Abstract
Recent evidence suggested an important role of matrix metalloproteinases 16 (MMP16) in the progression of several cancers. However, the contribution of MMP16 to colorectal cancer (CRC) remains elusive. In this study, we combined analyzed the MMP16 expression in The Cancer Genome Atlas (TCGA), GSE39582 database and in-house database. In TCGA and GSE39584 database, the log-rank test demonstrated that overall survival (OS) for patients with low MMP16 expression in tumor tissues was significantly higher than those with high expression (P < 0.05). In the validation cohort, high MMP16 expression was significantly correlated with N stage (P = 0.008) and lymphovascular invasion (P = 0.002). The 5-year OS and disease free survival (DFS) in high and low MMP16 expression groups were 66.0% and 80.6%, 54.3% and 72.8%, respectively. Univariate and multivariate analysis showed that high MMP16 expression was an independently prognosis factor for both OS and DFS (P < 0.05). Functional study found that silencing MMP16 expression could inhibit migration and invasion of colon cancer cells. In conclusion, high expression of MMP16 is associated with the aggressive malignant behavior and poor survival outcome of CRC patients. MMP16 can serve as an indicator of prognosis as well as a potential novel target for treatment of CRC patients.
Globally, colorectal cancer (CRC) is the third most frequently diagnosed cancer and one of the leading causes of cancer deaths1. Its incidence has been increasing in China in recent years. Although most patients at early stage can be successfully cured with surgery, about 20–45% of patients who underwent curative resection developed recurrence or metastasis2,3. Currently, tumor-node-metastasis (TNM) stage is the most powerful predictor for survival in CRC. However, the survival outcome is quite different even for patients at same TNM stage. Further understanding on the biological mechanisms of the metastasis and progression of CRC and developing effective measures to target this process are of vital importance. Much attention has been focused on the molecular-based prognostic markers, which are complementary to the data obtained by pathological diagnosis and can be used to give more information for clinical practice4,5,6.
Matrix metalloproteinases (MMPs) are a family of zinc dependent proteases capable of degrading most extracellular matrices (ECMs). MMPs participate in many physical and pathological processes such as morphogenesis, wound healing, tissue repair, and remodeling7. In addition, MMPs play a critical role in tumor progression through ECM turnover and cancer-cell migration, as well as regulating signaling pathways that control cell growth, inflammation, or angiogenesis8,9. As an important member of the MMPs family, MMP16 can also exhibit proteolytic activity against components of the extracellular matrix. MMP16 is frequently overexpressed in various human cancer tissues and help facilitate cancer metastasis and progression10,11,12,13. However, the clinical significance of MMP16 expression in CRC has rarely been investigated until now.
In the present study, we analyzed the MMP16 expression levels in public available databases, The Cancer Genome Atlas (TCGA) and GSE39582 in Gene Expression Omnibus (GEO), and then validated the results in in-house database to evaluate the correlations between the MMP16 level and clinicopathological features and survival outcomes. Functional studies were also conducted to figure out the role of MMP16 in oncogenesis.
Results
MMP16 expression in TCGA and GSE39582 database
A total of 579 eligible patients with CRC met the selection criteria in TCGA database, including 316 males and 263 females. The median age for all patients was 66 years (rang 31–90 years old). 987.9% (509/579) patients were at M0 stage. The median length of follow-up was 25 months (range, 0–142 months) and 123 (21.2%) patients had died at the end of follow-up. Table 1 showed the baseline characteristics of the two study cohorts.
