Abstract
Cotton leaf curl disease (CLCuD), the most complex disease of cotton, is a major limiting biotic factor to worldwide cotton productivity. Several whitefly-transmitted monopartite begomoviruses causing CLCuD have been characterized and designated as CLCuD-associated begomoviruses. Despite of being reported over 100 years ago in Africa, CLCuD became economically pandemic causing massive losses to cotton production in Pakistan and India during past couple of decades. In Asia, cotton has faced two major epidemics during this period viz. “Multan epidemic” and “Burewala epidemic.” The “Multan epidemic” era was 1988–1999 after which the virus remained calm until 2002 when “Burewala epidemic” broke into the cotton fields in Indo-Pak subcontinent, till 2013–2014. However, both the epidemics were caused by monopartite begomovirus complex. Similarly in Africa, Cotton leaf curl Gezira virus with associated DNA-satellites causes CLCuD. Quite recently, in the Old World (both Asia and Africa), bipartite begomoviruses have started appearing in the areas under cotton cultivation. Under such aggravated circumstances, it seems we are heading toward another epidemic of CLCuD in the Old World. Here we articulate the causes and potential emergence of the third epidemic of CLCuD in Asia. The current situation of CLCuD in Asia and Africa is also discussed.
Keywords: CLCuD, monopartite begomovirus, bipartite begomovirus, Multan epidemic, Burewala epidemic, SNPs
Cotton leaf curl disease (CLCuD) is a major threat to worldwide cotton productivity. Indo-Pak subcontinent is a paradigm where cotton production has been severely compromised in past few decades because of the onset of two CLCuD epidemics. India and Pakistan are third and fourth largest cotton producers in the world with 6.05 and 2.17 million ton annual production, respectively (FAOStat, 2013). The CLCuD, showing characteristic symptoms of upward or downward leaf curling, vein swelling, leaf enation, and growth stunting, is transmitted by a ubiquitous whitefly (Bemisia tabaci) and is caused by a complex of five single-stranded DNA viruses belonging to genus Begomovirus (family Geminiviridae) along with their DNA satellites, viz. betasatellite and alphasatellite (Briddon and Stanley, 2006; Zhou, 2013; Brown et al., 2015). Understanding the dynamics of this disease is essential in order to sustain the worldwide cotton productivity and socio-economic values of the people.
Two major CLCuD complexes are prevailing in the Old World, i.e., “the African complex” and “the Asian complex.” In Africa, CLCuD was reported, for the first time, in 1912 from Nigeria, later from Sudan in 1924 and Tanzania in 1926 (Farquharson, 1912; Kirkpatrick, 1931). In Africa, since 2002 a single begomovirus species Cotton leaf curl Gezira virus (CLCuGeV) associated with Cotton leaf curl Gezira betasatellite (CLCuGeB) and Cotton leaf curl Gezira alphasatellite (CLCuGeA) has been predominantly the causative agent of CLCuD (Idris and Brown, 2002; Idris et al., 2005). However, very recently a bipartite begomovirus Cotton yellow mosaic virus (CYMV) has been reported for the first time in cultivated cotton (Gossypium raimondii) from the west and central Africa (Leke et al., 2016). Unlike Africa, mapping the exact history of CLCuD is difficult in Asia, however, in Pakistan introgression of high yielding but highly susceptible varieties of cotton (Gossypium hirsutum) favored the onset of this disease (Ali, 1997). During 1967 leaf curl symptoms, as a minor sporadic nuisance, were observed in a small cotton field near Multan, Pakistan but remained un-noticed (Hussain and Ali, 1975); this was the first inception of CLCuD. The virus [most probably Cotton leaf curl Multan virus (CLCuMuV)] kept on proliferating and burst into an epidemic in 1988 at Moza Kokhran, Multan, Pakistan. The country-wide spread of CLCuD most likely started from this vicinity and later moved across the borders into the northwestern states of India. In India, the first evidence of CLCuD was reported near border area Sri Ganganagar in 1989, which later spread as an epidemic in 1994 (Monga et al., 2004). During 1990s, up to 80% yield losses were recorded in Pakistan that caused a shortfall of about US $5 billion to Pakistan’s economy (Briddon and Markham, 2000). Later in 1998, three different species of begomoviruses were found to be associated with the CLCuD-first epidemic (Figure 1A), which today are named: Cotton leaf curl Alabad virus (CLCuAlV), Cotton leaf curl Kokhran virus (CLCuKoV), and CLCuMuV (Zhou et al., 1998). This epidemic has been known as the “Multan epidemic” (Sattar et al., 2013). Later, other begomoviruses such as Tomato leaf curl Bangalore virus (ToLCBaV) and Papaya leaf curl virus (PaLCuV) were also identified infecting cotton (Zhou et al., 1998; Mansoor et al., 2003; Kirthi et al., 2004). Interestingly, all these three CLCuD-associated begomoviruses (CABs) were found associated with a single species of betasatellite Cotton leaf curl Multan betasatellite (CLCuMuB) and satellite-like component Cotton leaf curl Multan alphasatellite (CLCuMuA) (Figure 1A and Supplementary Table S1) (Mansoor et al., 1999). The major reason for Multan epidemic is unanimously considered to be the cultivation of highly susceptible G. hirsutum cultivar S-12, which was cultivated on 46% area in Punjab, Pakistan (Briddon and Markham, 2000).
FIGURE 1.
