Abstract
Acute undifferentiated febrile illnesses (AFIs) represent a significant health burden worldwide. AFIs can be caused by infection with a number of different pathogens including dengue (DENV) and Chikungunya viruses (CHIKV), and their differential diagnosis is critical to the proper patient management. While rapid diagnostic tests (RDTs) for the detection of IgG/IgM against a single pathogen have played a significant role in enabling the rapid diagnosis in the point-of-care settings, the state-of-the-art assay scheme is incompatible with the multiplex detection of IgG/IgM to more than one pathogen. In this paper, we present a novel assay scheme that uses two-color latex labels for rapid multiplex detection of IgG/IgM. Adapting this assay scheme, we show that 4-plex detection of the IgG/IgM antibodies to DENV and CHIKV is possible in 10 min by using it to correctly identify 12 different diagnostic scenarios. We also show that blue, mixed, and red colorimetric signals corresponding to IgG, IgG/IgM, and IgM positive cases, respectively, can be associated with distinct ranges of hue intensities, which could be exploited by analyzer systems in the future for making accurate, automated diagnosis. This represents the first steps toward the development of a single RDT-based system for the differential diagnosis of numerous AFIs of interest.
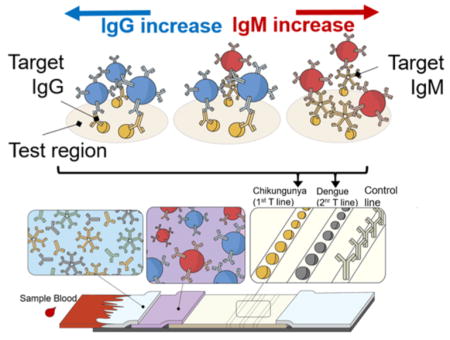
Graphical abstract
Acute undifferentiated febrile illnesses (AFIs) are responsible for substantial morbidity and mortality globally and impose a considerable economic cost, primarily in developing countries.1–3 In order to treat patients of AFIs appropriately and effectively, the AFIs need to be differentially diagnosed by identifying the causative infectious agents, examples of which include Chikungunya (CHIKV) and Dengue (DENV).4,5 CHIKV is a re-emerging mosquito-borne alphavirus responsible for a severe epidemic in countries of the Indian Ocean region with an estimated 7.5 million cases over five years6 and is now also widely prevalent in the Latin American region.7 DENV is the most rapidly spreading mosquito vector disease worldwide, with an estimated 50–100 million new dengue infections occurring every year in over 100 countries,8 and recent estimates suggest that this number may be as high as 390 million.9
In many settings, the initial diagnosis of patients presenting with febrile illness is done with the use of a standard rapid diagnostic test (RDT) based on lateral flow principles. These tests can be easily administered and yield quick results with relatively high sensitivities and specificities.10–13 The AFI diagnosis typically involves operating an antigen-specific test to detect the presence of infectious agents (e.g., nonstructural protein 1 of the DENV) which are present at high concentrations in human blood during the early clinical stage of infection.11 Because these concentrations fall below a detectable range in the later stages of infection, serological tests are necessarily performed in conjugation to the antigen-specific tests in order to make accurate AFI diagnosis. Most of the serological tests rely on detecting the Immunoglobulin M (IgM) antibodies to the infectious agent, which is supplemented by the Immunoglobulin G (IgG) antibody detection for the indication of a past infection.14 For these tests, the current state-of-the-art is a single RDT strip for the duplex IgG/IgM detection using two test lines (one for IgG and one for IgM detection). In such cases, detection is limited to that associated with a single infectious agent, and the detection of IgG/IgM to the other pathogens would require additional pathogen-specific strips to be used in parallel. Such practice of acquiring differential AFI diagnosis has major limitations as each test set would need to be prepared, operated, and interpreted separately by the user. Multiplexing the IgG/IgM detection of several AFIs on a single strip can have an immediate, significant impact by simplifying the operation process of the users, requiring less sample volume, and lowering the cost of the overall tests.15 These benefits could play a major role in facilitating the deployment of the rapid diagnostic tests (RDTs) and allowing much more rapid differential AFI diagnosis.
