Abstract
Growing evidence has demonstrated that autophagy plays important and paradoxical roles in carcinogenesis, while senescence is considered to be a crucial tumor-suppressor mechanism in cancer prevention and treatment. In the present study we demonstrated that both autophagy and senescence were induced in response to penta-1,2,3,4,6-O-galloyl-β-D-glucose (PGG), a chemopreventive polyphonolic compound, in multiple types of cancer cells. Analysis of these 2 events over the experimental time course indicated that autophagy and senescence occurred in parallel early in the process and dissociated later. The long-term culture study suggested that a subpopulation of senescent cells may have the capacity to reenter the cell cycle. Inhibition of autophagy by either a chemical inhibitor or RNA interference led to a significant reduction of PGG-induced senescence, followed by induction of apoptosis. These results suggested that autophagy promoted senescence induction by PGG and that PGG might exert its anticancer activity through autophagy-mediated senescence. For the first time, these findings uncovered the relationships among autophagy, senescence, and apoptosis induced by PGG. In addition, we identified that unfolded protein response signaling played a pivotal role in the autophagy-mediated senescence phenotype. Furthermore, our data showed that activation of MAPK8/9/10 (mitogen-activated protein kinase 8/9/10/c-Jun N-terminal kinases) was an essential upstream signal for PGG-induced autophagy. Finally, the key in vitro results were validated in vivo in a xenograft mouse model of human HepG2 liver cancer. Our findings provided novel insights into understanding the mechanisms and functions of PGG-induced autophagy and senescence in human cancer cells.
Keywords: PGG, autophagy, senescence, UPR, apoptosis, MAPK8/9/10
Introduction
Autophagy is the process by which cells break down their own long-lived proteins and damaged components, such as mitochondria. 1 Growing evidence has demonstrated that autophagy plays important and paradoxical roles in the regulation of carcinogenesis. Several studies have shown that autophagy functions as tumor suppressor. 2 - 5 In contrast, several other studies argue that autophagy could support tumor progression and survival. 6 - 9 Because of its contradictory functions, the current autophagy literature is often viewed as confusing. Therefore, further studies are clearly needed to better understand the functional role of autophagy in carcinogenesis.
Cellular senescence is an irreversible growth arrest. This phenomenon was first observed more than 50 years ago by Hayflick and Moorhead 10 as a special growth-arrest program that prevents normal fibroblasts from unrestricted proliferation and limits the life span of the cells in culture. This permanent growth-arrested state is referred to as replicative senescence in response to replicative aging. Recent evidence suggests that cellular senescence can also occur in cancer cells in response to stimuli such as chemopreventive agents or therapeutic drugs. 11 - 13 This response, now termed as senescence-like growth arrest (also known as premature senescence, stress-induced premature senescence, or accelerated senescence). Recent studies demonstrate that a small subpopulation of senescent cancer cells might be able to reenter the cell cycle. 14 , 15 Transformed or cancer cells possess unlimited mitotic division potential while senescence is characterized to be a permanent form of cell cycle arrest. Therefore, senescence-modulation is thought to be a very effective approach in preventing proliferation of cancer cells and a crucial tumor-suppressor mechanism in cancer prevention and treatment. 16 , 17
Penta-1,2,3,4,6-O-galloyl-β-D-glucose (PGG) is a naturally occurring gallotannin polyphenolic compound found in Oriental herbs such as Rhus chinensis Mill and Paeonia suffruticosa. 18 Previous studies show that PGG is effective against various types of cancer both in vitro and in vivo, suggesting its potential usefulness as a novel chemopreventive agent. 19 - 24 Our recent study has demonstrated that cellular features of both autophagy and senescence were induced in response to PGG in multiple types of cancer cells. 25 , 26 In the present study, we further investigated the crosstalk among these PGG-induced phenotypes and signal transduction mechanisms. We demonstrated here that autophagy-mediated activation of the unfolded protein response (UPR) contributed to PGG-induced senescence. Furthermore, these biological processes induced by PGG in vitro were indeed observed in vivo. These findings not only uncovered the relationships between PGG-induced autophagy and senescence, but also provided a novel insight into understanding the mechanisms involved in autophagy-mediated senescence.
Results
Both autophagy and the senescent phenotype were induced in response to PGG exposure in HepG2, MCF-7, and A549 cells
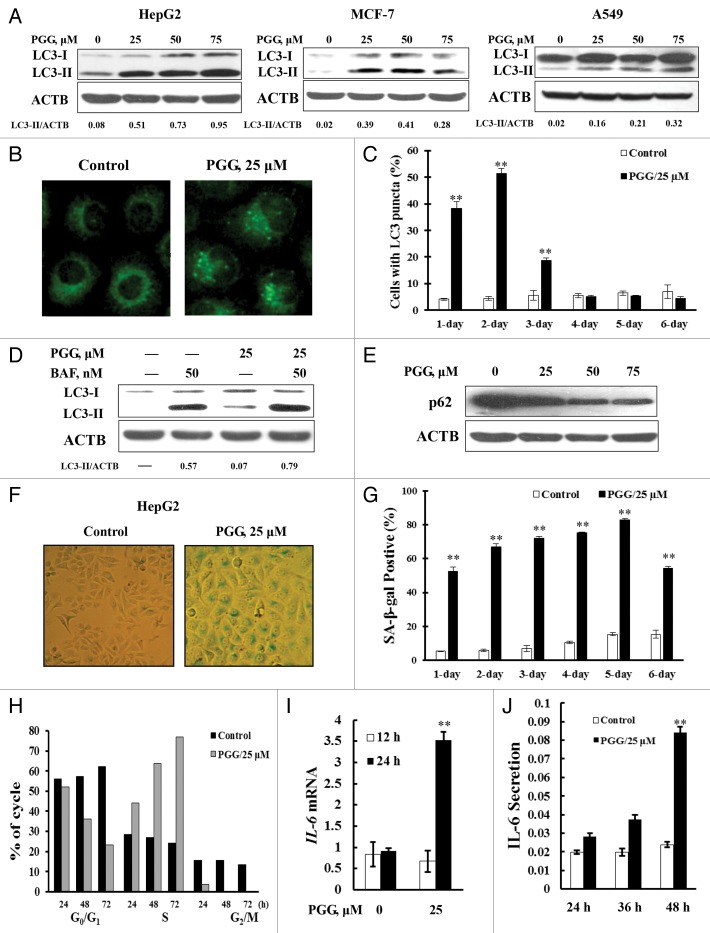
Our previous study showed that PGG induces autophagy in PC-3 human prostate cancer cells. 25 We asked if autophagy-induction by PGG also occurred in other types of human cancer cells. HepG2 human liver cancer cells, MCF-7 human breast cancer cells and A549 human lung cancer cells were treated with PGG at concentrations of 25 to 75 μM for 24 h and then a cleaved and lipidated form of LC3, LC3-II, an autophagic marker, was analyzed by western blotting. As shown in Figure 1A, treatment with PGG at concentrations of 25 to 75 μM led to a significant increase of LC3-II level in HepG2 (left), MCF-7 (middle), and A549 (right) cells. LC3 lipidation by PGG was further verified by immunofluorescence staining to analyze distribution patterns of LC3. As shown in Figure 1B, a diffuse localization of LC3 fluorescence was observed in control cells whereas a punctate pattern of LC3 fluorescence was detected in PGG-treated cells. Quantitative analysis of LC3-puncta-positive cells for the time-course experiment showed that the number of cells with LC3 puncta in PGG-treated cells was significantly increased compared with the untreated control at 24 h and reached a peak at 48 h and it then gradually diminished (Fig. 1C). We next measured autophagic flux to determine whether the elevated levels of LC3 cleavage and lipidation induced by PGG were due to increased formation of autophagosomes or to decreased degradation. As shown in Figure 1D, treatment with bafilomycin A1, an inhibitor of autophagosome degradation, was expected to increase accumulation of LC3-II, whereas PGG treatment significantly increased LC3-II levels either in the presence or absence of bafilomycin A1, suggesting that the increase of autophagosome formation contributed to PGG-induced autophagy. This notion was further supported by the evidence that PGG treatment led to a dose-dependent decrease of SQSTM1 (formerly also known as p62), the best-known mammalian autophagy-specific substrate 27 (Fig. 1E).
