Abstract
When yeast cells are challenged by a fluctuating environment, signaling networks activate differentiation programs that promote their individual or collective survival. These programs include the initiation of meiotic sporulation, the formation of filamentous growth structures and the activation of programmed cell death pathways. The establishment and maintenance of these distinct cell fates are driven by massive gene expression programs that promote the necessary changes in morphology and physiology. While these genomic reprogramming events depend on a specialized network of transcription factors, a diverse set of chromatin regulators, including histone-modifying enzymes, chromatin remodelers and histone variants, also play essential roles. Here, we review the broad functions of histone modifications in initiating cell fate transitions, with particular focus on their contribution to the control of expression of key genes required for the differentiation programs and chromatin reorganization that accompanies these cell fates.
Keywords: chromatin, post-translational modifications, meiosis, pseudohyphal growth, programmed cell death
Graphical Abstract
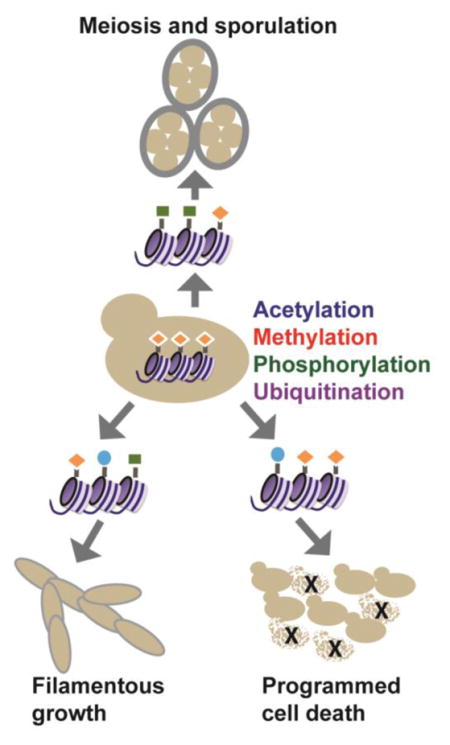
In yeast cells, a series of intrinsic and extrinsic signals initiate cell fate changes in a fluctuating environment. The signals are transduced through integrated networks that ultimately converge on highly-regulated gene expression programs, which are orchestrated by transcription factors, chromatin-modifying enzymes, chromatin remodelers and regulatory RNAs, such as long non-coding RNAs (lncRNAs) [1–3]. The well-characterized genome and tractable genetics of Sacchromyces cereviasie has yielded numerous insights into the genomic reprogramming that occurs in response to diverse environmental signals, and the key mechanisms discovered parallel similar reprogramming events in metazoan systems [4]. Here, we discuss our current understanding of histone modifications and modifying enzymes in S. cerevisiae and review the role of chromatin dynamics during key cell fate decisions, including the differentiation of diploids into haploid spores (meiosis and sporulation), the formation of filamentous growth structures and the initiation of programmed cell death pathways. Nutrient availability dictates the cell’s decision to enter either the sporulation or filamentous growth program [5, 6], and reprogramming depends on the tightly-executed activation of key genes, such as IME1 and FLO11, respectively, to drive the required cell cycle and morphological changes. Likewise, activation of programmed cell death pathways occurs in response to diverse cellular stresses in yeast and is accompanied by both transcriptional changes and massive changes to chromatin structure [7]. While these cell fate changes are orchestrated by a dedicated set of sequence-specific transcription factors, histone-modifying enzymes are critical to the reorganization of the chromatin landscape required for full execution of these fates.
Histone Modifications of Saccharomyces cerevisiae
The packaging of DNA into nucleosomes provides the structural foundation for genomic reprogramming events that underlie cell fate transitions. Nucleosomes are comprised of the four core histones H3, H4, H2A and H2B, and may also have one of two variant histones in budding yeast, Htz1 (H2A.Z), which marks heterochromatin-euchromatin boundaries and poised promoters [8, 9], or the centromere-specific H3 variant Cse4 (CENP-A) [10]. The role of individual nucleosomes in defining transcriptional states is determined by their level of histone modification, their position relative to defined sequence elements, the presence of histone variants and their interaction with the transcriptional machinery, all of which are regulated by the coordinated activity of chromatin-associated protein complexes and modifying enzymes [11, 12].
Histone-modifying enzymes, in particular, play a key role in integrating signals at chromatin through the dynamic process of post-translational modification (PTM), including methylation, acetylation, phosphorylation, and ubiquitination [1–3]. The enzymes that add histone PTMs are often referred to as writers and include histone methyltransferases (HMTs), histone acetyltransferases (HATs), kinases, and ubiquitin-protein transferases (UbTs). The marks are removed by erasers- histone demethylases (HDMs), histone deacetylases (HDACs), phosphatases, and deubiquitinases (DUbs)- allowing for dynamic regulation. Effector proteins, known as readers, are recruited to or stabilized at chromatin by these marks and act to transduce signals received at chromatin, governing such pathways as transcription and DNA repair. Reader proteins contain well-defined domains, such as bromodomains, chromodomains and PHD fingers, that recognize specific PTMs on histones [13–15]. Importantly, these domains are often found in writers and erasers, allowing for crosstalk between modifications and integrated signaling networks at chromatin.
As summarized in Table 1, a number of marks on the core histones have been extensively studied using proteomic and molecular techniques, and have well-characterized writers, erasers and readers. These marks, such as H3 and H4 acetylation, H3K4 and H3K36 methylation, are relatively abundant and have been implicated in diverse roles throughout the genome, especially in the realm of gene expression regulation. However, recent advances in proteomic techniques and other methods have revealed the presence of several previously unknown histone PTMs [16–20] that may act in a context-dependent manner. For example, newly-discovered acetylation at H4K44 has been identified in pre-meiotic and meiotic cells and is required for proper double strand break formation and recombination during meiosis [21]. Also, H4K5, K8 and K12 mono-methyl marks, catalyzed by Set5, which are present in approximately 1% of nucleosomes genome-wide [22], are likely to function in a context-dependent manner. Set5’s activity has been linked to stress responses and gene repression in coordination with Set1 [23], however the mechanism by which these methyl marks interpret and transduce signals at chromatin remains unclear.
