SUMMARY
Hepatic lipid droplet (LD) catabolism is thought to occur via cytosolic lipases such as adipose triglyceride lipase (ATGL) or through autophagy of LDs, a process known as lipophagy. We tested the potential interplay between these metabolic processes and its effects on hepatic lipid metabolism. We show that hepatic ATGL is both necessary and sufficient to induce both autophagy and lipophagy. Moreover, lipophagy is required for ATGL to promote LD catabolism and the subsequent oxidation of hydrolyzed fatty acids (FAs). Following previous work showing that ATGL promotes sirtuin 1 (SIRT1) activity, studies in liver-specific SIRT1−/− mice and in primary hepatocytes reveal that SIRT1 is required for ATGL-mediated induction of autophagy and lipophagy. Taken together, these studies show that ATGL-mediated signaling via SIRT1 promotes autophagy/lipophagy as a primary means to control hepatic LD catabolism and FA oxidation.
Graphical abstract
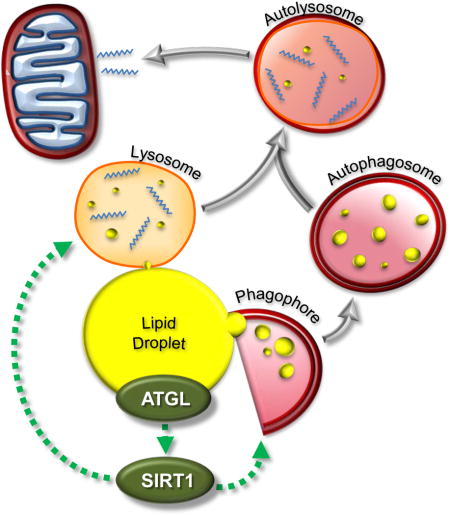
INTRODUCTION
LDs are the major storage organelle of intracellular triacylglycerol (TAG) and are present in nearly all cell types. Historically thought to be inert energy storage depots, LDs are increasingly recognized as dynamic organelles involved in many cellular processes beyond energy storage (Greenberg et al. 2011). Studies conducted by our laboratory and others have identified ATGL as a major hepatic lipase that regulates TAG turnover. Specifically, shRNA knockdown or genetic ablation of hepatic ATGL promotes steatosis (Ong et al. 2011; Wu et al. 2012), whereas overexpression of ATGL in the liver alleviates steatosis (Turpin et al. 2011). Additionally, ATGL selectively channels the liberated FAs to oxidative pathways and does not influence VLDL secretion (Ong et al. 2011; Wu et al. 2012). Thus, in addition to its prominent roles in tissues such as adipose and heart, ATGL also appears to play a critical role in regulating hepatic lipid metabolism.
Autophagy is a cellular recycling mechanism that provides cells with a source of energy during periods of nutrient insufficiency (He & Klionsky 2009). Autophagy can be classified into three categories based on lysosomal cargo delivery: macroautophagy (He & Klionsky 2009), chaperone-mediated autophagy (Cuervo & Wong 2014) and microautophagy (Santambrogio & Cuervo 2011). The role of autophagy in LD metabolism was unclear until Singh et al. showed that inhibiting autophagy in the liver promoted LD accumulation and attenuated oxidation of the hydrolyzed FAs (Singh et al. 2009). Since these findings of autophagy-mediated lipid mobilization, termed lipophagy, a plethora of studies have shown the existence of lipophagy in diverse cell types such as macrophages (Ouimet et al. 2011), neurons (Kaushik et al. 2011) and brown adipocytes (Cairó et al. 2016; Martinez-Lopez et al. 2016). Additional studies in which various autophagy genes have been manipulated further confirm the importance of autophagy/lipophagy in regulating hepatic TAG levels (Yang et al. 2010; Settembre et al. 2013; Schroeder et al. 2015).
Recent studies have provided some mechanistic insights into the potential crosstalk between ATGL-catalyzed lipolysis and autophagy/lipophagy in the liver. Chaperone-mediated autophagy has been shown to contribute to hepatic LD catabolism via its degradation of the LD proteins Perilipin 2 (PLIN2) and 3 (Kaushik & Cuervo 2015). The degradation of these proteins allows ATGL to access LDs and presumably facilitate lipolysis. A recent study demonstrated that ATGL contains a LC3 interacting region that facilitates its interaction with LC3 containing organelles (Martinez-Lopez et al. 2016). Mutating the LC3 interacting region prevents ATGL targeting to LDs suggesting an important crosstalk between autophagy and ATGL. Additionally, PNPLA5, a putative neutral lipid hydrolase and member of the patatin-like phospholipase domain-containing family, which includes ATGL/PNPLA2, promotes autophagy suggesting that cytosolic lipases could act upstream to regulate autophagy/lipophagy (Dupont et al. 2014).
