Abstract
Metastasis is a dynamic process in which cancer cells navigate the tumor microenvironment, largely guided by external chemical and mechanical cues. Our current understanding of metastatic cell migration has relied primarily on studies of single cell migration, most of which have been performed using two-dimensional (2D) cell culture techniques and, more recently, using three-dimensional (3D) scaffolds. However, the current paradigm focused on single cell movements is shifting toward the idea that collective migration is likely one of the primary modes of migration during metastasis of many solid tumors. Not surprisingly, the mechanics of collective migration differ significantly from single cell movements. As such, techniques must be developed that enable in-depth analysis of collective migration, and those for examining single cell migration should be adopted and modified to study collective migration to allow for accurate comparison of the two. In this review, we will describe engineering approaches for studying metastatic migration, both single cell and collective, and how these approaches have yielded significant insight into the mechanics governing each process.
Introduction
Tissue invasion by tumor cells is a critical, early step in metastasis, involving a breach of the basement membrane and subsequent migration through the stroma [1–3]. Following invasion, further steps in the metastatic cascade such as intravasation of tumor cells into the vasculature [4] and extravasation of cells in the circulation into surrounding tissue (Fig. 1) [5,6] can occur, ultimately resulting in the spread of tumor cells throughout the circulatory and/or lymphatic system and into distant secondary sites.
Fig. 1.
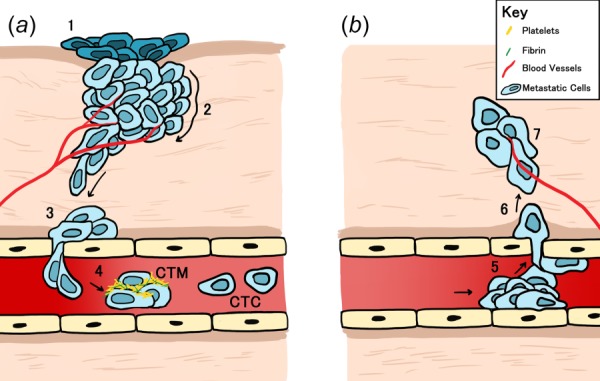
Cartoon depiction of the metastatic process. (a) To metastasize, cells in the (1) primary tumor, located at position 1; (2) separate and undergo EMT; (3) invading through local tissues surrounding the initial lesion before (4) intravasating from the basement membrane into the vasculature or lymphatic system. Metastatic cells then begin to travel as CTCs or CTM through the vasculature. (b) Cells in the metastatic cluster (5) adhere to the basement membrane and then (6) exit at a distal location in a process called extravasation to (7) form a tumor at a secondary site. Arrows indicate direction of migration.
Successful invasion is largely dependent on the migration of cancer cells through the extracellular matrix (ECM). In general, epithelial cells undergo a transition to a mesenchymal state to adopt a migratory phenotype, a process termed epithelial–mesenchymal transition [7–9]. The mechanism of migration employed by cells following this transition can be grouped into one of the two broad categories: single cell (Figs. 2(a) and 2(b)) and collective migration (Fig. 2(c)), where cells move individually or as an assembly of multiple cells, respectively [10]. Following entry into the vasculature, single cells can circulate individually as circulating tumor cells (CTCs) or in aggregates as circulating tumor microemboli (CTM) [11] (Fig. 1). Though recent evidence suggests that collective migration is the more dominant form of migration and CTM are the more aggressive form in circulation, often linked to poor clinical prognosis when seen in certain cancers [12,13], most published reports have focused on the mechanisms governing single cell migration and CTC dissemination [14,15]. Going forward, it is important to continue to broaden the scope of current research to include both modes of migration, with special consideration for the specific signaling and mechanical processes that govern each.
Fig. 2.
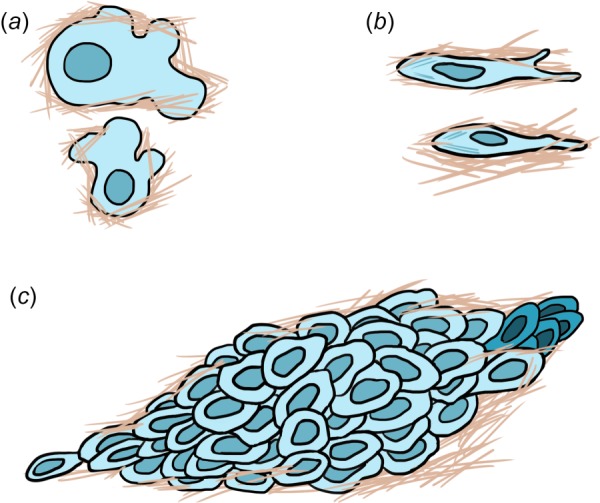
Mechanisms of cell migration. Modes of single cell migration include (a) amoeboid, characterized by blebbing, weak adhesions, and rapid polarity and (b) mesenchymal, characterized by strong stress fibers, polarization, and the presence of a leading and trailing edge. (c) Collective migration consists of a connected unit of cells, fronted by a select few leader cells (indicated by darker cells to the far right).
Single Cell Migration
Single cells utilize two main modes of migration, amoeboid (Fig. 2(a)) and mesenchymal (Fig. 2(b)). Amoeboid migration is characterized by blebbing, weak adhesions, and rapid motility whereas mesenchymal migration is characterized by strong stress fibers, polarization, and a leading and trailing edge [10,16] (Table 1). Amoeboid migration is primarily employed by cells moving through paths and pores in the ECM, as the cells deform through the pores and fibers. In contrast, mesenchymal migration results in the generation or expansion of paths as cells degrade the surrounding matrix [17]. Notably, certain cell types may alternate between migration modes at different points in time as they navigate the microenvironment [17,18]. The migration mode utilized by a cell is partly mediated by its adhesivity to its matrix, which is in turn controlled in part by the architecture, porosity, composition, and mechanical properties of the ECM. For example, actomyosin activity, intrinsic to the cell, is responsible for generating key mechanical signals from the ECM to the cell, and it results in the generation of contractile force within the cytoskeleton that is transmitted to adhesion complexes linking cells to their surroundings to facilitate movement along and within the matrix [19,20]. These adhesion complexes are essential to mesenchymal migration and are less critical during amoeboid migration. Multiple signaling pathways involving molecules such as Rho GTPases coordinate these intracellular force-generating processes and enable individual cells to migrate in a polarized, directed fashion [19]. The tuning of cell migration state is more thoroughly discussed by Friedl and Wolf, who described both intrinsic and extrinsic factors driving various migration modes [21].
Table 1.
Comparison of key features of single and collective migration modes
| Migration category | Circulating form | Mode of migration | Key features | References |
|---|---|---|---|---|
| Single | Circulating tumor cells (CTC) | Mesenchymal | Strong stress fibers, polarization, leading/trailing edges | [10,16] |
| Ameboid | Blebbing, weak adhesions, rapid motility | [17] | ||
| Collective | Circulating tumor microemboli (CTM) | Sheets, strands, tubes, clusters | Intact cell–cell junctions | [11–13] |
Two-Dimensional Versus 3D Models and Methods to Study Single Cell Migration.
In vitro analysis of single cell migration has consisted of various different two-dimensional (2D) and three-dimensional (3D) systems, ranging from platforms as straightforward as 2D glass or plastic to sophisticated 3D synthetic hydrogels and microfabricated models (Fig. 3(a)) [22–24]. Here, we briefly address frequently utilized systems for experimentally assessing single cell migration to lay the groundwork for comparison to more recent models of collective behaviors in metastasis.
Fig. 3.
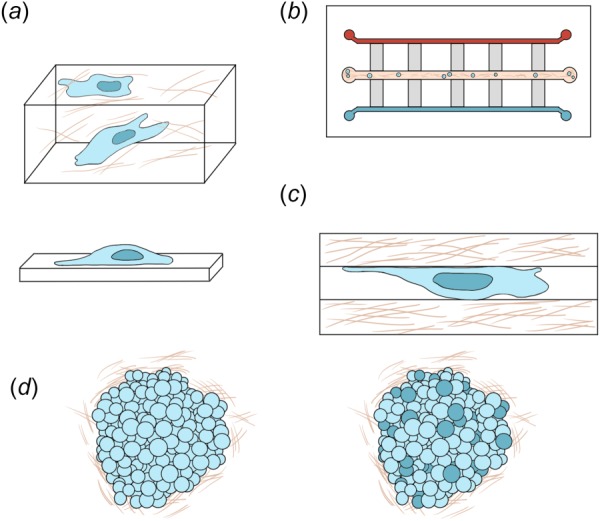
Cartoon representation of single and collective migration assays. (a) Matrix models for studying single cell migration, where cells are either seeded on 2D substrates (bottom) or embedded within 3D substrates (top). (b) Microfluidic model for studying single cell migration. Generally, the horizontal center channel is seeded with metastatically invasive cells in an ECM matrix and the outer horizontal channels are filled with varying types of media or chemogradients. While the vertical channels are used to study the migration characteristics of the cells. (c) Microtrack model for studying single cell migration. (d) Spheroid (left) and organoid (right) models for studying collective migration. Indicated by darker cells in the organoid model represent the heterogeneity of the cells taken from sources such as patient samples, mice, xenografts.
Studies of cell migration have traditionally been conducted using 2D systems in which cells are seeded on the surface of a substrate such as glass, plastic, or a gel [25–28]. Using these platforms, recent work has focused on the mechanical cues guiding cellular behavior [29–37] and the role of cellular mechanics in facilitating migration. Notably, measurements of cellular mechanical properties including cell stiffness and traction stress generation are much easier to perform using 2D platforms compared to 3D scaffolds [38]. However, measurements of cell deformability, as an example, are likely more important in 3D scaffolds because of the need for cells to squeeze through 3D spaces. As such, many of the mechanical probing techniques initially developed and utilized in 2D platforms have been modified for use in 3D scaffolds. However, one of the most significant remaining challenges in the field of cell mechanics is reconciling the results obtained using 2D platforms with the behaviors and properties of cells in 3D. As part of that challenge, measurement techniques that can be applied in both systems are required.
Two-Dimensional Platforms.
Despite the frequency with which they have and continue to be utilized for migration studies, 2D models have significant limitations, primarily owing to the fact that migration in vivo typically occurs in a 3D space. More recent work has shown that topography and dimensionality, among other parameters [25,39], play a critical role in determining migratory phenotype and the mechanical responses of cells. Micromolded and micromachined 2D surfaces [40] as well as tunably compliant 2D substrates such as polyacrylamide (PA) [39,41] or collagen gels [42–44] partially resolve the limitations of 2D systems by allowing for tuning of topography and stiffness, but still do not entirely account for dimensional cues from the environment. As a result, numerous 3D models have been developed to address these and other shortcomings of 2D systems. Confocal microscopy has been integral in the translation of 2D mechanics measurements into 3D as it allows for 3D imaging of the cell and its surroundings [45,46].
Three-Dimensional Collagen-Based Platforms.
Collagen is used frequently in in vitro models as it is very prevalent in many solid tumor microenvironments [47] and its fibrous architecture can be tuned to resemble that of the microenvironment present in many solid tumors [43,48]. It is also possible to tune the stiffness, porosity, and density of collagen by varying the polymerization kinetics or adding in crosslinking agents [49,50]. There are numerous other polymer systems that have been developed, including polyethylene glycol (PEG)-based hydrogels. These synthetic gels are inert [23] and possess tunable mechanical properties such as matrix stiffness [51], which enables independent assessment of various cellular processes [20].
Microfluidic Platforms.