For the detection of several antigens, as opposed to antibodies, multiplexing on a single strip has previously been achieved by introducing additional detection antibodies of the same label (e.g., gold nanoparticle labeled antibody for new target antigens) and adding the associated test lines that are spatially separated from each other for developing multiple test line signals.16,17 Unfortunately, such an approach cannot be adapted in multiplexing antibody RDTs whose test lines comprised of anti-IgG and anti-IgM and are not specific to a particular pathogen. This represents a challenge for multiplexing because, even if multiple detection probes with different pathogen specificity are used, they would all be captured on a single test line; as such, when a test line signal develops color, the information about the causative pathogen would still be unknown. Therefore, under this format, multiplexing requires the encoding of each detection probe with a different characteristic color (i.e., associating a distinct color with a certain pathogen) and subsequently analyzing the composition of the test region color as recently demonstrated by Yen et al.18 for a similar application in which multicolored silver nano-particles were used to detect several infectious agents. This approach has practical drawbacks including the requirements for distinctly different color labels (n-plex test would require n differently colored probes) and the complexity of analyzing the color development as n becomes large.
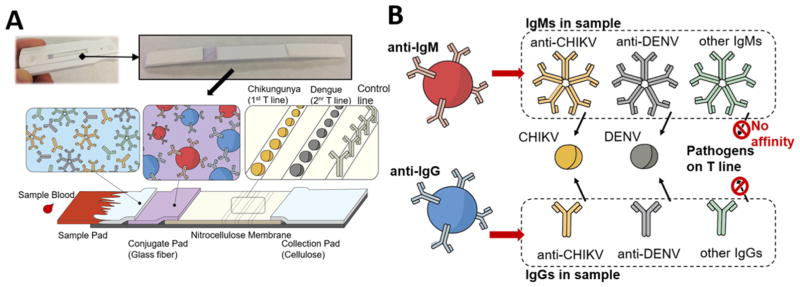
In this work, we present a novel lateral flow assay scheme for multiplexing the detection of several pathogen-specific IgGs/IgMs on a single strip using just two color labels. This method uses blue and red encodings to discriminate between the IgGs and IgMs, while the pathogen specificity is determined by observing the test line location of the color development. The principle of our two-color lateral flow assay is schematically illustrated in Figure 1. The assay consists of: the sample pad for accepting the sample, the conjugate pad for storing the detection probes, the nitrocellulose membrane for immobilizing the test and control region reagents, and the absorbent pad for collecting the waste mixtures (Figure 1a). To operate our test, the sample (which is envisioned to be blood/plasma/serum) and chase buffer solutions are added to the sample pad sequentially as is typically done for other traditional RDTs. Once wetted, the blue and red detection probes that have been dry-stored on the conjugate pad are released and become free to interact with all the sample IgGs and IgMs, respectively, including those associated with the AFIs of interest (Figure 1b). When the sample–probe mixture flows over the test regions, only the IgGs and IgMs to the targeted pathogens are captured at the respective test regions and develop color due to the accompanying blue and red detection probes from the earlier interaction. Further downstream, the control region immobilizes the secondary antibodies (i.e., antigoat IgG) that capture both the blue anti-IgG and red anti-IgM detection probes based on their host species (i.e., goat) and develops mixed colors to confirm that the test has completed successfully. Thus, after checking to find the mixed color development on the control region, the operator can obtain diagnostic AFI information from the two-color assay by (1) observing the color development on the test region where red, blue, and mixed colors indicate the presence of IgGs, IgMs, or both, respectively, and (2) determining the pathogen specificity of the detected IgG/IgM by noting the location of the test region under observation (e.g., two spatially separated test regions associated with DENV and CHIKV are shown as examples in Figure 1b). Here, such multiplexing is enabled by our use of anti-IgG and anti-IgM probes that serve as common agents in the IgG/IgM detection of different AFIs. This signifies that the two-color assay can be further multiplexed simply by introducing additional test regions (i.e., dispensing other recombinant pathogen molecules of interest such as Malaria, Chagas, Leptospirosis, and Typhoid fever) and ensuring that the anti-IgG and anti-IgM probes do not become limited during their interaction with the sample.
Figure 1.
Two-color lateral flow assay for multiplex IgG/IgM detection of several febrile illnesses. (A) Assay architecture comprised of sample, conjugate, nitrocellulose, and collection membranes. (B) Assay principles involving: Blue latex, anti-IgG, and Red latex, anti-IgM, detection probes for targeting the sample IgG and IgM, respectively, and the recombinant pathogen molecules on the test line for capturing the IgG/IgM to the respective pathogens.