ATGL activity promotes peroxisome proliferator activated receptor-α (PPAR-α) and PPAR-γ coactivator 1-α (PGC-1α) signaling as a means to increase FA oxidation and mitochondrial biogenesis (Haemmerle et al. 2011; Sapiro et al. 2009). Supplementation of ATGL knockout mice with the PPAR-α agonists Wy-14643 largely rescues PPAR-α/PGC-1 α signaling and normalizes heart TAG levels suggesting that ATGL mediates oxidative metabolism via the production of FA ligands for PPAR-α (Haemmerle et al. 2011). In contrast, we have shown that PPAR-α agonists are unable to normalize transcriptional signaling in response to hepatic ablation of ATGL indicating an alternate mechanism must exist through which ATGL influences PPAR-α (Ong et al. 2011). Subsequent studies have shown that ATGL promotes SIRT1 activity and that SIRT1 mediates the effects of ATGL on PPAR-α/PGC-1α signaling and the regulation of oxidative metabolism in the liver (Khan et al. 2015). Thus, ATGL plays an important role in cell signaling to facilitate pathways that promote fatty acid catabolism and oxidative metabolism.
Given that SIRT1, PGC-1α and PPAR-α are positive regulators of autophagy/lipophagy (Lee et al. 2008; Lee et al. 2014; Takikita et al. 2010) and are downstream of ATGL, we sought to further characterize the interplay between ATGL and autophagy/lipophagy. Herein, we show that ATGL acts as an important upstream signaling node that, via SIRT1, increases autophagy/lipophagy as a means to promote hepatic LD catabolism.
RESULTS
ATGL is required for autophagy/lipophagy induction
We have previously shown that hepatic ATGL plays an important role in TAG turnover and the subsequent channeling of hydrolyzed FAs to oxidative pathways as well as upregulating PPAR-α/PGC-1 a signaling to control oxidative metabolism (Ong et al. 2011; Khan et al. 2015). Given that lipophagy also contributes to hepatic LD catabolism (Schroeder et al. 2015; Settembre et al. 2013; Singh et al. 2009), we sought to dissect the links between hepatic ATGL-catalyzed lipolysis and autophagy/lipophagy. Inhibition of ATGL via shRNA-mediated knockdown in liver, as previously reported (Ong et al. 2011), significantly lowered the mRNA abundance of autophagy genes (Figure 1A). In agreement with the knockdown data, chemical inhibition of ATGL via ATGListatin also reduced autophagy gene expression in primary hepatocytes (Figure 1B) suggesting that ATGL activity rather than the protein itself is important for autophagy gene expression. ATGL knockdown decreased hepatic protein levels of LC3II and LAMP1, but increased p62 when compared to the scrambled controls (Figures 1C and S1A) consistent with reduced autophagy. ATGListatin also reduced the formation of LC3 punctae in MEFs (Figures 1D and S1B). To measure autophagic flux, we used the tandem GFP-RFP-LC3 sensor (Kimura et al.) that contains an acid-labile GFP and acid-resistant RFP to distinguish between autophagosome and autolysosome localization. We observed reduced RFP signal relative to GFP in response to chemical inhibition of ATGL indicating reduced autophagic flux (Figure 1E). ATGL knockdown in mouse livers also reduced LC3 staining and its colocalization with the LD marker PLIN2 indicating reduced lipophagy (Figure 1F). Similarly, ATGL knockdown reduced hepatic LD specific LC3 II levels (Figure S1C). Finally, ATGListatin reduced localization of lysosomes with LDs in primary hepatocytes (Figure 1G and S1D). Taken together, these data shown that ATGL promotes autophagy/lipophagy.
