Abstract
Early life neglect increases risk for the development of psychopathologies during childhood and adulthood, including depression and anxiety disorders. We recently reported epigenetic changes in DNA derived from saliva in three genes predicted depression in a cohort of maltreated children: DNA-binding protein inhibitor ID-3 (ID3), Glutamate NMDA Receptor (GRIN1), and Tubulin Polymerization Promoting Protein (TPPP). To validate the role of these genes in depression risk, secondary analyses were conducted of gene expression data obtained from medial prefrontal cortex (mPFC) tissue of mice subjected to a model of maternal neglect which included maternal separation and early weaning (MSEW). Anxiety and depression-like phenotype data derived using the elevated plus maze (EPM) and forced swimming test (FST), respectively, were also available for secondary analyses. Behavioral tests were conducted in MSEW and control adult male mice when they were between 65 and 80 days old. ID3, GRIN1 and TPPP gene expression in the mPFC were found to significantly predict behavioral differences in the EPM and FST. These results further support the role of these genes in the etiology of depressive and anxiety phenotypes following early life stress.
Keywords: Neglect, Depression, Methylation, DNA-binding protein inhibitor ID-3 (ID3), Glutamate NMDA receptor (GRIN1), Tubulin polymerization promoting protein (TPPP)
1. Introduction
Child maltreatment is highly prevalent worldwide; maternal neglect is the most common form of maltreatment reported in the United States [1]. Children who are victims of neglect often develop depression and anxiety disorders [2,3]. Epigenetic mechanisms, particularly DNA methylation in several candidate genes, have been associated with increased vulnerability for the development of these problems [4]. Specifically, methylation in the serotonin transporter (SLC6A4), brain derived neurotrophic factor (BDNF), glucocorticoid receptor (NR3C1), and FK506 binding protein (FKBP5) genes have been found to predict depression and anxiety in clinical samples [5], with the role of these genes validated in preclinical studies [6,7].
In a genome-wide methylation study, we recently showed that methylation in three genes (DNA Binding Protein Inhibitor ID-3 (ID3); Tubulin Polymerization Promoting Protein (TPPP); and Glutamate Receptor, Ionotropic N-methyl-d-aspartate (NMDA) 1 (GRIN1) [8]) were significant predictors of depression in a sample of maltreated children:. These genes are biologically relevant, involved in stress response, synaptic pasticity, and the development of white matter tracts [8]. While the role of GRIN1 in depression has been shown previously [9], the role of the other two genes implicated, ID3 and TPPP, needs to be further elucidated.
The child study findings were derived using DNA extracted from saliva samples, which may not be the best source to study epigenetic phenomenon in psychiatric disorders. There are two main things to consider: 1) whether changes observed in saliva-derived DNA are relevant in the brain, and 2) whether these epigenetic changes subsequently alter gene expression. Brain tissue is not accessible in studies with child populations, so interest in the utility of peripheral tissue specimens has increased. DNA methylation and gene expression patterns are tissue-specific [10-12], but nonetheless several studies have found that DNA methylation changes correlate between the brain and peripheral tissues, including saliva [13,14].
While human brain tissue is not available, animal models could be used to validate findings obtained from peripheral human samples. Towards this purpose, secondary analyses were conducted on data collected in a study of a mouse model of maternal neglect. In this model, mice are subjected to maternal separation with early weaning (MSEW) [15]. It was previously reported that MSEW mice display mild anxiety-like and depression phenotypes, as characterized by behavior examined using the elevated plus maze and forced swim test, respectively [15]. This paper reports the results of secondary analyses examining the association between anxiety and depression phenotype data and gene expression of ID3, GRIN1, and TPPP in the medial prefrontal cortex (mPFC) of mice subjected to MSEW.
2. Material and methods
2.1. Animals
As detailed elsewhere [15,16] experimentally naive C57 and DBA mice were obtained from Jackson Laboratories (Bar Harbor, ME), and subsequently bred in-house. Entire litters were randomized at birth to control or Maternal Separation with Early Weaning (MSEW) conditions, but only male mice were included in the research. Behavioral data were available from 38 C57 control animals, 35 C57 MSEW animals, 24 DBA control animals, and 29 DBA MSEW animals. From these behaviorally characterized animals, a total of 11 C57 control animals, 11 C57 MSEW animals, 10 DBA control animals, and 10 DBA MSEW animals were included in gene expression microarray analysis. Control animals were left undisturbed and weaned at postnatal day (P) 23. MSEW animals were separated from the dams for 4h per day on P2–P5, and 8h per day on P6–P16, and weaned on P17. The Yale University Institutional Animal Care and Use Committee approved all experimental procedures.
