Abstract
Systems Toxicology aims to change the basis of how adverse biological effects of xenobiotics are characterized from empirical end points to describing modes of action as adverse outcome pathways and perturbed networks. Toward this aim, Systems Toxicology entails the integration of in vitro and in vivo toxicity data with computational modeling. This evolving approach depends critically on data reliability and relevance, which in turn depends on the quality of experimental models and bioanalysis techniques used to generate toxicological data. Systems Toxicology involves the use of large-scale data streams (“big data”), such as those derived from omics measurements that require computational means for obtaining informative results. Thus, integrative analysis of multiple molecular measurements, particularly acquired by omics strategies, is a key approach in Systems Toxicology. In recent years, there have been significant advances centered on in vitro test systems and bioanalytical strategies, yet a frontier challenge concerns linking observed network perturbations to phenotypes, which will require understanding pathways and networks that give rise to adverse responses. This summary perspective from a 2016 Systems Toxicology meeting, an international conference held in the Alps of Switzerland, describes the limitations and opportunities of selected emerging applications in this rapidly advancing field. Systems Toxicology aims to change the basis of how adverse biological effects of xenobiotics are characterized, from empirical end points to pathways of toxicity. This requires the integration of in vitro and in vivo data with computational modeling. Test systems and bioanalytical technologies have made significant advances, but ensuring data reliability and relevance is an ongoing concern. The major challenge facing the new pathway approach is determining how to link observed network perturbations to phenotypic toxicity.
Introduction
As a subset of systems biology, systems toxicology aims to describe the resilience of biological systems to perturbation by toxicants, i.e., the ability (or lack thereof) to return to normal function. The toxicological community in the 21st century is repositioning from empirical, animal-based testing to a mechanistic understanding of chemical-induced biological perturbation in toxicity pathways and networks,1 ushering in a radical rethinking of safety assessment.
This repositioning has been driven by the revolution in genomics and a systems-oriented perspective on biology that aims to address biological processes as integrated systems of diverse interacting components.2 An understanding of biology from a systems perspective involves3 (1) collection of large sets of experimental data by high-content technologies and/or by mining molecular biology and biochemistry literature and databases; (2) proposal of mathematical models that might account for at least some significant aspects of this data set; (3) accurate computer simulation of the mathematical models to obtain numerical predictions; and (4) assessment of the quality of the models by comparing numerical simulations with the experimental data.
Systems Toxicology adds to this challenge a requirement to describe the perturbation of these systems and their resilience,4 in response to potential hazardous exposures. An overarching goal of Systems Toxicology is to relate complex exposures, via susceptibility factors and alterations of biological processes with impacts on a population level. A practical building block involves reliable experimental model systems to measure key events along pathways, which are really networks, and linking them to adverse outcomes. Addressing such adverse outcome pathways from a network perspective involves diverse strategies for the integrative analysis of omics measurements. Finally, observed network perturbations and the mathematical models that describe them need to be linked with particular phenotypes. This requires computational and empirical approaches for prediction and qualification.
Building on a highly successful Systems Toxicology conference held in Ascona, Switzerland in 2013, a second Systems Toxicology meeting was held in Les Diablerets, Switzerland, in early 2016.5 The 2013 meeting set out to evaluate how state-of-the-art systems biology tools can be used to elucidate toxicity pathways and provide realistic exposure and outcome assessments, as well as to establish a general framework for interpreting and applying Systems Toxicology data to inform chemical risk assessment policy and regulation.1 Three years later, the aim was to explore in greater detail specific applications of systems toxicology approaches. The objectives of the conference were to (1) illustrate real-world examples of how systems toxicology could be applied to elucidate toxic modes of action and contribute to realistic exposure and biological impact assessments; (2) learn how experimental and computational elements could be integrated in systems toxicology-based approaches; (3) reveal recent advances in complementary and multidisciplinary research with the potential to enhance further development and application of systems toxicology; and (4) bridge scientific approaches in systems toxicology with applications in human toxicological risk assessment.
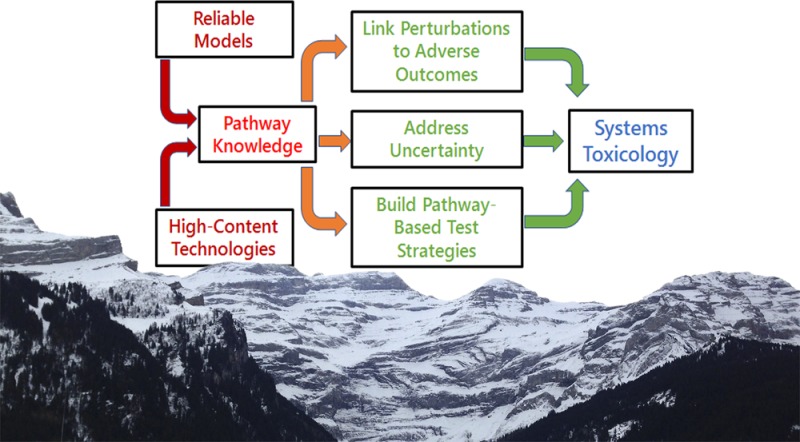
This perspective is based on presentations and discussions at the 2016 Systems Toxicology meeting. It is a simple and incomplete but we hope useful snapshot of current aspects of this rapidly developing field. The perspective addresses first the type of pathway information required for Systems Toxicology and the requirements the model systems and omics measurements have to satisfy to derive such information. Furthermore, it addresses three key challenges, i.e., how to link network perturbations to phenotypes, how to address uncertainty in computational models, and how to develop pathway-based testing strategies as a step toward systems toxicology risk assessment. Finally, a number of emerging examples toward systems toxicology are given, many of them recently highlighted in a Chemical Research in Toxicology virtual special issue on Pathway-Based Approaches for Environmental Monitoring and Risk Assessment.6
Toxicity Pathways and Networks
Pathway-based toxicology is at the core of the 21st Century Toxicity Testing vision (NRC 2007).7 Pathway-based mechanistic analysis is particularly important in interpreting the growing amount of chemical toxicity data based on high-content8,9 and high-throughput screening (HTS).10 For instance, how does one interpret the impact of toxicants on hundreds of proteins and genes in multiple pathways? What does it mean if multiple toxicants have shared mechanisms and pathways? Many toxicants actually simultaneously activate multiple pathways; this phenomenon has been termed promiscuity in chemical toxicity, i.e., most toxicants act via more than one discrete pathway. We need to consider mechanisms to make sense of observed perturbations.8
The term “Pathway of Toxicity” (PoT)11 was coined for this purpose. PoTs are defined on a molecular level, i.e., with a high level of detail, which typically requires the generation of new experimental information. They aim for quantitative relations and fluxes. PoTs rely mostly on the untargeted identification of molecular interactions, typically by omics technologies. Mechanistic validation, i.e., establishing whether identified mechanisms are relevant for human or environmental health effects, has been suggested.12
Pathway-oriented approaches are largely based on an assumed, principally linear sequence of events, i.e., A leads to B leads to C, and if we block B, A will not lead to C. Ultimately, however, there will be a need to combine linear pathways into network models.13 This is the basic goal of Systems Toxicology: Computational network models,14 e.g., virtual organ models,15 based on our pathway knowledge make predictions about organs or whole organism responses (such as the virtual embryo palate closure model).16 Comparison of model outputs with actual measurements will then show how close we are to understanding the characteristics of living networked systems.
Requirements for Test Systems to Derive Pathway Information
The pathway knowledge to build Systems Toxicology to a large extent still requires experimental models to derive such information, typically by omics technologies. The complexity of the organism, therefore, has to be reflected in the model. Test systems increasingly aim to reflect such aspects of the complexity of human physiology, for example, organotypic cultures or microphysiological systems. 3D organoid cultures17 and their bioengineered environments move closer to this goal18 by creating an organ-relevant model of multiple collaborating cell types, perfusion, organ architecture, and functionality. However, these elements are rarely combined in typical model systems.19
The data generated by test systems need to be both reliable and relevant. The OECD20 defines reliability in the context of formal validation as a measurement of “the extent to which a test method can be performed reproducibly within and between laboratories and over time, when performed by using the same protocol. It is assessed by calculating intra-laboratory and inter-laboratory reproducibility and intra-laboratory repeatability.” While it is obvious that such reproducibility is necessary in experimental test systems, it is astonishing how seldom this requirement is actually addressed in either in vivo or in vitro test systems (except for a small number of formal validation studies). This situation has undoubtedly contributed to the current reproducibility crisis in science.21 Recognizing this for in vitro testing, the Good Cell Culture Practice (GCCP) guidance22 has recently been revamped23 in an International GCCP collaboration.24 The upgrade includes work toward in vitro reporting standards.25 In short: 21st century toxicology starts with 21st century cell culture.26 The European Union Reference Laboratory for alternatives to animal testing (EURL ECVAM) is currently coordinating the development of an OECD guidance document on Good In Vitro Method Practice (GIVIMP) which may be adopted in 2017.27 GIVIMP should contribute to increased standardization and harmonization in the generation of in vitro information on test item safety.
What is measured must be not only reproducible but also relevant. Sometimes we reliably measure the wrong end point. The NIEHS (1997) definition of relevance indicates that it “describes whether a test method is meaningful and useful for a particular purpose. It is the extent to which the measurement result and uncertainty can accurately be interpreted as reflecting or predicting the biological effect of interest”.28 The way forward must focus our efforts on developing human cell-based in vitro systems,29 and the challenge is that such systems need to permit the reliable measurement of end points of relevant functional biological processes.
Thus, reliability and relevance are the cornerstones of formal validation and should be a basis for the test systems used to deduce pathways and networks.30,31 This assertion is not meant to suggest that only formally validated test systems should be used, only that it should be a goal. This is an iterative process, whereby the most mechanistically relevant test systems are used to further detail, challenge, and refine mechanistic understanding, which, in turn, can lead to improved test systems.
Requirements for Omics Data to Derive Pathway Information
Omics data can be a basis for the identification of pathways and networks,32 but many challenges remain, especially for metabolomics,33,34 which is important since it is closest to phenotypical changes.35,36
The progress in omics technologies is impressive; new generations of instruments and chips offer real advantages and enable measurement of an enormous number of signals. Each omics technology has its own advantages and limitations,37 as exemplified in Table 1 for some of the more common technologies.
