Summary
The recent advent of microphysiological systems – microfluidic biomimetic devices that aspire to emulate the biology of human tissues, organs and circulation in vitro – is envisaged to enable a global paradigm shift in drug development. An extraordinary US governmental initiative and various dedicated research programs in Europe and Asia have led recently to the first cutting-edge achievements of human single-organ and multi-organ engineering based on microphysiological systems. The expectation is that test systems established on this basis would model various disease stages, and predict toxicity, immunogenicity, ADME profiles and treatment efficacy prior to clinical testing. Consequently, this technology could significantly affect the way drug substances are developed in the future. Furthermore, microphysiological system-based assays may revolutionize our current global programs of prioritization of hazard characterization for any new substances to be used, for example, in agriculture, food, ecosystems or cosmetics, thus, replacing laboratory animal models used currently. Thirty-five experts from academia, industry and regulatory bodies present here the results of an intensive workshop (held in June 2015, Berlin, Germany). They review the status quo of microphysiological systems available today against industry needs, and assess the broad variety of approaches with fit-for-purpose potential in the drug development cycle. Feasible technical solutions to reach the next levels of human biology in vitro are proposed. Furthermore, key organ-on-a-chip case studies, as well as various national and international programs are highlighted. Finally, a roadmap into the future is outlined, to allow for more predictive and regulatory-accepted substance testing on a global scale.
Keywords: microphysiological systems, organ-on-a-chip, in vitro models, predictive toxicology, drug testing
1. The prediction dilemma of substance testing using laboratory animals
According to the most recent report from the European Commission (EC) to the Council and the European Parliament (EC, 2013), 11.5 million animals were used for experimental and other scientific purposes in the Member States of the European Union (EU) in 2011. The report indicates that the number of animals used for research and development for human medicine, dentistry and veterinary medicine has dropped since 2008 from 22.8 to 18.8 %. The number of animals used for toxicological and other safety evaluation, amounting to 8.75 % of the total, remained relatively unchanged. However, the percentage of animals used for fundamental biological research has increased sharply from 38 to 46 %. These three areas use by far the highest number of animals (8.7 million in 2011) for scientific purposes in the EU (Daneshian et al., 2015). Information and results generated from biological studies of a fundamental nature using animals are the groundwork for new medicine development in the pharmaceutical and biotech industries. The EC report highlights this fact by saying that ocular research, bone metabolism, fertility studies, potency testing, immunogenicity testing, studies in the areas of neuroscience and immunology, studies on pathophysiological mechanisms of tumors and research to gain experience for the determination of mechanisms of action of diseases for therapeutic purposes were the major causes for the increase in fundamental biological research. The last few decades of research and development have shown clearly that data from animal studies are often poorly indicative of the human situation (Hackam and Redelmeier, 2006; Leist and Hartung, 2013; Matthews, 2008; Olson et al., 2000; Perel et al., 2007; Hartung and Leist, 2008; Schnerch et al., 2010; Sena et al., 2010; Seok et al., 2013; van der Worp et al., 2010; Hartung, 2013). The latter is illustrated by repeated failures of drugs in clinical trials. Industry benchmarks for candidate success rates in a certain phase of clinical trials are 48 – 64 % for phase I, 29 – 32 % for phase II and 60 – 67 % for phase III, respectively (Cook et al., 2014; Hay et al., 2014). Hence, the report presented here focuses on the drug development aspects of the substance testing dilemma.
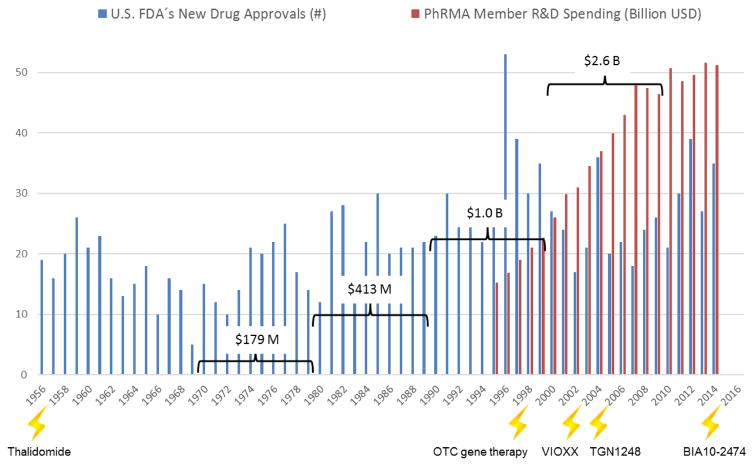
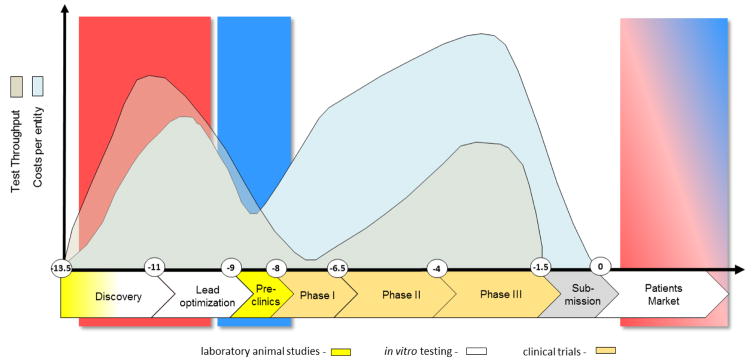
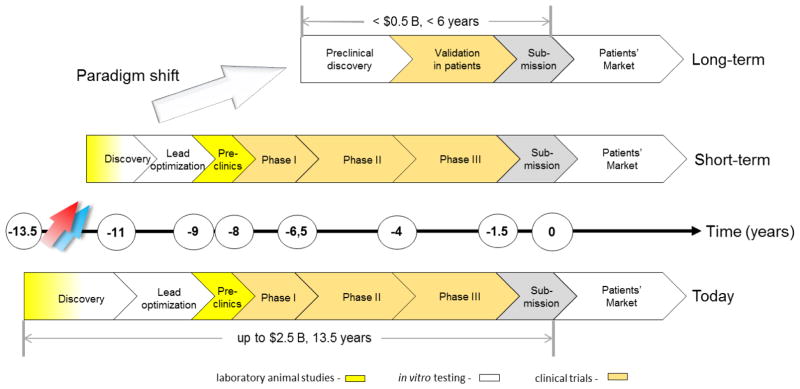
Development of new medicines is currently suffering from two major obstacles (Rovida et al., 2015a): The current approach in preclinical therapeutic drug discovery, which comes with a high candidate attrition before and even during clinical trials, and a decreasing R&D productivity at constantly increasing regulatory requirements regarding preclinical testing to avoid harm to human individuals (Fig. 1). The last seven decades hallmarked a significant increase of pharmaceutical drug development costs (spending of U.S. PhRMA members) while US Food and Drug Administration (FDA) new drug approval rates fluctuated in response to prime drug development disasters. R&D productivity has declined more than fifteen-fold, indicated by the respective increase of inflation-adjusted average spending from $179 million per successful drug (including the costs of failures) in the 1970s to $2.6 billion in the twenty-first century (Scannell et al., 2012; Tufts CSDD, 2014) (Fig. 1).
Fig. 1. Changes in drug development over the last seventy years.
Number of drugs approved by the U.S. FDA (FDA, 2013; FDA, 2014a) are plotted against the pharmaceutical research and development spending of the members of the Pharmaceutical Research and Manufacturers of America (PhARMA, 2015).
(lightening) – drug or substance failures with detrimental outcome for humans (see Table 1)
(brackets) – average costs to develop one new drug including costs of failures within the corresponding decade
The Thalidomide case was the first notorious drug failure with the resulting extreme handicaps for over 10,000 people. This event triggered the advent of pre-clinical teratogenicity testing. Each subsequent disaster naturally prompted an increase in relevant regulations for substance hazard identification, safety testing and efficacy evaluation. Some of these events, including the most recent fatty acid amide hydroxylase inhibitor failure from Bial-Portela & Ca. SA, are summarized in Table 1.
Tab. 1.
Prime examples of adverse outcome effects of drugs in human
| Date/Substance | Adverse outcome Pathway/Mode of Action | Reference |
|---|---|---|
| 1957 – 1961/Thalidomide | It was administered as a sleeping pill and an antidote to morning sickness suitable for pregnant women. Thalidomide or the metabolic breakdown products inhibit transaction of genes which interferes with normal angiogenesis resulting in truncation of the limb. | Fabro et al. (1965), Woollam, (1965), Bauer et al. (1998), Stephens and Fillmore (2000) |
| 1999/experimental gene therapy | Omithine transcarbamylase (OTC) deficiency is a genetic disease that prevents the body from breaking down ammonia, which is a metabolic waste product. The 18-year-old Jesse Gelsinger volunteered for a gene therapy experiment designed to test possible treatments for OTC. Four days after the injection of the adenovirus, the vector being used to deliver the OTC gene, he died. | Somia and Verma (2000), Edwards (2004) |
| 2004/VIOXX | Specific genetic predisposition is one of the key reasons for the now common and highly publicized drug withdrawals. This human genetic diversity is rarely addressed in preclinical and clinical safety studies at the present time. A sound hypothesis on the correlation of the morbidity with polymorphic genotypes for 5-LOX and 5-LOX activating proteins is one of many examples. | Dieppe et al. (2004), Arbor (2004) |
| 2006/TGN1412 | Super-agonist antibody TGN1412 was developed to direct the immune system to fight cancer cells or reduce arthritis pain. It triggered multiple organ failure m six healthy volunteers who participated in phase I clinical testing. The antibody, by binding to the CD28-receptor, overrides the basic control mechanisms of the whole immune system. | Clair (2008), Stebbings et al. (2012) |
| 2016/BIA 10-2474 | Clinical testing on an experimental fatty acid amide hydrolase inhibitor from Bial-Portela & Ca. SA, led to six patients being hospitalized on January 10, 2016 in France. Testing was stopped on January 11. One volunteer has died. The drug, aiming to treat mood and motor disorders, had previously been tested in mice, rats, dogs and monkeys with no adverse effects. Species differences in inhibitor activity have been discussed to be particularly problematic in drug discovery because in vivo efficacy models are largely based on rodent studies, but the candidate inhibitor is ultimately developed as a human drug. | Jouan (2016). Milem et al. (2008) |
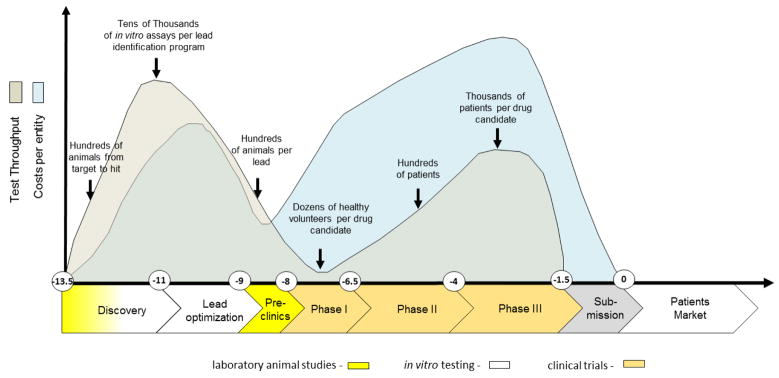
These historical drug failures with severe outcomes have led to the establishment of a development approach spanning several years of experimentation and involving animals, in vitro tests and healthy volunteers or patients (Fig. 2). A pool of more than 10,000 entities need to be fed into the drug development pipeline to finally arrive at one successful product (Kessel and Frank, 2007). A stringent regime of traditionally defined steps applies to that process. The goal for this extensive testing approach is to ensure patients’ safety. It has been the best approach possible to date. However, it is often inefficient, and would today subject blockbusters, such as Aspirin or Paracetamol, to regulatory disapproval (Hartung, 2009).
Fig. 2. Drug development cycle: test throughput and cost profile.
The vertical axis illustrates approximate numbers of tests performed (grey) and related spending (blue). The horizontal axis illustrates the development time in years (Paul et al., 2010). Animal tests are used in early discovery for mechanistic mode of action research and for toxicity and ADME profiling in the preclinical phase, while conventional in vitro assays are largely used in discovery for target validation, target-to-lead translation and lead optimization steps.
The smaller part of the investment during the cycle is spent on discovery and lead optimization (Paul et al., 2010), performing larger numbers of in vitro assays per lead identification and a significant number of laboratory animal tests per target or lead. Despite all these efforts, the phylogenetic distance between laboratory animals and humans and the discrepancy between current human in vitro tests and the human body have not improved attrition rates in clinical trials to a satisfactory level. Although animals represent systemic organisms, they are not human, and the in vitro tests on human cells are neither physiological nor systemic. Despite intense preclinical safety testing in a number of phase I trials, safety issues arise which may lead to the termination of a program (Cook et al., 2014; Schuster et al., 2005). Failure to predict efficacy and toxicity in the preclinical phases will lead to serious delays in the development of required drugs, exposure of subjects to inefficient substances and even unwanted side effects as well as initiating unsuccessful and expensive clinical programs, which are the largest investment points in the drug development process (Ledford, 2011).
In addition to this pharma prediction dilemma, a similar obstacle holds true for other industries, such as chemicals and consumer products, where animals are used for the toxicological hazard and risk assessment of substances. This situation has led to an ethical concern about the use of a large number of laboratory animals which demands justification with respect to predictive value.
Based on the above, it is obvious that new approaches are required to bridge the translational gap described and could eventually contribute to a higher productivity of the drug development process. We believe that biology-inspired microphysiological in vitro systems (MPS) will be a cornerstone of this bridge.
2. Microphysiological systems – an expanding toolbox for hazard, safety, disease and efficacy prediction of particulate matter, chemicals and drug candidates
2.1. Definition and terminology
Microphysiological systems are microfluidic devices capable of emulating human (or any other animal species’) biology in vitro at the smallest biologically acceptable scale, defined by purpose. The application of fluid flow (dynamic) for physiological nutrition of the tissues and for the creation of microenvironmental biomolecular gradients and relevant mechanical cues (e.g. shear stresses) is a major aspect of these systems, differentiating them from conventional (static) cell and tissue cultures.
The “system” component of MPS refers to devices which support human-like physiology of tissues and organ equivalents within the devices in vitro. This may include the maintenance of physical factors, such as temperature (e.g. 37 °C), relevant pH, and supply and control of the levels of oxygen and humidity required. Furthermore, it comprises mechanical coupling of organs by mimicking, for example, the flow of blood, urine, air in the lung, bile, pancreatic juice or cerebral fluid, shear stress regarding blood and lymphatic vessels, physical pressure on bone and cartilage, strain on skin, lung and stomach wall, intestinal peristaltic movement and muscle contraction. For more details on how microfluidic tools can be used to study mechanobiology we refer to a comprehensive review (Polacheck et al., 2013). In addition, the term “system” may incorporate readout of electrical activity of neuronal and cardiac tissues. The implementation of miniaturized relevant actuators and sensors into the devices enables necessary features for in vivo-like tissue-specific electro-mechano-biochemical signaling. They support expansion and compression forces, especially relevant for lung, bone and cartilage, and microelectrodes for the electrical stimulation and readout of muscle tissue (Ahadian et al., 2012; Dvir et al., 2012) or stimulation of cardiac cells or neurons (Bussek et al., 2009; Gramowski et al., 2011; Himmel et al., 2012; Johnstone et al., 2010, Koshferat Pakazad 2014). Moreover, such devices could be able to apply other technical means of measurement and control, such as noninvasive optical imaging. Finally, the “system” in an MPS implies a high degree of automation, which makes it robust and scalable. It is necessary to consider all of these in order to enable reproducible high throughput repeated substance exposure protocols mimicking human drug exposure as closely as possible. Further detailed information on these features are reviewed elsewhere (Andersson and van den Berg, 2004; Huh et al., 2011; Ingber and Whitesides, 2012; Kim et al., 2007; Park and Shuler, 2003; Wu et al., 2010).
The “physiological” component of MPS stands for the ambition to truly emulate human biology. In the current literature, the “true emulation” of human biology in vitro is described in different ways, depending on the background and prior experiences of the author(s). Adherence as much as possible to the emulation of human organ architecture, including proper cell-to-cell, cell-to-matrix, biochemical and mechanical signaling, is one of the cornerstones of our definition of “physiological.” Glass- and silicone-based devices complemented with polymers, textiles, ceramic or biological matrix entities are individually designed to match the well reviewed requirements regarding shape, surface pattern, stiffness, and microarchitecture of each specific organ microenvironment (Griffith and Swartz, 2006; Nelson and Gleghorn, 2011; Peyton et al., 2011). Another cornerstone encompasses the maintenance of healthy long-term organ homeostasis. The third cornerstone covers the ability to repair and regenerate organs which have been damaged or diseased. Thus, the latter cornerstone logically introduces one pathophysiological dimension of MPS. The final crucial cornerstone is to mimic human diseased tissue in the chip, this further introduces the pathophysiological dimension.
The “micro” component of MPS stands for miniaturization to the smallest biologically acceptable scale for each application. This miniaturization is driven by four major factors: mimicking organ function at laminar fluid flow in the cultured tissue, test throughput in the different phases of the drug development cycle (fit-for-purpose), minimum use of human cells and tissues, and, along with this, cost consideration. Scaling MPS-based organ equivalents down to an organoid level, as defined below, is one of the possible scaling strategies in the field. It is common knowledge today that almost all human organs consist of multiple, identical, functionally self-reliant, structural units, realizing the most relevant functions of the respective organ. Multiplication of these structures within a given organ is nature’s risk management tool to prevent a total loss of functionality during partial organ damage or disease for the majority of body organs. Regarding evolution, this concept has allowed the easy adjustment of organ size and shape to the needs of a given species – for example, liver in mice and men – still using nearly the same master plan to build up single functional organoids. The term “organoid” has been used as early as 1876 in different ways in life science (Bristowe, 1876, p. 73) and means the smallest functional organ or tissue unit. A selection of organoid histologies, all with a relevant functionality and highly variable conglomerate geometry, has been published for 15 key human organs (Marx et al. 2012). Ideally, the reactivity of organoids to drugs is representative of the reactivity of the organ. Therefore, a single organoid should reflect the smallest possible scale which can be used in MPS in order to emulate organ functionality. Selected functions may be emulated on a smaller scale, however, the investigation of disease emergence and progression, as well as tissue repair and regeneration processes requires more identical organoids to represent the organ response in an MPS. With regard to the scaling aspect of MPS, a computational methodology for design and operations for pharmacology applications was developed by Yu et al. (2015). Stokes et al. (2015) discuss on-platform scaling based on physiology and pharmacology and in vitro in vivo translation of results for complex MPS technologies. These different aspects reflect the current diversity of MPS within the scientific community.
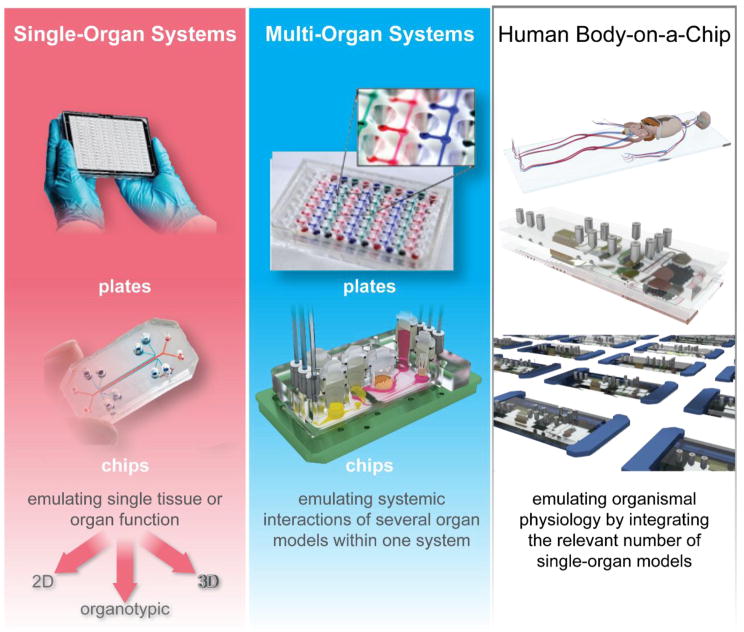
MPS devices exist in two categories: plates and chips, as illustrated in Fig. 3.
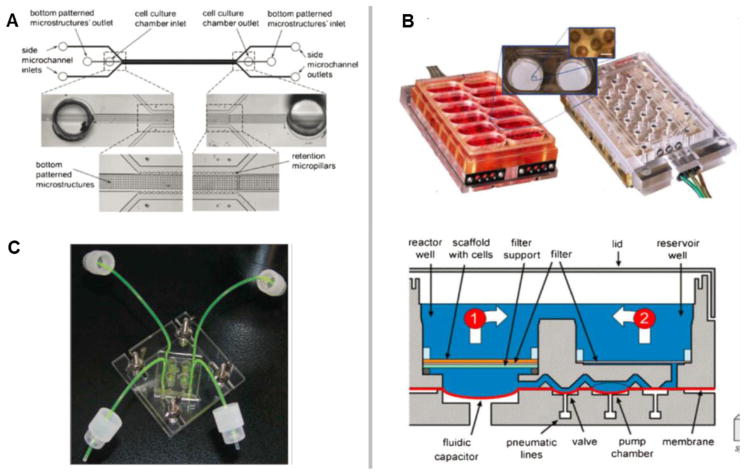
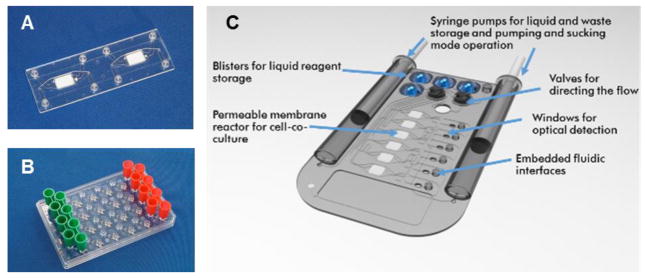
Fig. 3. Types of MPS used for emulation of human biology in vitro.