Table 1. Clinical characteristics of patients with colorectal cancer in the TCGA, GSE39582 and validation cohort.
| Variable | TCGA | GSE39582 | Validation Cohort | |||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| Sex | ||||||
| Male | 316 | 45.4 | 213 | 44.9 | 99 | 51.6 |
| Female | 263 | 54.6 | 261 | 55.1 | 93 | 48.4 |
| Age | 66 | 31–90 | 67 | 22–97 | 66 | 22–85 |
| Grade | ||||||
| G1 | / | / | / | / | 88 | 45.8 |
| G2 | / | / | / | / | 77 | 40.1 |
| G3 | / | / | / | / | 27 | 14.1 |
| T stage | ||||||
| T1/T2 | 115 | 19.9 | 52 | 11.0 | 46 | 24.0 |
| T3/T4 | 462 | 79.8 | 418 | 88.2 | 146 | 76.0 |
| TX | 2 | 0.3 | 4 | 0.8 | / | / |
| N stage | ||||||
| N0 | 323 | 55.8 | 283 | 59.7 | 110 | 57.3 |
| N1 | 144 | 24.9 | 108 | 22.8 | 55 | 28.6 |
| N2 | 108 | 18.7 | 78 | 16.5 | 27 | 14.1 |
| Nx | 4 | 0.7 | 1 | 1.1 | / | / |
| M stage | ||||||
| M0 | 426 | 73.6 | 474 | 100 | 192 | 100 |
| M1 | 83 | 14.3 | / | / | / | / |
| Mx | 70 | 12.1 | / | / | / | / |
| Lymphovascular invasion | ||||||
| Negative | 318 | 54.9 | / | / | 168 | 87.5 |
| Positive | 201 | 34.7 | / | / | 24 | 12.5 |
| Unknown | 60 | 10.4 | / | / | / | / |
| Perineural invasion | ||||||
| Negative | / | / | / | / | 164 | 85.4 |
| Positive | / | / | / | / | 28 | 14.6 |
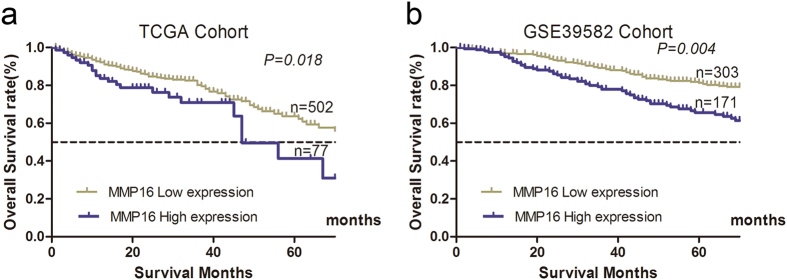
We then divided the patients in TCGA cohort into low or high risk subgroups according to the optimal cutoff value determined by ROC curve in terms of MMP16 expression levels. The log-rank test demonstrated that OS for patients with low MMP16 expression in tumor tissue was significantly higher than those in high group (P = 0.018; Fig. 1a). Then, we validated the results in GSE39582 database, the MMP16 was further confirmed as prognostic factor (P = 0.004, Fig. 1b).
Figure 1. Increased MMP16 expression was significantly associated with the overall survival of CRC patients in TCGA and GSE39582 database.
The data were analyzed using Kaplan-Meier survival analysis between patients with high MMP16 expression and low MMP16 expression in TCGA (a) and GSE39582 database.
Validation of MMP16 expression in in-house database
There were 192 eligible patients in the validation database, including 99 (51.6%) males and 93 (48.4%) females. The median follow-up period was 61 (12–89) months. Patient demographics and pathological features are summarized in Table 1.
We first studied MMP16 mRNA expression in 20 paired cases. As anticipated, the MMP16 mRNA expression levels in cancer tissues were significantly higher than their paired adjacent normal mucosa (P < 0.001, Fig. 2a). Then, we test MMP16 expression in 4 paired cancer tissues and their normal tissues by western blot, the results showed that there were higher MMP16 in cancer tissues than their controls’ (Fig. 2b). We further studied MMP16 mRNA and protein expression in 10 CRC tissues and found the MMP16 mRNA expressions were consisted with their protein expression levels (Data not shown).
Figure 2. Expression of MMP16 mRNA in colon cancer tumors and adjacent normal mucosa.
