Abstract
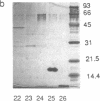
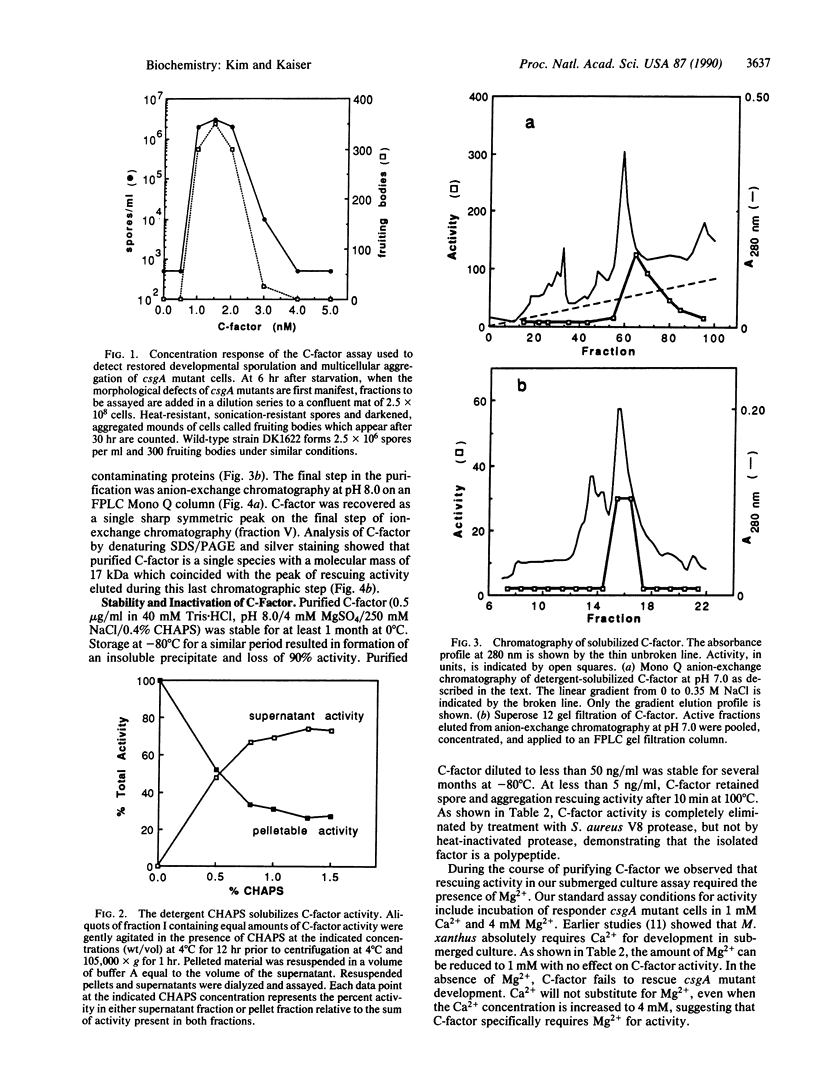
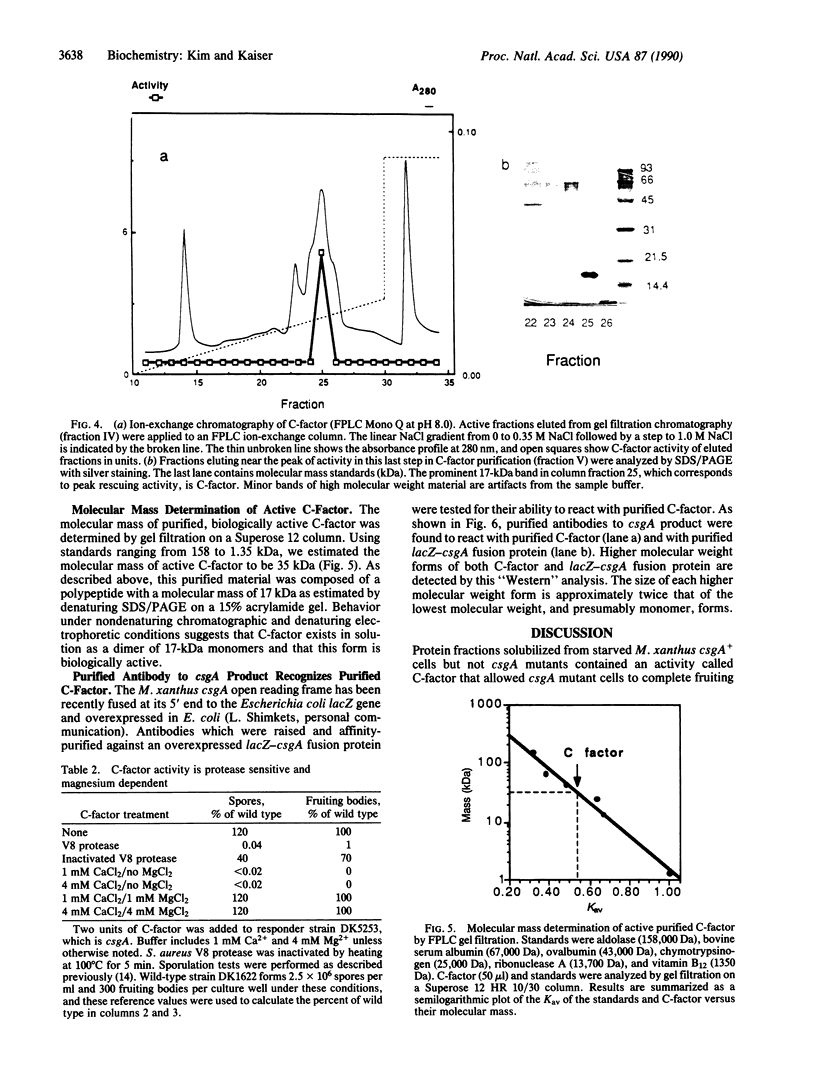
C-factor, a Myxococcus xanthus protein that restores the developmental defects of a class of nonautonomous mutants resulting from mutation of the csgA gene, has been purified approximately 1000-fold from starved wild-type cells. The monomeric form of C-factor is a single polypeptide with a molecular mass of 17 kDa that can be solubilized by detergent from membrane components. Characterization by gel filtration and denaturing gel electrophoresis suggests that biologically active C-factor is a dimer composed of two 17-kDa monomers. Antibodies against a form of the M. xanthus csgA gene product overexpressed in Escherichia coli react with purified C-factor.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Doe C. Q., Goodman C. S. Early events in insect neurogenesis. II. The role of cell interactions and cell lineage in the determination of neuronal precursor cells. Dev Biol. 1985 Sep;111(1):206–219. doi: 10.1016/0012-1606(85)90446-4. [DOI] [PubMed] [Google Scholar]
- Ey P. L., Ashman L. K. The use of alkaline phosphatase-conjugated anti-immunoglobulin with immunoblots for determining the specificity of monoclonal antibodies to protein mixtures. Methods Enzymol. 1986;121:497–509. doi: 10.1016/0076-6879(86)21050-2. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Hagen T. J., Shimkets L. J. Nucleotide sequence and transcriptional products of the csg locus of Myxococcus xanthus. J Bacteriol. 1990 Jan;172(1):15–23. doi: 10.1128/jb.172.1.15-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. R., Dworkin M. Cell-cell interactions in developmental lysis of Myxococcus xanthus. Dev Biol. 1985 Nov;112(1):194–202. doi: 10.1016/0012-1606(85)90133-2. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987 Oct;1(8):840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kuspa A., Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Kuner J. M., Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982 Jul;151(1):458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRossa R., Kuner J., Hagen D., Manoil C., Kaiser D. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J Bacteriol. 1983 Mar;153(3):1394–1404. doi: 10.1128/jb.153.3.1394-1404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J., Asher S. J. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol Gen Genet. 1988 Jan;211(1):63–71. doi: 10.1007/BF00338394. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J., Gill R. E., Kaiser D. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Kaiser D. Murein components rescue developmental sporulation of Myxococcus xanthus. J Bacteriol. 1982 Oct;152(1):462–470. doi: 10.1128/jb.152.1.462-470.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wireman J. W., Dworkin M. Morphogenesis and developmental interactions in myxobacteria. Science. 1975 Aug 15;189(4202):516–523. doi: 10.1126/science.806967. [DOI] [PubMed] [Google Scholar]