The latex-based detection probes were prepared by labeling the anti-IgG antibodies (goat antirabbit IgG secondary antibody, γ-chain specific; Jackson ImmunoResearch Laboratories, Inc.) with Blue latex beads (LATEX conjugation kit −400 nm Blue; Innova Biosciences Ltd.) and anti-IgM antibodies (goat antimouse IgM secondary antibody, μ-chain specific; Sigma-Aldrich Co.) with Red latex beads (LATEX conjugation kit −400 nm Red; Innova Biosciences Ltd.) based on antibody attachment to bead surfaces via lysine residues. The conjugates were subsequently diluted to 0.4% and dry-stored on Glass Fiber Conjugate Pads (EMD Millipore) with 10 cm × 5 mm dimensions. The details for preparation of the latex-based detection probe are provided in the Supporting Information.
The lateral flow assay was prepared by assembling the sample, conjugate, and absorbent pads onto the adhesive parts of the plastic-backed Hi-Flow Plus 90 Membrane Cards (EMD Millipore), which contained the nitrocellulose membrane that housed the test and control regions. The control regions of the test strip were prepared by dispensing 0.4 μL of 0.4 mg/mL antigoat IgG secondary antibody (LifeSpan Biosciences, Inc.) onto the center of the nitrocellulose membrane, 3 mm below the absorbent pads. The test regions were subsequently prepared 2 mm below the control regions by dispensing 0.5 μL of 1.0 mg/mL: recombinant CHIKV E2 protein (RaybioTech, Inc.) for the CHIKV test (Figure 2a) and/or recombinant dengue antigen (RaybioTech, Inc.) for the DENV test. To ensure proper wicking of the sample solution through the assembled assay, the components were assembled via the adhesive regions of the membrane card in the following order: (1) the latex-bead-treated conjugate pad was first attached below the nitrocellulose membrane with a 0.5 mm overlap, (2) the FR-1 Membrane (MDI Membrane Technologies) as the sample pad was attached below the conjugate pad with a 0.5 mm overlap, and (3) the Cellulose Fiber Membrane (EMD Millipore) as the absorbent pad was attached above the nitrocellulose membrane also with a 0.5 mm overlap. The assembled membrane card was cut manually into individual strips of 5 mm width (Figure 1a) using a rotary paper trimmer (Dahle North America, Inc.). Once prepared, the test could be run by inputting 10 μL of sample and 80 μL of chase buffer onto the sample pad of the assay (details for assay operation can be found in the Supporting Information).
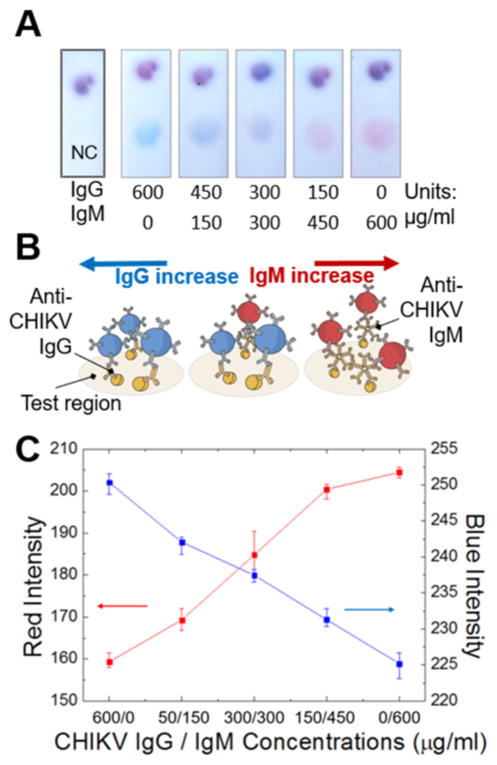
Figure 2.
Single-spot duplex IgG/IgM test for Chikungunya. (A) Color development on the test region of the two-color assay. (B) Schematic details of the test region interactions where blue and red colors correspond to the colored probes that have interacted with the anti-CHIKV IgG and IgM, respectively. (C) Image analysis for red and blue intensities on the test regions at different anti-CHIKV IgG/IgM concentrations.