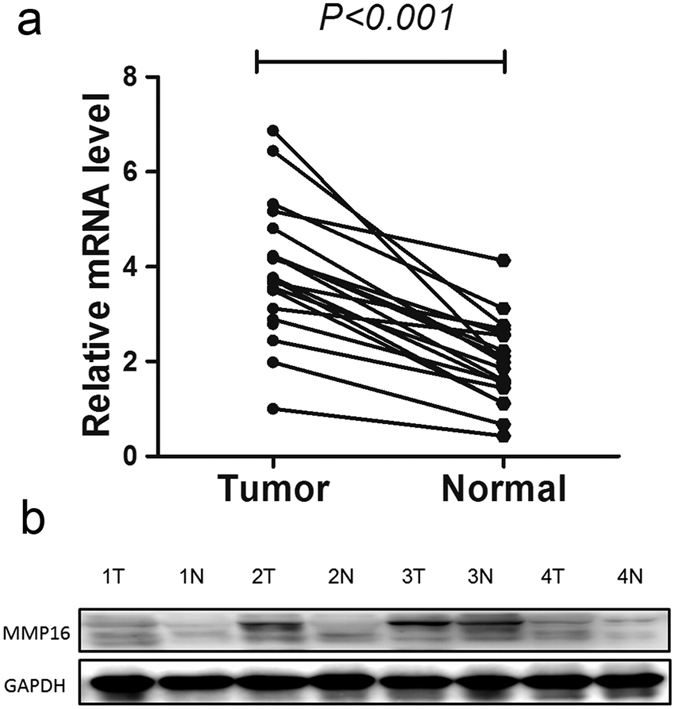
(a) Relative MMP16 mRNA levels in 20 matched colorectal tumors compared with the levels in normal mucosa specimens. The relative RQ value is used to represent the fold change in quantitative real-time polymerase chain reaction detection. (b) Evaluation of MMP16 in four paired cancer tissues and their normal control by western blot. The results showed that there were higher MMP16 expression in cancer tissues than their controls’.
Then, as mentioned previously, we divided patients into high and low MMP16 expression subgroups according to median MMP16 expression value. High MMP16 expression was significantly correlated with N stage (P = 0.008) and lymphovascular invasion (P = 0.002) (Table 2).
Table 2. Association between MMP16 expression and clinic pathological factors in the validation cohort.
| Variable | n | MMP16 Expression |
χ2 Value | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Gender | 0.021 | 0.885 | |||
| Male | 99 | 50 | 49 | ||
| Female | 93 | 46 | 47 | ||
| Age | 0.209 | 0.647 | |||
| ≦60 | 65 | 31 | 34 | ||
| >60 | 127 | 65 | 62 | ||
| T category | 0.457 | 0.499 | |||
| T1/2 | 46 | 21 | 25 | ||
| T3/4 | 146 | 75 | 71 | ||
| N stage | 9.546 | 0.008 | |||
| N0 | 110 | 64 | 46 | ||
| N1 | 55 | 18 | 37 | ||
| N2 | 27 | 14 | 13 | ||
| Pathological grading | 2.225 | 0.329 | |||
| High | 88 | 39 | 49 | ||
| Moderate | 77 | 43 | 34 | ||
| Poor | 27 | 14 | 13 | ||
| Lymphovascular invasion | 9.733 | 0.002 | |||
| Negative | 168 | 91 | 77 | ||
| Positive | 24 | 5 | 19 | ||
| Perineural invasion | 0.669 | 0.413 | |||
| Negative | 164 | 84 | 80 | ||
| Positive | 28 | 12 | 16 | ||
| Ki67 | 0.637 | 0.425 | |||
| Negative | 55 | 30 | 25 | ||
| Positive | 137 | 66 | 71 | ||
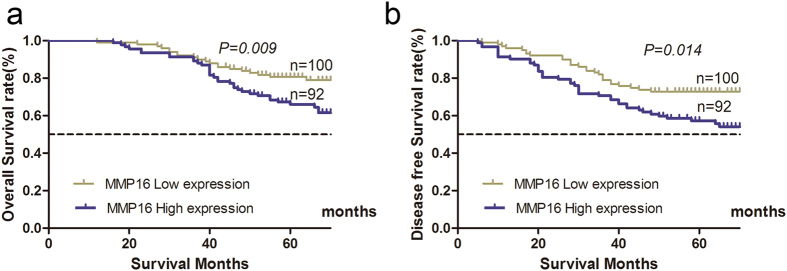
The 5-year OS and DFS in high and low MMP16 groups were 66.0% and 80.6%, 54.3% and 72.8%, respectively, both of which have statistically significant difference (P < 0.05, Fig. 3a,b).
