Abstract
Under conditions of nutrient shortage autophagy is the primary cellular mechanism ensuring availability of substrates for continuous biosynthesis. Subjecting cells to starvation or rapamycin efficiently induces autophagy by inhibiting the MTOR signaling pathway triggering increased autophagic flux. To elucidate the regulation of early signaling events upon autophagy induction, we applied quantitative phosphoproteomics characterizing the temporal phosphorylation dynamics after starvation and rapamycin treatment. We obtained a comprehensive atlas of phosphorylation kinetics within the first 30 min upon induction of autophagy with both treatments affecting widely different cellular processes. The identification of dynamic phosphorylation already after 2 min demonstrates that the earliest events in autophagy signaling occur rapidly after induction. The data was subjected to extensive bioinformatics analysis revealing regulated phosphorylation sites on proteins involved in a wide range of cellular processes and an impact of the treatments on the kinome. To approach the potential function of the identified phosphorylation sites we performed a screen for MAP1LC3-interacting proteins and identified a group of binding partners exhibiting dynamic phosphorylation patterns. The data presented here provide a valuable resource on phosphorylation events underlying early autophagy induction.
Keywords: autophagy, signal transduction, phosphorylation, proteomics, phosphoproteomics, mass spectrometry, bioinformatics
Introduction
To sustain a continuous cycle of biosynthesis it is critical that substrates are available to allow cells to respond to changes in the surroundings. The main source of anabolic substrates is nutrient uptake from the environment. But if the supply of nutrients falls scarce, cells are able to respond to starvation by degrading cellular components by autophagy. 1 Autophagy is a collective term for a range of processes by which cells can shuttle biomolecules, and even complete organelles, to lysosomes for degradation, generally, in vesicles termed autophagosomes. 2 , 3 To ensure that the correct amount of cytoplasm is degraded, autophagy is a tightly regulated process that integrates information about not only nutrient availability, but also hormone receptor stimulation and cellular energy levels, among others. 4 Autophagy was originally considered a largely unspecific process, but this view is changing as examples of specific degradation emerge. One of the mechanisms providing specificity to the selection of cargo for autophagosomal degradation is the interaction of proteins with the autophagosomal membrane protein MAP1LC3-II (LC3-II henceforth). 5
In addition to its critical role as a basic cellular process involved in nutrient availability and recycling of macromolecular complexes, autophagy has been described as a general stress response, its deregulation having important clinical implications. 6 - 8 In mammalian systems especially ULK (unc-51 like autophagy activating kinase), 9 , 10 AMPK (AMP-activated protein kinase), 11 phosphatidylinositol 3-kinase (whose catalytic subunit is PIK3C3, an ortholog of yeast Vps34) and MTOR [mechanistic target of rapamycin (serine/threonine kinase)] 12 are critical in the coordinated regulation of autophagy. When nutrients are abundant MTOR promotes cell growth. But upon starvation its activity is decreased resulting in the release of inhibitory phosphorylations on e.g., ULK1/2 and ATG13 leading to induction of autophagy. 13 Given the important roles of protein kinases in autophagy induction it is evident that protein phosphorylation serves as a main mechanism for relaying early signals to initiate autophagy.
To study cellular processes underlying autophagy, methodologies utilizing mass spectrometry (MS)-based proteomics have proven very powerful, 14 e.g., for establishing the protein composition of autophagosomes, 15 addressing protein degradation dynamics, 16 , 17 and studying protein-interaction networks. 18 In the past decade numerous technological advances have made quantitative phosphoproteomics the method of choice for studying protein phosphorylation in an unbiased manner. 19 Due to the versatility and robustness of phosphoproteomics strategies these have been applied to characterize the system-wide regulation of phosphorylation in many different settings and processes, thereby providing valuable information about crucial regulatory events. 20
Here we report the application of quantitative phosphoproteomics to characterize the early temporal dynamics of the phosphoproteome after withdrawal of amino acid supplements in the growth medium, or inhibiting the activity of MTOR by treatment with the pharmacological inhibitor rapamycin. Both of these treatments infer stress conditions on cells and are well-characterized inducers of autophagy. 21 The resulting catalog of phosphorylation dynamics provides a site-specific overview of the global impact of these treatments on the phosphoproteome. To complement this, we performed large-scale screens for identifying new LC3-interacting proteins, 41 of which we also identified as phosphorylated. In combination the data sets presented constitute comprehensive resources on the intricate phosphorylation dynamics accompanying induction of autophagy and, possibly, controlling autophagosomal targeting.
Results
Characterization of the phosphoproteome upon induction of autophagy
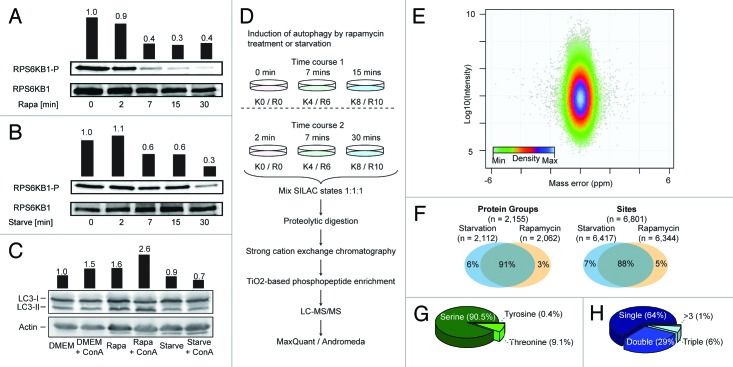
Before initiating large-scale phosphoproteomics screens to study autophagy induction we evaluated the efficiency and timing of starvation and rapamycin treatment by analyzing the phosphorylation of Thr389 on RPS6KB1, a well-established MTOR substrate site (Fig. 1A and B). From the results it is evident that within 30 min a decrease in phosphorylation is observed after both treatments. The level of this phosphorylation is reduced to 60% and 40% already after 7 min of starvation and rapamycin treatment, respectively, and after 30 min the phosphorylation is reduced below 40% in both treatments. The pronounced MTOR inhibition after 7 min of treatment indicates the need for a time-point prior to this to be able to identify the MTOR upstream events after starvation. Therefore we selected 2 min as the earliest time-point. We also noticed that the observed dynamics of RPS6KB1 phosphorylation were similar between the treatments, with only a short delay for starvation. To further evaluate the selected time-scale we performed western blots for the formation of LC3-II, a well-established marker of autophagosomes, and found that we could detect accumulation of LC3-II already after 30 min of rapamycin treatment (Fig. 1C). No increase in LC3-II formation was observed after starvation for 30 min. We ascribe this difference in the dynamics to the different modes of MTOR inhibition, since starvation depends on upstream signaling events to induce MTOR inhibition causing delayed autophagosome formation. 22
Figure 1. Characterization of the MCF7 phosphoproteome. (A) Levels of phosphorylation of Thr389 RPS6KB1 in response to rapamycin (Rapa) treatment and (B) starvation are shown. Bars indicate quantification of intensity normalized by total RPS6KB1 relative to the first lane. (C) Induction of autophagic flux analyzed by anti-GFP-LC3 blots. ConA was added to inhibit lysosomal degradation. Bars indicate LC3-II levels normalized to actin relative to the first lane. (D) Outline of experimental strategy for quantitative phosphoproteomics characterization of MCF7 cells upon induction of autophagy. Time courses 1 and 2 shared 7 min as common time point allowing the construction of 5 time points kinetics. (E) Mass error and intensity distribution of peptide identifications. (F) Overlap of identified phosphorylated protein groups and phosphorylation sites identified after starvation and rapamycin treatment. (G) Proportion of phosphorylated serine, threonine and tyrosine residues. (H) Distribution of single, double, and multiphosphorylated peptide identifications.
To perform stable isotope labeling by amino acids in cell culture (SILAC) labeling it is necessary to culture cells in dialyzed serum and to ensure that this did not have a negative impact on the obtained results we assayed LC3-II formation in dialyzed serum and obtained comparable results as for cells cultured in normal serum (compare Fig. 1C and Fig. S1). To reduce background of serum induced phosphorylations we serum-starved the cells for both, starvation and rapamycin, experiments before inducing autophagy, after ensuring that this did not interfere with the studied signaling pathways (Fig. S2). Having established optimal stimulation conditions and timing for the experiments, we continued to apply global quantitative phosphoproteomics to characterize the temporal dynamics of the phosphoproteome upon induction of autophagy.
To perform the quantitative phosphoproteomics experiments parallel MCF7 cell cultures were labeled by SILAC 23 , 24 and transferred to Hank’s balanced salt solution or serum-free medium containing rapamycin for 0, 2, 7, 15, or 30 min. Differentially labeled cells were then mixed in equal amounts, lysed, protein extracts were enzymatically digested followed by peptide fractionation by strong cation exchange (SCX)- and phosphopeptide enrichment by TiO2-chromatography, and the resulting samples were analyzed by LC-MS/MS 25 and processed with MaxQuant software (Fig. 1D). 26 The acquired data was stringently filtered for false discovery rates below 1% on the protein, peptide, and phosphorylation site level. Adding further credibility to the data is the very good precursor mass accuracy from high-accuracy MS analysis with a median absolute mass error of only 0.2 ppm (Fig. 1E). Combining the data from 2 biological replicates of the 2 treatments, each, we in total identified 6,801 phosphorylation sites on 2,155 proteins (Table S1) demonstrating good reproducibility (median relative standard deviation of site quantifications between replicates of 11.6%; Fig. S3). To evaluate the likelihood of the phosphorylation site localization inside the peptide sequence being correctly assigned, localization probabilities for all sites were calculated by MaxQuant. The median localization probability for all phosphorylation sites is 0.991 and of all identified phosphorylation sites 5,106 could be assigned to a specific residue with a probability above 0.75 (hereafter referred to as ‘class 1 sites’). Thus, the site localization is highly certain for the majority of the reported sites. 27
Despite the bias of shotgun proteomics for high abundant proteins 28 the identified phosphoproteins covered a wide range of functional groups, including proteins usually present at low copy-numbers. 29 , 30 As examples we found 213 class 1 phosphorylation sites on 86 protein kinases, 649 sites on 225 transcriptional regulators and 70 sites on 23 ubiquitin ligases (see Table S2 and Fig. S4).