Table 1. Advantages and Limitations of Common Omics Technologies.
| technology | advantages | limitations |
|---|---|---|
| RNASeq (including RASLseq and TempO seq) | less costly and getting continuously cheaper | number of transcripts (>27,000 genes plus others) |
| includes low abundance transcripts and noncoding RNA | RNA changes do not imply protein changes | |
| fully quantitative | large data sets posing bioinformatics challenges | |
| incomplete functional annotation | ||
| chip-based transcriptomics | best standardized as to performance, reporting, and functional annotation | number of transcripts (>27,000 genes plus others) |
| often strong signals (induction factors) | RNA changes do not imply protein changes | |
| interpretation by miRNA and transcription factor analysis advancing | semiquantitative at best | |
| proteomics | sensitivity and specificity | large number of (modified) proteins (up to 1 million) require extensive method development and multiple measurements |
| direct reflection of altered protein levels | ||
| may pick up direct compound–protein interactions | ||
| sensitivity of advanced mass-spectrometry-based methods gradually approaching that of chip-based transcriptomics | protein quantity changes do not imply functional changes | |
| only very few laboratories able to implement advanced proteomics methods | ||
| less costly per measurement | not all proteins in a sample can be identified and limited availability of antibodies | |
| metabolomics | actual phenotypic change | metabolite identification by MS |
| fewer number of substances (several thousands) | low sensitivity of NMR | |
| less costly per measurement | small effect strengths, i.e., often only slight changes over background activity | |
| NMR analysis is robust, noninvasive, and quantitative, and allows structural identification of metabolites | currently does not provide enough mechanistic information (this will improve in the near future) | |
| MS analysis is sensitive, quantitative, and detects a high number of metabolites | little standardized as to performance, reporting, and functional annotation | |
| differences between platforms | ||
| incomplete extraction of metabolites depending on extraction method |
Because of limited throughput and high cost, only a few repeat measurements and experimental conditions can typically be tested. Combined with considerable variability of the measurements, this makes it difficult to separate statistically significant and meaningful changes from measurement artifacts, and small perturbations often escape detection. Cherry-picking individual significant signals and simple clustering on pathways often produces irrelevant results (as has been recently shown for metabolomics34).
A promising approach is reduction in dimensionality of data, e.g., by transcription factor analysis38,39 or miRNA network analysis of transcriptomics data. A further promising approach is multiomics data analysis (as pursued, for example, in the Human Toxome project),40 whereby perturbations of the same pathways in several orthogonal omics technologies support greater confidence in reliability of pathway elucidation. One promising example of the result is proteogenomics, as well as the more challenging combination of transcriptomics with metabolomics.
It is often unclear whether the molecular changes and complex signatures detected by omics analyses actually represent adverse effects, and this lack of clarity hinders the broad application of omics measurements in safety assessments. To establish causal relationships, experimental challenges based on the Koch postulates or the Bradford-Hill criteria are necessary.41
The same issue of relevance also applies to the use of omics for exposome measurements42,43 involving the broad direct or indirect measurement of exogenous and endogenous substances in (human) biofluids as an indicator of the totality of exposure impacts. Thus, if we cannot interpret the patterns measured by omics, their use for exposome analysis is limited. Knowledge of mechanisms of toxicity could help identify meaningful signals.44 This approach, giving rise to candidate pathways of toxicity, could be expanded with targeted measurements of components of the pathway as a basis for explaining signatures in exposome measurements.
Requirements for High-Throughput and High-Content Imaging Data to Derive Pathway Information
The high-throughput screening (HTS) programs of ToxCast45 and Tox21 measure numerous cellular responses, and understanding the pathways by which such cellular responses can lead to adverse outcomes is central in the interpretation and validation of the HTS data46 and for designing future integrated testing strategies.47−49 Kleinstreuer et al. used computational clustering of ToxCast data from 641 environmental chemicals tested in primary human cell systems to identify potential chemical targets and mechanisms for elucidating toxicity pathways.50
Similarly, high-content imaging (HCI) provides data allowing the analysis of pathways. Shah et al. used HCI to simultaneously measure multiple cellular phenotypic changes in HepG2 cells induced by 967 chemicals in order to identify the “tipping point” at which the cells failed to show recovery toward a normal phenotypic state.51 The aim is to use such cellular tipping points to define points of departure for risk-based prioritization of environmental chemicals. These examples show that high-throughput and high-content imaging information can support pathway deductions, though experiences compared to omics approaches are rather limited.
Challenge of Linking Network Perturbations to Phenotypes
Large-scale data streams can be used to develop an integrative qualitative and quantitative view of complex networks operative in cells and organs. Linking these networks to in vivo adversity, however, remains a challenge.
Liu and colleagues used a machine-learning approach that integrates chemical structure, bioactivity, and toxicity data to classify rodent hepatotoxicants.52 While not directly aimed at elucidating pathways for liver toxicity, their data-driven approach empirically identifies putative biomarkers that could be key events involved in pathways to hepatic injury, inflammation, and neoplasia. Van den Hof et al. used gene expression profiles in vitro in HepG2 cells as a proof of principle for classifying known hepatotoxicants and nonhepatotoxicants.53
In order to link exposure and effect, quantitative high-content imaging of cellular adaptive stress response pathways for chemical toxicity visualizing pathway activation serves as an example. Garcia-Serna et al. computationally combined chemical, structural, and biological hazard data of bioactive small molecules to gain a better understanding of the mechanisms leading to adverse effects.54 Angrish et al. introduced a gas phase probe molecule into an in vitro system, observed normal steady state, added chemicals of interest, and quantitatively measured (from headspace gas) effects on metabolism that could be linked back to a well-defined corresponding in vivo effect.55 Similarly, Gonzalez-Suarez et al. selected three well-known harmful and potentially harmful constituents in tobacco smoke, established a high-content screening in normal human bronchial epithelial cells using 13 indicators of cellular toxicity complemented with a microarray-based whole-transcriptome analysis followed by a computational approach leveraging mechanistic network models, to identify and quantify perturbed molecular pathways.56
Challenge of Addressing Uncertainty in Computational Models for Systems Toxicology
Computational models in Systems Toxicology can involve multiple biological scales, from molecular signaling to tissue dynamics to whole organisms, as well as time scales from fractions of a second to human lifetimes. Small uncertainties at one scale could cause large errors in predictions at another scale. In building reliable predictive computer model systems, it is therefore important to consider uncertainties,57 including (at minimum): (1) Uncertainty in Systems Toxicology model structure: assessing whether the equations/network in use are appropriate. Would others fit the data equally well, but result in different predictions? (model selection). (2) Uncertainty in parameter values within the equations (minimization to fit data, inverse problems, parameter identifiability, dealing with variability): How sure are we that the numbers we are using in the simulation are accurate? Can we define probability distributions for them?. (3) Uncertainty propagation: how does the uncertainty in the model, parameters, and any inputs propagate through to uncertainty in our predictions of end points?
Assessing 1–3 is known as Uncertainty Quantification (UQ). UQ approaches are well developed, and often applied as standards in simple ADME compartmental concentration models, but extending UQ approaches to signaling pathway networks, adverse outcome pathways (AOP), and complex physiologically based pharmacokinetic (PBPK) models58 requires more attention.
Challenge of Pathway-Based Testing Strategies
Systems Toxicology can be seen as the ultimate goal of transitioning to a pathway-based approach in risk assessment, as it aims for the integration of our pathway knowledge into predictive models. This requires on the way, the generation of pathway-based information and the integrated use of such information to support risk assessment. By designing our testing strategies around the emerging pathway- and network-knowledge, we are converging with the Systems understanding and providing the data for its modeling.
Chemical risk assessment comprises hazard identification (adverse effects produced by a substance), hazard characterization (dose–response analysis of how much of a substance is required to produce adverse effects), and exposure assessment. Hazard characterization has traditionally used so-called “apical end points” (typically animal organ pathology) and has been plagued by interspecies and interindividual differences and the need for high-dose to low-dose and short-term to long-term extrapolations.59 Additional uncertainty in exposure assessments arises due to multiple kinetic factors that can affect target concentrations. As a consequence, uncertainty factors are added in risk assessments to err on the “safe” side. Still, concerns remain: Are we looking for the right adverse effects? Are those observed effects relevant for humans? Do kinetics differ between animals and humans? Are vulnerable human subpopulations not addressed in animal studies? Could kinetics and biological barrier functions differ in disease or at certain life stages? Could coexposure to other toxicants (mixture effects) alter exposure and hazard thresholds?
Some of these questions have been addressed by the World Health Organization International Program on Chemical Safety (WHO/IPCS) Mode of Action (MOA) framework, which introduced the concept of a chain of causal key events leading to toxicity.60−62 Using this framework, the question of human relevance of animal data has been addressed for many well-characterized toxicants.63,64
The MOA concept was subsequently adapted by the OECD Adverse Outcome Pathway (AOP) framework,65−67 which aims to identify key events in toxicity pathways between a molecular initiating event (MIE) and an adverse outcome (AO). In contrast to MOAs, AOPs are chemical-agnostic, i.e., they attempt to describe common key events triggered by multiple chemicals. Building a knowledgebase of these key events will assist in establishing the relevance of test systems.
It is important to note that AOPs are designed to be parsimonious descriptions that can be used for regulatory purposes, rather than detailed mechanistic descriptions of systems toxicity networks. The aim is to identify as many key events as necessary but as few as possible.68 The different levels of detail between AOPs and PoTs has led to the existence of different repositories, i.e., the AOP-Wiki69 and Effectopedia70 for AOPs, versus the Human Toxome Knowledgebase under construction.
In 2013, the Scientific Committees of the European Commission Directorate General for Health and Consumer Safety issued an opinion71 that “There is a trend/need to change the basis of risk assessment from the one based on standard tests to one that is centered on modes of action. In investigations using laboratory animals, increasing importance should be directed to characterizing the mode of action with less emphasis to end points based on histopathological criteria, body and organ weight, and blood chemistry.” This approach was adopted in the EU research program SEURAT-1 (Safety Evaluation Ultimately Replacing Animal Testing), funded by the European Community’s Seventh Framework Programme FP7,72 and subsequently in EUToxRisk,73,74 a large-scale, six-year EU project aiming to integrate in vitro and in silico toxicology, read-across methods, and adverse outcome pathways for the prediction of repeated dose systemic toxicity (liver, kidney, lung, and nervous system) and developmental/reproductive toxicity. It focuses on regulatory hazard and risk assessment rather than on individual test methods. AOPs and integrated approaches to testing and assessment (IATA) play a central role in the EU-ToxRisk project. The goal is to develop quantitative AOPs (qAOPs) for regulatory assessment of chemical safety in humans.73 This is all part of a systematic development of safety sciences.75
Many pieces of the puzzle are emerging from various places, e.g., Allen and colleagues76 proposed a unified approach to defining MIEs for risk assessment based on QSARs and receptor activation. Once chemicals can be accurately mapped to MIEs, potential AOPs can be inferred. This approach was implemented by Mellor and colleagues77 by computationally identifying structural alerts for nuclear receptor (NR) activators from 12,713 NR agonists in ChEMBL, a manually curated chemical database of bioactive molecules with drug-like properties by the European Bioinformatics Institute (EBI), of the European Molecular Biology Laboratory (EMBL). These structural alerts can be used to prioritize chemicals by NR-mediated pathways to hepatic steatosis. Similarly, Mekenyan and colleagues78 computationally linked chemicals to MIEs involved in respiratory sensitization based on absorption into epithelial membranes (physicochemical rules) and electrophilic reactivity (simulating metabolism). They further evaluated the predictive accuracy of their approach using known respiratory sensitizers. Similarly, Roberts and Aptula79 studied electrophilic reactivity in the context of skin sensitization potency.
Vinken80 provides an overview of constructing AOPs involved in drug-induced liver injury (DILI) with AOs steatosis, cholestasis, and fibrosis. Potential applications of such AOPs include read-across or chemical grouping based on MIEs using cheminformatics, as well as integrated approaches to testing and assessment (IATA).