The top left figure shows the MIMETAS OrganoPlate for 3D perfused cell culture in microtiter format, the middle left shows the Lung-on-a-Chip that was developed by the Wyss institute, top centre depicts hanging drop microtiter plate by ETH Basel with microfluidic channel connecting multiple spheroids, Centre bottom shows an artist’s impression of the four organ system developed by TissUse, Right shows artist’s impressions of a human Body-on-a-chip platform. (Courtesy of MIMETAS, The Netherlands; Wyss Institute, USA; ETH, Switzerland; and TissUse GmbH, Germany, respectively.)
Plates are usually based on the microtiter plate dimensions as defined by ANSI/SLAS and use passive gravity-based microfluidic flow or active on-board pumping (Ebrahimkhani et al., 2014). An advantage of plate-based systems, whether gravity-based or active flow-based, is that they can be used with standard liquid handling equipment, such as robotic pipettors and are fully compatible with automated microscopes and plate readers.
Chips of microscope slide size and other formats are operated by an active microfluidic flow (external or on-chip pumps are used). This supports the emulation of shear stress at physiological intra-capillary or interstitial rates mandatory to maintain stable protein and oxygen gradient-based microenvironments over long time periods. Typically, the single chip format is used when more complex tissue architectures need to be mimicked, while plate-based formats are preferred, when minimal amount of data points are needed.
Alternatively, microphysiological systems can be broken down into three distinct types: Single-organ systems, multi-organ systems and more complex systems which are often termed human “Body-on-a-chip” systems (see Fig. 3).
Single-organ systems are plates or chips emulating single tissue or organ function and are designed to improve early predictivity of human single-organ toxicity of a particulate matter, a chemical or a drug candidate. These tissues or organs can be modeled at a two-dimensional (2D), three dimensional (3D) or organotypic complexity: 2D stands for suspension or monolayer cell cultures; 3D involves multilayer cultures of different geometry, e.g. spheroids, strata and irregular tissue formations; and organotypic differs from 3D by capturing as many desirable features of the in vivo architecture as necessary to gain the appropriate organ function (Hartung, 2014). Single-organ formats range from those employing passive gravity-based flow in plates adapted from standard cell culture plates to a variety of plate-based and microfluidic chip-type approaches using active flow from on-board or external pumps. These approaches allow to generate various flow profiles, different fluid-to-tissue ratios and other physiologically relevant characteristics. Multi-organ systems emulate systemic interactions of two or more organ models within one system to enable adverse outcome pathway (AoP) and mode of action (MoA) data generation from their crosstalk. By contrast, human-on-a-chip (or body-on-a-chip) systems are envisioned to mimic the physiological interaction of a number of organs capable of emulating entire organismal functionality. A minimum of ten has been set by the Defense Advanced Research Projects Agency (DARPA)/National Institutes of Health (NIH) MPS program (Hartung and Zurlo, 2012), for example, as a means to identify the field of technical challenges. This number has yet to be defined by the scientific community and regulatory bodies.
The current literature on MPS describes different culture times: short-term cultures last hours or days, supporting single exposure acute substance testing, and long-term cultures last weeks and months, enabling disease modeling and repeated dose substance exposure.
2.2. Human cell supply for MPS
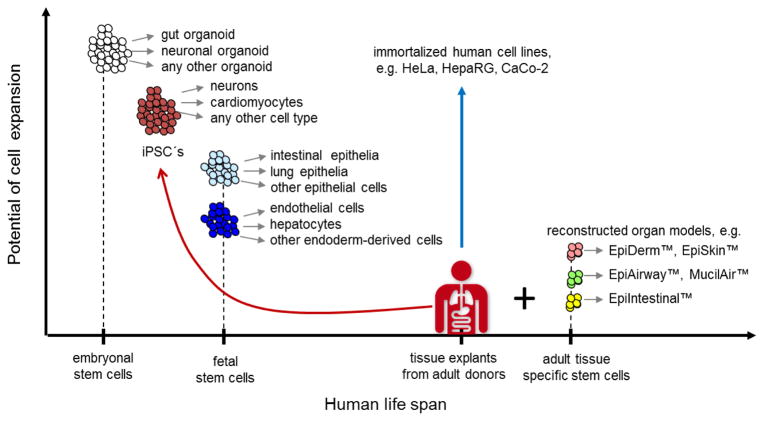
Human beings are the source for human cells and tissues used today in research and industrial applications in vitro (Fig. 4).
Fig. 4. Human cell sources for in vitro formation of organoids at a glance.
The expansion potential of different human cell and tissue sources used in MPS are plotted against their appearance in the human life-span
(Grey arrows) – indicate differentiation potential of the respective stem cell pool
(Red arrow) – illustrates induction of pluripotency using primary cells
(Blue arrow) – highlights unlimited expansion potential of immortalized cell lines
Human cell lines derived from adult donors (blue arrow, Fig. 4) have been the source of first choice for industrial in vitro assays historically, due to their unlimited expansion potential, relative stability and the ability for long-term cryostorage. The first human cell line – HeLa – was derived from the tumor of Henrietta Lacks, a female patient with a cervical adenocarcinoma, as early as 1951. Thousands of human cell lines more or less representative for different organ/tissue cell types are available nowadays, for example, patient-derived human cell lines like HepaRG, used in a number of toxicity assays, and the CaCo-2 cell line for intestinal permeation assays. With increasing knowledge of the genetic engineering of mammalian cells, an ever growing panel of designed human cell lines and human designer cells have been integrated into industrial and research applications (Sandig and Jordan, 2007). Existing well established human cell lines are a common source for MPS-based cell culture. The drawbacks of cell lines are the partial loss of functionality due to immortalization. These cells also display many physiological functions not representative for a healthy cell due to their cancer background in addition to an inability to form complex heterogenic tissue architecture. Primary tissue explants and biopsies from healthy donors for toxicity testing and adverse outcome pathway analysis or from patients for efficacy testing or disease modeling are the most complex and in vivo-like human tissue sources. These sources are used primarily in basic research and tissue engineering. Regarding use in MPS, the respective microsystem needs to be adapted for direct integration of such human explants and biopsies into the fluid flow. The drawbacks of tissue explants are their limited availability.
Another common approach to generate human tissue architecture in vitro is the de novo assembly or reconstruction of human organoids. Therefore, all primary tissues and/or their necessary adult tissue-specific stem cells are harvested from a donor or patient (Fig. 4) (Clevers 2015, Rookmaker 2015, Sato 2015). Consecutively, single cells are isolated from the tissues, expanded and reassembled in vitro. Prime targets for such reconstruction are the human barrier organs of skin, lung and intestine: EpiDerm™ from MatTek, EpiSkin™ from EpiSkin, EpiAirway™ and MucilAir™ from Epithelix and EpiIntestinal from MatTek, respectively, are prime reconstruction examples. See Gordon et al. (2015) for a comprehensive overview of non-animal models of epithelial skin, lung and intestinal barriers.
In order to integrate such models into MPS devices, the latter need to be adapted to the standard architecture of the respective reconstructed model. Optimized manufacturing procedures and a pool of standardized donors at the vendor site support a certain robustness of such reconstructed human tissue models. The drawbacks are limited architectural complexity, short-term stability and costs.
There are another three alternatives to derive necessary human cell sources for organoid engineering in MPS. All three are stem cell-based – embryonic, fetal and induced pluripotent – but only the induced pluripotent stem cell (iPSC) approach works with primary cells from an adult donor (red arrow, Fig. 4). Since the inception of the induced pluripotent stem cell technology by Takahashi and Yamanaka in 2006, induction of pluripotency in primary human cells from patients or healthy donors has become an experimental routine (Takahashi and Yamanaka, 2006). An impressive panel of companies, such as Axiogenesis, Axol, BioTalentum, Cellular Dynamics International, Pluriomics, Lonza, Takara Bio Europe AB, Viagene Biotech, iXCells Biotechnologies, Applied StemCell, Applied Biological Materials, Centre for Commercialization of Regenerative Medicine and Cedars-Sinai, have been established and are providing reprogramming and/or differentiation services. While reprogramming has become a professional industrial standard, leading to more than 1,500 cryopreserved and well characterized iPSC lines, differentiation into organ-specific human cells still remains a big challenge. See Bellin et al. (2012) for a comprehensive review. Within the last three years, there has been an unprecedented increase of experimental research into differentiation protocols, starting from induced pluripotency. Here we list a number papers trying to differentiate cell types relevant for MPS-based organoid engineering and subsequent assay development: hepatocytes (Szkolnicka et al., 2014), cardiomyocytes (Burridge et al., 2014), small intestine (Kauffman et al., 2013), neurons (Hoelting et al., 2013, Lippmann et al., 2014, Lancaster and Knoblich, 2014, Terrasso et al., 2015, Simao et al., 2016), lung (Ghaedi et al., 2013), gallbladder (Kauffman et al., 2013), pancreas (Pagliuca et al., 2014), kidney (Song et al., 2012), skin (Itoh et al., 2013), biliary ducts (Ogawa et al., 2015), bone-marrow (Hynes et al., 2014), ovaries (Leng et al., 2014) and vasculature (Patsch et al., 2014). To the best of our knowledge, none of the protocols or procedures have yet been adopted by pharmaceutical industry for robust reproducible assay establishment, but there are first data to hypothesize that MPS might be the right devices to drive the differentiation of pre-differentiated iPS cells toward a final stage by the physiological environment of the respective organoid (Moreno et al, 2015).
Fetal cells are rarely used in the engineering of human adult tissue, but recently, mesenchymal stem cells derived from fetal tissues could be differentiated into neuronal lineages (Zhang et al., 2011) and osteoblastic lineages (Zhang et al., 2010). The main advantage with fetal cells is that they retain their proliferative capacity, while being committed to an endpoint, and are not teratogenic in nature. However, their source is controversial.
Embryonic stem cells (ESC) are derived early after fertilization from the inner cell mass of human blastocysts. A variety of human embryonic stem cell lines has been created over the last few decades (Mummery 2014). These pluripotent progenitor cells lack the epigenetic modifications that may sometimes be seen in iPSC. Therefore, ESC might provide the broadest background for differentiation into any human organoid. The disadvantages of human embryonal cells or cell lines are the ethical debate surrounding their use and their propensity to give rise to teratomas, as shown in transplantation experiments (Lees et al., 2007, Leist et al., 2008)
Pluripotent stem cells of the origin described above have been used for the de novo assembly of human gastric (McCracken et al., 2011), cerebral (Lancaster et al., 2013), kidney (Xia et al., 2014) and lung organoids (Dye et al., 2015) in static culture systems. Physiological, mechanical and electrical coupling of such human organoids using MPS seem to be an attractive approach for their further differentiation into in vivo-like functional organ units. See Huch and Koo (2015) and Li et al. (2014) for a review on organoid cultures based on pluripotent human stem cells.
2.3. Current state of the art of MPS
2.3.1. Introduction
Technical drawbacks of any static cell culture-based assay include the limitation of oxygen and nutrients to a given 3D tissue size, or monolayer metabolism, and the lack of dynamic protein gradients. Oxygen restriction in cultures of primary human hepatocytes in static culture flasks, for example, was described as early as 1968 (McLimans et al., 1968). Microfabrication and microfluidics technologies, initiated as a scientific discipline in its own right as early as 1990 (Manz et al., 1990) and delivered commercially available lab-on-a-chip platforms end of 20th century (see El-Ali et al., 2006 and Whitesides, 2006 for reviews), and advances in bioengineering related to materials science have enabled the appearance of first plate- and chip-based microfluidic tissue culture systems aiming to solve the two aforementioned limitations of static cultures and to replicate the spatiotemporal, mechanical and biochemical cues that underlie the physiological behavior inherent in those tissues. MPS developers approached their development from two different angles, some of them targeting to integrate microfluidic principles of nutrient supply into well-established industrial high throughput compliant static cell culture microtiter well formats and some trying to miniaturize well established industrial tissue culture bioreactor systems.
Plate-based MPS developments accommodated conventional static cell culture microtiter plates with arrays of 96, 384 and 1536 wells which have become the standard of choice in any biological laboratory. Microtiter plates offer additional advantages in that they have been fully accepted as a standard across industries [SLAS, 2014]. These platforms appeared on the grounds that the sheer nature of experiments and inherent variability in biology requires a vast amount of control experiments, repetitions and dilutions. In addition, the trend in molecular and cell biological sciences has progressed to ever higher throughput approaches, testing more conditions in one experimental session. It is for these reasons that manual and automated liquid handling and readout equipment have been designed in accordance with this standard. Moreover, the high number of wells in a plate, such as 384, 1,536, and even 3,456, offer the additional advantage of reduced volumes of cells and reagents, leading to a reduction of cost per assay.
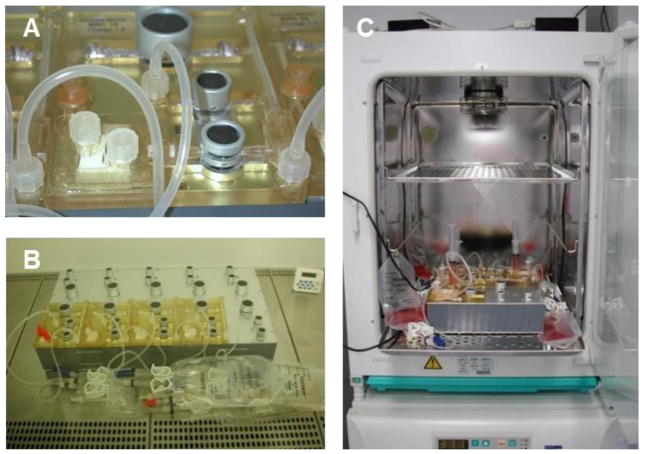
Chip-based microphysiological systems for tissue and organ modeling have usually been modified from conventional industrial cell-culture bioreactors by trying to downscale the fluidic and tissue culture compartments and to miniaturize the sensors and actuators necessary. Automated dynamic bioreactors, such as hollow-fiber bioreactors, invented in the early-1970s, solved the problem of oxygen and nutrient supply at large scale – up to 1 L tissue culture – by the introduction of artificial capillaries for media perfusion (Knazek, 1972). These dynamic perfusion tissue culture systems were advanced later into functional long-term in vitro organ equivalents introduced into medical practice as extracorporeal bioartificial liver devices in the late-1990s (Catapano and Gerlach, 2007; Gerlach, 1996; Wang et al., 2010). On the basis of this historical knowledge, numerous dynamic tissue-specific bioreactor devices, such as human artificial lymph nodes for immunogenicity testing, were miniaturized for in vitro testing purposes towards the lower mL per tissue culture scale (Giese et al., 2006). Figure 5 illustrates a miniaturized incubator-dependent dynamic bioreactor prototype for the simultaneous culture of human immune tissues at 0.5 ml each in operation.
Fig. 5. Single-organ microcassette bioreactor device with hollow-fiber-based perfusion.
Integration of electromechanically controlled peristaltic pumps (A). Controller for ten microcassettes (B). Micro-bioreactor in operation in an incubator for temperature, humidity and CO2 control (C). (Courtesy of ProBioGen AG, Germany.)
Unfortunately, it became obvious in the first decade of this century that the lower mL scale still required too much cell supply and was still far too large to enable simultaneous high content testing at reasonable costs. This triggered the advent of chip-based MPS, which adopted lab-on-a-chip platforms to decrease tissue culture volumes down from micro- to nanoliter scale per tissue culture compartment. These are aiming to apply automated pumping systems and electrical, optical and mechanical transduction to monitor and control complex minute tissue cultures in automated incubator-independent systems at relevant throughput.
In summary, plate- and chip-based MPS currently aim to reflect physiologically relevant parameters, including proper cell-to-cell, cell-to-matrix, and biochemical and mechanical signaling. These capabilities present unprecedented opportunities to create microphysiological systems with the potential of capturing the dynamics of disease appearance, of repair and regeneration processes and of drug effects in the human body. Recent advances in microphysiological system development have made it possible to initiate the engineering of cellular environments and/or functional units of lung, heart, blood vessels, muscles, bones, liver, reproductive system, nervous system (including eye), gut, skin, intestine and kidney. In this chapter, we report the status of the development of biology-inspired microphysiological single- and multi-organ systems and their use for substance testing. Furthermore, we introduce achievements in modeling diseases using MPS. Recent initiatives to advance multi-organ systems into human body-on-a-chip solutions are outlined in Chapter 4.
2.3.2. State of the art of microphysiological single-organ systems
Early microtiter plate-based microfluidic cell culture formats with passive gravity-based microfluidic flow approaches to in vitro tissue regeneration with application for human disease modeling and drug development were developed by CellAsic (Lee et al., 2007a). Inspired by the work of Luke Lee’s group (Lee et al., 2007b; Hung et al., 2005b), they developed a microtiter plate with eight independent flow units that did not require tubing for perfusion flow, but relied on passive leveling instead. The device consists of a polydimethylsiloxane (PDMS) molded microfluidic layer that is bonded to a glass bottom and an acrylic user interface. Cells were introduced with the help of a vacuum manifold and positioned by a filter consisting of densely packed micropillars. Perfusion flow was induced by gravity leveling in conjunction with a carefully tuned fluid resistance of the microfluidic channels. HP Medizintechnik GmbHs (Oberschleiβheim, Germany) intelligent microplate reader is an advanced 24-well-based microphysiological system incorporating an automated microscope, pipetting robot and microsensors for pH, dissolved oxygen and impedance, and is premounted in an incubator (Demmel et al., 2015). It was successfully used to monitor the viability of various cell types and primary tissue (Kleinhans et al., 2012) and to develop new toxicological endpoints (Wolf et al., 2011). The relatively high grade of parallelization and the automation of the pipetting robot and the microscope allow autonomous, label-free long-term experiments in the field of MPS. The PDMS material used, however, bears the potential of high unspecific binding of drug candidates to the device.
Dave Beebe’s group at the University of Wisconsin, Madison, US, developed a series of microtiter plate-based cell culture devices, including a 3D culture plate and a chemotaxis plate (Berthier et al., 2010). A total of 192 single channels on a microtiter plate were filled with cell-loaded gels (Meyvantsson, 2008). An interstitial flow through the gel was applied making use of differential meniscus pressure between two droplets of different size at the in- and outlet, respectively. A similar pumping concept was used for a 49-channel network chemotaxis device, consisting of a source and a sink channel that are connected to a very thin gradient channel.
The OrganoPlate was recently introduced by the company MIMETAS, Netherlands, consisting of 40 to 96 individually addressable microfluidic networks (Jang et al., 2015; Moreno et al., 2015; Trietsch et al., 2013). The OrganoPlate makes use of PhaseGuide technology to stratify extracellular matrix (ECM) gels in orderly layers, one next to the other. This enables a co-culture approach that is fully free of artificial membranes, while still enabling the engineering of tissues. The OrganoPlate is fully devoid of high absorbance materials, such as PDMS, does not require any other handling equipment apart from standard pipettes for operation, and provides continuous perfusion flow through passive leveling.
Plate-based microfluidic cell culture formats with active on-board pumping were introduced for liver models by the group of Linda Griffith at MIT, (Cambridge, USA) and commercialized by CN Bio Innovations (London, UK). The Liverchip™ system, such a plate-based system, is described in more detail below.
A wave of single-organ chip concepts covering a wide range of tissues were proposed during the last decade and were actively introduced into biological research. They began with single cell type cultures of different organ specificity, such as endothelial cells (Young and Simmons, 2010), myoblasts (Gu et al., 2004), hepatocytes (Carraro et al., 2008; Goral et al., 2010; Ho et al., 2006; Leclerc et al., 2004; Lee et al., 2007; Park et al., 2008; Powers et al., 2002; Toh et al., 2007; Toh et al., 2009), neurons (Rhee et al., 2005), mammary epithelial cells (Grafton et al., 2011), adipose cells (Nakayama et al., 2008), and human embryo cells (Chung et al., 2005; Hung at al., 2005a; Smith et al., 2012; Villa-Diaz et al., 2009).
Consequently, in a next phase, heterotypic microfluidic single-organ co-culture systems combining crucial cell types of a specific organoid into artificial functional units reflected aspects of the individual organs more realistically. Scientific activities in the field of organ-on-a-chip models have increased dramatically in the last five years (van de Stolpe et al., 2013). Therefore, we decided to exemplarily highlight the single-organ MPS development for human liver and lung modeling whilst referring the readers to the original MPS literature for other organs and systems.
MPS-based liver cultures have matured from dynamic random cell culture towards systems of higher tissue complexity. The unique importance of liver for organismal homeostasis (e.g. plasma protein synthesis, glucose biotransformation) and blood detoxification (e.g. urea, xenobiotic drug metabolism) has led to an evolutionary optimization of the human liver architecture at the scale of its smallest functional unit – the liver lobule. A precise zonal division of labor along a 500 μm long stretch of about 25 hepatocytes arranged in cords allows the management of blood detoxification at a blood contact time of only a few seconds. Not much is known about the fluid dynamics of plasma-hepatocyte contact. The nature of the plasma flow in the space of Disse and the mechanics applied to hepatocytes at these basolateral surfaces are under dispute. The high degree of fenestration may result in transmission of fluid shear stress to hepatocytes. Furthermore, the sinusoids in the periportal zone are of a very small diameter, the size of an erythrocyte. Blood cell passage through these tight sinusoids may additionally modulate shear stress. Four types of MPS-based liver equivalents have been developed so far: random dynamic hepatocyte monocultures, cord-like liver equivalents, endothelial sinusoid models and hepatic lobule “equivalents” (Materne et al., 2013). Figure 6 highlights prominent examples for the latter three types.
Fig. 6. Biology-inspired increasing complexity of MPS-based liver models.
Inspired by the lobulus architecture of human liver lobules, MPS have evolved from bile canaliculi forming cord-like liver cultures (A) through sinusoid-like arrangements supporting functional space Disse structures (B) towards tissue slice cultures maintaining functional organoid liver structures (C).