Table 1.
Histone modifications of Saccharomyces cerevisiae. Acetylation, methylation, phosphorylation and ubiquitination of the core histones from budding yeast are shown. For methylation, the extent of methylation is indicated in parentheses. All modifications listed have been detected using mass spectrometry and/or antibody-based methods and known writers and erasers are indicated.
| Histone | Residue | Modification | Writer | Eraser | References |
|---|---|---|---|---|---|
| H3 | K4 | Methylation (me1-3) | Set1 | Jhd2 | [102–107] |
| H3 | K4 | Acetylation | Gcn5, Rtt109 | Hst1, Sir2 | [108] |
| H3 | K9 | Acetylation | Gcn5 Rtt109 |
Rpd3 | [109–112] |
| H3 | S10 | Phosphorylation | Snf1, Ipl1 | Glc7 | [113, 114] |
| H3 | T11 | Phosphorylation | Mek1, Pkm2 | ? | [71, 100] |
| H3 | K14 | Acetylation | Gcn5 | Rpd3 | [115] |
| H3 | K18 | Acetylation | Gcn5 | Rpd3 | [111, 115] |
| H3 | K23 | Acetylation | Gcn5 | Rpd3 | [109, 111] |
| H3 | K27 | Acetylation | Gcn5 | Rpd3 | [111] |
| H3 | K36 | Methylation (me1-3) | Set2 | Rph1, Jhd1 | [116–118] |
| H3 | K36 | Acetylation | Gcn5 | ? | [119] |
| H3 | T45 | Phosphorylation | Cdc7 | ? | [120] |
| H3 | K56 | Acetylation | Rtt109 | Hst3, Hst4 | [121–124] |
| H3 | K79 | Methylation (me1-3) | Dot1 | ? | [125–128] |
| H3 | Y99 | Phosphorylation | Rad53 | ? | [129] |
| H4 | S1 | Phosphorylation | Cka1 | ? | [130] |
| H4 | R3 | Methylation (me1) | Rmt1 | ? | [127] |
| H4 | K5 | Methylation (me1) | Set5 | ? | [22] |
| H4 | K5 | Acetylation | Esa1, Hat1 | Hos2, Rpd3 | [111, 131–134] |
| H4 | K8 | Methylation (me1) | Set5 | ? | [22] |
| H4 | K8 | Acetylation | Esa1 | Hos2, Rpd3 | [111] [131, 132, 134] |
| H4 | K12 | Methylation (me1) | Set5 | ? | [22] |
| H4 | K12 | Acetylation | Esa1, Hat1 | Hos2, Rpd3 | [111, 131, 132, 134] |
| H4 | K16 | Acetylation | Sas2 | Sir2 | [135–138] |
| H4 | K20 | Methylation (me1) | ? | ? | [139] |
| H4 | K20 | Acetylation | ? | ? | [16] |
| H4 | K44 | Acetylation | ? | ? | [21] |
| H2A | K7 | Acetylation | Esa1 | Rpd3 | [111] |
| H2A | Y58 | Phosphorylation | Cka1 | ? | [140] |
| H2A | Q105 | Methylation | Nop1 | ? | [31] |
| H2A | S121 | Phosphorylation | ? | ? | [141] |
| H2A | S128 | Phosphorylation | Mec1 | ? | [142] |
| H2B | S10 | Phosphorylation | Ste20 | ? | [91, 92] |
| H2B | K11 | Acetylation | Gcn5 | Hda1, Hos3 | [93, 111] |
| H2B | K16 | Acetylation | Gcn5, Esa1 | Hda1, Rpd3 | [111, 143] |
| H2B | K37 | Methylation (me2) | ? | ? | [144] |
| H2B | K123 | Ubiquitination | Rad6, Bre1 | Ubp8, Ubp10 | [145–148] |
The genome reprogramming events underlying cell fate decisions rely on both widespread changes to the epigenome, as well as targeted events that may be specific to environmental stimuli or the regulation of individual genomic regions. Marks that appear to be lowly abundant or present only during specialized cellular programs or stress responses may have unique, context-dependent roles in the genome. Additionally, increasing evidence indicates an extensive role for so-called histone-modifying enzymes in the post-translational modification of non-histone proteins, which has been linked to critical signaling pathways [24–28]. The kinetochore protein Dam1 is methylated by Set1, supporting a role for Set1 in chromosome segregation independent of H3K4 methylation [29]. As another example, phosphoenolpyruvate carboxykinase Pck1 is acetylated by the HAT Esa1 and deacetylated by the HDAC Sir2 to dynamically regulate its activity in gluconeogenesis [30]. Furthermore, there are numerous potential histone-modifying enzymes for which no substrates have been identified, such as the candidate methyltransfersases Set4 and Set6, and demethylase Ecm5, and also novel modifications, such as glutamate methylation and lysine succinylation [19, 31], for which the functional consequences are largely unclear. Altogether, these observations suggest that we have only begun to scratch the surface regarding the potential roles of histone modifications and histone-modifying enzymes in signaling networks that govern cell fate decisions and genomic reprogramming. Further investigation of these pathways in yeast is expected to reveal principles common to similar events in metazoan systems.
Nutrient-responsive signaling and chromatin states
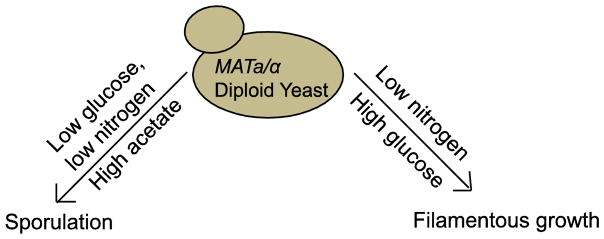
Changes to nutrient availability trigger multiple responses that promote survival of yeast cells under fluctuating environmental conditions (Figure 1). For example, nitrogen depletion, the absence of glucose and the presence of a nonfermentable carbon source, such as acetate, stimulates entry into sporulation [5, 32], a specialized cell fate in which MATa/α diploid cells undergo a meiotic division to produce haploid progeny (spores), which are packaged into a protective ascus. Sporulation is characterized by a genetic reprogramming event in which hundreds of genes are induced or repressed in a precisely timed synchrony that controls the progression of meiosis and the morphological changes required for spore formation [33, 34]. Alternatively, a change in environmental conditions in which glucose is abundant, but nitrogen is still limiting, promotes the dimorphic transition of diploid cells from the yeast state to a pseudohyphal state, in which filamentous structures (pseudohyphae) form, adhere to other cells and solid surfaces, such as agar [35]. Similar to sporulation, the filamentous state is also driven by a signaling cascade that activates a gene expression program required for the morphological and physiological changes associated with the new polarized growth structures. In both cases, the nutrient-responsive signaling pathways converge on genes that underlie these cell fate transitions, IME1 and FLO11, respectively. Below, we review the specific role of histone modifications in expression control of IME1 and FLO11, and highlight how the disruption of chromatin homeostasis alters the maintenance of the acquired cell fates.