Figure 1. PGG induces autophagy and senescence in HepG2, MCF-7, and A549 human cancer cells. (A) PGG caused a concentration-dependent increase of LC3-I to LC3-II conversion analyzed by western blotting in HepG2 (left), MCF-7 (middle), and A549 cells (right). (B) A punctate distribution of LC3 induced by PGG and detected by immunofluorescence staining. (C) Quantitative analysis of LC3-punctate cells for time-course experiments in PGG-treated and untreated cells. (D) Increase of autophagosome formation was involved in PGG-induced autophagy in HepG2 cells. The cells were treated with 25 μM PGG for 24 h in the presence or absence of 50 nM bafilomycin A1 (added 2 h before cells harvest) and then LC3 was analyzed by western blotting. (E) PGG caused a concentration-dependent decrease of SQSTM1 analyzed by western blotting in HepG2 cells. (F and G) PGG-induced senescence-like phenotype in HepG2 cells. The cells were treated with 25 μM PGG for 24 h and SA-β-gal activity was measured with senescence-associated β-galactosidase staining. The stained cells were viewed and photographed under an inverted microscope (F); Time-course analysis of β-gal-positive cells (G). (H) Cell cycle distribution induced by PGG. HepG2 cells were treated with PGG for the indicated times and cell cycle was analyzed by flow cytometry (the data are representative of 3 experiments). (I and J) PGG stimulated IL6 secretion in HepG2 cells. The cells were treated with 25 μM PGG for the indicated times and the levels of IL6 mRNA and its secreted gene product were measured by Real-time PCR and an ELISA Kit respectively. **P < 0.01.
To determine whether autophagy induction by PGG is accompanied by a senescent phenotype in these cancer cells, we employed the senescence-associated β-galactosidase (SA-β-gal) staining to detect acidic β-galactosidase (β-gal) activity at pH 6, a known characteristic of senescent cells not found in presenescent, quiescent, or immortal cells. As shown in Figure 1F and G, as well as Figure S1A and S1B, PGG treatment (25 μM, 24 h treatment) significantly increased the percentage of β-gal-positive cells in HepG2 (Fig. 1F and G), MCF-7, and A549 cells (Fig. S1A and S1B). Time-course analysis demonstrated that senescence induction by PGG reached a peak at 5 d and decreased thereafter (Fig. 1G) accompanied by a significantly increased apoptosis (data not shown). The cell cycle distribution analysis revealed that a progressive increase in the percentage of S-phase cells over time was observed in HepG2 (Fig. 1H), MCF-7, and A549 cells (Fig. S1C). Further BrdU incorporation assays showed that treatment with PGG caused a strong inhibition of DNA synthesis (Fig. S1D). Together, these data suggested that a persistent S-phase cell cycle arrest was induced in the PGG-treated cells. To further confirm PGG-induced senescent phenotype, we next measured the changes of senescence-associated secretory phenotype (SASP) key component IL6 using real-time PCR and an ELISA kit. As shown in Figure 1I and J, treatment with PGG caused a significant increased level of both IL6 mRNA and its secretory protein In line with the above changes, the typical morphological features of senescence such as an enlarged and flattened morphology with an increase of cytoplasmic vacuoles were observed in PGG-treated cells (data not shown). Consistent with the biochemical change, SASP and the typical morphological features of senescence, the majority of PGG-treated cells irreversibly lost their proliferative capacity after withdrawal of the treatment (Fig. S1E). Together, these results strongly supported that PGG was capable of inducing autophagy as well as a senescence-like phenotype in the cell lines tested.
Induction of autophagy contributed to PGG-induced senescent phenotype
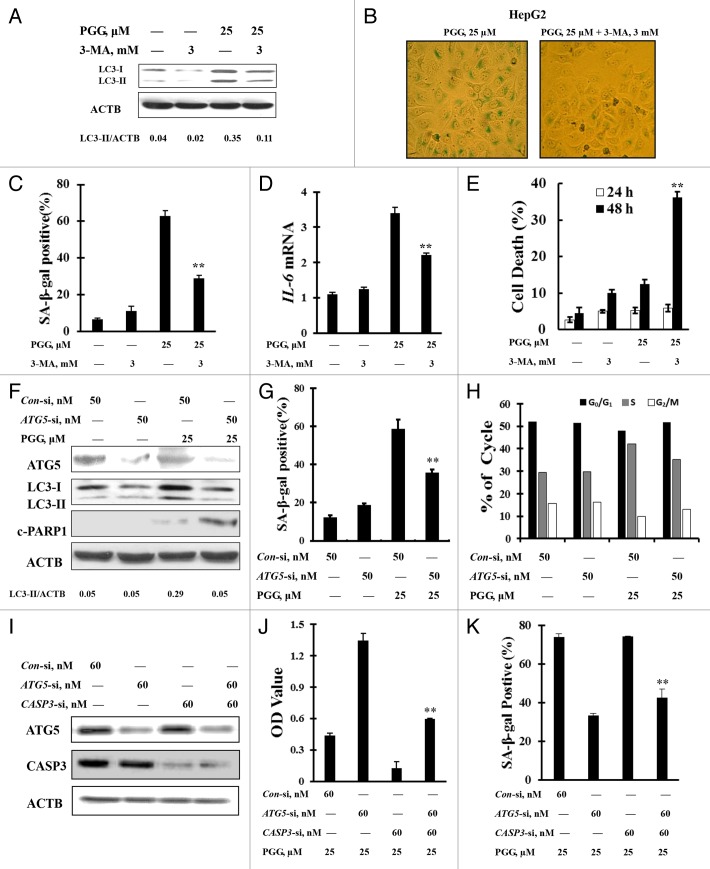
To determine if autophagy plays a role in the PGG-induced senescent phenotype, we first tested effects of autophagy inhibition by its inhibitor 3-MA on the PGG-induced senescent phenotype in HepG2, MCF-7, and A549 cells. The cells were treated with 25 μM PGG in the presence or absence of 3-MA for 24 h and then senescent phenotype was assessed using senescence-associated β-galactosidase staining. As shown in Figure 2A–C and Figure S2A and S2B, under the conditions in which autophagy was blocked by its inhibitor, the percentage of β-gal-positive cells induced by PGG was significantly decreased in HepG2 (Fig. 2B and C), MCF-7, and A549 cells (Fig. S2A and S2B) in comparison with PGG treatment without 3-MA (P < 0.01). Consistent with the reduction of β-gal-positive cells, PGG-induced IL6 mRNA was also significantly decreased in the presence of 3-MA (Fig. 2D). After the 48 h treatment, however, a dramatically increased apoptosis measured by ANXA5 staining was observed in PGG and/or 3-MA treated cells compared with PGG treatment alone in HepG2 (Fig. 2E), MCF-7, and A549 cells (Fig. S2C) and no significant changes of apoptosis were detected at 24 h (Fig. 2E; Fig. S2C) or 12 h (data not shown) by either PGG treatment alone or in combination with 3-MA. We next used a genetic approach to further validate the role of autophagy in the PGG-induced senescent phenotype. As shown in Figure 2F, ATG5, an essential gene for autophagy, 28 was silenced by RNA interference. Under this condition, the percentage of β-gal-positive cells induced by PGG was significantly decreased in HepG2 cells (Fig. 2G, 24 h), followed by an increased PARP1 cleavage (Fig. 2F, 48 h). We also assessed effects of autophagy inhibition on cycle distribution induced by PGG. As shown in Figure 2H, under the condition in which autophagy was inhibited by knocking down ATG5, the percentage of cells in S phase decreased, whereas the number of cells in G2/M phase increased, indicating that partial S-phase-arrested cells by PGG moved into G2/M phase. To critically determine the role of autophagy in the regulation of senescence induction by PGG, we further measured senescence induction by PGG in a system where apoptosis was suppressed. As shown in Figure 2I, simultaneous transfection of HepG2 cells with ATG5si and CASP3si led to a significant inhibition of ATG5 and CASP3 expression (Fig. 2I), and apoptosis induction by PGG was significantly suppressed (Fig. 2J). Under such conditions, a significant reduction of β-gal-positive cells induced by PGG was observed (Fig. 2K). Taken together, these results clearly indicated that autophagy induction was involved in the regulation of senescence and apoptosis induced by PGG.