In contrast to random dynamic hepatocyte cultures, the formation of 3D cord-like structures composed of polarized cells which form extended bile canalicular structures was presented by Goral and colleagues (2010). A series of retention pillars formed a microchannel centered between two side-channels (Fig. 6a). However, unlike other perfusion-based microdevices, the bottom of the cell culture chamber was patterned with microstructures, which provided additional control of hepatocyte polarity. After two weeks of perfusion culture, the cells remained viable and had formed a cord-like structure. An extended bile canalicular structure and the formation of gap junctions between the 3D structured cells could be shown. The system mentioned above allowed to arrange hepatocytes into an artificial, but cord-like assembly, restoring hepatocyte polarity and supporting sporadic bile segregation. Co-culture of hepatocytes with non-parenchymal cells in a spatially arranged 3D environment under constant perfusion might lead to the formation of a next level of architectural resemblance to the in vivo situation. A very interesting approach to modeling sinusoid-like structures in a microfluidic system has been developed by Linda Griffith’s group at MIT (Domansky et al., 2010). An array of multiple bioreactors was built into a multiwell plate comprising 12 autonomous microfluidic systems (Fig. 6b), each perfused by an integrated pneumatic micropump circulating a total volume of 3 ml. Each tissue culture scaffold contains 769 multichannels (0.24 mm deep, 0.34 mm diameter) and was seeded with 106 rat hepatocytes and endothelial cells at a 1:1 ratio. A continuous adjustable oxygen gradient could be established over long operating times. The scaffold supports near physiological tissue densities and the functional zonation of hepatocytes can be stipulated. The large channels of the scaffold support self-assembly of the two cell types in dynamic conditions. Liver sinusoid endothelial cells, known to lose their differentiated phenotype in vitro, maintained the expression of the functional marker SE-1 throughout the culture. The important features of adjustable flow rates on the basis of an oxygen consumption model, long-term steady gradient maintenance and the ability for co-culture of hepatocytes with different types of non-parenchymal cells made the system an interesting alternative for toxicity testing. It is one of the still rare cases where research MPS have been subsequently improved and transferred into industrial application. The scaffold size of this LiverChip™ system commercialized by CN Bio Innovations can be scaled for 10,000 – 1,000,000 cells, and plates are similarly scaled in 12- or 36-well formats. Active pumping to provide circulation allows flow rates through the tissue to be uncoupled from the morphology of the tissue. The system provides high-content data for drug metabolism and pharmacokinetics, toxicology and disease biology applications (Sarkar et al., 2015; Vivares et al., 2015). It has been applied in preclinical development (Dash et al., 2009; Vivare et al., 2015). Furthermore, a model of micrometastasis of cancer cells in liver has been developed on that platform and a demonstration of dormancy of the aggressive MDA-MB-231 cell line in this model has been established (Wheeler et al., 2014). A model of hepatitis B virus infection has also been developed using the same 3D perfused liver model. Infection of cryopreserved primary human hepatocytes is achieved through incubation with patient serum. The full viral life cycle is recapitulated making the model of utility (Wai et al., 2014) in both exploring the basic biology of hepatitis B virus and evaluating novel treatments. The highest level of architecture – the entire liver lobule – has not yet been reached, but a chip-based microfluidic approach to contain a major part of a liver lobule has been made by van Midwoud and colleagues (van Midwoud et al., 2010), who cultivated 3 mg of liver slice tissue in a microfluidic chip (Fig. 6c). The precision-cut liver slice (100 μm thick, 4 mm diameter) was perfused with a flow of 10 μl/min in an incubator chamber with constant pH and dissolved oxygen. Biotransformation activity was shown to be equal in control slices in static culture over three hours. The system does not demonstrate any advantage with regard to metabolic functionality over the three-hour measurement time. The authors highlight the advantage of continuous media perfusion at low tissue-to-fluid volume ratios in contrast to steady metabolite accumulation in static culture. Unfortunately, data on the culture performance over times longer than 72 hours are not yet presented (van Midwoud et al., 2011).
In addition to the examples described above, MPS-based single-organ liver modeling has led to a large variety of liver-on-a-chip devices (Baudoin et al., 2014; Hwa et al., 2007; Kane et al., 2006; Khetani and Bhatia, 2008; Leclerc et al., 2014; Leclerc et al., 2015; Legendre et al., 2013; Legendre et al., 2014; Pasirayi et al., 2014; Snouber et al., 2013) Research tools and assays have been developed to investigate liver metabolism and toxicity in single-organ chips (Baudoin et al., 2014; Leclerc et al., 2014; Leclerc et al., 2015; Legendre et al., 2013; Legendre et al., 2014; Pasirayi et al., 2014; Snouber et al., 2013). For further details on the development of liver-on-a-chip models readers are referred to respective reviews (Materne et al. 2013; van Midwoud et al., 2011).
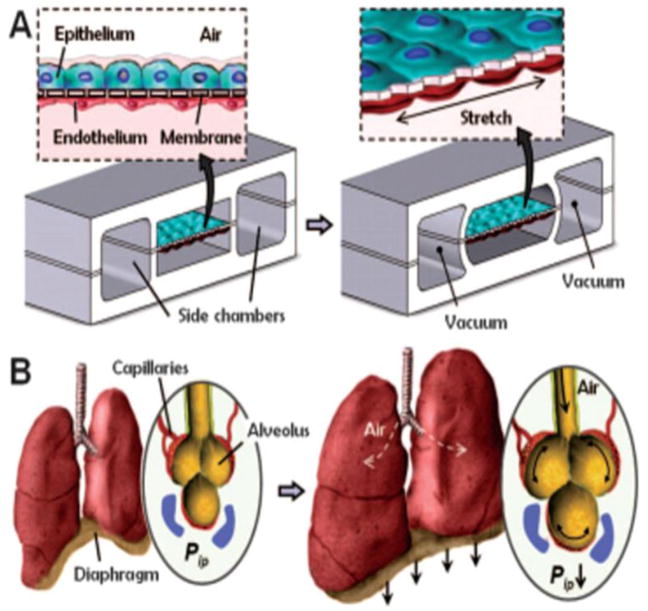
Another complex organ where biology-inspired MPS-based models are already making a difference regarding the understanding of human organ functionality, testing safety and modeling disease is the human lung. Human in vitro models of small lung airways and alveoli were developed historically in static air-liquid interface cultures on cell culture inserts. For a comprehensive overview on such models, readers are referred to the recent t4 report by Gordon and colleagues (2015). Until recently, only a few MPS trying to emulate functional parts of the lung have been developed (Nalayanda et al., 2007; Nalayanda et al., 2010), but in 2010, a microphysiological lung-on-a-chip system developed by Donald Ingber’s group at the Wyss Institute for Biologically Inspired Engineering at Harvard University Boston, MA, USA, made it for the first time into Science Magazine (Huh et al., 2010). Figure 7a shows the design and principles of this system which is mimicking the function of a lung alveola. This system is prepared by microfabricating a chamber with two microchannels separated by a thin (10 μm) porous PDMS membrane coated with an ECM (fibronectin or collagen). Human alveolar epithelial cells are cultivated on the top of the membrane (in the upper channel) and human pulmonary microvascular endothelial cells are cultivated on the lower side of the membrane (in the lower channel). Air is pumped through the top channel and a culture fluid is pumped through the lower channel as a substitute for blood. Thus, the system is biomimetic for the alveolar capillary barrier. By applying a vacuum to the side channels, it is possible to stretch the culture layer; release of vacuum causes elastic recoil of the membrane, returning it to its natural state, mimicking the stretching of the alveoli during breathing (Fig. 7b). Molecules present in the air cross the culture layer through a combination of passive diffusion and active transport, as in the alveoli in the lung. Huh and colleagues (2012) used their lung-on-a-chip system to develop a model of pulmonary edema. Pulmonary edema is a life-threatening disease associated with increased hydrostatic pressure or microvascular permeability caused by various diseases or dose-limiting drug toxicities (Willett, 2014). Huh and colleagues (2012) confirmed that administration of high doses of interleukin-2 (IL-2) caused a state consistent with pulmonary edema in their lung-on-a-chip system, similar to the effects of administering IL-2 to human cancer patients. They also found that angiopoietin-1 and GSK2193874, an inhibitor of the transient receptor potential vanilloid 4 ion channel, attenuated edema in vitro and may prevent this dose-limiting toxicity of IL-2 in clinical settings. The authors concluded that the system could be used to model diseases in other organs and predict the efficacies and toxicities of other drugs in humans. Emulate Inc. (Boston, USA) is commercializing tools based on this platform. The system has also been used to examine the production of intracellular reactive oxygen species (ROS) in response to nanoparticles (Huh, 2015; Huh et al., 2010). In these studies, silica nanoparticles with a diameter of 12 nm induced and amplified the acute toxic responses measured in terms of ROS generation. Cyclic stretching and relaxation of the porous membrane to mimic breathing further enhanced the absorption and translocation of the silica nanoparticles from the upper air channel to the lower fluid channel. Similar findings were observed in a ventilation perfusion model using whole mouse lung.
Fig. 7. Biology-inspired Microfluidic alveolar models applying mechanical stretch.
Design of a lung-on-a-chip system. (A) Cross-sectional view of the device in the native and stretched state. Human alveolar epithelial cells are cultivated on the top of the membrane and human pulmonary microvascular endothelial cells are cultivated on the bottom of the membrane. (B) View of the lung showing the stretch and resulting distribution of air during inhalation. Applying a cyclic vacuum through the side chambers causes the cell layer to stretch, mimicking the natural stretching during inhalation. Reprinted from Huh et al. (2010).
Other research groups have developed lung-on-a-chip systems using human primary pulmonary alveolar epithelial cells from patients who had undergone partial lung resection (Stucki et al. 2015) or primary airway epithelia (Bol et al., 2014; Sellgren et al., 2014) for selection of candidate drugs to treat pulmonary pathologies. Although the consensus is that lung-on-a-chip systems are valid tools for toxicological research, very few toxicology studies have been performed using these systems. Finally, a model for chemotherapy-induced pulmonary edema has been developed on the basis of a small-airway-on-a-chip model (Benam et al., 2015b).
The main biology-inspired aspect of the systems is the application of physiological cyclic strain placed on the cells, which not only closely mimics normal respiratory movements, but also influences the permeability of the cell layers. In vivo studies have shown that distention of the lung, as in inhalation, increases the permeability of lung to hydrophilic molecules (Marks et al., 1985). This is probably due to the stretching of the intercellular junctions, which increases the size of the pores and allows larger hydrophilic molecules to cross the junctions (Mason et al., 2001). Innovative approaches to develop microfluidic systems for the culture of other types of epithelia could potentially also be adapted to establish new lung-on-a-chip systems. For further information on MPS-based lung model development readers are referred to a recent review (Nichols et al., 2014).
Other organs models in single-organ MPS include the small artery (Günter et al., 2010), the nervous system (Booth and Kim, 2012; Brown et al., 2014; Kermanet al., 2015; Nery et al., 2015; Park et al., 2009; Robertson et al., 2014; Taylor et al., 2005), the pancreas (Silva et al., 2013; Jun et al., 2013; Lee et al., 2012), the kidney (Snouber et al., 2012; Snouber et al., 2011; Jang et al., 2013; Ferrell et al., 2012; Kim and Takayama, 2015; Huang et al., 2013; Mu et al., 2013), the bone-marrow (Cui et al., 2007), the skin and hair (Atac et al., 2013) and the intestine (Esch et al., 2012 ; Kim and Ingber, 2013; Kim et al., 2012; Kim et al., 2013; Kimura et al., 2008; Lahar et al., 2011; Mahler et al., 2009; McAuliffe et al., 2008; Ootani et al., 2010; Sato et al., 2009; Sung et al., 2011; Yu et al., 2012) at different levels of biological complexity. Gao and colleagues (2013) designed an integrated microfluidic device directly coupled to a mass spectrometer, for instance, to characterize drug permeability of the intestinal barrier in a real-time manner. They were able to measure the permeation of curcumin through a Caco-2 cell monolayer in a real-time and obtained results consistent with published in vivo data. Another integrated microfluidic platform, called the “NutriChip,” was set up to investigate the potential of the immune-modulatory function of dairy food (Ramadan et al., 2013). Ramadan and colleagues quantified the variation of pro-inflammatory cytokines expression directly online using magnetic beads and an optical detection device in their epithelial/immune cells co-culture model. These techniques offer useful tools for substance permeability studies or physiological response measurement. Finally, the lung-on-a-chip platform of the Wyss Institute at Harvard, Boston, USA, has recently been used for a gut-on-a-chip model for inflammatory bowel disease (Kim et al., 2016). This single-organ chip was used to co-culture multiple commensal microbes in contact with living human intestinal epithelial cells and to analyze how gut microbiome, inflammatory cells and peristalsis-associated mechanical deformations contribute independently to intestinal bacterial overgrowth and inflammation. This in vitro model replicated results from past animal and human studies, including a demonstration that probiotic and antibiotic therapies can suppress villus injury induced by pathogenic bacteria. By ceasing peristalsis-like motions while maintaining luminal flow, lack of epithelial deformation was shown to trigger bacterial overgrowth similar to that observed in patients with ileus and inflammatory bowel disease. Thus, this human gut-on-a-chip can be used to analyze contributions of the microbiome to intestinal pathophysiology and dissect disease mechanisms in a controlled manner that is not possible using existing in vitro systems or animal models.
For further information on single-organ systems modeling tissues of the respiratory, nervous, digestive and excretory systems, we refer the readers to a recent systematic review by Perestrelo and co-authors (2015), whilst the use of single-organ MPS to model diseases has been reviewed elsewhere (Benam et al., 2015a).
A recent survey of microfluidic and microengineered 3D cell culture systems conducted by Hankemeier and colleagues (van Duinen et al., 2015) at the Leiden University, Netherlands, found that most tissue modeling efforts published between 2012 and early-2015 in the field of MPS were focused on vasculature. Authors explained the striking dominance of efforts in vascular modeling by the fact that MPS is the only platform capable of perfusing such vessels, thereby including the vitally important flow and accompanying shear stress. MPS emulating microvessels to study angiogenesis (Bischel et al., 2013; Zheng et al., 2012), permeability (Lee et al., 2014), pattern diffusive gradients (Baker et al., 2013), micro-vascular environments (Hasenberg et al., 2015; Kim et al., 2013; Park et al., 2014; Tourovskaia et al., 2014; Wang et al., 2014) and vascular response to vessel geometries (Ye et al., 2014) have been developed within the last few years. Finally, vascular MPS are used as models for arterial thrombosis (Huh et al., 2007; Westein et al., 2013).
Another important finding of the survey of Hankemeier and colleagues (van Duinen et al., 2015) was the fact that microfluidic cancer models published between 2012 and early 2015 in the field hold approximately the same numeric share as the aforementioned microfluidic tissue models. Breast and lung cancer models comprise half of the cancer models published and many of the cancer models developed recently include a vasculature component. Here again, MPS technologies provide the only platform to model tumor cell intravasation into a surrogate blood stream or immune cell extravasation into the tumor by combining human microperfused 3D tumor models with human vasculature. These systems add to our understanding of tumor progression. Migration (Haessler et al., 2012; Hockemeyer et al., 2014), intravasation (Zervantonakis et al., 2012) extravasation (Bersini et al., 2014; Jeon et al., 2013) and metastasis (Griep et al., 2013) have been studied in such MPS-based cancer models.
2.3.3. Microphysiological multi-organ system developments
Combining single-organ models towards integrated multi-organ configurations lifts the degree of complexity to a systemic level of organ interaction. Human organ-to-organ crosstalk, ADME pathways and systemic regulatory circuits between organs are envisioned to be emulated using such multi-organ systems. Their development poses significant challenges both for plate- and chip-based formats. Different organ models have to be handled in the same device, remain fully functional at the same time and interact through the same circulating liquid phase. These challenges raise the qualification requirements for tools and approaches using multi-organ MPS to generate results which are reproducible and predictive to humans. Any technological plate- or chip-based MPS approaches chosen for these systemic models are a trade-off between complexity and in vivo resemblance, on the one hand, and ease of use, reproducibility and potential for parallelization, on the other hand (van Midwoud et al., 2011). Olivier Frey’s group at the ETH Zurich and InSphero AG, Switzerland, and collaborators pioneered the developed of multi-organ plate concepts using 3D micro-tissue spheroids as a 3D tissue model (Kim et al., 2015a; Kim et al., 2015b). The microfluidic platform is built in a way that micro-tissue development is completely decoupled from microfluidic culturing and the loading of the spheroids is realized in a modular way (Fig. 8A).
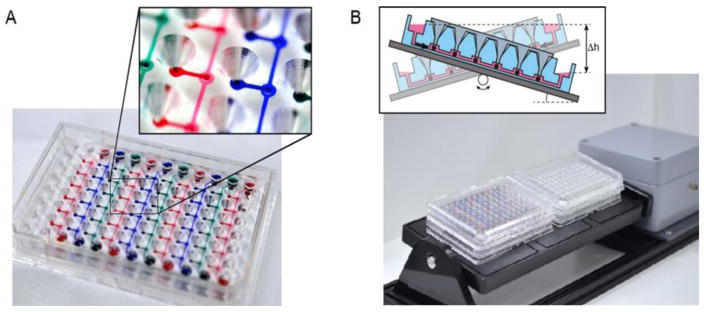
Fig. 8. A prime example of a plate-based multi-organ system.
(A) A 96-well format multi-tissue interaction testing chip (close-up shows spheroid compartment with loading port). Ten parallel microfluidic channels interconnect six culturing compartments, in which spheroids of different types can be loaded. (B) Platform operated in a standard incubator tilting the chips back and forth producing a gravity-induced flow between the different culturing compartments. (Courtesy of ETH, Switzerland.)
The plate consists of straight channels connecting up to 10 identical compartments in which spheroids can be loaded using simple pipetting. The medium is perfused between two lateral medium reservoirs using gravity-based flow through tilting the platform periodically. A single plate comprises up to 10 channels so that up 60 multi-organ conditions can be tested on a single tilting device (Fig. 8B). Human tissue spheroids generally possess inherent organotypic functionality and biomimetic morphology and can be manufactured precisely, reliably and flexibly in off-line automated systems. Their spherical shape makes them easy to handle and enabled the development of plate- or chip-based MPS that are simple to handle and robust to operate. Multi-organ arrangements using spheroids can be build up in a very flexible way regarding the large variety of cell types with which spheroids can be formed so as to represent different organ models and fluidically interconnected systems of many spheroid compartments. The spheroid model can, thereby, be improved and further developed continuously without the need to redesign the microfluidic test platform itself. While offering a large amount of flexibility in arranging different organ models in physiological order as well as tuning the different tissue volumes and ratios over the number of spheroids introduced into the system, the limitation of this approach lies within the spheroid model itself. Spheroids are frequently considered as the smallest functional tissue unit. However, it is clear that spheroids do not include mechanical cues (dynamic forces such as breathing strains) nor are they vascularized. Multi-organ models based on spheroids will, therefore, focus predominantly on biochemical and metabolic interactions between different tissue types (e.g. adding metabolic liver functionality to bioactivation applications). For proof of concept (Kim et al., 2015a; Kim et al., 2015b) primary rat liver tissues were cultured over eight days in combination with colorectal tumor micro-tissues (HCT-116). Interestingly, albumin secretion of the rat liver micro-tissues increased over the first few days in the fluidic devices compared to static culture conditions, which indicates further tissue maturation under flow conditions in the chip. The importance of interconnecting liver and tumor tissues was demonstrated by applying cyclophosphamide, a pro-drug which requires activation by the liver metabolism (mainly CYP2B6) to become effective. The impact of cyclophosphamide on tumor growth was simultaneously assessed under static culture conditions by discrete liquid transfer using a pipetting protocol and under perfusion conditions on the chip. Remarkably, hardly any effect on tumor growth was observed under static culture conditions, whereas a clear size decrease of the microtumor was detected after treatment with cyclophosphamide in the case of direct and continuous fluidic coupling in the chip. These findings illustrate the importance of continuous liquid and metabolite transfer between the different tissues or tissue compartments. Such transfer in isolated spheroid configurations could only be achieved with frequent discrete media exchanges. Suitable transfer timings and long enough incubation times to obtain the metabolic compounds desired are, however, difficult to estimate and optimize. Furthermore, the comparably large well volumes of conventional setups may entail a dilution of the active metabolites which is too large. Increasing the cell-to-medium volume ratio by co-culturing spheroids in the same well would lead to uncontrolled tissue fusion in a short time. The findings further demonstrate that the use of well formats with an already labor-intensive pipetting protocol cannot reproduce the results obtained with continuous media exchange, as has been exemplified for a setup including only two different tissue types. Pipetting approaches are no longer an option for experimental scenarios requiring more than two different tissue types, as the respective protocols become very complex, whereas microfluidic networks offer viable solutions. This plate-based MPS technology will be commercialized by InSphero AG, Switzerland (InSphero, 2012).
Another approach of the same group includes the use of the hanging drop itself – the primary technique to form spheroids – as on-chip culturing compartment (Frey et al., 2014). Arrays of hanging drops are linked to functional hanging-drop networks, in which the medium can be perfused between the drops and interconnects different spheroid types. The platform combines formation and culturing of spheroids of different cell types without any risk of adhesion and functional loss, as spheroids are located on the liquid-air interface. The array-based format enables parallel multi-tissue experiments and allowed to reproduce the cyclophosphamide bioactivation study described above. The integration of an on-chip pulsatile micropump, of which stroke rate has been synchronized with the beating of a human iPS-derived cardiac spheroid, recently “added the heart” to the hanging-drop network technology (Rismani et al., 2015).
Another plate-based system has been used to investigate intestinal first pass metabolism coupled with a secondary liver metabolism of paracetamol using a co-culture of CaCo-2 and HepG2/C3A cells over 24 hours. (Prot et al., 2014). The results demonstrated paracetamol transport through the intestinal unit followed by synergistic metabolism via the production of paracetamol sulfate in both the liver and intestinal units. Glucuronide paracetamol was detected when there was liver co-culture.
Michaels Shuler’s group at Cornell University, USA, pioneered the development of human multi-organ chips (MOCs). They developed a chip-based miniaturized microcell culture analog (μCCA) which was used for pharmacokinetic/pharmacodynamic (PK/PD) modeling as early as 2004 (Sin et al., 2004). The system supports physiologically based pharmacokinetic, quantitative structure activity relationship (QSAR) studies and quantitative in vitro to in vivo extrapolation (QIVIVE) modeling. It combines different human tissue cultures into a common media flow for the prediction of the time-dependent concentration of a parental compound and its metabolites. The principle design of the system is schematically illustrated in Figure 9. It relies on the recirculation of a common culture medium with around 10,000 cells per culture compartment and substance exposure times of up to four days.
Fig. 9. Scheme of the flow diagram of a μCCA.
The chip is 25 by 25 mm and flow channels are 20 – 100 μm wide. Flow is laminar and typically more than 10,000 cells populate each tissue culture compartment. The design is based on the Hagen Poiseulle Law which allows matching human-like fluid velocity in each channel and liquid residence time in each compartment with the respective PKPD model in silico. (Modified from Marx et al. 2012.)