Figure 1. Changes in nutrient availability promote alternative differentiation pathways in diploid yeast.
Low levels of nitrogen and glucose in the presence of a nonfermentable carbon source such as acetate promote meiotic division and sporulation. When glucose levels are high but nitrogen is limiting, filamentous growth is initiated, in which pseudohyphae form and can become invasive.
Chromatin-based control of IME1 expression, the meiotic master regulator
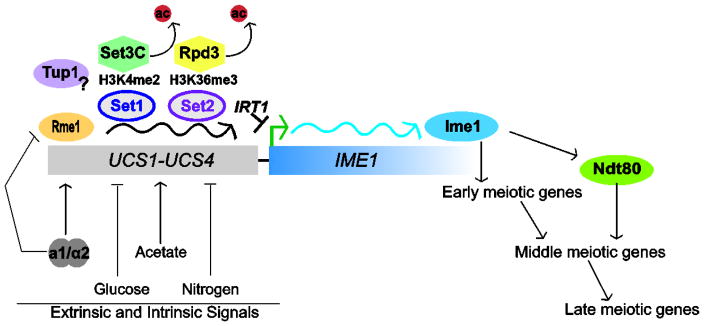
Sporulation occurs in three primary phases, defined as early, middle and late, which are each characterized by discrete events in meiosis and spore formation [5, 33, 36]. When the cell receives the necessary intrinsic (mating type and ploidy) and extrinsic (nutrient availability) signals (Figure 1), the transition from vegetative growth to the early phase of sporulation is marked by the rapid induction of IME1, the transcription factor which drives early gene expression required for entry into meiotic S phase [37–39]. The promoter of IME1 consists of four upstream control sequences (UCS) which serve as the integration platform for the positive and negative signals regulating sporulation (Figure 2) [40, 41]. The action of the transcription factor Rme1, which is uniquely expressed in haploids, establishes the chromatin landscape regulating IME1 expression. In haploid cells, IME1 is strongly repressed by the binding of Rme1 to the UCS elements to prevent the aberrant activation of the sporulation program [42–44]. The mechanism by which Rme1 repressed IME1 remained unknown for some time, however a breakthrough revealed that Rme1 promotes transcription of a long non-coding RNA (lncRNA). The transcription of this lncRNA, IRT1, from the upstream control sequences of IME1 generates a repressive environment by interfering with activator recruitment to the IME1 promoter and by the co-transcriptional recruitment of HMT and HDAC complexes (Figure 2) [45]. Specifically, IRT1 transcription promotes recruitment of Set2 and Set3, each of which generate repressive environments through interactions of histone methyl marks and chromatin reader domains [46–48]. vanWerven [45] and colleagues propose that co-transcriptional recruitment of Set1 during IRT1 induction deposits H3K4me2 at the 5′ end of the IME1 promoter, which stabilizes the Set3 complex and its HDAC activity [48], causing local repression. Likewise, co-transcriptional recruitment of Set2, which deposits H3K36me3, is postulated to promote association of the Rpd3(S) HDAC complex (as distinguished from Rpd3(L), a larger complex with distinct interactions) over the IRT1 coding region [45–47]. This work therefore uncovered a key mechanism of the combinatorial role of histone modifications in the control of a critical cell fate regulator.
Figure 2. Regulation of IME1 by signaling to chromatin modifiers.
Intrinsic and extrinsic signals received by the upstream control sequences 1 through 4 (UCS1-4) of the IME1 regulatory region. Nutrient signaling and the transcriptional regulators a1/α2 and Rme1 are essential to maintaining expression patterns of IME1. In diploids, the a1/α2 heterodimer, encoded by the MATa/MATα locus releases Rme1-mediated repression of IME1. In haploid cells, Rme1 promotes the expression of the lncRNA IRT1 from this region. Histone methyltransferases Set1 and Set2 are recruited co-transcriptionally and their respective methyl marks promote HDAC containing complexes Set3C and Rpd3C(L) chromatin association, repressing IME1 expression. Tup1 is also postulated to repress IME1, potentially as a parallel pathway promoting HDAC recruitment, however its precise role is unknown. IME1 expression initiates a transcriptional cascade, including inducing NDT80, which leads to the sporulation-specific gene expression program required for the meiotic divisions and spore formation.
A second pathway reported to repress IME1 in vegetatively-growing cells involves the Tup1/Ssn6 co-repressor complex [49, 50]. Cells lacking TUP1 show decreased nucleosome occupancy of the IME1 promoter, and increased recruitment of RNA pol II, consistent with constitutive expression of IME1 [50]. However, the mechanism by which Tup1 associates with IME1 is unclear, and whether it recruits other chromatin factors has not yet been investigated. Tup1 has been shown to play diverse roles at other nutrient-responsive promoters, such as driving the association of HDACs with FLO1 [51, 52] and cooperating with the SWR1 complex for deposition of H2A.Z (Htz1) at the GAL1 and SUC2 promoters [53]. As a repressor of IME1, Tup1 has been linked to nutrient-sensing through PKA and TOR complex signaling networks [50], whereas Rme1 levels are responsive to mating type and ploidy [42, 54, 55]. These two DNA-binding proteins therefore represent different nodes of signal integration which promote the required chromatin state at the IME1 promoter in response to both extrinsic and intrinsic signals.
