Abstract
Cells respond to the deprivation of nutrients by inducing autophagy. However, mechanisms through which cells coordinately regulate autophagy with metabolic state remain incompletely understood. We previously observed that prototrophic strains of yeast induce autophagy upon switch from a rich to minimal medium in the absence of severe nitrogen starvation. We determined that the sulfur-containing amino acid methionine and its downstream metabolite S-adenosylmethionine (SAM) are sufficient to strongly inhibit such autophagy. These metabolites function through Ppm1, an enzyme that methylates the catalytic subunit of the protein phosphatase PP2A. As such, methionine and SAM act as critical signals of amino acid sufficiency that reciprocally regulate autophagy and cell growth by modulating the methylation status of PP2A.
Keywords: methionine, SAM, methyltransferases, PP2A, TORC, autophagy
Autophagy is a cellular catabolic response to nutrient deprivation and other stresses. During autophagy, cellular components and organelles are delivered to the vacuole or lysosome for degradation and recycling. Thus, this process plays a critical role in maintaining cellular homeostasis. In eukaryotes, the TORC1 pathway is a major nutrient-responsive anabolic pathway that also regulates autophagy. The TORC1 pathway is inhibited during severe amino acid restriction, which in turn activates autophagy. In budding yeast, this occurs in part through the dephosphorylation of the TORC1 substrate Atg13. However, the mechanisms through which the TORC1 pathway senses specific nutrients to subsequently regulate autophagy are still poorly understood.
Genetic screens using budding yeast have enabled the identification of dozens of genes regulating autophagy that are conserved across eukaryotes. While these pioneering studies revealed many components of the core autophagy machinery, it should be noted that they were often performed using auxotrophic strains of yeast that cannot synthesize certain essential nutrients such as adenine, leucine, and uracil, which must consequently be supplemented into the growth medium. As such, autophagy was typically induced in these strains using severe nitrogen starvation conditions. It remained possible that autophagy might also be induced in response to less severe nutrient starvation and that the metabolic regulation of autophagy could be different in prototrophs vs. auxotrophs. We serendipitously observed that when prototrophic yeast strains were switched from nutrient-rich medium to a minimal medium still capable of supporting growth (albeit at slower rates), they still strongly induce autophagy as measured using multiple standard assays. Since a nitrogen source was not limiting under these conditions, we tentatively termed this form of autophagy “non-nitrogen-starvation” (NNS)-induced autophagy. We then conducted a visual genetic screen and identified a complex of 3 proteins (Iml1, Npr2, Npr3) that were required for NNS-autophagy (Fig. 1). Interestingly, these proteins were not required for nitrogen starvation-induced autophagy. We also showed that phosphorylation of the Npr2 protein seemed to be important to keep this complex intact and in a form critical for NNS-autophagy. While these proteins are conserved among eukaryotes, relatively little is known about them. Genetic studies have shown that they act as upstream negative regulators of TORC1 (Fig. 1) and more recent work suggests that this is at least in part due to their roles in regulating small G-proteins that activate this complex.
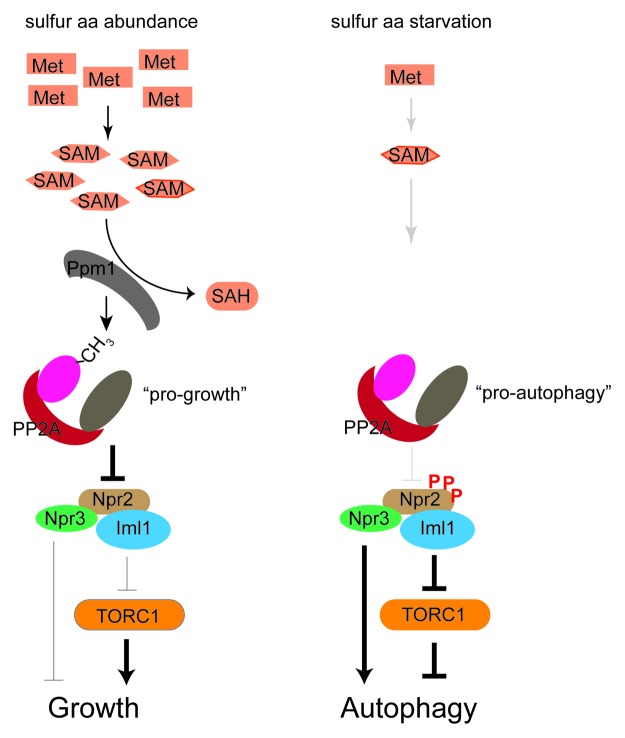
Figure 1. The influence of intracellular methionine and SAM on cell growth and autophagy. NNS-autophagy is inhibited by high intracellular concentrations of methionine and SAM. The methylation of the catalytic subunit of PP2A is responsive to SAM concentrations. Methylated PP2A can regulate the phosphorylation status of several substrates including Npr2, which is a component of a conserved complex that is required for NNS-autophagy. aa, amino acid; SAH, S-adenosylhomocysteine
However, there were 2 major unanswered questions. First, and perhaps most important, we did not know which metabolites or nutrients, when insufficient, could trigger NNS-autophagy. Second, we did not know the pathways through which this form of autophagy was regulated. The robust induction of autophagy in cells transferred to minimal growth media (from nutrient-rich media) allowed us to investigate whether the addition of specific metabolites could prevent autophagy. Surprisingly, we found that adding back a single amino acid, methionine, was sufficient to potently block such autophagy. Other amino acids, including leucine and glutamine, which have been linked to the regulation of TORC1 activity, had virtually no effect. We then determined that the downstream metabolite of methionine, S-adenosylmethionine (SAM), was responsible for this effect of methionine (Fig. 1). SAM is a central metabolite that serves as a methyl donor for a wide variety of metabolic transformations that result in the methylation of proteins, lipids, and nucleic acids. These methylation modifications are thought to play important roles in cell growth and regulation. We therefore screened non-essential yeast methyltransferase enzymes to determine if any of them were required for SAM-mediated inhibition of autophagy. Our screen identified Ppm1, a conserved methyltransferase enzyme that methylates the C-terminal leucine residue of a major protein phosphatase enzyme, PP2A (Fig. 1). Cells lacking Ppm1 (LCMT1 in mammals) were unable to respond to methionine addition and thus methionine could no longer inhibit autophagy in this mutant.
PP2A is a conserved protein phosphatase that regulates growth processes in response to nutrients by dephosphorylating specific target proteins, including several known TORC1 substrates. Indeed, PP2A has been linked to TORC1-dependent autophagy in both yeast and mammals. The specific role of methylation for PP2A function, however, is less clear. Our study revealed that mutation of the leucine residue in the PP2A catalytic subunit that is subject to methylation resulted in cells becoming completely unresponsive to methionine and SAM. We also identified the phosphorylation of Npr2 to be both methionine-dependent and PP2A-regulated (Fig. 1). Thus, our findings consequently suggest that TORC1 activity and signaling could be regulated by the methylation status of PP2A, which can dephosphorylate substrates that themselves are either regulators of TORC1 or TORC1 substrates. As such, PP2A itself may act as a sensor of amino acid sufficiency, through the ability of methionine to influence the methylation status of its catalytic subunit.
In summary, our studies demonstrate the importance of the sulfur-containing amino acid methionine in the regulation of autophagy and further define a pathway through which it is sensed. These studies also reveal that methionine and SAM function as sentinel metabolites that have a significant impact upon nutrient-responsive pathways that control cell growth and autophagy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.