The majority of phosphorylation sites were identified after both treatments with an overlap of 91% of the phosphoproteins and 88% of the individual phosphorylation sites (Fig. 1F). The amino acid distribution of the phosphorylation sites correlated well with previous reports (Fig. 1G) 25 , 27 and in accordance with the preference of SCX chromatography and TiO2 the majority of peptides contained only one phosphorylation (64%). 31 , 32 But a considerable proportion of double and multiphosphorylated peptides was also identified (Fig. 1H).
Temporal phosphorylation dynamics of starvation- and rapamycin-induced autophagy
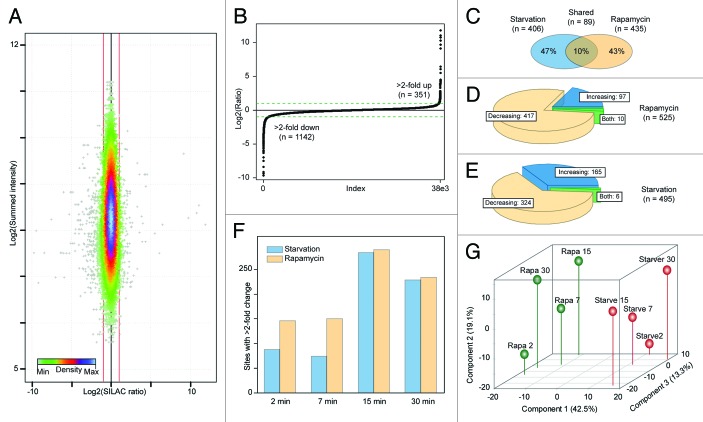
In total we recorded 38,154 individual phosphorylation site quantification ratios, the majority of which did not show considerable dynamics. Thus, the distribution of ratios clustered closely around 1:1 (Fig. 2A). Due to the presence of miss-cleavages, nonperfect fractionation prior to LC-MS/MS and the presence of phosphopeptides in variants with additional modifications, phosphosite quantification values are on average based on 12 independent ratio measurements. However, due to unavoidable measurement inaccuracies it is beneficial to assign a threshold for the minimum observed ratio required to regard a phosphosite as regulated. We and others have previously demonstrated that a 2-fold change is an appropriate value for a conservative cut-off and to consolidate this we calculated the MaxQuant significance values and found that a 2-fold change corresponds to an outlier with a P value of 0.009. 25 , 27 , 33 Furthermore, the 2-fold change cut-off is approximately equal to twice the average site variability of the individual site quantifications. Hence, we consider this a sufficiently robust threshold and based on this 1,493 ratios from 930 sites on 590 proteins showed significant dynamics with 351 ratios increasing and 1,142 decreasing (Fig. 2B). Within the 930 regulated sites 435 where specifically rapamycin sensitive, 406 specifically starvation sensitive and 89 sites on 74 proteins responded to both treatments and thus constitute a shortlist of potentially autophagy regulating phosphorylation events (Fig. 2C, Table S3). Despite the similar number of regulated sites after the treatments only 97 sites with increasing ratios were observed after rapamycin treatment (Fig. 2D) and 165 after starvation (Fig. 2E). The highest number of sites with dynamic phosphorylations was identified after 15 min for both treatments, but very early phosphorylation changes were also observed, since in total 230 sites showed above 2-fold dynamics already after 2 min treatment (Fig. 2F), demonstrating a rapid onset of potentially autophagy relevant signaling events.
Figure 2. Temporal dynamics of identified protein phosphorylations. (A) Density scatter plot of phosphorylation site quantification ratios vs. intensities, red lines indicating 2-fold dynamics (± 1 on Log2-scale). (B) Distribution of quantification ratios, dotted lines indicate regulation cut-off (± 1 on Log2-scale). (C) Venn diagram of regulated phosphorylation site identifications after the 2 treatments. (D) Distribution of sites with increasing or decreasing ratios after rapamycin treatment, sites indicated as both show both an increase and a decrease during the time-course. (E) Same as (D) for starvation. (F) Overview of the number of ratios with above 2-fold dynamics observed in each time point after the treatments. (G) Principal component analysis of log2 transformed phosphorylation-site ratios. Points corresponding to rapamycin and starvation treatment are shown in green and red, respectively. The numbering of the points indicates the time in minutes of the treatment duration.
To address the similarity between the observed phosphorylation dynamics at a global scale we performed a principal component analysis on the quantification ratios for each site and plotted the result in the dimensions of the first 3 components, which together explained 75% of the variability in the data (Fig. 2G). The primary observation from this analysis is that the variability between the effects of the treatments is larger than the variability between the time-points, which can be seen by the clear clustering of data points from the 2 treatments in the dimension of component 1. Furthermore, the late samples (15 and 30 min) in each treatment cluster closely together in the dimensions of component 1 and 2, whereas the earlier samples (2 and 7 min) are more distinct, illustrating bigger shifts after both treatments between 2, 7, and 15 min, probably due to a transition from early and intermediate signaling to more delayed events.
Serving as a consolidating observation we find threonine 70 of EIF4EBP1, a well-established MTOR substrate, within the group of shared sites with decreasing phosphorylation. In addition to this site we further identify 5 sites on EIF4EBP1 and 2 on EIF4EBP2. Of these, one site on each protein only responds to rapamaycin treatment indicating its direct influence on MTOR. Interestingly, Thr55 of the proteasomal subunit PSMA5 is one of the few sites, which displayed increased phosphorylation levels after both treatments indicating a crosstalk between both degradation pathways. As the site is solvent exposed in the assembled 26S proteasome (pdb code 4B4T), it might have regulatory function. A decrease in proteasome abundance and activity by functional autophagy has been shown recently. 15 Within the treatment-specific events we find e.g., downstream MTOR targets such as Thr9 on GSK3B, which decreases only after rapamycin treatment. As an example of a site responding only to starvation we find serine 2300 on HIVEP2, which exhibits a decrease in phosphorylation after 7 min starvation. Interestingly, HIVEP2 was recently shown to play a role in autophagy signaling in a large-scale siRNA screen. 34
Pathway analysis and inference of interactions between MTOR signaling pathway members
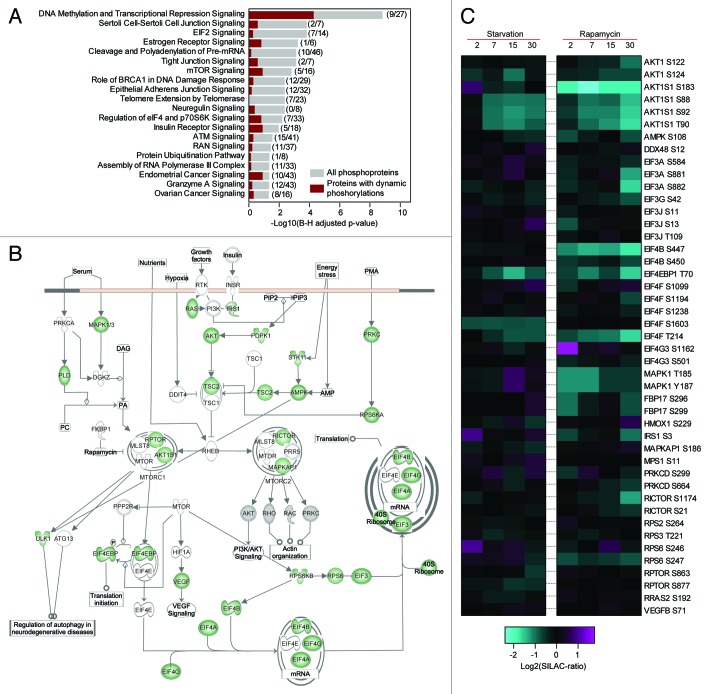
To investigate how different signaling pathways were covered by our data we mapped all identified phosphorylation sites to the curated Ingenuity protein pathways and identified from this analysis 20 signaling pathways as enriched (Fig. 3A). As a corroborating finding the MTOR signaling pathway was found among the enriched pathways with 97 phosphorylations on 39 proteins identified, corresponding to approximately 50% of the proteins in the pathway (Fig. 3B). Among the identified MTOR pathway proteins are 4 known MTOR complex proteins, including 2 sites on RPTOR and 5 sites on RICTOR. In addition, we also identified phosphorylation on many known downstream autophagy regulators such as ULK1, AMPK and MAPK1/3 (ERK1/2). In Figure 3C the ratios for the MTOR pathway sites for which we obtained quantifications in all time-points are visualized and it is clear that the majority of the ratios are decreasing. Furthermore, it is also clear that the impact on the pathway is faster by rapamycin treatment compared with starvation as we observe more ratios showing dynamics after the rapamycin treatment.
Figure 3. Pathway analysis of identified phosphorylation sites. (A) List of pathways from Ingenuity Pathway Analysis software found as enriched for the identified phosphoproteins, gray bars indicate P values for enrichment of all identified phosphoproteins and red bars for proteins with dynamically regulated phosphosites. Numbers in bracket indicate, first, the number of identified phosphoproteins with dynamic sites in the pathway and, second, all identified phosphoproteins in the pathway. Due to partial overlaps between pathways the same protein might occur in more than one pathway. (B) Graphical representation of the MTOR signaling pathway, proteins identified with phosphorylation are indicated in green. The connection between AMPK and ULK1 was added manually. (C) Dynamics of the sites identified on proteins in the MTOR signaling pathway shown in (B) for which quantification values in all time-points were obtained.
To complement this pathway analysis we set out to investigate if quantitative phosphoproteomics data could be used to extract putative protein-protein interactions. For this we applied the time-lagged correlation 35 strategy outlined in Figure S5 that uses a correlation coefficient to infer protein-protein interactions by determining the relationship between the temporal dynamics of 2 phosphosites. We applied this to all phosphosites on proteins in the MTOR signaling pathway and by combining the quantitative information after both treatments obtained a regulatory network for potential interactions between MTOR signaling proteins containing 15 protein-protein interactions, based on data from 39 sites (Table S4). In Figure S6 the complete inferred network is presented. Several well-described interactions are reconstructed. Furthermore, the inferred network also suggests new interactions between e.g., MTORC1 and proteins involved in translation via AKT1S1 36 indicating that this strategy might serve as an approach to extract new hypotheses about putative interactions.