Figure 2. Autophagy is required for the senescent phenotype induced by PGG. (A) Effects of autophagy inhibitor 3-MA on PGG-induced autophagy in HepG2 cells. (B) Effects of autophagy inhibition by 3-MA on PGG-induced senescence phenotype (SA-β-gal activity) in HepG2 cells. The cells were treated with PGG in the presence or absence of 3-MA for 24 h and senescence phenotype was measured by senescence-associated β-galactosidase staining. (C) Quantitative analysis of β-gal-positive cells induced by PGG in the presence or absence of 3-MA. (D) Effects of autophagy inhibition by 3-MA on IL6 mRNA induction by PGG analyzed by real-time PCR. (E) Effects of autophagy inhibition by 3-MA on PGG-induced apoptosis. The cells were treated with PGG in the presence or absence of 3-MA for the indicated times and apoptosis was analyzed by ANXA5 staining. (F and G) Effects of autophagy inhibition by knockdown of ATG5 on PGG-induced senescence phenotype and apoptosis. The cells were transfected with 50 nmol/L of ATG5 siRNA using siPORT™ NeoFX™ Transfection Agent. After 24 h transfection, the cells were treated with 25 μM PGG for 24 or 48 h. ATG5, LC3-I to LC3-II conversion and PARP1 cleavage were analyzed using western blotting (F) and senescence phenotype was measured by senescence-associated β-galactosidase staining (G). (n = 3, **P < 0.01). (H) Effects of autophagy inhibition by knocking down ATG5 on cell cycle distribution induced by PGG. The cells were transfected with 50 nmol/L of ATG5 siRNA using siPORT™ NeoFX™ Transfection Agent. After 24 h transfection, the cells were treated with 25 μM PGG for 24 h and cell cycle was assessed by flow cytometry. (I–K) Effects of autophagy inhibition by knocking down ATG5 on senescence induction by PGG in a system where apoptosis was suppressed by knocking down CASP3. HepG2 cells were simultaneously transfected with 50 nmol/L of ATG5 siRNA and 60 nmol/L of CASP3 siRNA (I). After 24 h transfection, the cells were treated with 25 μM PGG for 48 h, and then cell death and senescence phenotype (SA-β-gal activity) were measured by a Cell Death ELISA Kit (J) and senescence-associated β-galactosidase staining (K) respectively (n = 3, **P < 0.01).
PGG induced activation of the UPR
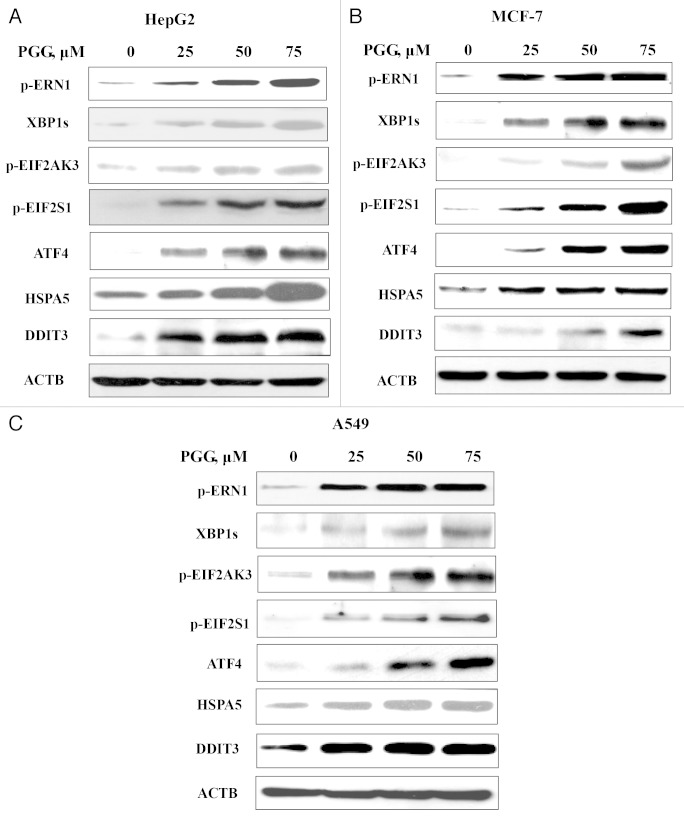
Our recent study has shown that stimulation of autophagosome formation could trigger activation of the UPR. 29 We then asked if the UPR was induced in response to PGG. HepG2 cells were treated with various concentrations of PGG for 24 h, and then the changes of endoplasmic reticulum (ER) stress markers after PGG treatment were analyzed by western blotting. As shown in Figure 3A, PGG treatment significantly increased protein abundance of ER chaperone HSPA5 as well as the phosphorylation levels of ER resident kinase EIF2AK3 and ERN1 in a concentration-dependent manner in HepG2 cells. In line with EIF2AK3 and ERN1 activation, phosphorylation of EIF2S1 and expression of ATF4, DDIT3, and XBP1s were significantly induced in response to PGG treatment. Furthermore, similar changes of these ER stress markers were also observed in MCF-7 (Fig. 3B) and A549 (Fig. 3C) cell lines. These results clearly indicated that PGG treatment led to activation of the UPR in all cell lines tested.
Figure 3. PGG induces the unfolded protein response (UPR) in HepG2, MCF-7, and A549 human cancer cells. The cells were treated with various concentrations of PGG for 24 h and ER stress associated markers were analyzed by western blotting. (A) HepG2 cells. (B) MCF-7 cells. (C) A549 cells.
Induction of autophagy contributed to PGG-induced activation of the UPR
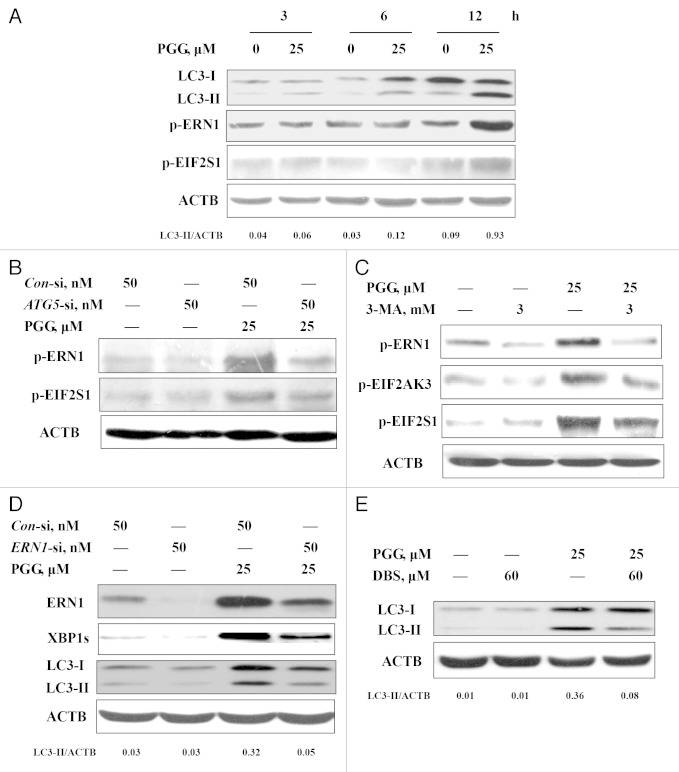
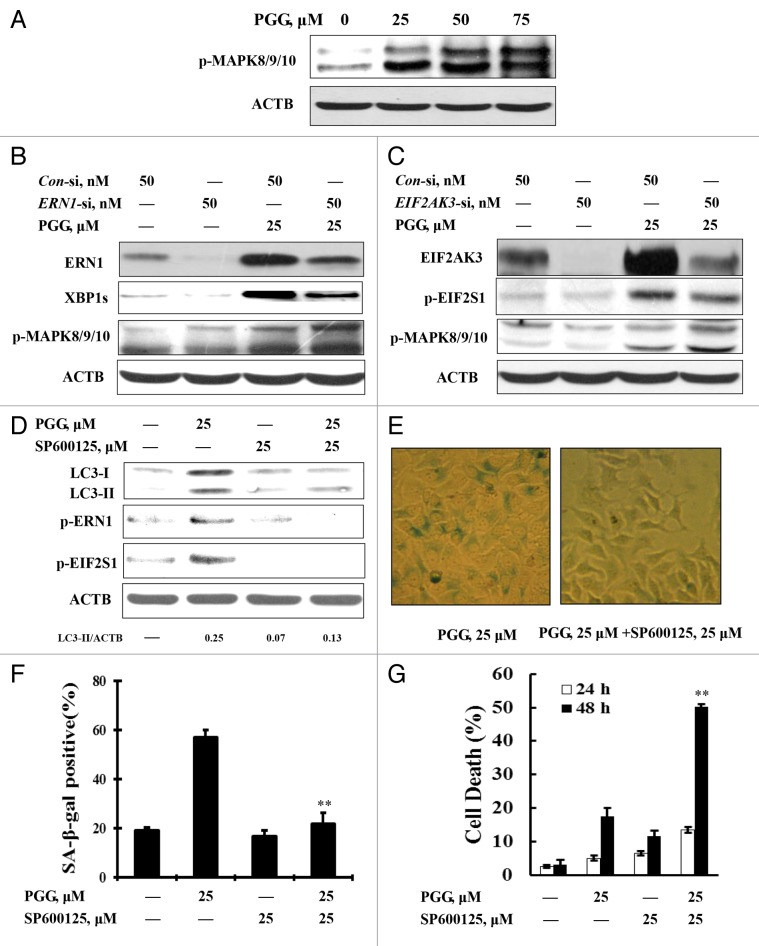
To determine the relationship between autophagosome formation and UPR activation, we first performed a time kinetic study assessing dynamic changes of key parameters of autophagy and ER stress. As shown in Figure 4A, PGG treatment caused a rapid LC3 cleavage and lipidation and a significantly increased LC3-II level was detected as early as after 6 h of PGG treatment, whereas the increase of ERN1 and EIF2S1 phosphorylation was observed at 12 h of the treatment. No cleavage of PARP1 was found during this time frame. These time-course results indicated that PGG induced autophagy preceding ER stress, which preceded apoptosis. To critically test the role of autophagosome formation in PGG-induced UPR activation, we next employed an RNA interference approach to assess effect of inhibition of autophagosome formation on UPR activation by PGG. As shown in Figure 4B, knockdown of ATG5 led to a significant attenuation of PGG-induced ERN1 and EIF2S1 phosphorylation. Similar results were also observed when autophagosome formation was blocked by a chemical inhibitor 3-MA (Fig. 4C). These results further supported that autophagy occurred upstream of UPR activation. However, inhibition of ERN1 by either RNAi (Fig. 4D) or a chemical inhibitor 3-ethoxy-5,6-dibromosalicylaldehyde 30 (Fig. 4E) led to a decrease of PGG-induced LC3-I to LC3-II conversion, suggesting that a positive feedback loop might exist between ER stress and autophagy induced by PGG.