Figure 1. Liver-specific inhibition of ATGL attenuates autophagy/lipophagy.
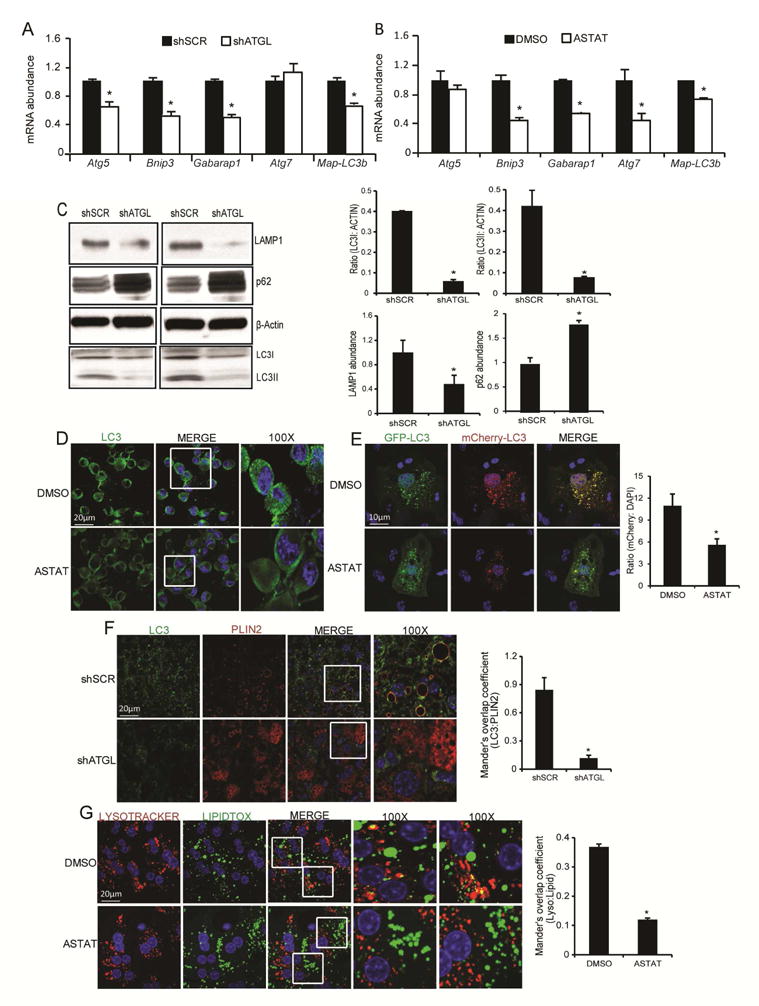
(A) In vivo ablation of hepatic ATGL via an ATGL shRNA adenovirus reduced the expression of autophagy genes (n=5); *P<0.05 vs. control shRNA. (B) Inhibition of ATGL in primary hepatocytes with ATGListatin (Astat, 30 μM for 36 hours) decreased autophagy gene expression (n=5); *P<0.05 vs. DMSO. (C) In vivo ATGL knockdown reduced the LC3II/LC3I ratio and protein levels of LAMP1, and increased p62 expression; a representative Western blot and densitometry from 3 mice is shown. (D) ATGLstatin decreased LC3 puncta in mouse embryonic fibroblasts (MEFs) (n=3). (E) Hepatocytes transfected with the dual RFP-GFP-LC3 plasmid were treated with ATGListatin to acutely inhibit ATGL. Increased red punctae is indicative of enhanced lysosomal activity as observed with the DMSO treatment when compared to ATGListatin treated cells (n=4). (F) Confocal imaging of liver sections show reduced LD localization (as measured with the LD protein perilipin 2, PLIN2) with LC3 in response to ATGL knockdown. (G) ATGListatin decreased lysosomal association with LDs in primary mouse hepatocytes (n=3 for F and G).
ATGL overexpression is sufficient to drive autophagy/lipophagy
In contrast to ATGL knockdown or inhibition, the livers of Ad-ATGL treated mice, described in (Ong et al. 2011), had increased expression of autophagy genes. Additionally, ATGL overexpression increased the protein abundance of LAMP1 and LC3II, and reduced p62 levels (Figures 2A–B, S1E). ATGL overexpression increased colocalization of LDs with LC3 and lysosomes suggesting increased lipophagy (Figures 2C–D). ATGL overexpression also enhanced autophagic flux in hepatocytes transfected with the dual sensor LC3 plasmid compared to the control (Figure 2E). Thus, these results show that ATGL overexpression is sufficient to increase autophagy and lipophagy in the liver.
Figure 2. ATGL overexpression is sufficient to promote autophagy/lipophagy.
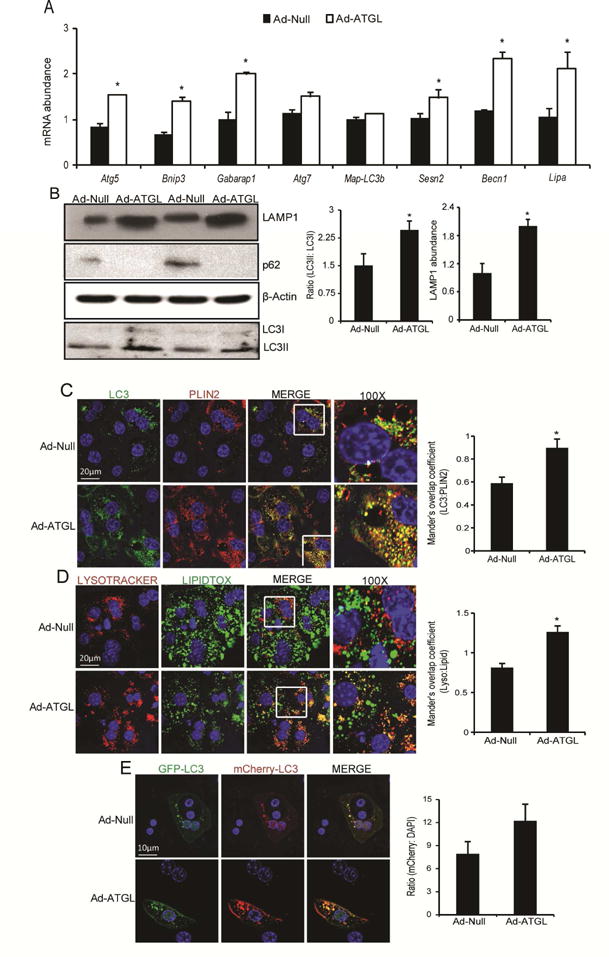
(A) In vivo overexpression of hepatic ATGL increased autophagy target gene expression (n=5); *P<0.05 vs. Ad-Null. (B) Hepatic ATGL overexpression increased the LC3II/LC3I ratio and LAMP1 protein levels as analyzed via western blotting. Livers overexpressing ATGL had no visible bands for p62 indicative of enhanced autophagosome clearance; a representative Western blot from 3 mice is shown. (C–D) In vivo overexpression of hepatic ATGL increased LC3 and lysosomal co-localization with LDs stained with PLIN2 or lipidtox. (E) The dual reporter RFP-GFP-LC3 was transfected in primary hepatocytes that were transduced with Ad-Null or Ad-ATGL adenoviruses; (n=3 for C–E).