Cluster analysis of phosphorylation site dynamics demonstrates widespread impact on biological processes
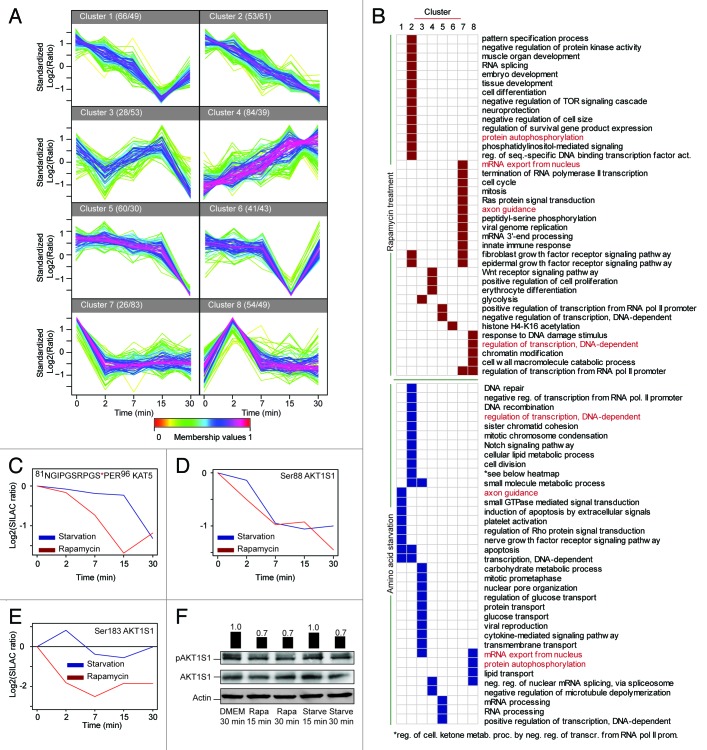
Given the size of contemporary phosphoproteomics data sets, the identified phosphorylation sites have a multitude of different temporal dynamic profiles making it challenging to summarize the complete data set. However, despite the large complexity present in the data this can often be reduced by grouping sites with similar patterns of regulation. To obtain an overview of the temporal dynamics we log2 transformed and standardized (z-scored) all the sites to enable clustering based on the overall pattern of the dynamics and not the absolute magnitude of the regulation. Subsequently, we submitted all sites that showed regulation in at least one time point to clustering by the fuzzy c-means algorithm 37 and found that the data could be partitioned into 8 clusters with roughly equal numbers of sites describing the most pronounced patterns of regulation within the data (Fig. 4A). From the cluster analysis it is evident that significant changes of phosphorylations are observed throughout the experimental time window and that very different modes of dynamics are seen. As an example the sites in clusters 2 and 4 show a gradual decrease and increase, respectively, which is in stark contrast to the sites in cluster 8 which increase rapidly after 2 min but return to the control level already after 7 min. In cluster 5 sites are found which respond only after 30 min treatment, indicating that the dynamics of these sites depend on upstream events to take place. In accordance, 2 times as many starvation responding sites are found in this cluster compared with rapamycin treatment.
Figure 4. Clustering and gene ontology enrichment analysis. (A) For cluster analysis ratios for sites with quantifications in all time-points were log2 transformed and standardized (z-scored) and submitted to clustering by fuzzy c-means. Numbers after cluster label indicate the number of sites after starvation (first) and rapamycin (second). (B) The phosphorylation sites found in each cluster in (A) were tested against the sites showing no dynamics for enrichment of GO biological process terms using Fisher’s exact test. The maps indicate the significantly enriched (P value < 0.05 after B-H correction) GO terms in the respective cluster(s) by rapamycin treatment (top, red) and starvation (bottom, blue). GO terms written in red represent terms that are found enriched after both treatments. Temporal profiles of (C) Ser90 of KAT5, (D) Ser88 of AKT1S1, and (E) Ser183 of AKT1S1. (F) Western blot of Ser183 on AKT1S1, bars indicate quantifications normalized to actin relative to the control (DMEM 30 min).
To explore whether phosphorylation of proteins with similar biological implications were observed in the different clusters we tested for enrichment of GO terms of source proteins within the sites in each of the clusters vs. the nonchanging, static sites (Fig. 4B). From this analysis we were able to extract panels of terms describing the global impact of phosphorylation dynamics on diverse biological processes. From Figure 4B it is apparent that the enriched GO terms are highly diverse ranging from e.g., DNA damage repair to glucose transport in the case of starvation, highlighting that both treatments have cellular consequences apart from the induction of autophagy. Another observation is that the majority of the GO terms are only found enriched after one of the treatments, illustrating that, although the 2 treatments both result in inactivation of MTOR, pronounced differences are observed between the impacts of these on the phosphoproteome. One of the terms that was enriched after both treatments, although in different clusters, is regulation of transcription, DNA-dependent, which attracted our attention since the understanding of connections between autophagy induction and transcriptional regulation is still sparse. Among the regulated sites on proteins involved in transcriptional regulation were sites on the histone acetyltransferase KAT5, which is intriguing, because KAT5 has been shown to acetylate one of the master regulators of autophagy, the kinase ULK. The activity of KAT5 itself is regulated by phosphorylation of Ser86 by GSK3 and as a consequence KAT5S86A mutant cells are resistant to serum starvation-induced autophagy. 38 We identified the peptide covering the amino acid residues 81 to 96, thus containing both Ser86 and Ser90, the latter being a potential priming site for Ser86. 38 Interestingly, we observed a marked dephosphorylation of the peptide containing only phosphorylation of Ser90 (Fig. 4C), whereas the peptide containing phosphorylation on both serine residues showed no significant change. The GO term analysis also identified the term negative regulation of TOR signaling cascade after rapamycin treatment, which is due to identification of dynamics of 4 sites on AKT1S1 (Fig. 4D and E), a member of the MTOR complex 1. We identified different dynamics for 2 sites on this protein, a decrease in phosphorylation of Ser88 after both starvation and rapamycin treatment, while the phosphorylation of Ser183 decreased only upon rapamycin treatment. To validate the regulation of Ser183 in AKT1S1 we performed western blot for this site and found again a pronounced dephosphorylation after rapamycin treatment (Fig. 4F). From the western blot results we also observed a delayed decrease in phosphorylation after starvation, which was not observed to the same extent by MS. This could be due to additional starvation-specific posttranslational modifications on the specific peptide interfering with either MS- or western blotting analysis.
Phosphorylation dynamics of protein kinases
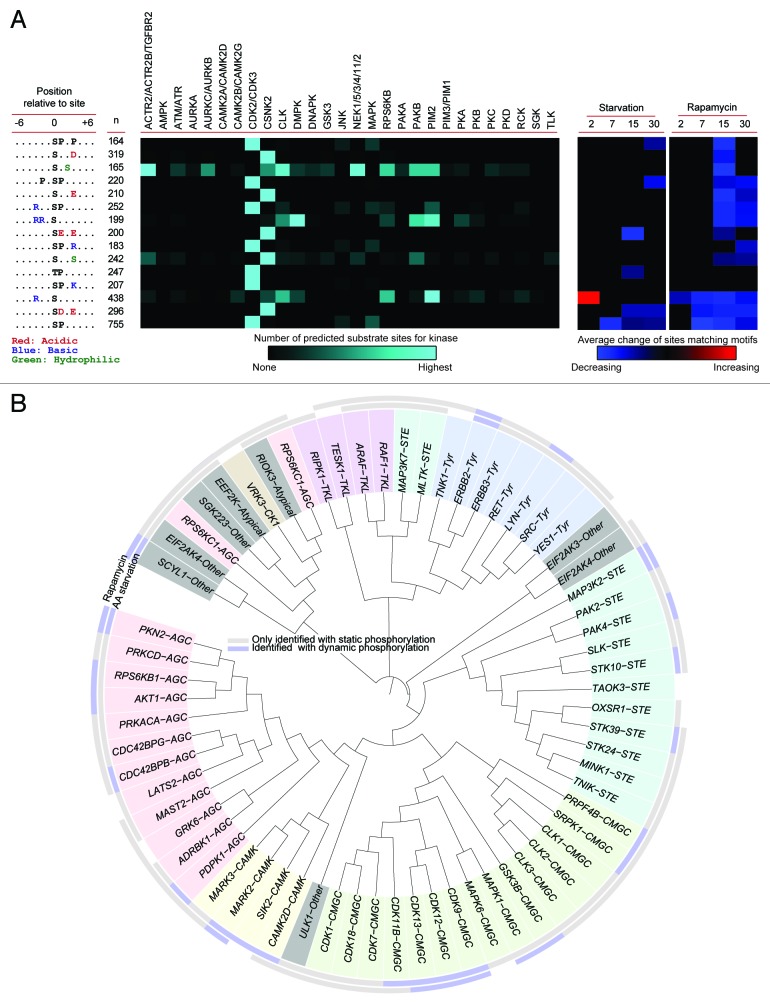
The specificity of serine and threonine kinases is largely controlled by the amino acid residues found proximal to substrate sites. 39 Thus, the presence or absence of specific amino acids serves to induce or prevent phosphorylation by different kinases. To identify potential phosphorylation motifs specific for particular kinases we looked for enrichment of amino acid residues proximal to all identified class 1 sites and found 15 distinct motifs (Fig. 5A). The identified phosphorylation motifs fell into 3 groups as they all contained a proline, a charged residue, or an additional serine. Some of the motifs are well known and have been shown to be the substrate of e.g., the MAPK family in the case of the PxSP members, or CSNK2 (casein kinase 2), which has a preference for acidic motifs. Several of the identified motifs have, however, not been connected with a specific kinase, to our knowledge. To obtain the identity of the kinases likely to target the extracted motifs we retrieved kinase predictions for the identified phosphorylation sites. From Figure 5A it is seen that the identified motifs were predicted to be substrates for a range of different kinases, particularly CDK2/3 and CSNK2. The prevalent identification of substrates for these kinases are, however, likely to be partly due to the loose requirements of the respective substrate motifs.