Histone modifications and meiotic progression
With the onset of the sporulation program, Ime1 activates a plethora of genes involved in meiotic DNA replication, chromosome remodeling and homologous recombination [5, 32]. A number of other chromatin-dependent mechanisms control the transcriptional dynamics of this cascade. Early and middle genes are also repressed by Ume6, a DNA binding protein that tethers Rpd3 and the chromatin remodeler Isw2 to promoter regions, resulting in histone deacetylation, chromatin compaction and occlusion of the TATA box [56, 57]. As Ime1 levels increase, Ume6 is targeted for degradation by the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase activity, in a manner partially dependent on Ime1, and early gene transcription is initiated [58]. Intriguingly, Ume6 is targeted by the HAT Gcn5 for acetylation, and can be deacetylated by Rpd3 [59]. Lysine acetylation of Ume6 appears to promote its destruction, revealing that a non-histone target of Gcn5 may be a critical substrate determining its role in early gene expression, and that Rpd3 has an additional role in stabilizing Ume6 prior to the onset of the meiotic program [59]. Acetylation at additional lysines of Ume6 also appears to release it from its DNA binding sites, providing another mechanism for the rapid inactivation of Ume6 and ensuring proper timing of the transcriptional cascade [60]. These observations highlight the potential contribution of non-histone targets of chromatin-modifying enzymes to the regulation of reprogramming events.
Middle genes are repressed by the Sum1 transcriptional repressor, which binds DNA and tethers the HDAC Hst1 to middle gene promoters through an interaction with the adaptor protein Rfm1 [61–63]. Sum1-Rfm1-Hst1 and Ume6-mediated mechanisms have been reported to repress the middle sporulation gene NDT80 [64], which encodes a master regulator transcription factor that activates a series of middle genes required for nuclear division and spore formation [63, 65]. Beyond this, we still have relatively little knowledge regarding the role of chromatin modifiers during middle, and also late-stage, gene expression. As an example, the methyltransferase Set1 has been implicated in repressing middle gene expression [66], however the mechanism by which it specifically represses this gene set has not been reported. While Set1’s activity promotes meiotic progression, this is primarily thought to be due to the role of H3K4 methylation in recombination and double strand break repair [67, 68], whereas the function or Set1 in meiotic gene expression control is unclear.
In addition to the large-scale changes in gene expression that accompany the sporulation program, massive chromosomal rearrangements, which rely on critical genome integrity pathways, also occur during meiosis. While the role of chromatin in these processes has been reviewed elsewhere [69], genetic and proteomic screens focused on meiotic chromatin and sporulation have uncovered previously-unknown modifications. These approaches have revealed that phosphorylation at H4S1 is required for post-meiotic chromatin compaction and spore maturation [70], H3T11ph has been implicated in meiotic progression [71] and H4K44ac is associated with meiotic recombination [21]. These studies not only elucidate key mechanisms of meiotic chromosomal reorganization, but also underscore the notion that novel, context-dependent histone modifications may be discovered through analysis of specialized cell fates.
Chromatin-based regulation of FLO11, initiator of filamentous growth
High expression levels of FLO11, a cell surface glycoprotein that mediates cell-cell and cell-surface adhesion, causes flocculation, haploid invasion, biofilm formation and diploid pseudohyphal, or filamentous, growth [6, 72, 73]. In S. cerevisiae, a family of related genes that encode adhesion proteins, FLO1, FLO5, FLO9 and FLO10, are not expressed due to their proximity to telomeres and position-dependent gene silencing [74]. This gene family is similar to the ALS and EPA adhesin gene families in the human pathogens Candida albicans and Candida glibrata, respectively. In particular, the EPA genes of C. glibrata are also adjacent to telomeres and subject to metastable gene silencing, with activation of individual EPA genes occurring in different host environments and generating cell surface variation for the pathogen [75]. Understanding the regulatory mechanisms controlling expression of these gene families is therefore of high clinical relevance due to their role in promoting adherence to host cells, such as human epithelial cells, and biofilm formation on abiotic surfaces, including implanted medical devices [76].
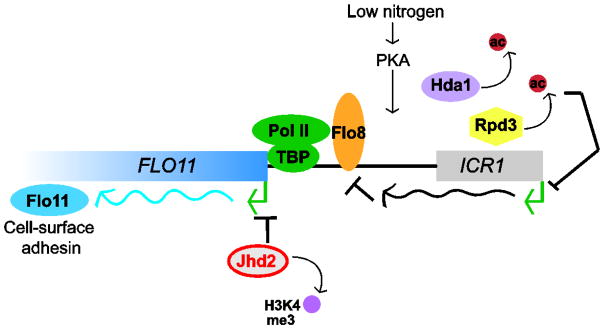
Similar to IME1, FLO11 expression is controlled by a very large, complex promoter that receives input from multiple nutrient-responsive signaling pathways, including the MAPK, SNF, TOR and RAS/cAMP-PKA pathways [6, 77]. Its expression levels are modulated through the coordinated activity of transcription factors, such as Flo8, which govern the contributions of histone-modifying enzymes, chromatin remodelers and lncRNAs to FLO11 expression (Figure 3). While genetic screens have revealed potential involvement of the HAT Gcn5, the histone variant Htz1 and chromatin remodelers RSC and SWI/SNF in FLO11 expression [78, 79], HDACs have the most well-documented roles in both repressing and activating FLO11 expression. Epigenetic silencing of FLO11 was first linked to the HDAC Hda1 [80], however other HDACs including Hos2 and Rpd3 have been implicated in its expression control. Interestingly, mutations of components of the Rpd3(L) complex (a larger Rpd3 complex than Rpd3(S) with unique interactions at chromatin) further repress FLO11 [79], and combined deletion of Rpd3(L) and Hda1 demonstrates their partially overlapping functions in nucleosome depletion at the promoter and FLO11 activation [81]. The paradoxical function of Rpd3(L) in FLO11 activation was found to be due to the upstream expression of the lncRNA ICR1, in a manner dependent on the competing activity of two transcription factors [82, 83] (Figure 3). In the absence of the repressive chromatin environment generated by Rpd3(L), ICR1 is transcribed and prevents expression of FLO11 by occluding key trans-acting factors, such as TBP and the Flo8 transcriptional activator, from the promoter region [82, 83]. While these studies revealed a critical link between histone deacetylation and lncRNA-mediated repression of FLO11, there remain many open questions regarding the roles of other HDACs in this regulatory network. As one example, the Set3 complex, of which the HDAC Hos2 is a member, promotes filamentous growth in Candida albicans [84] and is required for lncRNA-mediated repression of IME1, yet its potential role in FLO11 expression control is unclear.