2.2. Behavioral assessments
Male mice were between 65 and 80 days of age during behavioral testing, and scoring of all behavior was completed blind to rearing status. As described previously [15], all mice were administered the elevated plus maze task followed by the forced swim test.
2.2.1. Elevated plus maze (EPM)
Elevated plus maze testing was performed as previously described [15,17]. Mice were given 15minto explore a plus shaped maze, constructed of white Plexiglass according to the dimensions of most commercially available mouse mazes (e.g., San Diego Instruments, Panlab Harvard Apparatus). The maze, positioned 31.5 cm above the floor, contained two open and two closed arms, all 30 cm in length, connected by a 6 cm center square. Closed arms were encased in 28 cm high black walls. The maze was placed inside an opaque testing box (100 cm × 100 cm × 30 cm), which was positioned in the center of a dimly lit room devoid of any obvious visual cues. A video camera placed 3 ft above the maze acquired digital video, which was later viewed by an observer blind to experimental condition. Additionally, video tracking was performed offline using software written by the authors to determine time spent in each arm and speed at each time point. Behavior was scored for total open and closed arm entries (defined as all four limbs entering the arm), and total time spent in the open and closed arms.
2.2.2. Forced swim test
The forced swim test was performed according to published procedures [18,19] with minor modifications. Mice were placed in 4 I glass cylinders (16 cm diameter) filled to a depth of 10 cm with 25 °C water. Each mouse was tested for 15 min and the cylinder was cleaned and filled with fresh water following each test. At completion, mice were removed from the water, dried, placed in holding cages, and put under heat lamps for 30 min before being returned to their home cages. Digital video was acquired from above and later scored in 5-min intervals for 15 min by an observer blind to experimental condition. Behavior was classified as immobile (defined as the absence of movement with exception of what is necessary to keep the animal's head above water), or mobile (defined as active swim if there was movement of all four limbs, or paddling if engaged in low frequency movements involving only one or two limbs). Primary outcome measures included total time mobile and total time immobile in each of the three time periods assessed (i.e., 0–5, 5–10, 10–15 min).
2.3. Tissue collection and RNA extraction
As described previously [20], mice were anesthetized with chloral hydrate (1500mg/kg in sterile saline, IP) and rapidly decapitated. Whole brain was collected and placed in RNAlater (Qiagen) for 24 h, and then stored at −20 °C. Brains were removed from RNAlater, sectioned at 300 μm on a vibratory microtome (Vibratome), and sections were stored in 0.025% methylene blue in RNAlater at −20°C for 48h. Medial prefrontal cortex (mPFC), defined as anterior cingulate, pre-limbic cortex, and infra-limbic cortex, was dissected under a stereomicroscope, placed in RNAlater at 4°C, and processed within 24h. RNA and DNA were extracted using the Qiagen AllPrep mini kit. RNAlater was removed from the samples and 600 μL of buffer RLT (Qiagen) containing β-mercaptoethanol was added. Tissue was sonicated for 5 s at 10% power on ice, prior to RNA extraction. Protein was precipitated from the RNA column flow-through using the 2D Clean-Up Kit (GE) and pellets frozen at −80°C.
2.4. Microarray analysis
As detailed elsewhere [20], total RNA was collected and analyzed on a Bioanalyzer (Agilent). Samples with RNA integrity (RIN)>9 were used for the array analysis. Five hundred ng of RNA from each sample were hybridized to Illumina Mouse WG-6 arrays, whole genome expression arrays that profile more than 45,500 biologically relevant transcripts. The Lumi package in R/Bioconductor was used to perform variance stabilization and quantile normalization. The gene expression analyses examined in this manuscript were limited to ID3, TPPP, and GRIN1.
2.5. Statistics
Multiple linear regression analyses were conducted using the LME package in R. The linear regression model examined the following effects: gene expression (log2 transformed), treatment (control vs. MSEW), strain, treatment × strain, gene expression × treatment, and gene expression × strain. The three-way interaction term was dropped as it was found to be non-significant for all the probes tested.