Figure 5. Kinome and kinase-motif analysis. (A) Linear kinase motifs present in the data were extracted using Motif-X 40 (left panel) and predictions for kinase groups phosphorylating the sites matching the motifs were obtained from the NetworKIN algorithm (middle panel). 41 See Table S6 for individual kinase(s) most likely responsible for phosphorylation of respective motifs. The means of the regulated sites matching each motif were tested for difference from zero. If significant (P value < 0.05) the mean value is indicated in the right panel, with red indicating increasing and blue indicating decreasing phosphorylation levels. (B) All identified kinases were aligned and presented according to the homology of the kinase domains. Protein kinases belonging to the same family are colored in the same background color. The rims indicate if static, nonchanging sites (gray), or dynamic sites (blue) are identified on the respective kinase by starvation (inner rim), or rapamycin treatment (outer rim).
To evaluate if the activity of the kinases predicted to target the identified motifs appeared to be regulated we tested if the average of the regulated sites matching each motif were either above or below the control level. From the overview in Figure 5A it is observed that rapamycin induced the regulation of more motifs than starvation and for both treatments the regulation was predominantly observed at later time-points. To assess the value of this strategy we tested if this approach could pick up known kinase-substrate relationships identified from low throughput approaches and, indeed, found that we were able to reconstitute e.g., Ser13 on CDC37 as a CDK2 substrate and Ser239 on ATRIP as a CDK2 substrate, both showing significant dephosphorylations. 42 , 43
To complement the bioinformatics strategy for outlining kinase activity regulation we screened our data for experimental identifications of phosphorylation sites on kinases. Of the more than 80 kinases on which we identified phosphorylations, 23 showed phosphorylation dynamics within our time frame. Based on the hypothesis that homologous kinases might show co-regulation we ordered all identified kinases based on the similarity of their kinase domains and indicated if we observed regulated phosphorylation of these kinases after the 2 treatments (Fig. 5B). From this analysis it can be observed that, except for 2 cases, we did not see dynamic phosphorylation of tyrosine kinases or tyrosine-like kinase. From the kinases with dynamic phosphorylation it appears that the regulation after the treatments is not directed predominantly toward a specific family, since regulation is observed across the identified kinase families. One exception to this was, however, observed for the cyclin dependent-kinases as CDK9, CDK12 and CDK13 show dynamic phosphorylation after both treatments and CDK11B after rapamycin. Interestingly, the 2 analysis strategies could be connected since we found Ser249 on CDK12 to show an increase after 2 min starvation and this site also matched the identified RxxS motif. In addition, we observed also dynamic phosphorylation after both treatments for SCYL1, PKN2, MARK2, ERBB2, and MAP3K2 highlighting these as potentially autophagy relevant kinases.
Identification of LC3-interacting phosphoproteins
One of the predominant roles of protein phosphorylation in cellular systems is to alter the interactions in which proteins engage, thus regulating e.g., the recruitment of a protein to a protein complex. In the process of autophagy a key event is the targeting of proteins to autophagosomes. One way of attracting proteins to autophagosomes is the interaction with LC3-II located in autophagosomal membranes 5 and it was recently shown that phosphorylation can be an important regulator of LC3 mediated protein-protein interactions. 44 , 45
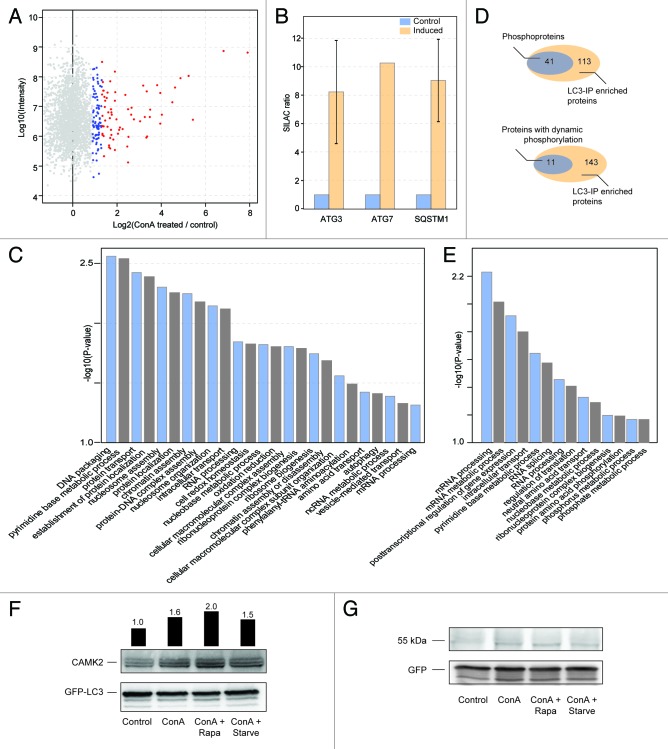
Based on this we hypothesized that the relevance of some of the observed phosphorylation events might be to affect the recruitment of proteins to autophagosomes. To evaluate this we performed pull-down experiments to identify proteins that showed an increased interaction with LC3 upon block of autophagosomal degradation by inhibiting lysosomal acidification using concanamycin A (conA). From 2 biological replicates we identified 154 proteins (Table S5) that showed a significantly increased interaction with LC3 upon treatment (P value < 0.05; Fig. 6A; Fig. S7A and S7B). Serving as a proof of principle for this strategy we identified 3 known LC3-interacting proteins ATG3, ATG7, and SQSTM1, all showing a strong increase upon autophagosomal accumulation (Fig. 6B). To obtain an overview about the potential biological function of LC3-interacting proteins we performed GO term enrichment analysis, which demonstrated a wide range of protein categories as LC3 interactors, many being involved in transport and trafficking (Fig. 6C). Recently, a particular sequence motif was identified as mediator of LC3 interaction and thus termed LC3-interacting region (LIR). 5 We screened our list of LC3-interacting proteins and found in total 716 potential LIR motifs, with 124 proteins containing LIR motifs and 113 of these containing more than one potential LIR motif (Fig. S7C and S7D). However, given the loose description of the motif it is likely that not all of the motifs are of functional relevance. In agreement with this, we did not find any correlation between number of LIR motifs in the sequence and extent of LC3 interaction.
Figure 6. Large-scale screen for phosphorylated LC3 interaction partners. (A) Using a rat eGFP-MAP1LC3-MCF7 cell line immunoprecipitations by anti-GFP antibodies were performed to enrich LC3-interacting proteins from untreated control cells and cells where autophagosomal degradation was blocked by the addition of 2 nM conA. From this analysis specific interactors could be identified as proteins exhibiting high ratios. Using the significance values provided by the MaxQuant software package significant interacting proteins were filtered (blue and red, P value below 0.05 and 0.01, respectively). (B) Enrichment ratios for the known LC3-interacting proteins ATG3, ATG7, and SQSTM1 are shown, error bars indicate standard error of 2 biological replicates. (C) Enriched GO terms for the identified LC3-interacting proteins were obtained from the DAVID resource 46 using default parameters. (D) Venn diagram of overlap between identified LC3-interacting proteins and all proteins which we identified as phosphorylated in this study (top) and the subset of the phosphoproteins identified which have one or more dynamic phosphorylation (bottom). (E) Same as (C) for the 43 proteins overlapping between phosphorylation and LC3-interaction screens. (F) CAMK2-LC3 interaction. Cells were left untreated or treated for 30 min as indicated. GFP-LC3 and interacting proteins were precipitated and blotted using a pan-CAMK2 antibody as readout. Accumulation of CAMK2 isoforms can be observed by a block of autophagy using conA. Bars indicate CAMK2 band intensities normalized to GFP-LC3 relative to the first lane. (G) Experiment performed as under (F) except for using GFP-only cells. GFP is shown as loading control.
Next we screened the 2 data sets covering the MCF7 phosphoproteome and the LC3 interactome against each other and found 41 of the 154 identified LC3-interacting proteins also in our list of phosphoproteins (Fig. 6D). This group covered a broad category of proteins with functions such as translation, intracellular transport, and regulation of transcription (Fig. 6E; Table S5), and included proteins which have already been linked to autophagy like SQSTM1, 47 major histocampatibility complex class I proteins, 48 and members of the ubiquitin-proteasome system. 15 Inspecting this group of proteins further we found regulated phosphorylation on 11 of these proteins within our 30 min time-frame (Fig. 6D), among these the kinase CAMK2 (calcium/calmodulin-dependent protein kinase II Δ). This was an interesting finding because other members of the CAMK family (CAMK4 and CAMK1) are involved in autophagy signaling. 49 , 50 To validate this approach as a strategy for identifying autophagosomal targeting of proteins we decided to perform GFP-LC3 immunoprecipitations and blotted the respective eluates against CAMK2 (Fig. 6F). Autophagy induction was performed for 30 min and conA was added to block autophagosomal degradation. An increase in interaction upon autophagy induction can be observed for all treatments, indicating that the interaction might be regulated by phosphorylation. As a control GFP immunoprecipitations were performed, which did not yield the respective signals (Fig. 6G).
Discussion
Due to numerous investigations the pivotal role of autophagy as a critical cellular process and stress response is becoming evident. It has been shown to have implications not only in energy and nutrient homeostasis, but also in human health and disease. Potential roles in clinical applications are emerging as an involvement of autophagy in i.e., immunity 6 , 7 and cancer 8 could be shown. Before autophagy can be exploited in a clinical setting there is a need to understand the highly complicated regulatory mechanisms underlying induction and progression of autophagy. To expand our knowledge of the initial signaling events in autophagy induction we have characterized phosphorylation dynamics within the first 30 min upon starvation and rapamycin treatment.