Lipophagy mediates the effects of ATGL on TAG catabolism and FA oxidation
The above data suggest lipophagy could either work in parallel with ATGL to promote LD degradation or act downstream of ATGL to mediate LD catabolism. Thus, we next explored if lipophagy was responsible for mediating the effects of ATGL on TAG catabolism and the subsequent oxidation of hydrolyzed FAs. Consistent with previous studies (Ong et al. 2011; Sapiro et al. 2009), overexpressing ATGL increased TAG turnover, but this effect was partially or completely abrogated when autophagy was blunted via exposure of hepatocytes to the autophagy inhibitor chloroquine or knockdown of ATG5 (Figures 3A–B). Moreover, blocking lysosomal lipid hydrolysis, via the lysosomal acid lipase inhibitor LAListat (Pearson et al. 2014), prevented the increase in TAG turnover in response to ATGL overexpression (Figure 3C). Consistent with the TAG turnover data, blocking autophagy or lysosomal lipid degradation attenuated the oxidation of hydrolyzed FAs following ATGL overexpression (Figures 3D–F). In further support of the above data, overexpressing ATGL reduced LD accumulation in lipid-loaded hepatocytes and this effect was completely abrogated when autophagy/lipophagy was blocked via chloroquine or LAListat (Figure 3G). Thus, these data show that autophagy/lipophagy acts downstream of ATGL to mediate hepatic LD catabolism and the subsequent oxidation of hydrolyzed FAs.
Figure 3. Lipophagy is required for ATGL-mediated effects on TAG catabolism.
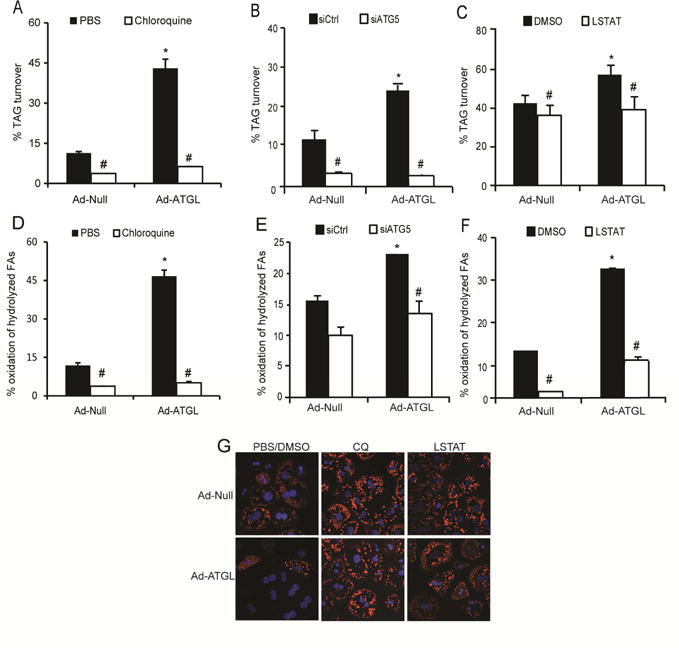
Isolated primary mouse hepatocytes were transduced with Ad-Null or Ad-ATGL adenoviruses and treated with 600 μM chloroquine (A), siATG5 (B), 10 μM LAListat (C) or their respective controls. Ad-ATGL increased TAG turnover, which was abolished with inhibition of autophagy/lipophagy. (D-F) Autophagy/lipophagy inhibitors impaired the ATGL-mediated induction in FA oxidation (n=6 for A-F); *P<0.05 vs. Ad-Null and #P<0.05 vs. siCtrl or vehicle. (G) Chloroquine and LAListat, added during the chase resulted in increased LD staining and blocked LD depletion in response to ATGL overexpression. Primary mouse hepatocytes were pulsed with a C-12 BODIPY FA (558/68) overnight, followed by an 8-hour chase in serum and insulin free M199 along with inhibitors (n=3).