3. Results
3.1. MSEW gene expression predicts depression and anxiety behavior
As depicted in Table 1, the gene expression data helped explain individual differences in the behavioral outcome measures reported previously [21]. A significant gene effect was found in the EPM with TPPP gene expression associated with greater total time in the closed arms (Beta = 0.0017, p = 0.0228), decreased total time in the open arms (Beta = −0.0021, p = 0.0066), and fewer open arm entries (Beta = −0.0105, p = 0.0112). With regard to the FST, a significant gene effect was found with gene expression of TPPP, GRIN1, and ID3, with mPFC expression of each of these genes associated with one or more of the FST behavioral measures. The results of these analyses are also depicted in Fig. 1.
Table 1.
Examination of mPFC Gene Expression Predictors of Depression and Anxiety Behaviors in Neglected and Control Mice.
| Behavioral Endpoint | Probe | Gene | SS | Df | F | Beta | P |
|---|---|---|---|---|---|---|---|
| EPM: Number of open arm entries | ILMN_2696110 | TPPP | 0.3369 | 1 | 7.1592 | −0.0105 | 0.0112 |
| EPM: Total time on closed arms | ILMN_2696110 | TPPP | 0.2356 | 1 | 5.6595 | 0.0017 | 0.0228 |
| EPM: Total time on open arms | ILMN_2696110 | TPPP | 0.3639 | 1 | 8.3025 | −0.0021 | 0.0066 |
| FST: Total time immobile 0–5 min | ILMN_1216667 | GRIN1 | 0.0384 | 1 | 4.6504 | −0.0018 | 0.0378 |
| FST: Total time mobile 0–5 min | ILMN_2687169 | ID3 | 0.0436 | 1 | 4.6629 | −0.0009 | 0.0376 |
| FST: Total time immobile 10–15 min | ILMN_1216667 | GRIN1 | 0.0729 | 1 | 10.4221 | 0.0084 | 0.0027 |
| FST: Mean time per immobile episode | ILMN_1253164 | GRIN1 | 0.0300 | 1 | 5.2240 | −0.0043 | 0.0283 |
| FST: Number of episodes of paddling | ILMN_2696110 | TPPP | 0.4180 | 1 | 9.9525 | v0.0158 | 0.0137 |
| FST: Number of mobile episodes | ILMN_2696110 | TPPP | 0.3286 | 1 | 6.7183 | −0.0158 | 0.0137 |
Legend. TPPP gene expression predicted anxiety-like behavior on the EPM task, and GRIN1, ID3, and TPPP predicted depression-like behavior on the FST. Note: EPM = Elevated Plus Maze; FST = Forced Swim Test.
Fig. 1.
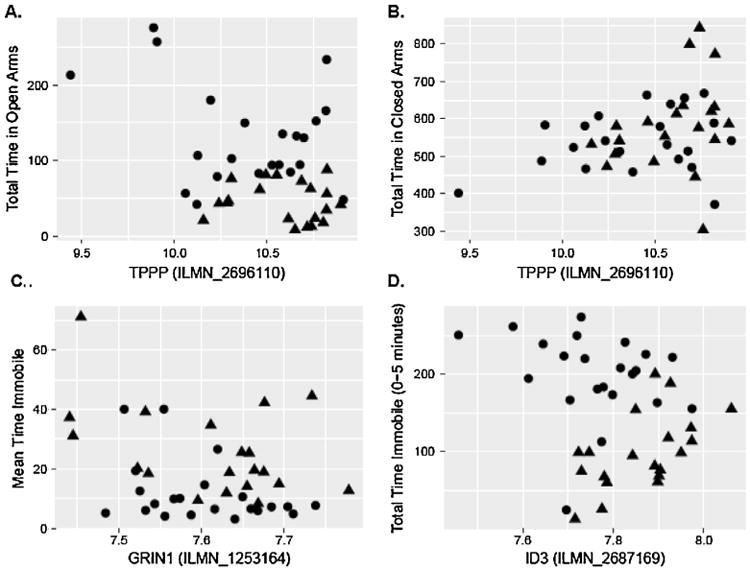
Association Between mPFC Gene Expression and Behavioral Data in Mice. Legend.
In mice, [A] increased TPPP gene expression in the mPFC was associated with reduced time in the open arm of the EPM and [B] increased time in the closed arm of the EPM. [C] Reduced GRIN1 expression was associated with more immobilization in the FST; and [D] Increased ID3 expression was associated with less immobilization in the FST. Code: EPM = Elevated Plus Maze; FST = Forced Swim Test; mPFC = medial prefrontal cortex. Circles represent C57 and the diamonds represent DBA mice.