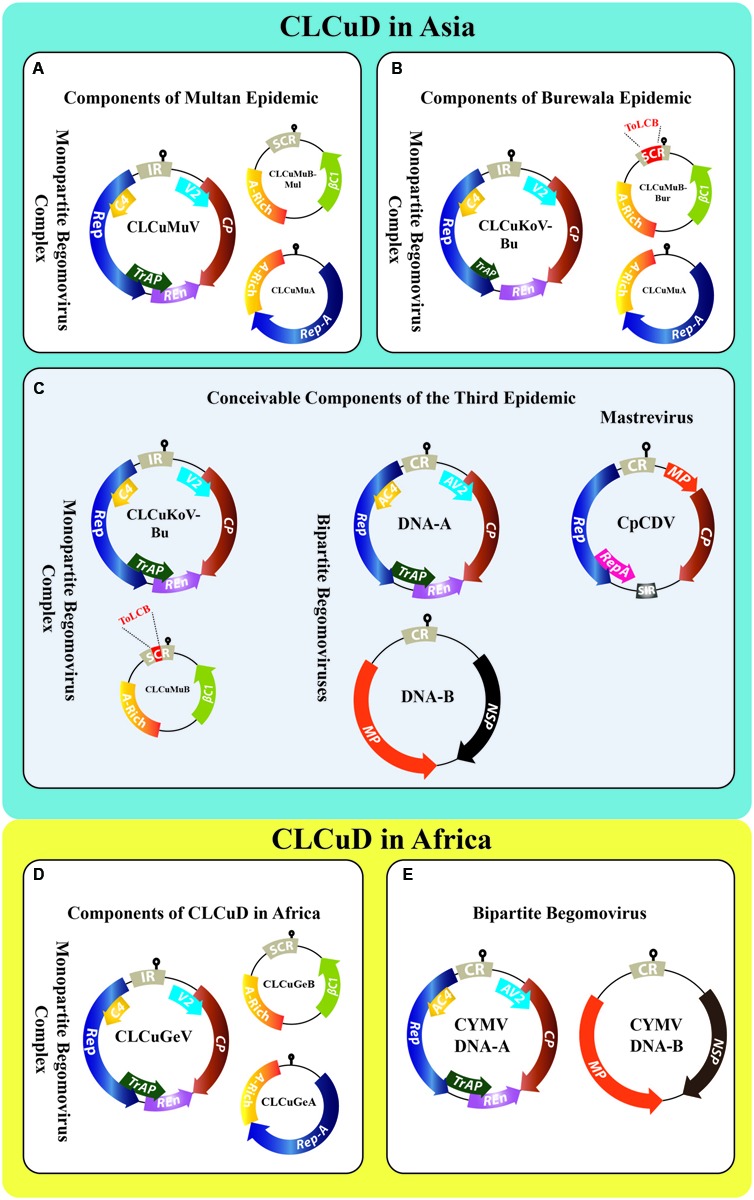
Genome organization of all the contributing viruses of CLCuD in Asia (A–C) and Africa (D,E). The key begomovirus along with DNA satellites of the “Multan epidemic” (A), “Burewala epidemic” (B), and of the proposed “third epidemic” (C) are shown. During the Multan epidemic era, several begomoviruses were reported concomitantly from cotton plants, i.e., CLCuAlV, CLCuKoV, PaLCuV, and ToLCBaV. However, the key begomovirus was CLCuMuV. The Burewala epidemic involved a truncated transcriptional activator protein (TrAP; short green arrow) and a recombinant CLCuMuBBur having a stretch of ∼98 nt in the SCR region (highlighted in red) acquired from ToLCB (B). The third epidemic in Asia may either involve CLCuKoV-Bu having intacted TrAP along with CLCuMuB having a stretch of ∼24 nt from ToLCB (highlighted in red); or ToLCNDV (DNA-A and DNA-B), ToLCV (DNA-A) together with ToLCNDV (DNA-B); or a Mastrevirus CpCDV (C). In parallel, the CLCuD situation in Africa involved monopartite begomovirus complex (D) and recently bipartite begomovirus CYMV (DNA-A and DNA-B) (E). The genomes of monopartite begomoviruses in the CLCuD-complex have single component DNA-A, which encodes two open reading frames (ORFs), i.e., V2 protein and coat protein (CP) in the virion-sense orientation whereas, four ORFs in the complementary-sense orientation are: replication associated protein (Rep), TrAP, replication enhancer protein (REn), and C4 protein (undermined functions). The bipartite begomovirus DNA-A resembles monopartite genome, however, they own a second genomic component DNA-B encoding two ORFs, i.e., nuclear shuttle protein (NSP) and movement protein (MP) in the virion- and complementary-sense orientation. The monopartite begomoviruses are also associated with DNA-satellites called alpha- and betasatellite. The alphasatellite encodes a single ORF (Rep-A) for its autonomous replication. Betasatellite encodes its single ORF βC1 in the complementary-sense orientation, which facilitates the helper begomovirus for its diverse functions.
In the late 1990s, after the Multan epidemic era (1989–1999), the cotton productivity was reinstated by developing resistant cultivars in Pakistan. However, theatrical appearance of a natural recombinant species that lacks a C2 gene (Figure 1B), named Cotton leaf curl Burewala virus [now known as CLCuKoV-Burewala strain (CLCuKoV-Bu)] in the year 2002–2003, broke this resistance and all the resistant germplasm became susceptible to this unique member of CLCuD (Amrao et al., 2010b). This was the start of the second epidemic of CLCuD in this region and here referred as “Burewala epidemic” (Sattar et al., 2013). It was speculated that dispense of C2 gene in CLCuKoV-Bu was a key player in resistance breaking (Amrao et al., 2010b; Briddon et al., 2014). However, the recent reports contradict this speculation as new field isolates of CLCuKoV-Bu with intact C2 gene have been reported from India (Kumar et al., 2015; Godara et al., 2016). Another interesting twist in this story was the appearance of “Burewala strain” of CLCuMuB (CLCuMuBBur), which was a recombinant of CLCuMuB and Tomato leaf curl betasatellite (ToLCB; Amin et al., 2006) (Figure 1B). The significance of both these invaders was the breakdown of established resistance in all the CLCuD-resistant cotton germplasm. The reasons for the ultimate selection of these recombinants through the use of resistant cultivars are not yet fully understood. According to Amin et al. (2006), the recombinant stretch of CLCuMuBBur may enhance the interaction between the betasatellite and the helper begomovirus.
