the growing size and dimensionality of the data sets. Precise sensors, rapid recording methods, and ease of expansion of storage capacities allow large amounts of data to be collected in a relatively short period of time (Keim et al., 2006). There is a tension between extracting meaningful information in an efficient way to assist with the discovery process and balancing the potential for cognitive overload. In reducing the data for more effective visualization, there is a concomitant risk of loss of information and a glossing over of the complexity of the phenomenon we seek to understand.
The cost of creating graphic visualizations has not decreased over the past decade. Scientists have often assumed an equal split between the time needed to generate and analyze data. However, we are entering an era in which the technologies developed to generate data are progressing and those used to visualize and analyze data lag behind and are requiring more time and effort (Fox & Hendler, 2011). Challenges also remain in effective visualizations for particular data types including textual data. Encoding of textual data is of primary importance as textual data are not pre-attentive.
Another ongoing challenge concerns data quality and veracity of the insights and findings generated. The value of visual representations is only as accurate as the data itself. While this is a challenge with any data analysis, given the large amount of complex, multidimensional data, it bears particularly close attention. Scientific visualization will only prove to be a valuable tool if the data quality is assured. As described earlier, undertaking a systematic evaluation of the visual tool’s merit, ability to support analysis tasks, decision making, and reasoning is critical to the demonstration that a rigorous and verifiable process has been used to draw inferences.
Innovations in health care research processes and technologies that support them have resulted in highly productive methods for gathering large, multidimensional data sets allowing nurse scientists to address vital questions in the field. The complexity of these data sets requires new approaches that harness the data and support the search for hidden patterns and trends. Scientific visualization will play a crucial role in helping nurse scientists make sense of the structure and underlying patterns that may be held in the data. This approach draws on the desire and ability to explore, discover patterns, and develop deeper insights into our areas of interest.
Acknowledgments
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Research reported in this publication was supported by the National Institute of Nursing Research of the National Institutes of Health under Award Numbers R01NR010548; R01NR010548-02S1; the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number R01HL118189-01; the National Library of Medicine of the National Institutes of Health under Award Number R21-LM010727; and the Renaissance Computing Institute, Duke University, Durham, North Carolina.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplemental Material
The online supplements are available at http://wjn.sagepub.com/supplemental.
References
- Abramson CM, Dohan D. Beyond text: Using arrays to represent and analyze ethnographic data. Sociological Methodology. 2015;45:272–319. doi: 10.1177/0081175015578740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D, Jones IG. John Snow, the broad street pump and modern epidemiology. International Journal of Epidemiology. 1983;12:393–396. doi: 10.1093/ije/12.4.393. [DOI] [PubMed] [Google Scholar]
- Card SK, Mackinlay JD, Shneiderman B, editors. Readings in information visualization: Using vision to think. Morgan Kaufman; San Francisco, CA: 1999. [Google Scholar]
- Chen C, Hou H, Hu Z, Liu S. An illuminated path: The impact of the work of Jim Thomas. In: Dill J, Earnshaw R, editors. Explanding the frontiers of visual analytics and visualization. Springer; New York, NY: 2012. pp. 9–20. [Google Scholar]
- Clark D. Four things you need to know in the Big Data era. Forbes. 2013 Aug; Retrieved from http://www.forbes.com/sites/dorieclark/2013/08/08/four-things-you-need-to-know-in-the-big-data-era/#6d712bc12e86.
- Cleveland WS. The elements of graphing data. Hobart Press; Summit, NJ: 1985. [Google Scholar]
- Cohen B. Florence nightingale. Scientific American. 1984;250:128–137. doi: 10.1038/scientificamerican0384-128. [DOI] [PubMed] [Google Scholar]
- Few S, editor. Data visualization for human perception. Aarhus, Denmark: 2009a. Retrieved from https://www.interaction-design.org/literature/book/the-encyclopedia-of-human-computer-interaction-2nd-ed/data-visualization-for-human-perception. [Google Scholar]
- Few S. Now you see it: Simple visualization techniques for quantitative analysis. Analytics Press; Oakland, CA: 2009b. [Google Scholar]
- Fox P, Hendler J. Changing the equation on scientific data visualization. Science. 2011;331:705–708. doi: 10.1126/science.1197654. [DOI] [PubMed] [Google Scholar]
- Friendly M. A brief history of data visualization. In: Chen C, Hardle W, Unwin A, editors. Handbook of computational statistics: Data visualization. Springer-Verlag; Heidelberg, Germany: 2006. pp. 16–48. [Google Scholar]
- Jin X, Wah BW, Cheng X, Wang Y. Significance and challenges of Big Data research. Big Data Research. 2015;2(2):59–64. [Google Scholar]