Figure 4. Induction of autophagy contributed to PGG-induced activation of the UPR. (A) Time-course analysis of key parameters of autophagy and ER stress induced by PGG in HepG2 cells. The cells were treated with PGG for the indicated time and then LC3 and phosphorylation of ERN1 and EIF2S1 were assessed by western blotting. (B) Effects of autophagy inhibition by ATG5 knockdown on PGG-induced UPR. The cells were transfected ATG5 siRNA using siPORT™ NeoFX™ Transfection Agent. After 24 h transfection, the cells were treated with 25 μM PGG for 24 h and ERN1 and EIF2S1 phosphorylation were assessed by western blotting. (C) Effects of autophagy inhibition by its inhibitor on PGG-induced UPR. The cells were treated with 25 μM PGG in the presence or absence of 3-MA for 24 h and ER markers were assessed by western blotting. (D) Effects of ERN1 knockdown on PGG-induced autophagy. The cells were transfected ERN1 siRNA using siPORT™ NeoFX™ Transfection Agent. After 24 h transfection, the cells were treated with 25 μM PGG for 24 h and ERN1 and LC3 were assessed by western blotting. (E) Effects of ERN1 inhibitor on PGG-induced autophagy. The cells were treated with PGG in the presence or absence of 3-ethoxy-5,6-dibromosalicylaldehyde for 24 h and LC3 was assessed by western blotting.
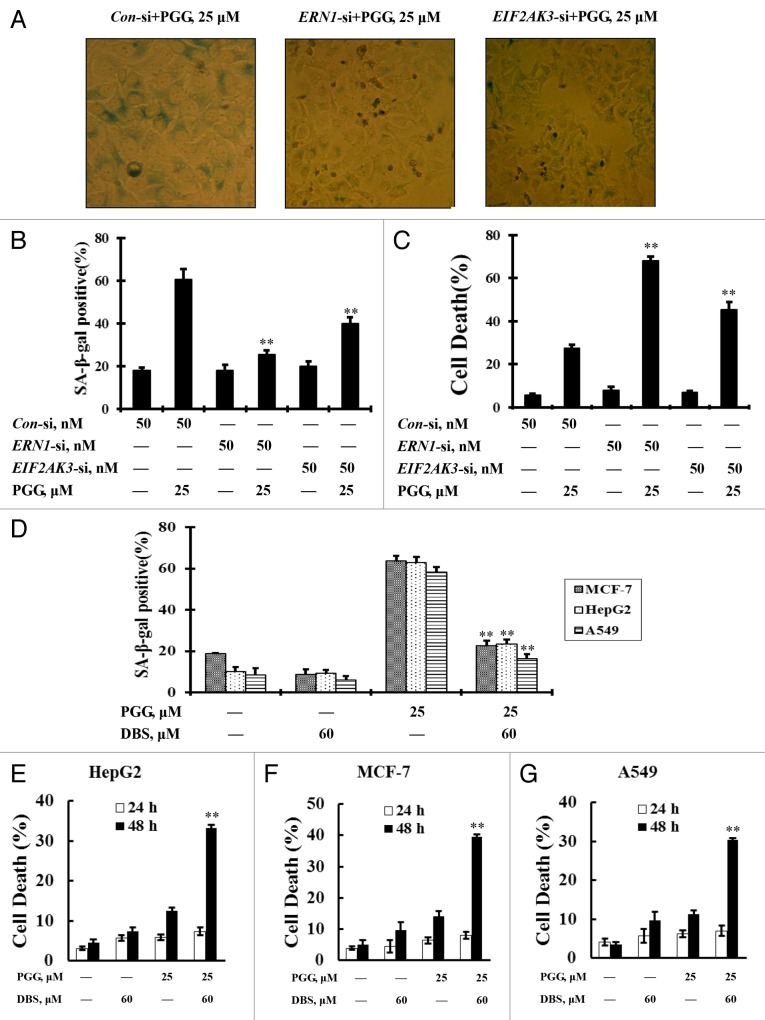
Both the ERN1 and EIF2AK3 arms of UPR signaling pathways were involved in the PGG-induced senescent phenotype
To investigate the role of UPR signaling pathways in the PGG-induced senescent phenotype, we first evaluated inhibition of the UPR by RNA interference on PGG-induced senescent markers. As shown in Figure 5A and B, when the UPR was inhibited by either knockdown of ERN1 or EIF2AK3, the population of β-gal-positive cells induced by PGG was significantly decreased (60.72% ± 3.76 in PGG and nontargeting siRNA-treated cells vs. 25.55% ± 1.95 in PGG and ERN1 siRNA-treated cells and 39.96% ± 2.75 in PGG and EIF2AK3-siRNA treated cells, P < 0.01). Following the senescence suppression, a dramatically increased apoptosis was induced when the treatment was extended over 48 h (Fig. 5C). We next used a chemical inhibitor to inactivate ERN1 to further validate the role of the UPR in PGG-induced senescent phenotype. HepG2, MCF-7, and A549 cells were treated with 25 μM PGG in the presence or absence of the inhibitor for 24 h and then β-gal activity was examined by senescence-associated β-galactosidase staining. As shown in Figure 5D, a decrease in β-gal-positive cell numbers, followed by an increase in apoptotic cell numbers in response to PGG was observed in the presence of the inhibitor compared with the absence of the inhibitor (Fig. 5E). Together, these results indicated that activation of both ERN1 and EIF2AK3 contributed to autophagy-mediated senescence by PGG.
Figure 5. Both the ERN1 and EIF2AK3 arms of UPR signaling pathways are involved in the PGG-induced senescent phenotype. (A and B) Effects of ERN1 or EIF2AK3 knockdown on PGG-induced senescence phenotype. HepG2 cells were transfected ERN1 or EIF2AK3 siRNA using siPORT™ NeoFX™ Transfection Agent. Twenty four h after transfection, the cells were treated with 25 μM PGG for 24 h and the senescence phenotype was measured by senescence-associated β-galactosidase staining. The stained cells were viewed and photographed under an inverted microscope (A); Quantitative analysis of β-gal-positive cells (B). (C) Effects of ERN1 or EIF2AK3 knockdown on PGG-induced apoptosis analyzed by ANXA5 staining after a 48 h treatment. (D) Effects of ERN1 inhibitor on the PGG-induced senescence phenotype. The cells were treated with PGG in the presence or absence of 3-ethoxy-5,6-dibromosalicylaldehyde for 24 h and the senescent phenotype was measured by senescence-associated β-galactosidase staining. (E) Effects of ERN1 inhibitor on PGG-induced apoptosis analyzed by ANXA5 staining after 24 and 48 h treatment.
MAPK8/9/10 activation lied upstream of PGG-induced autophagy
It has been shown that ER stress can induce autophagy via MAPK8/9/10 activation. 31 , 32 We asked if MAPK8/9/10 played a role in the feedback loop between PGG-induced ER stress and autophagy. We first analyzed MAPK8/9/10 (phosphorylation) activation in response to PGG in HepG2 cells. As shown in Figure 6A, treatment with PGG induced a concentration dependent increase of MAPK8/9/10 phosphorylation. We next tested effect of ER stress inhibition on PGG-induced MAPK8/9/10 phosphorylation. As shown in Figure 6B, under the condition that ERN1 was silenced by RNAi, phosphorylation of MAPK8/9/10 was further enhanced compared with PGG and control siRNA treatment. Similar results were also found when EIF2AK3 was inhibited by RNAi (Fig. 6C). These results suggested that MAPK8/9/10 activation was unlikely lying downstream of ERN1 or EIF2AK3 activation. We then further examined the effect of MAPK8/9/10 inactivation on PGG-induced autophagy and ER stress to determine if MAPK8/9/10 activation was signaling upstream. As shown in Figure 6D–F, MAPK8/9/10 inactivation by its inhibitor led to a near abolishment of PGG-induced LC3-I/LC3-II conversion, ERN1-EIF2S1 phosphorylation and β-gal-positive staining, followed by a dramatic increase of PGG-induced apoptosis (Fig. 6G). The results indicated that MAPK8/9/10 activation lied upstream of PGG-induced autophagy.