Figure 3. Chromatin-based regulation of FLO11.
Low nitrogen availability regulates the FLO11 promoter through PKA-mediated signaling to multiple transcription factors, including Flo8 (extensive signaling network regulation of FLO11 is reviewed elsewhere [6, 77]). Transcription of the lncRNA ICR1 approximately 3kb upstream of the FLO11 transcription start site (TSS) occludes binding of the activator Flo8 and TBP. ICR1 expression is repressed by Rpd3C(S) deacetylation, promoting FLO11 activation. Deacetylation by Hda1 also promotes FLO11 expression and dynamic regulation of H3K4me3 at the TSS by demethylase Jhd2 has been linked to FLO11 activation.
Recent genetic studies have implicated other types of chromatin-modifying enzymes in regulating FLO11 expression. The HAT Gcn5 has been reported to negatively regulate ICR1 expression, thereby promoting FLO11 transcriptional activation [85], most likely through regulation of a second ncRNA. Additionally, deletion of the demethylase JHD2 or a component of the CDK8 submodule of RNA pol II, SSN8, results in elevated H3K4me3 near the FLO11 transcription start site (TSS) and constitutive activation of FLO11 [86]. These data suggest a direct role for H3K4me3 in FLO11 activation, although its potential contribution to lncRNA-dependent regulation of FLO11 has not yet been investigated.
Chromatin dynamics during programmed cell death
Programmed cell death (PCD) in metazoans has key developmental roles, such as promoting proper tissue architecture and the destruction of damaged cells, and aberrant regulation of PCD pathways is implicated in diseases including cancer and neurodegenerative disorders [7, 87]. Based on the lack of some of the cell death machinery, it was not clear that S. cerevisiae exhibited PCD until apoptosis-related phenotypes were observed in a CDC48 mutant. Observed phenotypes included DNA fragmentation, chromatin compaction, increased reactive oxygen species and phosphatidylserine externalization [88]. It is now appreciated that budding yeast undergo multiple types of cell death, including caspase-dependent and caspase-independent intrinsic apoptosis, autophagic cell death and regulated necrosis [89]. These pathways are activated in response to a multitude of intrinsic and extrinsic signals, including oxidative and osmotic stress, acetic acid and aging, among others [7]. While PCD is clearly not required for tissue architecture in unicellular yeasts, it may act to eliminate old, damaged or nonreplicative cells, releasing nutrients to younger, replicative cells and promoting their survival [7, 89]. Interestingly, many filamentous fungi have more similar PCD mechanisms to animal cells than budding yeast, and PCD has frequently been observed to be important for pathogenic life cycles and the formation of multicellular-like structures in these organisms [90].
Histone modifications have the potential to be important for multiple components of PCD, including gene expression associated with stress responses, transcription of apoptotic regulators and the processes of chromatin condensation and DNA fragmentation. The Ste20 kinase phosphorylates H2BS10 in apoptotic chromatin in yeast [91], which is analogous to human H2BS14ph, catalyzed by the caspase-activated kinase Mst1 [92]. It was also found that deacetylation at H2BK11 by the HDAC Hos3 is a prerequisite for Ste20-mediated phosphorylation at H2BS10 [93]. This crosstalk between histone modifications appears to promote peroxide-mediated apoptosis, with H2BS10ph proposed to have a role in chromatin condensation [91, 93]. However, this mechanism has been debated [94], and it is unclear whether other substrates of Mst1 (or Ste20) may promote PCD-associated chromatin compaction [94, 95].
In a similar example of modification cross-talk, H2BK123Ub by Bre1 was reported to be an anti-apoptotic mark, with loss of the mark suspected to promote DNA damage or transcriptional deregulation of apoptotic factors [96]. H2BK123Ub precedes Set1-dependent H3K4 methylation, as well as H3K79 methylation catalyzed by Dot1 [97]. Subsequent investigation revealed that BRE1 mutants lacking H2BK123Ub undergo PCD due to the loss of H3K4 methylation by Set1 [98]. The loss of Set1 promotes PCD in a manner dependent on both Dot1/H3K79me and the checkpoint kinase Rad9. Increased PCD in SET1 mutants can be rescued by simultaneous inactivation of Nuc1, the yeast ortholog of enodonuclease G associated with DNA fragmentation [98]. Although further experiments are required, these data suggest that Set1 and H3K4 methylation may play a global role in promoting proper chromatin structure to prevent PCD-mediated condensation and fragmentation of DNA.
The studies conducted to date on histone modifications linked to yeast PCD have revealed that both program-specific (H2BS10ph) and general (H2BK123Ub, H3K4me) modifications make contributions to this developmental pathway and that cross-talk between modifications may serve to integrate multiple inputs from the signaling networks activating PCD. This highlights the need to comprehensively investigate the full complement of histone modifications participating in PCD. Additionally, acetylation of the autophagic protein Atg3 by the HAT Esa1 has been linked to the control of autophagy [99], indicating that there may be more diverse roles for typical chromatin-modifying enzymes that remain to be discovered in cell death pathways.
Concluding Remarks
We have reviewed a number of significant advances in our understanding of chromatin modifications critical to yeast cell fates. In the representative cases cited here, key themes emerge and areas for further exploration are highlighted. First, while chromatin-modifying enzymes such as Rpd3, Gcn5, Set1 and Set2 have broad functions throughout the genome, they also have critical context-dependent roles, responding to specific cues and acting within distinct genomic regions to advance cell fate programs. It is also evident that chromatin modifiers integrate signals from multiple sources, and their activity is governed by complex networks that signal to transcription factors, lncRNAs, and likely other factors, to promote the appropriate changes in the local chromatin landscape.