Until this period, the southern parts of Pakistan were disease free but following the resistance breakdown, CLCuKoV-Bu was extended to Sindh province as well as to northwestern India in 2005 (Rajagopalan et al., 2012). Moreover, CLCuGeV—one of the CABs causing CLCuD in Africa—trans-replicating CLCuMuB and Chili leaf curl betasatellite has been also found from cotton in Pakistan (Tahir et al., 2011). In these areas the CLCuD is characterized by CLCuKoV, CLCuKoV-Bu, CLCuKoV-Shahdadpur strain (CLCuKoV-Sha; old name Cotton leaf curl Shahdadpur virus), and CLCuGeV along with two strains of CLCuMuB, i.e., “Burewala strain” and “Shahdadpur strain” (Amrao et al., 2010a). The factors driving the rapid evolution of these new strains of CLCuMuB in this region are not yet clear (Akhtar et al., 2014).
Until the advent of Burewala epidemic, which remained active from 2002 until 2013–2014, only monopartite begomoviruses associated with DNA-satellites were found from the infected cotton plants. But quite recently, like in Africa, bipartite begomoviruses are also reportedly appearing in cotton plants including Tomato leaf curl virus (ToLCV) (Zaidi et al., 2015) and Tomato leaf curl New Delhi virus (ToLCNDV) (Zaidi et al., 2016) (Supplementary Table S1), in Pakistan. Moreover, the identification of Okra enation leaf curl virus (OELCuV) (Hameed et al., 2014) and a Mastrevirus Chickpea chlorotic dwarf virus (CpCDV) (Manzoor et al., 2014) has further aggravated the scenario. It has already been proven that ToLCNDV synergistically interacts with CLCuKoV-Bu thereby enhancing the accumulation of CLCuMuBBur (a member of the Burewala epidemic of CLCuD) in the infected plants (Zaidi et al., 2016). The DNA-A of ToLCNDV has been shown empirically to trans-replicate CLCuMuB in the absence of DNA-B (Saeed et al., 2007) and also can cause CLCuD symptoms in cotton (Saeed, 2010). During a most recent study, ToLCNDV has been found associated with CLCuD-infected cotton over a vast area in Punjab and Sindh provinces of Pakistan (Zaidi et al., 2016). ToLCNDV has been prevailing in Indo-Pak subcontinent from nearly three decades (Padidam et al., 1995). It has not only been established in Asia (Mizutani et al., 2011; Yazdani-Khameneh et al., 2016) but also spread to other continents including Africa (Mnari-Hattab et al., 2015) and Europe to Spain (Juarez et al., 2014; Ruiz et al., 2015) and Italy (Panno et al., 2016). If we have learnt some lesson from the previous epidemics, a wise interpretation could be that this is not just an accidental ephemeral intrusion of a bipartite begomovirus in cotton. Rather, it may be an alarm bell for the third epidemic of CLCuD in this area (Figure 1C). The ability of ToLCNDV to develop synergistic interactions with other begomoviruses has recently been shown to cause resistance breakdown in chilies (Singh et al., 2016). CLCuAlV has been found associated with and trans-replicating a recombinant DNA-B of a bipartite begomovirus in wild cotton germplasm, proving the ability of CABs to acquire components from bipartite begomoviruses (Nawaz-ul-Rehman et al., 2012). Thus, if we foresee correctly, the third epidemic might involve CLCuKoV-Bu (with intact C2) associated with CLCuMuB and in synergism with ToLCNDV in the Indo-Pak subcontinent. While in Sindh province, ToLCNDV, CLCuKoV-Bu, and CLCuGeV along with local DNA-satellites may represent the key players of the third epidemic.
The Asian and African CLCuD complexes are quite similar in having monopartite begomoviruses associated with DNA-satellites as causative agents (Figure 1). Yet this similarity has been extended to the possibility of another epidemic in both continents because of the identification of bipartite begomovirus from the field cotton in Africa (Leke et al., 2016) and Asia (Zaidi et al., 2015, 2016) (Figures 1C–E and Supplementary Table S1). The monopartite begomoviruses infection in cotton is the main cause of yield losses in Asia and Africa; however, the appearance of bipartite begomoviruses in the cotton crop fields is a stark reminder of another destructive epidemic. It seems that both the continents are going through a paradigm shift of CLCuD complex. We may speculate that a change in vector population, host plant–virus interactions, injudicious use of insecticides and other environmental factors could be the key players that are exacerbating the occurrence of CLCuD. Environmental changes concomitantly affect plant–virus–vector interactions and members of this complex consequently try to confront such fluctuations through differential expression of genes, mutation, and recombination. In such three-way interaction, virus induces multiple dramatic physiological changes, either in vector and/or host, to increase the transmission. This scenario has another level of complexity associated with particular species of whitefly. Studies show that higher incidence of CLCuD in Punjab (Pakistan) and northwestern India is associated with Asia II-1 species of B. tabaci, the single predominant whitefly specie in these regions (Ahmad et al., 2010; Hameed et al., 2012; Rana et al., 2012) which plays an important role in the transmission of CLCuD. The other factors include the increased, unwise and injudicious use of insecticides and higher adaption rate of Bacillus thuringiensis (BT)-cotton hybrids in Indo-Pak subcontinent. The cultivation of BT-crops not only affects the pest populations in an area but could also alter the weed flora in a particular region (Albajes et al., 2011) and thus indirectly may change the virus population in a particular niche. Among other problems, the higher incidence of CLCuD on unapproved BT varieties has become a major issue both in India and Pakistan. The rate of CABs infection was 54.24% in 2008, which increased to 83.1% in 2009 after BT-cotton cultivation (Kakakhel, 2009) in Pakistan. Similarly, the extensive cultivation of BT-cotton hybrids in north Indian states of Haryana, Punjab, and Rajasthan led to increase virus inoculum in the Indian ecosystem (Kranthi, 2014). The high rate of BT-cultivation in Indo-Pak subcontinent might have played an indirect role in proliferation of virus inoculum in this region because most of the BT varieties have higher susceptibility to CABs as compared to non-BT (Ali et al., 2010; Kranthi, 2014, 2015). Effects of BT crops on the insect vectors (and indirectly promoting virus proliferation) have been reported for Maize rough dwarf virus (MRDV) and Fijiviruses from maize in Spain (Pons et al., 2005; Grilli and Bruno, 2007; Achon and Alonso-Dueñas, 2009). Another alarming nuance is the hyper mutation rate of CABs (∼2.706 to 4.96 × 10-4), which is comparable to many RNA viruses (Saleem et al., 2016). It is thus conceivable that high mutation rate and environmental paradigm shift can lead to rapid evolution and emergence of more virulent species and/or strains of CABs, leading to a situation that we fear today, the “third epidemic” of CLCuD in Indo-Pak subcontinent.