SIRT1 mediates the effects of ATGL on autophagy induction
We have shown that hepatic ATGL regulates mitochondrial biogenesis and the expression of genes involved in β-oxidation through its regulation of SIRT1 (Khan et al. 2015). Additionally, SIRT1 is a well-established activator of autophagy through its deacetylation and activation of key components of the autophagy induction network such as the ATG protein family (Lee et al. 2008) and transcriptional regulators including FOXO1, CREB, TFEB and FXR (Brunet et al. 2004; Kemper et al. 2009; Qiang et al. 2011; Wang et al. 2010; Settembre et al. 2013). To examine the importance of SIRT1 in mediating the effects of ATGL on hepatic autophagy, we transduced liver-specific SIRT1−/− mice (L-SIRT1−/−) with an adenovirus harboring ATGL. Consistent with data from Figure 2A, Ad-ATGL increased the expression of autophagy genes in control SIRTflox/flox mice, but this effect was abolished in the absence of SIRT1 (Figure 4A). Western blot analysis revealed increased expression of LAMP1, ATG5 and LC3II with ATGL overexpression of SIRTflox/flox mice, but this effect was completely lost in the absence of hepatic SIRT1 (Figures 4B). We also used a dual adenovirus system to simultaneously overexpress ATGL and knockdown SIRT1 (shSIRT1) in the liver (Khan et al. 2015) to more acutely regulate SIRT1 (Figure S2A). Ad-ATGL treated livers had increased expression of autophagy genes, which was completely abolished when SIRT1 was knocked down (Figure S2B). To determine if the changes in autophagy/lipophagy genes translated to changes in lipid catabolism, we employed pulse-chase studies with FA tracers. ATGL promoted TAG turnover and the subsequent oxidation of the hydrolyzed FAs (Figures 3C and D) as we have shown previously (Ong et al. 2011). However, SIRT1 ablation abrogated the induction of TAG turnover and oxidation of hydrolyzed FAs in response to ATGL overexpression in primary hepatocytes (Figures 4C–D). The ability of ATGL overexpression to promote FA oxidation in vivo, as measured by serum ketone bodies, was also lost in L-SIRT1−/− mice (Figure 4E). ATGL overexpression enhanced LC3 colocalization with LDs in primary hepatocytes, which was blocked when cells were treated with the SIRT1 inhibitor EX527 (Figure 4F). Similar results were observed in the interaction between lysosomes and LDs in response to EX527 or SIRT1 knockdown (Figures 4G and S2C). Finally, the ATGL-mediated increase in autophagic flux was negated in the presence of the SIRT1 inhibitor EX527 (Figure S2D). Taken together, these data show that SIRT1 mediates the effects of ATGL on promoting autophagy/lipophagy.
Figure 4. SIRT1 mediates the effects of ATGL on autophagy/lipophagy.
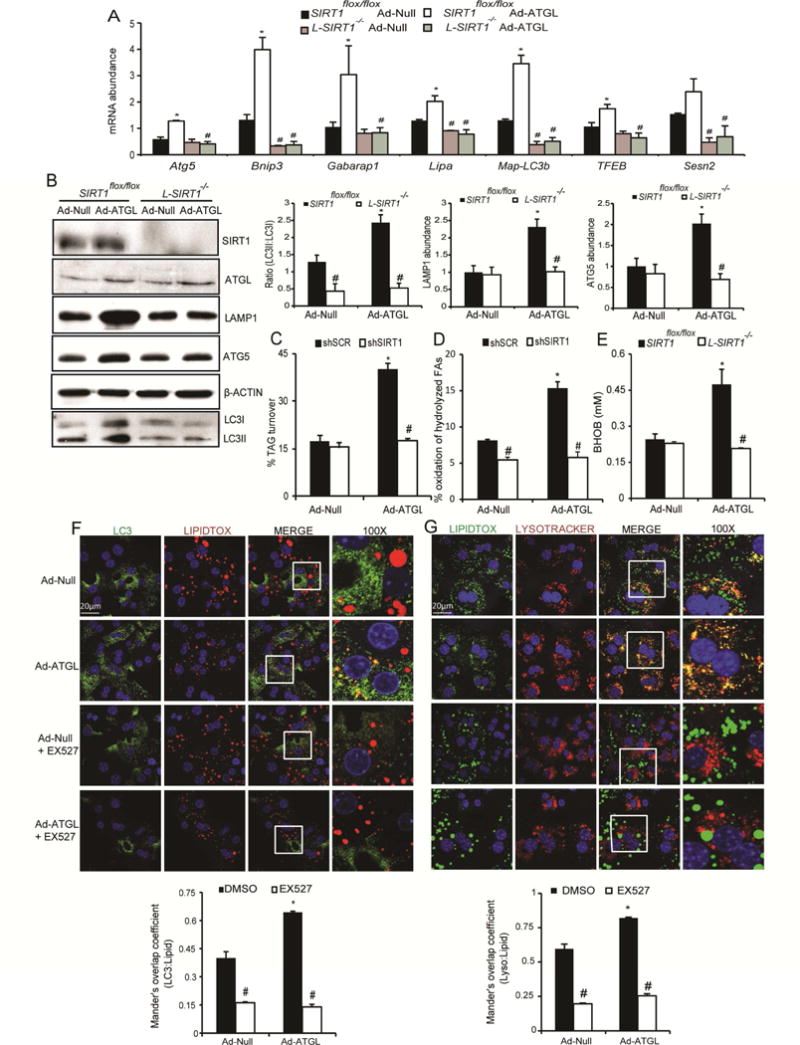
(A) The ATGL-mediated increase in autophagy gene expression was abolished in the absence of SIRT1. SIRT1flox/flox or L-SIRT1−/− mice were treated with Ad-Null or Ad-ATGL viruses for 7 days prior to sacrifice (n=6); *P<0.05 vs. Ad-Null and #P<0.05 vs. SIRT1+/+. (B) Protein expression of autophagy genes in response to ATGL overexpression and/or SIRT1 ablation; a representative Western blot and densitometry analysis from 4 mice is shown. (C-D) Primary mouse hepatocytes were treated with dual adenoviruses and TAG turnover and FA oxidation during the chase period were measured (n=5); *P<0.05 vs. Ad-Null and #P<0.05 vs. shSCR. (E) L-SIRT1−/− mice treated with Ad-ATGL had lower β-hydroxybutyrate compared to the floxed controls (n=6); *P<0.05 vs. Ad-Null and #P<0.05 vs. SIRT1+/+. (F–G) Confocal imaging revealed that Ad-ATGL enhanced LC3-LD and lysosome-LD co-localization, which was lost with 10 mM EX527, a SIRT1 inhibitor (n=2).
