Abstract
Background
Long-term PM2.5 exposure and aging have been implicated in multiple shared diseases; studying their relationship is a promising strategy to further understand the adverse impact of PM2.5 on human health.
Objective
We assessed the relationship of major PM2.5 component species (ammonium, elemental carbon, organic carbon, nitrate, and sulfate) with Horvath and Hannum DNA methylation (DNAm) age, two DNA methylation-based predictors of chronological age.
Methods
This analysis included 552 participants from the Normative Aging Study with multiple visits between 2000 and 2011 (n = 940 visits). We estimated 1-year PM2.5 species levels at participants’ addresses using the GEOS-chem transport model. Blood DNAm-age was calculated using CpG sites on the Illumina HumanMethylation450 BeadChip. We fit linear mixed-effects models, controlling for PM2.5 mass and lifestyle/environmental factors as fixed effects, with the adaptive LASSO penalty to identify PM2.5 species associated with DNAm-age.
Results
Sulfate and ammonium were selected by the LASSO in the Horvath DNAm-age models. In a fully-adjusted multiple-species model, interquartile range increases in both 1-year sulfate (95%CI: 0.28, 0.74, P < 0.0001) and ammonium (95%CI: 0.02, 0.70, P = 0.04) levels were associated with at least a 0.36-year increase in Horvath DNAm-age. No PM2.5 species were selected by the LASSO in the Hannum DNAm-age models. Our findings persisted in sensitivity analyses including only visits with 1-year PM2.5 levels within US EPA national ambient air quality standards.
Conclusion
Our results demonstrate that sulfate and ammonium were most associated with Horvath DNAm-age and suggest that DNAm-age measures differ in their sensitivity to ambient particle exposures and potentially disease.
Keywords: DNA methylation age, Particulate matter 2.5, Long-term exposure, Epigenetics
1. Introduction
Fine particulate matter (PM2.5) remains an inescapable environmental exposure and an enormous global public health concern (World Health, 2014). It is estimated that at least 2.1 million lives could be saved annually if PM2.5 guidelines were adhered to worldwide (Apte et al., 2015). For the millions of people exposed to PM2.5 daily, understanding the impact of PM2.5 on human health is critical for developing interventions aimed at reducing PM2.5-related morbidity and mortality globally. Researchers have consistently demonstrated that long-term PM2.5 exposure is a major contributor to cardiopulmonary disease (Künzli et al., 2005; Giorgini et al., 2015; Martinelli et al., 2013; Zhong et al., 2015; Kloog et al., 2015; Raaschou-Nielsen et al., 2016), and emerging evidence suggests that PM2.5 is a risk factor for previously unconsidered disease outcomes like cognitive decline (Terzano et al., 2010; Schikowski et al., 2015; Power et al., 2011). Nevertheless, much remains to be understood about how PM2.5 contributes to even its most well-documented disease outcomes. One promising strategy to better understand the adverse impact of PM2.5 on human health, is to study the relationship of PM2.5 with aging. Many studies have implicated PM2.5 as a contributor to accelerated aging (Chen and Schwartz, 2009; Weuve et al., 2012; Brown et al., 2015; Scheers et al., 2015; Shan et al., 2014; Wilker et al., 2015). Moreover, independent of PM2.5 exposures, aging is associated with cardiopulmonary disease, cognitive decline, and many other PM2.5-related disease outcomes (Rowe and Kahn, 1987, 2000, 2015; Chung et al., 2009). Thus, understanding how PM2.5 can contribute to aging, may provide additional insight into other adverse PM2.5-related health effects.
DNA methylation-based biomarkers of age have proved to be promising tools in understanding the relationship of PM2.5 with aging. These biomarkers have surpassed their initial utility of simply predicting chronological age, and have demonstrated remarkable usefulness in assessing individuals’ risk of mortality, malignancy, neurocognitive disease, and other biologically-relevant health endpoints (Marioni et al., 2015, 2016; Horvath et al., 2015; Horvath and Ritz, 2015; Levine et al., 2015a, 2015b, 2015c). Evidence also suggests that these biomarkers of age are reflective of individuals’ past environmental exposures (Horvath et al., 2014). One such study by our group demonstrated robust associations between PM2.5 exposure levels and Horvath DNA methylation (DNAm) age. Horvath DNAm-age is a tissue-independent predictor of chronological age that is calculated from DNA methylation values at 353 chronological age-correlated CpG dinucleotides in Illumina’s HumanMethylation450 BeadChip (Horvath, 2013). Specifically, in an elderly cohort and with fully-adjusted models, we observed that a 1 μg/m3 increase in 1-year PM2.5 exposure was associated with a 0.52-year increase in Horvath DNAm-age (Nwanaji-Enwerem et al., 2016).
Still, PM2.5 is a heterogeneous mixture of carbonaceous fractions, inorganics, and metals; and it is widely appreciated that PM2.5 component species often differ in their health effects (Zanobetti et al., 2009; Ito et al., 2011; Liu et al., 2016; Laurent et al., 2016). The present study builds upon our previous research and examines the relationships of PM2.5 component species with both Horvath and Hannum DNAm-age in elderly men. Hannum DNAm-age is also a DNA methylation-based predictor of chronological age, but it is based on measurements from 71 CpG dinucleotides (Hannum et al., 2013). Only 6 CpG dinucleotides are shared between the Horvath and Hannum metrics. By investigating the relationships of PM2.5 component species with these two forms of DNAm-age, we aim to (1) better understand how specific PM2.5 species are related to aging, and (2) demonstrate differences in the biological utility of different DNAm-age measures.
2. Materials and methods
2.1. Study population
The participants in this analysis were part of the U.S. Veterans Affairs Normative Aging Study (NAS), a longitudinal investigation of aging men established in Eastern Massachusetts in 1963 (Bell et al., 1966). The men were free of known chronic medical conditions at enrollment, and returned for onsite, follow-up visits every 3–5 years. During these visits, detailed physical examinations were performed, bio-specimens including blood were obtained, and questionnaire data pertaining to diet, smoking status, and additional lifestyle factors that may impact health were collected. All participants provided written informed consent to the VA Institutional Review Board (IRB), and both the Harvard T.H. Chan School of Public Health and VA IRBs granted human subjects approval.
All NAS men with continued study participation as of the year 2000, when PM2.5 component levels became available, were eligible for our study sample. After excluding participants with a diagnosis of leukemia (n = 11), due to its potential influence on the DNA methylation of blood cells (Horvath, 2013), and those incomplete for the covariates of interest (n = 16), we had a total of 552 participants with 940 observations between the years 2000 and 2011. Of the 552 participants, 249 (45%) had one visit, 218 (40%) had two visits, and 85 (15%) had three or more visits.
2.2. DNA methylation and calculation of DNAm-age
Laboratory staff extracted DNA from the buffy coat of whole blood collected from each participant at each NAS follow-up visit (QIAamp DNA Blood Kit, QIAGEN, Valencia, CA, USA). DNA samples were then treated with bisulfite conversion (EZ-96 DNA Methylation Kit, Zymo Research, Orange, CA, USA) and hybridized to the 12 sample Illumina HumanMethylation450 BeadChips (Infinium HD Methylation protocol, Illumina, San Diego, CA, USA). To ensure a similar age distribution and avoid confounding across chips and plates, study staff employed a two-stage age-stratified algorithm to randomize samples. For quality control, study staff removed samples where >5% of probes had a beadcount <3 or >1% of probes had a detection P-value >0.05. The Bioconductor minfi package Illumina-type background correction without normalization was used to preprocess the remaining samples and generate methylation beta values (Aryee et al., 2014). The beta values represent the percentage of methylation for each of the ~480,000 CpG sites in the BeadChip array. The 450 k arrays were run in the Genomics Core Facility at Northwestern University.