The correlation between various environmental factors and the vector-borne disease incidence has been a mechanistic determinant to assess the geographic spread of the disease risk (Chabot-Couture et al., 2014). Such correlations may also help beforehand disease assessments and to build models for their vector population (Grover-Kopec et al., 2005; White et al., 2011). A successful prevention of an emerging disease epidemic depends upon the early detection and timely control strategies. A number of plant disease epidemics are exemplary due to failure in timely detection and management, such as chestnut blight (Freinkel, 1997), white pine blister rust (Maloy, 1997) and citrus canker (Gottwald, 2007), etc. To validate our assumptions that “why this area (especially Multan city) has become such a ‘hot spot’ for new intrusions in cotton?” and to decipher the relation between CLCuD incidence and different meteorological parameters, “R-statistical and computing tool” was employed (R Development Core Team, 2016). A positive and highly significant correlation (0.85∗∗∗) between the disease severity and the disease incidence with respect to humidity and temperature was observed in all cotton growing areas of Pakistan (Figure 2A). Moreover, positive correlations between and among disease incidence, humidity (0.15), and temperature (0.25) support our hypothesis. Concomitant to this, disease incidence data of 19 years (1997–2015) along with the “Burewala epidemic” were mapped on a bar graph. Likewise to the Burewala epidemic during the years 1999–2002 when the disease incidence was dramatically at its lowest incidence (Figure 2B), similar disease scenario is prevailing in cotton growing areas of Pakistan (as shown in circle; Figure 2B) (2014–2015). Usually, during long hibernation/incubation periods, some pathogens remain cryptic while their infection continuously spreading (Tomlinson and Potter, 2010). This stage is pointing toward the likelihood of another epidemic of CLCuD in the cotton fields, which is supporting our hypothesis. Rapidly emerging begomoviruses bear huge numbers of single nucleotide polymorphism (SNP) in their genome to evolve and/or evade the host defense response. To assess the SNPs diversity in CABs, year-wise virus sequences were retrieved from the NCBI database, aligned by keeping the oldest known viral sequence as a master sequence and then SNPs were calculated semi-manually (NCBI-multiple sequence alignment tools) and by online web-based tool1. The SNPs with ≥75% frequency were mapped on the genomic structure of the respective virus (Figure 3). The present study demonstrates that an extensive SNPs diversity is associated with CABs genome and a highest SNPs diversity exists in CpCDV followed by ToLCNDV, CLCuGeV, and CLCuMuV, respectively, while least SNPs were evident in CLCuKoV-Bu genome (Figure 3). The occurrence of fewer SNPs in CLCuKoV-Bu may likely be due to recombination event from which this virus has emerged out. Further analysis revealed that >30% of the SNPs were non-synonymous, thus can have substantial role in the virus divergence and evolution.
FIGURE 2.
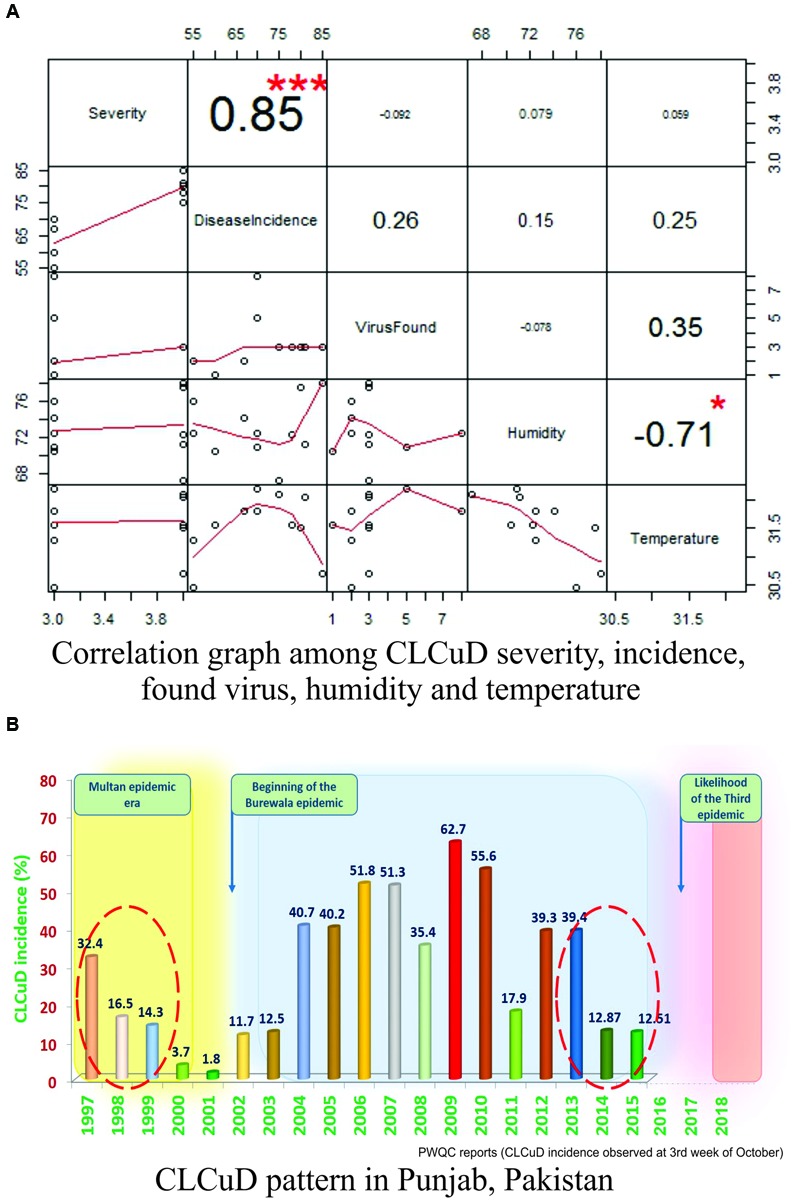
Statistical modeling of CLCuD in Pakistan. (A) The R-Plot represents the correlation structure in between disease severity, disease incidence, virus found, humidity, and temperature. Positive and significant correlations were calculated as numeric values among all the parameters. The numeric values with larger font and positive numbers denote higher correlation strength and vice versa. The significance of each correlation was represented by asterisks, i.e., “∗∗∗” represents very high significance and “∗” represents less significance (P-values <0.01), respectively. All the correlations were also represented in graphical forms (as line segments in red color) and their respective scales are given along x- and y-axis. (B) Graphical bar chart, representing 19 years data (1997–2015) of CLCuD incidence pattern in the Punjab province, Pakistan. The time scale of CLCuD was plotted against the CLCuD incidence (%). Each bar having numeric value and different color (green for least and red for highest incidence) represents the CLCuD incidence (%) in the respective year. The yellow shaded area represents the Multan epidemic era until the year 1999. Whereas, the gray shaded area represents the Burewala epidemic era starting from the year 2002 until 2013–2014. From here on, likely a new epidemic is being evolved (shaded area in pink color). The shaded area in between the epidemics represents the hibernation period of the virus(es). The red dotted circles are highlighting CLCuD incidence scenarios prior to the onset of a new disease outbreak.