To recreate more complicated 3D microenvironments, microfluidics have also been employed. In the tumor microenvironment, growth factors, cytokines, chemokine release, and matrix remodeling can drive irregular intercellular communication [52]. Microfluidic models, sometimes referred to as organ-on-a-chip devices [53], utilize 2D and 3D constructs in concert with channels, micromixers, and microwells to mimic the changing mechanical and chemical tumor microenvironment (Fig. 3(b)). As a result of recent advances in microtechnology and rapid prototyping, microfluidic devices can be fairly straightforward to design and construct [54]. Microfluidics, as opposed to conventional cell culture techniques, allows for the manipulation of fluids that are held within a device that can range from a few millimeters down to micron scale. Such small scales offer a number of advantages including cost effectiveness, low consumption of reagents, high sensitivity, high spatiotemporal resolution, and controlled laminar flow [55]. Because of these advantages, microfluidic devices have been highly valuable to the study of cellular migration and invasion.
Two-Dimensional Versus 3D Metastatic Movement Mechanics.
With the advent of more tractable 3D models for cell migration, subsequent studies have focused on describing the differences in migration in 2D versus 3D environments [56–59]. Though both types of migration involve the interaction of the cell with its environment and a heavy dependence on the cytoskeleton for efficient migration, the manner in which it occurs differs. For most cells migrating in 2D environments, protrusions at the leading edge are formed as a result of actin polymerization, whereas retraction of the trailing edge is controlled by contractile forces generated by myosin motors [20,34]. In 3D spaces, movement is generally more complicated, due in part to a significantly more complex cellular geometry and the ability to switch between amoeboid and mesenchymal modes of migration [18]. While Rho-mediated myosin contractility still plays a role in migration through 3D spaces by contributing to such tasks as matrix rearrangement and fiber alignment [60], cells can rely less on protrusions to navigate the matrix. For example, pharmacological inhibition of Rho-associated Protein Kinase (ROCK) activity does not necessarily halt migration in 3D matrices [61], pointing to a fundamentally different role of contractility in 2D versus 3D microenvironments. Although cell-matrix adhesions are still present in cells in a 3D matrix, they are thought to be more dynamic than those seen in 2D [58,62,63]. These data underscore the need to focus more on increasingly complex and more accurate models in studying cell migration mechanics.
Traction Stresses in 2D and 3D.
Cells utilize traction stresses to probe their environment, to move, and to remodel matrix [64,65]; as cells adhere to the matrix, they can deform it. There is significant interest within the biomechanics community on this cellular probing and cell-generated deformations, and how probing can generate signals internal to the cell that lead to behaviors like additional adhesion, focal adhesion maturation, and migration [40,66]. To better understand the role of contractility in migration, traction force microscopy (TFM) was first developed for use on 2D planar substrates [67] and has been widely used as such [68,69]. Microbeads embedded in the upper plane of a deformable substrate are tracked as a cell adheres and migrates, and these displacements are then translated into cell-mediated stresses. Our lab has used 2D systems to investigate differences in traction stresses exerted by metastatic and nonmetastatic cells [70] as well as other stromal cells in the tumor microenvironment [68]. More metastatic cells generate higher traction stresses, as do cells from a tumor stromal environment, compared to their counterparts from healthy tissue. These data point to an altered mechanosensitivity that is a function of metastatic and tumorigenic potential. Two-dimensional TFM methods have also been used to investigate collective motions on 2D substrates. For example, similar studies conducted on PA [71] have concluded that cellular migration is established as a cooperative endeavor that relies on cell–cell junctions across the monolayer, not just the leading edge [69,72].
The 2D traction force microscopy method has since been modified to be used in 3D microenvironments [73]. Confocal microscopy and its increasingly widespread use have facilitated the transition from studies of cell tractions in 2D to 3D. Microbeads embedded in PEG gels or collagen gels [45,74] have been used to map traction fields of embedded cells. Confocal microscopy allows for the cells, the matrix, and fiducial markers in the matrix to be imaged in real-time in 3D space. Additionally, matrix fiber movements can be imaged and tracked as cells exert force against the matrix as an overall metric of cell contractility [64]. While there has yet to be a thorough investigation of metastatic cell traction stresses in 3D matrices, 3D TFM studies indicate that the strongest forces and highest traction values are generated near long, thin cellular protrusions, which may correspond to invading regions [75]. Planar orientation of traction has also been investigated, with results indicating that the exertion of normal forces relative to focal adhesions is a uniquely 2D phenomena; cells embedded in 3D matrices do not appear to generate these rotational moments [73]. Notably, however, the primary focus of these 3D studies has been on single, isolated cells as opposed to cohorts of cells as seen in collective migration, discussed further below.
The Mechanics of Squeezing During Metastatic Cell Migration.
During cancer invasion, tumor cells have the ability to navigate around other cells and through the confines of the ECM, with pores that range from 0.1 to 30 μm [76–78]. To investigate how tumor cells are able to migrate through the ECM with relative ease, groups have developed microfluidic devices that utilize narrowing confined pillar arrays and channels to assess the ability of the tumor cells to deform [79–83]. The general consensus among these studies is that tumor cells have diminished rigidity and cell-surface frictional forces compared to healthy cells that facilitate their movements.
Through their extensive application in vitro, 2D systems have been used to uncover fundamental mechanical principles describing metastatic cell properties, including that metastatic cells are generally softer and more deformable than nonmetastatic cells. Numerous techniques have been used to probe the relationship between cellular deformability and metastatic ability including particle-tracking microrheology, atomic force microscopy (AFM), and cell stretching devices [84–88]. Overall, studies of cell deformability indicate that more metastatic cells are more compliant than their healthy counterparts and that deformability is predictive of cancer staging [89]. A more compliant cell may be better at navigating through a dense 3D environment; however, the molecular mechanisms controlling these rheological differences are not clear. Understanding these molecular mechanisms is critical in identifying therapeutic targets.
Particle-tracking microrheology of the cell cytoplasm, which was originally developed for use in cells plated on 2D substrates, has also been adapted for 3D systems [90–92]. Here, tracer beads are embedded within cells and tracked to determine various mechanical and viscoelastic properties [84] relatively noninvasively and over time [90]. Interestingly, rheological studies indicate that in 3D environments, cells are generally more solidlike whereas cells in 2D exhibit primarily fluidlike behavior [90]. Additionally, the cytoplasm is thought to be more compliant and less dense for cells migrating in 3D as opposed to 2D environments [92]. When investigated in concert with deformable matrices, for example, variations in matrix stiffness were shown to not significantly impact the intracellular rheology of probed cells in 2D but were shown to increase the compliance of cells in 3D matrices [84]. Together these data indicate that cellular deformability in 2D and 3D is not necessarily the same.
Two-dimensional platforms can be easily combined with techniques such as atomic force microscopy (AFM) (as an example) to not only measure the mechanical properties of migrating cells but also assess the cellular response to cellular and matrix deformations that are created using the AFM cantilever [93]. In metastasis research, AFM has been utilized in conjunction with 2D models of migration to study the mechanical properties of migrating cells by investigating and characterizing alterations to the cytoskeleton and ECM [94–98]. Such studies have demonstrated that the cytoskeleton of metastatic cells responds to mechanical loads differently than that of a nonmetastatic cell, as these cells are typically more compliant [99,100].
While there has been significant focus on the overall deformability of cells, work in 3D has suggested that the deformation of the nucleus is a rate-limiting step to a cell's ability to squeeze through pores in 3D matrices. Indeed, recent data show that the nucleus can undergo significant shape changes while cells migrate through confined spaces, and the nucleus moves through more slowly than the rest of the cell body [101–104]. The cells' ability to deform the nucleus is dependent on force generation of integrins and actomyosin [103]. Therefore, while data indicate the deformability of the cytoplasm scales with metastatic potential, it cannot necessarily be decoupled from the intricate network of cytoskeleton that connects it to the cell nucleus.
The Mechanics of Moving in and out of the Vasculature.
During metastasis, tumor cells migrate away from the primary tumor and can enter blood vessels [55,105] and exit at secondary sites through a process called extravasation. However, little is known about the mechanics governing intravasation and extravasation. Microfluidics platforms are ideal for mimicking intravasation and extravasation because they allow for the recreation of blood vessel mimics of varying size with well-defined flows [106–113]. The use of these devices has revealed numerous insights into the mechanical and chemical factors governing metastatic cell movements through the blood vessel wall. For example, a recent study created a microfluidic vascularized network to discover that breast cancer cells extravasate at a higher rate if adenosine receptors are blocked, showcasing the potential of the platform as a drug screening device [108]. Other studies suggest that macrophages are responsible for enabling intravasation by disrupting the integrity of the endothelium through the release of tumor necrosis factor alpha (TNF-α) [106]. The same group has also developed a highly tailored method to mimic bone and muscle microenvironments. The ultimate goal of these platforms is to investigate the seed-and-soil paradigm of extravasation that states certain organs have a propensity for metastatic cancer, suggesting that the spread of cancer to secondary sites depends on the preference and interaction of cancer cells with specific organs [109].
The Role of Matrix Metalloproteases (MMPs) and Metastatic Cell Movement in Microtracks.
In addition to squeezing through the matrix, cells can create paths in the matrix by secreting MMPs. MMPs likely do not play a significant role in 2D migration with regard to physically degrading the matrix to permit migration as they do in 3D, but they likely play a role in cleavage of cell-surface receptors [114–116]. The extent of remodeling and degradation of the matrix in 3D is dependent in part on matrix density and stiffness [117,118]. While it is somewhat intuitive that collagen density mediates cell migration in 3D (more dense gels restrict cell movement), recent data suggest that fiber architecture can dominate cell response [43,65,119]. More specifically, collagen constructs can be made with relatively high densities of collagen. However, if this collagen is polymerized in such a way to create large fibers with large pores, migration is enhanced compared to cells in less dense collagen containing smaller fibers and pores. Pore size and fiber size can be tuned based on temperature and pH at the time of polymerization [120,121]. These data underscore the importance of not only controlling collagen density in experiments but also characterizing the resulting fiber architecture.
In utilizing MMPs to degrade the matrix, the cells generate tunnels as they travel, which can facilitate the migration of subsequent cells through the matrix [19,25,122]. Similarly, cells have been shown to move through interstitial spaces in the matrix without the use of MMPs [115]; in these instances, cells rely on Rho kinases, integrins, and actomyosin to enable entry into pores via deformations to the cytoskeleton [19,104,123]. These tracks in the stroma have also been suggested to promote collective invasion of cohorts of cells by providing a path in which otherwise nonmetastatic cells can move [61]. Importantly, migration in tunnels is notably different than the behaviors observed in 2D, and it also distinct from migration in isotropic 3D microenvironments. Within these tunnels, cells have the ability to adhere to matrix on multiple sides of the cell (analogous to 3D migration), but unlike conventional 3D migration platforms, they do not need to clear a path to move through the matrix.
To mimic these tunnels in an in vitro platform, patterned microtracks (Fig. 3(c)) have been developed utilizing a variety of substrates such as polydimethylsiloxane (PDMS) and PA [81,102,124–128]. These platforms have been essential in understanding the effects of confinement on invasive migration, highlighting the reduced efficiency of myosin, ROCK/RhoA, and integrin inhibitors [81] and an increased dependence on microtubule rather than actin dynamics [19,81]. However, many of these mimetic systems do not fully recapitulate the in vivo microenvironment. PDMS and silicon exhibit stiffness ranges beyond what is detected in tissue [99], and PA, while softer, does not accurately reflect the architecture of the tumor microenvironment [124]. To address these issues, collagen has been utilized as a substrate to generate patterned microtracks of varying mechanical properties that more closely mimic tissue stiffness [126,128].