From a regulatory point of view, IATAs are essential for consistent and transparent evaluation of the relevance of AOP-based (frequently in vitro) data for specific end points and regulatory decisions.81−83 The first example of AOP-based IATA development is for in vitro skin sensitization testing.82 Two in vitro test guidelines have been adopted by the OECD, covering the molecular initiating event of the AOP for skin sensitization (covalent protein binding; OECD Test Guideline 442C)84 and the second key event (keratinocyte inflammation; OECD Test Guideline 442D).85 Combining the results of these two assays is more predictive than each assay alone,86 leading to an industry proposal for an Integrated Testing Strategy87 based on readouts from multiple key events, and an associated decision support system for the risk assessor. On the regulatory side, the U.S. Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) is developing integrated decision strategies based on in vitro, in chemico, and in silico information derived from the skin sensitization AOP.88 It is clear that much work needs to be done to provide simple, acceptable AOP-based IATAs for regulators.89
Outstanding regulatory challenges90,91 include the need for reliable data and data analysis, in addition to quantifying uncertainty due to genetic background, cell type and topography, life-stage, and exposure temporality in dose–response modeling.
Altogether, the AOP and PoT work is establishing the pathway-based approach to safety testing, which forms the basis of the adaption of Systems Toxicology at a later stage.
Emerging Building Blocks and Their Applications for Systems Toxicology
Mechanistic toxicology is flourishing in academic research and increasingly impacting additional or alternative evidence on the regulatory process. A lot of this is contributing to the development of Systems Toxicology. A few examples drawn from the congress and a recent virtual issue of this journal and Environmental Sciences and Technologies(92) are used to illustrate ongoing progress in the following.
Physiologically Based Pharmacokinetic (PBPK) Modeling
Computational PBPK models are used to estimate xenobiotic concentrations in various organs.93 Detailed permeability-limited PBPK models of the liver, kidney, lung, brain, intestine, and skin have been described.94−99 Both chemical properties and modeled physiology can be altered to investigate the effects of a given xenobiotic in individuals of different ethnicities, ages (e.g., pediatric and geriatric), or altered levels of organ function (e.g., renal and hepatic impairment).100 The combination of PBPK modeling with in vitro–in vivo extrapolation (IVIVE) permits bottom-up prediction of absorption, clearance, and distribution of xenobiotics.101,102
The combination of PBPK with pharmacodynamic or toxicodynamic models enables investigation of safety risks under conditions which are not amenable to clinical investigation.103,104 For example, using permeability-limited PBPK models, it was possible to predict the impact of a transporter genotype on the pharmacodynamics of rosuvastatin within the liver.105 PBPK models have also been coupled with information about the effects of xenobiotics on heart tissue (ion current disruption, contractility modification, and metabolic pathways disturbance leading to cell apoptosis) to simulate the cardiotoxicity of various agents. The verification of the simulation results against clinically observed end points (i.e., QT prolongation) demonstrates the usefulness of such combined modeling in drug safety assessment.106 Also, the risk of human nephrotoxicity can be estimated from animal studies by modeling drug-specific transporters to derive local kidney concentrations.107
Hepatic Toxicity
One example of network modeling is a large scale mechanistic simulation combining Flux Balance Analysis of Genome Scale Metabolic Network of human hepatocyte with a large-scale model of nuclear receptor signaling.108 This model can qualitatively link gene activity perturbation with bile acid homeostasis, thus permitting mechanistic assessment of the role of genetic polymorphism in toxicity and interpretation of omics data.
Cardiac Toxicity
Blockade of the hERG potassium channel by direct binding of a drug molecule causes QT prolongation and increases pro-arrhythmic risk. Since 2005, candidate drug compounds must be screened for hERG binding (ICH S7B guideline)109 and clinical long QT (ICH E14 guideline).110 These guidelines have been remarkably successful in preventing compounds with increased pro-arrhythmic risk reaching the market; this is a highly sensitive approach (few false negatives), but many safe compounds on the market since well before 2005 would fail to meet these guidelines, suggesting that they may have low specificity (many false positives). It has been proposed that multiple ion channel block may explain the discrepancy in sensitivity and specificity;111−113 put simply, blocking additional ion channels may compensate for blocking of hERG and reduce pro-arrhythmic risk.
In addition to multichannel effects, it is also important to consider drug kinetics at particular ion channels. Does the drug simply bind to the channel and block its current, or do we need to consider which conformational states the drug can bind, and how the channel behavior is affected by this?114−116
The development of improved test systems and bioanalytical methods has enabled a systems approach for measuring drug binding effects and simulating their consequences.117 First, new cell-line technologies (overexpression of ion channels of interest in immortal cell lines, together with automated patch-clamp ion current screening platforms) have made it possible to routinely screen compounds for blockade of multiple cardiac ion channels.118 Second, mathematical models of cardiac electrophysiology are very well established; they integrate the information on multiple ion channel effects by describing how action potentials are formed from a set of voltage-dependent ionic currents.119 Predicted and measured drug-induced action potential changes can be used as risk indicators. Notably, interference with one ion channel alters membrane voltage and thus the sensitivity to block of other ion channels. Modeling involves not just the biochemistry of ion channels and gene/protein networks but also the biophysics of membrane electrophysiology. These mathematical models are now in routine use to predict the results of safety tests120,121 and are an integral element of new initiatives to replace existing guidelines with more accurate preclinical assessments of pro-arrhythmic risk than the existing clinical studies.122,123
Renal Toxicity
In recent years, the toxicological community has shifted focus from animal studies to an in vitro molecular understanding of chemical-induced biological perturbations.124 The ability to map pathway activation at the mRNA level has uncovered networks of toxicologically relevant processes, including stress response pathways,125 that are involved at the early stages of many chemical-induced pathologies.126,127 Integrating transcriptomics with other omics information (such as proteomics and metabolomics) dramatically increases the depth of biological interpretation, allowing us to establish where breaks in cellular homeostatic regulation occur. We should be careful, however, to confirm the relevance of this information in real world situations and to normal human biology. Where possible, it is best practice to use human, stable, noncancerous, differentiated cells, rather than unstable, highly proliferating, glycolytic cells with practically alien karyotypes (such as A549 cells or MCF7 cells).128 It is also important to include adequate exposure data when interpreting high content toxicological data.
In the EU seventh Framework Project, Predict-IV, integrated omics analyses were combined with intracellular dosimetry in in vitro liver, kidney, and CNS test systems. Intracellular dosimetry analyses using inductively coupled plasma mass spectrometry (ICP-MS) and omics data (transcriptomics, metabolomics, and proteomics) were captured during the administration of renal toxicants for up to 14 days129−131 in the in vitro human proximal tubule RPTEC/TERT1 cell line, which has a close-to-normal cellular phenotype132−134 and can be maintained as a tissue monolayer for months.132,135 Cyclosporine A (CsA), an immunosuppressant agent and suspected nephrotoxin, is highly lipophilic and thus not readily soluble in aqueous solutions such as cell culture medium. At 15 but not 5 μM, CsA massively accumulated in the cells, likely due to the saturation of the ABC extrusion transporters. Integrated analysis of the exposure and omics data indicated that CsA at 15 μM floods the lipophilic compartments of the cell and causes both ER and mitochondrial disruption; the cells manage to stay alive by Nrf2 and ATF4 activation and by switching back to glycolysis for energy requirements. Increased glycolysis is reflected in increased lactate in the supernatant medium, which can thus be used as a surrogate marker for toxicity.136 The therapeutic mode of action of CsA is binding to cyclophilin and subsequent reduction of T cell activation and their immune response. In the RPTEC/TERT1 cells, CsA induced apical excretion of cyclophilin B (CyP-B) as a surrogate marker of clinical efficacy at both 5 and 15 μM.
From the lactate and CyP-B data, an in vitro therapeutic window is apparent for CsA, whereby strong CyP-B induction starts at 3 μM, but enhanced glycolysis does not begin until ≥10 μM (Figure 1). Human kinetic data indicate that CsA at therapeutic doses will not reach 10 μM in plasma, demonstrating a narrow therapeutic window between efficacy and toxicity.
Figure 1.
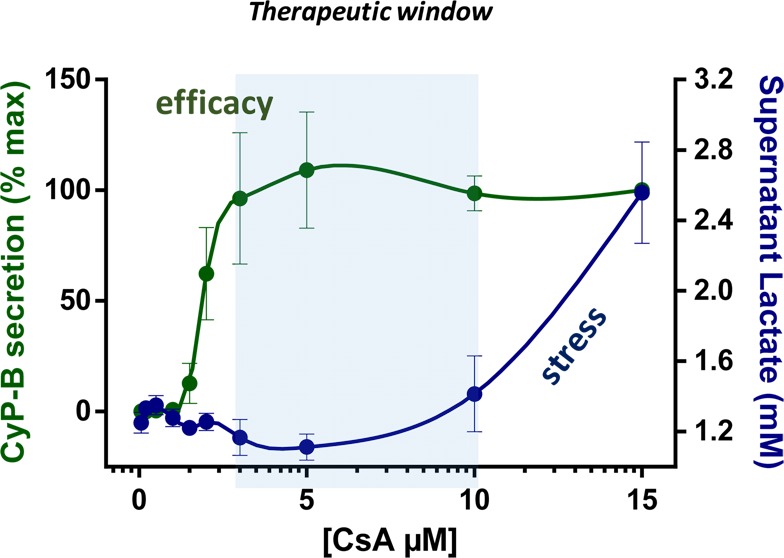
Effects of 24 h treatment with cyclosporine A (CsA) in RPTEC/TERT1 human kidney cells. Concentrations of cyclophilin B (CyP-B) and lactate in the supernatant medium were used as measures of efficacy and toxicity, respectively.131
It is understandable that there is some skepticism in accepting safety data derived solely in vitro, and thus, it is necessary to increase confidence by comparison with similar in vivo data where available. Because of species differences, comparing in vitro human cell data with in vivo animal data is unsatisfactory. A way around this is to verify in vitro biomarkers in clinical samples from diseased patients, which often have similar underlying etiology. We have shown, for example, that IL-19 is induced and secreted into the supernatant medium in RPTEC/TERT1 kidney cells exposed to several nephrotoxins, including zoledronate and antiviral compounds.137 IL-19 was found elevated in the urine of CKD patients and exhibited a negative correlation to the estimated glomerular filtration rate (a measure of renal function) and a positive correlation to urinary N-acetyl-beta-d-glucosaminidase and lipocalin 2 (biomarkers of proximal tubule injury). This type of in vitro to in vivo correlation increases the confidence in the relevance of specific biomarkers and their pathways.138
Conclusions
Three developments are enabling the progress of the Systems Toxicology approach: (1) reliable and relevant in vitro test systems, (2) high-throughput and high-content methods generating large-scale data streams, and (3) in silico model systems. All require delineation of pathways of toxicity. Development of pathway-based approaches in toxicology will require a mountain of work. The state-of-the-art approach summarized here is like a snapshot taken from the foothills. No single picture of a mountain can capture its complexity, but we believe that the current perspective will help us further plan the route to the top.
Acknowledgments
We are most grateful for the professional editing of this article provided by Mr. Michael Hughes, CAAT, Johns Hopkins University, Baltimore.