To explore potential differences in the relationship of PM2.5 and PM2.5 species with different forms of DNAm-age, we calculated both Horvath DNAm-age and Hannum DNAm-age using the 450 k beta values and Horvath’s publically available online calculator (http://labs.genetics.ucla.edu/horvath/dnamage/). Horvath DNAm-age was derived from an elastic net (penalized regression) using multiple data sets of varying tissue and cell types. 21,369 CpG probes, shared by the Illumina HumanMethylation27 and HumanMethylation450 BeadChip platforms were regressed on a calibrated version of chronological age. The elastic net selected 353 CpGs that correlate with age, and the resulting model coefficients are used by the calculator to predict the age of each DNA sample (DNAm-age) (Horvath, 2013). Hannum DNAm-age was also derived using an elastic net. However, Hannum DNAm-age was based on a single cohort where DNA methylation values were calculated from whole blood. This elastic net selected 71 CpG probes in the Illumina HumanMethylation450 BeadChip that are predictive of chronological age. Hannum DNAm-age was calculated as the sum of the beta values multiplied by the reported effect sizes for the Hannum predictor (Hannum et al., 2013). The Hannum and Horvath DNAm-ages only share 6 CpG probes (cg04474832, cg05442902, cg06493994, cg09809672, cg19722847, and cg22736354).
2.3. Assessment of environmental factors: ambient particles and temperature
We employed the widely used GEOS-chem chemical transport model (http://www.geoschem.org) to generate 1-year exposure estimates for PM2.5 and the following major PM2.5 component species: organic carbon (OC), elemental carbon (EC), sulfate, nitrate, and ammonium (van Donkelaar et al., 2010). These 5 component species were selected because they make up a large fraction of total PM2.5 mass (~ 88.6%) and were best predicted by the model. GEOS-chem incorporates nonlinear chemistry, meteorology, and detailed emissions inventories to simulate the formation and transportation of atmospheric components to give raw estimates of PM2.5 and its major chemical components. Ten-fold cross-validation demonstrated that the model performs well for PM2.5 mass and its component species with R2s ranging from 0.70 to 0.88 (Di et al., 2016). We generated daily estimates at the 1 × 1 km area resolution. Each participant’s residence was geocoded and linked to an area level grid-point. Time spent away from home (>7 days) and address changes were also accounted for as particle estimates were assigned to each participants’ address. Given that >90% of NAS participants are retired, home address exposures are expected to be a good proxy for their individual ambient exposures. We then generated 1-year total PM2.5 and PM2.5 component species exposure windows by averaging daily exposures for the 365 days prior to the day of each participants’ NAS visit. The 1-year PM2.5 exposure window was utilized because it has been previously reported to be robustly associated with DNAm-age (Nwanaji-Enwerem et al., 2016).
We used a spatiotemporal prediction multi-step approach to generate temperature (in Celsius) for each participant (Kloog et al., 2014). First, we obtained 1 × 1 km resolution daily physical surface temperature (Ts) data from NASA satellite measurements and daily near surface air (Ta) data from the Environmental Protection Agency, National Climatic Data Center, and Weather Underground Inc. We then used mixed model regression to calibrate Ts to Ta. The model was validated with a mean out of sample R2 of 0.95. To generate 1-year temperature measurements to complement 1-year particle exposures, we averaged daily temperature measurements over the 365 days prior to participants’ NAS visits.
2.4. Statistical analysis
We first used generalized linear mixed effects models to determine the relationship of DNAm-age (Horvath and Hannum independently) with 1-year PM2.5 exposure levels and 1-year PM2.5 component species exposure levels. All linear mixed effects models included a random participant-specific intercept to account for correlation between repeated measures (i.e. multiple visits for a participant).
We adjusted for confounders and covariates that have a priori biological/clinical relevance and/or are reported in the existing literature. Specifically, our previous publication was the first study examining associations of ambient particles and DNAm-age (Nwanaji-Enwerem et al., 2016). There, we used a tiered approach of adding confounders and covariates based on known relationships of ambient particles with DNA methylation and known relationships of ambient particles with older markers of aging (Horvath, 2013; Madrigano et al., 2011; Baccarelli et al., 2009; Bind et al., 2015; Peng et al., 2016). Tier one adjusted for chronological age and blood cell types. Tier two made additional adjustments for lifestyle and environmental factors. Tier three expanded on tier two by additionally adjusting for age-related diseases, and tier four expanded on tier two by additionally adjusting for medications of age-related diseases. After considering model fit (assessed via AIC) and considering biological factors that are known to be important, the tier two covariates were deemed to be most appropriate. Thus, in line with the previously published tier two framework (Nwanaji-Enwerem et al., 2016), the models for this analysis were adjusted for chronological age (continuous), six blood cell type estimates [i.e. plasma cells, CD4+ lymphocytes, CD8+ lymphocytes, natural killer (NK) cells, monocytes, and granulocytes] (continuous) determined via Houseman and Horvath methods (Horvath, 2013; Houseman et al., 2012), average 1-year temperature (continuous), cumulative cigarette pack years (continuous), smoking status (current, former, or never), season of visit (spring [March–May], Summer [June–August], Fall [September–November], and Winter [December–February]), body mass index (BMI) (lean [<25], overweight (Horvath and Ritz, 2015; Levine et al., 2015a, 2015b, 2015c; Horvath et al., 2014; Horvath, 2013), obese [>30]), alcohol intake (yes/no ≥2 drinks daily), and maximum years of education (continuous). All PM2.5 component species models were additionally adjusted for PM2.5 mass (Mostofsky et al., 2012).
To more rigorously identify the PM2.5 component species that may be associated with DNAm-age, we applied the adaptive LASSO (least absolute shrinkage and selection operator) (Schelldorfer et al., 2011). Given that PM2.5 component species are correlated, placing them together within the same standard linear regression model can result in unaccounted for stochastic errors. The LASSO is a regression shrinkage and selection approach that helps overcome such limitations. The LASSO applies an l1 penalty on the component regression coefficients, which minimizes the sum of squared errors subject to the sum of the absolute values of the coefficients being less than a given value (Tibshirani, 1996). The adaptive LASSO improves upon this procedure by utilizing weights for penalizing different coefficients in the l1 penalty to identify a subset of model predictors to achieve asymptotic normality (Zou, 2006). Furthermore, the adaptive LASSO has been successfully applied in air pollution and health research (Dai et al., 2016a, 2016b).
To identify and select PM2.5 component species associated with DNAm-age, we applied a penalty to all PM2.5 component species, but not to PM2.5 mass and the other covariates in the model. λ, the penalty parameter, determines how strongly the magnitude of the PM2.5 species regression coefficients is constrained. When λ is small, the regression coefficients are weakly penalized and mirror those that would be given from a standard linear mixed effects model. When λ is large, the coefficients are strongly penalized, shrinkage is maximized, and all coefficients tend towards zero such that the resulting model includes fixed covariates only. When λ takes a value in between the extremes, the result is a penalized model where some PM2.5 component species will have coefficients of zero and others will be non-zero. PM2.5 component species with non-zero coefficients are considered as “selected” by the adaptive LASSO. We ran the model across a range of λs, beginning with a λ of 0, and selected the λ resulting in the model with the smallest Bayesian Information Criterion (BIC) (Schwarz et al., 1978). Following LASSO selection, we fit a final multiple-species linear mixed effects model using the selected PM2.5 component species and our fixed covariates. From this final model, we were able to estimate component species effect sizes and their corresponding 95% confidence intervals.