4. Discussion
4.1. Convergent findings in a mouse model
This study examined GRIN1, IP3 and TPPP genes, previously reported to be associated with depression in maltreated children, in an animal model of maternal neglect. The data show that mice exposed to maternal neglect exhibit anxiety and depressive-like behavior, which are associated with gene expression changes in these three genes within the mPFC.
The medial prefrontal cortex (mPFC) is a forebrain structure known to play a role in cognitive and emotional processes [22] and to regulate the stress response [23]. Clinical studies have consistently shown reduced gray matter volume, and decreased synapses and glial cells in the mPFC of patients with anxiety and mood disorders [24,25]. In addition, preclinical studies have shown that early life stress induces changes in gene expression and structural abnormalities in the mPFC [26], as well as alterations in DNA methylation in the Brain-derived neurotrophic factor (Bdnf) gene, reelin gene, and genes involved in the DNA methylation regulation in the mPFC [26]. Here, we found that decreased ID3 and increased GRIN1 mRNA expression in the mPFC are associated with lower depression-like behavior as evaluated in the forced swim test. We also found that increased TPPP is associated with increased anxiety-like behavior ascertained via the elevated plus maze. Although the role of GRIN1 in depression has been suggested previously [27], these findings further support a role for ID3 and TPPP in the pathogenesis of depression following early life stress.
The available data, however, do not allow us to determine if the depression and anxiety phenotypes observed in children who are victims of neglect and animals subjected to MSEW are associated with ID3, GRIN1, and TPPP gene expression changes in the same direction. In our child epigenome study, we found that decreased methylation at CpG sites in ID3, TPPP, and GRIN1 were associated with higher depression in maltreated children [8]. These CpG sites, however, were located on the gene bodies, where the effects of methylation are variable. Methylation on the gene body may be associated with either increased or decreased gene expression, depending on the tissue and cell type where the methylation occurs [28]. Since gene expression data are not available for the clinical cohort, this study is limited by its inability to compare the direction of gene expression changes and methylation findings across species. Nevertheless, this study further validates the role of these genes in the etiology of depressive and anxiety-like behaviors following experiences of early life stress.
Overall, the present study provides additional support for roles for ID3, TPPP, and GRIN1 in the etiology of depression following early adversity. Ongoing iterative clinical and translational studies will help to further elucidate the mechanisms by which early life stress confers risk for depression and other adverse consequences later in life.
Highlights.
mPFC gene expression examined in mouse model of maternal neglect. (Characters with spaces: 67).
Stress (ID3), synaptic plasticity (GRIN1) and myelin related (TPPP) genes explored. (Characters with spaces: 85).
Genes selected to validate results obtained from peripheral samples in children. (Characters with spaces: 82).
ID3, GRIN1 and TPPP expression predict depression and anxiety behaviors in mice. (Characters with spaces: 82).
Acknowledgments
This work was supported by the NIH R01MH098073 (JK); the National Center for Posttraumatic Stress Disorder-Veterans Affairs Connecticut (JG,JK); the VA Cooperative Study #575B, Genomics of Posttraumatic Stress Disorder in Veterans (JG, JK) and the Biological Sciences Training Program through Grant Number 5T32 MH14276 (JLMO).