The proof of concept was provided in studies with naphtalene as a model toxicant (Viravaidya et al., 2004). Furthermore, two combination therapies using doxorubicin (Tatosian and Shuler, 2009) and tegafur (Sung and Shuler, 2009) have been tested on human cancer cell lines in μCCA layouts, specifically developed for substance testing applications. The system at its selected scale provides in vivo-like tissue mass ratios among the culture compartments, a media inflow split, equivalent to the respective blood flow split in humans, and relevant residence times in tissue compartments. Furthermore, the microfluidic chip design is claimed to support physiological shear stress and liquid to cell ratios, mimicking those of the respective organ. It can be operated over periods of up to four days. The platform is commercialized by Hμrel Corporation, USA. The layout of the μCCA has progressed over the years (Mahler et al., 2009a; Mahler et al., 2012; Mahler et al., 2009b; Sung et al., 2010; Sung and Shuler, 2010). The following issues to advance the μCCA technology into a routine test assay platform remained until 2012 (Shuler, 2012):
Avoidance of any pump system by design of a “pumpless” chip with a gravity-based media flow;
improvement of the monitoring and control periphery;
maintenance of mechanical forces or electrical coupling, where relevant;
improvement of the common culture medium into a more realistic blood surrogate; and
improvement of organotypicness of cell culture compartments from 3D human cell line constructs towards primary human organoids, addressing their specific ECM architecture, epithelial barriers, stromal tissue impact, physiological absorption, and secretion or evaporation kinetics of water where relevant.
A μCCA platform-based two-organ chip mimicking the gastrointestinal tract by a co-culture of CaCo-2 cells and the mucin producing cell line TH29-MTX and the liver by a HepG2/C3A cell line culture has recently been presented by Shuler’s team (Esch et al., 2014). This intestine-liver chip was exposed to nanoparticles over a period of 24 hours. The results suggest that ingested carboxylated polystyrene nanoparticles have the potential to cause liver injury in the system. In a next step Michael Shulers group developed a four-organ chip for toxicity testing (Oleago et al., 2016). A low-cost pumpless gravity driven flow system for maintaining viable and functional human cardiac, liver, skeletal muscle and neuronal cultures within a common defined medium for two weeks has been recently presented. These cell types were chosen to provide insight into important metabolic and functional changes in human tissues in response to challenge with Doxorubicin, Atorvastatin, Valproic Acid, Acetaminophen and N-Acetyl-aminophenol with well-defined toxicological properties. The presented data demonstrates the survival and continued functionality of all 4 human cell types over a 14-day culture period under flow and their response to a 48 h single dose exposure to the drugs. The results of all drug treatments were in general agreement with published toxicity results from human and animal data. The presented phenotypic culture model exhibits a multi-organ toxicity response and constitutes a step towards an in vitro “human-on-a-chip” assay for systemic toxicity screening. The latest prototype of the development in Shulers lab is a 10-organ prototype illustrated in chapter 4 of this report.
A range of other MOCs targeting systemic arrangements of different human tissues on a chip have emerged. A customizable Lego-like plug & play system, μOrgano, which enables initial individual culture of single organ-on-a-chip systems and subsequent connection to create integrated multi-organ microphysiological systems has been constructed by Loskill and colleagues (2015). As a proof of concept, the μOrgano system was used to connect multiple heart chips in series with excellent cell viability and spontaneously physiological beat rates. Experiments with multi-tissue arrangements are ongoing. Zhang and co-workers from the Institute of Bioengineering and Nanotechnology, Singapore, have developed a multichannel 3D micro-fluidic cell culture system providing four separate channel-based cell culture spaces which can be loaded with tens of thousands of cells each (Zhang et al., 2009). Human liver, lung and kidney cell lines and primary human adipose cells were arranged in microchannels shielded from direct shear stress to mimic in vivo shear stress protection by endothelial walls. Recirculation at flow rates of 0.2 mL/min and exposure times of two days for transforming growth factor beta 1 led to a discrete biological response of the human lung cells, whilst other compartments remain uncompromised. Another integrated systemic solution for the emulation of intestinal absorption, hepatic metabolism and responsiveness of human breast carcinoma cells applying four drugs used commonly has been reported by Imura and co-workers from the Department of Applied Biological Chemistry, School of Agricultural and Life Sciences of the University of Tokyo (Imura et al., 2010). Their system provides a unidirectional flow in a chip format, the area of a microscope slide. It supports the constant perfusion of tens of thousands of human HEPG2 cell line-based hepatocytes and human MCF-7 cell line-based human breast carcinoma cells, consecutively arranged in a single microchannel. Drugs are provided through a tightly closed monolayer of human CaCo2 cell line-based intestinal epithelial cells into the media flow of the channel before the liver compartment, emulating the absorptive properties of the human intestines. Exposure time reached two days at a flow rate of 0.4 μL/min and led to discrete biological effects on the human breast carcinoma cells. Ifosfamide nephrotoxicity has been investigated over 72 hours in a liver-kidney co-culture MPS in comparison to static (Snouber et al., 2013). The results suggest that ifosfamide nephrotoxicity in a liver-kidney microfluidic co-culture model using HepaRG-MDCK cells is induced by the metabolism of ifosfamide into chloroacetaldehyde. This study demonstrated the capability of multi-organ systems to detect secondary metabolite toxicity, combining a human cell line-based liver model with a second target organ, in this case, kidney.
Gordana Vunjak-Novakovic and her colleagues are developing a HeLiVa multi-organ platform with functionally connected vascular, liver and cardiac microtissues derived from a single line of human pluripotent stem cells (Vunjak-Novakovic et al., 2013). The ability to derive all necessary cell types for multi-organ systems from one donor source is a very important prerequisite for advancement in to immunocompetent multi-organ systems to avoid organ rejection in that systems due to donor incompatibility. The platform enables functional representation of human physiology in conjunction with real-time biological readouts (via imaging and homologous reporters for all three cell phenotypes) and compatibility with high-throughput/high-content analysis. The first prototypes of microfluidic platforms for the formation and cultivation of vascular, cardiac and hepatic microtissues are currently under evaluation to demonstrate utility of human iPS cell derived microtissues for physiological and pharmacological studies.
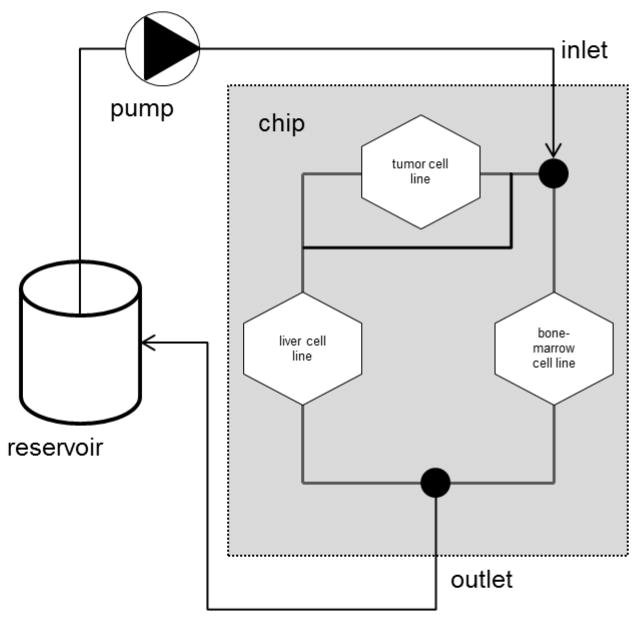
In order to overcome the non-physiological fluid-to-tissue ratio caused by the use of external pumps and reservoirs in all pump-based aforementioned MOCs, the group of Uwe Marx at the University of Technology Berlin and their collaborators designed and prototyped a MOC platform (Fig. 10) at a standardized chip format of the area of a microscopic slide, equipped with a robust peristaltic on-chip micropump, modified from Wu and colleagues (2008).
Fig. 10. Multi-organ chip platform.
(A) A PDMS chip (yellow), 3 mm high, bonded onto a microscopic slide hosts two independent microcircuits with a circulation channel of 100 × 500 μm. Each channel connects two tissue culture compartments, supporting the integration of 3D tissues, such as cell spheroids, and standard 96-well inserts for reconstructed barrier organ models. A peristaltic on-chip micropump (black) enables pulsatile unidirectional fluid flow at physiological frequencies. (B) Represents a worm’s-eye view of two blood-perfused circuits. (Reprinted from Marx et al. 2012.)
It supports pulsatile recirculation of media flow through one, two or more tissue culture compartments at near in vivo-like rates and pulse frequencies. Furthermore, the organ culture compartments of this MOC platform universally support the culture of 3D cell spheroids, reconstructed tissue equivalents, such as insert-based skin or intestine and donor-derived tissue explants or biopsies. The platform aims at homeostatic long-term performance of co-cultured human organ models over at least 28 days to enable repeated dose systemic substance exposure. A robust and reproducible 28-day co-culture process of human HepaRG-Stellate cell spheroids interconnected with insert-based human skin biopsies could be established, each a 1/100,000 of the biomass of their original human organ counterparts (Wagner et al., 2013). Skin could be maintained functional in an air-liquid interface during on-chip co-culture. Furthermore, the total fluid-to-tissue ratio in the system supports tissue crosstalk, indicated by the consumption by the skin of albumin produced by liver tissue. Finally, the co-cultures revealed a dose-dependent toxic response to a 6-day repeated dose exposure to troglitazone, a diabetes drug withdrawn from the market due drug-induced liver injury. Another two-organ co-culture using the MOC platform was aiming to combine the 3D liver model described with a commercially available reconstructed human 3D small intestine model (Maschmeyer et al., 2015a). This aimed to increase human organotypicness of the barrier compared to the CaCo-2 barriers and to improve robustness and industrial applicability. A reproducible and easy to perform oral substance administration protocol was applied over a stretch of 11 days after a three-day administration-free adaptation period, resulting in a continuous two-week MOC performance at repeated dose testing conditions. Immunohistochemistry of transporters, transepithelial electrical resistance measurement of barrier integrity and gene expression analysis revealed stable intestinal barrier functionality, whilst the troglitazone treatment decreased the albumin mRNA expression in the liver spheroids.
The same platform was used to repeatedly expose a long-term co-culture of human liver and neuronal spheroids to hexanedione (Materne et al., 2015). Therefore, 14-day co-cultures of the aforementioned liver spheroids with human neuronal spheroids, preformed and differentiated in a stirred tank bioreactor using the NT2 cell line, were exposed daily to different doses of hexanedione over an 8-day period. Sensitivity profiles of the co-culture could be compared with the responses of the single organ chips and revealed higher levels of sensitivity. Finally, a robust procedure to integrate biological vasculature into the microfluidic channels of the MOC platform has been established under physiological fluid flow conditions (Schimek et al., 2013). Thus, the MOC platform qualified for long-term repeated dose substance evaluation at homeostatic conditions combining human primary or cell line-based 3D micro-tissues. It is commercialized by TissUse GmbH, Berlin, Germany. Rapid prototyping tools allow the flexible adjustment of MOCs to any new format. Thus, 28-day four-organ co-culture has recently been established in the MOC format aiming for a human organotypic ADMET test chip. (Maschmeyer et al., 2015b). Reconstructed human small intestine was interconnected with a liver spheroid model, a skin biopsy at air-liquid interface and a human proximal tubules cell monolayer barrier. All human tissues maintained a high cell viability and discrete physiological tissue architecture over the entire co-culture period of 28 days. The human small intestine epithelia formed stable 3D villi-like structures of up to 270 μm height and expressed an apical brush border. Skin biopsies developed a stratified stratum corneum at the air liquid interface. The human proximal tubule epithelia maintained a functional polarized monolayer barrier. Small intestine and kidney proximal tubule cell barriers were able to keep a continuously stable glucose gradient balance between the intestinal lumen, the surrogate blood circuit and the excretory circuit of the system. Moreover, in-depth metabolic and gene analysis revealed the establishment of a reproducible homeostasis between all four tissues over at least 28 days, independent of the individual human cell line or tissue donor background used for each organ equivalent. The four-organ chip has been designed to support ADME profiling of substances along with repeated dose systemic toxicity testing of drug candidates. Evaluation of physiological absorption, first pass metabolism and pharmacokinetic and pharmacodynamics parameters such as effective concentration, maximum tolerable dose, time course, and intensity of therapeutic and adverse effects are envisioned. Finally, the system might support investigation of damage and regeneration of the intestine and skin compartments due to their stable physiological turnover in the system. The next development step of the MOC at Uwe Marx lab is a 10-organ prototype illustrated in chapter 4 of this report.
Other multi-organ systems integrating two or more organ equivalents are under development in the Linda Griffith lab at MIT and in Donald Ingber’s lab at the Wyss Institute at Harvard financed by the US DARPA/NIH/FDA initiative. Both groups have successfully demonstrated several two-organ chip variations (e.g. MIT liver/immune, gut/immune, endometrium/epithelial and lung/airway MPS) to the project evaluation committee. In addition, the progress of a four-organ platform with these MPS, where two-week viability and functionality was achieved, was recently presented to DARPA/NIH/FDA by the MIT team. The DARPA/NIH/FDA initiative is described in more detail in chapter 4 of this report in the context of the development of human body-on-a-chip platforms aiming to emulate organismal complexity of human biology in vitro.
2.4. Computational models, in-process controls and readouts for MPS
The use of computational and experimental paradigms for single and multi-MPS is critical to build physiologically and pharmacologically relevant MPS and interactomes. On-platform scaling describes where tissue weights, media volumes and inter-MPS flow rates can be based on either allometric scaling or functional scaling (Stokes et al., 2015). Computational fluid dynamics simulations calculate flow and oxygen/drug distributions, and shear stresses (Domansky et al., 2009). Physiologically based pharmacokinetic (PBPK) models investigate platform design and data interpretation via predicting/analyzing drug concentrations and exposure for given operational parameters, such as inter-MPS flow rates and MPS volumes. (Yu et al., 2015). Correlation, extrapolation and translation of results in such MPS systems can only be performed with the proper use of various computational models. In vitro/in vivo correlation of pharmacokinetic parameters is widely used for simpler systems (Brown et al., 2007; Obach, 1999). The superior advantage of MPS technologies comes into play to extrapolate drug and organ interaction where the biology is less well understood. Combining systems biology and pharmacology models with high-content experimental data from MPS technologies will lead to in vitro to in vivo translation advancing clinical outcomes (Stokes et al., 2015).
MPS-based assays aim for comparability with standard tests and approaches currently in use. Therefore, process controls and endpoint analysis should at least match that required in current gold standards. For laboratory animal experiments – the gold standard in the field – daily laboratory analysis of blood and urine, body temperature and visual monitoring of animal behavior are the most common in-process controls. These can be matched by daily medium analysis and viability testing in MPS-based assays. Instrumentation of MPS should be able to monitor and control tissue culture temperature, pH and oxygen consumption. This, in combination with, for example, LDH measurement and glucose consumption analyses in media, provides a good match to the visual inspection of animals for general behavior. A critical aspect of these fundamental in-process controls is the limited amount of medium (blood surrogate) available for analysis due to miniature size of the MPS. Prime endpoints for toxicity studies in animals are dose-dependent death and the histological analysis of all relevant organs after necropsy. Viability assessment in MPS has already been discussed. Organ histology in animals, to a certain degree, can be matched with histological analysis of the tissues similarly fixed, paraffin-embedded and stained, including immunofluorescent staining for proteins and other markers. Disease modeling and efficacy testing in animals applies disease-specific in-process controls (e.g. serum biomarkers, leucocyte monitoring, biopsies, neuronal electrodes, EEG, ECG) and endpoint analysis (e.g. pathohistology, restoration of organ function). Not all of them are easily applicable to MPS-based disease models. However, in contrast to readouts for laboratory animals and patients, MPS have much more to offer.
Due to their minute size, often real-time optical access at high spatial resolution and the tight instrumentation-based control of their environment, biological events which cannot be monitored in animals or patients can eventually be analyzed in MPS-based organ models. In principle, all modern tissue and cell analysis methods, such as PCR, (methylated) DNA and RNA sequencing, mRNA expression profiling, proteomics, metabolomics, and multiplex protein analysis (e.g. Luminex), can be used on (single) cells, tissue slices or flow through solutions to characterize cells, intracellular processes and tissue architecture. Model-specific assays can also be developed, including electrical monitoring by microelectrode arrays, for example, for MPS-based heart or brain models. Optical access to the micro-tissues of MPS allow for the use of 3D microscopic imaging of cell morphology and ECM architecture, as well as fluorescent imaging, both at high resolution (at least around 1 micron) to distinguish nuclear or cytoplasmic localization in a cell sufficiently. The main challenge is to combine sufficient focus depth with high resolution. Optical coherence tomography to a depth of 1 – 2 mm into the tissue, with space- and time-resolutions of micrometers and tenths of a second, is an attractive option. Other imaging techniques, such as confocal laser scanning microscopy, dual or multi-photon fluorescent microscopy, Raman spectroscopy or Fluorescence-lifetime imaging microscopy, can be applied.
A variety of label-free, nonoptical methods exist and have proven to be valuable tools for the monitoring of living systems (Fang, 2015; Wang and Liu, 2010). Extracellular acidification can be monitored with pH sensors, such as light addressable potentiometric sensors, ion sensitive field effect transistors, opto-chemically or simple potentiometric sensors (Brischwein et al., 2009). Dissolved oxygen can be measured with amperometric sensors or optochemically. Impedance sensors, quartz crystal microbalance systems, surface plasmon resonance systems and surface acoustic wave systems are used for monitoring morphological changes. The activity of electrophysiologically active cells (e.g. cardiomyocytes or nerves) is monitored with microelectrode arrays (Liu et al., 2014). These label-free methods allow real-time, time-resolved monitoring of cellular parameters which may be a great advantage in the field of MPS, since automated label-free monitoring of the living system for hours and days is achievable. However, the usage of, for example, fluorescent markers, bears the risk of activating receptors and altering the behavior of the MPS (Shinawi et al., 2013)
2.5. Instrumentation, prototyping and manufacturing of MPS
The MPS are composed of a cell culture part – a plate or a chip – and the necessary instrumentation to control and operate the cell culture part: incubators, pumps, tilting devices, control units with sensors and actuators, optical imaging devices and automated pipettors. The developmental challenges of instrumentation of MPS have been reviewed elsewhere (Wikswo et al., 2013). On-chip pumps have been one of the most challenging hurdles. Electromechanical or gas pressure controlled peristaltic micropumps are integrated into the microchannel circuits for a maximum of autonomous operation and to ensure low perfusion rates for circulation and exchange of culture media (Busek et al., 2013; Schimek et al., 2013). Miniaturized sensors based on thin-film or microbead-based fluorescent indicators (Liebsch et al., 2000; Schmälzlin et al., 2005) are increasingly implemented into MPS to monitor and control dO2, dCO2 or pH. Several reviews have summarized the current status of technologies for electrical coupling of neuronal and cardiac cell cultures to record physiological signals and stimulate the cells (Gosselin, 2011; Hierlemann et al., 2011; Simmons et al., 2012; Spira and Hai, 2013). Electrical coupling of neuronal cells in a dynamic microfluidic environment using microelectrodes has been described as early as 2005 (Pearce et al., 2005a; Pearce et al., 2005b).
Electrical coupling of muscle cells cultured in MPS have also been described (Agarwal et al., 2013; Ahadian et al., 2012; Cheng et al., 2006; Dvir et al., 2012; Grosberg et al., 2012; Klauke et al., 2003, 2006, 2007; Werdich et al., 2004). A prime example for a feasible instrumentation for MPS is the cellasys (Kronburg, Germany) technology (Fig. 11), which employs BioChips with label-free microsensors for pH, dissolved oxygen, impedance and temperature, and was used to monitor living cells in suspension, adherent monolayers, primary tissue and 3D spheroids for more than one month (Weiss et al., 2013; Wiest, 2015). The 6xIMOLA-IVD consists of six measurement modules which can be operated in parallel or serial mode. A computer-controlled fluidic system with peristaltic pump, valves and air bubble detectors allows a flexible arrangement of the MPS with 24 channels. This modular approach can be used to select different units out of a toolbox for building various organ-on-a-chip setups.
Fig. 11. A prime example of MPS instrumentation.
(A) BioChip-D (24 × 24 mm2) with microsensors for pH, dissolved oxygen, impedance and temperature. (B) The sensors are manufactured on a transparent substrate allowing additional optoanalytical methods. Computer-controlled 6xIMOLA-IVD arrangement with six measurement units and an automated fluidic system pre-mounted in an incubator (B). (Courtesy of cellasys GmbH, Germany.)
Prototyping and manufacturing of the (mostly disposable) cell culture part of MPS is another challenge in the field. Affordable, easy to handle, reliable and scalable manufacturing approaches applicable for volume fabrication in an aseptic environment are required to meet the demand in ready-to-use chips, eventually combining polymers, metallic sensors and immobilized biological molecules, cells or tissues. For these reasons, fabrication of MPS requires a multidisciplinary approach combining the engineering aspect with the biological functionality. General principles of relevant microsystems for biological applications have been described in detail (Ziaie et al., 2004). For most applications in the life sciences polymer devices are in use. In industrial applications the thermoplastic materials are dominating since this is a scalable fabrication technology, whilst in the academic world the soft polymer PDMS is a widely used material. The particular challenges of fabrication of microfluidic cell culture systems and the pros and cons of soft PDMS-based systems versus thermoplastic polymer systems in the field of MPS have been reviewed elsewhere (Alrifaiy et al., 2012; Andersson & van den Berg 2004; Berthier et al., 2012; Huh et al., 2013; Kim et al. 2007; Park et al. 2003; Wu et al. 2010). A replication-based technology, such as injection molding or injection compression molding, is a valid option for scalable industrial fabrication with a long track record in the industry of life science consumables (Becker and Gaertner, 2008, 2012). The technology chain for the realization of MPS in thermoplastic polymers is summarized in Fig. 12.
Fig. 12.