Additionally, we considered that the LASSO may not select the PM2.5 species that are most correlated with total PM2.5 mass. Thus, we conducted a sensitivity analysis where we performed LASSO selection without adjusting for PM2.5 mass. From this sensitivity analysis model, we fit a multiple-species linear mixed effects model using the selected PM2.5 component species and estimated component species effect sizes and their corresponding 95% confidence intervals.
After finding that Horvath DNAm-age alone was significantly associated with PM2.5 component species, we evaluated the relationships of the DNA methylation values of each of the 353 Horvath CpG probes with the particles in the aforementioned LASSO-selected multiple-species linear mixed effects model. In addition to the already described covariates, we included technical covariates (450 k plate, chip, row, and column) to this analysis. To account for multiple hypothesis testing, we also performed FDR correction in this analysis. We then performed gene ontology analysis on the list of significant CpGs (FDR P-value < 0.05) using the publically available GoTermFinder tool (http://go.princeton.edu/cgi-bin/GOTermFinder).
In an additional sensitivity analyses, we re-ran our models excluding participant visits with PM2.5 exposures > 12 μg/m3. This allowed us to assess if our findings persisted even at the PM2.5 levels currently deemed acceptable by the U.S. Environmental Protection Agency (EPA) National Ambient Air Quality Standards (NAAQS) (US EPA, O.A.R., n.d.).
All statistical analyses were performed using R Version 3.1.1 (R Core Team, Vienna, Austria) and we considered a P-value <0.05 to be statistically significant.
3. Results
3.1. Descriptive results
Table 1 summarizes the characteristics of the study population. All study participants were Caucasian males with a mean ± SD age of 74.7 ± 6.99 years across all study visits. Average Horvath DNAm-age and Hannum DNAm-age were 74.0 ± 7.92 years and 75.1 ± 8.95 years respectively. Horvath DNAm-age (r = 0.59, p < 0.0001) and Hannum DNAm-age (r = 0.77, p < 0.0001) were both strongly correlated with chronological age in the study population. Both measures of DNAm-age were also strongly correlated to each other (r = 0.69, p < 0.0001).
Table 1.
Characteristics of study subjects (2000–2011).
| Variable | First visit (N = 552) | All visits (N = 940) |
|---|---|---|
| Age (years), mean (SD) | 73.3 (6.82) | 74.7 (6.99) |
| Horvath DNAm-age (years), mean (SD) | 73.7 (7.77) | 74.0 (7.92) |
| Hannum DNAm-age (years), mean (SD) | 73.8 (8.80) | 75.1 (8.95) |
| Temperature (°C), mean (SD) | 11.5 (1.12) | 11.3 (1.00) |
| Pack years, mean (SD) | 20.7 (24.7) | 20.5 (24.4) |
| Smoking status, N (%) | ||
| Current | 25 (4) | 40 (4) |
| Former | 355 (64) | 614 (65) |
| Never | 172 (32) | 286 (31) |
| Season, N (%) | ||
| Spring | 145 (26) | 241 (26) |
| Summer | 115 (21) | 199 (21) |
| Fall | 177 (32) | 313 (33) |
| Winter | 115 (21) | 187 (20) |
| BMI, N (%) | ||
| Healthy/lean | 119 (21) | 216 (23) |
| Overweight | 302 (55) | 493 (52) |
| Obese | 131 (24) | 231 (25) |
| Alcohol consumption, N (%) | ||
| <2 drinks/day | 441 (80) | 761 (81) |
| ≥2 drinks/day | 111 (20) | 179 (19) |
| Education, N (%) | ||
| ≤12 years | 146 (27) | 242 (26) |
| 12–16 years | 262 (47) | 434 (46) |
| >16 years | 144 (26) | 264 (28) |
Table 2 reports 1-year PM2.5 and PM2.5 component species exposure levels across all study visits. The participants had a mean ± SD 1-year PM2.5 exposure level of 10.3 ± 1.60 μg/m3, with an interquartile range (IQR) of 2.16 μg/m3. Of the measured PM2.5 component species, sulfate accounted for the largest proportion of total PM2.5 mass (33%), followed by organic carbon (28.6%), nitrate (11.5%), ammonium (10.1%), and elemental carbon (5.4%). OC was the PM2.5 species most correlated with total PM2.5 mass (r = 0.67). 1-year PM2.5 and PM2.5 species Pearson correlations across all visits are reported in Table S1. Moreover, 1-year PM2.5 and PM2.5 species exposure distributions across first visits are reported in Table S2.
Table 2.
Mean 1-year particulate matter 2.5 (PM2.5) and component species concentrations across all study visits.
| Particle (μg/m3) | Mean (SD) | IQR | Proportion of PM2.5 (%) | Pearson correlation with PM2.5 | N |
|---|---|---|---|---|---|
| PM2.5 | 10.3 (1.60) | 2.16 | – | – | 940 |
| PM2.5 Component Species | |||||
| EC | 0.56 (0.17) | 0.23 | 5.4 | 0.62 | 940 |
| OC | 2.94 (0.91) | 1.28 | 28.6 | 0.67 | 940 |
| Sulfate | 3.40 (1.23) | 0.82 | 33.0 | 0.30 | 940 |
| Nitrate | 1.18 (0.32) | 0.42 | 11.5 | 0.46 | 940 |
| Ammonium | 1.04 (0.31) | 0.3 | 10.1 | 0.53 | 940 |
3.2. 1-year PM2.5 and PM2.5 component species as predictors of DNAm-age
Table 3 summarizes the results of three model frameworks where PM2.5 and its component species were modeled as predictors of both Horvath and Hannum DNAm-age. Residuals from all models appeared normally distributed. In the model framework 1, PM2.5 was modeled as a predictor of Horvath and Hannum DNAm-age independently. In the fully adjusted model, an IQR increase in 1-year PM2.5 exposure was significantly associated with a 0.64-year increase in Horvath DNAm-age (p = 0.005). However, an IQR increase in 1-year PM2.5 exposure was not significantly associated with Hannum DNAm-age (β = 0.06, p = 0.74).
Table 3.