Micromolded systems have highlighted the impact of cell confinement on properties such as cell stiffness and the deformability of the cytoskeleton and how these in turn affect a cell's ability to migrate [81,124–126]. Cells with lower stiffness and higher deformability exhibit greater velocities and therefore enhanced migration when moving through confined channels [124]. Increasing channel adhesiveness by coating channels with proteins such as fibronectin results in enhanced cell migration [126]. The effects of parameters such as channel dimensions and stiffness of the channel wall have also been investigated in these systems; results show that once the nucleus has deformed sufficiently to enable the cell to enter a channel, migration is enhanced in confined as opposed to unconfined channels, where the width of the channel exceeds the diameter of the cell [124,126,127]. Increasing matrix stiffness also led to enhanced migration in PDMS channels [127], whereas no effect was seen with collagen microtracks when stiffness was manipulated through alterations in density [1]. This relative independence of microtrack migration on collagen density, though significant in 3D isotropic collagen matrices, may simply be a result of steric effects. Preformed tracks involve less physical constraint to invading cells as compared to migration through a matrix, and as such they provide a path where less resistance is imposed on the cells as they migrate [1,104].
In vitro microtrack platforms have been primarily utilized for studying single cell migration, specifically to study how alterations in external properties of the matrix or internal cellular cues affect the ability of individual cells to navigate existing channels. However, it has been suggested that collective cell movements are also facilitated by microtracks [61,129], and as such, adapting these systems for use in studies of collective migration is an important next step in our understanding of metastatic cell movements (Table 2).
Table 2.
Comparison of single and collective migration models and their applications
|
Single Cell Migration Models | ||||
|---|---|---|---|---|
| Migration assay | Key investigative topic | Dimensionality | Substrate | References |
| Matrix model | Single cell migration, polarity, tumor microenvironment | 2D | Glass, plastic, collagen, PA | [25–37,39,40,66] |
| Single cell migration, tumor microenvironment | 3D | Collagen, PEG | [18,45,58,60–63,74] | |
| Microfluidic model | Tumor microenvironment, chemical cues | 2D | PDMS, PA, ECM proteins | [79–83,105–108,110–113] |
| Microtrack model | Single cell migration | Pseudo-3D | Collagen, PA, PDMS, silicon | [81,101,102,124–129] |
Collective Cell Migration
While significant work has been performed in the study of single cell movements, collective migration in cancer has been suggested to potentially be the clinically predominant form of metastatic migration [148]. Recent studies have shown that collectively migrating tumor cells are more aggressive and have higher survivability in response to chemotherapeutics [138,149,150]. Importantly, collective migration is not exclusive to metastatic cancer invasion. It occurs in a variety of contexts in vivo including embryogenesis and wound healing [130,151].
Cells can migrate collectively using a variety of modes including sheets, strands, tubes, and clusters [10]. Collective migration is distinct from single cell migration primarily in that cells remain connected throughout the process, maintaining intact cell–cell junctions during movement and ultimately displaying migratory patterns that are fundamentally different from the migratory patterns of single cells [130,152]. In vivo, collections of tumor cells outgrow, depositing matrix, realigning the matrix fibers, and excreting growth factors to cause mechanical stress and enhanced cellular proliferation [47,153,154]. These multifaceted changes in the ECM cannot be recreated in a 2D model system, and as a result, systems to investigate cellular interactions in 3D are needed. Given the numerous modes of collective migration that occur in vivo, several in vitro platforms have been developed including spheroid models and organoid models (Fig. 3(d)).
Spheroid Model of Collective Migration.
The tumor spheroid model allows for a level of complexity to be recreated that bridges the gap between 2D cellular techniques and tumors in vivo. Tumor spheroids are aggregates of cells that are made in vitro from cell lines or primary cells. Typically, tumor spheroids are made by aggregating and compacting a cell suspension cultured in growth media and Methocult (StemCell Technologies, Vancouver, BC, Canada) in nonadherent 96-well round bottom plates [155]. The spheroid protocol is fairly straightforward and allows for single or multiple cancerous cell lines or primary cells to be incorporated [64]. When embedded in 3D scaffolds, the dynamics of collective metastatic cell migration can be observed and studied.
In the current practice, there are four different classifications of cancer spheroid models commonly described in literature: multicellular tumor spheroids, tumorspheres, tissue-derived tumor spheres, and organotypic multicellular organ spheres [156]. Briefly, multicellular tumor spheroids are derived from cell suspension and used to study cellular migration through the matrix. Tumorspheres, meanwhile, are derived from cancer stem cells and are often used for investigating cancer stemness. Organotypic multicellular organ spheres and tissue-derived tumor spheres are both derived from primary cancer tissue and are often used for characterizing the primary tumor and for drug screening.
Numerous studies have used multicellular tumor spheroids for invasion and migration assays [130–133]. A major benefit of the spheroid model is its capacity to be implanted and incorporated into a number of different types of matrices. Moreover, spheroids have been shown to exhibit similar growth kinetics to those measured in vivo [157]. Spheroid models have been used to show that ECM density and reorganization, in addition to high cell–cell adhesion and MMP secretion rates, are important factors mediating collective cell migration and migration plasticity [135]. During collective migration, cohorts of cells move together, requiring significantly more remodeling of the matrix to accommodate the large size of the aggregate compared to the free space in the stroma.
Interestingly, in comparing the physical mechanisms of single cell migration to those of collective migration, it has been suggested that collective migration may be thermodynamically favorable compared to single cell migration [136]. Collective invasion minimizes energy costs to the cells associated with entering the stroma and coordinating movement with other cells, among other functions. To facilitate movement of the cohort of cells, a dynamic invasive front is utilized, led by a leader cell with various cells in the aggregate assuming the role of the leader during the process of migration [12,136]. The mechanics governing leader–follower cell dynamics are being investigated, but recent work has proposed that migration involves an energy cost to the leader as compared to the followers, due to the energy required to create paths for the collective unit to utilize to navigate the ECM [136]. A switching of leader cells is speculated to occur to minimize the energy costs to any one cell. Leader cells are thought to be the most invasive cells arising from a tumor; as such, being able to study and distinguish leader cells from the surrounding follower cells is critical to our understanding of the drivers of the metastatic process. Importantly, they may also be important clinical targets to disrupt collective movements [137].
Organoid Model of Collective Migration.
In addition to spheroid models, several labs have developed organoid models to investigate collective migration and leader–follower behavior in invasive cancers [137–139]. In the organoid model, tumors are extracted from patient samples, mice, or xenografts and either mechanically or chemically digested into smaller aggregates that are then embedded in extracellular matrix. This methodology offers several advantages over spheroid models, most notably that many of the native tumor features and cellular heterogeneities are retained. These models may be useful for pharmacological testing and have clear implications for personalized medicine.
In the context of migration, organoid models have been very useful in revealing important dynamics about leader–follower behaviors. For example, recent data suggest that leader cells share a molecular phenotype distinct from that of followers [137,139], expressing a number of molecules including keratin-14 [138,158–160], which regulates cell–cell adhesion, cell-ECM adhesion, and cellular invasion [138]. These results suggest that leader cells contain specific markers identifying them as such and that these identifying properties may then be responsible for the ability of a leader cell to invade the surrounding tissue and/or function as a leader.
Previous studies have utilized TFM to examine the role of the leader cell in migration; these have determined that a majority of the traction forces originate from the leading edge of advancing cells [140–145]. However, recent traction force research also shows that however pivotal the leader cells are in migration, a large proportion of the migration forces are originating from the region between the tumor and the leader cell [69,72].
Both the spheroid and organoid models have also been vital in understanding the impact of cell–cell interactions in the tumor microenvironment [147,156]. Cocultured spheroids consisting of MCF-10A mammary epithelial cells and MDA-MB-231 malignant breast cells have been used to study the leader and follower roles often observed in collective migration. Interestingly, in this model, malignant cells induce the collective invasion of epithelial cells considered to not be invasive [61]. The transformation from noninvasive to invasive phenotype was also observed in cells exposed to senescence-associated secretory phenotype (SASP), which inhibits ROCK/RhoA and promotes migration [161].
In many ways, the spheroid and organoid models come closer to mimicking the in vivo environment than do systems with single cells embedded in collagen. They allow for collective or single cell migration to occur and enable investigation of the role of the microenvironment in dictating the mode of migration rather than prescribing the mode a priori based on the experimental platform. To create even more in vivo-like environments, additional recent work has focused on the interaction between malignant cells and the adjacent vasculature by creating an in vitro model containing a model blood vessel and spheroid embedded in collagen [147]. These more sophisticated systems will enable a deeper understanding of the mechanical and chemical communication occurring across cell types in the tumor microenvironment.
Concluding Thoughts and Future Directions
Mechanics apparently plays a clear role in cancer progression, but there is still much to be understood about how mechanical changes contribute to cancer and how therapeutic interventions may be designed to target mechanics. Numerous in vitro models and mechanical measurement tools have emerged for studying single cell and collective cell movements in cancer. By improving these techniques and more accurately modeling migration, we can further develop our hypotheses on the mechanical forces and properties that govern migration, potentially leading to the development of tools that more effectively inhibit metastatic cancer.
Using these current models, engineers and biologists have uncovered critical roles for cell and matrix mechanics in guiding cell movements. It is becoming clear that collective migration is a major, if not the dominating, aspect of the dissemination of cancer cells throughout the body and that the advent of 3D cell culture techniques and advanced imaging will continue to produce additional insights into the molecular and mechanical mechanisms governing collective migration in metastasis. Three-dimensional cell culture techniques for studying collective migration provide a level of control that cannot be obtained with in vivo models, and they recapitulate cellular behaviors that closely mimic in vivo tumors [134,157].
Since collective motion occurs in development and wound healing and has been widely studied in these contexts, it is likely that there is much we can learn about collective migration in metastasis from these other fields. The role of cell–cell adhesions, mechanical cues, cell deformation, and so-called “tissue tectonics” are likely features that translate across both fields [162–164]. At the same time, however, it is important to note that numerous distinct differences exist between development and cancer which necessitate that collective migration in cancer and development each be studied independently (as opposed to assuming that because a certain finding is true in collective movement in development that it is also true in cancer).
The transition of collective movements into widespread dissemination in the metastatic cascade remains an open question. The role of follower cells as opposed to leader cells is unclear. Moreover, it is not yet clear whether the leader cell is predetermined to be a leader or simply changes its phenotype into that of a leader due to its location in the migrating strand. Microenvironmental context becomes key in answering these questions since one might expect that the leader cell's contact with a certain ECM type, architecture, composition, or stiffness may alter its ability to lead or, alternatively, induce its leader phenotype.
Ultimately, our understanding of single cells and collective movements will help guide the development of therapeutics to prevent metastasis. Based on emerging data, it is clear that the same molecular pathways may not be equally important in both modes, and suggests that it may be important clinically to distinguish between the modes of dissemination in a given patient for effective targeting of the relevant pathways. Additionally, our understanding of the role of cell and matrix mechanics in collective migration in metastasis is continuing to increase and it is not impossible to conceive that one could ultimately develop targets against mechanics and mechanical properties that promote disease in an emerging field of mechanomedicine.
Acknowledgment
This work was supported in part by grants from the National Science Foundation (Award numbers 1055502 and 1233827) and the National Heart, Lung and Blood Institute (Award number HL 127499) to CAR, a Sloan Foundation Fellowship to ML and an NIH diversity supplement (HL127499 02S) to AM.
Contributor Information
Marianne Lintz, The Nancy E. and Peter C. Meinig , School of Biomedical Engineering, , Cornell University, , 309 Weill Hall, , Ithaca, NY 14853.
Adam Muñoz, The Nancy E. and Peter C. Meinig , School of Biomedical Engineering, , Cornell University, , 309 Weill Hall, , Ithaca, NY 14853.
Cynthia A. Reinhart-King, The Nancy E. and Peter C. Meinig , School of Biomedical Engineering, , Cornell University, , 302 Weill Hall, , Ithaca, NY 14853 , e-mail: cak57@cornell.edu.