Technology chain for scalable industrial manufacturing of polymer-based MPS
The starting point is the design of the system, followed by the realization of a mold insert with the respective microstructures that will, subsequently, be replicated. The mold insert fabrication is the critical process, since all follow-up fabrication steps rely on the quality of the template generated at this point. After mold insert generation, the master will be replicated in polymers by injection or injection-compression molding allowing for a high volume fabrication. Keeping in mind that these first fabrication steps consume 80 % of the cost of prototyping or low-volume manufacturing, at a higher-volume fabrication scale, the so-called back-end processes consume up to 80 % of the overall cost. Back-end processes summarize steps, such as surface functionalization, bonding, assembly, integration of hybrid elements, e.g. membranes, sensors, valves, and dry and liquid reagents. The final step is quality control, which requires, in addition to physical inspection, technologies such as optical evaluation, in particular a functional control in the case of biological reagents, and finally, cells or pre-assembled tissues, particularly, can be shipped. Taking the cost-structure into account, a design for manufacturing is a crucial part for a later commercial success and the penetration of the new technology into standard laboratory life.
It is beneficial for any product development to start directly with the material that is intended to be used for the later product. Envisioning using thermoplastic polymers, such as cyclo-olefinic polymers (trade name “Zeonor”), cyclo-olefinic co-polymers (tradename “Topas”) or, in particular, polystyrene, which is a standard material for petri dishes for cell culture or microtiter plates; these materials should also be used for prototyping. Similar to the microfluidic area of the life science world, from where the MPS approach originates, the same standard formats are used: the format of a microscopy slide (75.5 × 25.5 mm) and that of a microtiter plate. This leads to two major benefits: family tools for fabrication can be used for a large variety of application-specific designs, therefore, not requiring larger investments even at medium scale production. Furthermore, the same fabrication chain for the back-end processes can be applied, making the realization of the MPS through scalable fabrication technologies affordable from the beginning. Regarding applications, the standardization of MPS in those formats allows for the use of the complete instrumentation from the lab automation and screening world, namely pipetting robots, imaging systems, incubation and storage elements and standard microscope setups. Fig. 13 illustrates two different MPS formats – a chip and a plate – fabricated using the technology chain outlined above from microfluidic ChipShop.
Fig. 13. Hard micro-machining of microfluidic plates and chips.
MPS examples of a chip and a plate manufactured using thermoplastic polymers. Microscopy slide format chip with integrated fluidic interfaces (A) with cross-flow operating membranes for on-chip co-culture of cells. A microtiter plate-sized platform with on-plate reservoirs for passive hydrodynamic driven fluid flow (design HepaChip project, FKZ 031A121D) (B). Animation of an integrated MPS with parallel on-chip co-culture of different organ equivalents and on-chip liquid supply, including windows optical imaging of metabolites (C). (Courtesy of Microfluidic ChipShop, Germany.)
The chip in the format of a microscopic slide requires a microstructured molded part, integration of a membrane, assembly of top and bottom with a cover lid and surface functionalization. With this straightforward scalable fabrication technology, simple devices for cell co-cultures on-chip can be realized. The lower left picture illustrates a microtiter plate-sized device combining fabrication steps such as injection molding, thin film technology for electrode integration, surface functionalization, biological reagent patterning and assembly with a cover film. The design of an integrated device fulfilling multiple tasks is shown on the right-hand side: Injection molding, integration of membranes and valves, liquid reagent implementation and pressure driven flow through embedded syringe pumps are some of the challenges towards a fully integrated MPS illustrated.
3. Industries test standards, unmet needs and potential for MPS adoption
Although there are only a small number of young companies, e.g. AIM Biotech, Bionas, cellasys, CN Bio Innovations, Emulate, Hesperos, Hμrel, InSphero, Kirkstall, micronit, Mimetas and TissUse, providing MPS devices, they are already advanced enough to provide exploratory platforms for feasibility studies. Certain members of the pharma and cosmetics industries have already been investigating the use of MPS for hazard potential identification, and safety and efficacy testing. It was common sense among the workshop experts that none of the MPS-based approaches and tools existing currently have yet been adopted by the industries. In this chapter, we report the results of the workshop discussions and subsequent surveys with regard to current standards of substance testing in the pharmaceutical, chemical/cosmetics and tobacco industries. On the basis of this information, experts from each industry segment summarize the unmet needs in the field. Finally, potential directions for demand-driven industrial adoption of relevant MPS-based approaches in the industries are highlighted in this chapter.
3.1. Pharmaceutical industry – test standards and potential for MPS adoption
3.1.1. Aspects of pharmaceutical safety assessment
Drug safety assessment is an ever evolving process which takes into account both historical data and newer findings, together with changing regulations from authorities and new experimental and technical capabilities. Drug safety assessment today spans a large battery of in silico, in vitro and in vivo tools which aim at selecting and optimizing the best candidates for safe dosing in humans. To a significant extent, a portion of those tests are required to get approval by regulatory authorities, while many of the additional tests being conducted are used for internal decision-making and for mechanistic understanding of adverse effects. Tests required include, for example, assessment of the genotoxic potential by MNT (micronucleus test) or AMES (Bruce Ames’ test), an indication of risk of cardiac arrhythmias (torsades de pointes, assessed by measuring the inhibition of the human ether-a-go-go-related gene channel) or the estimation of a drug drug interaction potential in liver using in vitro assays. On an in vivo level, the indication of target organs of toxicity, types of toxicities, plasma exposure, and distribution and excretion patterns on a quantitative basis in both a rodent and a non-rodent species, taking into account comparable treatment duration, are expected to be demonstrated as part of a package for entry into human trials. These are just a few examples and the requirements vary depending on the type of drug modality (small molecule, peptide, large molecule, other), drug administration (orally, by inhalation), disease indication and existing knowledge in a particular area. That said, drug safety assessment is an iterative process with subsequent learning depending on the stage, however, the extent of testing can vary greatly – from minimal test data to large packages including all possible screening, investigational or accepted tests.
Despite continuous improvements, there are failures in drug programs, both in preclinical and in clinical stages (e.g. Arrowsmith and Miller, 2013). Drug programs stopped at preclinical stages usually face issues with unacceptable, acute or severe toxicities, a drop of exposure or similar types of effects, which would make successful development of a drug unlikely. As highlighted in Chapter 1 of this report, industry benchmarks for candidate success rates in phase I clinical trials are 48 – 64 %, indicating a still significant share of safety-related candidate failures (Cook et al., 2014: Hay et al., 2014). Cardiac and liver toxicity are the prime contributors to these withdrawals, followed by kidney toxicity. Thus, there is still an unmet need to develop in vitro systems mimicking these human organs in a near to in vivo environment. Here, MPS-based single-organ systems might contribute with long-term in vitro stability for repeated dose substance testing over weeks. A significant improvement of existing MPS-based liver, neuronal, cardiac or kidney systems would be desirable. For easier industrial adoption, we recommend the generation of a biological baseline for each chip-based single organ equivalent. Exemplarily, it is necessary, therefore, for MPS-based liver systems to determine the architectural and functional correlation of the chip-based liver equivalent with the in vivo liver based on histology, analysis of gene expression, protein levels, such as albumin secretion, CYP450 (particularly selected protein activities such as CYPs 1A, CYP2C and, CYP3A) ATP, GSH, LDH, AST, ALT, and metabolite analysis. We further recommend the generation of an extensive time course of both early and late time points and to compare it with data of standard hepatocyte cultures. In a next step, well-known hepatotoxicants should be assessed by the chip-based liver equivalents to prove that additional data can be derived at tissue level in contrast to static hepatocyte models. We would advise the use of early and late time points and, if possible, pharmacologically relevant doses for assessment. Improvement of the MPS-based liver models reported would be a segregation of bile from long-term cultures and the integration of immunocompetent cells, e.g. Kupffer cells. This scheme of improvement of single organ MPS can be universally applied to the other human organ models.
Another important added value for the pharma safety testing could be the qualification and validation of the liver+X two-organ systems presented in Chapter 2.3.3. Such organ arrangements might lead to reliable assays, allowing the elimination of candidates from nonclinical pipelines, which are not toxic per se, but are toxic through their liver metabolites. Finally, immunogenicity evaluation of biopharmaceuticals is another unmet safety need. The majority of large biomolecules in the pipeline are of human origin. This makes immunogenicity testing of such candidates in animals obsolete. Human immune organ cultures able to predict immunogenicity of candidates are under development, but have not yet been adopted industrially (Giese and Marx, 2014). MPS approaches can play a leading role in the establishment of such test tools.
A big challenge in the pharmaceutical safety assessment space are drug-induced idiosyncratic injuries of individual organs, e.g. liver, based on genetic predisposition of a tiny subpopulation of patients and the properties of a given drug. Drugs inducing this type of toxicity often fail much later in clinical stages or after market approval. In many cases, the program is stopped if the mechanism cannot be fully elucidated, albeit only a very small proportion of patients showed toxicity. These effects usually arise due to a potential liability coming from the drug which, under normal conditions, does not become apparent and a patient-derived factor (genetic predisposition, disease-specific). A toxicity arises only when these two aspects occur together. This so-called idiosyncratic type of clinical toxicity poses a significant challenge, because if the mechanism is not known, tens of thousands of MPS-based tissue models need to be tested from tens of thousands of different donors to get a statistical chance of identifying the human subpopulation at risk prior to exposure of the drug in humans. However, some of the idiosyncratic drug-induced liver mechanisms have been found to be dependent on specific HLA types and possibly on specific factors of the adaptive immune system. For these cases, the development of efficient MPS-based screening platforms might be worth trying. Furthermore, the reported extension of the multi-tissue systems towards systems physiologically arranging an intestine and a kidney equivalent with a liver equivalent on a chip may enable a new application for multi-tissue liver equivalents – the generation of ADME data and PK/PD analysis within one experimental setting, provided quantitative aspects of drug uptake, distribution and excretion can be recapitulated in vitro. All the desirable MPS developments mentioned previously can be implemented both in screening strategies and in mechanistic investigations.
There are other major fundamental challenges when working in vitro; one being the lack of a quantitative translation from an in vitro to an in vivo effect. The concept of investigating AoPs, for example, which has arisen from the cosmetics and chemical industry in their effort to seek alternatives to systemic toxicity studies, cannot really be linked to the concept of defining a “No Observed Adverse Effect Level” established for clinical drug development: What does a toxic effect at 10 μM influencing an AoP mean for the clinic if we do not know the kinetic parameters in humans; for example, what are the exposures, metabolism, distribution and elimination of the drug likely to be in humans? Therefore, information on AoPs may not be directly usable for the in vivo situation where doses have to be defined. In addition, such quantitative estimates are generally not available in the early stages of drug development when in vitro tests are applied, and this makes firm statements on the relevance of an in vitro flag difficult. There are historical data to validate some of those endpoints, but not for all the endpoints needed, and a drawback of historical datasets is that the chemical space of, for example, 1970 drug development may be very different from the one in 2016. Therefore, MPS-based assays, mimicking human multi-organ arrangements with physiological blood flow and at scale down parameters, matching basic physiological principles in patients are highly desirable for pharmaceutical safety testing.
3.1.2 Test standards, unmet needs and new approaches in efficacy testing
Efficacy of drug candidates is assessed along the drug development cycle. Drugs no longer fail in the clinic because of undesirable physicochemical or pharmacokinetic properties. Instead, a major reason for failure is lack of efficacy. In vivo efficacy and activity testing in laboratory animals are a major cornerstone and requirement in preclinical phases. These tests are accompanied by in vitro bioassays, such as target binding and potency assays. A compilation of attritions for oral small molecules from four large pharmaceutical companies revealed a lower attrition of 35 % of such compounds in phase II due to efficacy concerns (Waring et al., 2014). Thus, an average attrition rate of 35 – 71 % in phase II trials indicates the limited knowledge gained in preclinical studies on the efficacy and MoA of a drug candidate. The clinical relevance of most targets pursued in drug discovery is often not completely understood. Since the advent of the rational design concept in the late 1970th, the focus on most screening programs in pharmaceutical R&D has been on single drug targets, which are typically proteins that appear to have a role in disease pathogenesis. Target-based screening platforms also fit well into high throughput screening strategies and very potent and selective compounds can often be found and specific chemical and molecular knowledge can be applied to modify the chemical entities. However, the low success rates indicate that alternative preclinical strategies are needed that focus on relevant biological pathways and their clinical relevance. Swinney and Anthony (2011) analyzed the productivity of R&D and especially the innovative medicines including new molecular entities and new biologics that were approved by the US FDA between 1999 and 2008. The majority of first-in-class small molecules that were approved came from phenotypic screening and not from target-based approaches, and that is in an era in which the major focus was on target-centric approaches. One major reason why the reductionist screening strategies using a single receptor/protein often fail could be that most diseases are multifactorial events with a systemic impact on many organs and, consequently, a response needs to be observed at the level of an intact organ and its interaction with the other organs in the right pathophysiological context.
There is, thus, an urgent need to develop preclinical assays that translate more effectively into clinical effects in patients with a specific disease. Metabolic syndrome is a good example of a multi-factorial and multi-organ disease collecting a number of risks associated with cardiovascular disease and diabetes. Metabolic syndrome is becoming increasingly common largely as a result of the increased prevalence of obesity. It is an insidious and progressive disorder that can culminate in type 2 diabetes which carries a high risk of cardiovascular disorders and is also prevalent in patients with chronic kidney diseases. There are no approved drugs that can reliably reduce all the metabolic risk factors. To avoid the problem of polypharmacy, these patients would be helped with drugs that target several or all risk factors with single therapies.
A human relevant preclinical MPS-based multi-organ model useful for a disease such as metabolic syndrome is urgently needed due to the low predictive power of animal models in the field. It needs to incorporate the key organs, such as pancreas, liver, adipose, muscle vasculature and the kidney. In such a model, the key steps in the progression of the disease from abdominal obesity and insulin resistance to Type 2 diabetes and diabetic complications could be investigated. The model could be build up by healthy tissues where the progression of metabolic disorder can be studied by, for example, medium, nutritional or hormonal manipulation. Another strategy would be to use tissues or cells from diseased subjects. Such a model could be used to unravel new biological pathways and targets and their clinical relevance at various stages of disease progression in the model. The ultimate aim for microphysiological systems in drug development is to recapitulate the various stages of a disease and even understand the stages before the disease is clinically manifested, which may open the way for new treatment paradigms.
The optimization of drug candidates can include target-based screening if a specific target has been selected. The optimized compound can, in this case, be investigated in the disease model for treatment effects. Eventually the selected compounds can be evaluated in the disease model and an in vitro dose-finding study to guide the clinical program. This strategy integrates early drug discovery and translational discovery. In addition to treatment effects in a disease relevant model, the MPS-based model can also be used to investigate useful in vivo treatment biomarkers.
The multi-organ model can use either material from diseased individuals or the disease can be induced in the system. The disease pathogenesis can be investigated and biological networks unraveled by using the latest multi-parameter read-out technologies and bioinformatics. The insight into the disease pathology at the molecular level can indicate whether a single target approach or a more complex strategy pursuing multiple targets is warranted. Bioinformatics and conventional structural biology tools combined with chemoinformatics and computational analysis can guide the choice of intervention and aid in identifying the types of chemistry needed for pathway perturbation. A schematic overview of a possible patient-centered drug discovery model using the MOC as a focal point to evaluate intervention strategies, evaluate targets and dose setting is described in Fig. 14.
Fig. 14. Impact of MPS-based models on drug discovery screening strategies.
A model of how to transform drug discovery screening strategies by including disease mimicking multi-organ system approaches and tools is proposed. The model can be used to analyze and unravel disease mechanisms and define treatment interventions.
Another important opportunity for advanced multi-organ MPS in current pharmaceutical new drug candidate assessment is combined toxicity and efficacy testing in the oncology space. After the identification of a valuable cancer target, compound libraries are screened for inhibitors (or sometimes activators) of this target. Subsequently, compounds which have passed this first step are tested for in vitro efficacy by determining the inhibitory effect of these so-called hits on tumor cell lines. In a next step, compounds with promising in vitro inhibitory potential and an acceptable pharmacokinetic profile are administered to tumor-bearing rodents.
During these early phases, safety aspects can be investigated in non-cancer cell lines or primary cell cultures (Tab. 2). The endpoints in these in vitro toxicity assays are rather simple and determine the inhibition of proliferation in short-term assays lasting from 24 to 72 hours, for example, in cultures of proliferating keratinocytes (HaCaT(p)) or overt cytotoxicity in non-proliferating cells (HaCaT(q)). Primary rat hepatocytes are frequently used in order to reflect the potential metabolism of a compound. Such an isolated in vitro test battery is able to identify those compounds which lack specificity towards the tumor target by comparing the EC50 obtained in the tumor cell line test with the cytotoxic concentrations in the quiescent culture or the IC50 in the proliferating non-cancer cell. An IC50 in the range of the EC50 clearly indicates that the specificity to the tumor target is too low and adverse effects on healthy, particularly on proliferating tissues have to be expected in patients. Compound 2 shows cytotoxicity in a non-proliferating cell at concentrations less than 10× above the effective concentrations, as determined in HeLa cells, and is, therefore, regarded as problematic. Compound 1 is less toxic towards non-proliferating cells, but still shows anti-proliferative effects in a concentration lower than the EC50, and HeLa cells are also inhibited, i.e. the anti-proliferative specificity of this compound is questionable.
Tab. 2. Illustration of a conventional early in vitro safety assessment of oncology drug candidates.
The test compounds are tested in three different safety screens and the results are compared to the results of the tumor cell line inhibition (Hepas: primary rat hepatocytes; HaCaT: human keratinocytes, non-proliferating quiescent (q) or proliferating (p)). Efficacy of both compounds is in the same range; compound 2 is ranked as critical (category 2) because of its cytotoxicity in a non-proliferating cell at concentrations less than 10-fold above the effective concentrations.
However, this comparative approach is not able to reflect effects which might be caused by the interaction between tumor and healthy tissues, as it occurs in the whole organism. In addition, effects which are elicited only upon chronic exposure cannot be depicted due to the short-term nature of the test systems described.
It would, therefore, be desirable to obtain an MPS where cancer cells could be cultivated over several days or even weeks as spheroids and co-cultured with a panel of non-cancer organoids, such as liver, heart, skin and/or kidney, raised from human cells or tissue explants. Such a model would allow to obtain the readout for both efficacy and first safety parameters in just one system. In addition, it would be able to reflect changes in the microenvironment and display the effect of drug metabolites on both healthy and cancerous tissue. In theory, such a model residing on human material should also offer a better translatability of the effects observed to the clinical situation both in terms of safety and efficacy.
In summary, MPS could be used in the pharma industry as screening tools for both efficacy and toxicity testing of new chemical or biological molecules. The resulting test information can contribute to the selection of targets and lead structures, as well as for the prioritizing and selection of drug candidates for further development. Scientific information or data arising from such assays may also be used for internal decision-making and guidance in the course of the development process. In addition, as part of mechanistic investigations, it may also be attached as supplementary state-of-the art scientific information to IMPD and/or IND documentation. Figure 15 illustrates the potential application segments for single- or multi-organ systems within the drug development life cycle.
Fig. 15. Target segments of various categories of MPS along the drug development cycle.
Single-organ systems are envisioned to add significant value from target selection toward lead optimization (red box) with plate-based systems serving primarily the high throughput demand of earlier discovery and chip-based systems serving primarily the demand of lead identification and early lead optimization. Multi-organ systems are supposed to enable predictive measurements in the late lead optimization phase and early preclinical evaluation (blue box). All categories of MPS might add relevant data to the post-approval extension or restriction of indication to specific genotypic subpopulations (red-blue box). The vertical axis illustrates approximate numbers of tests performed (grey) and related spending (blue). The horizontal axis illustrates the development time in years.
Single-organ systems are envisioned to add significant value in earlier phases at higher throughput, whilst multi-organ systems are supposed to enable predictive measurements later in the preclinical stage. The latter could provide more systemic in vivo-like assays where the therapeutic impact is more readily translated. However, a disadvantage is the challenge to optimize the molecular properties of candidate drugs without the design parameters provided by the exact knowledge of the target(s) and the lower throughput. Although preclinical discovery and qualitative clinical data generation will become the prime application of MPS-based tools, their use for post-approval evaluation could be of further added value. MPS-based tools bear the potential to compete with animal models in early discovery and the preclinical stage in the future. It may also be possible to use information from MPS testing throughout the entire drug development process, even during clinical development. Finally, during this process, mechanistic information on findings occurring may support clinical use. Notably, the pharmaceutical industry will only adopt such systems when compelling data has shown that MPS-based tools provide a clear added value, for example, in the areas of unmet needs. Subsequent validation of MPS-based tools will be essential for complete adoption within the entire pharmaceutical industry. Information on a validation strategy for MPS-based tools are provided in chapter 6 of this report.
3.2. Chemical and cosmetics industries – how MPS can make a difference
Substance testing in chemical and consumer products, e.g. the cosmetics industry, aims at the identification of any hazard potential a substance might entail for humans. In contrast to the pharmaceutical industry, where a risk-benefit analysis may lead to the acceptance of a certain drug toxicity level of a candidate drug if its efficacy benefit for the patient outweighs the safety downsides, the regulatory requirement for chemicals and consumer products do not accept any meaningful hazard potential of substances used for humans. Often it is skin that these substances are exposed to. Contact with the skin is either wanted or inevitable for many daily life products, such as cosmetics, household products, perfumes, volatile organic compounds and fertilizers. Tests and strategies have been developed and validated for the in vitro assessment of skin irritation, corrosion and penetration. The risk of skin sensitization towards specific components and, consequently, allergic responses to products is another important toxicological concern. Allergic responses to consumer products can be explained by occupational skin sensitization, cross-reactivity towards structurally similar antigens, interindustry use of ingredients and sensitization of customers by the product itself. Animal test methods, such as the Local Lymph Node Assay (LLNA, Gerberick et al., 2007) and the Guinea Pig Maximization Test (Basketter and Scholes, 1992), have been the gold standards and enabled risk assessors to identify both the hazard potential and the potency of a compound, the latter parameter referring to the concentration range sufficient to evoke skin sensitization. The respective OECD guideline for skin sensitization testing is number 406 (OECD, 1992). By complementing this information with additional read-across information and exposure scenarios, risk assessors were able to make judgments on safe concentrations of specific compounds in the final product. In order to minimize risk, default safety factors are applied to give consideration to the interspecies differences in penetration, metabolism and immunology, as well as the inherent fallibility of toxicity testing (Basketter et al., 2014; Dean et al., 2001).