Figure 6. MAPK8/9/10 activation lies upstream of PGG-induced autophagy-dependent senescence in HepG2 cells. (A) PGG induced a concentration-dependent MAPK8/9/10 phosphorylation (activation) analyzed by western blotting. (B and C) Effects of ERN1 or EIF2AK3 knockdown on PGG-induced MAPK8/9/10 phosphorylation. The cells were transfected with ERN1 (B) or EIF2AK3 (C) siRNA using siPORT™ NeoFX™ Transfection Agent. After 24 h transfection, the cells were treated with 25 μM PGG for 24 h and MAPK8/9/10 phosphorylation was assessed by western blotting. (D) Effects of MAPK8/9/10 inactivation by its inhibitor on PGG-induced autophagy and UPR. The cells were treated with 25 μM PGG in the presence or absence of SP600125 for 24 h. LC3 and phosphorylation of ERN1 and EIF2S1 were analyzed by western blotting. (E and F) Effects of MAPK8/9/10 inactivation by its inhibitor on the PGG-induced senescence phenotype. The cells were treated with PGG in the presence or absence of SP600125 for 24 h and the senescence phenotype was measured by senescence-associated β-galactosidase staining. (G) Effects of MAPK8/9/10 inactivation by its inhibitor on PGG-induced apoptosis. The cells were treated with PGG in the presence or absence of SP600125 for 48 h and apoptosis was assessed by ANXA5 staining.
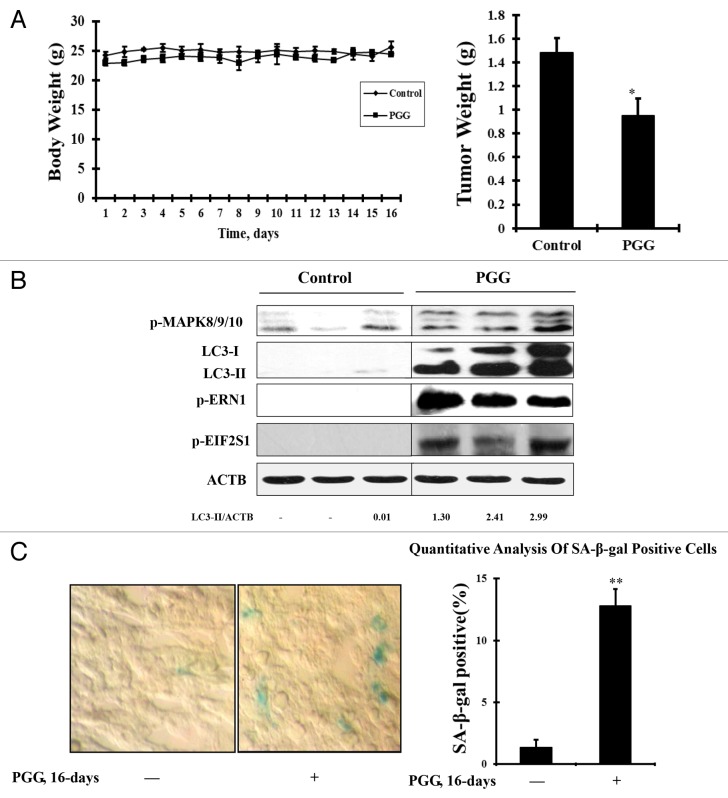
PGG induced autophagy and senescence in vivo
Lastly, the above in vitro findings were validated in vivo in a xenograft mouse model of human HepG2 liver cancer. PGG was given after one week inoculation of cancer cells by i.p injection for 16 days and then the tumors were harvested for western blot analysis of proteins of interest and senescence-associated β-galactosidase staining. As shown in Figure 7A, treatment with PGG resulted in a significant reduction of tumor weight without affecting body weight obviously. Moreover, dramatically enhanced LC3-I/LC3-II conversion and ERN1 or EIF2S1 phosphorylation were detected in the tumors of PGG-treated mice in comparison with that found in the untreated control. Consistent with the changes of these autophagy and ER stress markers, MAPK8/9/10 was activated by PGG treatment in the tumors (Fig. 7B). The senescence assay showed that PGG treatment caused a significant increase of β-gal-positive cell numbers in the tumors of PGG-treated mice (Fig. 7C. P < 0.05, compared with the untreated control). Together, these data suggested that treatment with PGG indeed caused autophagy and senescence in vivo, which were possibly associated with activation of MAPK8/9/10 and ER stress signaling pathways.
Figure 7. PGG induces autophagy and senescence in vivo. PGG was given daily by i.p. injection (20 mg/kg body weight) starting 7 d after s.c. inoculation of cancer cells for 16 d. (A) Effects of PGG on tumor and bodyweight. (B) Western blotting analysis of the markers of autophagy and ER stress. (C) Senescence-associated β-galactosidase staining assay for detecting senescence phenotype.
Discussion
The implication of autophagy in carcinogenesis still remains controversial. Autophagy functions either as prooncogenic or antioncogenic signaling. Our current study demonstrated that autophagy was induced by PGG, followed by senescence induction not only in vitro, but also in vivo. Moreover, we identified autophagy as an important mechanism to promote PGG-induced senescence phenotype in all 3 cell lines tested. Given the important role of senescence in tumor suppression, our findings supported that PGG-induced autophagy might act as a tumor suppressor mechanism through induction of senescence.
Autophagy and senescence are 2 important cellular processes that have been shown to play critical role in regulation of carcinogenesis. Recently, a number of studies have shown that autophagy and senescence were closely related and autophagy can promote senescence induction. 33 - 38 In the current study, we found that both autophagy and senescence were induced in response to PGG treatment. Inhibition of autophagy by either chemical inhibitor or genetic approach led to a significant decrease of PGG-induced senescence, followed by an increased induction of apoptosis. We further revealed that the reduction of senescence by autophagy inhibition was not due to apoptosis induction. These data indicated that autophagy facilitated the senescence phenotype in response to PGG, which is in agreement with the previous studies showing that inhibition of autophagy can reduce and/or delay the senescence response by therapeutic drugs. 34 , 38 The time-course analysis indicated that autophagy was rapidly induced with a peak at 48 h after PGG exposure and it then gradually diminished, whereas senescence induction by PGG reached a peak at 5 d and decreased thereafter accompanied by a significant increase in apoptosis, suggesting these 2 events occurred in parallel only at the early stage of senescence and then autophagy was progressively inactivated in the senescent cells. The possible reasons for the decreased percentage of senescent cells over time (after 6 d) include senescent cells undergoing apoptosis, nonsenescent cell proliferation, and senescent cells reentering proliferation. This was supported by significant apoptosis, which was detected starting from 6 d (data not shown), and a certain amount of nonsenescent cells that were observed in the PGG-treated cell population during the time-course analysis. Having found the involvement of autophagy in PGG-induced senescence, we next questioned how autophagy promoted senescence. Our previous study has shown that induction of autophagy can activate the UPR in response to a chemopreventive agent ursolic acid, 29 we then hypothesized that the UPR could be activated by PGG which might mediate the link between the senescence and autophagy induced by PGG. Our data demonstrated that PGG treatment led to an increased protein abundance of ER chaperone HSPA5 and activation of ER resident kinase EIF2AK3 and ERN1 in all 3 cell lines tested. Inhibition of autophagy by either RNA interference or a chemical inhibitor resulted in a significant attenuation of these UPR signals. Furthermore, inhibition of EIF2AK3 or ERN1 activation by both genetic and chemical approaches caused a dramatic decrease of PGG-induced senescent cell populations. Together, these data strongly indicated that autophagy-mediated UPR signaling played a critical role in the PGG-induced senescence of cancer cells, supporting our hypothesis. Our current study established a critical role of UPR signaling in the autophagy-mediated senescence phenotype. The detailed mechanisms involved in UPR modulation of the autophagy-mediated senescence phenotype clearly need to be further investigated.