1-year particulate matter 2.5 (PM2.5) and component species as predictors of DNA methylation (DNAm) age.
| Particle | Difference in Horvath DNAm-age for IQR (95% CI) | P | Difference in Hannum DNAm-age for IQR (95% CI) | P | N |
|---|---|---|---|---|---|
| Model framework 1 | |||||
| PM2.5 | 0.64 (0.20, 1.09) | 0.005 | 0.06 (−0.28, 0.40) | 0.74 | 940 |
| Model framework 2 | |||||
| EC | 0.27 (−0.25, 0.80) | 0.30 | −0.09 (−0.48, 0.29) | 0.64 | 940 |
| OC | 0.93 (0.37, 1.50) | 0.001 | 0.35 (−0.05, 0.77) | 0.09 | 940 |
| Sulfate | 0.59 (0.37, 0.81) | <0.0001 | 0.08 (−0.09, 0.25) | 0.36 | 940 |
| Nitrate | 0.58 (0.11, 1.04) | 0.01 | 0.30 (−0.04, 0.65) | 0.08 | 940 |
| Ammonium | 0.59 (0.26, 0.92) | 0.0004 | 0.06 (−0.18, 0.30) | 0.63 | 940 |
| Model framework 3 | |||||
| PM2.5 | 0.18 (−0.30, 0.66) | 0.45 | – | – | 940 |
| Sulfate | 0.51 (0.28, 0.74) | <0.0001 | – | – | 940 |
| Ammonium | 0.36 (0.02, 0.70) | 0.04 | – | – | 940 |
Model framework 1: adjusted for chronological age, blood cell types, temperature, pack years, smoking status, season, BMI, alcohol consumption, and education. Model frame-work 2: PM2.5 component species as independent predictors of DNAm-age adjusted for PM2.5 in addition to model 1 covariates. Model framework 3: PM2.5, sulfate, and ammonium as joint predictors of DNAm-age (given selection of sulfate and ammonium by the adaptive LASSO) adjusted for model 1 covariates. No species were selected as predictors of Hannum DNAm-age. Bold values indicate significance at P < 0.05.
Under the model framework 2, each PM2.5 component species was modeled as an independent predictor of Horvath and Hannum DNAm-age adjusting for all covariates and total PM2.5 mass. 1-year IQR increases in OC (β = 0.93, p = 0.001), sulfate (β = 0.59, p < 0.0001), nitrate (β = 0.58, p = 0.01), and ammonium (β = 0.59, p = 0.0004) were all significantly associated with increases in Horvath DNAm-age of at least 0.58 years. No PM2.5 component species were significantly associated with Hannum DNAm-age (Table 3).
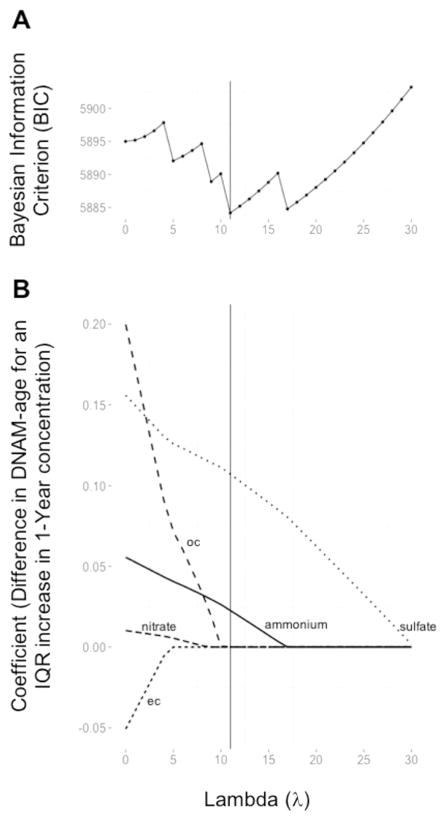
The model 3 framework reflects the results of the multiple-species fully-adjusted linear mixed effects models with the PM2.5 component species selected by the adaptive LASSO. The adaptive LASSO selected sulfate and ammonium as important predictors of Horvath DNAm-age. Fig. 1A depicts the relationship between BIC, the model selection criterion, and λ, the adaptive LASSO penalty parameter. The model with the smallest BIC had λ = 11. Fig. 1B shows the LASSO coefficient paths for the PM2.5 component species. Each component species coefficient is expressed as the difference in mean Horvath DNAm-age per an IQR increase in the 1-year component species exposure level. Each curve depicts the rate at which the component species coefficient shrinks towards zero as λ increases. At λ = 0, all components species have a non-zero coefficient.
Fig. 1.
A) The relationship between BIC, a criterion for model selection and λ (lambda), the adaptive LASSO penalty parameter, for DNAm-age. The vertical line at λ = 11 denotes the penalty parameter with the lowest BIC. B) LASSO coefficient paths: plot of coefficient profiles for PM2.5 components as a function of λ. At λ = 11, sulfate and ammonium are the only PM2.5 components with a non-zero coefficient.
In the multiple-species fully-adjusted linear mixed effects model, both sulfate (β = 0.51, p < 0.0001) and ammonium (β = 0.36, p = 0.04) remain significant positive predictors of Horvath DNAm-age. The adaptive LASSO did not select any PM2.5 component species as important predictors of Hannum DNAm-age.
In our sensitivity analysis – where LASSO selection was performed without adjusting for total PM2.5 mass – sulfate, ammonium, and OC were selected as important predictors of DNAm-age (Fig. S1). Nonetheless, in a multiple-species fully-adjusted linear mixed effects model, both sulfate (β = 0.45, p = 0.0003) and ammonium (β = 0.34, p < 0.05) remained significant positive predictors of Horvath DNAm-age, but OC (β = 0.42, p = 0.16) was not a significant predictor of Horvath DNAm-age (Table S3). Again, the sensitivity analysis adaptive LASSO did not select any PM2.5 component species as important predictors of Hannum DNAm-age.
Significant findings from the main analysis multiple-species fully-adjusted linear mixed effects model persisted in the second sensitivity analyses excluding participant visits with PM2.5 exposures > 12 μg/m3, the annual PM2.5 exposure level currently deemed acceptable by the U.S. Environmental Protection Agency (EPA) National Ambient Air Quality Standards (NAAQS) (Table S4).
3.3. Associations between 1-year PM2.5 and PM2.5 component species levels and methylation values at Horvath DNAm-age CpG sites
After FDR correction, 47 out of 353 Horvath DNAm-age CpG sites had methylation values that were significantly associated with total PM2.5 levels in the fully-adjusted multiple-species linear mixed effects model. PM2.5 levels were positively or negatively associated with CpG methylation values depending on the CpG site (Table 4). 46 of the 47 CpG sites mapped to known genes. 9 of these 46 genes (ENPP2, FAM50B, LZTFL1, SGCE, C14orf105, ZBTB5, TMEM132E, CATSPERG, and NDUFA13) were previously reported in a similar, previously published PM2.5 Horvath CpG analysis (Nwanaji-Enwerem et al., 2016). Gene ontology of our 46 genes combined with the genes in the previously reported study returned the GO term “regulation of translational initiation” (Table S5).
Table 4.