After the color was allowed to develop for 10 min on the test and control regions, the images of the test and control regions were taken using a 8 megapixel CMOS camera of iPhone 5s under the same room lighting conditions. The images were transferred to a standard laptop computer and red (R), green (G), and blue (B) intensities were obtained using RGB Measure function of ImageJ (National Institutes of Health). Within the test regions which typically measured 120 pixels by 120 pixels, three different areas of 40 pixels by 40 pixels were analyzed in order to examine the effect of nonuniformities of the color development. The R, G, and B values were then averaged and used to calculate the hue (H) intensities (Tables S.1 and S.2) based on the following equation:
where M = max(R, G, B) and m = min(R, G, B). Here, the use of hue channel to investigate the test region color is convenient; as in the range of our interest, hue intensity is found to increase monotonically as the ratio of red to blue color increases. Furthermore, previous studies have found hue to be less affected by illumination and more precise than RGB for quantitative analysis.19 In our analysis, the test regions that did not develop any color were not considered.
In Figure 2a, we first demonstrate our assay scheme for the duplex detection of anti-CHIKV IgG/IgM. As expected, only the blue color can be observed on the CHIKV test region when the sample contains the significant concentrations of anti-CHIKV IgG only. The blue color on the CHIKV test region is indicative of the blue latex–anti-IgG probes that have bound to the anti-CHIKV IgG in the sample, which in turn has been captured by the CHIKV molecules on the test region (Figure 2b). As the sample concentration of anti-CHIKV IgG is decreased and that of anti-CHIKV IgM is increased, the blue color diminishes and the red color begins to emerge which correlates to the increasing number of red latex–anti-IgM probes accumulating on the CHIKV test region. This color trend continues to the case where only the significant concentration of anti-CHIKV IgM is found in the sample, and the red color dominates the CHIKV test region. In Figure 2c, we confirm this trend by acquiring the R and B intensities from the CHIKV test regions. In addition, we show that hue intensity of the test region increases as expected when red color shows a stronger presence over blue (Figure S.1). These results suggest that, as currently optimized, our two-color lateral flow assay develops distinguished blue/red colors when the concentrations of anti-CHIKV IgG/IgM, respectively, are above 300 μg/mL. As the concentrations of AFI related IgG/IgM in human blood are elevated significantly beyond 300 μg/mL for those infected and found in much lower concentrations otherwise,20 this version of our assay has the potential to diagnose the presence of anti-CHIKV IgG/IgM in point-of-care settings. Our novel assay scheme, in comparison to the commercial RDTs for duplex anti-CHIKV IgG/IgM detection that requires two separate test regions, is able to achieve the same level of multiplexing on just a single test region, which could lead to the reductions in the strip dimensions, material cost, and required sample volume.18
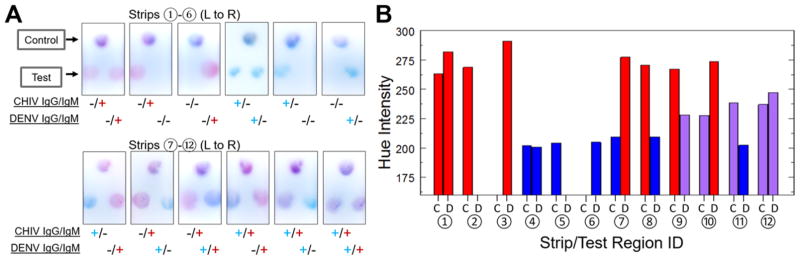
In Figure 3a, we demonstrate the multiplexing capabilities of our novel assay scheme that enables the 4-plex detection of the IgG/IgM to the two distinct AFIs, CHIKV and DENV, on a single strip. The colorimetric development on the CHIKV test region (left) can be observed to indicate the presence of anti-CHIKV IgG and/or anti-CHIKV IgM above 300 μg/mL, where the concentrations above this cutoff are highly suggestive of current (IgM) or past (IgG) infection. In parallel, the DENV test region (right) on the same strip provides information about the presence of anti-DENV IgG and/or anti-DENV IgM. On both test regions, we show that blue, red, or mixed colors develop when the sample contains the pathogen-specific IgGs, IgMs, or both, respectively, which could be used to correctly identify 12 different diagnostic scenarios involving anti-CHIV and anti-DENV IgG/IgM. In Figure 3b, we quantitatively confirm these observations by evaluating the hue intensity of the test regions. The results indicate that the presence of IgG, IgG/IgM, and IgM can be associated with three distinct ranges of hue values at 200–209, 227–247, and 262–290, respectively (summarized in Figure S.2).
Figure 3.