DISCUSSION
The past decade has seen significant advances in our knowledge of autophagy and its role in the pathophysiology of many diseases. Despite a long known linkage between LAL deficiency and steatosis in humans (Burke & Schubert 1972), lipophagy has only recently been shown to contribute directly to hepatic lipid catabolism (Singh et al. 2009). Since this seminal discovery, numerous studies show that lipophagy is a major regulator of hepatic LD accumulation (Cingolani & Czaja 2016). Despite the recognized importance of lipophagy in LD catabolism, the relative contribution of lipophagy versus cytosolic lipases such as ATGL has not been fully elucidated. Previous studies have highlighted the importance of chaperone-mediated autophagy and LC3 in targeting ATGL to the surface of LDs (Kaushik & Cuervo 2015; Martinez-Lopez et al. 2016). While these studies further highlight upstream regulation of ATGL, they did not explore the mechanism through which ATGL promotes LD catabolism once it is present on the LD surface. The findings in the current study extend these data and show that ATGL activity promotes autophagy/lipophagy to facilitate catabolism of LDs. Thus, rather than ATGL being the primary enzyme responsible for TAG hydrolysis and LD catabolism in the liver, our data suggest that ATGL acts as an inducer of autophagy/lipophagy, the latter of which is responsible for bulk LD degradation. The above studies showing that ATGListatin impairs autophagy/lipophagy suggest that ATGL could influence lipophagy by providing a signal that allows for recognition of LDs by autophagosomes and lysosomes in addition to increasing lipophagic/autophagic machinery. Additionally, we can’t rule out the possibility that LD accumulation itself, as would occur with ATGL inhibition, could act to suppresses autophagy/lipophagy. Regardless, the mechanistic details through which the LDs are recognized and targeted for lipophagy is still an intense area of investigation.
LDs are commonly found in association with various organelles including mitochondria and autophagy organelles such as autophagosomes and lysosomes (Pu et al. 2011). The direct interaction with mitochondria is postulated to provide a bridge for the transfer of hydrolyzed FAs to mitochondria for oxidation (Herms et al. 2015; Rambold et al. 2015). Despite the intimacy between these organelles, direct transfer of FAs from the LD to the mitochondria has yet to be demonstrated. Although the current data do not rule out that direct transfer of FAs between LDs and mitochondria in the liver occurs, the data in Figure 3, which shows that lipophagy largely mediates the effects of ATGL on oxidation of hydrolyzed FAs, suggests that this pathway is of less importance relative to the supply of FAs generated from lipophagy.
SIRT1 is well recognized as a major regulator of autophagy and directly regulates autophagy through the deacetylation of several mediator proteins of autophagy such as ATG5, ATG7 and ATG8/LC3 (Lee et al. 2008). In addition, SIRT1 regulates the transcriptional control of autophagy via numerous transcription factors and coactivators including PGC-1α and PPARα. Our laboratory has shown that liver-specific deletion of ATGL results in lipid accumulation with a concomitant decrease in SIRT1 activity, PPAR-α/PGC-1 α target gene expression and downstream mitochondrial biogenesis and β-oxidation (Khan et al. 2015; Ong et al. 2011). Consistent with its role as mediator of ATGL signaling, the above data shows that the effects of ATGL on autophagy/lipophagy are completely abrogated following SIRT1 ablation (genetic, shRNA-mediated or chemical). Although the mechanism through which ATGL regulates SIRT1 is still under investigation, the current studies highlight the importance of SIRT1 as a critical signaling node that links ATGL-catalyzed lipolysis to autophagy/lipophagy induction.
In summary, our data suggest that ATGL is not directly responsible for the mass breakdown of LDs in the liver. Rather, ATGL acts as in important signaling node that promotes autophagy/lipophagy, which in turn catalyzes the majority of LD catabolism. This study also highlights the importance of SIRT1 as a key mediator downstream of ATGL that acts to promote autophagy/lipophagy. Overall, these data further clarify the roles of ATGL and autophagy/lipophagy in LD catabolism and provide a new framework in our understanding of hepatic LD catabolism and FA trafficking.
EXPERIMENTAL PROCEDURES
Animals, diets and adenovirus administration
All animal protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee. Eight to ten week old C57Bl/6J male mice were housed and acclimatized as previously described (Ong et al. 2011). Adenoviruses to manipulate ATGL and SIRT1 expression were provided by Drs. Andrew Greenberg and X.C. Dong, respectively, and were administered via the tail vein as previously described (Khan et al. 2015). All mice had free access to water and were fed a purified control diet (TD 94045; Harlan Teklad Premier Laboratory) after adenoviral administration. One week following adenovirus injection, the mice were sacrificed for liver tissue and serum collection after a 16 hour overnight fast (shATGL treatment) or a 4 hour fast (Ad-ATGL treatment). Homozygous SIRT1 floxed mice were purchased from JAX Labs and were cross-bred to Alb-Cre transgenic mice to generate liver specific SIRT1−/− mice.