FIGURE 3.
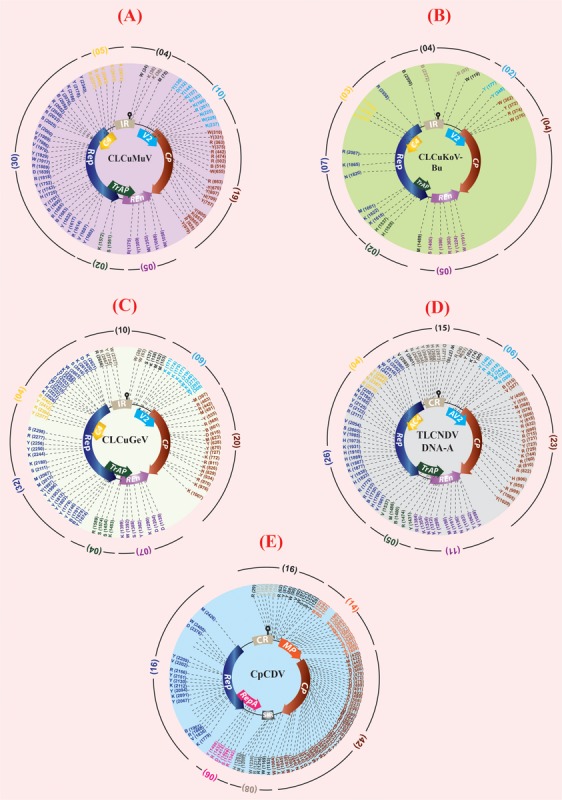
Graphical representation of the location and type(s) of single nucleotide polymorphisms (SNPs) in CLCuD-associated begomoviruses (CABs) DNA-A (A–D) and Mastrevirus (E). Maximum available genome sequences (∼40–100 for each candidate virus) were retrieved from NCBI GenBank database. Semi-manual (NCBI based multiple sequence alignment tools) and web-based tools were used for SNPs identification and classification following the International Union of Pure and Applied Chemistry (IUPAC). For CLCuMuV, CLCuKoV-Bu, and CLCuGeV the oldest reported virus sequence was set as reference sequence for alignment and to locate the viral SNPs. Whereas, in case of ToLCNDV and CpCDV, the sequences reported from cotton crop were set as reference sequence. In the second method, a web-based tool (www.fludb.org/brc/snpAnalysis) was used and all the aligned sequences of the CABs were subjected to SNPs calculation. Only those nucleotide positions were marked as SNPs, which had a quality score ≥40% in both the methods used. The SNPs in the non-coding regions are shown in black and gray text whereas, the SNPs in the coding regions are shown in the respective colors of their ORFs. The total number of SNPs found in the ORFs of each molecule is mentioned separately along-sides (in brackets), respectively.
Thus, we may speculate that today we come across a similar situation of early 2000s in terms of virus incubation period (Figure 2B). Based upon our statistical modeling and the currently available data, we may infer that the Multan, Pakistan (the place for Multan- and Burewala-epidemic origin) may face a serious eruption of CLCuD with new arsenals, which may spread across the borders.
Various strategies encompassing classical breeding and biotechnological techniques have been employed to control the CLCuD (Iqbal et al., 2016). All these techniques showed a laconic control over CLCuD. In a three-way interaction of cotton–whitefly–CABs (along with DNA-satellites), none (either breeding or biotechnological techniques) have ever been tried to target this complex simultaneously. Recently, a CRISPR/Cas9-based multiplexing technique has been proposed (Iqbal et al., 2016) as an ultimate strategy to target whole CLCuD complex, i.e., the CABs as well as associated DNA-satellites. However, authors hereby would like to propose that the third epidemic could be obstructed by targeting more than one component of the cotton–whitefly–CABs complex through multiplexing CRISPR/Cas9 system.
Concluding Remarks
Whitefly populations responsible for transmitting CLCuD appear to be favored by the humid and high temperature conditions in the Indo-Pak subcontinent, where cotton is a key crop. Due to expansion in the geographic range of CABs from South Asia to China, a dynamic change in CABs in South Asia is an emerging threat to cotton production (Cai et al., 2010). The key factor is whitefly being a vector for both monopartite and bipartite begomoviruses. The anticipated global warming, poor social practices, unwise use of insecticides, extensive cultivation of unapproved CLCuD susceptible BT-cotton hybrids and non-BT germplasm, mono-cropping system, transportation of infected plants (especially ornamental plants) and higher mutation and recombination rate in the virus genome will exacerbate the whitefly-transmitted CABs. Ultimately, it may enhance the likelihood of another epidemic to occur due to the emergence of more virulent species/strains. It is therefore of a dire need to devise more effective and sustainable approaches to achieve control over host plant–virus–vector complexes. The best way to control CLCuD is the exploitation of natural resistance by stacking multiple resistance sources together, i.e., through modern genetic tools. Understanding of various aspects of pathosystem like host–vector interaction, insect vector alternative hosts, biological role of new begomoviruses involvement in the CLCuD and environmental factors will also help to manage unpleasant surprises in the future.