While substantial progress has been made in understanding the functions of diverse histone modifications in meiosis and sporulation, pseudohyphal differentiation and programmed cell death pathways in yeast, it is clear that there are still many open questions. Continued investigation into the new modifications revealed by high-resolution proteomics may elucidate state-specific roles for these marks, particularly for those that are lowly abundant or enriched in certain environmental conditions. Moreover, further understanding of the role of non-histone PTMs catalyzed by enzymes classified as chromatin modifiers may uncover additional regulatory nodes critical to these cell fate programs and diversify the biological functions of these enzymes. And finally, given the role of nutritional cues in these cell fate signaling networks, it will be critical to delineate how metabolic changes within the cell affect chromatin-modifying enzymes. There is increasing evidence that metabolic changes can signal directly to these enzymes, potentially regulating their activity, localization or co-factor availability to drive cell fate changes [100, 101]. The high conservation of chromatin-modifying pathways and the breadth of knowledge regarding the genome and epigenome suggest that continued investigation of histone modifications in yeast will yield key insights into cell fate decisions in diverse organisms.
Highlights.
Diverse histone modifications in S. cerevisiae regulate cell fate decisions
Histone modifying enzymes are targeted by intrinsic and extrinsic signaling cues to regulate transcription and chromatin reorganization
Histone modifications play critical roles in genomic reprogramming during meiotic sporulation, filamentous growth and programmed cell death
Acknowledgments
The authors acknowledge members of the Green lab for fruitful discussions and comments on the manuscript. This work was supported in part by NIH R03AG052018 and a UMBC START award to EMG.
Abbreviations
- lncRNA
long non-coding ribonucleic acid
- DNA
deoxyribonucleic acid
- PTM
post-translational modification
- HAT
histone acetyltransferase
- HDAC
histone deacteylase
- HMT
histone methyltransferase
- HDM
histone demethylase
- UbT
ubiquitin-protein transferase
- DUB
deubiquinase
- me
methylation
- ac
acetylation
- ph
phosphorylation
- MAPK
mitogen activated protein kinase
- TOR
target of rapamycin
- PKA
protein kinase A
- UCS
upstream control sequence
- APC/C
anaphase-promoting complex/cyclosome
- PCD
programmed cell death
- kb
kilobases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–99. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 2.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–24. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 5.Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189:737–65. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen PJ, Sprague GF., Jr The regulation of filamentous growth in yeast. Genetics. 2012;190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strich R. Programmed Cell Death Initiation and Execution in Budding Yeast. Genetics. 2015;200:1003–14. doi: 10.1534/genetics.115.179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–36. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–31. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–13. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 11.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–6. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tee WW, Reinberg D. Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development. 2014;141:2376–90. doi: 10.1242/dev.096982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musselman CA, Lalonde ME, Côté J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–27. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–55. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 17.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnaudo AM, Garcia BA. Proteomic characterization of novel histone post–translational modifications. Epigenetics Chromatin. 2013;6:24. doi: 10.1186/1756-8935-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–7. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Garcia BA. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb Perspect Biol. 2015;7:a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, Donahue G, Dorsey J, Govin J, Yuan Z, Garcia BA, et al. H4K44 Acetylation Facilitates Chromatin Accessibility during Meiosis. Cell Rep. 2015;13:1772–80. doi: 10.1016/j.celrep.2015.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green EM, Mas G, Young NL, Garcia BA, Gozani O. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat Struct Mol Biol. 2012;19:361–3. doi: 10.1038/nsmb.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martín GM, King DA, Green EM, Garcia-Nieto PE, Alexander R, Collins SR, et al. Set5 and Set1 cooperate to repress gene expression at telomeres and retrotransposons. Epigenetics. 2014:9. doi: 10.4161/epi.27645. [DOI] [PMC free article] [PubMed]
- 24.Zhang X, Huang Y, Shi X. Emerging roles of lysine methylation on non-histone proteins. Cell Mol Life Sci. 2015;72:4257–72. doi: 10.1007/s00018-015-2001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 26.Moore KE, Gozani O. An unexpected journey: lysine methylation across the proteome. Biochim Biophys Acta. 2014;1839:1395–403. doi: 10.1016/j.bbagrm.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore KE, Carlson SM, Camp ND, Cheung P, James RG, Chua KF, et al. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol Cell. 2013;50:444–56. doi: 10.1016/j.molcel.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazur PK, Reynoird N, Khatri P, Jansen PW, Wilkinson AW, Liu S, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014 doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–34. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–84. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tessarz P, Santos-Rosa H, Robson SC, Sylvestersen KB, Nelson CJ, Nielsen ML, et al. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature. 2014;505:564–8. doi: 10.1038/nature12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honigberg SM, Purnapatre K. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J Cell Sci. 2003;116:2137–47. doi: 10.1242/jcs.00460. [DOI] [PubMed] [Google Scholar]
- 33.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 34.Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, et al. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–23. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 35.Shively CA, Eckwahl MJ, Dobry CJ, Mellacheruvu D, Nesvizhskii A, Kumar A. Genetic networks inducing invasive growth in Saccharomyces cerevisiae identified through systematic genome-wide overexpression. Genetics. 2013;193:1297–310. doi: 10.1534/genetics.112.147876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith HE, Driscoll SE, Sia RA, Yuan HE, Mitchell AP. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics. 1993;133:775–84. doi: 10.1093/genetics/133.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kassir Y, Granot D, Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell. 1988;52:853–62. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 39.Mandel S, Robzyk K, Kassir Y. IME1 gene encodes a transcription factor which is required to induce meiosis in Saccharomyces cerevisiae. Dev Genet. 1994;15:139–47. doi: 10.1002/dvg.1020150204. [DOI] [PubMed] [Google Scholar]
- 40.Granot D, Margolskee JP, Simchen G. A long region upstream of the IME1 gene regulates meiosis in yeast. Mol Gen Genet. 1989;218:308–14. doi: 10.1007/BF00331283. [DOI] [PubMed] [Google Scholar]
- 41.Sagee S, Sherman A, Shenhar G, Robzyk K, Ben-Doy N, Simchen G, et al. Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1985–95. doi: 10.1128/mcb.18.4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Covitz PA, Mitchell AP. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 1993;7:1598–608. doi: 10.1101/gad.7.8.1598. [DOI] [PubMed] [Google Scholar]
- 43.Shimizu M, Hara M, Murase A, Shindo H, Mitchell AP. Dissection of the DNA binding domain of yeast Zn-finger protein Rme1p, a repressor of meiotic activator IME1. Nucleic Acids Symp Ser. 1997:175–6. [PubMed] [Google Scholar]