4-plex IgG/IgM test for Chikungunya and Dengue on a single strip. (A) CHIKV test region (C; left) and DENV test region (D; right) developing red, blue, or mixed colors for different diagnostic scenarios. (B) Hue intensity of CHIKV and DENV test regions for the 4-plex strips.
This ability to multiplex IgG/IgM tests of more than one pathogen on a single strip is unprecedented and has the potential to facilitate the differential diagnosis of AFIs in the point-of-care settings. As the number of test regions increases and to accommodate the users with color-perception difficulties, the need for an analyzer system becomes apparent. For the intended point-of-care applications, this requirement can be fulfilled by the use of smartphone-based analyzer systems that have been previously developed by our group21,22 and others23,24 for similar applications. Besides the analytical capabilities, these systems could facilitate the AFI management by allowing quick communication of results via e-mail and collecting/tracking epidemiological data.25,26
The choice of our anti-IgG and anti-IgM probes that have affinities toward rabbit IgGs and mouse IgMs, respectively, has been to demonstrate our assay scheme using the commercially available IgGs and IgMs of DENV and CHIKV. In order for this assay to be useful in the diagnosis of AFIs in humans, the current anti-IgG and anti-IgM probes would need to be replaced with those with human IgG/IgM affinities; however, the probe preparation steps would remain the same. In the near future, once our two-color assay is adapted for human IgG/IgM, the assay performance needs to be validated in human samples of AFI positivity. Notably, we hope to investigate the effect of other IgG/IgMs present in human samples on our assay signals and establish reliable methods to interpret our color signals to determine the presence of target IgG/IgMs as well as provide other clinically useful information such as differentiation of primary and secondary infections. Our assay can also be improved in terms of its ability to diagnose AFI in the beginning days of infection by incorporating the infectious agent detection on the same strip, where antibody against a specific AFI agent would be introduced as a testline on the strip.
For this initial prototype version of the assay, pipetting of test and control regions provided convenient and cost-effective means to prepare the small batch of test strips (<50). For mass production of the test strips, a lateral flow dispenser (e.g., Lateral Flow Reagent Dispenser; Claremont Biosolution) could be adapted to yield higher repeatability.
In conclusion, we present a novel lateral flow assay scheme that relies on two-color detection probes to multiplex the detection of IgG/IgM to CHIKV and DENV on a single lateral flow assay strip. We show that, by observing the color and location of the CHIKV and DENV test regions, our assay can indicate the presence of anti-CHIKV IgG/IgM and anti-DENV IgG/IgM. As our choice of the detection probes is commonly applicable in the detection schemes of different AFI-associated IgG/IgM tests, our assay could be easily multiplexed beyond the two AFIs presented here. Thus, this represents the significant step toward the development of a single point-of-care rapid test for the differential diagnosis of multiple AFIs, which could be integrated with smartphone reader systems to facilitate the results analysis and communication of diagnostic findings.
Supplementary Material
Acknowledgments
D.E. and S.M. acknowledge support from the US National Institutes of Health through award R01EB021331 and the US National Science Foundation through award 1343058. In addition, S.L. acknowledges the support of the National Science and Engineering Research Council of Canada (NSERC) through a Postgraduate scholarship. S.L would like to thank Jess Hohenstein, Ruisheng Wang, Aadhar Jain, and Perry Schein for the helpful discussions on the assay development.
Footnotes
Author Contributions
S.L., S.M., and D.E. designed the study. S.L. developed the assay and performed the experiments. S.L., S.M., and D.E wrote and edited the paper.
The authors declare the following competing financial interest(s): D.E. and S.M. are unpaid board members for a diagnostic start up focused on measurement of nutritional biomarkers at the point-of-care utilizing the results from their research.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.6b01828.