Author Contributions
MS conceived the idea, created the figures and drafted the first draft. ZI wrote part of the manuscript, collected meteorological and CLCuD incidence data, and helped in preparation of the first draft. MT helped in retrieving all the data and prepared the tables. SU performed all statistical analysis. Final draft was checked and approved by all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Thomas Martin, Uppsala University Sweden, for critically reading the article and the valuable suggestions. The authors are also thankful to Dr. Shahiq Iqbal to help in collecting disease incidence data for CLCuD in Punjab, Pakistan. The authors hereby acknowledge Pakistan Meteorological Department, Islamabad for the provision of meteorological data, and Ayub Agriculture Research Institute, Faisalabad and Central Cotton Research Institute, Multan, Pakistan for providing CLCuD incidence data.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00631/full#supplementary-material
References
- Achon M., Alonso-Dueñas N. (2009). Impact of 9 years of Bt-maize cultivation on the distribution of maize viruses. Transgenic Res. 18 387–397. 10.1007/s11248-008-9231-2 [DOI] [PubMed] [Google Scholar]
- Ahmad S., Islam N., Mahmood A., Ashraf F., Hayat K., Hanif M. (2010). Screening of cotton germplasm against Cotton leaf curl virus. Pak. J. Bot. 42 3327–3342. [Google Scholar]
- Akhtar S., Tahir M. N., Baloch G. R., Javaid S., Khan A. Q., Amin I., et al. (2014). Regional changes in the sequence of cotton leaf curl multan betasatellite. Viruses 6 2186–2203. 10.3390/v6052186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albajes R., Lumbierres B., Pons X. (2011). Two heteropteran predators in relation to weed management in herbicide-tolerant corn. Biol. Control 59 30–36. 10.1016/j.biocontrol.2011.03.008 [DOI] [Google Scholar]
- Ali M. (1997). Breeding of cotton varieties for resistance to cotton leaf curl virus. Pak. J. Phytopathol. 9 1–7. [Google Scholar]
- Ali N., Datta S. K., Datta K. (2010). RNA interference in designing transgenic crops. GM Crops 1 207–213. 10.4161/gmcr.1.4.13344 [DOI] [PubMed] [Google Scholar]
- Amin I., Mansoor S., Amrao L., Hussain M., Irum S., Zafar Y., et al. (2006). Mobilisation into cotton and spread of a recombinant cotton leaf curl disease satellite. Arch. Virol. 151 2055–2065. 10.1007/s00705-006-0773-4 [DOI] [PubMed] [Google Scholar]
- Amrao L., Akhter S., Tahir M. N., Amin I., Briddon R. W., Mansoor S. (2010a). Cotton leaf curl disease in Sindh province of Pakistan is associated with recombinant begomovirus components. Virus Res. 153 161–165. 10.1007/s00705-010-0760-7 [DOI] [PubMed] [Google Scholar]
- Amrao L., Amin I., Shahid S. M., Briddon R. W., Mansoor S. (2010b). Cotton leaf curl disease in resistant cotton is associated with a single begomovirus that lacks an intact transcriptional activator protein. Virus Res. 152 153–163. 10.1016/j.virusres.2010.06.019 [DOI] [PubMed] [Google Scholar]
- Briddon R. W., Akbar F., Iqbal Z., Amrao L., Amin I., Saeed M., et al. (2014). Effects of genetic changes to the begomovirus/betasatellite complex causing cotton leaf curl disease in South Asia post-resistance breaking. Virus Res. 186 114–119. 10.1016/j.virusres.2013.12.008 [DOI] [PubMed] [Google Scholar]
- Briddon R. W., Markham P. G. (2000). Cotton leaf curl virus disease. Virus Res. 71 151–159. 10.1016/S0168-1702(00)00195-7 [DOI] [PubMed] [Google Scholar]
- Briddon R. W., Stanley J. (2006). Sub-viral agents associated with plant single-stranded DNA viruses. Virology 344 198–210. 10.1016/j.virol.2005.09.042 [DOI] [PubMed] [Google Scholar]
- Brown J. K., Zerbini F. M., Navas-Castillo J., Moriones E., Ramos-Sobrinho R., Silva J. F., et al. (2015). Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 160 1593–1619. 10.1007/s00705-015-2398-y [DOI] [PubMed] [Google Scholar]
- Cai J. H., Xie K., Lin L., Qin B. X., Chen B. S., Meng J. R., et al. (2010). Cotton leaf curl Multan virus newly reported to be associated with cotton leaf curl disease in China. Plant Pathol. 59 794–795. 10.1111/j.1365-3059.2010.02266.x [DOI] [Google Scholar]
- Chabot-Couture G., Nigmatulina K., Eckhoff P. (2014). An environmental data set for vector-borne disease modeling and epidemiology. PLoS ONE 9:e94741 10.1371/journal.pone.0094741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOStat.(2013). Food and Agriculture Organization of the United Nations Statistics Division. Available: http://faostat3.fao.org/home/E [Google Scholar]
- Farquharson C. O. (1912). “Report of mycologist. Annual report of the agricultural department of Nigeria,” in Cotton Growth in Gezira Environment, eds Siddique M. A., Hungus L. C. (Cambridge: W. Haffer and Sons Ltd; ) 106. [Google Scholar]
- Freinkel S. (1997). American Chestnut: The Life, Death, and Rebirth of a Perfect Tree. Oakland, CA: University of California Press. [Google Scholar]
- Godara S., Paul Khurana S. M., Biswas K. K. (2016). Three variants of cotton leaf curl begomoviruses with their satellite molecules are associated with cotton leaf curl disease aggravation in New Delhi. J. Plant Biochem. Biotechnol. 26 97–105. 10.1007/s13562-016-0370-x [DOI] [Google Scholar]
- Gottwald T. R. (2007). Citrus canker and citrus huanglongbing, two exotic bacterial diseases threatening the citrus industries of the Western Hemisphere. Outlooks Pest Manage. 18 274–279. 10.1564/18dec09 [DOI] [Google Scholar]