Glossary
Abbreviations
- AOP
adverse outcome pathway
- ChEMBL
chemical database by the EMBL
- CsA
cyclosporine A
- EBI
European Bioinformatics Institute
- EMBL
European Molecular Biology Laboratory
- EURL ECVAM
European Union Reference Laboratory for Alternatives to Animal Testing
- GIVIMP
Good In Vitro Method Practice
- GCCP
Good Cell Culture Practice
- HCI
high-content imaging
- HTS
high-throughput screening
- IATA
integrated approaches to testing and assessment
- ICP-MS
inductively coupled plasma mass spectrometry
- IVIVE
in vitro–in vivo extrapolation
- MOA
mode of action
- NIEHS
US National Institute for Environmental Health Sciences
- NR
nuclear receptor
- PBPK
physiologically based pharmacokinetic
- PoT
pathway of toxicity
- qAOPs
quantitative AOPs
- SEURAT-1
EU project Safety Evaluation Ultimately Replacing Animal Testing
- UQ
uncertainty quantification
- WHO/IPCS
World Health Organization International Program on Chemical Safety
Biographies
Professor Thomas Hartung, M.D., Ph.D., is Professor of Toxicology and Chair for Evidence-based Toxicology, Pharmacology, Molecular Microbiology and Immunology at Johns Hopkins Bloomberg School of Public Health, Baltimore, and University of Konstanz, Germany; he also is Director of their Centers for Alternatives to Animal Testing (CAAT). CAAT organizes the Evidence-based Toxicology, Good Cell Culture Practice, and Green Toxicology Collaborations. He heads the Human Toxome project and is the former Head of the European Commission’s Center for the Validation of Alternative Methods. He was a scientific advisor and key note speaker at Systems Toxicology 2016 in Switzerland.
Dr. Rex FitzGerald, Ph.D., is a Regulatory Toxicologist at the Swiss Centre for Applied Human Toxicology (SCAHT). Prior to joining SCAHT in 2010, he was the head of reproduction toxicology and a medical advisor for gynecological endocrinology at Ciba-Geigy. In 1996, he founded FitzGerald Toxicology & Clinical Services, providing safety documentation and regulatory advice to the chemical–pharmaceutical industry. He is a Diplomate of the American Board of Toxicology (DABT). His research and professional interests include developmental neurotoxicity and human risk assessment.
Professor Paul Jennings, Ph.D., is an Assistant Professor in the Division of Physiology at the Medical University of Innsbruck. His research is centered on understanding molecular mechanisms of chemical induced nephrotoxicity with a strong focus on stress response pathways including but not limited to Nrf2, unfolded protein response, and p53. This research involves the integration of multiomic data streams with biokinetic data with the aim to improve chemical safety prediction utilising human-based in vitro models.
Dr. Gary R. Mirams, Ph.D., is the Wellcome Trust & Royal Society Sir Henry Dale Fellow in the Faculty of Science at the University of Nottingham, where he is part of the Centre for Mathematical Medicine & Biology in the School of Mathematical Sciences. He is an applied mathematician working on problems in biology, particularly in the area of cardiac electrophysiology and drug safety. He is working with a number of pharmaceutical companies to embed simulation in their safety work and to develop free open source software to perform these simulations.
Professor Manuel C. Peitsch, Ph.D., is Vice President of Research and Development and Chief Scientific Officer at Philip Morris International in Switzerland. He is Titular Professor of Bioinformatics at the University of Basel and cofounder and chairman of the board of the Swiss Institute of Bioinformatics. At PMI he leads the department responsible for the assessment of candidate Reduced Risk Tobacco Products through pre-clinical toxicology, systems toxicology, and clinical studies, as well as for their regulatory submissions.
Professor Amin Rostami-Hodjegan, Ph.D., is Professor of Systems Pharmacology at the Centre for Applied Pharmacokinetic Research (CAPKR) at the University of Manchester and was previously Professor of Systems Pharmacology at the University of Sheffield. Professor Rostami has authored/coauthored over 200 peer-reviewed full articles and serves on the Editorial Boards of several journals. He has been an invited speaker at over 170 conferences and has led a number of hands on workshops in the area of in vitro–in vivo extrapolation as applied to ADME in Drug Development. Professor Rostami is also the Senior Vice President of Research & Development and Chief Scientific Officer at Certara, a company with a scientific team which includes almost 100 Ph.D.s or M.D.s. His mission is to ensure that the latest science is incorporated into all of the company’s products and activities across Certara’s Business Units.
Dr. Imran Shah, Ph.D., is a computational systems biologist in EPA’s National Center for Computational Toxicology (NCCT). Dr. Shah provides NCCT leadership in innovative computational approaches to rapidly evaluate health implications for thousands of environmental stressors. His research focuses on predicting chemical-induced toxicity from complex large-scale molecular data sets using novel machine learning and systems biology methods.
Professor Martin F. Wilks, M.D., Ph.D., is the Director of the Swiss Centre for Applied Human Toxicology (SCAHT) and in 2012 was appointed Professeur titulaire at the Medical Faculty of the University of Geneva. He is a EUROTOX Registered Toxicologist, a Fellow of the British Toxicology Society, and Honorary Consultant Medical Toxicologist at Guy’s & St. Thomas’ Hospital Trust, London. His research and professional interests include clinical toxicology, toxico-epidemiology, and risk assessment of chemical exposures.
Professor Shana J. Sturla, Ph.D., is Professor of Toxicology at the ETH Zurich. Researchers in her laboratory (www.toxicology.ethz.ch) address the chemical basis of disease incidence and treatment by investigating relationships among chemical structure, biotransformation, and cellular responses. She received a European Research Council Grant, the American Chemical Society Young Investigator Award for Chemical Research in Toxicology, is Vice President of the Swiss Society of Toxicology, and Chair Elect of the American Chemical Society Division of Toxicology.
G.R.M. gratefully acknowledges support from a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 101222/Z/13/Z). P.J. acknowledges support of the European Union’s seventh Framework Programme (FP7/2007-2013) under grant agreement no 202222, Predict-IV. S.J.S. acknowledges support of the Swiss National Science Foundation (Grant Number 156280), the European Research Council (Grant Number 680920), and the European Union’s Horizon 2020 Program (Grant Number 633172). R.E.F. and M.F.W. are at the Swiss Centre for Applied Human Toxicology, which is funded by the Swiss Confederation and the Universities of Basel, Geneva, and Lausanne; T.H. is funded by the NIH transformative research project on “Mapping the Human Toxome by Systems Toxicology” (R01ES020750).
The funding bodies had no involvement in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the perspective for publication.
The authors declare the following competing financial interest(s): T.H. is the founder of Organome LLC, Baltimore, share-holder and chief scientific advisor of Atheralabs, Luxembourg, and consults AstraZeneca, Cambridge, UK, all in the field of organo-typic cultures.
References
- Sturla S. J.; Boobis A. R.; FitzGerald R. E.; Hoeng J.; Kavlock R. J.; Schirmer K.; Whelan M.; Wilks M. F.; Peitsch M. C. (2014) Systems toxicology: from basic research to risk assessment. Chem. Res. Toxicol. 27, 314–329. 10.1021/tx400410s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario D.; Hartung T. (2014) Glossary of reference terms for alternative test methods and their validation. ALTEX 31, 319–335. 10.14573/altex.140331. [DOI] [PubMed] [Google Scholar]
- Duffus J. H.; Nordberg M.; Templeton D. M. (2007) Glossary of terms used in toxicology, 2nd edition. Pure Appl. Chem. 79, 1153–1344. 10.1351/pac200779071153. [DOI] [Google Scholar]
- Smirnova L.; Harris G.; Leist M.; Hartung T. (2015) Cellular Resilience. ALTEX 32, 247–260. 10.14573/altex.1509271. [DOI] [PubMed] [Google Scholar]
- Systems Toxicology (2016) https://systox.ch (last accessed 21 Mar, 2017).
- Ankley G.; Escher B.; Hartung T.; Shah I. (2016) Pathway-based approaches for environmental monitoring and risk assessment. Chem. Res. Toxicol. 29, 1789–1790. 10.1021/acs.chemrestox.6b00321. [DOI] [PubMed] [Google Scholar]
- National Research Council/National Academy of Sciences (2007) Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press, Washington, DC. DOI: 10.17226/11970. [DOI]
- Hartung T.; McBride M. (2011) Food for thought··· on mapping the human toxome. ALTEX 28, 83–93. 10.14573/altex.2011.2.083. [DOI] [PubMed] [Google Scholar]
- van Vliet E.; Danesian M.; Beilmann M.; Davies A.; Fava E.; Fleck R.; Julé Y.; Kansy M.; Kustermann S.; Macko P.; Mundy W.; Roth A.; Shah I.; Uteng M.; van de Water B.; Hartung T.; Leist M. (2014) Current approaches and future role of high content imaging in safety sciences and drug discovery. ALTEX 31, 479–493. [DOI] [PubMed] [Google Scholar]
- Judson R. S.; Mortensen H. M.; Shah I.; Knudsen T. B.; Elloumi F. (2012) Using pathway modules as targets for assay development in xenobiotic screening. Mol. BioSyst. 8, 531–542. 10.1039/C1MB05303E. [DOI] [PubMed] [Google Scholar]