1-Year Particle Exposures as Predictors of Horvath CpG Probe.
| CpG | Gene | Process/function | Difference in methylation per SD (%) | Direction of association | FDR adjusted P |
|---|---|---|---|---|---|
| PM2.5 | |||||
| cg15262928 | TIMM17A | Mitochondrial protein import | 24.18 | + | 0.001 |
| cg14409958 | ENPP2* | Nucleic acid binding | 19.85 | + | 0.001 |
| cg01570885 | FAM50B* | Protein binding | 19.64 | − | 0.004 |
| cg08186124 | LZTFL1* | Protein binding: cytoplasm | 19.59 | + | 0.004 |
| cg18139769 | SGCE* | Calcium binding | 19.25 | − | 0.004 |
| cg15547534 | PPP1R35 | Phosphatase binding | 18.79 | + | 0.004 |
| cg26456957 | PPP1R12C | Protein kinase binding | 18.74 | + | 0.001 |
| cg05847778 | BBS5 | Transcription initiation | 18.01 | + | 0.006 |
| cg15661409 | C14orf105* | Uncharacterized | 17.52 | − | <0.001 |
| cg02335441 | NEK11 | DNA replication | 17.32 | + | 0.008 |
| cg17285325 | TYMP | Phosphorylase activity | 17.23 | + | 0.007 |
| cg04094160 | ZBTB5* | Transcriptional regulation | 16.92 | + | 0.003 |
| cg03682823 | SGCE | Calcium binding | 16.80 | − | 0.008 |
| cg07663789 | NPR3 | Hormone binding/blood volume | 16.13 | + | 0.003 |
| cg15703512 | PDZD9 | Uncharacterized | 15.80 | + | 0.015 |
| cg22190114 | NLRP8 | ATP binding | 15.60 | + | 0.015 |
| cg19008809 | SFMBT1 | Transcription corepressor activity | 15.48 | + | 0.013 |
| cg00374717 | ARSG | Sulfatase enzyme activity | 15.29 | − | 0.004 |
| cg12985418 | MIB1 | Protein binding | 15.14 | + | 0.018 |
| cg03588357 | GPR68 | G-protein coupled receptor activity | 15.09 | + | 0.020 |
| cg14424579 | AGBL5 | Metallocarboxypeptidase | 15.07 | + | 0.007 |
| cg14597908 | GNAS | G-protein binding | 15.04 | − | 0.015 |
| cg19044674 | LEPRE1 | Oxidoreductase activity | 14.88 | + | 0.023 |
| cg09441152 | PQLC1 | Membrane component | 14.87 | + | 0.027 |
| cg07849904 | MN1 | Transcriptional activator | 14.85 | + | 0.015 |
| cg19273182 | PAPOLG | Polynucleotide adenylyltransferase activity | 14.81 | + | 0.025 |
| cg17063929 | NOX4 | Nucleotide binding | 14.56 | − | 0.015 |
| cg24116886 | DEFB127 | Immunologic response | 14.40 | − | 0.015 |
| cg09191327 | PRDM12 | Methyltransferase activity | 14.30 | + | 0.027 |
| cg23662675 | ZMYND8 | Transcription cofactor activity | 14.16 | + | 0.014 |
| cg14992253 | EIF3I | Translation initiation | 13.12 | − | 0.018 |
| cg05442902 | P2RX6 | Channel activity | 12.82 | − | 0.026 |
| cg06557358 | TMEM132E* | Integral component of membrane | 12.60 | + | 0.031 |
| cg11932564 | TNFRSF13C | Immunologic response | 12.51 | + | 0.037 |
| cg18031008 | MRPS21 | Mitochondrial ribosome | 12.10 | + | 0.030 |
| cg19945840 | SDF4 | Calcium binding | 12.08 | − | 0.023 |
| cg19167673 | PDGFB | Protein homodimerization activity | 12.03 | + | 0.031 |
| cg25159610 | PLK2 | Cell division | 11.96 | + | 0.038 |
| cg22006386 | CATSPERG* | Ion channel activity | 11.89 | + | 0.026 |
| cg27377450 | unknown | unknown | 11.85 | − | 0.026 |
| cg20100381 | NAE1 | Protein heterodimerization activity | 11.84 | + | 0.046 |
| cg04268405 | CHST3 | Sulfotransferase | 11.47 | − | 0.026 |
| cg07595943 | ADAD2 | RNA binding | 11.36 | − | 0.023 |
| cg25505610 | EIF3M | Translation initiation | 10.67 | + | 0.042 |
| cg16744741 | PRKG2 | Protein kinase activity | 10.17 | − | 0.038 |
| cg21395782 | NDUFA13* | NADH dehydrogenase activity | 8.27 | + | 0.027 |
| cg01459453 | SELP | Oligosaccharide binding | 8.05 | − | 0.023 |
| Ammonium | |||||
| cg02275294 | SOAT1 | Fatty-acyl-CoA binding | 10.81 | + | 0.036 |
All models are fully adjusted.
CpGs associated with PM2.5 levels in a prior publication.
Only 1 out 353 CpG sites (cg02275294) had methylation values that were significantly associated with ammonium levels in the fully-adjusted multiple-species linear mixed effects model. No individual CpG sites had methylation values that were significantly associated with sulfate levels after FDR correction.
4. Discussion
In this study, we report positive associations of 1-year PM2.5 exposure levels with Horvath DNAm-age in a population of community-dwelling, elderly men. Additionally, we utilized the adaptive LASSO to identify 1-year sulfate and ammonium levels as the PM2.5 components most robustly associated with Horvath DNAm-age. To our knowledge, this is the first report of associations of multiple PM2.5 component species with DNAm-age and the second time that satellite-derived PM2.5 exposure levels have been found to be associated with Horvath DNAm-age. In addition to being consistent with the existing literature (Nwanaji-Enwerem et al., 2016), our findings also demonstrate important public health relevance as they persist in sensitivity analyses including only participant visits with 1-year PM2.5 levels within current US EPA national ambient air quality standards (US EPA, O.A.R., n.d.). Our study also extends the literature by exploring PM2.5 relationships with Hannum DNAm-age although these relationships were found to be null. Furthermore, we identified 47 CpG sites, 9 of which were previously reported, whose methylation values were significantly associated with PM2.5 levels in fully-adjusted linear mixed effects models. Only 1 CpG was associated with ammonium levels and 0 were associated with sulfate levels.
Given our prior report of robust associations of PM2.5 levels from satellite-based spatiotemporal models with Horvath DNAm-age, we expected to observe a similar positive relationship using PM2.5 levels from the GEOS-chem chemical transport model. As expected, we observed that an IQR increase in 1-year PM2.5 exposure was associated with a 0.64-year increase in Horvath DNAm-age. Since PM2.5 component species are highly related to total PM2.5, we also expected that PM2.5 component species would be associated with Horvath DNAm-age, even when adjusting for PM2.5 mass. Given the existing literature concerning the differential health effects of PM2.5 component species, we speculated that some component species may be more robustly associated with Horvath DNAm-age than others. In particular, we expected the carbonaceous fractions to be among the species most robustly associated with DNAm-age due to the extensive literature (including work from our group) on the adverse nature of carbonaceous fraction exposures on health (Nwanaji-Enwerem et al., 2016; Baccarelli et al., 2009; Colicino et al., 2014; Zanobetti et al., 2014; McCracken et al., 2010). In our fully adjusted one-species linear mixed effects models, we observed strong positive associations of 4 out of the 5 component species examined with Horvath DNAm-age. IQR range increases in organic carbon, sulfate, nitrate, and ammonium were all significantly associated with at least a 0.58-year increase in Horvath DNAm-age.