- Grilli M. P., Bruno M. (2007). Regional abundance of a planthopper pest: the effect of host patch area and configuration. Entomol. Exp. Appl. 122 133–143. 10.1111/j.1570-7458.2006.00498.x [DOI] [Google Scholar]
- Grover-Kopec E., Kawano M., Klaver R., Blumenthal B., Ceccato P. (2005). An online operational rainfall-monitoring resource for epidemic malaria early warning systems in Africa. Malar. J. 4:6 10.1186/1475-2875-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed S., Hameed S., Sadia M., Malik S. A. (2012). Genetic diversity analysis of Bemisia tabaci populations in Pakistan using RAPD markers. Electron J. Biotechnol. 15 1–9. 10.2225/vol15-issue6-fulltext-5 [DOI] [Google Scholar]
- Hameed U., Zia-Ur-Rehman M., Herrmann H.-W., Haider M. S., Brown J. K. (2014). First report of Okra enation leaf curl virus and associated cotton leaf curl Multan betasatellite and cotton leaf curl Multan alphasatellite infecting cotton in Pakistan: a new member of the cotton leaf curl disease complex. Plant Dis. 98:1447 10.1094/pdis-10-13-1067-pdn [DOI] [PubMed] [Google Scholar]
- Hussain T., Ali M. (1975). A review of cotton diseases in Pakistan. Pak. Cottons 19 71–86. [Google Scholar]
- Idris A. M., Briddon R. W., Bull S. E., Brown J. K. (2005). Cotton leaf curl Gezira virus-satellite DNAs represent a divergent, geographically isolated Nile Basin lineage: predictive identification of a satDNA REP-binding motif. Virus Res. 109 19–32. 10.1016/j.virusres.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Idris A. M., Brown J. K. (2002). Molecular analysis of cotton leaf curl virus-Sudan reveals an evolutionary history of recombination. Virus Genes 24 249–256. 10.1023/A:1015380600089 [DOI] [PubMed] [Google Scholar]
- Iqbal Z., Sattar M. N., Shafiq M. (2016). CRISPR/Cas9: a tool to circumscribe cotton leaf curl disease. Front. Plant Sci. 7:475 10.3389/fpls.2016.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M. A., Tovar R., Fiallo-Olivé E., Aranda M. A., Gosálvez B., Castillo P., et al. (2014). First detection of Tomato leaf curl New Delhi virus infecting zucchini in Spain. Plant Dis. 98 857 10.1094/pdis-10-13-1050-pdn [DOI] [PubMed] [Google Scholar]
- Kakakhel I. (2009). Govt. decides to use CLCV-tolerant BT cotton varieties. Daily Times, Lahore: Available at: http://www.dailytimes.com.pk/default.asp?page=2009\%5C09\%5C30\%5Cstory_30-9-2009_pg5_17 [Google Scholar]
- Kirkpatrick T. W. (1931). Further studies on leaf-curl of cotton in the Sudan. Bull. Entomol. Res. 22 323–363. 10.1017/S0007485300029862 [DOI] [Google Scholar]
- Kirthi N., Priyadarshini C. G. P., Sharma P., Maiya S. P., Hemalatha V., Sivaraman P., et al. (2004). Genetic variability of begomoviruses associated with cotton leaf curl disease originating from India. Arch. Virol. 149 2047–2057. 10.1007/s00705-004-0352-5 [DOI] [PubMed] [Google Scholar]
- Kranthi K. R. (2014). “Cotton leaf curl virus time bomb,” in Cotton Statistics and News, ed. Singh A. (Mumbai: Cotton Association of India; ). [Google Scholar]
- Kranthi K. R. (2015). Technologies are breaking down -What next? Cotton Statistics and News. [Google Scholar]
- Kumar J., Gunapati S., Alok A., Lalit A., Gadre R., Sharma N. C., et al. (2015). Cotton leaf curl Burewala virus with intact or mutant transcriptional activator proteins: complexity of cotton leaf curl disease. Arch. Virol. 160 1219–1228. 10.1007/s00705-015-2384-4 [DOI] [PubMed] [Google Scholar]
- Leke W. N., Khatabi B., Mignouna D. B., Brown J. K., Fondong V. N. (2016). Complete genome sequence of a new bipartite begomovirus infecting cotton in the Republic of Benin in West Africa. Arch. Virol. 161 2329–2333. 10.1007/s00705-016-2894-8 [DOI] [PubMed] [Google Scholar]
- Maloy O. C. (1997). White pine blister rust control in North America: a case history. Annu. Rev. Phytopathol. 35 87–109. 10.1146/annurev.phyto.35.1.87 [DOI] [PubMed] [Google Scholar]
- Mansoor S., Briddon R. W., Bull S. E., Bedford I. D., Bashir A., Hussain M., et al. (2003). Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA β. Arch. Virol. 148 1969–1986. 10.1007/s00705-003-0149-y [DOI] [PubMed] [Google Scholar]
- Mansoor S., Khan S. H., Bashir A., Saeed M., Zafar Y., Malik K. A., et al. (1999). Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology 259 190–199. 10.1006/viro.1999.9766 [DOI] [PubMed] [Google Scholar]
- Manzoor M. T., Ilyas M., Shafiq M., Haider M. S., Shahid A. A., Briddon R. W. (2014). A distinct strain of chickpea chlorotic dwarf virus (genus Mastrevirus, family Geminiviridae) identified in cotton plants affected by leaf curl disease. Arch. Virol. 159 1217–1221. 10.1007/s00705-013-1911-4 [DOI] [PubMed] [Google Scholar]
- Mizutani T., Daryono B. S., Ikegami M., Natsuaki K. T. (2011). First report of Tomato leaf curl New Delhi virus infecting cucumber in central java, Indonesia. Plant Dis. 95 1485 10.1094/pdis-03-11-0196 [DOI] [PubMed] [Google Scholar]
- Mnari-Hattab M., Zammouri S., Belkadhi M. S., Doña D. B., ben Nahia E., Hajlaoui M. R. (2015). First report of Tomato leaf curl New Delhi virus infecting cucurbits in Tunisia. New Dis. Rep. 31:21 10.5197/j.2044-0588.2015.031.021 [DOI] [Google Scholar]