Figure 3.
Influence of MMP16 expression patterns on overall survival (a) and disease free survival (b) by Kaplan-Meier analyses in the validation cohort.
In a standardized way using Cox regression model, all factors that were statistically significant in the univariate were tested in multivariate Cox regression analysis for association with OS and DFS. Multivariate analysis demonstrated that high MMP16 expression level, poor tumor grade, and advanced T and N stage were independently associated with both OS and DFS (P < 0.05) (Tables 3 and 4).
Table 3. Univariate and multivariate Cox proportional hazards analysis of MMP16 expression and overall survival for patients with colorectal cancer in the validation cohort.
| Factor | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Gender | 0.677 (0.403–1.136) | 0.139 | ||
| Age | 1.148 (0.659–2.000) | 0.625 | ||
| Grade | 1.909 (1.356–2.687) | <0.001 | 1.518 (1.045–2.205) | 0.028 |
| T category | 7.152 (2.237–22.863) | 0.001 | 4.273 (1.305–13.992) | 0.016 |
| N stage | 3.765 (2.677–5.293) | <0.001 | 3.114 (2.069–4.685) | <0.001 |
| Lymphovascular invasion | 2.193 (1.182–4.069) | 0.013 | 1.109 (0.585–2.103) | 0.750 |
| Perineural invasion | 2.390 (1.329–4.299) | 0.004 | 0.781 (0.406–1.503) | 0.459 |
| Tumor location | 0.837 (0.491–1.427) | 0.514 | ||
| MMP16 | 1.992 (1.168–3.395) | 0.011 | 1.938 (1.129–3.372) | 0.038 |
Abbreviation: CI, confidence interval; HR, hazard ratio.
Bold type indicates statistical significance.
Table 4. Univariate and multivariate Cox proportional hazards analysis of MMP16 expression and disease free survival for patients with colorectal cancer in the validation cohort.
| Factor | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Gender | 0.787 (0.488–1.268) | 0.325 | ||
| Age | 1.065 (0.641–1.770) | 0.809 | ||
| T category | 4.054 (1.752–9.377) | 0.001 | 2.736 (1.155–6.483) | 0.022 |
| N stage | 3.259 (2.377–4.469) | <0.001 | 2.711 (1.881–3.907) | <0.001 |
| Grade | 1.941 (1.411–2.671) | <0.001 | 1.650 (1.171–2.325) | 0.004 |
| Lymphovascular invasion | 2.165 (1.202–3.900) | 0.010 | 1.139 (0.616–2.106) | 0.677 |
| Perineural invasion | 1.969 (1.108–3.491) | 0.021 | 0.771 (0.412–1.443) | 0.416 |
| Tumor location | 0.726 (0.439–1.200) | 0.211 | ||
| MMP16 | 1.818 (1.118–2.955) | 0.022 | 1.839 (1.122–3.016) | 0.023 |
Abbreviation: CI, confidence interval; HR, hazard ratio.
Bold type indicates statistical significance.