The role of MAPK8/9/10 pathway in autophagy has been well established in a number of models, including ER stress, 31 serum deprivation, 39 and insulin-like growth factor-1 exposure, 40 possibly through regulation of several autophagic genes, such as ATG5, 41 ATG7, 42 SQSTM1, 43 and DRAM. 44 In the present study, we found that MAPK8/9/10 was activated in response to PGG and inhibition of MAPK8/9/10 activation significantly prevented autophagy induction by PGG. Consistent with autophagy inhibition by MAPK8/9/10 inactivation, the downstream events including ER stress and senescence were dramatically attenuated, whereas apoptosis was significantly increased when MAPK8/9/10 activation was blocked by its inhibitor. Together, these results suggested that MAPK8/9/10 activation played an apical role in PGG-induced autophagy and its downstream events. Initially, we hypothesized that MAPK8/9/10 activation was due to induction of the UPR based, on the literature. 31 , 32 Therefore we tested effect of ER stress inhibition on PGG-induced MAPK8/9/10 activation. Unexpectedly, knockdown of ERN1 or EIF2AK3 resulted in an enhanced MAPK8/9/10 activation (phosphorylation), suggesting that ER stress is unlikely the mediator of MAPK8/9/10 activation. Our current study established a critical role of MAPK8/9/10 signaling in PGG-induced autophagy, the detailed mechanisms involved in MAPK8/9/10-mediated autophagy in response to PGG exposure are being investigated.
Finally, we translated our cell culture results into an animal model. The key markers of autophagy and senescence were observed in the animal study. Moreover, the signaling pathways have been implicated in PGG-induced autophagy-dependent senescence in vitro including MAPK8/9/10, ERN1, and EIF2S1 were indeed activated in the tumors of PGG-treated mice. These results indicated that the findings of the cell culture study were well consistent with those found in the animal study. Our findings provided the first evidence that PGG was capable of inducing autophagy and senescence in vivo.
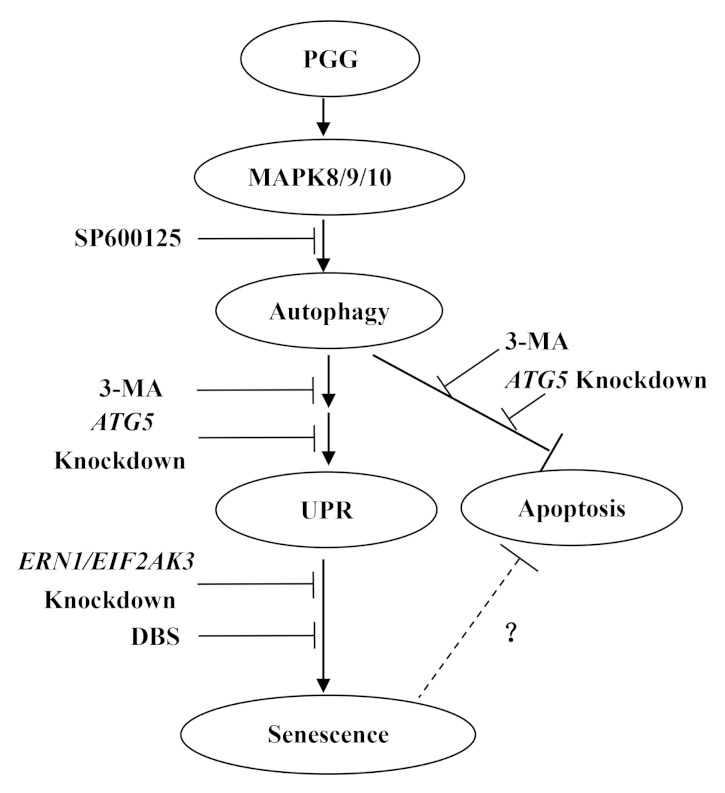
In summary (Fig. 8), PGG induced autophagy-dependent senescence through UPR activation. MAPK8/9/10 activation was an essential upstream signal to trigger autophagy. Our findings provided novel insights into understanding the mechanisms and functions of PGG-induced autophagy and senescence.
Figure 8. Potential signaling pathways involved in PGG-induced autophagy and senescence in human cancer cells. PGG induced autophagy-dependent senescence through UPR (ERN1 and EIF2AK3) activation. MAPK8/9/10 activation was an essential upstream signal to trigger autophagy.
Materials and Methods
Chemicals and reagents
PGG was prepared by a protocol refined and scaled up by Lu lab 45 through acidic methanolysis of tannic acid (Fischer Chemical, batch A310-500) and purification. The chemical identity and purity were established by proton NMR, specific rotation, and high-performance liquid chromatography (HPLC). The typical purity is 99% or higher.
3-methyladenine (3-MA) (M9281), bafilomycin A1 (B1793), 5-Bromo-2′-deoxyuridine (BrdU) (B5002) and 3-ethoxy-5,6-dibromosalicylaldehyde (DBS) (SML0149) were purchased from Sigma-Aldrich. Antibody for BrdU (11903800) was purchased from Roche. Antibodies specific for phospho-EIF2S1 (3597), phospho-MAPK8/9/10 (4668), HSPA5 (3177), DDIT3 (2895), ACTB (4970), CASP3 (9665) and cleaved poly (ADP-ribose) polymerase (PARP1; p89) (9548) were purchased from Cell Signaling Technology. Antibodies for phospho-EIF2AK3 (SC-35277) and ATF4 (SC-22800) were purchased from Santa Cruz Biotecnology. The antibody for phospho-ERN1 (ab124945) was purchased from Abcam. The antibody for XBP1s (647502) was purchased from Biolegend. Antibodies for LC3 (PM036) and ATG5 (M153-3) were purchased from MBL International Corporation. The MAPK8/9/10 inhibitor SP600125 (420119) were purchased from Calbiochem. Human IL6 Quantikine HS ELISA Kit (HS600B) was purchased from R&D. RNeasy Mini Kit (74104) and QuantiFast Probe RT-PCR Kit (204443) were purchased from Qiagen. The High Capacity cDNA Reverse Transcription Kit (4368814) was purchased from Invitrogen.
Cell culture and treatments
HepG2, MCF-7, and A549 cell lines were obtained from the American Type Culture Collection. The cells were grown in Dulbecco’s Modification of Eagle’s Medium (HyClone, SH30022.01B) supplemented with 10% fetal bovine serum without antibiotics. At 24 to 48 h after plating when cells were at 50% to 60% confluence, the medium was changed before staring the treatment with PGG and/or other agents.
Cell proliferative capacity assay
For the evaluation of the proliferative capacity of PGG-treated cells, HepG2 cells were treated with 25 µM PGG for 48 h and then trypsinized, counted, and sparsely seeded into six-well plate at 1 × 104 cells/well with fresh complete medium. The cells were continually cultured for 2, 4, 6, 8, 10, 12, 14, and 16 d with the medium change every 2 d. The cell number was counted by hemocytometer.
Assessment of senescence-associated β-galactosidase (SA-β-gal) activity
SA-β-gal activity at pH 6.0 was cytochemically detected according to the manufacturer’s instructions (Cell Signaling Technology, 9860). In brief, after the treatment, the cells were fixed with the fixative solution and stained with the staining solution provided with the kit. The stained cells were viewed and photographed under microscope. Total cell numbers and SA-β-gal-positive cell numbers were counted randomly for 5 to 10 fields per slide/well. SA-β-gal-positive cells were calculated as the percentage of positive cells per area.
DNA synthesis measurement by BrdU labeling
HepG2 cells were treated with 25 µM PGG for 24, 48, and 72 h. After the treatment, fresh media with 1µM BrdU was given to the cells for 1 h. The cells were collected, fixed, and processed for binding with a primary anti-BrdU monoclonal antibody (Roche, 11903800) and a secondary FITC-conjugated antibody (Sigma, F2883). Finally, the cells were stained with propidium iodide (MP Biomedicals, 195458) for flow cytometry analyses.
Reverse transcription-PCR
Total RNA was extracted at 12 h and 24 h after PGG treatment using an RNeasy Mini kit (Qiagen, 74104) and was used for reverse transcription according to the manufacturer's instruction. The primers used for PCR analysis were synthesized by Sangon Biotech. The sequences of the primers were as follows: for IL6, 5′ AAAGAGGCAC TGGCAGAAAA 3′ (forward) and 5′ TTTCACCAGG CAAGTCTCCT 3′ (reverse), for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′ CAGCCTCAAG ATCATCAG CA 3′ (forward) and 5′ GTCTTCTGGG TGGCAGTGAT 3′ (reverse). The quantification of real-time PCR products was performed using QuantiFast Probe RT-PCR Kit (Qiagen, 204443). The housekeeping gene GAPDH was used as internal control for RNA integrity and loading normalization.
Quantitative determination of human IL6 secretion
HepG2 cells were treated with 25 µM PGG for 24 h and conditioned medium was collected at various time points. Secretory IL6 was measured using an ELISA kit (R&D, HS600B) according to the manufacturer’s protocol.