The cosmetics and fragrance industry is particularly challenged due to the European Commission Regulation (EU) No 655/2013 (EC, 2013c), also called the European cosmetics directive (Hartung, 2008) and, consequently, is very active in the field of non-animal skin sensitization assessment. The chemical industry is also actively working on strategies for in vitro skin sensitization testing, aiming to avoid unnecessary reassessment of chemicals due to the Registration, Evaluation, Authorisation and Restrictions of Chemicals (REACH) regulation (Rovida and Hartung, 2009; Hartung and Rovida, 2009). The legal framework for human risk assessment for chemicals in the EU is detailed in Chapter 5.5. of this report (Hartung, 2010a). Here, we focus on test standards for hazard potential identification in skin.
In the field of skin sensitization, the European animal testing ban spurred the development of a multitude of in vitro assays and, in parallel, strategies how to facilitate the development and validation of those methods for regulatory acceptance (Basketter et al., 2012, 2015; Leist et al., 2014). Activities in this field are intellectually and financially supported by associations such as Cosmetics Europe and the Research Institute for Fragrance Materials (RIFM), as well as institutions such as the European Centre for the Validation of Alternative Methods (EURL-ECVAM), the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM), the Centre for Alternatives to Animal Testing (CAAT) and the European Partnership for Alternative Approaches to Animal Testing (EPAA).
The development of non-animal in vitro methods for skin sensitization benefited from extensive data gathered by academic and clinical research, enabling scientific insights into the complex mechanisms of sensitization. Four key steps in an adverse outcome pathway (AoP) for skin sensitization were identified and published by the OECD in 2012 (EC, 2012b): 1) covalent binding of compound to skin proteins, 2) activation of keratinocytes, 3) activation and migration of skin-residing dendritic cells (DCs) to the draining lymph nodes, and 4) DC-dependent, allergen-specific priming of naïve T-cells in the lymph node, resulting in T-cell proliferation and T-memory cell formation. These insights specified appropriate targets for in vitro methods and, in addition, alluded to AoP-based testing strategies.
To date, EURL-ECVAM lists 18 skin sensitization testing options in different formal validation phases, most of them addressing single AoP events (ECVAM, 2015). Strategic approaches to systematically evaluate different non-animal testing options by comparing different assays have also been reported from the cosmetics industry (Reisinger et al., 2015). A recent major breakthrough for in vitro testing was the formal validation and regulatory acceptance of the direct peptide reactivity assay (DPRA; Gerberick et al., 2004; Gerberick et al., 2007) and the KeratinoSens™ (Emter et al., 2010; Natsch et al., 2013) in 2015. Following an endorsement by EURL ECVAM, OECD test guidelines were published for both assays (OECD TG 442C and OECD TG 442D, respectively). A third assay, the human cell line activation test (h-CLAT; Ashikaga et al., 2006), is currently rated as provisionally accepted by the OECD. The three assays conveniently encompass the first three AoP events; i) the DPRA assessing the haptenization of cysteine or lysine-containing artificial peptides; ii) the KeratinoSens™ analyzing the Nrf2-dependent stress response based on ARE promoter-driven luciferase reporter gene expression in HaCat cells, and, iii) the hCLAT analyzing the activation of the dendritic-like cell line THP-1 based on the induction of CD54 and CD86 co-stimulatory cell surface markers. Examples for integrated testing strategies are provided in Table 3.
Tab. 3. Examples for Integrated Testing Strategies and Integrated Approaches to Testing and Assessment.
Current in vitro skin sensitization assessment follows a pathophysiology-inspired chain of adverse effects from covalent binding to proteins to the immunogenic activation of dendritic cells. Several methods are used to integrate and interpret the corresponding complex in vitro information in combination with (Q)SAR methods for subsequent risk assessment.
| IATA | (Q)SAR / in silico methods | Covalent binding to proteins | Keratinocyte response | Dendritic cell activation | Prediction model | Reference |
|---|---|---|---|---|---|---|
| P&G | TIMES-M, Bioavailability variables | DPRA | KeratinoSens | h-CLAT | Bayesian Network | Jaworska et al., (2013) |
| Kao | DPRA | KeratinoSens | h-CLAT | Tiered system | Nukada et al., (2013) | |
| Shiseido | Molecular-orbital descriptors | DPRA | AREc32-cell-based assay | h-CLAT | artificial neural network (iSENS) | Tsujita-Inoue et al. (2014) |
| BASF | OECD QSAR Toolbox | DPRA, SH Test | ARE/Nrf2-based cell activation assays (KeratinoSens, LuSens) | h-CLAT or mMUSST | 2 of 3 approach | Bauch et al. (2012); Natsch et al. (2013) |
| RIVM, Givaudan | DEREK, CAESAR, MultiCASE, OECD toolbox | DPRA | KeratinoSens or Gene signature assay | h-CLAT | Tiered strategy | van der Veen (2013) |
Though DPRA, KeratinoSensTM and the h-CLAT may be ahead in terms of validation, other alternative assays, addressing similar endpoints, are actively advanced by method developers. The PPRA may introduce partial metabolism to peptide reactivity assessment to improve in chemico evaluation of pro-haptens (Gerberick et al., 2009), whereas the LuSens assay (Ramirez et al., 2014) and other Nrf-2-reporter cell line-based assays present alternatives to KeratinoSensTM testing. Regarding DC activation, several tests besides the h-CLAT exist, most prominently the Myeloid U937 Skin Sensitization Test (U-SENS, formerly known as “MUSST” assay, Ade et al., 2006), the Modified myeloid U937 skin sensitisation test modified MUSST assay and the PBMDC (Reuter et al. 2011). Despite substantial efforts, the development of assays for the fourth AoP key event is not solved, yet. First promising steps towards a T cell-based assay were published recently (Richter et al., 2013). However, the complex “organ response” in the lymph node, where activated dendritic cells from the periphery present haptenized peptides to naïve T cells, thereby priming the biologically important proliferation of antigen-specific and skin-homing T effector and T memory cells is difficult to emulate in vitro using suspension cell-based test designs.
There is general agreement that the results of the AoP-aligned tests are more complementary than exclusive (Basketter et al., 2015). However, the interpretation of testing results is challenging and the need for transparent and reproducible decision frameworks stimulated the generation of Integrated Testing Strategies (ITS, Hartung et al., 2013) and Integrated Approaches to Testing and Assessment (IATAs, Tollefsen et al., 2014; Bal-Price et al., 2015). These strategies have been recently reviewed in a t4 report on Integrated Testing Strategies for safety assessment (Rovida et al., 2015b). ITS, according to a perspicuous explanation by Jaworska and Hoffmann, represent a guiding framework for the generation of comprehensive, non-redundant in vitro data by means of a reasonable and logical combination of tests that cover relevant mechanistic steps (Jaworska and Hoffmann, 2010; Jaworska et al., 2010; JRC, 2005; Luechtefeld et al., 2015). To allow for an optimized risk assessment, the comprehensive data generated by ITS approaches should be interpreted according to thoroughly evaluated decision schemes and should consider applicability domains of singular assays as well as the consistency of assay results, especially if the ITS contains assays with partial mechanistic overlap.
The IATAs extend the ITS concept by strategically integrating and weighing all relevant data to inform regulatory decision on potential hazard and/or risk and/or the need for further targeted and, thus, minimal testing (Barroso, 2014). Information sources for IATA are not restricted to in vitro results, but include physico-chemical data, in silico predictions and weight of evidence considerations. Several skin sensitization IATAs have been developed, often based on combinations of AoP-key event assays enriched by additional assays, e.g. for surface thiol concentration, and toxicogenomics. Six IATAs were presented and discussed in a recent workshop organized by the EPAA and Cosmetics Europe. The ITS and IATA approaches generate highly complex data. Pragmatic approaches, such as the 2-of-3 concept (Bauch et al., 2012) are delivered promising results. Machine learning systems, such as Bayesian models, artificial neural networks and support vector machines, turned out to be valuable tools to establish prediction models for hazard (and partially potency) information for specific compounds (Hirota et al., 2013; Jaworska et al., 2015; Tsujita-Inoue et al., 2015). Furthermore, those systems are able to refine and improve predictive parameters easily by virtue of extended training sets. However, However, it should be noticed that the “black box” character of hidden layers in the computational models does not provide mechanistic insights for the ensuing risk assessment.
Though current IATA case studies for prediction of skin sensitization look promising, quantitative approaches for potency prediction are only recently beginning to gain traction. Potency determination is a keystone in risk assessment and is still a major gap for most non-animal skin sensitization assessment strategies. While singular assays exist that aim to provide potency information, for example, the toxicogenomic-based Genomic Allergen Rapid Detection test (GARD; Johansson et al., 2013) and Sens-Is assay (Cottrez et al., 2015), IATA approaches are also now closing in to categorize the potency of test compounds.
Three-dimensional skin models, such as MatTek Epiderm, Phenion and Episkin, have proved to be valuable tools for toxicity testing and have been extensively reviewed in a recent t4 report on Non-Animal Models of Epithelial Barriers (Skin, Intestine and Lung) in Research, Industrial Applications and Regulatory Toxicology (Gordon et al., 2015). Characterization of skin models showed, in some cases, high similarity to skin metabolism in vivo. Though difficult, progress has been made in the creation of immune-competent skin models. In parallel, skin sensitization assays based on co-cultures of HaCat cells and DCs were reported to show increased sensitivity towards pro-haptens, explained by increased metabolic capacity. Recent publications report on successful integration of dendritic-like Mutz-3 cells into skin models (Ouwehand et al., 2011) and showed the responsiveness towards irritants and sensitizers (Kosten et al., 2015).
In summary, promising advances have been made over the last few years in the field of skin sensitization assessment in vitro. The predictive parameters of current ITS and IATAs for hazard are comparable to, or even superior to, the gold standard LLNA. The AoP-based ITS and IATA strategies split the biological complexity of animal testing into practicable pieces for testing. However, the corresponding pieces of information need to be assembled by complex computational models. In addition, the applicability of cell culture-based assays can be often restricted by compound solubility and application form. Only a few skin sensitization assays are based on skin models which can deal with water-insoluble compounds or formulations. Moreover, the metabolic capacity of the test system is important for the detection of pro-haptens and needs to be considered for the interpretation of results.
The potential of MPS being used in hazard identification and toxicity testing has been recognized by consumer products industries and first feasibility studies are ongoing to evaluate the potential avenues of adoption of MPS by the industry segments. The MPS may facilitate a comprehensive testing approach by integrating the four aforementioned biologically and AoP-relevant key events and multiple readouts in a single immunocompetent skin MPS. Skin explants or 3D skin models are suitable for topical application of compounds and, for example, cosmetic formulations. In addition, they would (partially) add bioavailability parameters directly influencing the skin sensitizing effect of a compound. Skin penetration for sensitizing compounds, is probably not only influenced by molecular weight and solubility, but also by its chemical reactivity, which may result in absorption of a compound by, for example, stratum corneum proteins. First approaches to combine primary DCs and epidermal models in the microfluidic MOC platform described in Chapter 2.3. were carried out and revealed parameters that need to be adjusted for future successful integration of both cell types (unpublished data). A further step towards MPS approaches integrating all relevant parameters would be the combination of immunocompetent human skin models with human lymph nodes in a two-organ chip. Such systems might allow the detection and quantification of lymph node responses towards topically applied chemicals, which in a way mirrors the readouts of the LLNA, the animal-based gold standard in the industry. Such MPS approaches have another advantage over testing strategies comprising several assays. Combinations of assays increase the overall sensitivity at the expense of false positive results. False positive results complicate not only the interpretation of inconsistent results, but also the previous generation of computational prediction models.
Apart from skin sensitization testing the chemicals testing landscape uses a broad repertoire of toxicity tests, again, historically primarily based on laboratory animal tests. There are possibilities to apply in vitro methods, but no accepted alternative methods are available yet for studies on systemic toxicity (e.g. acute toxicity, repeated dose toxicity and reproduction toxicity). This is another unmet need in the domain of cosmetics in Europe, where animal tests have been banned completely. As of yet, there have not been any applications for market admittance for cosmetics which are based entirely on alternative methods. Needless to point out, there is a great demand for alternative methods for systemic toxicity testing which can be accepted for regulatory use.
Acute toxicity tests focus on organism death, an effect that cannot be easily mimicked with conventional static in vitro tests, as it can involve a single-organ failure and also interplay between organs. Current efforts to replace acute toxicity tests involve cytotoxicity tests in vitro with cell lines, such as the 3T3 NRU cytotoxicity assay, which need complementation with kinetic data and in vitro target organ assays (Prieto et al., 2013). Improved target organ models, such as the MPS models, in which cytotoxicity may also be analyzed could improve the predictive value of these in vitro tests for acute toxicity. The potential of MPS to substantially improve kinetic measurement and systemic toxicity testing, described in Chapter 3.1. for pharmaceutical safety assessment are generally equally applicable for chemicals and cosmetics testing of hazard potential. Thus, MPS tools described in Chapter 2.3. for kinetics (e.g. gut-on-a-chip; gut+liver-on-a-chip and gut+liver+skin+kidney-on-a-chip) are envisaged to add value for kinetic endpoints, as the different kinetic processes are reasonably separated with a different organ for each process. While the regulatory need for kinetic data does not seem large, according to Table 3 of Chapter 5.5. of this report, when using in vitro tests for hazard endpoints, the addition of kinetic data becomes essential in combination with in vitro tests or strategies. The reason is that kinetics are included in whole animal tests, but not in tests with only a small part of the organism, such as in vitro tests. Thus, kinetic data must be added to the result of an in vitro hazard test to make this result more predictive of an in vivo response. As outlined above, coupling MPS-based testing systems to physiologically based toxicokinetic modeling would be advisable. These models ideally allow an estimation of local substance and metabolite concentrations in different tissues and organs. Together with a given exposure, they can, thus, potentially identify target organs together with the relevant organ doses for subsequent testing in vitro. However, there is little experience with regard to the applicability and reliability of such a “tox-test dummy” (Tralau et al., 2012). In addition, the data concerning the substance absorption, distribution and metabolism required is often not available (Caldwell et al., 2012). It would, thus, be warranted to conduct blinded pilot studies with established and already assessed substances before any routine use.
Finally, the achievement of stable functional homeostasis in MOCs combining human intestine, liver, skin and kidney equivalents over time frames of at least four weeks (Maschmeyer et al., 2015b) opens possibilities for MPS-based 28-day repeated dose testing, which, after improvement and validation, might become competitive with gold standard animal testing used according to OECD guidelines 407, 410, 412 and 419 where appropriate. The human origin of MPS provides additional competitive advantages versus animal testing. When combined with AoPs, which connect early biochemical effects with clinical outcomes, and kinetic models, which can extrapolate accumulating doses, responses seen in MPS could become predictive of ultimate toxicological endpoints after prolonged exposures. However, the difference between four weeks of culture and 90-day repeated dose testing or 1- or 2-year animal tests which are required for certain toxicity evaluation in animals according to OECD guidelines 408, 411, 413 and 453, respectively, in geographies and segments where there is no ban, will still remain a significant technical challenge for MPS to overcome.
In summary, MPS-based advanced immunocompetent human skin models and their combination with human lymph nodes can have an extraordinary added value for hazard potential identification and potency assessment in the chemical and cosmetics industries. Moreover, human MOCs enabling stable physiological organ homeostasis and crosstalk of universal combinations of human organs over periods of 28 days and longer can mature towards valuable approaches for predictive systemic acute and repeated dose toxicity testing. In contrast to the pharmaceutical industry, where qualified assays can immediately improve the drug development process and internal decision-making, MPS-based assay adoption in the chemical and cosmetics industries requires much more formal validation efforts prior to full industrial adoption. Information on a validation strategy for MPS-based tools is given in Chapter 6 of this report.
3.3. The tobacco industry, the human respiratory system and MPS approaches
The tobacco industry is under immense pressure to significantly increase the safety of their product portfolio due to the proven high hazard potential of tobacco smoke for the human respiratory system. The US FDA Center for Tobacco Products (CTP) was set up in 2009 when the FDA was authorized to regulate the manufacture, marketing and distribution of cigarettes and smokeless tobacco products in the Family Smoking Prevention and Tobacco Control Act. The FDA recommends that applicants conduct nonclinical studies to address the known clinical toxicities of tobacco products and evaluate a range of potential toxicities of the product compared to other tobacco products on the market. Nonclinical toxicology studies should use methods that are sufficiently sensitive to assess the actual differences between the use of the product and the use of other tobacco products, or between the use of the product and the nonuse of tobacco products. Animal models are the industry standard to assess the effects of tobacco smoke on the respiratory system. Limitations associated with human or animal studies require alternative tools to align with the goals of the 21st century toxicology framework, and develop a “systems approach that fully integrates exposure and toxicity information in a holistic framework for improved public health decision-making” (Sheldon and Cohen Hubel, 2009). The FDA noted commitment to the three Rs and the inclusion of “modernizing toxicology” as one of the priority areas to address (FDA, 2011a; Fitzpatrick) and, thus, has expressed interest in learning of current assays that could meet their goals (IMNA, 2012).
In addition to the regulatory aspects, the industry’s efforts to develop new approaches and tools for assessment of smoke-related adverse effects on the human respiratory system are driven by ethical considerations and the aim of developing new safer products. The prime effect of tobacco smoke is targeting airways and alveoli in the lung. A comprehensive recent review on lung models in vitro has been provided in the t4 report on Non-Animal Models of Epithelial Barriers (Skin, Intestine and Lung) in Research, Industrial Applications and Regulatory Toxicology (Gordon et al., 2015). Another recent review by Manupello and Sullivan (2015) specifically emphasises the technological advances in the assessment of tobacco products in vitro, including reconstituted human lung tissues cultures that facilitate the exposure at the air-liquid interface (ALI) to test the toxicity of tobacco smoke, E-cig aerosols, constituents and particulates. The community of experts from the industry and academia together with regulatory stakeholders are currently working on addressing the challenges pertaining to the standardization of methods and endpoints for the new in vitro toxicity testing approaches. Regarding the assessment of potential modified risks of tobacco products, animal use should be minimized and mechanistic data should be acquired based on human cell-based in vitro systems that have the potential to provide specific, actionable and human relevant insights more quickly.
Existing in vitro models to study the biological impact of inhaled toxicants on cells of the human airway epithelia are currently based on 2D or 3D cell culture systems. Some of the drawbacks of using epithelial cell lines for in vitro toxicity studies include a limited metabolic competency which may affect their responsiveness to toxic stimuli (Garcia-Canton et al., 2013), and an absence or reduced formation of tight junctions or refractoriness to differentiation (Stewart et al., 2012). One of the most suitable in vitro models to characterize the impact of inhaled toxicants in current use is the 3D culture system, where fully differentiated primary epithelial cells grow at the ALI. This model system features the full cellular complexity of a pseudo-stratified epithelia found in human airways, containing ciliated/non-ciliated epithelial cells and basal (progenitor) cells (Prytherch et al., 2011). Furthermore, it resembles epithelial functionality most closely by apical mucus secretion, expression of ion channels and the presence of tight junctions, which warrant biological barrier function (BéruBé at al., 2009). Despite the fact that 3D cell cultures resemble the native tissue closely, they usually lack cellular components of the immune system; a gap that is being addressed by a number of co-culture systems which combine the co-culturing of immune cells with epithelial cell layers as well as utilizing ALI conditions (Klein at al., 2013). A static system comprising three cell types: epithelial cells, human blood monocyte-derived macrophages and DCs was designed as an in vitro model of the alveolar wall to examine the physiological responses of these cells to environmental particles (Lehmann et al., 2011; Rothen-Rutishauser et al., 2008). It was used to examine the role of surfactant protein D in modulating allergen particle uptake and inflammatory responses (Schleh et al., 2012), and the physiological effects and uptake of gold nanoparticles (Brandenberger et al., 2010). In these systems, the investigators used A549 cells, a lung adenocarcinoma cell line that may not be representative of healthy epithelial cells.
An urgent unmet need for the tobacco industry are approaches to investigate the onset of lung diseases. Smoking induces chronic obstructive pulmonary disease (COPD) and accounts for as many as 8 out of 10 COPD-related deaths in the US (DHHS, 2014). The title COPD refers to a group of diseases that cause airflow blockage and breathing-related problems: COPD includes emphysema, chronic bronchitis and, in some cases, asthma. With COPD, less air flows through the airways because of the destruction of alveoli, inflammation of airways, increased excretion of mucus in the airways and loss of ability of the lung tissue to stretch and shrink. Modeling such complex behavior of the human lungs is impossible in the static in vitro lung models described above. The MPS-based Lung-on-a-chip model systems, outlined in detail in Chapter 2.2. of this report, resemble additional aspects of the human respiratory tract, such as the mechanobiology of breathing (Huh et al., 2010; Stucki et al., 2015) and, therefore, provide great opportunities to model aspects of COPD, investigating toxicity-induced lung injury and repair following exposure to smoke. As outlined in Chapter 2.2. a model for chemotherapy-induced pulmonary edema has already been developed on the basis of a small-airway-on-a-chip model (Benam et al., 2015b). The authors established a human lung “small airway-on-a-chip” containing a differentiated, mucociliary bronchiolar epithelium and an underlying microvascular endothelium that experiences fluid flow, which allows for the analysis of organ-level lung pathophysiology in vitro with culture times of 4 to 6 weeks. Exposure of the epithelium to interleukin 13 reconstituted the goblet cell hyperplasia, cytokine hypersecretion and decreased ciliary function of asthmatics. Small airway chips lined with epithelial cells from individuals with chronic obstructive pulmonary disease recapitulated features of the disease, such as selective cytokine hypersecretion, increased neutrophil recruitment and clinical exacerbation by exposure to viral and bacterial infections. The improvement of such a model towards smoke-induced pulmonary edema might be one avenue for MPS-based assay adoption by the tobacco industry. Another aspect of smoke-induced disease onset is malignant transformation. First single-organ MPS developments are focusing on the malignant transformation of bronchial epithelial cells due to tobacco (Li et al., 2014). A single-organ MPS has been applied to investigate the role of macrophages in promoting benzopyrene (BaP)-induced malignant transformation of human bronchial epithelial cells in comparison to animal models. The MPS data showed that macrophages promoted BaP-induced malignant transformation of human bronchial epithelial cells, which was mediated by nuclear factor (NF)-κB and STAT3 pathways to induce cell proliferation, colony formation in chip culture and tumorigenicity in nude mice. Blockage of interleukin 6 or tumor necrosis factor α signaling or inhibition of NF-κB, STAT3 or cyclinD1 expression abrogated the effect of macrophages on malignant transformation in the lung airway chip, which was in line with results obtained in comparable induction experiments in animals. This indicates that MPS-based lung models are capable of modeling key molecular events involved in COPD progression in humans. This makes MPS-based lung models an attractive target for industrial adoption by the tobacco industry.