Autophagy can be induced by a wide range of different cellular stimuli, including amino acid 51 or glucose starvation, 52 MTOR inhibition by rapamycin, 12 growth factor deprivation, 53 SIRT1 (sirtuin 1) activation, 54 spermidine-treatment, 55 mitochondrial damage, 56 elevated ammonia concentrations, 57 oxidative stress, 58 and hypoxia. 59 We selected starvation and MTOR inhibition by rapamycin as the treatments for induction of autophagy, because these are among the best described, thus providing a framework for the analysis of the acquired data. Applying 2 different treatments allows discrimination between treatment specific regulatory events, not necessarily related to autophagy, and shared events observed after both treatments likely to be related to autophagy. Furthermore, since one of the consequences of starvation is inhibition of MTOR, the use of these treatments allows an identification of signaling events upstream of MTOR and also events independent of MTOR. The initiation of autophagy occurs rapidly. 60 , 61 Thus, we hypothesized that phosphorylation would be important in the earliest steps after induction and focused on the events happening within the first half-hour of treatment. Nevertheless, it is clear that phosphorylation continues to play a role throughout the process of autophagy exemplified by a recent study by Bennetzen et al. 62 that applied a similar experimental setup to characterize the phosphorylation response after 2 h treatment, 62 focusing on events taking place after the first wave of phosphorylation signaling. In this study autophagy was induced by treating cells with the compounds resveratrol or spermidine. A direct comparison with this study is challenging as both, resveratrol and spermidine, target the acetylation machinery, and thus function most likely by different signaling routes than starvation and rapamycin. 55 Nevertheless, it is interesting to note that in agreement with this study we also identified regulation of the most apparent kinase motifs identified by Bennetzen et al., 62 namely RxxS and SP motifs.
Recently, the MTOR-responsive phosphoproteome has been the focus of at least 3 studies focusing on the role of MTOR in cancer and growth factor signaling. 33 , 63 , 64 Furthermore, in this issue of Autophagy, Harder et al. present data from a phosphoproteomics analysis of the induction of autophagy by rapamycin and ammonia. To evaluate the robustness of the data presented here we have examined the similarity between the data published by us and by Harder et al. In this comparison we found it relevant to also include data published by Hsu et al. 64 and Yu et al., 33 which also combined the use of rapamycin and phosphoproteomics to study MTOR-dependent signaling. 33 , 64 Figure S8A summarizes the cellular treatment regime used in the four studies and it can be noticed that there are considerable experimental differences between the data presented here and the data from Hsu et al. 64 and Yu et al. 33 In particular the length of the treatment is different as rapamycin is applied for 1 h 20 min or 2 h before cell harvest in these studies. Furthermore, Hsu et al. 64 stimulate cells with insulin 20 min before harvest, thus inducing MTOR-independent events and also increasing MTOR activity in nonrapamycin treated controls resulting in larger phosphosite ratios. Also the differences in cellular systems complicate a direct comparison since Hsu et al. 64 used HEK293 cells and Yu et el. 33 performed their studies in mouse embryonic fibroblast with nonfunctional TSC2, resulting in hyperactive MTOR signaling. Despite these differences all studies identify a substantially overlapping part of the phosphoproteome (Fig. S8B) and more importantly 98 shared proteins with dynamic phosphorylation are identified in the 4 studies (Fig. S8C). Despite the large overlap of the pairwise comparisons only 3 proteins were identified as rapamycin sensitive in all 4 studies, RPS6KB1, FOXK1 and AKT1S1. As noted from Figure S8A our data and the data from Harder et al. have a direct intersect at 15 min rapamycin treatment and, furthermore, both studies used the same cell line, MCF7 cells. Therefore, a direct site-by-site comparison is feasible. The observed ratios in both data sets are very similar (Fig. S8D) and the most robust rapamycin sensitive sites correlate well exhibiting in some cases nearly identical ratios (Fig. S8E). Comparing our data with the results of a genome-wide siRNA screen for genes regulating autophagy by Lipinski et al. 34 we identified phosphorylation on 33 of the proposed autophagy regulators, including dynamic phosphorylation of e.g., HIVEP2, PDCD5, TPR, heterogeneous ribonucleoprotein particles and STIM1/GOK after starvation and TACC2 after rapamycin treatment.
To complement the value of the data as resource we have applied bioinformatics strategies to extract information about the global regulation of the phosphoproteome. From a general analysis of the dynamic sites it is clear that, despite the fact that both treatments inhibit MTOR, there are 841 phosphorylation events that are specific to a single treatment, as only 10% of the dynamic sites are affected by both treatments. Furthermore, the majority of dynamic sites showed a decrease correlating well with the inhibition of MTOR signaling and downstream kinases by the 2 treatments. Another important result is the observation that 230 dynamic events take place as early as after 2 min of treatments. From a PCA analysis it was evident that there is a shift in signaling between 7 and 15 min, and, thus, sampling only later events observed at an hour timescale will likely cause early signaling events to be overlooked. On the contrary, by focusing on the earliest events in autophagy we are likely to miss later phosphorylated events important for autophagy. In addition, mass spectrometry has a bias toward high abundant phosphosites/proteins, which might be one reason why we do not observe any dynamics for sites on the pivotal autophagy regulator ULK1, or why ATG13 is not detected at all. In return however, quantitative phosphoproteomics allows a nonhypothesis-driven approach, which has allowed us to identify potential autophagy relevant phosphorylation events that would not be identified by more targeted strategies.
Combining unsupervised clustering and GO term enrichment analysis allowed us to extract the terms that best describe the sites affected by the treatments, such as the term regulation of transcription, DNA-dependent, which was found significantly enriched in part due to the dynamics of KAT5. The regulation of serine 90 of KAT5, which exhibited a marked decrease in phosphorylation after both treatments, is intriguing as it may be a critical point of crosstalk between the phosphorylation and acetylation machinery. The exact consequence of the phosphorylation pattern of KAT5 is difficult to predict, but it seems likely that phosphorylation of this particular region of KAT5 is a general feature in autophagy induction since this is observed after distinct treatments. 38
From an analysis of phosphorylation of protein kinases we identified regulated phosphorylation sites on 23 kinases including the cyclin-dependent kinases (CDK) 9, 12, and 13 that responded to both treatments, indicating that this class of kinases may play an important role in the early events of autophagy. Recent studies support a proposed role of cyclin-dependent kinases in autophagy. Wilkinson et al. show that CDK11 is required for functional autophagy, 65 Capparelli et al. outline that CDK inhibition induces autophagy, 66 and Furuya et al. highlight that CDK1 and CDK5 phosphorylate PIK3C3/VPS34, negatively regulating its interaction with BECN1/Beclin 1 in mitosis. 67 Starvation leads to a cell cycle arrest and rapamycin treatment to a marked increase in doubling time (own unpublished data) and therefore it is not farfetched to envisage an active crosstalk between autophagy and cell cycle regulation. Another kinase responsive to both treatments was SCYL1, which has been shown to be inhibitory for autophagic flux functioning upstream of MTOR. 68 The finding that phosphorylation on this kinase is responsive to MTOR inhibition by rapamycin as well as by starvation indicates that the regulation of the kinase circuitry of these 2 proteins might be complex, since SCYL1 activity might be dependent on MTOR itself.
Unarguably, one of the biggest challenges in contemporary phophoproteomics is the immediate lack of high-throughput assays to assess the consequences of phosphorylation. As a result the function of the vast majority of phosphorylation sites identified by large scale screens is unknown. To address whether a subset of the phosphorylations we identified might have a role in the targeting of proteins for autophagosomal degradation, we performed a proteomics based screen of LC3-interacting proteins. Recently, Behrends et al. applied a partially similar experimental setup 18 and identified 67 LC3-interacting proteins under basal conditions, of which 5 were also identified in our screen (SQSTM1, NIPSNAP1, HADHA, ATG3, ATG7). By cross-referencing our interactome and phosphorylation data we could extract a group of proteins where the function of the sites might regulate LC3 interaction. Clearly, this does not provide a final proof, but combining these 2 experiments allows us to propose a potential role for a subset of the identified phosphorylation sites. E.g. we identified 4 phosphosites on SQSTM1, among them Ser332 in close vicinity of the LIR motif. These sites were not regulated in the observed timeframe. However, this does not exclude that they are regulated at later time points, possibly influencing SQSTM1-LC3 interaction. As we do not detect phosphorylation of Ser403, a site that was previously shown to be important for the function of SQSTM1, we cannot rule out that the observed behavior is due to sites not identified in this study. 69 Of the 11 LC3 interactors exhibiting dynamic phosphorylations, the kinase calcium/calmodulin-dependent protein kinase type II subunit delta might be of special interest, as kinases of this class have already been linked to autophagy. 49 , 70 This might indicate that the autophagosome functions as a scaffold orchestrating signal transduction by bringing together relevant kinases.
Taken together, by combining time-resolved phosphoproteomics and LC3 interaction data, potential autophagosomal target mechanisms are highlighted. Signaling events in autophagy induction have been linked to cell cycle regulation, among others, and the impact on the kinome is described. Thus, this study serves to illustrate the value of integrating data from high-throughput studies complementing and supporting each other. Clearly, as information continues to accumulate, this is bound to point out new directions in autophagy research by generating new hypotheses for future functional studies.
Materials and Methods
Cell culture and sample preparation
For SILAC labeling MCF7 cells were cultured in Dulbecco's modified Eagle's medium (GE Healthcare, E15-085) supplemented with penicillin/streptomycin (100 units/ml, 100 μg/ml), glutamine, and 10% dialyzed fetal calf serum (Gibco, Invitrogen, 26400). To differentially label parallel cultures these were grown in media containing l-arginine (Arg0, Sigma-Adrich, 11039) and l-lysine (Lys0, Sigma-Adrich, 62929), l-lysine–2H4 (Lys4, Silantes, 211104113), and l-arginine–U-13C6 (Arg6, Silantes, 201204102), or l-lysine–U-13C6-15N2 (Lys8, Silantes, 211604102) and l-arginine–U-13C6-15N4 (Arg10, Silantes, 201604102) to generate “light-,” “medium-,” and “heavy-” labeled cells, respectively. Fully labeled cells were grown to 80 to 90% confluence, serum-starved overnight and either washed 3 times in PBS and transferred to Hank’s buffered salt solution containing glucose (1 g/l) or treated with rapamycin (Sigma-Aldrich, R0395) at a final concentration of 100 nM. Light-labeled cells were subjected to the treatments for either 0 or 2 min, medium-labeled cells were treated for 7 min in all experiments to serve as a common time point and heavy-labeled cells were treated for 15 or 30 min.