Silencing of MMP16 expression inhibits the migration and invasion of human colon cancer cells
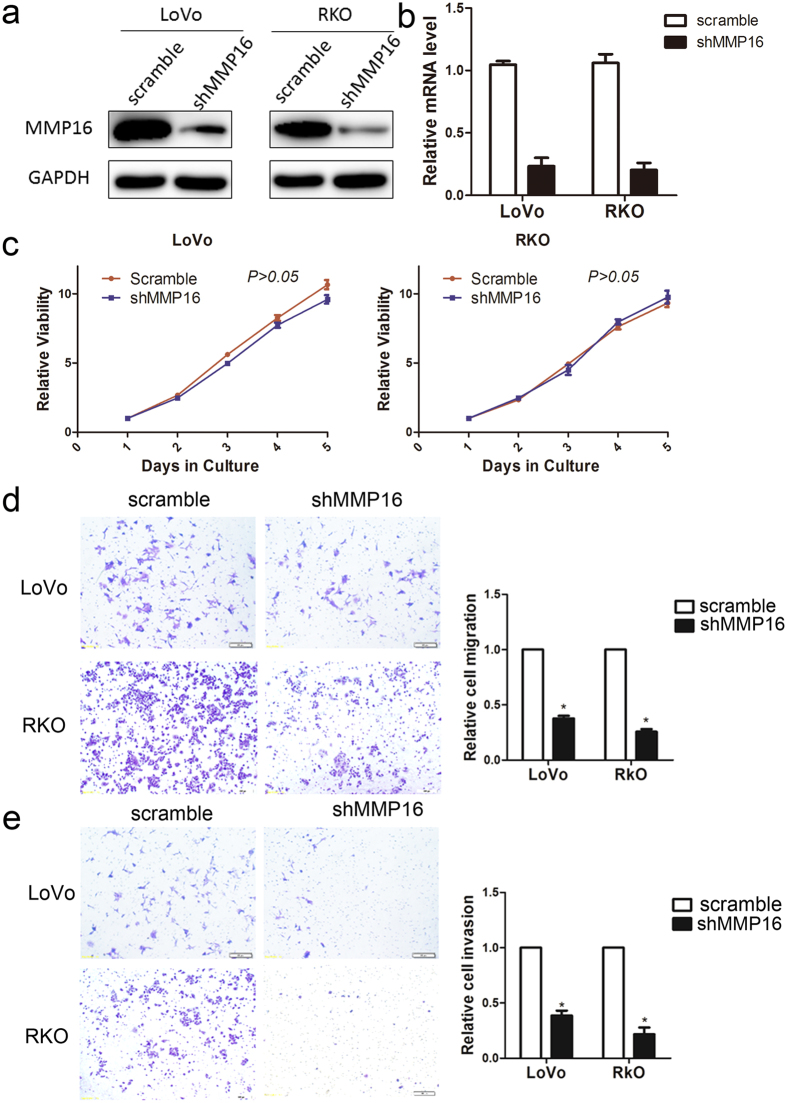
To determine the role of MMP16 in colon cancer cells viability and progression, we used lentivirus-mediated method to establishe stable MMP16-knockdown in LoVo and RKO cells, and the knockdown efficiency were v determined by RT-PCR and western blotting (Fig. 4a,b). CCK8 assay showed that no significantly different cell growth rates between MMP16-knockdown cells and their control cells was found. (P > 0.05, Fig. 4c). The effect of MMP16 on tumor cell migration and invasion were then measured by Transwell analysis without (migration) and with (invasion) matrigel, and the results demonstrated that there were significantly decreased in cell motility and invasion abilities in MMP16 knockdown cells, as compared with control cells. (P < 0.05, Fig. 4d,e).
Figure 4. Influence of MMP16 in colon cancer cell proliferation and invasion.
The MMP16 in LoVo and RKO cells after transfection of shRNA against MMP16 or scramble sequence was analyzed by Western blot (a) and RT-PCR (b). (c) Growth curves of LoVo and RKO cells with transfected shRNAs or scramble sequence. Cell growth was determined by CCK-8. Representative images were shown of migration (d) or invasion (e) of LoVo and RKO cells via transwells without or with matrigel, measured by direct counting of trespassing cells.