References
- [1]. Carey, S. P. , Rahman, A. , Kraning-Rush, C. M. , Romero, B. , Somasegar, S. , Torre, O. M. , Williams, R. M. , and Reinhart-King, C. A. , 2015, “ Comparative Mechanisms of Cancer Cell Migration Through 3D Matrix and Physiological Microtracks,” Am. J. Physiol.: Cell Physiol., 308(6), pp. C436–447. 10.1152/ajpcell.00225.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Carey, S. P. , D'Alfonso, T. M. , Shin, S. J. , and Reinhart-King, C. A. , 2012, “ Mechanobiology of Tumor Invasion: Engineering Meets Oncology,” Crit. Rev. Oncol. Hematol., 83(2), pp. 170–183. 10.1016/j.critrevonc.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Liotta, L. A. , and Stetler-Stevenson, W. G. , 1991, “ Tumor Invasion and Metastasis: An Imbalance of Positive and Negative Regulation,” Cancer Res., 51(Suppl. 18), pp. 5054s–5059s.http://cancerres.aacrjournals.org/content/51/18_Supplement/5054s.full-text.pdf [PubMed] [Google Scholar]
- [4]. Chiang, S. P. H. , Cabrera, R. M. , and Segall, J. E. , 2016, “ Tumor Cell Intravasation. A Review in the Theme: Cell and Molecular Processes in Cancer Metastasis,” Am. J. Physiol.: Cell Physiol., 311(1), pp. C1–C14. 10.1152/ajpcell.00238.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Bersini, S. , Jeon, J. S. , Moretti, M. , and Kamm, R. D. , 2014, “ In Vitro Models of the Metastatic Cascade: From Local Invasion to Extravasation,” Drug Discovery Today, 19(6), pp. 735–742. 10.1016/j.drudis.2013.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Koop, S. , Schmidt, E. E. , MacDonald, I. C. , Morris, V. L. , Khokha, R. , Grattan, M. , Leone, J. , Chambers, A. F. , and Groom, A. C. , 1996, “ Independence of Metastatic Ability and Extravasation: Metastatic Ras-Transformed and Control Fibroblasts Extravasate Equally Well,” Proc. Natl. Acad. Sci. U.S.A., 93(20), pp. 11080–11084. 10.1073/pnas.93.20.11080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Thiery, J. P. , Acloque, H. , Huang, R. Y. J. , and Nieto, M. A. , 2009, “ Epithelial-Mesenchymal Transitions in Development and Disease,” Cell, 139(5), pp. 871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- [8]. Thiery, J. P. , and Lim, C. T. , 2013, “ Tumor Dissemination: An EMT Affair,” Cancer Cell, 23(3), pp. 272–273. 10.1016/j.ccr.2013.03.004 [DOI] [PubMed] [Google Scholar]
- [9]. Nieto, M. A. , Huang, R. Y.-J. , Jackson, R. A. , and Thiery, J. P. , 2016, “ EMT: 2016,” Cell, 166(1), pp. 21–45. 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- [10]. Friedl, P. , 2004, “ Prespecification and Plasticity: Shifting Mechanisms of Cell Migration,” Curr. Opin. Cell Biol., 16(1), pp. 14–23. 10.1016/j.ceb.2003.11.001 [DOI] [PubMed] [Google Scholar]
- [11]. Duda, D. G. , Duyverman, A. M. M. J. , Kohno, M. , Snuderl, M. , Steller, E. J. A. , Fukumura, D. , and Jain, R. K. , 2010, “ Malignant Cells Facilitate Lung Metastasis by Bringing Their Own Soil,” Proc. Natl. Acad. Sci. U.S.A., 107(50), pp. 21677–21682. 10.1073/pnas.1016234107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Gaggioli, C. , Hooper, S. , Hidalgo-Carcedo, C. , Grosse, R. , Marshall, J. F. , Harrington, K. , and Sahai, E. , 2007, “ Fibroblast-Led Collective Invasion of Carcinoma Cells With Differing Roles for RhoGTPases in Leading and Following Cells,” Nat. Cell Biol., 9(12), pp. 1392–1400. 10.1038/ncb1658 [DOI] [PubMed] [Google Scholar]
- [13]. Hou, J. M. , Krebs, M. G. , Lancashire, L. , Sloane, R. , Backen, A. , Swain, R. K. , Priest, L. J. C. , Greystoke, A. , Zhou, C. , Morris, K. , Ward, T. , Blackhall, F. H. , and Dive, C. , 2012, “ Clinical Significance and Molecular Characteristics of Circulating Tumor Cells and Circulating Tumor Microemboli in Patients With Small-Cell Lung Cancer,” J. Clin. Oncol., 30(5), pp. 525–532. 10.1200/JCO.2010.33.3716 [DOI] [PubMed] [Google Scholar]
- [14]. Kang, Y. , and Pantel, K. , 2013, “ Tumor Cell Dissemination: Emerging Biological Insights From Animal Models and Cancer Patients,” Cancer Cell, 23(5), pp. 573–581. 10.1016/j.ccr.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Braun, S. , and Naume, B. , 2005, “ Circulating and Disseminated Tumor Cells,” J. Clin. Oncol., 23(8), pp. 1623–1626. 10.1200/JCO.2005.10.073 [DOI] [PubMed] [Google Scholar]
- [16]. Huttenlocher, A. , and Horwitz, A. R. , 2011, “ Integrins in Cell Migration,” Cold Spring Harbor Perspect. Biol., 3(9), p. a005074. 10.1101/cshperspect.a005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Hecht, I. , Bar-El, Y. , Balmer, F. , Natan, S. , Tsarfaty, I. , Schweitzer, F. , and Ben-Jacob, E. , 2015, “ Tumor Invasion Optimization by Mesenchymal-Amoeboid Heterogeneity,” Sci. Rep., 5, p. 10622. 10.1038/srep10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Panková, K. , Rösel, D. , Novotný, M. , and Brábek, J. , 2010, “ The Molecular Mechanisms of Transition Between Mesenchymal and Amoeboid Invasiveness in Tumor Cells,” Cell. Mol. Life Sci., 67(1), pp. 63–71. 10.1007/s00018-009-0132-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Mak, M. , Spill, F. , Kamm, R. D. , and Zaman, M. H. , 2016, “ Single-Cell Migration in Complex Microenvironments: Mechanics and Signaling Dynamics,” ASME J. Biomech. Eng., 138(2), p. 21004. 10.1115/1.4032188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Chi, Q. , Yin, T. , Gregersen, H. , Deng, X. , Fan, Y. , Zhao, J. , Liao, D. , and Wang, G. , 2014, “ Rear Actomyosin Contractility-Driven Directional Cell Migration in Three-Dimensional Matrices: A Mechano-Chemical Coupling Mechanism,” J. R. Soc., Interface, 11(95), p. 20131072. 10.1038/nrm2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Friedl, P. , and Wolf, K. , 2010, “ Plasticity of Cell Migration: A Multiscale Tuning Model,” J. Cell Biol., 188(1), pp. 11–19. 10.1007/s00418-008-0529-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Gupton, S. L. , and Waterman-Storer, C. M. , 2006, “ Spatiotemporal Feedback Between Actomyosin and Focal-Adhesion Systems Optimizes Rapid Cell Migration,” Cell, 125(7), pp. 1361–1374. 10.1016/j.cell.2006.05.029 [DOI] [PubMed] [Google Scholar]
- [23]. Raeber, G. P. , Lutolf, M. P. , and Hubbell, J. A. , 2005, “ Molecularly Engineered PEG Hydrogels: A Novel Model System for Proteolytically Mediated Cell Migration,” Biophys. J., 89(2), pp. 1374–1388. 10.1529/biophysj.104.050682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Peela, N. , Sam, F. S. , Christenson, W. , Truong, D. , Watson, A. W. , Mouneimne, G. , Ros, R. , and Nikkhah, M. , 2016, “ A Three Dimensional Micropatterned Tumor Model for Breast Cancer Cell Migration Studies,” Biomaterials, 81, pp. 72–83. 10.1016/j.biomaterials.2015.11.039 [DOI] [PubMed] [Google Scholar]
- [25]. Stroka, K. M. , Gu, Z. , Sun, S. X. , and Konstantopoulos, K. , 2014, “ Bioengineering Paradigms for Cell Migration in Confined Microenvironments,” Curr. Opin. Cell Biol., 30, pp. 41–50. 10.1016/j.ceb.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Mogilner, A. , and Oster, G. , 1996, “ Cell Motility Driven by Actin Polymerization,” Biophys. J., 71(6), pp. 3030–3045. 10.1016/S0006-3495(96)79496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Palecek, S. P. , Huttenlocher, A. , Horwitz, A. F. , and Lauffenburger, D. A. , 1998, “ Physical and Biochemical Regulation of Integrin Release During Rear Detachment of Migrating Cells,” J. Cell Sci., 111(7), pp. 929–940.http://jcs.biologists.org/content/111/7/929.long [DOI] [PubMed] [Google Scholar]
- [28]. Smith, J. T. , Elkin, J. T. , and Reichert, W. M. , 2006, “ Directed Cell Migration on Fibronectin Gradients: Effect of Gradient Slope,” Exp. Cell Res., 312(13), pp. 2424–2432. 10.1016/j.yexcr.2006.04.005 [DOI] [PubMed] [Google Scholar]
- [29]. Lo, C. M. , Wang, H. B. , Dembo, M. , and Wang, Y. L. , 2000, “ Cell Movement is Guided by the Rigidity of the Substrate,” Biophys. J., 79(1), pp. 144–152. 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Discher, D. E. , Janmey, P. , and Wang, Y.-L. , 2005, “ Tissue Cells Feel and Respond to the Stiffness of Their Substrate,” Science, 310(5751), pp. 1139–1143. 10.1126/science.1116995 [DOI] [PubMed] [Google Scholar]
- [31]. Stroka, K. M. , and Aranda-Espinoza, H. , 2009, “ Neutrophils Display Biphasic Relationship Between Migration and Substrate Stiffness,” Cell Motil. Cytoskeleton, 66(6), pp. 328–341. 10.1002/cm.20363 [DOI] [PubMed] [Google Scholar]
- [32]. Zhang, J. , Guo, W.-H. , Rape, A. , and Wang, Y.-L. , 2013, “ Micropatterning Cell Adhesion on Polyacrylamide Hydrogels,” Methods Mol. Biol., 1066, pp. 147–156. 10.1007/978-1-62703-604-7_13 [DOI] [PubMed] [Google Scholar]
- [33]. Moore, S. W. , Roca-Cusachs, P. , and Sheetz, M. P. , 2010, “ Stretchy Proteins on Stretchy Substrates: The Important Elements of Integrin-Mediated Rigidity Sensing,” Dev. Cell, 19(2), pp. 194–206. 10.1016/j.devcel.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Ridley, A. J. , Schwartz, M. A. , Burridge, K. , Firtel, R. A. , Ginsberg, M. H. , Borisy, G. , Parsons, J. T. , and Horwitz, A. R. , 2003, “ Cell Migration: Integrating Signals From Front to Back,” Science, 302(5651), pp. 1704–1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- [35]. Kim, D.-H. , and Wirtz, D. , 2013, “ Focal Adhesion Size Uniquely Predicts Cell Migration,” FASEB J., 27(4), pp. 1351–1361. 10.1096/fj.12-220160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Goldfinger, L. E. , Han, J. , Kiosses, W. B. , Howe, A. K. , and Ginsberg, M. H. , 2003, “ Spatial Restriction of Alpha4 Integrin Phosphorylation Regulates Lamellipodial Stability and Alpha4beta1-Dependent Cell Migration,” J. Cell Biol., 162(4), pp. 731–741. 10.1016/S0092-8674(00)81301-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Nishiya, N. , Kiosses, W. B. , Han, J. , and Ginsberg, M. H. , 2005, “ An Alpha4 Integrin-Paxillin-Arf-GAP Complex Restricts Rac Activation to the Leading Edge of Migrating Cells,” Nat. Cell Biol., 7(4), pp. 343–352. 10.1038/ncb1234 [DOI] [PubMed] [Google Scholar]