- Kleensang A.; Maertens A.; Rosenberg M.; Fitzpatrick S.; Lamb J.; Auerbach S.; Brennan R.; Crofton K. M.; Gordon B.; Fornace A. J. Jr.; Gaido K.; Gerhold D.; Haw R.; Henney A.; Ma’ayan A.; McBride M.; Monti S.; Ochs M. F.; Pandey A.; Sharan R.; Stierum R.; Tugendreich S.; Willett C.; Wittwehr C.; Xia J.; Patton G. W.; Arvidson K.; Bouhifd M.; Hogberg H. T.; Luechtefeld T.; Smirnova L.; Zhao L.; Adeleye Y.; Kanehisa M.; Carmichael P.; Andersen M. E.; Hartung T. (2014) Pathways of toxicity. ALTEX 31, 53–61. 10.14573/altex.1309261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T.; Stephens M.; Hoffmann S. (2013) Food for Thought...Mechanistic validation. ALTEX 30, 119–130. 10.14573/altex.2013.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boue S., Fields B., Hoeng J., Park J., Peitsch M. C., Schlage W. K., Talikka M., Binenbaum I., Bondarenko V., Bulgakov O. V., Cherkasova V., Diaz-Diaz N., Fedorova L., Guryanova S., Guzova J., Igorevna Koroleva G., Kozhemyakina E., Kumar R., Lavid N., Lu Q., Menon S., Ouliel Y., Peterson S. C., Prokhorov A., Sanders E., Schrier S., Schwaitzer Neta G., Shvydchenko I., Tallam A., Villa-Fombuena G., Wu J., Yudkevich I., and Zelikman M.. Enhancement of COPD biological networks using a web-based collaboration interface, DOI: 10.12688/f1000research.5984.2. [DOI] [PMC free article] [PubMed]
- Hartung T.; van Vliet E.; Jaworska J.; Bonilla L.; Skinner N.; Thomas R. (2012) Systems toxicology. ALTEX 29, 119–128. 10.14573/altex.2012.2.119. [DOI] [PubMed] [Google Scholar]
- Shah I.; Wambaugh J. (2010) Virtual tissues in toxicology. J. Toxicol. Environ. Health, Part B 13 (2–4), 314–328. 10.1080/10937404.2010.483948. [DOI] [PubMed] [Google Scholar]
- Hutson M. S., Leung M. C., Baker N. C., Spencer R. M., and Knudsen T. B. (2017) Computational Model of Secondary Palate Fusion and Disruption. Chem. Res. Toxicol., DOI: 10.1021/acs.chemrestox.6b00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alépée N.; Bahinski T.; Daneshian M.; De Wever B.; Fritsche E.; Goldberg A.; Hansmann J.; Hartung T.; Haycock J.; Hogberg H.; Hoelting L.; Kelm J. M.; Kadereit S.; McVey E.; Landsiedel R.; Leist M.; Lübberstedt M.; Noor F.; Pellevoisin C.; Petersohn D.; Pfannenbecker U.; Reisinger K.; Ramirez T.; Rothen-Rutishauser B.; Schäfer-Korting M.; Zeilinger K.; Zurich M.-G. (2008) State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology – a t4 report. ALTEX 31, 441–477. 10.14573/altex1406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx U.; Andersson T. B.; Bahinski A.; Beilmann M.; Beken S.; Cassee F. R.; Cirit M.; Daneshian M.; Fitzpatrick S.; Frey O.; Gaertner C.; Giese C.; Griffith L.; Hartung T.; Heringa M. B.; Hoeng J.; de Jong W. H.; Kojima H.; Kuehnl J.; Luch A.; Maschmeyer I.; Sakharov D.; Sips A. J. A. M.; Steger-Hartmann T.; Tagle D. A.; Tonevitsky A.; Tralau T.; Tsyb S.; van de Stolpe A.; Vandebriel R.; Vulto P.; Wang J.; Wiest J.; Rodenburg M.; Roth A. (2016) Biology-inspired microphysiological system approaches to solve the prediction dilemma of substance testing using animals. ALTEX 33, 272–321. 10.14573/altex.1603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T. (2014) 3D - a new dimension of in vitro research. Adv. Drug Delivery Rev. 69-70, vi. 10.1016/j.addr.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (2005) Guidance Document on the Validation and International Acceptance of New or Updated Test Methods for Hazard Assessment, OECD Series on Testing and Assessment, Guidance Document 34, Environment Directorate, Organisation for Economic Cooperation and Development, Paris, France, ENV/JM/MONO(2005)14. [Google Scholar]
- Baker M. (2016) 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454. 10.1038/533452a. [DOI] [PubMed] [Google Scholar]
- Coecke S.; Balls M.; Bowe G.; Davis J.; Gstraunthaler G.; Hartung T.; Hay R.; Merten O.-W.; Price A.; Schechtman L.; Stacey G.; Stokes W. (2005) Guidance on Good Cell Culture Practice. Altern. Lab. Anim. 33, 261–287. [DOI] [PubMed] [Google Scholar]
- Pamies D.; Bal-Price A.; Simeonov A.; Tagle D.; Allen D.; Gerhold D.; Yin D.; Pistollato F.; Inutsuka T.; Sullivan K.; Stacey G.; Salem H.; Leist M.; Daneshian M.; Vemuri M. C.; McFarland R.; Coecke S.; Fitzpatrick S. C.; Lakshmipathy U.; Mack A.; Wang W. B.; Daiju Y.; Sekino Y.; Kanda Y.; Smirnova L.; Hartung T. (2016) Good Cell Culture Practice for stem cells and stem-cell-derived models. ALTEX 34, 95–132. 10.14573/altex.1607121. [DOI] [PubMed] [Google Scholar]
- Pamies D., Barreras P., Block K., Makri G., Kumar A., Wiersma D., Smirnova L., Zang C., Bressler J., Christian K. M., Harris G., Ming G.-L., Kyro K., Berlinicke C., Song H., Pardo C. A., Hartung T., and Hogberg H. T. (2016) A human brain microphysiological system derived from iPSC to study central nervous system toxicity and disease. ALTEX, DOI: 10.14573/altex.1609122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M.; Efremova L.; Karreman C. (2010) Food for thought··· considerations and guidelines for basic test method descriptions in toxicology. ALTEX 27, 309–317. 10.14573/altex.2010.4.309. [DOI] [PubMed] [Google Scholar]
- Pamies D.; Hartung T. (2017) 21st century cell culture for 21st century toxicology. Chem. Res. Toxicol. 30, 43–52. 10.1021/acs.chemrestox.6b00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (2016) http://www.oecd.org/env/ehs/testing/OECD_Draft_GIVIMP_in_Human_Safety_Assessment.pdf (last accessed 14 Dec 2016).
- National Institute of Environmental Health Sciences (1997) Validation and Regulatory Acceptance of Toxicological Test Methods: A Report of the ad Hoc Interagency Coordinating Committee on the Validation of Alternative Methods, NIH Publication No. 97-3981, NIEHS, Research Triangle Park, NC. [Google Scholar]
- Jennings P. (2015) The future of in vitro toxicology. Toxicol. In Vitro 29, 1217–1221. 10.1016/j.tiv.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Hartung T.; Bremer S.; Casati S.; Coecke S.; Corvi R.; Fortaner S.; Gribaldo L.; Halder M.; Hoffmann S.; Roi A. J.; Prieto P.; Sabbioni E.; Scott L.; Worth A.; Zuang V. A. (2004) Modular approach to the ECVAM principles on test validity. Altern. Lab. Anim. 32, 467–472. [DOI] [PubMed] [Google Scholar]
- Leist M.; Hasiwa M.; Daneshian M.; Hartung T. (2012) Validation and quality control of replacement alternatives – current status and future challenges. Toxicol. Res. 1, 8. 10.1039/c2tx20011b. [DOI] [Google Scholar]
- Pendse S. N.; Maertens A.; Rosenberg M.; Roy D.; Fasani R.; Vantangoli M.; Madnick S.; Boekelheide K.; Fornace A.; Odwin S.-A.; Yager J.; Hartung T.; Andersen M. E.; McMullen P. D. (2017) Information-dependent enrichment analysis reveals time-dependent transcriptional regulation of the estrogen pathway of toxicity. Arch. Toxicol. 91, 1749. 10.1007/s00204-016-1824-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhifd M.; Beger R.; Flynn T.; Guo L.; Harris G.; Hogberg H. T.; Kaddurah-Daouk R.; Kamp H.; Kleensang A.; Maertens A.; Odwin-DaCosta S.; Pamies D.; Robertson D.; Smirnova L.; Sun J.; Zhao L.; Hartung T. (2015) Quality assurance of metabolomics. ALTEX 32, 319–326. 10.14573/altex.1509161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens A.; Bouhifd M.; Zhao L.; Odwin-DaCosta S.; Kleensang A.; Yager J. D.; Hartung T. (2017) Metabolomic network analysis of estrogen-stimulated MCF-7 cells: a comparison of over-representation analysis, quantitative enrichment analysis and pathway analysis versus metabolite network analysis. Arch. Toxicol. 91, 217–230. 10.1007/s00204-016-1695-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez T.; Daneshian M.; Kamp H.; Bois F. Y.; Clench M. R.; Coen M.; Donley B.; Fischer S. M.; Ekman D. R.; Fabian E.; Guillou C.; Heuer J.; Hogberg H. T.; Jungnickel H.; Keun H. C.; Krennrich G.; Krupp E.; Luch A.; Noor F.; Peter E.; Riefke B.; Seymour M.; Skinner N.; Smirnova L.; Verheij E.; Wagner S.; Hartung T.; van Ravenzwaay B.; Leist M. (2013) Metabolomics in toxicology and preclinical research. ALTEX 30, 209–225. 10.14573/altex.2013.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhifd M.; Hartung T.; Hogberg H. T.; Kleensang A.; Zhao L. (2013) Review: toxicometabolomics. J. Appl. Toxicol. 33, 1365–1383. 10.1002/jat.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet E. (2011) Current standing and future prospects for the technologies proposed to transform toxicity testing in the 21st century. ALTEX 28, 17–44. 10.14573/altex.2011.1.017. [DOI] [PubMed] [Google Scholar]