Despite the results from our fully adjusted one-species linear mixed effects models, we desired a method to more comprehensively identify the component species most associated with DNAm-age. Nevertheless, we were aware that simply modeling highly-correlated PM2.5 species together would result in unaccounted for stochastic errors. Thus, we employed the adaptive LASSO as a penalized regression method to help overcome this difficulty. The literature has shown that carbonaceous fractions are robustly associated with age-related health outcomes (Nwanaji-Enwerem et al., 2016; Baccarelli et al., 2009; Colicino et al., 2014; Zanobetti et al., 2014; McCracken et al., 2010); however, neither elemental or organic carbon were selected in our models. Rather, sulfate and ammonium were selected. This difference may be explained by the fact that a majority of the aforementioned studies did not consider other PM2.5 component species in addition to the carbonaceous fractions. Even in our single-species linear mixed effects models, we note that organic carbon was among the four species significantly associated with Horvath DNAm-age (Table 3). However, when all five component species are considered together in the adaptive LASSO, only sulfate and ammonium were selected. It is also possible that the LASSO did not select the carbonaceous fractions because the selection was performed under PM2.5 adjustment and PM2.5 may be capturing most of the variability of organic and elemental carbon. Thus, we performed LASSO selection not adjusting for total PM2.5 mass as a sensitivity analysis. This time LASSO did select organic carbon along with sulfate and ammonium. However, when these three component species were modeled with PM2.5 in a multiple-species fully-adjusted linear mixed effects model, organic carbon was the only species that was not a significant predictor of DNAm-age. This suggests that organic carbon was selected in the sensitivity analysis because of its strong correlation with PM2.5 mass and not because organic carbon itself is a good predictor of DNAm-age. This finding also reiterates the notion that adjustment for PM2.5 mass in component species models is very important as PM2.5 mass often confounds the relationship between the outcome and species (Mostofsky et al., 2012). Failing to include PM2.5 mass may lead to misleading findings about species. In all, our data suggests that of the considered species, sulfate and ammonium have the most important relationships with DNAm-age. Furthermore, existing studies that do consider a range of PM2.5 components demonstrate that other non-carbonaceous components are important to age-related outcomes (Dai et al., 2016b; Wu et al., 2013; Wu et al., 2015). These data, together with our findings, also suggests the important need to consider a range of PM2.5 components, rather than one or two species, in air pollution and health studies.
Both sulfate and ammonium are classified in the inorganic fraction of PM2.5. Sulfates are often produced from oxidation or photochemical reactions involving primary gases derived from sources like coal-burning power plants (Huang et al., 2014). Additionally, ammonia from organic sources including animal feeds and fertilizers can contribute to the existence of sulfates in the form of atmospheric ammonium sulfate (Frank, n.d.). As far as direct ambient sulfate and ammonium toxicity to human health is concerned, existing studies are limited. Yet, there has been extensive evidence describing the ability of acidic sulfates, like ammonium sulfate, to increase the number and toxicity of biologically harmful secondary particles (Mostofsky et al., 2012; Popovicheva et al., 2011; Rubasinghege et al., 2010; Li et al., 2011; Lepeule et al., 2012; Schwartz and Lepeule, 2012). For instance, ammonium sulfate aerosols have been shown to influence the photo-chemical reactions of nitrogen oxides and toluene hastening the production of secondary organic aerosols (Wu et al., 2007). Moreover, sulfur concentrations have been found to be directly proportional to the ability of soluble particle extracts to generate biologically damaging oxidants (Ghio et al., 1999). Furthermore, a prior study in the NAS has reported a 27% decrease in long interspersed nucleotide element-1 methylation per every IQR increase in 90-day sulfate exposure. This study provides evidence for the influence of sulfates on DNA methylation, which may be a potential pathway for sulfate toxicity (Madrigano et al., 2011). It is still unclear what the molecular relevance of Horvath DNAm-age is, but our findings along with the existing literature will be helpful in providing additional insight for future work.
Following the selection of sulfate and ammonium by the adaptive LASSO, we constructed a final multiple-species linear mixed effects model adjusted for PM2.5 mass and all covariates. Even in this model, sulfate and ammonium remained significant positive predictors of Horvath DNAm-age. We then looked to see if there were specific Horvath DNAm-age component CpG sites with methylation values that were associated with PM2.5, sulfate, and/or ammonium in our fully-adjusted multiple-species linear model. From this analysis, we identified 47 significant CpG sites after FDR adjustment. These sites mapped to 46 genes, and 9 of them were reported in a previous CpG-level analysis of the same 353 sites in the Horvath algorithm that we conducted using PM2.5 levels from a satellite-based spatiotemporal model. To better grasp the impact of PM2.5 levels on methylation, we divided the coefficients for each significant CpG site (i.e. difference in methylation per IQR increase in particle level) by the standard deviation of the respective particle level. We were pleased to see that 5 of the 9 CpGs that were shared between both PM2.5 prediction models were in the top 20% of our gene list. We then combined the gene lists from both PM2.5 prediction models (removing any duplicates) and performed a gene ontology (GO) analysis. The GO analysis returned the term “regulation of translational initiation” with the following genes from our list falling into this category: RXRA, EIF3M, EIF31. Though the GO term itself is not highly specific, combining this pathway with what is known about the toxicity of PM2.5 will be useful in further understanding how PM2.5 may contribute to aging and disease. Only 1 CpG was associated with ammonium levels and it mapped to the gene SOAT1, which is involved in fatty-acyl-CoA binding. SOAT1 has been implicated in a number of diseases including familial hypercholesterolemia (Peters et al., 2011). No CpG sites were specifically associated with sulfate levels. The finding that almost no CpGs sites were associated with ammonium and sulfate further demonstrates that Horvath DNAm-age is simply not a reflection of its 353 component CpGs, and reiterates the need for work focused on defining the molecular relevance of DNAm-age.
Finally, our study demonstrates that all DNAm-age measures are not the same. In the literature there is evidence of both Horvath and Hannum DNAm-age reflecting the same disease outcome and evidence where they differ in their reporting ability. For instance, both Horvath and Hannum DNAm-age appear to be useful in predicting mortality (Chen et al., 2016; Wolf et al., 2016). However, in a study of male and female veterans, Hannum DNAm-age was associated with post-traumatic stress disorder and neural integrity, but Horvath DNAm-age was not (Perna et al., 2016). The differences in these two DNAm-age measures may stem from the fact that they are derived from almost entirely different CpG sites or from the fact that Horvath DNAm-age was constructed using many datasets of multiple tissue types and the Hannum DNAm-age was based only on blood from one dataset (Horvath, 2013; Hannum et al., 2013). Our results suggest that Hannum DNAm-age is not sensitive to exposure levels of PM2.5 and its component species. Additional studies in different populations will be necessary to confirm these findings more broadly. Nonetheless, continued research exploring the specific sensitivity of DNAm-age measures will be a crucial next step in the growth of this field of research. Once more is known about the profiles of these markers, we can begin to use them more effectively in answering questions concerning human health.
Strengths of our study include rigorous statistical methods and access to a large cohort with extensive and repeated information regarding pollutant exposures, potential confounders, and DNA methylation data from multiple study visits. However, our study does have several limitations. First, although we used a validated chemical transport model to estimate the levels of ambient PM2.5 and its component species at participants’ addresses, we recognize that these estimates may differ from personal exposures. Nonetheless, we know that a majority of NAS participants are retired and spend most of their time at home. Moreover, our approach is expected to result in non-differential misclassification that is likely to underestimate the observed associations rather than bias them away from the null (Kioumourtzoglou et al., 2014). Secondly, it is known that LASSO regression is limited to linear relationships. Given the linear relationship of our particle exposures with DNAm-age and the scope of this paper, the adaptive LASSO was a good tool for identifying PM2.5 components that are independently important to DNAm-age. However, for future studies potentially interested in the interactions between PM2.5 components, another technique may be necessary as PM2.5 species interactions that are important for the prediction of DNAm-age may be more complex (i.e. not linear). Third, we note that our findings are based on an elderly cohort of Caucasian males that reside in a lightly-polluted environment. Hence, additional studies involving other demographic groups and in different environments will be necessary to confirm our findings more broadly. Finally, we used the existing literature and a priori knowledge of biological/clinical relevance to adjust for potential confounders. Nonetheless, we cannot rule out the possibility of unknown or residual confounding in our analyses.