RNA and protein analysis
mRNA and protein analyses were performed as described previously (Khan et al. 2015). The LAMP1 antibody was obtained from Abcam (catalog #ab24170) and LC3 and ATG5 antibodies were purchased from Novus Biologicals (catalog #NB100-2220 and NB110-53818, respectively). SIRT1, ATGL and p62 antibodies were purchased from Cell Signaling Technology (catalog #8469S, 2439S and 5114S, respectively). The β-actin antibody was obtained from Bio-Rad (catalog #MCA5775GA).
Cell culture, adenoviral infection and radiolabeling
Hepatocytes were isolated as described previously (Ong et al. 2011) and were transduced with adenoviruses 4 hours after plating. Experiments measuring lipid incorporation (pulse period) to measure TAG turnover or FA oxidation were performed in M199 media (Invitrogen) prepared as described previously (Khan et al. 2015). The following morning the cells were pulsed with 500 μM oleate and trace [1-14C]oleate for 2 hours. Some cells were harvested at the end of the pulse period to measure radiolabel incorporation into cellular lipid fractions. Unless noted otherwise, the remaining cells were washed with PBS and media were replaced with fresh complete M199 (as described above) lacking insulin and serum for an additional 6–8 hours (chase period) followed by collection of media and cells for lipid extraction. FAs oxidized during the chase period are expressed as a percentage of the pulse [14C]TAG. Lipids were extracted and separated into different fractions by thin layer chromatography and analyzed as described previously (Ong et al. 2014).
Lipid droplet isolation
Intact LDs were isolated from livers harboring adenoviruses against scrambled control or ATGL knockdown as described previously (Storey et al. 2011) and quantified for protein levels of LC3 and PLIN2.
Chemical reagents/vectors
The ATGL-specific inhibitor ATGListatin was purchased from Xcess Biosciences (catalog #MC0150-2s) and used in cells at 30 μM for 4 hours (for imaging studies) and up to 36 hours for the gene expression studies. Chloroquine was purchased from Sigma-Aldrich (catalog #c6628). All autophagy inhibitors were used at the specified concentrations for the duration of the chase. siATG5 was custom synthesized from Qiagen. The dual reporter plasmid RFP-GFP-LC3 was a kind donation from Dr. Mark McNiven (Mayo clinic). All vectors were administered using Qiagen’s Effectene Transfection Reagent. LAListat was a kind donation from Dr. Paul Helquist (Notre Dame University).
Confocal imaging
For immunofluorescence microscopy, cells were grown on coverslips and fixed with 4% paraformaldehyde for 30 minutes and blocked with 1% BSA, 10% donkey serum in PBS. Incubation with primary (overnight at 4°C) and secondary (1 hour at room temperature) antibodies was done in the above-mentioned blocking buffer. The secondary antibody conjugated to Alexa Fluor 488 or Fluor 567 was purchased from Invitrogen. For LD staining, cells were incubated with LipidTOX™ Deep Red or Green Neutral Lipid Stain (1 μM; Invitrogen) for 30 minutes at 37°C after fixatio n. For lysosomal staining, cells were incubated with 20 nM Lysotracker Red DND-99 (Invitrogen) for 30 minutes at 37°C before fixation. Nuclei were stained with DAPI (4′, 6-diamidino-2-phenylindole) for 10 minutes followed by mounting onto slides for visualization. All images were acquired with a Nikon A1 Spectral Confocal Microscope (Nikon Ti2000E inverted fluorescence microscope with DIC optics) with a 60X oil objective and 0.6 numerical aperture, and prepared using ImageJ (NIH). Images from 5 different fields per well were captured, and experiments were performed in triplicate.
For imaging tissues, paraffin blocks were generated from formaldehyde fixed livers. Slides from these blocks were de-paraffinized using xylene and ethanol washes alternatively. This was followed by an antigen retrieval step which included boiling the slides in 10 mM sodium citrate buffer with 0.5% Tween-20 and subsequent permeabilization in a buffer containing 0.2% Triton X-100 in PBS. Finally, the slides were blocked using 10% donkey serum in 1% BSA for 1 hour at room temperature followed by overnight incubation with primary antibodies at a dilution of 1:100. The following day, slides were washed and incubated with fluorophore-conjugated secondary antibodies at 1:100 dilutions for 1 hour at room temperature, followed by addition of DAPI and subsequent mounting with coverslips.
Statistical analysis
Statistical comparisons were made using analysis of variance or student t-test. All data are presented as means ± SEM and statistical significance was declared at P<0.05.
Acknowledgments
The authors declare no conflict of interest regarding the publication of this paper. This work was supported by grants from the National Institutes of Health (DK090364) to D.G. M. and the Minnesota Obesity Center (NIH DK050456). The authors express their gratitude to Charles Najt for his expertise with lipid droplet isolations as well as Codruta Vizoli and Mary McCourt for their excellent technical support. They also extend their gratitude to the staff at the University of Minnesota’s Imaging Center, in particular Grant Barthel and Alexander Cramer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
Conceptualization, A.S and D.G.M; Methodology, A.S; Investigation, A.S, M.T.M; Validation, A.S; Writing-Original Draft, A.S; Writing-Review & Editing, A.S. and D.G.M; Visualization, A.S; Project and Funding Acquisition, D.G.M; Supervision, D.G.M.