Subsequent to the induction of autophagy cells were mixed in 1:1:1 ratio and lysed in 0.1% deoxycholate (Fluka, 30970; Replicate 1) or modified RIPA buffer (1% NP-40, 0.1% deoxycholate, 150 mM NaCl, 1mM EDTA, 50 mM Tris, pH 7.5) followed by acetone precipitation and reconstitution in 8 M urea (Replicate 2) and proteins were reduced by addition of dithiothreitol to a final concentration of 1 mM followed by alkylation of cysteines by addition of iodoacetamide (Sigma-Aldrich, I1149) at a final concentration of 5.5 mM. Proteins were digested with trypsin (Sequencing-grade, Promega, V5113 at an enzyme to protein ratio 1:50 overnight, trifluoroacetic acid (TFA; Promochem, SO-9668-B001 catalog number) was added to a concentration of 0.1% and the sample was cleared by centrifugation. Thus, 2 biological replicates were performed for each treatment resulting in a total of 4 quantitative phosphoproteomics experiments.
Fractionation and phosphopeptide enrichment
SCX chromatographic separations were performed on an Akta System using a 1-mL column (Resourse S, GE Healthcare, 17-1178-01). Tryptic digests of MCF7 cells were acidified by formic acid to a pH under 3, diluted by acetonitrile (ACN; Promochem, SO-9340-B025) to 30%, and loaded to an equilibrated column with 30% ACN containing 5 mM KH2PO4, pH 2.7 (Solvent A). After 10 min in 100% A, peptides were eluted with 30% ACN containing 5 mM KH2PO4 and 300 mM KCl, pH 2.7 (Solvent B) using a gradient from 0% to 30% in 30 min followed by 30% to 100% B in 5 min and then maintained at 100% B for 10 min at a flow rate of 1 mL/min.
For TiO2 based enrichment 1 to 10 microliters of a 50% slurry of TiO2 (GL Sciences, 5020-75010) in 30 mg/mL dihydroxybenzoic acid (Sigma-Aldrich, 149357) were added to each fraction and flow-through (first replicate) or whole cell lysate (second replicate) and incubated for 30 min at room temperature. Beads were washed with 100 μL of 10% ACN and 1% TFA followed by 100 μL of 80% ACN and 1% TFA and finally 100 μL of water. Phosphopeptides were eluted using 25% ammonium hydroxide in 20% and 40% ACN, respectively. Eluted phosphopeptides were dried to less than 5 μL and resuspended in 15 μL of 0.5% acetic acid for analysis. Five microliters of each fraction were used for MS analysis. For consecutive incubations, the peptide beads slurry was centrifuged and the supernatant was incubated with another aliquot of freshly prepared TiO2 beads.
eGFP-LC3 immunoprecipitation, western blots, and antibodies
For autophagosome accumulation cells were treated with 2 nM conA (Sigma-Aldrich, 27689) for 7 h. Anti-GFP (Santa Cruz Biotechnology, sc-9996) immunoprecipitations of SILAC-labeled MCF7 cells expressing rat eGFP-MAP1LC3 71 were performed in modified RIPA buffer containing 1% NP-40, 150 mM NaCl, 0.25% Na deoxycholate, 50 mM Tris pH 7.5 and Complete Protease Inhibitor tablets (Roche, 11836145001). Cells were lysed for 10 min on ice followed by centrifugation (17,000 × g/15 min) to remove nuclei. The supernatant fraction was precleared with 20 µl protein G sepharose beads slurry (Sigma-Aldrich, P3296) for 30 min followed by an incubation with 80 µl bead-conjugated anti-GFP-antibody for 5 h at 4 °C. Beads were washed 3 times and proteins were eluted by incubation with 1× SDS loading buffer with 1 mM DTT for 10 min at 95 °C.
All western blots were performed minimally 2 times. Additional antibodies used for western blots were: anti-pan-CaMKII (Cell Signaling, 4436), anti-actin (Sigma-Aldrich, A3853); RPS6KB1 (Cell Signaling, 9202, 9206 anti-P-Thr389), anti-AKT1S1 (Cell Signaling, 2691, 936 anti-p-Ser183)
LC-MS/MS and data processing
Samples for LC-MS/MS were fractionated by nanoscale–HPLC on either an Agilent 1200 or an Eksigent NanoLC-ultra connected online to a LTQ-Orbitrap XL (Thermo Scientific). Peptides were separated over a linear gradient from 10% to 30% ACN in 0.5% acetic acid with a flow rate of 250 nl/min. All full-scan acquisition was done in the FT-MS part of the mass spectrometers in the range from m/z 350 to 2000 with an automatic gain control target value of 106 and at resolution 60,000 at m/z 400. MS acquisition was done in data-dependent mode to sequentially perform MS/MS on the 5 (first replicate) or 10 (second replicate) most intense ions in the full scan (Top10) in the LTQ using the following parameters. AGC target value: 5,000. Ion selection thresholds: 1000 counts (first replicate) or 100 (second replicate) and a maximum fill time of 100 ms. Wide-band activation was enabled with an activation q = 0.25 applied for 30 ms at a normalized collision energy of 35%. Singly charged and ions with unassigned charge state were excluded from MS/MS. Dynamic exclusion was applied to reject ions from repeated MS/MS selection for 45 s. All MS/MS scans were recorded with multistage activation enabled with a neutral loss mass list of m/z 195.95, m/z 97.97, m/z 48.99 and m/z 32.66.
All recorded LC-MS/MS raw files (359) were processed together in MaxQuant 26 version 1.2.2.5 with default parameters using the March 2012 UniProt database, which contains 81,470 protein sequences. For databases searching parameters were mass accuracy thresholds of 0.5 (MS/MS) and 6 ppm (precursor), maximum two missed cleavages, carbamidomethylation (C) as fixed modification and deamidation (NQ), oxidation (M), phosphorylation(STY) and protein N-terminal acetylation as variable modifications. MaxQuant was used to filter the identifications for a false discovery rate below 1% for peptides, sites and proteins using forward-decoy searching. Match between runs were enabled with a retention time window of 2 min.
Data analysis and bioinformatics
For fuzzy c-means clustering quantification ratios for sites with above 2-fold dynamics in at least one time point were Log2 transformed and z-scored by subtracting the mean and dividing with the standard deviation, clustered and plotted using GProX 1.1.8 72 with a fuzzification parameter of 2 and 100 algorithm iterations. To test for enriched Gene Ontologies within the sites found in each cluster GO biological process terms were retrieved from UniProt for all proteins using GProX and Fisher exact test was used to test for over-representation at α = 0.05 after Benjamini and Hochberg correction, requiring at least 3 observations in the test cluster.
For motif analysis sequence windows of ±6 amino acids of all class 1 sites were submitted to Motif-X 40 requested a maximum P value of 0.000001 and at least 150 occurrences.
Kinase predictions were extracted and filtered using default settings with PhosphoSiteAnalyzer 73 and phosphorylation motifs were scored as regulated if the average was different from zero at α 0.05. To analyze homology of detected kinases, the kinase domain(s) of these were aligned using ClustalW2 74 and visualized using iTOL. 75
Calculation of the time-lagged correlation coefficients and measurement of the inferred interaction networks
In the data used for network interface, each phosphosites xi has 5 time points of fold changes data with the experimental conditions of after starvation and rampacyin treatments, which can be represented as [xi(t11), xi(t12), xi(t13), xi(t14), xi(t15), xi(t21), xi(t22), xi(t23), xi(t24), xi(t25)]. We calculated the time-lagged correlation coefficients based on the Pearson correlation coefficients between two phosphosites xi and xj with one time-interval lag:
where X = [xi(t11), xi(t12), xi(t13), xi(t14), xi(t21), xi(t22), xi(t23), xi(t24)] and Y = [xj(t12), xj(t13), xj(t14), xj(t15), xj(t22), xj(t23), xj(t24), xj(t25)].
To assess the performance of the inferred interaction networks, we calculated the precision-recall curves of the inferred networks by comparing the inferred networks to the corresponding protein-protein interactions present in STRING database (v9.0, active prediction methods of experiments plus databases with confidence scores > 0.7). Precision is the fraction of predicted interactions that are shown in STRING database [TP/(TP + FP)], and recall is the fraction of all known interactions that are predicted by the inferred network [TP/(TP + FN)], where TP is the number of true positives, FP is the number of false positives, and FN is the number of false negatives. Precision and recall were estimated over a range of thresholds. Interactions with ranking scores below the optimal threshold were removed from the final inferred network. To increase the precision of the prediction, we used a prior knowledge that translation proteins downstream of MTOR cannot directly regulate other phosphosites of upstream proteins. We used a P value formula introduced by Husmeier 76 to evaluate the significance of the predicted regulatory interaction network:
where n is the number of predicted interactions (TP+FP), p is the probability of obtaining a true link by random selection.
Supplementary Material
Acknowledgments
This work was supported by the Excellence Initiative of the German Federal and State Governments through Freiburg Institute for Advanced Studies (FRIAS), School of Life Sciences—LifeNet (JD) and the Center for Biological Signalling Studies (BIOSS, ZZ, and JD), by grants DE 1757/2-1 from the German Research Foundation, DFG, and through GerontoSys II—NephAge (031 5896 A) from the German Ministry for Education and Research, BMBF (ZZ and JD), and by a grant from the Danish Natural Sciences Research Council (KR).