- 44.Shimizu M, Li W, Covitz PA, Hara M, Shindo H, Mitchell AP. Genomic footprinting of the yeast zinc finger protein Rme1p and its roles in repression of the meiotic activator IME1. Nucleic Acids Res. 1998;26:2329–36. doi: 10.1093/nar/26.10.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, et al. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–81. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 47.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–72. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuno T, Nakazawa N, Remgsamrarn P, Kunoh T, Oshima Y, Harashima S. The Tup1-Ssn6 general repressor is involved in repression of IME1 encoding a transcriptional activator of meiosis in Saccharomyces cerevisiae. Curr Genet. 1998;33:239–47. doi: 10.1007/s002940050332. [DOI] [PubMed] [Google Scholar]
- 50.Weidberg H, Moretto F, Spedale G, Amon A, van Werven FJ. Nutrient Control of Yeast Gametogenesis Is Mediated by TORC1, PKA and Energy Availability. PLoS Genet. 2016;12:e1006075. doi: 10.1371/journal.pgen.1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davie JK, Edmondson DG, Coco CB, Dent SY. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J Biol Chem. 2003;278:50158–62. doi: 10.1074/jbc.M309753200. [DOI] [PubMed] [Google Scholar]
- 52.Fleming AB, Beggs S, Church M, Tsukihashi Y, Pennings S. The yeast Cyc8-Tup1 complex cooperates with Hda1p and Rpd3p histone deacetylases to robustly repress transcription of the subtelomeric FLO1 gene. Biochim Biophys Acta. 2014;1839:1242–55. doi: 10.1016/j.bbagrm.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gligoris T, Thireos G, Tzamarias D. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol Cell Biol. 2007;27:4198–205. doi: 10.1128/MCB.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitchell AP, Herskowitz I. Activation of meiosis and sporulation by repression of the RME1 product in yeast. Nature. 1986;319:738–42. doi: 10.1038/319738a0. [DOI] [PubMed] [Google Scholar]
- 55.Covitz PA, Herskowitz I, Mitchell AP. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1-alpha 2. Genes Dev. 1991;5:1982–9. doi: 10.1101/gad.5.11.1982. [DOI] [PubMed] [Google Scholar]
- 56.Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–33. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 57.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–71. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 58.Mallory MJ, Cooper KF, Strich R. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-directed APC/C. Mol Cell. 2007;27:951–61. doi: 10.1016/j.molcel.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mallory MJ, Law MJ, Sterner DE, Berger SL, Strich R. Gcn5p-dependent acetylation induces degradation of the meiotic transcriptional repressor Ume6p. Mol Biol Cell. 2012;23:1609–17. doi: 10.1091/mbc.E11-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Law MJ, Mallory MJ, Dunbrack RL, Strich R. Acetylation of the transcriptional repressor Ume6p allows efficient promoter release and timely induction of the meiotic transient transcription program in yeast. Mol Cell Biol. 2014;34:631–42. doi: 10.1128/MCB.00256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–54. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCord R, Pierce M, Xie J, Wonkatal S, Mickel C, Vershon AK. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol Cell Biol. 2003;23:2009–16. doi: 10.1128/MCB.23.6.2009-2016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winter E. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2012;76:1–15. doi: 10.1128/MMBR.05010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pierce M, Benjamin KR, Montano SP, Georgiadis MM, Winter E, Vershon AK. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol Cell Biol. 2003;23:4814–25. doi: 10.1128/MCB.23.14.4814-4825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–96. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- 66.Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Géli V, et al. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004;23:1957–67. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sommermeyer V, Béneut C, Chaplais E, Serrentino ME, Borde V. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell. 2013;49:43–54. doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 69.Govin J, Berger SL. Genome reprogramming during sporulation. Int J Dev Biol. 2009;53:425–32. doi: 10.1387/ijdb.082687jg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krishnamoorthy T, Chen X, Govin J, Cheung WL, Dorsey J, Schindler K, et al. Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis. Genes Dev. 2006;20:2580–92. doi: 10.1101/gad.1457006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger SL. Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev. 2010;24:1772–86. doi: 10.1101/gad.1954910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lo WS, Dranginis AM. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–71. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–81. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 74.Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A. 2000;97:12158–63. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Las Peñas A, Juárez-Cepeda J, López-Fuentes E, Briones-Martín-Del-Campo M, Gutiérrez-Escobedo G, Castaño I. Local and regional chromatin silencing in Candida glabrata: consequences for adhesion and the response to stress. FEMS Yeast Res. 2015:15. doi: 10.1093/femsyr/fov056. [DOI] [PubMed] [Google Scholar]
- 76.Nobile CJ, Johnson AD. Candida albicans Biofilms and Human Disease. Annu Rev Microbiol. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brückner S, Mösch HU. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2012;36:25–58. doi: 10.1111/j.1574-6976.2011.00275.x. [DOI] [PubMed] [Google Scholar]
- 78.Fischer C, Valerius O, Rupprecht H, Dumkow M, Krappmann S, Braus GH. Posttranscriptional regulation of FLO11 upon amino acid starvation in Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8:225–36. doi: 10.1111/j.1567-1364.2007.00331.x. [DOI] [PubMed] [Google Scholar]
- 79.Barrales RR, Jimenez J, Ibeas JI. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics. 2008;178:145–56. doi: 10.1534/genetics.107.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–15. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- 81.Barrales RR, Korber P, Jimenez J, Ibeas JI. Chromatin modulation at the FLO11 promoter of Saccharomyces cerevisiae by HDAC and Swi/Snf complexes. Genetics. 2012;191:791–803. doi: 10.1534/genetics.112.140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A. 2009;106:18321–6. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, et al. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 2012;45:470–82. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hnisz D, Majer O, Frohner IE, Komnenovic V, Kuchler K. The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 2010;6:e1000889. doi: 10.1371/journal.ppat.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang LC, Montalvo-Munoz F, Tsai YC, Liang CY, Chang CC, Lo WS. The Histone Acetyltransferase Gcn5 Regulates ncRNA-ICR1 and FLO11 Expression during Pseudohyphal Development in Saccharomyces cerevisiae. Biomed Res Int. 2015;2015:284692. doi: 10.1155/2015/284692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Law MJ, Ciccaglione K. Fine-tuning of histone H3 Lys4 methylation during pseudohyphal differentiation by the CDK submodule of RNA polymerase II. Genetics. 2015;199:435–53. doi: 10.1534/genetics.114.172841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiraz Y, Adan A, Kartal Yandim M, Baran Y. Major apoptotic mechanisms and genes involved in apoptosis. Tumour Biol. 2016 doi: 10.1007/s13277-016-5035-9. [DOI] [PubMed] [Google Scholar]