Discussion
Local recurrence and distant metastasis are suggested to be the key reasons for poor prognosis and cancer related death in tumor patients. Previous studies have shown that MMP16 is overexpressed in gastric cancer, glioma cancer and melanoma and has implications for tumor invasion and prognosis10,11,12,13. However, little is known regarding its expression pattern and clinical value in CRC. In this study, we first studied MMP16 expression in TCGA database and GSE39582 database, and found that its expression was correlated with poor OS. For TCGA and GSE39582 database lacks some important clinicopathological features (eg. lymphovascular invasion and perineural invasion) and therapy information (eg. radical resection or palliative resection), we then validated clinical value of MMP16 in in-house database and confirmed that high MMP16 expression in CRC was negatively correlated with both OS and DFS. Furthermore, functional study found knockdown of MMP16 expression could inhibit the migration and invasion of colon cancer cells.
MMP16 is one number of the important MMP family. MMP16 functions in activating pro-MMP2 (gelatinase A) into its active form as the zymogen is excreted out of the cell14. Therefore, activating MMP2 would be an indirect mechanism of determining the activity of MMP1610,11. The activated MMP2 can promote the migration and invasion of tumor cells13 by denaturing type IV collagen and partially degrading type I collagen and other ECM proteins in basement membrane10,15,16. Therefore, it is not surprising that high MMP16 expression promoted the invasion and metastasis abilities and led to poor survival outcomes in CRC. In the validation database, we demonstrated that MMP16 expression was significantly correlated with N stage and lymphovascular invasion, both of which were indicated of high invasive abilities of CRC. However, our results seems contradiction with the results from Moon et al. who demonstrated that the MMP16 promoter is frequently hypermethylated in CRC and that downregulation of MMP16 may increase cell migration in CRC17. Our results were first got from public available TCGA database and then validated in in-house database, which made our results more reliable and convincible. Xu et al. also confirmed MMP16 as oncogene in CRC18. MMP16 can promote the invasion and metastasis of melanoma cells by decreasing cell adhesion, inhibiting collagen alignment and inducing lymphatic invasion12. Overexpression of MMP16 can promote migration and invasion of gastric cancer cells and then cause worse long time survival in gastric cancer10. MMP16 is a downstream of β-catenin target gene in human gastric cancer, induction of the MMP16 protein expression is vital to the Wnt-mediated invasion and metastasis in gastric cancer cells12,19, all of which indicated that MMP16 acts as an oncogene by facilitating metastasis in solid tumor.
In summary, we combined analysis the public available database and in-house cohort firmly and demonstrated that overexpression of MMP16 was closely correlated with poor OS and DFS. Therefore, MMP16 can serve as an indicator of prognosis as well as a potential novel target for treatment in CRC patients.
Materials and Methods
Patients in TCGA and GSE39582 database
Gene expression (RNA-Seq) data and corresponding clinical data of CRC samples were retrieved from TCGA database ((https://genome-cancer.ucsc.edu/) and GSE39582 database (https://www.ncbi.nlm.nih.gov/geo/). All patients included in the study should be pathological diagnosed with adenocarcinoma, have no pretreatment, and with intact OS information. Patients who died within one months were excluded from this study. Patients who died with tumor at last follow-up were defined as the clinical endpoint for tumor specific survival. Follow-up was completed on Apr 27, 2016 in TCGA database on Feb 24, 2017 in GSE39582 database.
Patients in the validation database
CRC specimens from patients who underwent intentionally curative surgical resection from January 2004 to December 2009 were obtained to validate the conclusions from TCGA database. Tumor tissues were histopathologically verified as adenocarcinoma and noncancerous tissues were confirmed as negative. Tissue fragments were immediately put in RNA-later and stored at −80 °C. Specimens and data were anonymized, and the need for ethical consent was obtained from the institutional ethics committee of The Affiliated Yancheng Hospital of Southeast University Medical College, Yancheng Third People’s Hospital. The methods were carried out in accordance with the approved guidelines. Written informed consent was obtained from all subjects. Inclusion criteria were patients with pathological confirmed colorectal adenocarcinomas, absence of distant metastasis (M0) at the time of surgery and without neoadjuvant chemotherapies. All patients were restaged according to 7th edition TNM stage system. For OS analysis, patients who died at the last follow-up were defined as clinical endpoints. For analysis of DFS, tumor progression after surgical resection was the clinical endpoint, documented as either tumor recurrence or metastasis. Follow-up data were recorded by phone or medical records.