- [38]. Baker, B. M. , and Chen, C. S. , 2012, “ Deconstructing the Third Dimension: How 3D Culture Microenvironments Alter Cellular Cues,” J. Cell Sci., 125(Pt 13), pp. 3015–3024. 10.1242/jcs.079509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Mousavi, S. J. , and Doweidar, M. H. , 2015, “ Three-Dimensional Numerical Model of Cell Morphology During Migration in Multi-Signaling Substrates,” PLoS One, 10(3), p. e0122094. 10.1371/journal.pone.0122094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Galbraith, C. G. , and Sheetz, M. P. , 1997, “ A Micromachined Device Provides a New Bend on Fibroblast Traction Forces,” Proc. Natl. Acad. Sci. U.S.A., 94(17), pp. 9114–9118. 10.1073/pnas.94.17.9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Mao, A. S. , Shin, J.-W. , and Mooney, D. J. , 2016, “ Effects of Substrate Stiffness and Cell-Cell Contact on Mesenchymal Stem Cell Differentiation,” Biomaterials, 98, pp. 184–191. 10.1016/j.biomaterials.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Wu, P.-H. , Giri, A. , Sun, S. X. , and Wirtz, D. , 2014, “ Three-Dimensional Cell Migration Does Not Follow a Random Walk,” Proc. Natl. Acad. Sci. U.S.A., 111(11), pp. 3949–3954. 10.1073/pnas.1318967111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43]. Carey, S. P. , Kraning-Rush, C. M. , Williams, R. M. , and Reinhart-King, C. A. , 2012, “ Biophysical Control of Invasive Tumor Cell Behavior by Extracellular Matrix Microarchitecture,” Biomaterials, 33(16), pp. 4157–4165. 10.1016/j.biomaterials.2012.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Starke, J. , Wehrle-Haller, B. , and Friedl, P. , 2014, “ Plasticity of the Actin Cytoskeleton in Response to Extracellular Matrix Nanostructure and Dimensionality,” Biochem. Soc. Trans., 42(5), pp. 1356–1366. 10.1007/PL00000864 [DOI] [PubMed] [Google Scholar]
- [45]. Hall, M. S. , Long, R. , Feng, X. , Huang, Y. , Hui, C.-Y. , and Wu, M. , 2013, “ Toward Single Cell Traction Microscopy Within 3D Collagen Matrices,” Exp. Cell Res., 319(16), pp. 2396–2408. 10.1016/j.yexcr.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Starke, J. , Maaser, K. , Wehrle-Haller, B. , and Friedl, P. , 2013, “ Mechanotransduction of Mesenchymal Melanoma Cell Invasion Into 3D Collagen Lattices: Filopod-Mediated Extension–Relaxation Cycles and Force Anisotropy,” Exp. Cell Res., 319(16), pp. 2424–2433. 10.1016/j.yexcr.2013.04.003 [DOI] [PubMed] [Google Scholar]
- [47]. Provenzano, P. P. , Eliceiri, K. W. , Campbell, J. M. , Inman, D. R. , White, J. G. , and Keely, P. J. , 2006, “ Collagen Reorganization at the Tumor-Stromal Interface Facilitates Local Invasion,” BMC Med., 4(1), p. 38. 10.1016/0955-0674(95)80117-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Polacheck, W. J. , Zervantonakis, I. K. , and Kamm, R. D. , 2013, “ Tumor Cell Migration in Complex Microenvironments,” Cell. Mol. Life Sci., 70(8), pp. 1335–1356. 10.1007/s00018-012-1115-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Kreger, S. T. , Bell, B. J. , Bailey, J. , Stites, E. , Kuske, J. , Waisner, B. , and Voytik-Harbin, S. L. , 2010, “ Polymerization and Matrix Physical Properties as Important Design Considerations for Soluble Collagen Formulations,” Biopolymers, 93(8), pp. 690–707. 10.1002/bip.21431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Charulatha, V. , and Rajaram, A. , 2003, “ Influence of Different Crosslinking Treatments on the Physical Properties of Collagen Membranes,” Biomaterials, 24(5), pp. 759–767. 10.1016/S0142-9612(02)00412-X [DOI] [PubMed] [Google Scholar]
- [51]. Schultz, K. M. , Kyburz, K. A. , and Anseth, K. S. , 2015, “ Measuring Dynamic Cell-Material Interactions and Remodeling During 3D Human Mesenchymal Stem Cell Migration in Hydrogels,” Proc. Natl. Acad. Sci. U.S.A., 112(29), pp. E3757–3764. 10.1073/pnas.1511304112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Li, L.-L. , Shu, X.-S. , Wang, Z.-H. , Cao, Y. , and Tao, Q. , 2011, “ Epigenetic Disruption of Cell Signaling in Nasopharyngeal Carcinoma,” Chin. J. Cancer, 30(4), pp. 231–239. 10.5732/cjc.011.10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Young, E. W. K. , 2013, “ Cells, Tissues, and Organs on Chips: Challenges and Opportunities for the Cancer Tumor Microenvironment,” Integr. Biol. (Cambridge), 5(9), pp. 1096–1109. 10.1039/c3ib40076j [DOI] [PubMed] [Google Scholar]
- [54]. Xia, Y. , and Whitesides, G. M. , 2015, “ Soft Lithography,” Annu. Rev. Mater. Sci., 28, pp. 153–184. 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- [55]. Friedl, P. , and Alexander, S. , 2011, “ Cancer Invasion and the Microenvironment: Plasticity and Reciprocity,” Cell, 147(5), pp. 992–1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- [56]. Fraley, S. I. , Feng, Y. , Giri, A. , Longmore, G. D. , and Wirtz, D. , 2012, “ Dimensional and Temporal Controls of Three-Dimensional Cell Migration by Zyxin and Binding Partners,” Nat. Commun., 3, p. 719. 10.1038/ncomms1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Tang, H. , Li, A. , Bi, J. , Veltman, D. M. , Zech, T. , Spence, H. J. , Yu, X. , Timpson, P. , Insall, R. H. , Frame, M. C. , and MacHesky, L. M. , 2013, “ Loss of Scar/WAVE Complex Promotes N-WASP-and FAK-Dependent Invasion,” Curr. Biol., 23(2), pp. 107–117. 10.1016/j.cub.2012.11.059 [DOI] [PubMed] [Google Scholar]
- [58]. Yu, X. , and Machesky, L. M. , 2012, “ Cells Assemble Invadopodia-Like Structures and Invade Into Matrigel in a Matrix Metalloprotease Dependent Manner in the Circular Invasion Assay,” PLoS One, 7(2), p. e30605. 10.1371/journal.pone.0030605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59]. Zaman, M. H. , Trapani, L. M. , Sieminski, A. L. , Siemeski, A. , Mackellar, D. , Gong, H. , Kamm, R. D. , Wells, A. , Lauffenburger, D. , and Matsudaira, P. , 2006, “ Migration of Tumor Cells in 3D Matrices is Governed by Matrix Stiffness Along With Cell-Matrix Adhesion and Proteolysis,” Proc. Natl. Acad. Sci. U.S.A., 103(29), pp. 10889–10894. 10.1073/pnas.0604460103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60]. Provenzano, P. P. , Inman, D. R. , Eliceiri, K. W. , Trier, S. M. , and Keely, P. J. , 2008, “ Contact Guidance Mediated Three-Dimensional Cell Migration is Regulated by Rho/ROCK-Dependent Matrix Reorganization,” Biophys. J., 95(11), pp. 5374–5384. 10.1529/biophysj.108.133116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Carey, S. P. , Starchenko, A. , McGregor, A. L. , and Reinhart-King, C. A. , 2013, “ Leading Malignant Cells Initiate Collective Epithelial Cell Invasion in a Three-Dimensional Heterotypic Tumor Spheroid Model,” Clin. Exp. Metastasis, 30(5), pp. 615–630. 10.1007/s10585-013-9565-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62]. Hung, W.-C. , Chen, S.-H. , Paul, C. D. , Stroka, K. M. , Lo, Y.-C. , Yang, J. T. , and Konstantopoulos, K. , 2013, “ Distinct Signaling Mechanisms Regulate Migration in Unconfined Versus Confined Spaces,” J. Cell Biol., 202(5), pp. 807–824. 10.1007/s00418-008-0529-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63]. Kubow, K. E. , and Horwitz, A. R. , 2011, “ Reducing Background Fluorescence Reveals Adhesions in 3D Matrices,” Nat. Cell Biol., 13(1), pp. 3–5. 10.1038/ncb0111-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64]. Kraning-Rush, C. M. , Carey, S. P. , Califano, J. P. , Smith, B. N. , and Reinhart-King, C. A. , 2011, “ The Role of the Cytoskeleton in Cellular Force Generation in 2D and 3D Environments,” Phys. Biol., 8(1), p. 15009. 10.1088/1478-3975/8/1/015009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65]. Kraning-Rush, C. M. , Carey, S. P. , Califano, J. P. , and Reinhart-King, C. A. , 2012, “ Quantifying Traction Stresses in Adherent Cells,” Methods Cell Biol., 110, pp. 139–178. 10.1016/B978-0-12-388403-9.00006-0 [DOI] [PubMed] [Google Scholar]
- [66]. Ben-Yaakov, D. , Golkov, R. , Shokef, Y. , and Safran, S. A. , 2015, “ Response of Adherent Cells to Mechanical Perturbations of the Surrounding Matrix,” Soft Matter, 11(7), pp. 1412–1424. 10.1039/C4SM01817F [DOI] [PubMed] [Google Scholar]
- [67]. Dembo, M. , and Wang, Y. L. , 1999, “ Stresses at the Cell-to-Substrate Interface During Locomotion of Fibroblasts,” Biophys. J., 76(4), pp. 2307–2316. 10.1016/S0006-3495(99)77386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68]. Alcoser, T. A. , Bordeleau, F. , Carey, S. P. , Lampi, M. C. , Kowal, D. R. , Somasegar, S. , Varma, S. , Shin, S. J. , and Reinhart-King, C. A. , 2015, “ Probing the Biophysical Properties of Primary Breast Tumor-Derived Fibroblasts,” Cell. Mol. Bioeng., 8(1), pp. 76–85. 10.1007/s12195-014-0360-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69]. Trepat, X. , Wasserman, M. R. , Angelini, T. E. , Millet, E. , Weitz, D. A. , Butler, J. P. , and Fredberg, J. J. , 2009, “ Physical Forces During Collective Cell Migration,” Nat. Phys., 5(6), pp. 426–430. 10.1038/nphys1269 [DOI] [Google Scholar]
- [70]. Kraning-Rush, C. M. , Califano, J. P. , and Reinhart-King, C. A. , 2012, “ Cellular Traction Stresses Increase With Increasing Metastatic Potential,” PLoS One, 7(2), p. e32572. 10.1371/journal.pone.0032572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71]. Serra-Picamal, X. , Conte, V. , Sunyer, R. , Muñoz, J. J. , and Trepat, X. , 2015, “ Mapping Forces and Kinematics During Collective Cell Migration,” Methods Cell Biol., 125, pp. 309–330. 10.1016/bs.mcb.2014.11.003 [DOI] [PubMed] [Google Scholar]