LITERATURE CITED
- Ahmadian M, et al. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes. 2009;58(4):855–66. doi: 10.2337/db08-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science (New York, NY) 2004;303(5666):2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Burke JA, Schubert WK. Deficient activity of hepatic acid lipase in cholesterol ester storage disease. Science (New York, NY) 1972;176(4032):309–10. doi: 10.1126/science.176.4032.309. [DOI] [PubMed] [Google Scholar]
- Cairó M, et al. Thermogenic activation represses autophagy in brown adipose tissue. International journal of obesity (2005) 2016;40(10):1591–1599. doi: 10.1038/ijo.2016.115. [DOI] [PubMed] [Google Scholar]
- Cingolani F, Czaja MJ. Regulation and Functions of Autophagic Lipolysis. Trends in endocrinology and metabolism: TEM. 2016;27(10):696–705. doi: 10.1016/j.tem.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell research. 2014;24(1):92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Current biology: CB. 2014;24(6):609–20. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AS, et al. The role of lipid droplets in metabolic disease in rodents and humans. The Journal of clinical investigation. 2011;121(6):2102–10. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nature medicine. 2011;17(9):1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms A, et al. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nature communications. 2015;6:7176. doi: 10.1038/ncomms8176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell metabolism. 2011;14(2):173–83. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nature cell biology. 2015;17(6):759–70. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper JK, et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell metabolism. 2009;10(5):392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, et al. ATGL-catalyzed lipolysis regulates SIRT1 to control PGC-1α/PPAR-α signaling. Diabetes. 2015;64(2):418–26. doi: 10.2337/db14-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3(5):452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(9):3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516(7529):112–5. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, et al. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell metabolism. 2016;23(1):113–27. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KT, et al. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology (Baltimore, Md) 2011;53(1):116–26. doi: 10.1002/hep.24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KT, et al. Hepatic ATGL mediates PPAR-α signaling and fatty acid channeling through an L-FABP independent mechanism. Journal of lipid research. 2014;55(5):808–15. doi: 10.1194/jlr.M039867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M, et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell metabolism. 2011;13(6):655–67. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson GL, et al. Lysosomal acid lipase and lipophagy are constitutive negative regulators of glucose-stimulated insulin secretion from pancreatic beta cells. Diabetologia. 2014;57(1):129–39. doi: 10.1007/s00125-013-3083-x. [DOI] [PubMed] [Google Scholar]
- Pu J, et al. Interactomic study on interaction between lipid droplets and mitochondria. Protein & Cell. 2011;2(6):487–496. doi: 10.1007/s13238-011-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, et al. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through Creb deacetylation. Cell metabolism. 2011;14(6):758–67. doi: 10.1016/j.cmet.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Developmental cell. 2015;32(6):678–92. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santambrogio L, Cuervo AM. Chasing the elusive mammalian microautophagy. Autophagy. 2011;7(6):652–4. doi: 10.4161/auto.7.6.15287. [DOI] [PubMed] [Google Scholar]
- Sapiro JM, et al. Hepatic triacylglycerol hydrolysis regulates peroxisome proliferator-activated receptor alpha activity. Journal of lipid research. 2009;50(8):1621–9. doi: 10.1194/jlr.M800614-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B, et al. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology (Baltimore, Md) 2015;61(6):1896–907. doi: 10.1002/hep.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nature cell biology. 2013;15(6):647–58. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey SM, et al. The phospholipid monolayer associated with perilipin-enriched lipid droplets is a highly organized rigid membrane structure. American journal of physiology Endocrinology and metabolism. 2011;301(5):E991–E1003. doi: 10.1152/ajpendo.00109.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikita S, et al. Fiber type conversion by PGC-1α activates lysosomal and autophagosomal biogenesis in both unaffected and Pompe skeletal muscle. PloS one. 2010;5(12):e15239. doi: 10.1371/journal.pone.0015239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin SM, et al. Adipose triacylglycerol lipase is a major regulator of hepatic lipid metabolism but not insulin sensitivity in mice. Diabetologia. 2011;54(1):146–56. doi: 10.1007/s00125-010-1895-5. [DOI] [PubMed] [Google Scholar]
- Wang RH, et al. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. The Journal of clinical investigation. 2011;121(11):4477–90. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Li C, Deng CX. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. International journal of biological sciences. 2010;6(7):682–90. doi: 10.7150/ijbs.6.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, et al. Fasting energy homeostasis in mice with adipose deficiency of desnutrin/adipose triglyceride lipase. Endocrinology. 2012;153(5):2198–207. doi: 10.1210/en.2011-1518. [DOI] [PubMed] [Google Scholar]
- Xiong X, et al. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. The Journal of biological chemistry. 2012;287(46):39107–14. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell metabolism. 2010;11(6):467–78. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