Apoptosis evaluation
Apoptosis was assessed by 3 methods. The first was ANXA5 staining (MBL International Corporation, BV-K101-3) of externalized phosphatidylserine. The second method was conducted with a Cell Death ELISA kit (Roche, 11544675001) to detect cytoplasmic histone-associated-DNA-fragments purchased from Roche Diagnostics. The third one was immunoblot analysis of PARP1 cleavage.
Autophagy detection
Autophagy induction was determined by 2 methods. The first method was western blotting for conversion of the microtubule-associated protein 1 light chain 3 (LC3) -I to LC3-II. The second one was immunofluorescence staining for LC3 localization as reported previously. 46
Western blotting
The cell lysate was prepared in ice-cold radioimmunoprecipitation assay buffer. Cell lysate proteins were resolved by electrophoresis and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, IPVH00010). The blot was then probed with a primary antibody followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies. The signal was visualized by enhanced chemiluminescence (Fisher/Pierce, 34080) and recorded on an X-ray film (Estman Kodak Company, XBT-1).
RNA interference
siRNAs targeting ERN1(sc-40705), EIF2AK3 (sc-36213), and CASP3 (sc-29237) were purchased from Santa Cruz Biotechnologies. siRNAs targeting ATG5 and nontargeting siRNAs were synthesized by Integrated DNA Technologies. The cells were transfected with 50 nmol/L of specific or non-targeting siRNA using siPORT™ NeoFX™ Transfection Agent (Life Technologies, AM4510) for 24 h and then were used for subsequent experiments.
Animal study
To establish the cancer xenograft, 2 × 106 HepG2 cells were mixed with Matrigel (50%) (Becton Dickinson, 354234) and injected subcutaneous (s.c.) into the right flank of 7- to 8-wk-old BALB/c athymic nude mice (Charles River Laboratories). PGG was given daily by i.p. injection (20 mg/kg body weight) starting 7 d after s.c. inoculation of cancer cells for 16 d. This dose was chosen based on our earlier work with DU145 prostate cancer xenografts. 19 After the treatment, the tumors were harvested and the markers of autophagy, senescence, and ER stress were analyzed by western blotting or senescence-associated β-galactosidase staining.
Statistical analysis
Data are presented as mean ± SD. These data were analyzed with ANOVA with appropriate post-hoc comparisons among means. P < 0.05 was considered statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (NSFC, 31071533, 31371752, 30972172), Ministry of Science and Technology of China (2012BAD33B09), and the US. National Cancer Institute (R01 CA136953).
Glossary
Abbreviations:
- 3-MA
3-methyladenine
- ATG5
autophagy-related 5
- ATG7
autophagy-related 7
- ATF4
activating transcription factor 4
- ACTB
actin, beta
- β-gal
β-galactosidase
- BAF
bafilomycin A1
- BrdU
5-bromo-2’-deoxyuridine
- CASP3
caspase 3, apoptosis-related cysteine peptidase
- DBS
3-ethoxy-5,6-dibromosalicylaldehyde
- DDIT3
DNA-damage-inducible transcript 3
- EIF2AK3
eukaryotic translation initiation factor 2-alpha kinase 3
- EIF2S1
eukaryotic translation initiation factor 2, subunit 1 alpha
- ER
endoplasmic reticulum
- ERN1
endoplasmic reticulum to nucleus signaling 1
- HSPA5
heat shock 70 kDa protein 5 (glucose-regulated protein, 78kDa)
- IL6
interleukin 6
- MAP1LC3 (in the text
LC3), microtubule-associated protein 1 light chain 3
- MAPKs
mitogen-activated protein kinases
- PARP1
poly (ADP-ribose) polymerase 1
- PGG
penta-1,2,3,4,6-O-galloyl-beta-D-glucose
- s.c.
subcutaneous
- SQSTM1/p62
sequestosome 1
- UPR
unfolded protein response
- XBP1s
spliced X-box binding protein 1
References
- 1. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. . Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069 - 75; http://dx.doi.org/ 10.1038/nature06639; PMID: 18305538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jin S, White E. . Role of autophagy in cancer: management of metabolic stress. Autophagy 2007; 3:28 - 31; PMID: 16969128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. . Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137:1062 - 75; http://dx.doi.org/ 10.1016/j.cell.2009.03.048; PMID: 19524509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. . Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011; 25:795 - 800; http://dx.doi.org/ 10.1101/gad.2016211; PMID: 21498569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koren I, Kimchi A. . Cell biology. Promoting tumorigenesis by suppressing autophagy. Science 2012; 338:889 - 90; http://dx.doi.org/ 10.1126/science.1230577; PMID: 23161981 [DOI] [PubMed] [Google Scholar]
- 6. Levine B, Kroemer G. . Autophagy in the pathogenesis of disease. Cell 2008; 132:27 - 42; http://dx.doi.org/ 10.1016/j.cell.2007.12.018; PMID: 18191218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen N, Debnath J. . Autophagy and tumorigenesis. FEBS Lett 2010; 584:1427 - 35; http://dx.doi.org/ 10.1016/j.febslet.2009.12.034; PMID: 20035753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Y, Iglehart JD, Richardson AL, Wang ZC. . The amplified cancer gene LAPTM4B promotes tumor growth and tolerance to stress through the induction of autophagy. Autophagy 2012; 8:273 - 4; http://dx.doi.org/ 10.4161/auto.8.2.18941; PMID: 22301992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, Qiu B, Zhang H, Cerniglia G, Bi M, et al. . ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest 2012; 122:4621 - 34; http://dx.doi.org/ 10.1172/JCI62973; PMID: 23143306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayflick L, Moorhead PS. . The serial cultivation of human diploid cell strains. Exp Cell Res 1961; 25:585 - 621; http://dx.doi.org/ 10.1016/0014-4827(61)90192-6; PMID: 13905658 [DOI] [PubMed] [Google Scholar]
- 11. Gewirtz DA, Holt SE, Elmore LW. . Accelerated senescence: an emerging role in tumor cell response to chemotherapy and radiation. Biochem Pharmacol 2008; 76:947 - 57; http://dx.doi.org/ 10.1016/j.bcp.2008.06.024; PMID: 18657518 [DOI] [PubMed] [Google Scholar]
- 12. Christov KT, Shilkaitis AL, Kim ES, Steele VE, Lubet RA. . Chemopreventive agents induce a senescence-like phenotype in rat mammary tumours. Eur J Cancer 2003; 39:230 - 9; http://dx.doi.org/ 10.1016/S0959-8049(02)00497-5; PMID: 12509956 [DOI] [PubMed] [Google Scholar]
- 13. Heiss EH, Schilder YD, Dirsch VM. . Chronic treatment with resveratrol induces redox stress- and ataxia telangiectasia-mutated (ATM)-dependent senescence in p53-positive cancer cells. J Biol Chem 2007; 282:26759 - 66; http://dx.doi.org/ 10.1074/jbc.M703229200; PMID: 17626009 [DOI] [PubMed] [Google Scholar]
- 14. Wang Q, Wu PC, Roberson RS, Luk BV, Ivanova I, Chu E, Wu DY. . Survivin and escaping in therapy-induced cellular senescence. Int J Cancer 2011; 128:1546 - 58; http://dx.doi.org/ 10.1002/ijc.25482; PMID: 20503268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roberson RS, Kussick SJ, Vallieres E, Chen SY, Wu DY. . Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res 2005; 65:2795 - 803; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-1270; PMID: 15805280 [DOI] [PubMed] [Google Scholar]
- 16. Campisi J. . Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 2005; 120:513 - 22; http://dx.doi.org/ 10.1016/j.cell.2005.02.003; PMID: 15734683 [DOI] [PubMed] [Google Scholar]
- 17. Hornsby PJ. . Senescence as an anticancer mechanism. J Clin Oncol 2007; 25:1852 - 7; http://dx.doi.org/ 10.1200/JCO.2006.10.3101; PMID: 17488983 [DOI] [PubMed] [Google Scholar]
- 18. Zhang J, Li L, Kim SH, Hagerman AE, Lü J. . Anti-cancer, anti-diabetic and other pharmacologic and biological activities of penta-galloyl-glucose. Pharm Res 2009; 26:2066 - 80; http://dx.doi.org/ 10.1007/s11095-009-9932-0; PMID: 19575286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oh GS, Pae HO, Oh H, Hong SG, Kim IK, Chai KY, Yun YG, Kwon TO, Chung HT. . In vitro anti-proliferative effect of 1,2,3,4,6-penta-O-galloyl-beta-D-glucose on human hepatocellular carcinoma cell line, SK-HEP-1 cells. Cancer Lett 2001; 174:17 - 24; http://dx.doi.org/ 10.1016/S0304-3835(01)00680-2; PMID: 11675148 [DOI] [PubMed] [Google Scholar]