Finally, onset of both chronic obstructive and malignant disease in human lung is intimately related to the entire organismal homeostasis. Uptake of smoke-induced substances by the liver, lymph node-based immunological responses toward the induced tumor cells and interactions of the diseased lung with the cardiovascular system requires more complex multi-organ systems. The potential of MOCs to provide tools for systemic lung+X (e.g. X = liver or lymph node) on-chip assays are of high interest to the industry. Adoption of MPS-based assays by the tobacco industry needs robust qualification, but does not need regulatory acceptance yet. Value-adding MPS-based tools and approaches, once qualified, can immediately be used for internal decision-making in product development.
4. Initiatives to advance MPS developments into human Body-on-a-chip solutions
4.1. Human Body-on-a-chip – a courageous promise to reduce animal testing and provide a solution to unmet needs
We have reported the status quo and potential aspects for the adoption of MPS emulating human single- and multi-organ models in different industries. These MPS may contribute to the reduction of laboratory animal testing methods due to the early effective elimination of toxic drug candidates from further testing and better preselection of effective drugs. However, these types of MPS are incapable of emulating human biology at an entire organismal level. Some research groups have advanced their MPS platforms towards concepts to emulate a functional human-like mini-organism in vitro (Hartung and Zurlo, 2012). These concepts are called “Human-on-a-chip” or “Body-on-a-chip” approaches. Teams have designed and constructed first prototypes (Fig. 16) which are in experimental evaluation now. Current human Body-on-a-chip developments focus on the emulation of non-pregnant adulthood, as this time span has the largest numeric relevance for safety and efficacy testing in humans. It is postulated that organs from at least the following ten systems should be interconnected in a biological manner to gain minimal human organismal homeostasis in vitro: circulatory, endocrine, gastrointestinal, immune, integumentary, musculoskeletal, nervous, reproductive, respiratory, and urinary.
Fig. 16. First prototypes towards a human body-on-a-chip.
Images illustrate four MPS prototypes in experimental evaluation aiming to advance multi-organ systems into miniature organisms on a chip. (A) The German “GO-Bio-MOC” program-derived 10-organ prototype of TissUse GmbH, Germany (Courtesy of TissUse GmbH, Germany); (B) The Russian “Homunculus” program-derived 6-organ prototype design of Bioclinicum, Russia (Courtesy of Bioclinicum, Russia); (C) The US 10-organ prototype of Cornell University, USA (Courtesy of Cornell University, USA); and (D) a US 4-way PhysioMimetics prototype of the Human Physiome on a Chip Program, MIT, USA (Courtesy of MIT, USA), developed within the framework of the US DARPA/NIH/FDA MPS initiative.
With regard to the scale of chip-based organisms, the first approaches towards a biologically representative scale-down of human organs have been published (Marx et al., 2012; Moraes et al., 2013; Wikswo et al., 2013). In the following, we glance at the most prominent initiatives, all at a very early stage, aiming at human Body-on-a-chip solutions. The initiatives are highlighted in this chapter in the chronological order of their inception.
4.2. March 2010 – the German multi-organ chip GO-Bio initiative
The German Human-on-a-chip initiative was launched under a GO-Bio grant from the German Federal Ministry of Education and Research in March 2010. The grant was awarded to Uwe Marx’s group at the Technische Universitat Berlin, in collaboration with its spinoff TissUse GmbH, and the Fraunhofer IWS in Dresden and the Fraunhofer IGB in Stuttgart. This initiative represented a reduced biphasic version of a larger VAMOS grant proposal to develop a human-on-a-chip platform under the FP7 Health 2010 Program on alternatives to laboratory animals aiming at the development of in vitro solutions for systemic substance testing at a European level; the VAMOS proposal was not funded. The eight-year goal of the German GO-Bio program is the development of a human body-on-a-chip consisting of a relevant number of organs to allow for self-contained organismal long-term homeostasis. This includes the chip-based utilization of physiological organ functions for functional maintenance of the human body-on-a-chip: the intestine absorbing digested food, the liver supplying albumin for all organs and metabolizing xenobiotics, the bone-marrow realizing adequate hematopoiesis for the mini-organism, the lung providing oxygen through red blood cells to each and every organ, the kidney excreting waste products from blood plasma, the immune system guarding health status, etc. The final human Body-on-a-chip platform should be able to emulate the key pathophysiological behavior of patient groups in clinical trials. A first five-year period with an investment of about 6.1 million Euro was dedicated to establishing a MOC platform capable of hosting a relevant number of organs for such a human-on-a-chip solution; an organ miniaturization algorithm which reflects the organoid structure of a human was applied. A “man-on-a-chip” at 0.00001 scale was designed which fitted the format of a microscopic slide (Marx et al., 2012). A first proof of concept milestone was reached in 2013 when a reproducible 28-day co-culture of human 3D liver and skin models equivalents and long-term repeated dose substance could be established in a two-organ version of the MOC platform (Wagner et al., 2013). Subsequently, the second milestone – human two-organ chip vasculature – was reached (Schimek et al., 2013). Results and further platform improvements towards a four-organ ADMET chip for long-term testing (Maschmeyer et al., 2015b) have been described in Chapter 2.3.3. of this report. Since the market launch of the two-organ chip in 2013, a number of successful industrial feasibility studies have proved the commercial viability of the concept, which has been the last milestone of the first phase. Therefore, the second phase, a three-year program at TissUse GmbH under the auspices of the GO-Bio grant (031B0062) dedicated to providing the proof of concept for a human Body-on-a-chip solution was initiated in 2015 and continues until 2018. The researchers’ “man-on-a-chip” design has been illustrated in a video (http://www.tissuse.com/technology.html) and has recently been converted into a first chip prototype of a mini-organism (Fig. 16A) which is now under evaluation.
4.3. October 2011 – the Russian “Homunculus” program
The Russian Ministry of Education and Science launched a program aiming at the development of an “organ-on-a-chip” and “human-on-a-chip” technology in October 2011. Dmitri Sakharov and his team at the Scientific Research Center “Bioclinicum” in Moscow has been awarded grant money from the Russian Ministry of Education and Sciences and the Ministry of Production and Trade for platform development, totaling 5.7 million Euro. The resulting “Homunculus” microbioreactor is a MPS platform that provides prolonged cell culture viability in vitro and emulates physiological processes (Senyavina et al., 2013). The system includes a control unit connected to a maximum of four microfluidic chips. The disposable microchip is designed for co-culturing of up to six cell micromodels in a single closed circuit with automatic medium exchange (Samatov et al., 2015). The system is adapted for the cultivation of cellular monolayers in standard transwell inserts, chip vascularization by seeding cells in the microchannels or work with any other cellular objects, including 3D models and human tissue samples in cell chambers. The chip contains a peristaltic micropump and cell culture wells connected by microchannels with circulating culture medium. The microfluidic channels imitate blood vessels. The control unit sustains the desired temperature of the chip and provides constant circulation of culture medium, allowing the variation of pressure on the micropump valves and their switching frequency. The chip construction allows the integration of various sensors to monitor cell culture parameters in real-time mode: at the moment, cover-embedded electrodes provide the impedance-based measurement of barrier models during cultivation. The microscopic glass at the bottom of the chip provides the opportunity for noninvasive cell viability observation by the use of fluorescent vital dyes and optical fibers (Sergachev et al., 2013). The microcirculation introduces convective mass transfer to the cellular microenvironment, which mimics physiological conditions, accelerating transport fluxes of nutrients and metabolites compared to mass transfer limited to diffusion in static culture. Thus, the permeability values predicted become more accurate. Various microcirculation regimes promote in vivo-like mechanical influences, including physiological values of shear stress and interstitial pressure. The “Homunculus” device is commercialized by Bioclinicum and is currently passing the national registration for medical use to certify the in vitro trials of drugs and chemical compounds with regard to bioavailability, biotransformation and drug drug interaction on specialized cellular models. The aim of the “Homunculus” is to be used to determine dose-dependent and integrated toxicity of chemical compounds to target cells, given the toxicity of the metabolites produced in human liver. The class of danger of hazardous chemical waste and mixtures thereof can be determined using this device applied to environmental monitoring, and sanitary and hygiene regulation. The six-organ chip prototype designed, as shown in Figure 16B, is currently in experimental evaluation.
4.4. March 2012 – the European “Body on a chip” consortium
Back in March 2012, a so-called “Body on a chip” consortium consisting of four European sites (http://cordis.europa.eu/project/rcn/104027_en.html) was awarded 1.4 million Euro by the EC for a “Body on a Chip” development under a 7th Framework program FET Open project (STREP #296257) and included InSphero AG, Switzerland, ETH Zurich, Switzerland, KU Leuven, Belgium. Technical University Dortmund, Germany, and Astra Zeneca Pharmaceuticals, United Kingdom. The “Body on a chip” consortium directive was to create a comprehensive, next-generation in vitro drug-testing device which interconnects multi-organ tissue models to identify toxicity and/or decreased efficacy due to metabolic activity (http://www.insphero.com/company/eu-project-the-body-on-a-chip). The partnership developed a plate-based multi-tissue device within a timeframe of three years (Kim et al., 2015a; Kim et al., 2015b; Frey et al., 2014) which is described in part 2.3.3. of this report.
4.5. June 2012 – the US NIH/DARPA/FDA Microphysiological (Tissues-on-a-chip) Systems program
The FDA/National Center for Advancing Translational Sciences (NCATS)/DARPA Microphysiological Systems collaboration, which started in 2011, was initiated to support the development of human microsystems, or organ “chips,” to screen for safe and effective drugs more swiftly and efficiently than current methods. The NCATS and DARPA, in coordination with the FDA, solicited proposals from industry, government labs, academic institutions and other research organizations on how best to develop the chip by bringing the latest advances in engineering, biology and toxicology to bear on this complex problem. A joint NIH/DARPA/FDA tissue-on-a-chip five-year program at a total of around $140 million was launched in June 2012. The DARPA awarded two contracts, one to Donald Ingber’s group at the Wyss Institute for Biologically Inspired Engineering at Harvard University Boston, and one to the Human Physiome on a Chip program led by Linda Griffith at the Massachusetts Institute of Technology (MIT; Cambridge), for the development of engineering platforms and biological proof-of-principle for a human body-on-a-chip. Each research team was required to develop a human body-on-a-chip with the following characteristics: (1) Reconfigurable platform; (2) Ten or more in vitro physiological systems; (3) Ability to monitor resident tissues for up to 4 weeks; (4) Uses human cells; (5) Commercial availability; and (6) Includes plan for validating integrated platform performance. Concurrently, NCATS awarded several grants in two complimentary areas: (1) Stem/Progenitor Cell Grants to develop stem- and progenitor-derived cell resources to seed circulatory, endocrine, gastrointestinal, immune, integumentary, musculoskeletal, nervous (including eye), reproductive, respiratory and urinary microsystems, and (2) Integrated Microphysiological Systems for Drug Efficacy and Toxicity Grants to develop in vitro microphysiological systems representative of major organs/tissues in the human body that will facilitate the assessment of biomarkers, bioavailability, efficacy and toxicity of therapeutic agents prior to clinical trials. Throughout the five-year research plan, NCATS, FDA and DARPA meet biannually with all the researchers to ensure progress and collaboration for all the projects. Moreover, through this program, the development of well characterized multicellular models that represent the disease pathogenesis, cellular diversity, genomic complexity, sex, microenvironment and functional output of human organ systems is being supported. Essential characteristics of advanced multicellular models of human physiology should include all of the following features:
Multicellular architecture that represents characteristics of the target tissue;
Functional representation of normal human physiology;
3D culture modeling of multicellular microenvironments;
Reproducible and viable operation under physiological conditions maintained up to four weeks in culture;
Representation of normal and disease phenotypes;
Representation of population diversity; and
Use of patient-derived cell lines where patient consent allows commercialization and broad sharing of cells and cellular derivatives with academic and industry researchers.
A critical challenge in engineering an integrated network of microsystems will be the ability to develop and assemble 3D tissue modules that are physiologically relevant, reproducible and compatible with high content screening platforms that include multiple molecular readouts, such as gene expression, proteomic, metabolomic or epigenomic analyses. The in vitro microsystems should provide spatial and temporal control of the cellular microenvironment, while enabling continuous monitoring (sensing), probing (direct in-cell measurements) and sampling (testing and continuous data collection and analysis) of the system in a nondestructive, real-time and label-free assay. The integrated platform should ideally support a network of microsystems that can monitor epithelial and endothelial barrier functions, exposure to environmental and infectious agents, and absorption and metabolism of drugs in real-time and at high resolution inflammatory responses. Platform challenges include: 1) ease of use and portability; 2) cost-effective designs; 3) optimization of the full range of possible physiological functions in each organ system; 4) replacement of PDMS with materials that do not absorb/bind hydrophobic molecules and exhibit physiologically relevant stiffness; 5) implementation of iPS-derived cells, when possible, to harness heterogeneities of responses in humans; 6) increase throughput with automation; 7) creation of an integrated set of organ systems with physiological flow, shear, optimal metabolic scaling between organ systems, and spatiotemporal scaling (adult male ~ 4.5 liters of blood; 4.5 μl for μHuman); 8) integration of dynamic mechanical, chemical and electrical cues including key contributions of missing organ systems; 9) optimization of analytical chemistry in nl to μl volumes for pH, O2 and glucose; 10) implementation of a combination of real-time biosensors of key cell functions; 11) implementation of a universal medium with media to cell ratios reflecting physiological values; 12) implementation of designs with minimal dead volumes in tubing and reservoirs; 13) implementation of the MPS data base for integrated systems; 14) development of modeling methods to harness the rich datasets; and 15) integration of disease models. It is such program designs that prompted collaboration between DARPA and NIH investigators to engineer their platforms in anticipation of integration and to work towards common requirements.
A draft summary of the human body-on-a-chip programs within the US initiative has been provided recently (Dance, 2015).
Figure 16 shows the MPS-based human “Body-on-a-chip” prototypes currently evaluated in biological experiments in the US: one of them in the lab of Michael Shuler at Cornell University (Ithaca, NY, Fig. 16C) and another one in the lab of Linda Griffith at the MIT (Cambridge, MA, Fig. 16D).
4.6. February 2015 – the Netherlands Institute for Human Organ and Disease Model Technologies
The Institute for Human Organ and Disease Model technologies (hDMT) is a precompetitive non-profit technological R&D institute, initiated in the Netherlands. This institute, established on 6 February, 2015, integrates state of the art human stem cell technologies with top level engineering and a variety of other expertise from academia and industry to develop, implement and valorize human organ and disease models-on-a-chip. These miniature models in chip format closely mimic human tissue and organs in vivo. This will primarily meet the need of pharma, clinicians and society for representative human test model systems for drug development, and an active dialogue is encouraged to achieve this central aim. As an additional advantage, the use of animal experiments might be reduced. The hDMT has a flat, efficient organizational structure and combines technologies, facilities and expertise from different disciplines and multiple organizations and locations, thereby forming a virtual institute, “a laboratory without walls.” The hDMT Consortium, coordinated by hDMT, consists of internationally known scientists with backgrounds in biology, physics, chemistry, pharmacology, medicine and engineering. The hDMT researchers share their knowledge, expertise and facilities to develop the organ-on-chip models. A clinical expert team of medical specialists is also present. The Consortium aims at further expanding their expertise in MPS technologies, applying for research and infrastructure grants, and making results available to the global MPS research community by open access publication. The current partners are the Leiden University and University Medical Centre, Erasmus Medical Centre in Rotterdam, the Hubrecht Institute in Utrecht, the three Universities of Technology (Twente, Eindhoven and Delft), and two companies – Genmab and Galapagos. The academic partners carry out the research and the private partners provide input about the validation and application of the models. The “scalability and manufacturability” of the technologies will also be addressed within the hDMT Institute. The hDMT will valorize the MPS-based models developed through technology and pharma companies both in The Netherlands and internationally. The hDMT also seeks collaboration with other researchers in the field of human organ and disease modeling worldwide to develop complementary international programs that will lead to new insights in human disease, and to new treatments and diagnostic tests.
Although hDMT has only recently been established, it has already organized highly interdisciplinary research teams. Its research projects fall within two research lines that reinforce and complement each other: development of innovative human organ and disease models, and development of organ-on-chip technology platforms. Four themes are currently prioritized in both healthy and disease-format: vessels on chip, heart on chip, cancer on chip and brain on chip. Other models will be added in the near future, including immune system on a chip, lifestyle models (skin and hair) and environmental models (nutrition and infection). All hDMT research programs are interdisciplinary programs to which each partner contributes with a specific technology focus embedded in ongoing research, clinical interest and expertise. For more information about the hDMT Institute, see www.hDMT.technology.
4.7. January 2016 - EU-ToxRisk – a H2020 project
In a large (30 Million €) H2020-supported integrated European ‘Flagship’ program driving mechanism-based toxicity testing and risk assessment for the 21st century called EU-ToxRisk, academia joins forces with small and medium-sized enterprises (SMEs), large industry, contract research organisations (CROs) and regulatory bodies to achieve a paradigm shift in toxicology towards a more efficient and animal-free chemical safety assessment. An international consortium of 39 partner organisations is funded by the European Commission to work on the integration of new concepts for regulatory chemical safety assessment. These new concepts involve cutting-edge human-relevant in vitro non-animal methods such as MPS-based approaches and in silico computational technologies to translate molecular mechanistic understanding of toxicity into safety testing strategies. The ultimate goal is to deliver reliable, animal-free hazard and risk assessment of chemicals. Coordinated by Bob van de Water, Professor of Toxicology at Leiden University (The Netherlands), EU-ToxRisk intends to become the European flagship for animal-free chemical safety assessment. The project will integrate advancements in cell biology, omics technologies, systems biology and computational modelling to define the complex chains of events that link chemical exposure to toxic outcome. The consortium will provide proof of concept for a new mechanism-based chemical safety testing strategy with a focus on repeated-dose systemic toxicity as well as developmental and reproductive toxicity. Importantly, novel mechanism-based test methods will be integrated in fit-for-purpose testing batteries that are in line with the regulatory framework and will meet industrial implementation.
5. Selected legal frameworks, regulatory science and acceptance criteria
5.1. General remarks
A stringent regulatory network of laws, regulations and guidelines currently covers substance development, registration and approval for all areas of potential use, such as medicine, food, agriculture, chemistry and cosmetics.
To the best of our knowledge, MPS-based test assays for the evaluation of substances have not yet been introduced into any regulatory assessment procedure in any country. This is due to the early development status at the research level and a lack of practical experience of both industry and regulatory bodies. However, experience and mutual acceptance are prerequisites for new technologies, as was experienced during the recent history of the ban of laboratory animal testing for the evaluation of cosmetics in the EU in 2013. The ban has thrown the global cosmetics industry into a dilemma: Cosmetics containing new ingredients tested on laboratory animals are no longer allowed to be sold in the EU, while the same cosmetics are prohibited from being sold in the rest of the world if they have not been tested on laboratory animals.
Two different approaches are being investigated to avoid a similar situation in drug development. Firstly, the FDA has launched a regulatory science-based approach, and the EU European Medicines Agency seems to be following their lead. Secondly, key academic and industrial players are actively disseminating the same approach, but to regulatory bodies in other countries, such as China and Russia.
At a regulatory science panel discussion at the 9th World Congress on Alternatives and Animal Use in the Life Sciences (WC9) in Prague, 2014, the representations of regulatory bodies from the US, EU, China, Russia and Japan issued a statement of intent: “Taking into account the large-scale projects ongoing on a global scale with regard to the human-on-a-chip technologies and the potential interest for global regulatory authorities of different sectors, it is considered important that a systematic mechanism for exchange of information is being set up. The latter should take advantage of the collaborative initiatives already established for different sectors (e.g. FDA/EMA qualification exercise). Such a forum could also allow for cross-sectorial discussions on qualification criteria and performance standards in order to foster possible qualification of human-on-a-chip technologies.”
In this chapter, some of the panel members summarize the status quo of substance testing regulations subject to major regulatory bodies.
5.2. US FDA drug regulation and regulatory science approach
Recent breakthroughs in science and technology have the potential to transform the FDA’s ability to prevent, diagnose and treat disease. Recognizing this, the US FDA Advancing Regulatory Science Strategic Plan was designed to advance regulatory science to speed innovation, improve regulatory decision-making, and get safe and effective products to market in a timely manner. This strategic plan meets both today’s public health needs and the challenges and opportunities of tomorrow (FDA, 2011b). One of the key elements of this plan is to improve the prediction of product safety and efficacy by developing better models of human adverse response, identifying and evaluating biomarkers and endpoints that can be used in nonclinical and clinical evaluations and developing and using computational methods and in silico modeling. By addressing these needs, the FDA will be better able to identify and accurately predict and reduce the magnitude and likelihood of risks associated with the products that it regulates. The FDA, the NCATS and the DARPA partnership for the development of in vitro microphysiological systems, described in detail in Chapter 4.5. of this report, is a groundbreaking example of the types of partnerships that are needed to bring innovative new technologies into the regulatory paradigm. The challenge is to develop an integrated microsystem platform that can incorporate several different modular organs on a chip and to incorporate it into the regulatory paradigm. The FDA’s role in this partnership is to advice on regulatory requirements, validation and qualification of the organ-on-a-chip or human body-on-a-chip models. Qualification is a formal process that enables the FDA to advise product sponsors that a tool is available for use in a regulatory application. Qualification is a conclusion that, within the stated context of use, the results of assessment with a tool can be relied upon to have a specific interpretation and application in product development and regulatory decision-making.
Context of use is the key concept in the qualification process of drug development tools (DDT). It refers to a clearly articulated description delineating the manner and purpose of use for the tool (when and how will it be used?) including its limitations. Once qualification is granted, any product sponsor can submit data obtained with the qualified tool without being asked for further evidence in support of its suitability. Three DDT qualification programs are currently being implemented in the FDA Center for Drug Development and Research: (1) biomarkers; (2) clinical outcome assessments; and (3) animal models (FDA, 2014b). The context of use qualification concept can be applicable to any tool proposed for use in regulatory decision-making, including in vitro microphysiological systems.