- Johnson C, Feinglos M, Pereira K, Hassell N, Blascovich J, Nicollerat J, Vorderstrasse A. Feasibility and preliminary effects of a virtual environment for adults with type 2 diabetes: Pilot study. JMIR Research Protocols. 2014;3(2):e23. doi: 10.2196/resprot.3045. doi:10.2196/resprot.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer J, Hauser H. Visualization and visual analysis of multifaceted scientific data: A survey. IEEE Transactions on Visualization and Computer Graphics. 2013;19:495–513. doi: 10.1109/TVCG.2012.110. [DOI] [PubMed] [Google Scholar]
- Keim DA, Andrienko G, Fekete JD, Gorg C, Kohlhammer J, Melancon G. Visual analytics: Definition, process, and challenges. In: Kerren A, editor. Information visualization. Springer-Verlag; Berlin, Germany: 2008. pp. 154–175. [Google Scholar]
- Keim DA, Mansmann F, Schneidewind J, Ziegler H. IV ’06 Proceedings of the Tenth International Conference on Information Visualization. IEEE Computer Society; Washington, DC: 2006. Challenges in visual data analysis; pp. 9–16. [Google Scholar]
- Kerren A, Stasko JT, Dykes J. Teaching information visualization. In: Kerren A, Stasko JT, Fekete JD, North C, editors. Information visualization. Springer; New York, NY: 2008. pp. 65–90. [Google Scholar]
- Kitchin R, McArdle G. What makes Big Data, Big Data? Exploring the ontological characteristics of 26 datasets. Big Data & Society. 2016;3:1–10. doi:10.1177/2053951716631130. [Google Scholar]
- Knigge L, Cope M. Grounded visualization: Integrating the analysis of qualitative and quantitative data through grounded theory and visualization. Environment and Planning. 2006;38:2021–2037. [Google Scholar]
- Lam H, Bertini E, Isenberg P, Plaisant C, Carpendale S. Empirical studies in information visualization: Seven scenarios. IEEE Transactions on Visualization and Computer Graphics, Institute of Electrical and Electronics Engineers. 2012;18:1520–1536. doi: 10.1109/TVCG.2011.279. [DOI] [PubMed] [Google Scholar]
- Leventhal R. Survey: Variety, not volume, the biggest challenge to analyzing Big Data. 2014 Jul 14; [Web log post]. Retrieved from http://www.healthcare-informatics.com/news-item/survey-variety-not-volume-biggest-challenge-analyzing-big-data.
- Lim AK, Thuemmler C. Opportunities and challenges of internet-based health interventions in the future internet; 12th International Conference on Information Technology; Las Vegas, NV. Apr, 2015. [Google Scholar]
- Lynch M. Discipline and the material form of images: An analysis of scientific visibility. Social Studies of Science. 1985;15:37–66. [Google Scholar]
- Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, Green ED. The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: Capitalizing on biomedical Big Data. Journal of the American Medical Informatics Association. 2014;21:957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North C. Toward measuring visualization insight. IEEE Computer Graphics and Applications. 2006;26(3):6–9. doi: 10.1109/mcg.2006.70. [DOI] [PubMed] [Google Scholar]
- Onwuegbuzie AJ, Dickinson WB. Mixed methods analysis and information visualization: Graphical display for effective communication of research results. The Qualitative Report. 2008;13:204–225. [Google Scholar]
- Shamir A, Stolpnik A. Multivariate graphs: Exploration using interactive visual queries. Computers & Graphics. 2012;36:257–264. [Google Scholar]
- Sharts-Hopko NC. Tackling complex problems, building evidence for practice, and educating doctoral nursing students to manage the tension. Nursing Outlook. 2013;61:102–108. doi: 10.1016/j.outlook.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Shaw J. Why “Big Data” is a big deal. Harvard Magazine. 2014 Mar-Apr; Retrieved from http://harvardmagazine.com/2014/03/why-big-data-is-a-big-deal.
- Tominski C, Abello J, Schumann H. CGV—An interactive graph visualization system. Computers & Graphics. 2009;33:660–678. [Google Scholar]
- Tufte ER. The visual display of quantitative information. Graphics Press; Cheshire, CT: 1983. [Google Scholar]
- Tufte ER. Envisioning information. Graphics Press; Cheshire, CT: 1990. [Google Scholar]
- Tufte ER. Visual explanations: Images and quantities, evidence and narrative. Graphics Press; Cheshire, CT: 1997. [Google Scholar]
- Tufte ER. The visual display of quantitative information. 2nd Graphics Press; Cheshire, CT: 2001. [Google Scholar]
- Tufte ER. Beautiful evidence. Graphics Press; Cheshire, CT: 2006. [Google Scholar]
- Tukey JW. Exploratory data analysis. Pearson; New York, NY: 1977. [Google Scholar]
- Vorderstrasse A, Melkus G, Lewinski A, Johnson CM. Diabetes learning in virtual environments: Testing the efficacy of self-management training and support in virtual environments (Randomized controlled trial protocol) Nursing Research. 2015;64:485–494. doi: 10.1097/NNR.0000000000000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GH, Hauser H. Interactive visual exploration and analysis. 2014 Retrieved from https://www.semanticscholar.org/paper/Interactive-Visual-Exploration-and-Analysis-Weber-Hauser/17b65ea9dc2cbf9844df55572ef66ef3 32c18d23.
- Wong PC, Bergeron RD. Proceedings in scientific visualization, overviews, methodologies, and techniques. IEEE Computer Society; Washington, DC: 1994. 30 years of multidimensional multivariate visualization; pp. 3–33. [Google Scholar]
- Wu LH, Hsu PY. An interactive and flexible information visualization method. Information Sciences. 2013;221:306–305. [Google Scholar]