- 88.Madeo F, Fröhlich E, Fröhlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–34. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin SJ, Austriaco N. Aging and cell death in the other yeasts, Schizosaccharomyces pombe and Candida albicans. FEMS Yeast Res. 2014;14:119–35. doi: 10.1111/1567-1364.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shlezinger N, Goldfinger N, Sharon A. Apoptotic-like programed cell death in fungi: the benefits in filamentous species. Front Oncol. 2012;2:97. doi: 10.3389/fonc.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 92.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–17. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 93.Ahn SH, Diaz RL, Grunstein M, Allis CD. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol Cell. 2006;24:211–20. doi: 10.1016/j.molcel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 94.Ura S, Nishina H, Gotoh Y, Katada T. Activation of the c-Jun N-terminal kinase pathway by MST1 is essential and sufficient for the induction of chromatin condensation during apoptosis. Mol Cell Biol. 2007;27:5514–22. doi: 10.1128/MCB.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wen W, Zhu F, Zhang J, Keum YS, Zykova T, Yao K, et al. MST1 promotes apoptosis through phosphorylation of histone H2AX. J Biol Chem. 2010;285:39108–16. doi: 10.1074/jbc.M110.151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Walter D, Matter A, Fahrenkrog B. Bre1p-mediated histone H2B ubiquitylation regulates apoptosis in Saccharomyces cerevisiae. J Cell Sci. 2010;123:1931–9. doi: 10.1242/jcs.065938. [DOI] [PubMed] [Google Scholar]
- 97.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–96. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 98.Walter D, Matter A, Fahrenkrog B. Loss of histone H3 methylation at lysine 4 triggers apoptosis in Saccharomyces cerevisiae. PLoS Genet. 2014;10:e1004095. doi: 10.1371/journal.pgen.1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–7. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 100.Li S, Swanson SK, Gogol M, Florens L, Washburn MP, Workman JL, et al. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Mol Cell. 2015;60:408–21. doi: 10.1016/j.molcel.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 101.Etchegaray JP, Mostoslavsky R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol Cell. 2016;62:695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–95. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–48. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–4. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, et al. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–5. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 106.Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, et al. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol. 2007;14:240–2. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 107.Liang G, Klose RJ, Gardner KE, Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat Struct Mol Biol. 2007;14:243–5. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- 108.Guillemette B, Drogaris P, Lin HH, Armstrong H, Hiragami-Hamada K, Imhof A, et al. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011;7:e1001354. doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 110.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–7. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–9. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 112.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, et al. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–53. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, et al. Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–6. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 114.Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–91. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 115.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–72. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 116.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, et al. Identification of histone demethylases in Saccharomyces cerevisiae. J Biol Chem. 2007;282:14262–71. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim T, Buratowski S. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. J Biol Chem. 2007;282:20827–35. doi: 10.1074/jbc.M703034200. [DOI] [PubMed] [Google Scholar]
- 119.Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, et al. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem. 2007;282:7632–40. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baker SP, Phillips J, Anderson S, Qiu Q, Shabanowitz J, Smith MM, et al. Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat Cell Biol. 2010;12:294–8. doi: 10.1038/ncb2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–5. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 122.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, et al. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–9. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 123.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, et al. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–12. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang B, Miller A, Kirchmaier AL. HST3/HST4-dependent deacetylation of lysine 56 of histone H3 in silent chromatin. Mol Biol Cell. 2008;19:4993–5005. doi: 10.1091/mbc.E08-05-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–56. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 126.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–8. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 127.Lacoste N, Utley RT, Hunter JM, Poirier GG, Côte J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–4. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 128.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–27. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–33. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheung WL, Turner FB, Krishnamoorthy T, Wolner B, Ahn SH, Foley M, et al. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol. 2005;15:656–60. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 131.Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–19. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–26. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 134.Sharma VM, Tomar RS, Dempsey AE, Reese JC. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol Cell Biol. 2007;27:3199–210. doi: 10.1128/MCB.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shia WJ, Osada S, Florens L, Swanson SK, Washburn MP, Workman JL. Characterization of the yeast trimeric-SAS acetyltransferase complex. J Biol Chem. 2005;280:11987–94. doi: 10.1074/jbc.M500276200. [DOI] [PubMed] [Google Scholar]
- 136.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–7. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 137.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–83. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 138.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 139.Edwards CR, Dang W, Berger SL. Histone H4 lysine 20 of Saccharomyces cerevisiae is monomethylated and functions in subtelomeric silencing. Biochemistry. 2011;50:10473–83. doi: 10.1021/bi201120q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Basnet H, Su XB, Tan Y, Meisenhelder J, Merkurjev D, Ohgi KA, et al. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature. 2014;516:267–71. doi: 10.1038/nature13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moore JD, Yazgan O, Ataian Y, Krebs JE. Diverse roles for histone H2A modifications in DNA damage response pathways in yeast. Genetics. 2007;176:15–25. doi: 10.1534/genetics.106.063792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–4. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 143.Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL. Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail. J Biol Chem. 2007;282:27923–34. doi: 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- 144.Gardner KE, Zhou L, Parra MA, Chen X, Strahl BD. Identification of lysine 37 of histone H2B as a novel site of methylation. PLoS One. 2011;6:e16244. doi: 10.1371/journal.pone.0016244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–4. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 146.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–74. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 147.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–63. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell. 2005;17:585–94. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]