- Maertens A.; Luechtefeld T.; Kleensang A.; Hartung T. (2015) MPTP’s pathway of toxicity indicates central role of transcription factor SP1. Arch. Toxicol. 89, 743–755. 10.1007/s00204-015-1509-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendse S. N.; Maertens A.; Rosenberg M.; Roy D.; Fasani R.; Vantangoli M.; Madnick S.; Boekelheide K.; Fornace A.; Odwin S.-A.; Yager J.; Hartung T.; Andersen M. E.; McMullen P. D. (2017) Information-dependent enrichment analysis reveals time-dependent transcriptional regulation of the estrogen pathway of toxicity. Arch. Toxicol. 91, 1749. 10.1007/s00204-016-1824-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhifd M.; Andersen M. E.; Baghdikian C.; Boekelheide K.; Crofton K. M.; Fornace A. J. Jr.; Kleensang A.; Li H.; Livi C. B.; Maertens A.; McMullen P. D.; Rosenberg M.; Thomas R.; Vantangoli M.; Yager J. D.; Zhao L.; Hartung T. (2015) The Human Toxome project. ALTEX 32, 112–124. 10.14573/altex.1502091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez T.; Daneshian M.; Kamp H.; Bois F. Y.; Clench M. R.; Coen M.; Donley B.; Fischer S. M.; Ekman D. R.; Fabian E.; Guillou C.; Heuer J.; Hogberg H. T.; Jungnickel H.; Keun H. C.; Krennrich G.; Krupp E.; Luch A.; Noor F.; Peter E.; Riefke B.; Seymour M.; Skinner N.; Smirnova L.; Verheij E.; Wagner S.; Hartung T.; van Ravenzwaay B.; Leist M. (2013) Metabolomics in toxicology and preclinical research. ALTEX 30, 209–225. 10.14573/altex.2013.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport S. M.; Smith M. T. (2010) Environment and disease risks. Science 330, 460–461. 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. W.; Jones D. P. (2014) The nature of nurture: refining the definition of the exposome. Toxicol. Sci. 137, 1–2. 10.1093/toxsci/kft251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher B. I.; Hackermüller J.; Polte T.; Scholz S.; Aigner A.; Altenburger R.; Böhme A.; Bopp S. K.; Brack W.; Busch W.; Chadeau-Hyam M.; Covaci A.; Eisenträger A.; Galligan J.; Garcia-Reyero N.; Hartung T.; Hein M.; Herberth G.; Jahnke A.; Kleinjans J.; Kluever N.; Krauss M.; Lamoree M.; Lehmann I.; Luckenbach T.; Miller G. W.; Mueller A.; Phillips D. H.; Rappaport S. M.; Reemtsma T.; Rolle-Kampczyk U.; Schüürmann G.; Schwikowski B.; Tan Y.-M.; Trump S.; Walter-Rohde S.; Wambaugh J. F. (2017) From the exposome to mechanistic understanding of chemical-induced adverse effects. Environ. Int. 99, 97–106. 10.1016/j.envint.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R.; Dix D. (2010) Computational toxicology as implemented by the U.S. EPA: providing high throughput decision support tools for screening and assessing chemical exposure, hazard and risk. J. Toxicol. Environ. Health, Part B 13, 197–217. 10.1080/10937404.2010.483935. [DOI] [PubMed] [Google Scholar]
- Judson R.; Kavlock R.; Martin M.; Reif D.; Houck K.; Knudsen T.; Richard A.; Tice R.; Whelan M.; Xia M.; Huang R.; Austin C.; Daston G.; Hartung T.; Fowle J. R. III; Wooge W.; Tong W.; Dix D. (2013) Perspectives on validation of high-throughput pathway-based assays supporting the 21st century toxicity testing vision. ALTEX 30, 51–66. 10.14573/altex.2013.1.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T.; Luechtefeld T.; Maertens A.; Kleensang A. (2013) Integrated testing strategies for safety assessments. ALTEX 30, 3–18. 10.14573/altex.2013.1.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida C.; Alépée N.; Api A. M.; Basketter D. A.; Bois F. Y.; Caloni F.; Corsini E.; Daneshian M.; Eskes C.; Ezendam J.; Fuchs H.; Hayden P.; Hegele-Hartung C.; Hoffmann S.; Hubesch B.; Jacobs M. N.; Jaworska J.; Kleensang A.; Kleinstreuer N.; Lalko J.; Landsiedel R.; Lebreux F.; Luechtefeld T.; Locatelli M.; Mehling A.; Natsch A.; Pitchford J. W.; Prater D.; Prieto P.; Schepky A.; Schuurmann G.; Smirnova L.; Toole C.; van Vliet E.; Weisensee D.; Hartung T. (2015) Integrated testing strategies (ITS) for safety assessment. ALTEX 32, 171–181. 10.14573/altex.1411011. [DOI] [PubMed] [Google Scholar]
- Tollefsen K. E.; Scholz S.; Cronin M. T.; Edwards S. W.; de Knecht J.; Crofton K.; Garcia-Reyero N.; Hartung T.; Worth A.; Patlewicz G. (2014) Applying adverse outcome pathways (AOPs) to support integrated approaches to testing and assessment (IATA). Regul. Toxicol. Pharmacol. 70, 629–640. 10.1016/j.yrtph.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer N. C.; Yang J.; Berg E. L.; Knudsen T. B.; Richard A. M.; Martin M. T.; Reif D. M.; Judson R. S.; Polokoff M.; Dix D. J.; Kavlock R. J.; Houck K. A. (2014) Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol. 32, 583–591. 10.1038/nbt.2914. [DOI] [PubMed] [Google Scholar]
- Shah I.; Setzer R. W.; Jack J.; Houck K. A.; Judson R. S.; Knudsen T. B.; Liu J.; Martin M. T.; Reif D. M.; Richard A. M.; Thomas R. S.; Crofton K. M.; Dix D. J.; Kavlock R. J. (2016) Using ToxCast Data to Reconstruct Dynamic Cell State Trajectories and Estimate Toxicological Points of Departure. Environ. Health Perspect. 124, 910–919. 10.1289/ehp.1409029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Mansouri K.; Judson R. S.; Martin M. T.; Hong H.; Chen M.; Xu X.; Thomas R. S.; Shah I. (2015) Predicting Hepatotoxicity Using ToxCast in Vitro Bioactivity and Chemical Structure. Chem. Res. Toxicol. 28, 738–751. 10.1021/tx500501h. [DOI] [PubMed] [Google Scholar]
- Van den Hof W. F.; Coonen M. L.; van Herwijnen M.; Brauers K.; Wodzig W. K.; van Delft J. H.; Kleinjans J. C. (2014) Classification of hepatotoxicants using HepG2 cells: A proof of principle study. Chem. Res. Toxicol. 27, 433–442. 10.1021/tx4004165. [DOI] [PubMed] [Google Scholar]
- Garcia-Serna R.; Vidal D.; Remez N.; Mestres J. (2015) Large-scale predictive drug safety: from structural alerts to biological mechanisms. Chem. Res. Toxicol. 28, 1875–1887. 10.1021/acs.chemrestox.5b00260. [DOI] [PubMed] [Google Scholar]
- Angrish M. M.; Madden M. C.; Pleil J. D. (2015) Probe molecule (PrM) approach in adverse outcome pathway (AOP) based high-throughput screening (HTS): in vivo discovery for developing in vitro target methods. Chem. Res. Toxicol. 28, 551–559. 10.1021/acs.chemrestox.5b00024. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez I.; Sewer A.; Walker P.; Mathis C.; Ellis S.; Woodhouse H.; Guedj E.; Dulize R.; Marescotti D.; Acali S.; Martin F.; Ivanov N. V.; Hoeng J.; Peitsch M. C. (2014) Systems biology approach for evaluating the biological impact of environmental toxicants in vitro. Chem. Res. Toxicol. 27, 367–376. 10.1021/tx400405s. [DOI] [PubMed] [Google Scholar]
- Mirams G. R.; Pathmanathan P.; Gray R. A.; Challenor P.; Clayton R. H. (2016) Uncertainty and variability in computational and mathematical models of cardiac physiology. J. Physiol. 594, 6833–6847. 10.1113/JP271671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaioun K.; Blaauboer B. J.; Hartung T. (2016) Evidence-based absorption, distribution, metabolism, excretion and toxicity (ADMET) and the role of alternative methods. ALTEX 33, 343–358. 10.14573/altex.1610101. [DOI] [PubMed] [Google Scholar]
- Hartung T. (2009) Toxicology for the twenty-first century. Nature 460, 208–212. 10.1038/460208a. [DOI] [PubMed] [Google Scholar]
- Boobis A. R.; Daston G. P.; Preston R. J.; Olin S. S. (2009) Application of key events analysis to chemical carcinogens and noncarcinogens. Crit. Rev. Food Sci. Nutr. 49, 690–707. 10.1080/10408390903098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek M. E.; Boobis A.; Cote I.; Dellarco V.; Fotakis G.; Munn S.; Seed J.; Vickers C. (2014) New developments in the evolution and application of the WHO/IPCS framework on mode of action/species concordance analysis. J. Appl. Toxicol. 34, 1–18. 10.1002/jat.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek M. E.; Palermo C. M.; Bachman A. N.; North C. M.; Jeffrey Lewis R. (2014) Mode of action human relevance (species concordance) framework: Evolution of the Bradford Hill considerations and comparative analysis of weight of evidence. J. Appl. Toxicol. 34, 595–606. 10.1002/jat.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boobis A. R.; Cohen S. M.; Dellarco V.; McGregor D.; Meek M. E.; Vickers C.; Willcocks D.; Farland W. (2006) IPCS framework for analyzing the relevance of a cancer mode of action for humans. Crit. Rev. Toxicol. 36, 781–792. 10.1080/10408440600977677. [DOI] [PubMed] [Google Scholar]
- Boobis A. R.; Doe J. E.; Heinrich-Hirsch B.; Meek M. E.; Munn S.; Ruchirawat M.; Schlatter J.; Seed J.; Vickers C. (2008) IPCS framework for analyzing the relevance of a noncancer mode of action for humans. Crit. Rev. Toxicol. 38, 87–96. 10.1080/10408440701749421. [DOI] [PubMed] [Google Scholar]
- Villeneuve D. L.; Crump D.; Garcia-Reyero N.; Hecker M.; Hutchinson T. H.; LaLone C. A.; Landesmann B.; Lettieri T.; Munn S.; Nepelska M.; Ottinger M. A.; Vergauwen L.; Whelan M. (2014) Adverse outcome pathway development II: best practices. Toxicol. Sci. 142, 321–330. 10.1093/toxsci/kfu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings P. (2013) Stress response pathways, toxicity pathways and adverse outcome pathways. Arch. Toxicol. 87, 13–14. 10.1007/s00204-012-0974-4. [DOI] [PubMed] [Google Scholar]
- Yoon M.; Adeleye Y.; Clewell R.; Jennings P.; Whelan M. (2016) Moving beyond prioritization toward true in vitro safety assessment. Appl. In Vitro Toxicol. 2, 67–73. 10.1089/aivt.2016.29005.rtl. [DOI] [Google Scholar]
- Hartung T. (2017) Utility of the Adverse Outcome Pathway concept in drug development. Expert Opin. Drug Metab. Toxicol. 13, 1–3. 10.1080/17425255.2017.1246535. [DOI] [PubMed] [Google Scholar]
- https://aopwiki.org/wiki/index.php/Main_Page (last accessed 21 Mar, 2017).
- https://www.effectopedia.org (last accessed 21 Mar, 2017).
- SCCS (2013) Addressing the New Challenges for Risk Assessment, Opinion adopted March 2013, European Union Scientific Committee on Health and Environmental Risks (SCHER), Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR), Scientific Committee on Consumer Safety (SCCS). http://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_131.pdf (accessed 26 Aug, 2016).
- Daston G.; Knight D. J.; Schwarz M.; Gocht T.; Thomas R. S.; Mahony C.; Whelan M. (2015) SEURAT: Safety Evaluation Ultimately Replacing Animal Testing--recommendations for future research in the field of predictive toxicology. Arch. Toxicol. 89, 15–23. 10.1007/s00204-014-1421-5. [DOI] [PubMed] [Google Scholar]
- Daneshian M.; Kamp H.; Hengstler J.; Leist M.; van de Water B. (2016) Highlight report: Launch of a large integrated European in vitro toxicology project: EU-ToxRisk. Arch. Toxicol. 90, 1021–1024. 10.1007/s00204-016-1698-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.eu-toxrisk.eu (last accessed 21 Mar, 2017).