Glossary
Abbreviations:
- ACN
acetonitrile
- AKT1S1
AKT1 substrate 1 (proline-rich)
- AMPK
AMP-activated protein kinase
- ATG
autophagy-related
- ATRIP
ATR-interacting protein
- BECN1
Beclin 1, autophagy-related
- CAMK1/2/4
calcium/calmodulin-dependent protein kinase I/II Δ/IV
- CDC37
cell division cycle 37
- CDK2/3/9/11B/12/13
cyclin-dependent kinase 2/3/9/11B/12/13
- conA
concanamycin A
- CSNK2
casein kinase 2
- EIF4EBP1
eukaryotic translation initiation factor 4E binding protein 1
- ERBB2
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2
- FOXK1
forkhead box K1
- GO
gene ontology
- GSK3
glycogen synthase kinase 3
- HADHA
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit
- HIVEP2
human immunodeficiency virus type I enhancer binding protein 2
- KAT5
K(lysine) acetyltransferase 5
- LIR
LC3-interacting region
- MAP1LC3/LC3
microtubule-associated protein 1 light chain 3
- MAP3K2
mitogen-activated protein kinase kinase kinase 2
- MAPK1/3
mitogen-activated protein kinase 1/3
- MARK2
MAP/microtubule affinity-regulating kinase 2
- MS
mass spectrometry
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- NIPSNAP1
nipsnap homolog 1 (C. elegans)
- PDCD5
programmed cell death 5
- PIK3C3
phosphatidylinositol 3-kinase, catalytic subunit type 3
- PKN2
protein kinase N2
- PSMA5
proteasome (prosome, macropain) subunit, alpha type, 5
- RICTOR
RPTOR independent companion of MTOR, complex 2
- RPS6KB1
ribosomal protein S6 kinase, 70 kDa, polypeptide 1
- RPTOR
regulatory associated protein of MTOR, complex 1
- SCX
strong cation exchange
- SCYL1
SCY1-like 1 (S. cerevisiae)
- SILAC
stable isotope labeling by amino acids in cell culture
- SIRT1
sirtuin 1
- SQSTM1
sequestosome 1
- STIM1
stromal interaction molecule 1
- TACC2
transforming acidic coiled-coil containing protein 2
- TFA
trifluoroacetic acid
- TPR
translocated promoter region, nuclear basket protein
- ULK
unc-51 like autophagy activating kinase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Rabinowitz JD, White E. . Autophagy and metabolism. Science 2010; 330:1344 - 8; http://dx.doi.org/ 10.1126/science.1193497; PMID: 21127245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. De Duve C, Wattiaux R. . Functions of lysosomes. Annu Rev Physiol 1966; 28:435 - 92; http://dx.doi.org/ 10.1146/annurev.ph.28.030166.002251; PMID: 5322983 [DOI] [PubMed] [Google Scholar]
- 3. Klionsky DJ. . Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8:931 - 7; http://dx.doi.org/ 10.1038/nrm2245; PMID: 17712358 [DOI] [PubMed] [Google Scholar]
- 4. He C, Klionsky DJ. . Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67 - 93; http://dx.doi.org/ 10.1146/annurev-genet-102808-114910; PMID: 19653858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johansen T, Lamark T. . Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011; 7:279 - 96; http://dx.doi.org/ 10.4161/auto.7.3.14487; PMID: 21189453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Randow F, Münz C. . Autophagy in the regulation of pathogen replication and adaptive immunity. Trends Immunol 2012; 33:475 - 87; http://dx.doi.org/ 10.1016/j.it.2012.06.003; PMID: 22796170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dumit VI, Dengjel J. . Autophagosomal protein dynamics and influenza virus infection. Front Immunol 2012; 3:43; http://dx.doi.org/ 10.3389/fimmu.2012.00043; PMID: 22566925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. White E. . Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12:401 - 10; http://dx.doi.org/ 10.1038/nrc3262; PMID: 22534666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mizushima N. . The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 2010; 22:132 - 9; http://dx.doi.org/ 10.1016/j.ceb.2009.12.004; PMID: 20056399 [DOI] [PubMed] [Google Scholar]
- 10. Chan EY. . Regulation and function of uncoordinated-51 like kinase proteins. Antioxid Redox Signal 2012; 17:775 - 85; http://dx.doi.org/ 10.1089/ars.2011.4396; PMID: 22074133 [DOI] [PubMed] [Google Scholar]
- 11. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. . Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456 - 61; http://dx.doi.org/ 10.1126/science.1196371; PMID: 21205641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. . Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 1995; 270:2320 - 6; http://dx.doi.org/ 10.1074/jbc.270.5.2320; PMID: 7836465 [DOI] [PubMed] [Google Scholar]
- 13. Neufeld TP. . TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol 2010; 22:157 - 68; http://dx.doi.org/ 10.1016/j.ceb.2009.11.005; PMID: 20006481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zimmermann AC, Zarei M, Eiselein S, Dengjel J. . Quantitative proteomics for the analysis of spatio-temporal protein dynamics during autophagy. Autophagy 2010; 6:1009 - 16; http://dx.doi.org/ 10.4161/auto.6.8.12786; PMID: 20603599 [DOI] [PubMed] [Google Scholar]
- 15. Dengjel J, Høyer-Hansen M, Nielsen MO, Eisenberg T, Harder LM, Schandorff S, Farkas T, Kirkegaard T, Becker AC, Schroeder S, et al. . Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol Cell Proteomics 2012; 11:014035; http://dx.doi.org/ 10.1074/mcp.M111.014035; PMID: 22311637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engelke R, Becker AC, Dengjel J. . The degradative inventory of the cell: proteomic insights. Antioxid Redox Signal 2012; 17:803 - 12; http://dx.doi.org/ 10.1089/ars.2011.4393; PMID: 22074050 [DOI] [PubMed] [Google Scholar]
- 17. Kristensen AR, Schandorff S, Høyer-Hansen M, Nielsen MO, Jäättelä M, Dengjel J, Andersen JS. . Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics 2008; 7:2419 - 28; http://dx.doi.org/ 10.1074/mcp.M800184-MCP200; PMID: 18687634 [DOI] [PubMed] [Google Scholar]
- 18. Behrends C, Sowa ME, Gygi SP, Harper JW. . Network organization of the human autophagy system. Nature 2010; 466:68 - 76; http://dx.doi.org/ 10.1038/nature09204; PMID: 20562859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Macek B, Mann M, Olsen JV. . Global and site-specific quantitative phosphoproteomics: principles and applications. Annu Rev Pharmacol Toxicol 2009; 49:199 - 221; http://dx.doi.org/ 10.1146/annurev.pharmtox.011008.145606; PMID: 18834307 [DOI] [PubMed] [Google Scholar]
- 20. Rigbolt K, Blagoev B. . Quantitative phosphoproteomics to characterize signaling networks. Sem Cell Dev Biol 2012; http://dx.doi.org/ 10.1016/j.semcdb.2012.05.006; PMID: 22677334 [DOI] [PubMed] [Google Scholar]
- 21. Becker AC, Bunkenborg J, Eisenberg T, Harder LM, Schroeder S, Madeo F, Andersen JS, Dengjel J. . Friend or food: different cues to the autophagosomal proteome. Autophagy 2012; 8:995 - 6; http://dx.doi.org/ 10.4161/auto.20286; PMID: 22572990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. . Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012; 149:410 - 24; http://dx.doi.org/ 10.1016/j.cell.2012.02.044; PMID: 22424946 [DOI] [PubMed] [Google Scholar]
- 23. Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. . Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002; 1:376 - 86; http://dx.doi.org/ 10.1074/mcp.M200025-MCP200; PMID: 12118079 [DOI] [PubMed] [Google Scholar]
- 24. Rigbolt KT, Blagoev B. . Proteome-wide quantitation by SILAC. Methods Mol Biol 2010; 658:187 - 204; http://dx.doi.org/ 10.1007/978-1-60761-780-8_11; PMID: 20839105 [DOI] [PubMed] [Google Scholar]
- 25. Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. . System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 2011; 4:rs3; http://dx.doi.org/ 10.1126/scisignal.2001570; PMID: 21406692 [DOI] [PubMed] [Google Scholar]
- 26. Cox J, Mann M. . MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26:1367 - 72; http://dx.doi.org/ 10.1038/nbt.1511; PMID: 19029910 [DOI] [PubMed] [Google Scholar]
- 27. Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. . Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006; 127:635 - 48; http://dx.doi.org/ 10.1016/j.cell.2006.09.026; PMID: 17081983 [DOI] [PubMed] [Google Scholar]
- 28. Prakash A, Piening B, Whiteaker J, Zhang H, Shaffer SA, Martin D, Hohmann L, Cooke K, Olson JM, Hansen S, et al. . Assessing bias in experiment design for large scale mass spectrometry-based quantitative proteomics. Mol Cell Proteomics 2007; 6:1741 - 8; http://dx.doi.org/ 10.1074/mcp.M600470-MCP200; PMID: 17617667 [DOI] [PubMed] [Google Scholar]
- 29. Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. . Global quantification of mammalian gene expression control. Nature 2011; 473:337 - 42; http://dx.doi.org/ 10.1038/nature10098; PMID: 21593866 [DOI] [PubMed] [Google Scholar]
- 30. Boisvert F-M, Ahmad Y, Gierliński M, Charrière F, Lamont D, Scott M, Barton G, Lamond AI. . A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteomics 2012; 11:011429; http://dx.doi.org/ 10.1074/mcp.M111.011429; PMID: 21937730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zarei M, Sprenger A, Metzger F, Gretzmeier C, Dengjel J. . Comparison of ERLIC-TiO2, HILIC-TiO2, and SCX-TiO2 for global phosphoproteomics approaches. J Proteome Res 2011; 10:3474 - 83; http://dx.doi.org/ 10.1021/pr200092z; PMID: 21682340 [DOI] [PubMed] [Google Scholar]
- 32. Zarei M, Sprenger A, Gretzmeier C, Dengjel J. . Combinatorial use of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) and strong cation exchange (SCX) chromatography for in-depth phosphoproteome analysis. J Proteome Res 2012; 11:4269 - 76; http://dx.doi.org/ 10.1021/pr300375d; PMID: 22768876 [DOI] [PubMed] [Google Scholar]
- 33. Yu Y, Yoon S-O, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. . Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011; 332:1322 - 6; http://dx.doi.org/ 10.1126/science.1199484; PMID: 21659605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, et al. . A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell 2010; 18:1041 - 52; http://dx.doi.org/ 10.1016/j.devcel.2010.05.005; PMID: 20627085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmitt WA Jr., Raab RM, Stephanopoulos G. . Elucidation of gene interaction networks through time-lagged correlation analysis of transcriptional data. Genome Res 2004; 14:1654 - 63; http://dx.doi.org/ 10.1101/gr.2439804; PMID: 15289483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kazi AA, Lang CH. . PRAS40 regulates protein synthesis and cell cycle in C2C12 myoblasts. Mol Med 2010; 16:359 - 71; http://dx.doi.org/ 10.2119/molmed.2009-00168; PMID: 20464060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Futschik ME, Carlisle B. . Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol 2005; 3:965 - 88; http://dx.doi.org/ 10.1142/S0219720005001375; PMID: 16078370 [DOI] [PubMed] [Google Scholar]