5. Conclusion
Our study utilizes the GEOS-chem chemical transport model to validate novel positive associations between long-term PM2.5 exposure levels and Horvath DNAm-age. For the first time, we demonstrate that sulfate and ammonium are among the PM2.5 component species most associated with Horvath DNAm-age in this population of elderly men. In contrast, we observed no relationships of long-term PM2.5 and PM2.5 component species exposure levels with Hannum DNAm-age. These results suggest that DNA methylation-based biomarkers of age differ in their sensitivity to ambient particle exposures and potentially disease outcomes. Future studies in other populations will be critical for defining the environmental and disease sensitivity profiles of DNAm-age measures.
Supplementary Material
Acknowledgments
This study is supported by grants from the National Institute of Environmental Health Sciences (NIEHS) (R01ES021733 and R01ES025225-01A1). Other support comes from NIEHS grants ES015172, ES014663 and ES020010, and Environmental Protection Agency (EPA) grant RD832416. The US Department of Veterans Affairs (VA) Normative Aging Study (NAS) is supported by the Cooperative Studies Program/ERIC, US Department of Veterans Affairs, and is a research component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). The views expressed in this paper are those of the authors and do not necessarily represent the views of the US Department of Veterans Affairs. Additional support was provided by the US Department of Agriculture, Agricultural Research Service (contract 53-K06-510).
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.envint.2016.12.024.
Footnotes
Contributors
JCN and JDS conceived and designed the study. EC, QD, IK, ACJ, LH, and PV gathered data. JCN performed the data analyses and drafted the manuscript. LD, JDS, EC, YO, MGW, and AAB contributed to the analyses. All authors revised and approved the manuscript.
Conflict of interest statement
None declared.
Ethics approval
Boston VA Medical Center, Harvard T.H. Chan School of Public Health (protocol 14027-102).
Data availability
Data are from the Normative Aging Study, from which restricted data are available for researchers who meet the criteria. A subset of the methylation data is deposited at NCBI dbGaP (study accession number: phs000853.v1.p1).
Competing financial interests related to this research
None.
References
- Apte JS, et al. Addressing global mortality from ambient PM2.5. Environ Sci Technol. 2015;49(13):8057–8066. doi: 10.1021/acs.est.5b01236. [DOI] [PubMed] [Google Scholar]
- Aryee MJ, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179(7):572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B, Rose CL, Damon A. The veterans administration longitudinal study of healthy aging. The Gerontologist. 1966;6(4):179–184. doi: 10.1093/geront/6.4.179. [DOI] [PubMed] [Google Scholar]
- Bind MA, et al. Beyond the mean: quantile regression to explore the association of air pollution with gene-specific methylation in the normative aging study. Environ Health Perspect. 2015;123(8):759–765. doi: 10.1289/ehp.1307824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, et al. Occupational exposure to PM2.5 and incidence of ischemic heart disease: longitudinal targeted minimum loss-based estimation. Epidemiology. 2015;26(6):806–814. doi: 10.1097/EDE.0000000000000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BH, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8(9):1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Schwartz J. Neurobehavioral effects of ambient air pollution on cognitive performance in US adults. Neurotoxicology. 2009;30(2):231–239. doi: 10.1016/j.neuro.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Chung HY, et al. Molecular inflammation: underpinnings of aging and age-related diseases. Ageing Res Rev. 2009;8(1):18–30. doi: 10.1016/j.arr.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicino E, et al. Mitochondrial haplogroups modify the effect of black carbon on age-related cognitive impairment. Environ Health. 2014;13(1):42. doi: 10.1186/1476-069X-13-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, et al. Fine particles, genetic pathways, and markers of inflammation and endothelial dysfunction: analysis on particulate species and sources. J Expo Sci Environ Epidemiol. 2016a;26(4):415–421. doi: 10.1038/jes.2015.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, et al. Use of the adaptive LASSO method to identify PM2.5 components associated with blood pressure in elderly men: the veterans affairs normative aging study. Environ Health Perspect. 2016b;124(1):120–125. doi: 10.1289/ehp.1409021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Q, Koutrakis P, Schwartz J. A hybrid prediction model for PM 2.5 mass and components using a chemical transport model and land use regression. Atmos Environ. 2016;131:390–399. [Google Scholar]
- Frank N. The Chemical Composition of PM2.5 to Support PM Implementation 2007 [Google Scholar]
- Ghio AJ, et al. Sulfate content correlates with iron concentrations in ambient air pollution particles. Inhal Toxicol. 1999;11(4):293–307. doi: 10.1080/089583799197104. [DOI] [PubMed] [Google Scholar]
- Giorgini P, et al. Air pollution exposure and blood pressure: an updated review of the literature. Curr Pharm Des. 2016;22(1):28–51. doi: 10.2174/1381612822666151109111712. [DOI] [PubMed] [Google Scholar]
- Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging. 2015;7(12):1130–1142. doi: 10.18632/aging.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci. 2014;111(43):15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, et al. The cerebellum ages slowly according to the epigenetic clock. Aging. 2015;7(5):294–306. doi: 10.18632/aging.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XHH, et al. Characterization of PM2.5 major components and source investigation in suburban Hong Kong: a one year monitoring study. Aerosol Air Qual Res. 2014;14(1):237–250. [Google Scholar]
- Ito K, et al. Fine particulate matter constituents associated with cardiovascular hospitalizations and mortality in New York City. Environ Health Perspect. 2011;119(4):467–473. doi: 10.1289/ehp.1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioumourtzoglou MA, et al. Exposure measurement error in PM2.5 health effects studies: a pooled analysis of eight personal exposure validation studies. Environ Health. 2014;13(1):2. doi: 10.1186/1476-069X-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, et al. Predicting spatiotemporal mean air temperature using MODIS satellite surface temperature measurements across the Northeastern USA. Remote Sens Environ. 2014;150:132–139. [Google Scholar]
- Kloog I, et al. Effects of airborne fine particles (PM2.5) on deep vein thrombosis admissions in the northeastern United States. J Thromb Haemost. 2015;13(5):768–774. doi: 10.1111/jth.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzli N, et al. Ambient air pollution and atherosclerosis in Los Angeles. Environ Health Perspect. 2005;113(2):201–206. doi: 10.1289/ehp.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent O, et al. Low birth weight and air pollution in California: which sources and components drive the risk? Environ Int. 2016;92–93:471–477. doi: 10.1016/j.envint.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepeule J, et al. Chronic exposure to fine particles and mortality: an extended follow-up of the Harvard Six Cities study from 1974 to 2009. Environ Health Perspect. 2012;120(7):965–970. doi: 10.1289/ehp.1104660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, et al. Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol. 2016a;22(3):366–375. doi: 10.1007/s13365-015-0406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, et al. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging. 2015b;7(9):690–700. doi: 10.18632/aging.100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, et al. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging. 2015c;7(12):1198–1211. doi: 10.18632/aging.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, et al. Internally mixed sea salt, soot, and sulfates at Macao, a coastal city in South China. J Air Waste Manage Assoc. 2011;61(11):1166–1173. doi: 10.1080/10473289.2011.603996. [DOI] [PubMed] [Google Scholar]