- 20. Huh JE, Lee EO, Kim MS, Kang KS, Kim CH, Cha BC, Surh YJ, Kim SH. . Penta-O-galloyl-β-D-glucose suppresses tumor growth via inhibition of angiogenesis and stimulation of apoptosis: roles of cyclooxygenase-2 and mitogen-activated protein kinase pathways. Carcinogenesis 2005; 26:1436 - 45; http://dx.doi.org/ 10.1093/carcin/bgi097; PMID: 15845650 [DOI] [PubMed] [Google Scholar]
- 21. Hu H, Lee HJ, Jiang C, Zhang J, Wang L, Zhao Y, Xiang Q, Lee EO, Kim SH, Lü J. . Penta-1,2,3,4,6-O-galloyl-beta-D-glucose induces p53 and inhibits STAT3 in prostate cancer cells in vitro and suppresses prostate xenograft tumor growth in vivo. Mol Cancer Ther 2008; 7:2681 - 91; http://dx.doi.org/ 10.1158/1535-7163.MCT-08-0456; PMID: 18790750 [DOI] [PubMed] [Google Scholar]
- 22. Kuo PT, Lin TP, Liu LC, Huang CH, Lin JK, Kao JY, Way TD. . Penta-O-galloyl-beta-D-glucose suppresses prostate cancer bone metastasis by transcriptionally repressing EGF-induced MMP-9 expression. J Agric Food Chem 2009; 57:3331 - 9; http://dx.doi.org/ 10.1021/jf803725h; PMID: 19320436 [DOI] [PubMed] [Google Scholar]
- 23. Hu H, Zhang J, Lee HJ, Kim SH, Lü J. . Penta-O-galloyl-beta-D-glucose induces S- and G(1)-cell cycle arrests in prostate cancer cells targeting DNA replication and cyclin D1. Carcinogenesis 2009; 30:818 - 23; http://dx.doi.org/ 10.1093/carcin/bgp059; PMID: 19269999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee HJ, Seo NJ, Jeong SJ, Park Y, Jung DB, Koh W, Lee HJ, Lee EO, Ahn KS, Ahn KS, et al. . Oral administration of penta-O-galloyl-β-D-glucose suppresses triple-negative breast cancer xenograft growth and metastasis in strong association with JAK1-STAT3 inhibition. Carcinogenesis 2011; 32:804 - 11; http://dx.doi.org/ 10.1093/carcin/bgr015; PMID: 21289371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu H, Chai Y, Wang L, Zhang J, Lee HJ, Kim SH, Lü J. . Pentagalloylglucose induces autophagy and caspase-independent programmed deaths in human PC-3 and mouse TRAMP-C2 prostate cancer cells. Mol Cancer Ther 2009; 8:2833 - 43; http://dx.doi.org/ 10.1158/1535-7163.MCT-09-0288; PMID: 19825802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yin S, Dong Y, Li J, Lü J, Hu H. . Penta-1,2,3,4,6-O-galloyl-beta-D-glucose induces senescence-like terminal S-phase arrest in human hepatoma and breast cancer cells. Mol Carcinog 2011; 50:592 - 600; http://dx.doi.org/ 10.1002/mc.20743; PMID: 21319227 [DOI] [PubMed] [Google Scholar]
- 27. Johansen T, Lamark T. . Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011; 7:279 - 96; http://dx.doi.org/ 10.4161/auto.7.3.14487; PMID: 21189453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie Z, Klionsky DJ. . Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102 - 9; http://dx.doi.org/ 10.1038/ncb1007-1102; PMID: 17909521 [DOI] [PubMed] [Google Scholar]
- 29. Zhao C, Yin S, Dong Y, Guo X, Fan L, Ye M, Hu H. . Autophagy-dependent EIF2AK3 activation compromises ursolic acid-induced apoptosis through upregulation of MCL1 in MCF-7 human breast cancer cells. Autophagy 2013; 9:196 - 207; http://dx.doi.org/ 10.4161/auto.22805; PMID: 23182854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Volkmann K, Lucas JL, Vuga D, Wang X, Brumm D, Stiles C, Kriebel D, Der-Sarkissian A, Krishnan K, Schweitzer C, et al. . Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem 2011; 286:12743 - 55; http://dx.doi.org/ 10.1074/jbc.M110.199737; PMID: 21303903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. . Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26:9220 - 31; http://dx.doi.org/ 10.1128/MCB.01453-06; PMID: 17030611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moretti L, Cha YI, Niermann KJ, Lu B. . Switch between apoptosis and autophagy: radiation-induced endoplasmic reticulum stress?. Cell Cycle 2007; 6:793 - 8; http://dx.doi.org/ 10.4161/cc.6.7.4036; PMID: 17377498 [DOI] [PubMed] [Google Scholar]
- 33. Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavaré S, Arakawa S, Shimizu S, Watt FM, et al. . Autophagy mediates the mitotic senescence transition. Genes Dev 2009; 23:798 - 803; http://dx.doi.org/ 10.1101/gad.519709; PMID: 19279323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh K, Matsuyama S, Drazba JA, Almasan A. . Autophagy-dependent senescence in response to DNA damage and chronic apoptotic stress. Autophagy 2012; 8:236 - 51; http://dx.doi.org/ 10.4161/auto.8.2.18600; PMID: 22240589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goehe RW, Di X, Sharma K, Bristol ML, Henderson SC, Valerie K, Rodier F, Davalos AR, Gewirtz DA. . The autophagy-senescence connection in chemotherapy: must tumor cells (self) eat before they sleep?. J Pharmacol Exp Ther 2012; 343:763 - 78; http://dx.doi.org/ 10.1124/jpet.112.197590; PMID: 22927544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gewirtz DA. . Autophagy and senescence: a partnership in search of definition. Autophagy 2013; 9:808 - 12; http://dx.doi.org/ 10.4161/auto.23922; PMID: 23422284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gewirtz DA. . Autophagy and senescence in cancer therapy. J Cell Physiol 2014; 229:6 - 9; PMID: 23794221 [DOI] [PubMed] [Google Scholar]
- 38. Knizhnik AV, Roos WP, Nikolova T, Quiros S, Tomaszowski KH, Christmann M, Kaina B. . Survival and death strategies in glioma cells: autophagy, senescence and apoptosis triggered by a single type of temozolomide-induced DNA damage. PLoS One 2013; 8:e55665; http://dx.doi.org/ 10.1371/journal.pone.0055665; PMID: 23383259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. . JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008; 30:678 - 88; http://dx.doi.org/ 10.1016/j.molcel.2008.06.001; PMID: 18570871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jia G, Cheng G, Gangahar DM, Agrawal DK. . Insulin-like growth factor-1 and TNF-alpha regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol Cell Biol 2006; 84:448 - 54; http://dx.doi.org/ 10.1111/j.1440-1711.2006.01454.x; PMID: 16942488 [DOI] [PubMed] [Google Scholar]
- 41. Byun JY, Yoon CH, An S, Park IC, Kang CM, Kim MJ, Lee SJ. . The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis 2009; 30:1880 - 8; http://dx.doi.org/ 10.1093/carcin/bgp235; PMID: 19783847 [DOI] [PubMed] [Google Scholar]
- 42. Wong CH, Iskandar KB, Yadav SK, Hirpara JL, Loh T, Pervaiz S. . Simultaneous induction of non-canonical autophagy and apoptosis in cancer cells by ROS-dependent ERK and JNK activation. PLoS One 2010; 5:e9996; http://dx.doi.org/ 10.1371/journal.pone.0009996; PMID: 20368806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, Auberger P. . Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res 2010; 70:1042 - 52; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-3537; PMID: 20103647 [DOI] [PubMed] [Google Scholar]
- 44. Lorin S, Borges A, Ribeiro Dos Santos L, Souquère S, Pierron G, Ryan KM, Codogno P, Djavaheri-Mergny M. . c-Jun NH2-terminal kinase activation is essential for DRAM-dependent induction of autophagy and apoptosis in 2-methoxyestradiol-treated Ewing sarcoma cells. Cancer Res 2009; 69:6924 - 31; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-1270; PMID: 19706754 [DOI] [PubMed] [Google Scholar]
- 45. Li L, Shaik AA, Zhang J, Nhkata K, Wang L, Zhang Y, Xing C, Kim SH, Lü J. . Preparation of penta-O-galloyl-β-D-glucose from tannic acid and plasma pharmacokinetic analyses by liquid-liquid extraction and reverse-phase HPLC. J Pharm Biomed Anal 2011; 54:545 - 50; http://dx.doi.org/ 10.1016/j.jpba.2010.09.028; PMID: 20970943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Herman-Antosiewicz A, Johnson DE, Singh SV. . Sulforaphane causes autophagy to inhibit release of cytochrome C and apoptosis in human prostate cancer cells. Cancer Res 2006; 66:5828 - 35; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-0139; PMID: 16740722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.