- 38. Lin S-Y, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang S-M, Lian G, Liu Q, Ruan K, et al. . GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 2012; 336:477 - 81; http://dx.doi.org/ 10.1126/science.1217032; PMID: 22539723 [DOI] [PubMed] [Google Scholar]
- 39. Miller ML, Jensen LJ, Diella F, Jørgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, et al. . Linear motif atlas for phosphorylation-dependent signaling. Sci Signal 2008; 1:ra2; http://dx.doi.org/ 10.1126/scisignal.1159433; PMID: 18765831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwartz D, Gygi SP. . An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 2005; 23:1391 - 8; http://dx.doi.org/ 10.1038/nbt1146; PMID: 16273072 [DOI] [PubMed] [Google Scholar]
- 41. Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. . Systematic discovery of in vivo phosphorylation networks. Cell 2007; 129:1415 - 26; http://dx.doi.org/ 10.1016/j.cell.2007.05.052; PMID: 17570479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shao J, Prince T, Hartson SD, Matts RL. . Phosphorylation of serine 13 is required for the proper function of the Hsp90 co-chaperone, Cdc37. J Biol Chem 2003; 278:38117 - 20; http://dx.doi.org/ 10.1074/jbc.C300330200; PMID: 12930845 [DOI] [PubMed] [Google Scholar]
- 43. Myers JS, Zhao R, Xu X, Ham A-J, Cortez D. . Cyclin-dependent kinase 2 dependent phosphorylation of ATRIP regulates the G2-M checkpoint response to DNA damage. Cancer Res 2007; 67:6685 - 90; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-0495; PMID: 17638878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. . Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011; 333:228 - 33; http://dx.doi.org/ 10.1126/science.1205405; PMID: 21617041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu Y, Massen S, Terenzio M, Lang V, Chen-Lindner S, Eils R, Novak I, Dikic I, Hamacher-Brady A, Brady NR. . Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem 2013; 288:1099 - 113; http://dx.doi.org/ 10.1074/jbc.M112.399345; PMID: 23209295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang W, Sherman BT, Lempicki RA. . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44 - 57; http://dx.doi.org/ 10.1038/nprot.2008.211; PMID: 19131956 [DOI] [PubMed] [Google Scholar]
- 47. Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, Johansen T. . p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 2010; 285:22576 - 91; http://dx.doi.org/ 10.1074/jbc.M110.118976; PMID: 20452972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippé R, et al. . Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 2009; 10:480 - 7; http://dx.doi.org/ 10.1038/ni.1720; PMID: 19305394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Evankovich J, Zhang R, Cardinal JS, Zhang L, Chen J, Huang H, Beer-Stolz D, Billiar TR, Rosengart MR, Tsung A. . Calcium/calmodulin-dependent protein kinase IV limits organ damage in hepatic ischemia-reperfusion injury through induction of autophagy. Am J Physiol Gastrointest Liver Physiol 2012; 303:G189 - 98; http://dx.doi.org/ 10.1152/ajpgi.00051.2012; PMID: 22575222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pfisterer SG, Mauthe M, Codogno P, Proikas-Cezanne T. . Ca2+/calmodulin-dependent kinase (CaMK) signaling via CaMKI and AMP-activated protein kinase contributes to the regulation of WIPI-1 at the onset of autophagy. Mol Pharmacol 2011; 80:1066 - 75; http://dx.doi.org/ 10.1124/mol.111.071761; PMID: 21896713 [DOI] [PubMed] [Google Scholar]
- 51. Munafó DB, Colombo MI. . A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci 2001; 114:3619 - 29; PMID: 11707514 [DOI] [PubMed] [Google Scholar]
- 52. Marambio P, Toro B, Sanhueza C, Troncoso R, Parra V, Verdejo H, García L, Quiroga C, Munafo D, Díaz-Elizondo J, et al. . Glucose deprivation causes oxidative stress and stimulates aggresome formation and autophagy in cultured cardiac myocytes. Biochim Biophys Acta 2010; 1802:509 - 18; http://dx.doi.org/ 10.1016/j.bbadis.2010.02.002; PMID: 20176105 [DOI] [PubMed] [Google Scholar]
- 53. Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. . Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005; 120:237 - 48; http://dx.doi.org/ 10.1016/j.cell.2004.11.046; PMID: 15680329 [DOI] [PubMed] [Google Scholar]
- 54. Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. . A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 2008; 105:3374 - 9; http://dx.doi.org/ 10.1073/pnas.0712145105; PMID: 18296641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. . Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 2009; 11:1305 - 14; http://dx.doi.org/ 10.1038/ncb1975; PMID: 19801973 [DOI] [PubMed] [Google Scholar]
- 56. Dai D-F, Rabinovitch P. . Mitochondrial oxidative stress mediates induction of autophagy and hypertrophy in angiotensin-II treated mouse hearts. Autophagy 2011; 7:917 - 8; http://dx.doi.org/ 10.4161/auto.7.8.15813; PMID: 21505274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eng CH, Yu K, Lucas J, White E, Abraham RT. . Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal 2010; 3:ra31; http://dx.doi.org/ 10.1126/scisignal.2000911; PMID: 20424262 [DOI] [PubMed] [Google Scholar]
- 58. Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. . Oxidative stress induces autophagic cell death independent of apoptosis in transformed and cancer cells. Cell Death Differ 2008; 15:171 - 82; http://dx.doi.org/ 10.1038/sj.cdd.4402233; PMID: 17917680 [DOI] [PubMed] [Google Scholar]
- 59. Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. . Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 2008; 283:10892 - 903; http://dx.doi.org/ 10.1074/jbc.M800102200; PMID: 18281291 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60. Høyvik H, Gordon PB, Berg TO, Strømhaug PE, Seglen PO. . Inhibition of autophagic-lysosomal delivery and autophagic lactolysis by asparagine. J Cell Biol 1991; 113:1305 - 12; http://dx.doi.org/ 10.1083/jcb.113.6.1305; PMID: 1904444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Seglen PO, Gordon PB. . 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A 1982; 79:1889 - 92; http://dx.doi.org/ 10.1073/pnas.79.6.1889; PMID: 6952238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bennetzen MV, Mariño G, Pultz D, Morselli E, Færgeman NJ, Kroemer G, Andersen JS. . Phosphoproteomic analysis of cells treated with longevity-related autophagy inducers. Cell Cycle 2012; 11:1827 - 40; http://dx.doi.org/ 10.4161/cc.20233; PMID: 22517431 [DOI] [PubMed] [Google Scholar]
- 63. Chen R-Q, Yang Q-K, Lu B-W, Yi W, Cantin G, Chen Y-L, Fearns C, Yates JR 3rd, Lee J-D. . CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res 2009; 69:2663 - 8; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-3222; PMID: 19276368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. . The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011; 332:1317 - 22; http://dx.doi.org/ 10.1126/science.1199498; PMID: 21659604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilkinson S, Croft DR, O’Prey J, Meedendorp A, O’Prey M, Dufès C, Ryan KM. . The cyclin-dependent kinase PITSLRE/CDK11 is required for successful autophagy. Autophagy 2011; 7:1295 - 301; http://dx.doi.org/ 10.4161/auto.7.11.16646; PMID: 21808150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Capparelli C, Chiavarina B, Whitaker-Menezes D, Pestell TG, Pestell RG, Hulit J, Andò S, Howell A, Martinez-Outschoorn UE, Sotgia F, et al. . CDK inhibitors (p16/p19/p21) induce senescence and autophagy in cancer-associated fibroblasts, “fueling” tumor growth via paracrine interactions, without an increase in neo-angiogenesis. Cell Cycle 2012; 11:3599 - 610; http://dx.doi.org/ 10.4161/cc.21884; PMID: 22935696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T, Shen Y, Rameh L, Yankner B, Tsai L-H, et al. . Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell 2010; 38:500 - 11; http://dx.doi.org/ 10.1016/j.molcel.2010.05.009; PMID: 20513426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Szyniarowski P, Corcelle-Termeau E, Farkas T, Høyer-Hansen M, Nylandsted J, Kallunki T, Jäättelä M. . A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy 2011; 7:892 - 903; http://dx.doi.org/ 10.4161/auto.7.8.15770; PMID: 21508686 [DOI] [PubMed] [Google Scholar]
- 69. Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. . Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell 2011; 44:279 - 89; http://dx.doi.org/ 10.1016/j.molcel.2011.07.039; PMID: 22017874 [DOI] [PubMed] [Google Scholar]
- 70. Dagda RK, Zhu J, Kulich SM, Chu CT. . Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy 2008; 4:770 - 82; PMID: 18594198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. . LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720 - 8; http://dx.doi.org/ 10.1093/emboj/19.21.5720; PMID: 11060023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rigbolt KT, Vanselow JT, Blagoev B. . GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol Cell Proteomics 2011; 10:007450; http://dx.doi.org/ 10.1074/mcp.O110.007450; PMID: 21602510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bennetzen MV, Cox J, Mann M, Andersen JS. . PhosphoSiteAnalyzer: A Bioinformatic Platform for Deciphering Phospho Proteomes Using Kinase Predictions Retrieved from NetworKIN. J Proteome Res 2012; 11:3480 - 6; http://dx.doi.org/ 10.1021/pr300016e; PMID: 22471441 [DOI] [PubMed] [Google Scholar]
- 74. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. . Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947 - 8; http://dx.doi.org/ 10.1093/bioinformatics/btm404; PMID: 17846036 [DOI] [PubMed] [Google Scholar]
- 75. Letunic I, Bork P. . Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2007; 23:127 - 8; http://dx.doi.org/ 10.1093/bioinformatics/btl529; PMID: 17050570 [DOI] [PubMed] [Google Scholar]
- 76. Husmeier D. . Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics 2003; 19:2271 - 82; http://dx.doi.org/ 10.1093/bioinformatics/btg313; PMID: 14630656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.