FIG 7 .
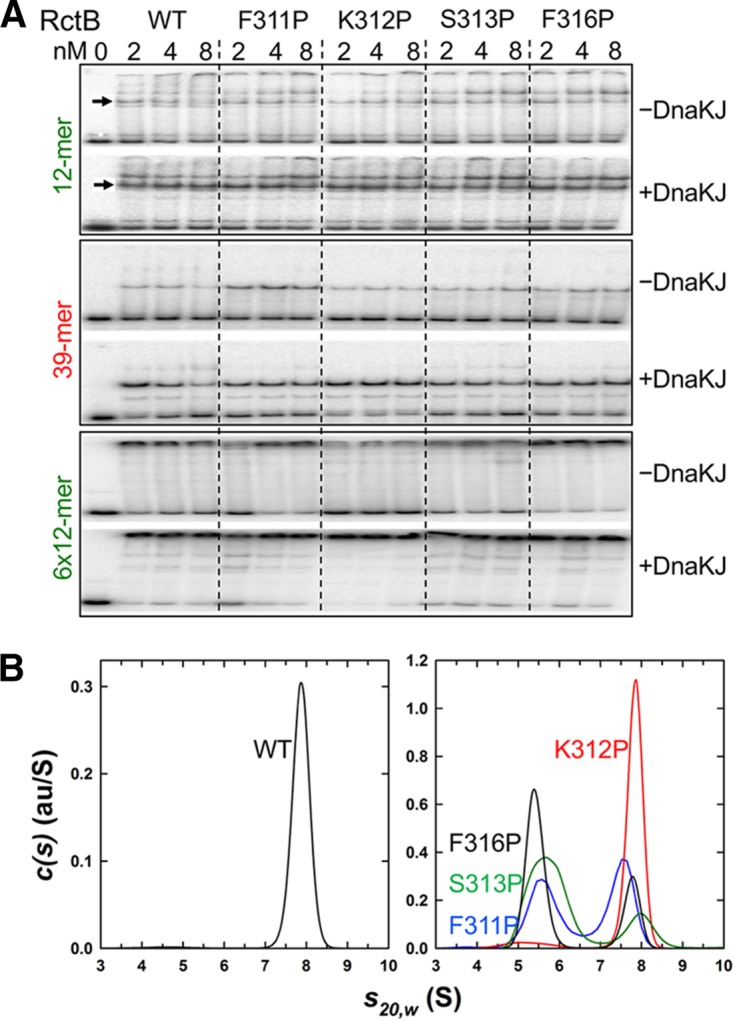
DNA binding and dimerization of representative RctB mutants with residues changed at the dimerization interface. (A) EMSA. Four mutants, F311P, K312P, S313P, and F316P, were studied for binding to a single 12-mer, a single 39-mer, and an array of 6×12-mers in the absence and presence of DnaKJ. (B) Sedimentation velocity absorbance c(s) profile for WT MBP-RctB at 0.9 μM based on data collected at 280 nm (left panel) and for its derivatives F311P (blue), K312P (red), S313P (green), and F316P (black) at 0.4 μM based on data collected at 230 nm (right panel). The c(s) profiles of WT MBP-RctB at multiple concentrations show the presence of a single dimeric species at 7.90 S having an estimated molar mass of 215 kDa. At loading concentrations of 0.4 (shown) and 1.2 μM, the c(s) profiles for the samples represented in the right panel indicate the presence of a reversible monomer-dimer equilibrium with the presence of both the MBP-RctB monomer and dimer. au/S, absorbance units per Svedberg.