- Busquet F.; Hartung T. (2008) The need for strategic development of safety sciences. ALTEX 34, 3–21. 10.14573/altex.1701031. [DOI] [PubMed] [Google Scholar]
- Allen T. E. H.; Goodman J. M.; Gutsell S.; Russell P. J. (2014) Defining molecular initiating events in the adverse outcome pathway framework for risk assessment. Chem. Res. Toxicol. 27, 2100–2112. 10.1021/tx500345j. [DOI] [PubMed] [Google Scholar]
- Mellor C. L.; Steinmetz F. P.; Cronin M. T. D. (2016) Using molecular initiating events to develop a structural alert based screening workflow for nuclear receptor ligands associated with hepatic steatosis. Chem. Res. Toxicol. 29, 203–212. 10.1021/acs.chemrestox.5b00480. [DOI] [PubMed] [Google Scholar]
- Mekenyan O.; Patlewicz G.; Kuseva C.; Popova I.; Mehmed A.; Kotov S.; Zhechev T.; Pavlov T.; Temelkov S.; Roberts D. W. (2014) A mechanistic approach to modeling respiratory sensitization. Chem. Res. Toxicol. 27, 219–239. 10.1021/tx400345b. [DOI] [PubMed] [Google Scholar]
- Roberts D. W.; Aptula A. O. (2014) Electrophilic reactivity and skin sensitization potency of SNAr electrophiles. Chem. Res. Toxicol. 27, 240–246. 10.1021/tx400355n. [DOI] [PubMed] [Google Scholar]
- Vinken M. (2015) Adverse Outcome Pathways and drug-induced liver injury testing. Chem. Res. Toxicol. 28, 1391–1397. 10.1021/acs.chemrestox.5b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (2013) Guidance Document on Developing and Assessing Adverse Outcome Pathways, OECD Environment Directorate, Joint Meeting of the Chemicals Committee and the Working Party on Chemicals, Pesticides and Biotechnology, Organisation for Economic Co-operation and Development, Series on Testing and Assessment No. 184, ENV/JM/MONO(2013)6, 17-Apr-2013. http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono(2013)6&docLanguage=en. [Google Scholar]
- OECD (2015) Report of the Workshop on a Framework for the Development and Use of Integrated Approaches to Testing and Assessment (17–19 Nov, 2014, Crystal City VA, USA), Organisation for Economic Co-operation and Development, Series on Testing and Assessment No. 215. ENV/JM/MONO(2015)22, 22-Jul-2015. http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=ENV/JM/MONO(2015)22&docLanguage=En. [Google Scholar]
- Benfenati E.; Berggren E.; Fritsche E.; Hartung T.; Slikker W. Jr.; Spielmann H.; Testai E.; Tice R. R.; Tiramani M.; Villenave R. (2016) Novel chemical hazard characterisation approaches. EFSA J. 14, s0506. 10.2903/j.efsa.2016.s0506. [DOI] [Google Scholar]
- OECD (2015) Test Guideline 442C. In Chemico Skin Sensitisation. Direct Peptide Reactivity Assay (DPRA), OECD Guidelines for the Testing of Chemicals, Section 4: Health Effects, published 05 February 2015. http://www.oecd-ilibrary.org/environment/test-no-442c-in-chemico-skin-sensitisation_9789264229709-en.
- OECD (2015) Test Guideline 442D, In Vitro Skin Sensitisation, ARE-Nrf2 Luciferase Test Method, OECD Guidelines for the Testing of Chemicals, Section 4: Health Effects, published 05 Feb, 2015. http://www.oecd-ilibrary.org/environment/test-no-442d-in-vitro-skin-sensitisation_9789264229822-en.
- Natsch A.; Emter R. (2015) Reporter cell lines for skin sensitization testing. Arch. Toxicol. 89, 1645–1468. 10.1007/s00204-015-1555-0. [DOI] [PubMed] [Google Scholar]
- Jaworska J. S.; Natsch A.; Ryan C.; Strickland J.; Ashikaga T.; Miyazawa M. (2015) Bayesian integrated testing strategy (ITS) for skin sensitization potency assessment: a decision support system for quantitative weight of evidence and adaptive testing strategy. Arch. Toxicol. 89, 2355–2383. 10.1007/s00204-015-1634-2. [DOI] [PubMed] [Google Scholar]
- Strickland J.; Zang Q.; Kleinstreuer N.; Paris M.; Lehmann D. M.; Choksi N.; Matheson J.; Jacobs A.; Lowit A.; Allen D.; Casey W. (2016) Integrated decision strategies for skin sensitization hazard. J. Appl. Toxicol. 36, 1150–1162. 10.1002/jat.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsen K. E.; Scholz S.; Cronin M. T.; Edwards S. W.; de Knecht J.; Crofton K.; Garcia-Reyero N.; Hartung T.; Worth A.; Patlewicz G. (2014) Applying Adverse Outcome Pathways (AOPs) to support Integrated Approaches to Testing and Assessment (IATA). Regul. Toxicol. Pharmacol. 70, 629–640. 10.1016/j.yrtph.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Tralau T.; Oelgeschläger M.; Gürtler R.; Heinemeyer G.; Herzler M.; Höfer T.; Itter H.; Kuhl T.; Lange N.; Lorenz N.; Müller-Graf C.; Pabel U.; Pirow R.; Ritz V.; Schafft H.; Schneider H.; Schulz T.; Schumacher D.; Zellmer S.; Fleur-Böl G.; Greiner M.; Lahrssen-Wiederholt M.; Lampen A.; Luch A.; Schönfelder G.; Solecki R.; Wittkowski R.; Hensel A. (2015) Regulatory toxicology in the twenty-first century: challenges, perspectives and possible solutions. Arch. Toxicol. 89, 823–850. 10.1007/s00204-015-1510-0. [DOI] [PubMed] [Google Scholar]
- Tralau T.; Luch A. (2015) Moving from rats to cellular omics in regulatory toxicology: great challenge toward sustainability or ″up-shit-creek without a paddle″?. Arch. Toxicol. 89, 819–821. 10.1007/s00204-015-1511-z. [DOI] [PubMed] [Google Scholar]
- Ankley G.; Escher B.; Hartung T.; Shah I. (2016) Pathway-based approaches for environmental monitoring and risk assessment. Environ. Sci. Technol. 50, 10295–10296. 10.1021/acs.est.6b04425. [DOI] [PubMed] [Google Scholar]
- Rostami-Hodjegan A.; Tucker G. T. (2007) Simulation and prediction of in vivo drug metabolism in human populations from in vitro data. Nat. Rev. Drug Discovery 6, 140–148. 10.1038/nrd2173. [DOI] [PubMed] [Google Scholar]
- Gaohua L.; Wedagedera J.; Small B. G.; Almond L.; Romero K.; Hermann D.; Hanna D.; Jamei M.; Gardner I. (2015) Development of a multicompartment permeability-limited lung PBPK model and its application in predicting pulmonary pharmacokinetics of antituberculosis drugs. CPT: Pharmacometrics Syst. Pharmacol. 4, 605–613. 10.1002/psp4.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff S., Gaohua L., Burt H., Jamei M., Li L., Tucker G. T., and Rostami-Hodjegan A. (2013) Accounting for Transporters in Renal Clearance: Towards a Mechanistic Kidney Model (Mech KiM), in Transporters in Drug Development (Steffansen B., and Sugiyama Y., Eds.) pp 155–177, Springer, New York, DOI: 10.1007/978-1-4614-8229-1_7. [DOI] [Google Scholar]
- Neuhoff S.; Yeo K. R.; Barter Z.; Jamei M.; Turner D. B.; Rostami-Hodjegan A. (2013) Application of permeability-limited physiologically-based pharmacokinetic models: part I-digoxin pharmacokinetics incorporating P-glycoprotein-mediated efflux. J. Pharm. Sci. 102, 3145–3160. 10.1002/jps.23594. [DOI] [PubMed] [Google Scholar]
- Jamei M.; Bajot F.; Neuhoff S.; Barter Z.; Yang J.; Rostami-Hodjegan A.; Rowland-Yeo K. (2014) A mechanistic framework for in vitro-in vivo extrapolation of liver membrane transporters: prediction of drug-drug interaction between rosuvastatin and cyclosporine. Clin. Pharmacokinet. 53, 73–87. 10.1007/s40262-013-0097-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak S.; Ghobadi C.; Mishra H.; Ahamadi M.; Patel N.; Jamei M.; Rostami-Hodjegan A. (2012) Prediction of concentration-time profile and its inter-individual variability following the dermal drug absorption. J. Pharm. Sci. 101, 2584–2595. 10.1002/jps.23155. [DOI] [PubMed] [Google Scholar]
- Gaohua L.; Neuhoff S.; Johnson T. N.; Rostami-Hodjegan A.; Jamei M. (2016) Development of a permeability-limited model of the human brain and cerebrospinal fluid (CSF) to integrate known physiological and biological knowledge: Estimating time varying CSF drug concentrations and their variability using in vitro data. Drug Metab. Pharmacokinet. 31, 224–233. 10.1016/j.dmpk.2016.03.005. [DOI] [PubMed] [Google Scholar]
- EMA (2016) Reflection Paper on Extrapolation of Efficacy and Safety in Paediatric Medicine Development, European Medicines Agency, Draft 31-Mar-2016, EMA/199678/2016. http://www.ema.europa.eu/docs/en_GB/document_library/Regulatory_and_procedural_guideline/2016/04/WC500204187.pdf (accessed 19 Sep, 2016).
- Rostami-Hodjegan A. (2012) Physiologically based pharmacokinetics joined with in vitro-in vivo extrapolation of ADME: a marriage under the arch of systems pharmacology. Clin. Pharmacol. Ther. 92, 50–61. 10.1038/clpt.2012.65. [DOI] [PubMed] [Google Scholar]
- Tsaioun K.; Blaauboer B. J.; Hartung T. (2016) Evidence-based absorption, distribution, metabolism, excretion and toxicity (ADMET) and the role of alternative methods. ALTEX 33, 343–358. 10.14573/altex.1610101. [DOI] [PubMed] [Google Scholar]
- Zhao P.; Zhang L.; Grillo J. A.; Liu Q.; Bullock J. M.; Moon Y. J.; Song P.; Brar S. S.; Madabushi R.; Wu T. C.; Booth B. P.; Rahman N. A.; Reynolds K. S.; Gil Berglund E.; Lesko L. J.; Huang S. M. (2011) Applications of physiologically based pharmacokinetic (PBPK) modeling and simulation during regulatory review. Clin. Pharmacol. Ther. 89, 259–267. 10.1038/clpt.2010.298. [DOI] [PubMed] [Google Scholar]
- Salem F.; Rostami-Hodjegan A.; Johnson T. N. (2013) Do children have the same vulnerability to metabolic drug–drug interactions as adults? A critical analysis of the literature. J. Clin. Pharmacol. 53, 559–566. 10.1002/jcph.13. [DOI] [PubMed] [Google Scholar]
- Rose R. H.; Neuhoff S.; Abduljalil K.; Chetty M.; Rostami-Hodjegan A.; Jamei M. (2014) Application of a physiologically based pharmacokinetic model to predict OATP1B1-related variability in pharmacodynamics of rosuvastatin. CPT: Pharmacometrics Syst. Pharmacol. 3, e124. 10.1038/psp.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniowska B.; Polak S. (2016) Virtual clinical trial toward polytherapy safety assessment: combination of physiologically based pharmacokinetic/pharmacodynamic-based modeling and simulation approach with drug-drug interactions involving terfenadine as an example. J. Pharm. Sci. 105, 3415–3424. 10.1016/j.xphs.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Scotcher D.; Jones C.; Posada M.; Galetin A.; Rostami-Hodjegan A. (2016) Key to Opening Kidney for In Vitro-In Vivo Extrapolation Entrance in Health and Disease: Part II: Mechanistic Models and In Vitro-In Vivo Extrapolation. AAPS J. 18, 1082–1094. 10.1208/s12248-016-9959-1. [DOI] [PubMed] [Google Scholar]
- Fisher C. P.; Plant N. J.; Moore J. B.; Kierzek A. M. (2013) QSSPN: dynamic simulation of molecular interaction networks describing gene regulation, signalling and whole-cell metabolism in human cells. Bioinformatics 29, 3181–3190. 10.1093/bioinformatics/btt552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICH S7B (2005) The Non-clinical Evaluation of the Potential for Delayed Ventricular Repolarization (QT Interval Prolongation) by human Pharmaceuticals, ICH Finalised Guideline, May 2005. http://www.ich.org/products/guidelines/safety/article/safety-guidelines.html.