Real-time PCR
MMP16 mRNA levels were analyzed using a real-time PCR assay. The total RNA from tissues was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and reversely transcripted to cDNA with PrimeScript™ RT Master Mix (Perfect Real Time) kit (RR036A, Takara) based on the manufacturer’s instruction. RT-PCR was performed using the SYBR Green Master Mix (Roche, Mannheim, Germany) on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) in triplicate, and non-template controls were run for each assay under the same conditions. Primers were as follows: GAPDH-F, 5′-GCACCGTCAAGGCTGAGAAC-3′, GAPDH-R, 5′-TGGTGAAGACGCCAGTGGA-3′; MMP16-F, 5′-GGACAGAAATGGCAGCACAAGC-3′, MMP16-R, 5′-CATCAAAGGCACGGCGAATAGC-3′10.
The cycling conditions were as follows: initial denaturation for 10 min at 95 °C followed by 35 cycles of denaturation (15 s at 95 °C), annealing and elongation (30 s at 60 °C). The relative expression of MMP16 was calculated and normalized using the RQ value method relative to GAPDH.
Western blotting
The MMP16 expression was assessed by western blotting analysis and samples were normalized to GAPDH. Total proteins were extracted from the cultured cells solubilized in lysis buffer (RIPA Lysis Buffer, Thermo Scientific Pierce). The protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes (Bio-rad). The membranes were blocked within 5% Bovine Serum Albumin (BSA) at room temperature for 2 h and incubated overnight at 4 °C with primary anti-MMP16 (1:500, Abgent) and anti-GAPDH (1:5000, Santa Cruz), respectively. The membranes subsequently washed and incubated with appropriate secondary antibodies. After being incubated with ECL, the protein bands were visualized.
Cell culture
The human CRC cell lines (LoVo and RKO) were originally purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Invitrogen).
Stable transfection of colon cancer cells
Biologically active short hairpin RNAs (shRNA) were generated using the lentiviral expression vector pLKO.1-puro. The shRNA target sequence for human MMP16 was 5′-CGTGATGTGGATATAACCATT-3′. PLKO.1-scramble shRNA with limited homology with any known sequences in the human was used as a negative control. LoVo and RKO cells were transfected with the pLKO.1-shMMP16 expression vector or pLKO.1-scramble. The cells stably transfected were isolated using puromycin selection to obtain stable MMP16 knockdown cells.
Cell proliferation assays
Cell proliferation Reagent Kit (CCK-8, Dojindo, Japan) was used to assess cell proliferation. Transfected cells were plated in each well of a 96-well plate and assessed every 24 h according to the manufacturer’s instructions. The cell viability of different groups at each measuring time point was compared.
Cell migration and invasion assay
The migration and invasion ability of LoVo and RKO cells after different transfection was measured by Transwell assay (without or with matrigel). Approximately 105 cells were seeded on the upper chamber of the transwell with 200 μl serum-free growth medium (105 cells per well of 8.0 μm Pore Polycarbonate Membrane Insert). Complete medium containing 10% FBS was added to the lower chamber as a chemo-attractant. After 48 h of incubation at 37 °C, non-migratory cells on the upper surface of upper chamber were removed slightly by cotton swabs, and cells that migrated to the bottom of the membrane were fixed and stained. The number of invaded cells was counted under light microscope. To minimize the bias, five randomly selected fields with 200× magnification were counted, then the average number was calculated.
Statistical Analysis
Two-tailed χ2 test was used to evaluate the expression difference between theclinicopathological features and MMP16 expression. The survival curves were estimated by Kaplan-Meier analysis, and P values were calculated by log rank test. Univariate Cox proportional hazards regressions were applied to estimate the individual hazard ratio (HR) for the DFS and OS. The HR with 95% confidence interval (CI) was measured to estimate the hazard risk of individual factors. All experiments were performed independently a minimum of three times. All P values were two-sided, and P < 0.05 was considered statistically significant. Statistical calculations were all performed using SPSS 17.0.