- [72]. Tambe, D. T. , Hardin, C. C. , Angelini, T. E. , Rajendran, K. , Park, C. Y. , Serra-Picamal, X. , Zhou, E. H. , Zaman, M. H. , Butler, J. P. , Weitz, D. A. , Fredberg, J. J. , and Trepat, X. , 2011, “ Collective Cell Guidance by Cooperative Intercellular Forces,” Nat. Mater., 10(6), pp. 469–475. 10.1038/nmat3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Legant, W. R. , Choi, C. K. , Miller, J. S. , Shao, L. , Gao, L. , Betzig, E. , and Chen, C. S. , 2013, “ Multidimensional Traction Force Microscopy Reveals Out-of-Plane Rotational Moments About Focal Adhesions,” Proc. Natl. Acad. Sci. U.S.A., 110(3), pp. 881–886. 10.1073/pnas.1207997110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]. Meseke, M. , and Förster, E. , 2013, “ A 3D-Matrigel/Microbead Assay for the Visualization of Mechanical Tractive Forces at the Neurite-Substrate Interface of Cultured Neurons,” J. Biomed. Mater. Res., Part A, 101A(6), pp. 1726–1733. 10.1002/jbm.a.34477 [DOI] [PubMed] [Google Scholar]
- [75]. Legant, W. R. , Miller, J. S. , Blakely, B. L. , Cohen, D. M. , Genin, G. M. , and Chen, C. S. , 2010, “ Measurement of Mechanical Tractions Exerted by Cells in Three-Dimensional Matrices,” Nat. Methods, 7(12), pp. 969–971. 10.1038/nmeth.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76]. Doerschuk, C. M. , Beyers, N. , Coxson, H. O. , Wiggs, B. , and Hogg, J. C. , 1993, “ Comparison of Neutrophil and Capillary Diameters and Their Relation to Neutrophil Sequestration in the Lung,” J. Appl. Physiol., 74(6), pp. 3040–3045.http://jap.physiology.org/content/74/6/3040.long [DOI] [PubMed] [Google Scholar]
- [77]. Stoitzner, P. , Stössel, H. , Romani, N. , and Pfaller, K. , 2002, “ A Close-Up View of Migrating Langerhans Cells in the Skin,” J. Invest. Dermatol., 118(1), pp. 117–125. 10.1046/j.0022-202x.2001.01631.x [DOI] [PubMed] [Google Scholar]
- [78]. Weigelin, B. , Bakker, G.-J. , and Friedl, P. , 2012, “ Intravital Third Harmonic Generation Microscopy of Collective Melanoma Cell Invasion,” IntraVital, 1(1), pp. 32–43. 10.4161/intv.21223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Liu, Z. , Lee, Y. , Jang, J. H. , Li, Y. , Han, X. , Yokoi, K. , Ferrari, M. , Zhou, L. , and Qin, L. , 2015, “ Microfluidic Cytometric Analysis of Cancer Cell Transportability and Invasiveness,” Sci. Rep., 5, p. 14272. 10.1038/srep14272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Fu, Y. , Chin, L. K. , Bourouina, T. , Liu, A. Q. , and VanDongen, A. M. J. , 2012, “ Nuclear Deformation During Breast Cancer Cell Transmigration,” Lab Chip, 12(19), pp. 3774–3778. 10.1039/c2lc40477j [DOI] [PubMed] [Google Scholar]
- [81]. Balzer, E. M. , Tong, Z. , Paul, C. D. , Hung, W.-C. , Stroka, K. M. , Boggs, A. E. , Martin, S. S. , and Konstantopoulos, K. , 2012, “ Physical Confinement Alters Tumor Cell Adhesion and Migration Phenotypes,” FASEB J., 26(10), pp. 4045–4056. 10.1096/fj.12-211441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82]. Liu, Y.-J. , Le Berre, M. , Lautenschlaeger, F. , Maiuri, P. , Callan-Jones, A. , Heuzé, M. , Takaki, T. , Voituriez, R. , and Piel, M. , 2015, “ Confinement and Low Adhesion Induce Fast Amoeboid Migration of Slow Mesenchymal Cells,” Cell, 160(4), pp. 659–672. 10.1016/j.cell.2015.01.007 [DOI] [PubMed] [Google Scholar]
- [83]. Davidson, P. M. , Denais, C. , Bakshi, M. C. , and Lammerding, J. , 2014, “ Nuclear Deformability Constitutes a Rate-Limiting Step During Cell Migration in 3-D Environments,” Cell. Mol. Bioeng., 7(3), pp. 293–306. 10.1007/s12195-014-0342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84]. Baker, E. L. , Bonnecaze, R. T. , and Zaman, M. H. , 2009, “ Extracellular Matrix Stiffness and Architecture Govern Intracellular Rheology in Cancer,” Biophys. J., 97(4), pp. 1013–1021. 10.1016/j.bpj.2009.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85]. Tseng, Y. , Kole, T. P. , and Wirtz, D. , 2002, “ Micromechanical Mapping of Live Cells by Multiple-Particle-Tracking Microrheology,” Biophys. J., 83(6), pp. 3162–3176. 10.1016/S0006-3495(02)75319-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86]. Xu, W. , Mezencev, R. , Kim, B. , Wang, L. , McDonald, J. , and Sulchek, T. , 2012, “ Cell Stiffness is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells,” PLoS One, 7(10), p. e46609. 10.1371/journal.pone.0046609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87]. Swaminathan, V. , Mythreye, K. , O'Brien, E. T. , Berchuck, A. , Blobe, G. C. , and Superfine, R. , 2011, “ Mechanical Stiffness Grades Metastatic Potential in Patient Tumor Cells and in Cancer Cell Lines,” Cancer Res., 71(15), pp. 5075–5080. 10.1158/0008-5472.CAN-11-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88]. Guck, J. , Ananthakrishnan, R. , Mahmood, H. , Moon, T. J. , Cunningham, C. C. , and Käs, J. , 2001, “ The Optical Stretcher: A Novel Laser Tool to Micromanipulate Cells,” Biophys. J., 81(2), pp. 767–784. 10.1016/S0006-3495(01)75740-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89]. Ochalek, T. , Nordt, F. J. , Tullberg, K. , Variants, M. C. , and Burger, M. M. , 1988, “ Correlation Between Cell Deformability and Metastatic Potential in B16-F1 Melanoma Cell Variants Correlation Between Cell Deformability and Metastatic Potential in B16-F1,” Cancer Res., 48(18), pp. 5124–5128.http://cancerres.aacrjournals.org/content/47/14/3835.long [PubMed] [Google Scholar]
- [90]. Mak, M. , Kamm, R. D. , and Zaman, M. H. , 2014, “ Impact of Dimensionality and Network Disruption on Microrheology of Cancer Cells in 3D Environments,” PLoS Comput. Biol., 10(11), p. e1003959. 10.1371/journal.pcbi.1003959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91]. Jones, D. P. , Hanna, W. , El-Hamidi, H. , and Celli, J. P. , 2014, “ Longitudinal Measurement of Extracellular Matrix Rigidity in 3D Tumor Models Using Particle-Tracking Microrheology,” J. Visualized Exp., (88), pp. e51302. 10.3791/51302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92]. Gal, N. , and Weihs, D. , 2012, “ Intracellular Mechanics and Activity of Breast Cancer Cells Correlate With Metastatic Potential,” Cell Biochem. Biophys., 63(3), pp. 199–209. 10.1007/s12013-012-9356-z [DOI] [PubMed] [Google Scholar]
- [93]. Haase, K. , and Pelling, A. E. , 2015, “ Investigating Cell Mechanics With Atomic Force Microscopy,” J. R. Soc., Interface, 12(104), p. 20140970. 10.1038/nmat1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94]. Alenghat, F. J. , and Ingber, D. E. , 2002, “ Mechanotransduction: All Signals Point to Cytoskeleton, Matrix, and Integrins,” Sci. STKE, 2002(119), p. pe6.http://stke.sciencemag.org/content/2002/119/pe6.long [DOI] [PubMed] [Google Scholar]
- [95]. Volakis, L. I. , Li, R. , Ackerman, W. E., IV , Mihai, C. , Bechel, M. , Summerfield, T. L. , Ahn, C. S. , Powell, H. M. , Zielinski, R. , Rosol, T. J. , Ghadiali, S. N. , and Kniss, D. A. , 2014, “ Loss of Myoferlin Redirects Breast Cancer Cell Motility Towards Collective Migration,” PLoS One, 9(2), p. e86110. 10.1371/journal.pone.0086110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96]. Mihai, C. , Bao, S. , Lai, J.-P. , Ghadiali, S. N. , and Knoell, D. L. , 2012, “ PTEN Inhibition Improves Wound Healing in Lung Epithelia Through Changes in Cellular Mechanics That Enhance Migration,” Am. J. Physiol.: Lung Cell. Mol. Physiol., 302(3), pp. L287–299. 10.1152/ajplung.00037.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97]. Staunton, J. R. , Doss, B. L. , Lindsay, S. , and Ros, R. , 2016, “ Correlating Confocal Microscopy and Atomic Force Indentation Reveals Metastatic Cancer Cells Stiffen During Invasion Into Collagen I Matrices,” Sci. Rep., 6, p. 19686. 10.1038/srep19686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98]. Tartibi, M. , Liu, Y. X. , Liu, G.-Y. , and Komvopoulos, K. , 2015, “ Single-Cell Mechanics—An Experimental–Computational Method for Quantifying the Membrane–Cytoskeleton Elasticity of Cells,” Acta Biomater., 27, pp. 224–235. 10.1016/j.actbio.2015.08.028 [DOI] [PubMed] [Google Scholar]
- [99].Physical Sciences—Oncology Centers Network, Agus, D. B. , Alexander, J. F. , Arap, W. , Ashili, S. , Aslan, J. E. , Austin, R. H. , Backman, V. , Bethel, K. J. , Bonneau, R. , Chen, W.-C. , Chen-Tanyolac, C. , Choi, N. C. , Curley, S. A. , Dallas, M. , Damania, D. , Davies, P. C. W. , Decuzzi, P. , Dickinson, L. , Estevez-Salmeron, L. , Estrella, V. , Ferrari, M. , Fischbach, C. , Foo, J. , Fraley, S. I. , Frantz, C. , Fuhrmann, A. , Gascard, P. , Gatenby, R. A. , Geng, Y. , Gerecht, S. , Gillies, R. J. , Godin, B. , Grady, W. M. , Greenfield, A. , Hemphill, C. , Hempstead, B. L. , Hielscher, A. , Hillis, W. D. , Holland, E. C. , Ibrahim-Hashim, A. , Jacks, T. , Johnson, R. H. , Joo, A. , Katz, J. E. , Kelbauskas, L. , Kesselman, C. , King, M. R. , Konstantopoulos, K. , Kraning-Rush, C. M. , Kuhn, P. , Kung, K. , Kwee, B. , Lakins, J. N. , Lambert, G. , Liao, D. , Licht, J. D. , Liphardt, J. T. , Liu, L. , Lloyd, M. C. , Lyubimova, A. , Mallick, P. , Marko, J. , McCarty, O. J. T. , Meldrum, D. R. , Michor, F. , Mumenthaler, S. M. , Nandakumar, V. , O'Halloran, T. V. , Oh, S. , Pasqualini, R. , Paszek, M. J. , Philips, K. G. , Poultney, C. S. , Rana, K. , Reinhart-King, C. A. , Ros, R. , Semenza, G. L. , Senechal, P. , Shuler, M. L. , Srinivasan, S. , Staunton, J. R. , Stypula, Y. , Subramanian, H. , Tlsty, T. D. , Tormoen, G. W. , Tseng, Y. , van Oudenaarden, A. , Verbridge, S. S. , Wan, J. C. , Weaver, V. M. , Widom, J. , Will, C. , Wirtz, D. , Wojtkowiak, J. , and Wu, P.-H. , 2013, “ A Physical Sciences Network Characterization of Non-Tumorigenic and Metastatic Cells,” Sci. Rep., 3, p. 1449. 10.1038/srep01449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100]. Plodinec, M. , Loparic, M. , Monnier, C. A. , Obermann, E. C. , Zanetti-Dallenbach, R. , Oertle, P. , Hyotyla, J. T. , Aebi, U. , Bentires-Alj, M. , Lim, R. Y. H. , and Schoenenberger, C.-A. , 2012, “ The Nanomechanical Signature of Breast Cancer,” Nat. Nanotechnol., 7(11), pp. 757–765. 10.1038/nnano.2012.167 [DOI] [PubMed] [Google Scholar]