- ICH-E14 (2005) The Clinical Evaluation of QT/QTc Interval Prolongation and Proarrhythmic Potential for Non-antiarrhythmic Drugs, ICH Finalised Guideline, May 2005. http://www.ich.org/products/guidelines/efficacy/article/efficacy-guidelines.html.
- Mirams G. R.; Cui Y.; Sher A.; Fink M.; Cooper J.; Heath B. M.; Mcmahon N. C.; Gavaghan D. J.; Noble D. (2011) Simulation of multiple ion channel block provides improved early prediction of compounds’ clinical torsadogenic risk. Cardiovasc. Res. 91, 53–61. 10.1093/cvr/cvr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J.; Obejero-Paz C.; Myatt G.; Kuryshev Y.; Bruening-Wright A.; Verducci J. S.; Brown A. M. (2013) MICE models: superior to the HERG model in predicting Torsade de Pointes. Sci. Rep. 3, 2100. 10.1038/srep02100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J. D.; Yang P.-C.; Bankston J. R.; Grandi E.; Bers; Kass R. S.; Clancy C. E. (2013) Ranolazine for congenital and acquired late INa linked arrhythmias: in silico pharmacologic screening. Circ. Res. 113, e50–61. 10.1161/CIRCRESAHA.113.301971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Veroli G. Y.; Davies M. R.; Zhang H.; Abi-Gerges N.; Boyett M. R. (2013) High-throughput screening of drug-binding dynamics to HERG improves early drug safety assessment. AJP Hear. Circ. Physiol. 304, H104–H117. 10.1152/ajpheart.00511.2012. [DOI] [PubMed] [Google Scholar]
- Di Veroli G. Y.; Davies M. R.; Zhang H.; Abi-Gerges N.; Boyett M. R. (2014) hERG Inhibitors With Similar Potency But Different Binding Kinetics Do Not Pose the Same Proarrhythmic Risk: Implications for Drug Safety Assessment. J. Cardiovasc. Electrophysiol. 25, 197–207. 10.1111/jce.12289. [DOI] [PubMed] [Google Scholar]
- Moreno J. D.; Zhu Z. I.; Yang P. C.; Bankston J. R.; Jeng M. T.; Kang C.; Wang L.; Bayer J. D.; Christini D. J.; Trayanova N. A.; Ripplinger C. M.; Kass R. S.; Clancy C. E. (2011) A computational model to predict the effects of class I anti-arrhythmic drugs on ventricular rhythms. Sci. Transl. Med. 3, 98ra83. 10.1126/scitranslmed.3002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirams G. R.; Noble D. (2011) Is it time for in silico simulation of drug cardiac side effects?. Ann. N. Y. Acad. Sci. 1245, 44–47. 10.1111/j.1749-6632.2011.06324.x. [DOI] [PubMed] [Google Scholar]
- Elkins R. C.; Davies M. R.; Brough S. J.; Gavaghan D. J.; Cui Y.; Abi-Gerges N.; Mirams G. R. (2013) Variability in high-throughput ion-channel screening data and consequences for cardiac safety assessment. J. Pharmacol. Toxicol. Methods 68, 112–122. 10.1016/j.vascn.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D.; Garny A.; Noble P. J. (2012) How the Hodgkin – Huxley equations inspired the Cardiac Physiome Project. J. Physiol. 590, 2613–2628. 10.1113/jphysiol.2011.224238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie K. A.; Luscombe C.; Williams G.; Munoz-Muriedas J.; Gavaghan D. J.; Cui Y.; Mirams G. R. (2013) Evaluation of an in silico cardiac safety assay: using ion channel screening data to predict QT interval changes in the rabbit ventricular wedge. J. Pharmacol. Toxicol. Methods 68, 88–96. 10.1016/j.vascn.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. R.; Mistry H. B.; Hussein L.; Pollard C. E.; Swinton J.; Valentin J.-P.; Abi-Gerges N. (2012) An in silico canine cardiac midmyocardial action potential duration model as a tool for early drug safety assessment. Am. J. Physiol. Hear. Circ. Phys. 302, H1466–H1480. 10.1152/ajpheart.00808.2011. [DOI] [PubMed] [Google Scholar]
- Sager P. T.; Gintant G.; Turner J. R.; Pettit S.; Stockbridge N. (2014) Rechanneling the cardiac proarrhythmia safety paradigm: a meeting report from the Cardiac Safety Research Consortium. Am. Heart J. 167, 292–300. 10.1016/j.ahj.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Fermini B.; Hancox J. C.; Abi-Gerges N.; Bridgland-Taylor M.; Chaudhary K. W.; Colatsky T.; Correll K.; Crumb W.; Damiano B.; Erdemli G.; Gintant G.; Imredy J.; Koerner J.; Kramer J.; Levesque P.; Li Z.; Lindqvist A.; Obejero-Paz C. A.; Rampe D.; Sawada K.; Strauss D. G.; Vandenberg J. I. (2016) A new perspective in the field of cardiac safety testing through the comprehensive in vitro proarrhythmia assay paradigm. J. Biomol. Screening 21, 1–11. 10.1177/1087057115594589. [DOI] [PubMed] [Google Scholar]
- Jennings P. (2015) The future of in vitro toxicology. Toxicol. In Vitro 29, 1217–1221. 10.1016/j.tiv.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Judson R.; Houck K.; Martin M.; Richard A. M.; Knudsen T. B.; Shah I.; Little S.; Wambaugh J.; Setzer R. W.; Kothiya P.; Phuong J.; Filer D.; Smith D.; Reif D.; Rotroff D.; Kleinstreuer N.; Sipes N.; Xia M.; Huang R.; Crofton K.; Thomas R. S. (2016) Analysis of the Effects of Cell Stress and Cytotoxicity on In Vitro Assay Activity Across a Diverse Chemical and Assay Space. Toxicol. Sci. 153, 409–409. 10.1093/toxsci/kfw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings P.; Limonciel A.; Felice L.; Leonard M. O. (2013) An overview of transcriptional regulation in response to toxicological insult. Arch. Toxicol. 87, 49–72. 10.1007/s00204-012-0919-y. [DOI] [PubMed] [Google Scholar]
- Limonciel A.; Moenks K.; Stanzel S.; Truisi G. L.; Parmentier C.; Aschauer L.; Wilmes A.; Richert L.; Hewitt P.; Mueller S. O.; Lukas A.; Kopp-Schneider A.; Leonard M. O.; Jennings P. (2015) Transcriptomics hit the target: Monitoring of ligand-activated and stress response pathways for chemical testing. Toxicol. In Vitro 30, 7–18. 10.1016/j.tiv.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Peng K. J.; Wang J. H.; Su W. T.; Wang X. C.; Yang F. T.; Nie W. H. (2010) Characterization of two human lung adenocarcinoma cell lines by reciprocal chromosome painting. Dongwuxue Yanjiu 31, 113–121. 10.3724/SP.J.1141.2010.02113. [DOI] [PubMed] [Google Scholar]
- Crean D.; Bellwon P.; Aschauer L.; Limonciel A.; Moenks K.; Hewitt P.; Schmidt T.; Herrgen K.; Dekant W.; Lukas A.; Bois F.; Wilmes A.; Jennings P.; Leonard M. O. (2015) Development of an in vitro renal epithelial disease state model for xenobiotic toxicity testing. Toxicol. In Vitro 30, 128–137. 10.1016/j.tiv.2014.11.015. [DOI] [PubMed] [Google Scholar]
- Wilmes A.; Bielow C.; Ranninger C.; Bellwon P.; Aschauer L.; Limonciel A.; Chassaigne H.; Kristl T.; Aiche S.; Huber C. G.; Guillou C.; Hewitt P.; Leonard M. O.; Dekant W.; Bois F.; Jennings P. (2015) Mechanism of cisplatin proximal tubule toxicity revealed by integrating transcriptomics, proteomics, metabolomics and biokinetics. Toxicol. In Vitro 30, 117–127. 10.1016/j.tiv.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Wilmes A.; Limonciel A.; Aschauer L.; Moenks K.; Bielow C.; Leonard M. O.; Hamon J.; Carpi D.; Ruzek S.; Handler A.; Schmal O.; Herrgen K.; Bellwon P.; Burek C.; Truisi G. L.; Hewitt P.; Di Consiglio E.; Testai E.; Blaauboer B. J.; Guillou C.; Huber C. G.; Lukas A.; Pfaller W.; Mueller S. O.; Bois F. Y.; Dekant W.; Jennings P. (2013) Application of integrated transcriptomic, proteomic and metabolomic profiling for the delineation of mechanisms of drug induced cell stress. J. Proteomics 79, 180–194. 10.1016/j.jprot.2012.11.022. [DOI] [PubMed] [Google Scholar]
- Aschauer L.; Gruber L. N.; Pfaller W.; Limonciel A.; Athersuch T. J.; Cavill R.; Khan A.; Gstraunthaler G.; Grillari J.; Grillari R.; Hewitt P.; Leonard M. O.; Wilmes A.; Jennings P. (2013) Delineation of the key aspects in the regulation of epithelial monolayer formation. Mol. Cell. Biol. 33, 2535–2550. 10.1128/MCB.01435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser M.; Stadler G.; Jennings P.; Streubel B.; Pfaller W.; Ambros P.; Riedl C.; Katinger H.; Grillari J.; Grillari-Voglauer R. (2008) hTERT alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am. J. Physiol. Renal Physiol. 295, F1365–1375. 10.1152/ajprenal.90405.2008. [DOI] [PubMed] [Google Scholar]
- Wilmes A.; Aschauer L.; Limonciel A.; Pfaller W.; Jennings P. (2014) Evidence for a role of claudin 2 as a proximal tubular stress responsive paracellular water channel. Toxicol. Appl. Pharmacol. 279, 163–172. 10.1016/j.taap.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Aschauer L.; Limonciel A.; Wilmes A.; Stanzel S.; Kopp-Schneider A.; Hewitt P.; Lukas A.; Leonard M. O.; Pfaller W.; Jennings P. (2015) Application of RPTEC/TERT1 cells for investigation of repeat dose nephrotoxicity: A transcriptomic study. Toxicol. In Vitro 30, 106–116. 10.1016/j.tiv.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Limonciel A.; Aschauer L.; Wilmes A.; Prajczer S.; Leonard M. O.; Pfaller W.; Jennings P. (2011) Lactate is an ideal non-invasive marker for evaluating temporal alterations in cell stress and toxicity in repeat dose testing regimes. Toxicol. In Vitro 25, 1855–1862. 10.1016/j.tiv.2011.05.018. [DOI] [PubMed] [Google Scholar]
- Jennings P.; Crean D.; Aschauer L.; Limonciel A.; Moenks K.; Kern G.; Hewitt P.; Lhotta K.; Lukas A.; Wilmes A.; Leonard M. O. (2015) Interleukin-19 as a translational indicator of renal injury. Arch. Toxicol. 89, 101–106. 10.1007/s00204-014-1237-3. [DOI] [PubMed] [Google Scholar]
- Wilmes A., Limonciel A., Leonard M., and Jennings P. (2014) Translational Biomarkers, in Vitro and in Vivo, in In Vitro Toxicology Systems. Methods in Pharmacology and Toxicology (Bal-Price A., and Jennings P., Eds.) pp 459–478, Springer, New York. [Google Scholar]