- Monga D., Raj S., Mayee C. D. (2004). “Strategies for cotton leaf curl virus disease management,” in Proceedings of the National Symposium on Changing(World )Order-Cotton Research Development and Policy in Context, (Hisar: CCS Haryana Agricultural University; ), 205–213. [Google Scholar]
- Nawaz-ul-Rehman M. S., Briddon R. W., Fauquet C. M. (2012). A melting pot of old world begomoviruses and their satellites infecting a collection of Gossypium species in Pakistan. PLoS ONE 7:e40050 10.1371/journal.pone.0040050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam M., Beachy R. N., Fauquet C. M. (1995). Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J. Gen. Virol. 76 25–35. 10.1099/0022-1317-76-1-25 [DOI] [PubMed] [Google Scholar]
- Panno S., Iacono G., Davino M., Marchione S., Zappardo V., Bella P., et al. (2016). First report of Tomato leaf curl New Delhi virus affecting zucchini squash in an important horticultural area of southern Italy. New Dis. Rep. 33:6 10.5197/j.2044-0588.2016.033.006 [DOI] [Google Scholar]
- Pons X., Lumbierres B., Lopez C., Albajes R. (2005). Abundance of non-target pests in transgenic Bt-maize: a farm scale study. Eur. J. Entomol. 102 73–79. 10.14411/eje.2005.010 [DOI] [Google Scholar]
- R Development Core Team. (2016). R: A Language and Environment for Statistical Computing. Vienna: R foundation for statistical computing. [Google Scholar]
- Rajagopalan P. A., Naik A., Katturi P., Kurulekar M., Kankanallu R. S., Anandalakshmi R. (2012). Dominance of resistance-breaking cotton leaf curl Burewala virus (CLCuBuV) in northwestern India. Arch. Virol. 157 855–868. 10.1007/s00705-012-1225-y [DOI] [PubMed] [Google Scholar]
- Rana V. S., Singh S. T., Priya N. G., Kumar J., Rajagopal R. (2012). Arsenophonus GroEL interacts with CLCuV and is localized in midgut and salivary gland of whitefly B. tabaci. PLoS ONE 7:e42168 10.1371/journal.pone.0042168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M. L., Simón A., Velasco L., García M. C., Janssen D. (2015). First report of Tomato leaf curl New Delhi virus infecting tomato in Spain. Plant Dis. 99:894 10.1094/pdis-10-14-1072-pdn [DOI] [Google Scholar]
- Saeed M. (2010). Tomato leaf curl virus and Cotton leaf curl Multan betasatellite can cause mild transient symptoms in cotton. Australas. Plant Dis. Notes 5 58–60. 10.1071/DN100211833-928X/10/010058 [DOI] [PubMed] [Google Scholar]
- Saeed M., Zafar Y., Randles J. W., Rezaian M. A. (2007). A monopartite begomovirus-associated DNA b satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 88 2881–2889. 10.1099/vir.0.83049-0 [DOI] [PubMed] [Google Scholar]
- Saleem H., Nahid N., Shakir S., Ijaz S., Murtaza G., Khan A. A., et al. (2016). Diversity, mutation and recombination analysis of cotton leaf curl geminiviruses. PLoS ONE 11:e0151161 10.1371/journal.pone.0151161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar M. N., Kvarnheden A., Saeed M., Briddon R. W. (2013). Cotton leaf curl disease – an emerging threat to cotton production worldwide. J. Gen. Virol. 94 695–710. 10.1099/vir.0.049627-0 [DOI] [PubMed] [Google Scholar]
- Singh A. K., Kushwaha N., Chakraborty S. (2016). Synergistic interaction among begomoviruses leads to the suppression of host defense-related gene expression and breakdown of resistance in chilli. Appl. Microbiol. Biotechnol. 100 4035–4049. 10.1007/s00253-015-7279-5 [DOI] [PubMed] [Google Scholar]
- Tahir M. N., Amin I., Briddon R. W., Mansoor S. (2011). The merging of two dynasties - identification of an African cotton leaf curl disease-associated begomovirus with cotton in Pakistan. PLoS ONE 6:e20366 10.1371/journal.pone.0020366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I., Potter C. (2010). ‘Too little, too late’? Science, policy and Dutch elm disease in the UK. J. Hist. Geogr. 36 121–131. 10.1016/j.jhg.2009.07.003 [DOI] [Google Scholar]
- White M. T., Griffin J. T., Churcher T. S., Ferguson N. M., Basanez M. G. (2011). Modelling the impact of vector control interventions on Anopheles gambiae population dynamics. Parasit. Vectors 4:153 10.1186/1756-3305-4-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani-Khameneh S., Aboutorabi S., Shoori M., Aghazadeh A., Jahanshahi P., Golnaraghi A., et al. (2016). Natural occurrence of Tomato leaf curl New Delhi virus in Iranian cucurbit crops. Plant Pathol. J. 32 201–208. 10.5423/ppj.oa.10.2015.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S.-E.-A., Iqbal Z., Amin I., Mansoor S. (2015). First report of Tomato leaf curl Gujarat virus, a bipartite begomovirus on cotton showing leaf curl symptoms in Pakistan. Plant Dis. 99 1655 10.1094/PDIS-02-15-0195-PDN [DOI] [Google Scholar]
- Zaidi S.-E.-A., Shafiq M., Amin I., Scheffler B. E., Scheffler J. A., Briddon R. W., et al. (2016). Frequent occurrence of Tomato Leaf Curl New Delhi Virus in cotton leaf curl disease affected cotton in Pakistan. PLoS ONE 11:e0155520 10.1371/journal.pone.0155520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. (2013). Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 51 357–381. 10.1146/annurev-phyto-082712-102234 [DOI] [PubMed] [Google Scholar]
- Zhou X., Liu Y., Robinson D. J., Harrison B. D. (1998). Four DNA-A variants among Pakistani isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J. Gen. Virol. 79 915–923. 10.1099/0022-1317-79-4-915 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