References
- 1.DHHS. The AFCARS Report, in Adoption and Foster Care Analysis and Reporting System (AFCARS) FY 2013 data. U.S. Department of Health and Human Services, Administration for Children and Families, Administration on Children, Youth and Families, Children's Bureau, programs/cb; 2014. [Google Scholar]
- 2.Bhatia SK, Bhatia SC. Childhood and adolescent depression. Am Fam Physician. 2007;75(1):73–80. [PubMed] [Google Scholar]
- 3.Widom CS, DuMont K, Czaja SJ. A prospective investigation of major depressive disorder and comorbidity in abused and neglected children grown up. Arch Gen Psychiatry. 2007;64(1):49–56. doi: 10.1001/archpsyc.64.1.49. [DOI] [PubMed] [Google Scholar]
- 4.Yang BZ, et al. Child abuse and epigenetic mechanisms of disease risk. Am J Prev Med. 2013;44(2):101–107. doi: 10.1016/j.amepre.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagot RC, et al. Epigenetic signaling in psychiatric disorders: stress and depression. Dialogues Clin Neurosci. 2014;16(3):281–295. doi: 10.31887/DCNS.2014.16.3/rbagot. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kundakovic M, et al. DNA methylation of BDNF as a biomarker of early-life adversity. Proc Natl Acad Sci U S A. 2015;112(22):6807–6813. doi: 10.1073/pnas.1408355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klengel T, et al. The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology. 2014;80:115–132. doi: 10.1016/j.neuropharm.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Weder N, et al. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J Am Acad Child Adolesc Psychiatry. 2014;53(4):417–424 (e5). doi: 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanacora G, Treccani G, Popoli M. Towards a glutamate hypothesis of depression: an emerging frontier of neuropsychopharmacology for mood disorders. Neuropharmacology. 2011;3:p3. doi: 10.1016/j.neuropharm.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith AK, et al. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(6):700–708. doi: 10.1002/ajmg.b.31212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta D, et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110(20):8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong CC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2014;19(4):495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(3):261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- 14.Smith AK, et al. DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(1):36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George ED, et al. Maternal separation with early weaning: a novel mouse model of early life neglect. BMC Neurosci. 2010;11:123. doi: 10.1186/1471-2202-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bordner KA, et al. Functional genomic and proteomic analysis reveals disruption of myelin-related genes and translation in a mouse model of early life neglect. Front Psychiatry. 2011;2:18. doi: 10.3389/fpsyt.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simen BB, et al. TNFalpha signaling in depression and anxiety: behavioral consequences of individual receptor targeting. Biol Psychiatry. 2006;59(9):775–785. doi: 10.1016/j.biopsych.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Porsolt RD, et al. Behavioural despair in rats: a new model sensitive to antidepressant treatments. Eur J Pharmacol. 1978;47(4):379–391. doi: 10.1016/0014-2999(78)90118-8. [DOI] [PubMed] [Google Scholar]
- 19.Porsolt RD, Bertin A, Jalfre M. Behavioral despair in mice: a primary screening test for antidepressants. Arch Int Pharmacodyn Ther. 1977;229(2):327–336. [PubMed] [Google Scholar]
- 20.Bordner KA, et al. Functional genomic and proteomic analysis reveals disruption of myelin-related genes and translation in a mouse model of early life neglect. Front Psychiatry. 2011;2(18):18. doi: 10.3389/fpsyt.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George ED, et al. Maternal separation with early weaning: a novel mouse model of early life neglect. BMC Neurosci. 2010;11(123):123. doi: 10.1186/1471-2202-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heidbreder CA, Groenewegen HJ. The medial prefrontal cortex in the rat: evidence for a dorso-ventral distinction based upon functional and anatomical characteristics. Neurosci Biobehav Rev. 2003;27(6):555–579. doi: 10.1016/j.neubiorev.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Diorio D, Viau V, Meaney MJ. The role of the medial prefrontal cortex (cingulate gyrus) in the regulation of hypothalamic-pituitary-adrenal responses to stress. J Neurosci. 1993;13(9):3839–3847. doi: 10.1523/JNEUROSCI.13-09-03839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drevets WC, Savitz J, Trimble M. The subgenual anterior cingulate cortex in mood disorders. CNS Spectr. 2008;13(8):663–681. doi: 10.1017/s1092852900013754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chocyk A, et al. Early-life stress affects the structural and functional plasticity of the medial prefrontal cortex in adolescent rats. Eur J Neurosci. 2013;38(1):2089–2107. doi: 10.1111/ejn.12208. [DOI] [PubMed] [Google Scholar]
- 26.Montalvo-Ortiz JL, et al. RDoC and translational perspectives on the genetics of trauma-related psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2016;171(1):81–91. doi: 10.1002/ajmg.b.32395. http://dx.doi.org/10.1002/ajmg.b.32395 (Epub 2015 Nov 22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tordera RM, et al. Chronic stress and impaired glutamate function elicit a depressive-like phenotype and common changes in gene expression in the mouse frontal cortex. Eur Neuropsychopharmacol. 2011;21(1):23–32. doi: 10.1016/j.euroneuro.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 28.Xu H, et al. Sex-biased methylome and transcriptome in human prefrontal cortex. Hum Mol Genet. 2014;23(5):1260–1270. doi: 10.1093/hmg/ddt516. [DOI] [PMC free article] [PubMed] [Google Scholar]