- [101]. Mak, M. , and Erickson, D. , 2013, “ A Serial Micropipette Microfluidic Device With Applications to Cancer Cell Repeated Deformation Studies,” Integr. Biol. (Cambridge), 5(11), pp. 1374–1384. 10.1039/c3ib40128f [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102]. Mak, M. , Reinhart-King, C. A. , and Erickson, D. , 2013, “ Elucidating Mechanical Transition Effects of Invading Cancer Cells With a Subnucleus-Scaled Microfluidic Serial Dimensional Modulation Device,” Lab Chip, 13(3), pp. 340–348. 10.1039/C2LC41117B [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103]. McGregor, A. L. , Hsia, C.-R. , and Lammerding, J. , 2016, “ Squish and Squeeze—The Nucleus as a Physical Barrier During Migration in Confined Environments,” Curr. Opin. Cell Biol., 40, pp. 32–40. 10.1016/j.ceb.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104]. Wolf, K. , te Lindert, M. , Krause, M. , Alexander, S. , te Riet, J. , Willis, A. L. , Hoffman, R. M. , Figdor, C. G. , Weiss, S. J. , and Friedl, P. , 2013, “ Physical Limits of Cell Migration: Control by ECM Space and Nuclear Deformation and Tuning by Proteolysis and Traction Force,” J. Cell Biol., 201(7), pp. 1069–1084. 10.1007/s00418-008-0529-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105]. Quigley, J. P. , and Armstrong, P. B. , 1998, “ Tumor Cell Intravasation Alu-Cidated: The Chick Embryo Opens the Window,” Cell, 94(3), pp. 281–284. 10.1016/S0092-8674(00)81470-1 [DOI] [PubMed] [Google Scholar]
- [106]. Zervantonakis, I. K. , Hughes-Alford, S. K. , Charest, J. L. , Condeelis, J. S. , Gertler, F. B. , and Kamm, R. D. , 2012, “ Three-Dimensional Microfluidic Model for Tumor Cell Intravasation and Endothelial Barrier Function,” Proc. Natl. Acad. Sci. U.S.A., 109(34), pp. 13515–13520. 10.1073/pnas.1210182109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107]. Lee, H. , Park, W. , Ryu, H. , and Jeon, N. L. , 2014, “ A Microfluidic Platform for Quantitative Analysis of Cancer Angiogenesis and Intravasation,” Biomicrofluidics, 8(5), p. 54102. 10.1063/1.4894595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108]. Jeon, J. S. , Bersini, S. , Gilardi, M. , Dubini, G. , Charest, J. L. , Moretti, M. , and Kamm, R. D. , 2015, “ Human 3D Vascularized Organotypic Microfluidic Assays to Study Breast Cancer Cell Extravasation,” Proc. Natl. Acad. Sci. U.S.A., 112(1), pp. 214–219. 10.1073/pnas.1417115112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109]. Qian, C.-N. , and Teh, B. T. , 2011, “ ‘Seed and Soil’ Theory of Metastasis,” Encyclopedia of Cancer, Springer, Berlin, pp. 3354–3355. [Google Scholar]
- [110]. Zhang, Q. , Liu, T. , and Qin, J. , 2012, “ A Microfluidic-Based Device for Study of Transendothelial Invasion of Tumor Aggregates in Realtime,” Lab Chip, 12(16), pp. 2837–2842. 10.1039/c2lc00030j [DOI] [PubMed] [Google Scholar]
- [111]. Jeon, J. S. , Zervantonakis, I. K. , Chung, S. , Kamm, R. D. , and Charest, J. L. , 2013, “ In Vitro Model of Tumor Cell Extravasation,” PLoS One, 8(2), p. e56910. 10.1371/journal.pone.0056910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112]. Chen, M. B. , Whisler, J. A. , Jeon, J. S. , and Kamm, R. D. , 2013, “ Mechanisms of Tumor Cell Extravasation in an In Vitro Microvascular Network Platform,” Integr. Biol. (Cambridge), 5(10), pp. 1262–1271. 10.1039/c3ib40149a [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113]. Riahi, R. , Yang, Y. L. , Kim, H. , Jiang, L. , Wong, P. K. , and Zohar, Y. , 2014, “ A Microfluidic Model for Organ-Specific Extravasation of Circulating Tumor Cells,” Biomicrofluidics, 8(2), p. 24103. 10.1063/1.4868301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114]. Fraley, S. I. , Feng, Y. , Krishnamurthy, R. , Kim, D.-H. , Celedon, A. , Longmore, G. D. , and Wirtz, D. , 2010, “ A Distinctive Role for Focal Adhesion Proteins in Three-Dimensional Cell Motility,” Nat. Cell Biol., 12(6), pp. 598–604. 10.1038/ncb2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115]. Chiru, A. A. Z. , Popescu, C. R. , and Gheorghe, D. C. , 2014, “ Enzymatic Aspects in ENT Cancer-Matrix Metalloproteinases,” J. Med. Life, 7(3), pp. 379–380.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4233443/ [PMC free article] [PubMed] [Google Scholar]
- [116]. Sabeh, F. , Shimizu-Hirota, R. , and Weiss, S. J. , 2009, “ Protease-Dependent Versus-Independent Cancer Cell Invasion Programs: Three-Dimensional Amoeboid Movement Revisited,” J. Cell Biol., 185(1), p. 11. 10.1007/s00418-008-0529-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117]. Ehrbar, M. , Sala, A. , Lienemann, P. , Ranga, A. , Mosiewicz, K. , Bittermann, A. , Rizzi, S. C. , Weber, F. E. , and Lutolf, M. P. , 2011, “ Elucidating the Role of Matrix Stiffness in 3D Cell Migration and Remodeling,” Biophys. J., 100(2), pp. 284–293. 10.1016/j.bpj.2010.11.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118]. Reinhardt, J. W. , Krakauer, D. A. , and Gooch, K. J. , 2013, “ Complex Matrix Remodeling and Durotaxis Can Emerge From Simple Rules for Cell-Matrix Interaction in Agent-Based Models,” ASME J. Biomech. Eng., 135(7), p. 71003. 10.1115/1.4024463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119]. Pizzo, A. M. , Kokini, K. , Vaughn, L. C. , Waisner, B. Z. , and Voytik-Harbin, S. L. , 2005, “ Extracellular Matrix (ECM) Microstructural Composition Regulates Local Cell-ECM Biomechanics and Fundamental Fibroblast Behavior: A Multidimensional Perspective,” J. Appl. Physiol., 98(5), pp. 1909–1921. 10.1152/japplphysiol.01137.2004 [DOI] [PubMed] [Google Scholar]
- [120]. Yang, Y. , Motte, S. , and Kaufman, L. J. , 2010, “ Pore Size Variable Type I Collagen Gels and Their Interaction With Glioma Cells,” Biomaterials, 31(21), pp. 5678–5688. 10.1016/j.biomaterials.2010.03.039 [DOI] [PubMed] [Google Scholar]
- [121]. Roeder, B. A. , Kokini, K. , Sturgis, J. E. , Robinson, J. P. , and Voytik-Harbin, S. L. , 2002, “ Tensile Mechanical Properties of Three-Dimensional Type I Collagen Extracellular Matrices With Varied Microstructure,” ASME J. Biomech. Eng., 124(2), p. 214. 10.1115/1.1449904 [DOI] [PubMed] [Google Scholar]
- [122]. Wolf, K. , Wu, Y. I. , Liu, Y. , Geiger, J. , Tam, E. , Overall, C. , Stack, M. S. , and Friedl, P. , 2007, “ Multi-Step Pericellular Proteolysis Controls the Transition From Individual to Collective Cancer Cell Invasion,” Nat. Cell Biol., 9(8), pp. 893–904. 10.1038/ncb1616 [DOI] [PubMed] [Google Scholar]
- [123]. Wyckoff, J. B. , Pinner, S. E. , Gschmeissner, S. , Condeelis, J. S. , and Sahai, E. , 2006, “ ROCK- and Myosin-Dependent Matrix Deformation Enables Protease-Independent Tumor-Cell Invasion In Vivo,” Curr. Biol., 16(15), pp. 1515–1523. 10.1016/j.cub.2006.05.065 [DOI] [PubMed] [Google Scholar]
- [124]. Rolli, C. G. , Seufferlein, T. , Kemkemer, R. , and Spatz, J. P. , 2010, “ Impact of Tumor Cell Cytoskeleton Organization on Invasiveness and Migration: A Microchannel-Based Approach,” PLoS One, 5(1), p. e8726. 10.1371/journal.pone.0008726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125]. Mak, M. , Reinhart-King, C. A. , and Erickson, D. , 2011, “ Microfabricated Physical Spatial Gradients for Investigating Cell Migration and Invasion Dynamics,” PLoS One, 6(6), p. e20825. 10.1371/journal.pone.0020825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126]. Lautscham, L. A. , Kämmerer, C. , Lange, J. R. , Kolb, T. , Mark, C. , Schilling, A. , Strissel, P. L. , Strick, R. , Gluth, C. , Rowat, A. C. , Metzner, C. , and Fabry, B. , 2015, “ Migration in Confined 3D Environments is Determined by a Combination of Adhesiveness, Nuclear Volume, Contractility, and Cell Stiffness,” Biophys. J., 109(5), pp. 900–913. 10.1016/j.bpj.2015.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127]. Pathak, A. , and Kumar, S. , 2012, “ Independent Regulation of Tumor Cell Migration by Matrix Stiffness and Confinement,” Proc. Natl. Acad. Sci., 109(26), pp. 10334–10339. 10.1073/pnas.1118073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128]. Kraning-Rush, C. M. , Carey, S. P. , Lampi, M. C. , and Reinhart-King, C. A. , 2013, “ Microfabricated Collagen Tracks Facilitate Single Cell Metastatic Invasion in 3D,” Integr. Biol. (Cambridge), 5(3), pp. 606–616. 10.1039/c3ib20196a [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129]. Ilina, O. , Bakker, G.-J. , Vasaturo, A. , Hofmann, R. M. , and Friedl, P. , 2011, “ Two-Photon Laser-Generated Microtracks in 3D Collagen Lattices: Principles of MMP-Dependent and -Independent Collective Cancer Cell Invasion,” Phys. Biol., 8(1), p. 15010. 10.1088/1478-3975/8/1/015010 [DOI] [PubMed] [Google Scholar]
- [130]. Friedl, P. , and Gilmour, D. , 2009, “ Collective Cell Migration in Morphogenesis, Regeneration and Cancer,” Nat. Rev. Mol. Cell Biol., 10(7), pp. 445–457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- [131]. Sodek, K. L. , Ringuette, M. J. , and Brown, T. J. , 2009, “ Compact Spheroid Formation by Ovarian Cancer Cells is Associated With Contractile Behavior and an Invasive Phenotype,” Int. J. Cancer, 124(9), pp. 2060–2070. 10.1002/ijc.24188 [DOI] [PubMed] [Google Scholar]