Blue and red latex-based detection probe preparation, assay procedures, and RGB and Hue values and charts for the duplex and 4-plex testing (PDF)
References
- 1.Chrispal A, Boorugu H, Gopinath KG, Chandy S, Prakash JAJ, Thomas EM, Abraham AM, Abraham O, Thomas K. Trop Doct. 2010;40:230–234. doi: 10.1258/td.2010.100132. [DOI] [PubMed] [Google Scholar]
- 2.Suaya JA, Shepard DS, Beatty M. Dengue burden of disease and cost of illness. World Health Organization Scientific Working Group; 2007. Dengue: Burden of Disease and Costs of Illness. Report on Dengue Vol. TDR/SWG/08. [Google Scholar]
- 3.Lau C, Smythe L, Weinstein P. Travel Medicine and Infectious Disease. 2010;8:33–39. doi: 10.1016/j.tmaid.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Laoprasopwattana K, Kaewjungwad L, Jarumanokul R, Geater A. Pediatric Infectious Disease Journal. 2012;31:459–463. doi: 10.1097/INF.0b013e31824bb06d. [DOI] [PubMed] [Google Scholar]
- 5.Hofer M, Mahlaoui N, Prieur A-M. Best Practice & Research Clinical Rheumatology. 2006;20:627–640. doi: 10.1016/j.berh.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz O, Albert ML. Nat Rev Microbiol. 2010;8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 7.Vega-Rúa A, Zouache K, Girod R, Failloux A-B, Lourenço-de-Oliveira R. Journal of Virology. 2014;88:6294–6306. doi: 10.1128/JVI.00370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO. Dengue and severe dengue. World Health Organization; Geneva: 2015. [Factsheet no. 117, revised May 2015] [Google Scholar]
- 9.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chappuis F, Alirol E, d’Acremont V, Bottieau E, Yansouni CP. Clin Microbiol Infect. 2013;19:422–431. doi: 10.1111/1469-0691.12154. [DOI] [PubMed] [Google Scholar]
- 11.Andries AC, Duong V, Ngan C, Ong S, Huy R, Sroin KK, Te V, YB, Try PL, Buchy P. PLoS Neglected Trop Dis. 2012;6:e1993. doi: 10.1371/journal.pntd.0001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajani MD, Ashford DA, Bragg SL, Woods CW, Aye T, Spiegel RA, Plikaytis BD, Perkins BA, Phelan M, Levett PN, Weyant RS. Journal of Clinical Microbiology. 2003;41:803–809. doi: 10.1128/JCM.41.2.803-809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson JL, May HT, Horne BD, Bair TL, Hall NL, Carlquist JF, Lappe DL, Muhlestein JB. Am J Cardiol. 2010;106:963–968. doi: 10.1016/j.amjcard.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Blacksell SD, Jarman RG, Bailey MS, Tanganuchitcharnchai A, Jenjaroen K, Gibbons RV, Paris DH, Premaratna R, de Silva HJ, Lalloo DG, Day NPJ. Clinical and Vaccine Immunology. 2011;18:2095–2101. doi: 10.1128/CVI.05285-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Acremont V, Bosman A. WHO informal consultation on fever management in peripheral health care settings: a global review of evidence and practice. Malaria Policy Advisory Committee; Geneva: 2013. [Google Scholar]
- 16.Song S, Liu N, Zhao Z, Njumbe Ediage E, Wu S, Sun C, De Saeger S, Wu A. Anal Chem. 2014;86:4995–5001. doi: 10.1021/ac500540z. [DOI] [PubMed] [Google Scholar]
- 17.Yang M, Caterer NR, Xu W, Goolia M. J Virol Methods. 2015;221:119–126. doi: 10.1016/j.jviromet.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Yen CW, de Puig H, Tam JO, Gomez-Marquez J, Bosch I, Hamad-Schifferli K, Gehrke L. Lab Chip. 2015;15:1638–1641. doi: 10.1039/c5lc00055f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oncescu V, O’Dell D, Erickson D. Lab Chip. 2013;13:3232–3238. doi: 10.1039/c3lc50431j. [DOI] [PubMed] [Google Scholar]
- 20.Parry JV, Perry KR, Mortimer PP. Lancet. 1987;330:72–75. doi: 10.1016/s0140-6736(87)92737-1. [DOI] [PubMed] [Google Scholar]
- 21.Lee S, Oncescu V, Mancuso M, Mehta S, Erickson D. Lab Chip. 2014;14:1437–1442. doi: 10.1039/c3lc51375k. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, O’Dell D, Hohenstein J, Colt S, Mehta S, Erickson D. Sci Rep. 2016;6:28237. doi: 10.1038/srep28237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A. Lab Chip. 2012;12:2678–2686. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK. Sci Transl Med. 2015;7:273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 25.Erickson D, O’Dell D, Jiang L, Oncescu V, Gumus A, Lee S, Mancuso M, Mehta S. Lab Chip. 2014;14:3159–3164. doi: 10.1039/c4lc00142g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tucker C. Nation’s Health. 2011;41(1):14–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.