Additional Information
How to cite this article: Wu, S. et al. High expression of matrix metalloproteinases 16 is associated with the aggressive malignant behavior and poor survival outcome in colorectal carcinoma. Sci. Rep. 7, 46531; doi: 10.1038/srep46531 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
The authors acknowledge the efforts of the National Cancer Institute’s Center for Cancer Genomics and the National Human Genome Research Institute in the creation of the TCGA database. We also thanked the public available of GSE39582 database. The authors thank Prof. Shenglin Huang and Weixing Dai at Fudan University Shanghai Cancer Center for technical help in analyzing the TCGA database.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.W. and W.L. conceived of and designed the study. C.M., S.S. and L.Z. performed the analyses. C.M. and S.S. prepared all figures and tables. S.W. and W.L. wrote the main manuscript. All authors reviewed the manuscript.
References
- Siegel R. L., Miller K. D. & Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- Winawer S. et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology 124, 544–560 (2003). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Nomograms for predicting prognostic value of inflammatory biomarkers in colorectal cancer patients after radical resection. International journal of cancer 139, 220–231 (2016). [DOI] [PubMed] [Google Scholar]
- Eschrich S. et al. Molecular staging for survival prediction of colorectal cancer patients. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 23, 3526–3535 (2005). [DOI] [PubMed] [Google Scholar]
- McLeod H. L. & Murray G. I. Tumour markers of prognosis in colorectal cancer. British journal of cancer 79, 191–203 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Overexpression of forkhead Box C2 promotes tumor metastasis and indicates poor prognosis in colon cancer via regulating epithelial-mesenchymal transition. American journal of cancer research 5, 2022–2034 (2015). [PMC free article] [PubMed] [Google Scholar]
- Nagase H., Visse R. & Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovascular research 69, 562–573 (2006). [DOI] [PubMed] [Google Scholar]
- Kessenbrock K., Plaks V. & Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D. W., Zhang Y. Y., Qi Y., Zhou X. T. & Lv G. Y. Prognostic significance of MMP-7 expression in colorectal cancer: a meta-analysis. Cancer epidemiology 39, 135–142 (2015). [DOI] [PubMed] [Google Scholar]
- Cao L. et al. MMP16 is a marker of poor prognosis in gastric cancer promoting proliferation and invasion. Oncotarget (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. miR-132 can inhibit glioma cells invasion and migration by target MMP16 in vitro. OncoTargets and therapy 8, 3211–3218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tatti O. et al. MMP16 Mediates a Proteolytic Switch to Promote Cell-Cell Adhesion, Collagen Alignment, and Lymphatic Invasion in Melanoma. Cancer research 75, 2083–2094 (2015). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16. Cancer letters 339, 260–269 (2013). [DOI] [PubMed] [Google Scholar]
- Nakada M. et al. Expression and tissue localization of membrane-type 1, 2, and 3 matrix metalloproteinases in human astrocytic tumors. The American journal of pathology 154, 417–428 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe R. G. & Weiss S. J. Breaching the basement membrane: who, when and how? Trends in cell biology 18, 560–574 (2008). [DOI] [PubMed] [Google Scholar]
- Arslantas A. et al. Genomic alterations in low-grade, anaplastic astrocytomas and glioblastomas. Pathology oncology research: POR 13, 39–46 (2007). [DOI] [PubMed] [Google Scholar]
- Moon J. W. et al. Promoter hypermethylation of membrane type 3 matrix metalloproteinase is associated with cell migration in colorectal adenocarcinoma. Cancer genetics 208, 261–270 (2015). [DOI] [PubMed] [Google Scholar]
- Xu X. T. et al. MicroRNA expression profiling identifies miR-328 regulates cancer stem cell-like SP cells in colorectal cancer. British journal of cancer 106, 1320–1330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy A. M. et al. beta-Catenin/Wnt signaling regulates expression of the membrane type 3 matrix metalloproteinase in gastric cancer. Cancer research 66, 4734–4741 (2006). [DOI] [PubMed] [Google Scholar]