- [132]. Gole, B. , Durán Alonso, M. B. , Dolenc, V. , and Lah, T. , 2009, “ Post-Translational Regulation of Cathepsin B, But Not of Other Cysteine Cathepsins, Contributes to Increased Glioblastoma Cell Invasiveness In Vitro,” Pathol. Oncol. Res., 15(4), pp. 711–723. 10.1007/s12253-009-9175-8 [DOI] [PubMed] [Google Scholar]
- [133]. Hauptmann, S. , Budianto, D. , Denkert, C. , Köbel, M. , Borsi, L. , and Siri, A. , 2003, “ Adhesion and Migration of HRT-18 Colorectal Carcinoma Cells on Extracellular Matrix Components Typical for the Desmoplastic Stroma of Colorectal Adenocarcinomas,” Oncology, 65(2), pp. 174–181. 10.1159/000072344 [DOI] [PubMed] [Google Scholar]
- [134]. Sutherland, R. M. , 1986, “ Importance of Critical Metabolites and Cellular Interactions in the Biology of Microregions of Tumors,” Cancer, 58(8), pp. 1668–1680. [DOI] [PubMed] [Google Scholar]
- [135]. Kumar, S. , Kapoor, A. , Desai, S. , Inamdar, M. M. , and Sen, S. , 2016, “ Proteolytic and Non-Proteolytic Regulation of Collective Cell Invasion: Tuning by ECM Density and Organization,” Sci. Rep., 6, p. 19905. 10.1038/srep19905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136]. Liu, L. , Duclos, G. , Sun, B. , Lee, J. , Wu, A. , Kam, Y. , Sontag, E. D. , Stone, H. A. , Sturm, J. C. , Gatenby, R. A. , and Austin, R. H. , 2013, “ Minimization of Thermodynamic Costs in Cancer Cell Invasion,” Proc. Natl. Acad. Sci., 110(5), pp. 1686–1691. 10.1073/pnas.1221147110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137]. Cheung, K. J. , and Ewald, A. J. , 2014, “ Invasive Leader Cells: Metastatic Oncotarget,” Oncotarget, 5(6), pp. 1390–1391. 10.18632/oncotarget.1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138]. Cheung, K. J. , Padmanaban, V. , Silvestri, V. , Schipper, K. , Cohen, J. D. , Fairchild, A. N. , Gorin, M. A. , Verdone, J. E. , Pienta, K. J. , Bader, J. S. , and Ewald, A. J. , 2016, “ Polyclonal Breast Cancer Metastases Arise From Collective Dissemination of Keratin 14-Expressing Tumor Cell Clusters,” Proc. Natl. Acad. Sci., 113(7), p. 201508541. 10.1073/pnas.1508541113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139]. Cheung, K. J. , Gabrielson, E. , Werb, Z. , and Ewald, A. J. , 2013, “ Collective Invasion in Breast Cancer Requires a Conserved Basal Epithelial Program,” Cell, 155(7), pp. 1639–1651. 10.1016/j.cell.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140]. du Roure, O. , Saez, A. , Buguin, A. , Austin, R. H. , Chavrier, P. , Silberzan, P. , Siberzan, P. , and Ladoux, B. , 2005, “ Force Mapping in Epithelial Cell Migration,” Proc. Natl. Acad. Sci. U.S.A., 102(7), pp. 2390–2395. 10.1073/pnas.0408482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141]. Vaughan, R. B. , and Trinkaus, J. P. , 1966, “ Movements of Epithelial Cell Sheets In Vitro,” J. Cell Sci., 1(4), pp. 407–413.http://jcs.biologists.org/content/1/4/407.long [DOI] [PubMed] [Google Scholar]
- [142]. Omelchenko, T. , Vasiliev, J. M. , Gelfand, I. M. , Feder, H. H. , and Bonder, E. M. , 2003, “ Rho-Dependent Formation of Epithelial ‘Leader’ Cells During Wound Healing,” Proc. Natl. Acad. Sci., 100(19), pp. 10788–10793. 10.1073/pnas.1834401100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143]. Friedl, P. , Hegerfeldt, Y. , and Tusch, M. , 2004, “ Collective Cell Migration in Morphogenesis and Cancer,” Int. J. Dev. Biol., 48(5–6), pp. 441–449. 10.1387/ijdb.041821pf [DOI] [PubMed] [Google Scholar]
- [144]. Gov, N. S. , 2007, “ Collective Cell Migration Patterns: Follow the Leader,” Proc. Natl. Acad. Sci., 104(41), pp. 15970–15971. 10.1073/pnas.0708037104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145]. Poujade, M. , Grasland-Mongrain, E. , Hertzog, A. , Jouanneau, J. , Chavrier, P. , Ladoux, B. , Buguin, A. , and Silberzan, P. , 2007, “ Collective Migration of an Epithelial Monolayer in Response to a Model Wound,” Proc. Natl. Acad. Sci. U.S.A., 104(41), pp. 15988–15993. 10.1073/pnas.0705062104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146]. Casey, R. C. , Burleson, K. M. , Skubitz, K. M. , Pambuccian, S. E. , Oegema, T. R. , Ruff, L. E. , and Skubitz, A. P. , 2001, “ Beta 1-Integrins Regulate the Formation and Adhesion of Ovarian Carcinoma Multicellular Spheroids,” Am. J. Pathol., 159(6), pp. 2071–2080. 10.1016/S0002-9440(10)63058-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147]. Ehsan, S. M. , Welch-Reardon, K. M. , Waterman, M. L. , Hughes, C. C. W. , and George, S. C. , 2014, “ A Three-Dimensional In Vitro Model of Tumor Cell Intravasation,” Integr. Biol. (Cambridge), 6(6), pp. 603–610. 10.1039/c3ib40170g [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148]. Cheung, K. J. , and Ewald, A. J. , 2016, “ A Collective Route to Metastasis: Seeding by Tumor Cell Clusters,” Science, 352(6282), pp. 167–169. 10.1126/science.aaf6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149]. Aceto, N. , Bardia, A. , Miyamoto, D. T. , Donaldson, M. C. , Wittner, B. S. , Spencer, J. A. , Yu, M. , Pely, A. , Engstrom, A. , Zhu, H. , Brannigan, B. W. , Kapur, R. , Stott, S. L. , Shioda, T. , Ramaswamy, S. , Ting, D. T. , Lin, C. P. , Toner, M. , Haber, D. A. , and Maheswaran, S. , 2014, “ Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis,” Cell, 158(5), pp. 1110–1122. 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150]. Maddipati, R. , and Stanger, B. Z. , 2015, “ Pancreatic Cancer Metastases Harbor Evidence of Polyclonality,” Cancer Discovery, 5(10), pp. 1086–1097. 10.1158/2159-8290.CD-15-0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151]. Montell, D. J. , 2008, “ Morphogenetic Cell Movements: Diversity From Modular Mechanical Properties,” Science, 322(5907), pp. 1502–1505. 10.1126/science.1164073 [DOI] [PubMed] [Google Scholar]
- [152]. Valencia, A. M. J. , Wu, P.-H. , Yogurtcu, O. N. , Rao, P. , DiGiacomo, J. , Godet, I. , He, L. , Lee, M.-H. , Gilkes, D. , Sun, S. X. , and Wirtz, D. , 2015, “ Collective Cancer Cell Invasion Induced by Coordinated Contractile Stresses,” Oncotarget, 6(41), pp. 43438–43451. 10.18632/oncotarget.5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153]. Rizwan, A. , Bulte, C. , Kalaichelvan, A. , Cheng, M. , Krishnamachary, B. , Bhujwalla, Z. M. , Jiang, L. , and Glunde, K. , 2015, “ Metastatic Breast Cancer Cells in Lymph Nodes Increase Nodal Collagen Density,” Sci. Rep., 5, p. 10002. 10.1038/srep10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154]. Jain, R. K. , 2005, “ Normalization of Tumor Vasculature: An Emerging Concept in Antiangiogenic Therapy,” Science, 307(5706), pp. 58–62. 10.1126/science.1104819 [DOI] [PubMed] [Google Scholar]
- [155]. Mason, B. N. , Starchenko, A. , Williams, R. M. , Bonassar, L. J. , and Reinhart-King, C. A. , 2013, “ Tuning Three-Dimensional Collagen Matrix Stiffness Independently of Collagen Concentration Modulates Endothelial Cell Behavior,” Acta Biomater., 9(1), pp. 4635–4644. 10.1016/j.actbio.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156]. Weiswald, L.-B. , Bellet, D. , and Dangles-Marie, V. , 2015, “ Spherical Cancer Models in Tumor Biology,” Neoplasia, 17(1), pp. 1–15. 10.1016/j.neo.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157]. Sutherland, R. M. , 1988, “ Cell and Environment Interactions in Tumor Microregions: The Multicell Spheroid Model,” Science, 240(4849), pp. 177–184. 10.1126/science.2451290 [DOI] [PubMed] [Google Scholar]
- [158]. Dang, T. T. , Westcott, J. M. , Maine, E. A. , Kanchwala, M. , Xing, C. , and Pearson, G. W. , 2016, “ ΔNp63α Induces the Expression of FAT2 and Slug to Promote Tumor Invasion,” Oncotarget, 7(19), pp. 28592–28611. 10.18632/oncotarget.8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159]. Maine, E. A. , Westcott, J. M. , Prechtl, A. M. , Dang, T. T. , Whitehurst, A. W. , and Pearson, G. W. , 2016, “ The Cancer-Testis Antigens SPANX-A/C/D and CTAG2 Promote Breast Cancer Invasion,” Oncotarget, 7(12), pp. 14708–14726. 10.18632/oncotarget.7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160]. Westcott, J. M. , Prechtl, A. M. , Maine, E. A. , Dang, T. T. , Esparza, M. A. , Sun, H. , Zhou, Y. , Xie, Y. , and Pearson, G. W. , 2015, “ An Epigenetically Distinct Breast Cancer Cell Subpopulation Promotes Collective Invasion,” J. Clin. Invest., 125(5), pp. 1927–1943. 10.1172/JCI77767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161]. Aifuwa, I. , Giri, A. , Longe, N. , Lee, S. H. , An, S. S. , and Wirtz, D. , 2015, “ Senescent Stromal Cells Induce Cancer Cell Migration Via Inhibition of RhoA/ROCK/Myosin-Based Cell Contractility,” Oncotarget, 6(31), pp. 30516–30531. 10.18632/oncotarget.5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162]. Okegawa, T. , Pong, R.-C. , Li, Y. , and Hsieh, J.-T. , 2004, “ The Role of Cell Adhesion Molecule in Cancer Progression and Its Application in Cancer Therapy,” Acta Biochim. Pol., 51(2), pp. 445–457.http://www.actabp.pl/pdf/2_2004/445.pdf [PubMed] [Google Scholar]
- [163]. Branco da Cunha, C. , Klumpers, D. D. , Koshy, S. T. , Weaver, J. C. , Chaudhuri, O. , Seruca, R. , Carneiro, F. , Granja, P. L. , and Mooney, D. J. , 2016, “ CD44 Alternative Splicing in Gastric Cancer Cells is Regulated by Culture Dimensionality and Matrix Stiffness,” Biomaterials, 98, pp. 152–162. 10.1016/j.biomaterials.2016.04.016 [DOI] [PubMed] [Google Scholar]
- [164]. Blanchard, G. B. , Kabla, A. J. , Schultz, N. L. , Butler, L. C. , Sanson, B. , Gorfinkiel, N. , Mahadevan, L. , and Adams, R. J. , 2009, “ Tissue Tectonics: Morphogenetic Strain Rates, Cell Shape Change and Intercalation,” Nat. Methods, 6(6), pp. 458–464. 10.1038/nmeth.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]