- Liu S, et al. Fine particulate matter components and emergency department visits among a privately insured population in Greater Houston. Sci Total Environ. 2016;566–567:521–527. doi: 10.1016/j.scitotenv.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Madrigano J, et al. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect. 2011;119(7):977–982. doi: 10.1289/ehp.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44(4):1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Harris SE, Shah S, McRae AF, von Zglinicki T, Martin-Ruiz C, … Deary IJ. The epigenetic clock and telomere length are independently associated with chronological age and mortality. International journal of epidemiology. 2016;45(2):424–432. doi: 10.1093/ije/dyw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Olivieri O, Girelli D. Air particulate matter and cardiovascular disease: a narrative review. Eur J Intern Med. 2013;24(4):295–302. doi: 10.1016/j.ejim.2013.04.001. [DOI] [PubMed] [Google Scholar]
- McCracken J, et al. Annual ambient black carbon associated with shorter telomeres in elderly men: veterans affairs normative aging study. Environ Health Perspect. 2010;118(11):1564–1570. doi: 10.1289/ehp.0901831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostofsky E, et al. Modeling the association between particle constituents of air pollution and health outcomes. Am J Epidemiol. 2012;176(4):317–326. doi: 10.1093/aje/kws018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwanaji-Enwerem JC, et al. Long-term ambient particle exposures and blood DNA methylation age: findings from the VA normative aging study. Environ Epigenet. 2016;2(2) doi: 10.1093/eep/dvw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, et al. Particulate air pollution and fasting blood glucose in nondiabetic individuals: associations and epigenetic mediation in the normative aging study, 2000–2011. Environ Health Perspect. 2016;124(11):1715–1721. doi: 10.1289/EHP183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna L, et al. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8:64. doi: 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BJ, et al. Genetic variability within the cholesterol lowering pathway and the effectiveness of statins in reducing the risk of MI. Atherosclerosis. 2011;217(2):458–464. doi: 10.1016/j.atherosclerosis.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovicheva OB, et al. Quantification of the hygroscopic effect of soot aging in the atmosphere: laboratory simulations. J Phys Chem A. 2011;115(3):298–306. doi: 10.1021/jp109238x. [DOI] [PubMed] [Google Scholar]
- Power MC, et al. Traffic-related air pollution and cognitive function in a cohort of older men. Environ Health Perspect. 2011;119(5):682–687. doi: 10.1289/ehp.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaschou-Nielsen O, et al. Particulate matter air pollution components and risk for lung cancer. Environ Int. 2016;87:66–73. doi: 10.1016/j.envint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- Rowe JW, Kahn RL. Human aging: usual and successful. Science. 1987;237(4811):143–149. doi: 10.1126/science.3299702. [DOI] [PubMed] [Google Scholar]
- Rowe JW, Kahn RL. Successful aging and disease prevention. Adv Ren Replace Ther. 2000;7(1):70–77. doi: 10.1016/s1073-4449(00)70008-2. [DOI] [PubMed] [Google Scholar]
- Rowe JW, Kahn RL. Successful aging 2.0: conceptual expansions for the 21st century. J Gerontol B Psychol Sci Soc Sci. 2015;70(4):593–596. doi: 10.1093/geronb/gbv025. [DOI] [PubMed] [Google Scholar]
- Rubasinghege G, et al. Simulated atmospheric processing of iron oxyhydroxide minerals at low pH: roles of particle size and acid anion in iron dissolution. Proc Natl Acad Sci U S A. 2010;107(15):6628–6633. doi: 10.1073/pnas.0910809107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheers H, et al. Long-term exposure to particulate matter air pollution is a risk factor for stroke: meta-analytical evidence. Stroke. 2015;46(11):3058–3066. doi: 10.1161/STROKEAHA.115.009913. [DOI] [PubMed] [Google Scholar]
- Schelldorfer J, et al. Estimation for high-dimensional linear mixed-effects models using 31-penalization. Scand J Stat. 2011;38(2):197–214. [Google Scholar]
- Schikowski T, et al. Association of air pollution with cognitive functions and its modification by APOE gene variants in elderly women. Environ Res. 2015;142:10–16. doi: 10.1016/j.envres.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Schwartz J, Lepeule J. Is ambient PM2.5 sulfate harmful? Schwartz and Lepeule respond. Environ Health Perspect. 2012;120(12):a454–a455. doi: 10.1289/ehp.1205873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G, et al. Estimating the dimension of a model. Ann Stat. 1978;6(2):461–464. [Google Scholar]
- Shan M, et al. A feasibility study of the association of exposure to biomass smoke with vascular function, inflammation, and cellular aging. Environ Res. 2014;135:165–172. doi: 10.1016/j.envres.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Terzano C, et al. Air pollution ultrafine particles: toxicity beyond the lung. Eur Rev Med Pharmacol Sci. 2010;14(10):809–821. [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B Methodol. 1996;58(1):267–288. [Google Scholar]
- US, E.P.A. O.A.R, NAAQS Table. 2015. [Google Scholar]
- van Donkelaar A, et al. Global estimates of ambient fine particulate matter concentrations from satellite-based aerosol optical depth: development and application. Environ Health Perspect. 2010;118(6):847–855. doi: 10.1289/ehp.0901623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weuve J, et al. Exposure to particulate air pollution and cognitive decline in older women. Arch Intern Med. 2012;172(3):219–227. doi: 10.1001/archinternmed.2011.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EH, et al. Long-term exposure to fine particulate matter, residential proximity to major roads and measures of brain structure. Stroke. 2015;46(5):1161–1166. doi: 10.1161/STROKEAHA.114.008348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, et al. Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–162. doi: 10.1016/j.psyneuen.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health, O. Burden of Disease from Household Air Pollution for 2012. WHO; Geneva: 2014. [Google Scholar]
- Wu S, et al. Effect of ammonium sulfate aerosol on the photochemical reaction of toluene/NO(x)/air mixture. Huan Jing Ke Xue. 2007;28(6):1183–1187. [PubMed] [Google Scholar]
- Wu S, et al. Blood pressure changes and chemical constituents of particulate air pollution: results from the healthy volunteer natural relocation (HVNR) study. Environ Health Perspect. 2013;121(1):66–72. doi: 10.1289/ehp.1104812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, et al. Association of chemical constituents and pollution sources of ambient fine particulate air pollution and biomarkers of oxidative stress associated with atherosclerosis: a panel study among young adults in Beijing, China. Chemosphere. 2015;135:347–353. doi: 10.1016/j.chemosphere.2015.04.096. [DOI] [PubMed] [Google Scholar]
- Zanobetti A, et al. Fine particulate air pollution and its components in association with cause-specific emergency admissions. Environ Health. 2009;8:58. doi: 10.1186/1476-069X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanobetti A, et al. Associations between arrhythmia episodes and temporally and spatially resolved black carbon and particulate matter in elderly patients. Occup Environ Med. 2014;71(3):201–207. doi: 10.1136/oemed-2013-101526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, et al. Cardiac autonomic dysfunction: particulate air pollution effects are modulated by epigenetic immunoregulation of toll-like receptor 2 and dietary flavonoid intake. J Am Heart Assoc. 2015;4(1):e001423. doi: 10.1161/JAHA.114.001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. J Am Stat Assoc. 2006;101(476):1418–1429. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.