Developing confidence in the predictive ability of in vitro microphysiological systems begins with identifying the critical regulatory gaps which can be addressed with these new tools, the consequences of an incorrect or incomplete answer, and an assessment of the level of confidence in each of the integral parts of an in vitro microphysiological system. A determination needs to be made on the essential biological and functional features that are needed in the test organ on a chip to address the context of use. The test or reference library of compounds for qualification should be tied directly to the regulatory question that is being addressed in the context of use. This means that a different context of use may require a different set of test chemicals. Additionally, how to use the data to make/add or refine a regulatory decision should be decided up front. Combining the information above for a “context of use” evaluation of the overall predictive ability of a new tool to answer critical regulatory questions can be used as a framework for qualifying in vitro microphysiological systems in a transparent and scientifically defensible manner. A potential qualification and validation strategy for MPS is described in Chapter 7 of this report.
5.3. Chinese State Food and Drug Administration (SFDA) drug regulation and MPS
As part of the government restructuring announced in March 1998, the Chinese Ministry of Health’s Department of Drug Administration merged with the State Pharmaceutical Administration of China (SPAC) to become the State Drug Administration (SDA). As a result, SDA oversees all drug manufacturing, trade, and registration. In 2003, the SDA was restructured to become the State Food and Drug Administration (SFDA). The Chinese government’s establishment of a single drug regulatory authority was an important step toward foreign access, because it eliminated the conflicting standards that prevailed among provincial government agencies, centralized the Chinese healthcare regulatory system, and made it more transparent. SFDA’s new regulations follow US FDA’s model. In July 1999, as part of medical insurance reform, SFDA released its first list of over-the-counter (OTC) medications, and in 2000, the state began to regulate OTC and prescription drugs separately. SFDA did so to encourage patients to purchase OTC medicines for less serious diseases, thereby reducing government medication expenditures and hospital visits. SFDA required all pharmaceutical companies in China to obtain GMP certificates from SFDA by June 30, 2004 to be licensed to sell their drug products in China. In 2005, SFDA launched a regulation on drug research and supervision management aimed at enforcing GLP to investigative drugs, traditional Chinese medicine injections and biotechnology products. The regulation aims to help China’s drug research and development gain international recognition. Founded in 1950, the National Institutes for food and drug Control (NIFDC) is a subordinate agency of the SFDA. The main professional areas of NIFDC cover pharmaceutical products, biological products, medical devices, food, healthy food, cosmetics, reference standards, laboratory animals, drug safety evaluation and others. A prime mission of the NIFDC is to organize the international collaboration and cooperation in the field of testing and analysis of drug, medical devices, healthy food, cosmetics, and restaurant food safety. On the basis of that mission, a representative of the NIFDC actively participated in the panel discussion on regulatory science and Human-on-a-chip approaches at the 9th World Congress on Alternatives and Animal Use in the Life Sciences in Prague 2014. In order to get hands on experiences with MPS-based approaches the NIFDC signed a memorandum of understanding with the Technische Universität Berlin, Germany, for a joint 3-year collaboration in the field of MPS. A NIFDC team has been trained to operate MOC-based equipment. Next steps include MPS-based assay evaluation in comparison to test assays currently in use at the institute.
5.4. Russian drug regulation and considerations on MPS impact
The Russian Federation has been taking steps to modernize the healthcare system by stimulating the development of pharmaceutical and medical products (Russian Government, 2014). As a part of these measures, a national program “Development of pharmacological and medical industries 2013 – 2020” was approved. The program is monitored by the Ministry of Health and supported by the Ministry of Industry and the Ministry of Education and Science. The main goal of this program is the introduction of novel pharmaceutical compounds and new methods of testing them. This implies the progressive transition of Russian pharmaceutical companies and research laboratories to the international Good Manufacturing Practice (GMP), Good Laboratory Practice (GLP) and Good Clinical Practice (GCP) standards and the development of corresponding educational programs.
The Ministry of Health approved the procedure for the evaluation of medical devices, as well as the regulations for their registration for the technical testing, toxicological studies and clinical trials, in January 2014. In the meeting of the presidium of the Council for Economic Modernization and Innovative Development of Russia on 16 May, 2014, the Prime Minister Dmitry Medvedev emphasized that the introduction of innovative approaches should rely on domestic R&D programs and products, referring to the development of modern biopreparations, cell and tissue products, genomic technology, and biocompatible materials (Russian Government, 2014). New standards governing the testing of chemical impacts on the human body involving the use of an in vitro model of human skin (GOST 32634-2014), micronucleus test on mammalian cells (GOST 32635-2014) and the evaluation of gene mutations (GOST 32638-2014) were introduced in June, 2015.
5.5. Legal framework for human risk assessment for chemicals in the EU
Although animal use in biological research has surpassed that required for regulatory and chemical safety testing, the latter are major drivers for alternative method development and testing systems. With the advent of “omics,” high-throughput testing, 3D cell cultures and, lately, MPS, the pressure to bring such methods into regulatory use has increased tremendously, as has the debate about the usability and application domain of such methods (Tralau et al., 2015). However, regulators continue to take a cautious stand on this matter. This is reflected not least by the comparably modest number of validated and internationally accepted alternative methods, none of which harbors a MPS yet. The main obstacles to the implementation of any such methods are, apart from the often slow progress of legal implementation, method validation and an inherent limitation of cellular and MPS to reflect established toxicological endpoints and organism plasticity (Tralau and Luch, 2015). Without changes to long established practices, this makes it difficult to readily adapt such systems for the purpose of quantifiable risk assessments or to transpose the results obtained to the classifications required by the Globally Harmonized System of Classification and Labelling of Chemicals (GHS), Classification, Labelling and Packaging (CLP) or REACH. Unsurprisingly, a lot of the regulatory testing in the EU is still carried out in vivo, the exceptions to the rule being skin and eye irritation, corrosion and genotoxicity.
A legal ban on the use of laboratory animals for cosmetic ingredient/substance testing has existed in the EU since 2013. Prior to that ban, significant efforts were made to develop validated in vitro alternatives to the animal tests with moderate success (Adler et al., 2011). This specific EU-wide situation encouraged us to report on the different legal frameworks existing for different domains of chemical substance evaluation (e.g. pesticides, cosmetics and food additives) apart from the legal frameworks for drug candidate testing in the EU. Each regulatory framework involves different requirements for information that need to be submitted in the dossier for market acceptance, with guidelines or guidance documents usually describing which tests are acceptable for providing this information. Table 4 below gives an overview of the information required for some well-established EU frameworks for chemicals and the OECD Test Guidelines (TGs) that can provide this information. The table shows how animal tests still predominate (in dark blue).
Tab. 4. Overview of toxicity tests/information required for human risk assessment in the different regulations for the use of chemicals in the EU1.
Requirements based on in vivo tests are marked in dark blue, while requirements for which in vitro alternatives are included are marked in light blue, and white boxes indicate no data are required.
| EU Regulation | Types of toxicity data (i.e. endpoints) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Kinetics | Acute toxicity | Irritation/Corrosion | Sensitization | Chronic | Mutagenicity | Carcinogenicity | Reproduction toxicity | Other | |
| REACH (EASHW, 2006) • 1–10 t/y (Annex VII) |
|||||||||
| • 10–100 t/y (Annex VIII)2 | |||||||||
| • 100–1000 t/y (Annex IX)2 | |||||||||
| • >1000 t/y (Annex X)2 | |||||||||
| Cosmetics Regulation (EC, 2009) | |||||||||
| Pesticides • Active substances (EC, 2013a) |
|||||||||
| • Plant protection products (EC, 2013b) | |||||||||
| Biocides (EC, 2012) | |||||||||
| Food additives (EC, 2008) | |||||||||
| Food contact materials (EC, 2004; + SCF Guidelines) • Migration <0.05 mg/kg food |
|||||||||
| • Migration 0.05 – 5 mg/kg food | |||||||||
| • Migration > 5 mg/kg food | |||||||||
| Pharmaceuticals (EC, 2001; + ICH Guidelines) | |||||||||
Many regulations include waivers and conditions for the tests required or may be replaced by alternatives, such as QSAR predictions or in vitro systems, which are not included in this table for the sake of keeping a clear overview. For these waivers, conditions, and alternatives, one is referred to the specific regulation texts; optional tests, i.e. tests that are only necessary in case of certain outcomes of previous tests, are given a footnote with an explanation or are put in parentheses.
The required tests shown for this tonnage/migration level are additional to the tests required at lower tonnage/migration levels; the tests required at the lower tonnage/migration levels are also required here.
The highest numbers of experimental animals and resources are required for systemic and repeated dose toxicity (accounting for more than 65 % of all experimental animals used; see ECHA, 2011; van der Jagt et al., 2004; Rovida and Hartung, 2009). The respective toxicological endpoints are challenging to address, in vivo as well as in vitro, and it is in this context that MPS would ideally put their strength to the test, especially since they allow the breakdown of the species barrier. Moreover, they could provide solutions for some of the open issues and regulatory challenges in the field of consumer protection, i.e. endocrine disruptors beyond the sex and thyroid hormone systems, low-dose effects, epigenetically active substances and the influence of background exposures.
Therefore, much of this will constitute rather mid- to long-term options. The more immediate regulatory use of MPS comprises the OECD’s “adverse outcome pathways” initiative (OECD, 2013), integrated testing strategies, adverse biomarker identification and testing prioritization (e.g. similar to the Tox21-programme; Krewski et al., 2010). Concomitantly MPS should be part of preliminary test runs for the aforementioned tox-test dummy (Tralau et al., 2012). Such blinded cross-validation trials would not only help to assess their already existing regulatory potential but help to identify shortfalls hands on.
A special situation exists with regard to cosmetics, which are subject to a ban on animal testing (EC, 2003; EC, 2009). With effect of 2013, the ban not only prohibits live animal testing of cosmetic products, but also the marketing of products tested on animals or containing animal-tested ingredients (the only exception being chemicals that are within their scope of use also subject to REACH). It should be noted that, as of yet, there has been no applications for market admittance for cosmetics which are based entirely on alternative methods of testing. Consequently, the requirement to adapt alternative toxicity testing for cosmetics is pressing. The possible areas of adoption of MPS-based systems by the chemical and cosmetics industries are described in Chapter 3.2. whereas a potential tiered approach for qualification and validation is provided in Chapter 6.
6. How to qualify and validate MPS-based tools?
Qualification and validation of 3D in vitro assays in general and MPS-based assays in particular is a major hurdle for industrial adoption and regulatory acceptance (Astashkina et al., 2012). Several challenges remain to be addressed before MPS-based assays can be adopted by the different industries. In particular, validation of MPS-based assays requires initial qualification of the equipment and tissue sources used. Furthermore, adherence to the well-established rules of Good Cell Culture Practice (Coecke et al., 2005) and the recommendations for improving the quality and utility of organotypic models (Alépée et al., 2014) are mandatory requirements to ensure that MPS-based assays generate reliable, reproducible and robust data. Noteworthy, a concept of mechanistic validation (Hartung et al., 2013a) has been put forward out of the context of Evidence-based Toxicology (Hartung, 2010b) that promises to support MPS validation. As outlined in Chapter 2, only a few studies to date have sought to qualify the readouts of MPS-based assays against relevant in vivo models. For easier industrial adoption, we recommend the generation of a biological baseline for each and every MPS-based single organ equivalent, as exemplarily described for single-liver MPS in Chapter 3.1.1 of this report. We recommend a two-tiered approach for the validation of a MPS-based test assay within the current regulatory landscape in US and Europe (Fig. 17).
Fig. 17.
A possible stepwise validation roadmap for upcoming MPS-based tools
Validation is a process, which needs to be continuously adapted to the technologies assessed for the purpose of their use (Leist et al., 2012). In a first step, we advise the voluntary “fit-for-purpose” or “context of use” qualification of a MPS-based test tool through the aforementioned FDA DDT Qualification Program. Here, collaboration between MPS-providers and pharmaceuticals companies adopting the particular assay would be advisable. This qualification involves MPS providers, pharma industry and FDA officers and eventually leads to a MPS tool validation independent from the drug candidates used for validation. This, in turn, supports an easier adoption of the tool for further use in drug assessment. In addition, such a first validation step for a given MPS-based tool generates a validation algorithm and tool-specific data, which then, in a second step, for example, can be fed into a validation program for the use of the same tool for chemical testing. For MPS-based models aiming to replace the animal or existing in vitro models used in OECD guidelines in chemical safety assessment, the adherence to the existing OECD guidance document on the validation and international acceptance of new or updated test methods for hazard assessment (OECD, 2005), the EMA guideline on regulatory acceptance of 3R methods and qualification of novel methodologies for drug development (EMEA/CHMP/SAWP/72894/2008 Corr1), via the EMA Scientific Advice Working Party (SAWP) and the recommendation of the ICH Safety Topic Recommendation Working Group could be instrumental. However, given that MPS are usually based on human cell material it might be important to re-assess the suitability of animal data as the gold-standard during validation. As of today, a subsequent combination of the two qualification and validation steps, schematically illustrated in Fig. 17, seems to be the most efficient road map toward validation of upcoming MPS-based drug development tools arising from single-organ, multi-organ or human “body-on-a-chip” developments.
7. Visions are becoming reality
Single-organ MPS have been developed to establish physiological substance testing assays. Human organ models of the lung, heart, blood vessels, muscles, bones, liver, reproductive system, nervous system, gut, skin, intestine and kidney have been established and more models are to come. This development was followed by multi-organ MPS allowing the co-culture of different human organ equivalents in integrated microphysiological arrangements. First success in 28-day human intestine/liver/skin/kidney co-culture at homeostasis, for example, underlines the ability to move the technology towards systemic modeling of a human Body-on-a-chip – a miniature organism. The future development of these three types of MPS will, we think, eventually lead towards the reduction and replacement of animal and human testing (Fig. 18). Prime challenges to improve single- and multi-organ systems over the next five years include the integration of biological vasculature, immunocompetence, electrical coupling and mechanical stress (where relevant). Physiological skin, intestine and lung barriers and predictive drug-induced organ toxicity assays or lung disease models may, for example, result from such single-organ improvements. Improved multi-organ systems may be suitable to enable ADMET profiling, physiological multi-organ-crosstalk and multi-organ failure models. Subsequently, the advanced knowledge and experience acquired from these successful improvements will logically enable the development of a human Body-on-a-chip, providing systemic solutions to disease modeling and repeated dose toxicity assays (see Fig. 18 for overview).
Fig. 18. Next MPS developments meeting industrial needs.
The next hurdles to overcome in order to achieve significant reduction and replacement of animal testing are further integration of vasculature, immunocompetence, electrical signaling and mechanical forces into the respective organ models. These developments are expected to lead to a further reduction of industrial substance testing in animals and will strongly support the development of human “Body-on-a-chip” solutions aiming to replace systemic testing and disease modeling in laboratory animals.
The experts at the workshop recognized a cyclic pattern of phases and levels of innovation in the field of MPS. It took the scientific community more than five years to take first single- and multi-organ systems towards initial industrial evaluation (see Fig. 19). Future development breaks down into three distinct phases: short-term, mid-term and long-term, over a total time period of more than 25 years. Each and every phase of innovative solutions triggers a consecutive phase of industrial adoption, which, in turn, instigates regulatory acceptance in a consecutive phase (see diagonal arrows in Fig. 19). The proof of concept of single-organ MPS, for example, may lead to their use for MoA assessment and/or toxicity evaluation in industry within the next five years. We think that this should result in first validated regulatory safety assays between 2020 and 2030.
Fig. 19. A roadmap toward the reduction and replacement of animals and patients.
Red blue and white arrows – translational impact of single-organ, multi-organ and human Body-on-a-chip MPS-based approaches, respectively.
The individual future phases are highlighted in the roadmap (Fig. 19). The vision of the development of MPS in the long-term after 2030 could be, for example, the adoption of precision medicine tools (“You-on-a-chip”) by industry for personalized disease models. Another target envisioned would be the on-chip modeling of aspects of clinical trials at an industrial level. All these developments could result in the eventual reduction and curtailing of tests on both animals and humans.
We expect MPS-based MoA assessment and toxicity testing adopted by the pharmaceutical industry in the short-term to lead to an initial reduction of the first two phases of the drug discovery life cycle (discovery and lead optimization) due to faster and more precise selection of targets and relevant leads (Fig. 20).
Fig. 20. MPS-based approaches and tools can shift the current drug testing paradigm.
The substance testing paradigm shift envisioned (top) compares the drug development phases and related costs with the current situation (below)
Once human Body-on-a-chip platforms in the mid-term enter the pharmaceutical space with tools for predictive systemic testing and on-chip clinical trial data generation (Fig. 20), this may nearly obviate the necessity of involving animals in preclinical development and healthy volunteers in phase I testing in the long-term. This combined with expected high predictivity and, therefore, significant reduction of clinical attrition rates, may create a new drug development paradigm. The latter would change drug development from a stepwise approach to an ultrafast discovery and validation approach with an enormous economic impact and value for society (see Figure 20).
The authors of this report are fully aware that this challenging roadmap, if successful, not only leads to a paradigm shift of drug development and chemical safety assessment but that it can only be successfully realized with all the parties involved (academia, industry and regulatory bodies) collaborating on a global scale.
Acknowledgments
We thank Philip Saunders for linguistic help and Peter Mangel for his artistic work on many of the illustrations.
1TissUse GmbH, Berlin, Germany
2AstraZeneca, Cardiovascular and Metabolic Diseases, Innovative Medicines and Early Development Biotech Unit, Mölndal, Sweden
3Section of Pharmacogenetics, Department of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden
4Wyss Institute for Biologically Inspired Engineering at Harvard University Boston, USA
5Boehringer Ingelheim Pharma GmbH & Co. KG, Non-clinical Drug Safety, Biberach, Germany
6Federal Agency for Medicines and Health Products, Brussels, Belgium
7National Institute for Public Health & the Environment, Bilthoven, The Netherlands
8Institute for Risc Assessment Science, Utrecht University, The Netherlands
9Massachusetts Institute of Technology, Cambridge, USA
10Center for Alternatives to Animal Testing-Europe, University of Konstanz, Konstanz, Germany
11US Food and Drugs Administration, Center for Food Safety and Applied Nutrition, College Park, USA
12ETH Zurich, Dept. Biosystems Science and Engineering, Bio Engineering Laboratory, Basel, Switzerland
13microfluidic ChipShop GmbH, Jena, Germany
14ProBioGen AG, Berlin, Germany
15Center for Alternatives to Animal Testing, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, USA
16Philip Morris International R&D, Neuchatal, Switzerland
17Japanese Center for Validation of Animal Methods, Tokyo, Japan
18Beiersdorf, Hamburg, Germany
19German Federal Institute for Risk Assessment, Department of Chemicals and Product Safety, Berlin, Germany
20Scientific Research Centre Bioclinicum, Moscow, Russia
21Bayer, Investigational Toxicology, Berlin, Germany
22National Center for Advancing Translational Sciences, National Institutes of Health, Bethesda, USA
23National Center of Medical Radiological Research, Moscow, Russia
24Russian Ministry of Production and Trade, Moscow, Russia
25The Institute for Human Organ and Disease Model Technologies, Leiden, The Netherlands
26MIMETAS BV, Leiden, The Netherlands
27Chinese National Center for Safety Evaluation for Drugs, Beijing, China
28cellasys GmbH, Kronburg, Germany
29F. Hoffmann-La Roche Ltd, Roche Innovation Centre Basel, Switzerland
Footnotes
Conflict of Interest Statement
Uwe Marx is founder of TissUse GmbH which commercializes MPS platforms.
Thomas Hartung is cofounder of Organome LLC which aims to make mini-brains and other organotypic organ models commercially available.
References
- Adler S, Basketter D, Creton S, et al. Alternative (non-animal) methods for cosmetics testing: current status and future prospects – 2010. Arch Toxicol. 2011;85:367–485. doi: 10.1007/s00204-011-0693-2. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Goss JA, Cho A, et al. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip. 2013;13:3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahadian S, Ramón-Azcón J, Ostrovidov S, et al. Interdigitated array of Pt. electrodes for electrical stimulation and engineering of aligned muscle tissue. Lab Chip. 2012;12:3491–3503. doi: 10.1039/c2lc40479f. [DOI] [PubMed] [Google Scholar]
- Alépée N, Bahinski A, Daneshian M, et al. State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology. ALTEX. 2014;31:441–477. doi: 10.14573/altex1406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alrifaiy A, Lindahl O, Ramser K. Polymer-based microfluidic devices for pharmacy, biology and tissue engineering. Polymers. 2012;4:1349–1398. [Google Scholar]
- Andersson H, van den Berg A. Microfabrication and microfluidics for tissue engineering: State of the art and future opportunities. Lab Chip. 2004;4:98–103. doi: 10.1039/b314469k. [DOI] [PubMed] [Google Scholar]
- Arbor A. Withdrawal of Vioxx casts a shadow over Cox-2 inhibitors. Science. 2004;306:384–385. doi: 10.1126/science.306.5695.384. [DOI] [PubMed] [Google Scholar]
- Arrowsmith J, Miller P. Trial watch: phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov. 2013;12:569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- Ashikaga T, Yoshida Y, Hirota M, et al. Development of an in vitro skin sensitization test using human cell lines: the human Cell Line Activation Test (h-CLAT). I. Optimization of the h-CLAT protocol. Toxicol In Vitro. 2006;20:767–773. doi: 10.1016/j.tiv.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Astashkina A, Mann B, Grainger DW. A critical evaluation of in vitro cell culture models for high-throughput drug screening and toxicity. Pharmacol Ther. 2012;134:82–106. doi: 10.1016/j.pharmthera.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Ataç B, Wagner I, Horland R, et al. Skin and hair on-a-chip: in vitro skin models versus ex vivo tissue maintenance with dynamic perfusion. Lab Chip. 2013;13:3555–3561. doi: 10.1039/c3lc50227a. [DOI] [PubMed] [Google Scholar]
- Bal-Price A, Crofton KM, Leist M, et al. International STakeholder NETwork (ISTNET): creating a developmental neurotoxicity (DNT) testing road map for regulatory purposes. Arch Toxicol. 2015;89:269–287. doi: 10.1007/s00204-015-1464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Trappmann B, Stapleton SC, et al. Microfluidics embedded within extracellular matrix to define vascular architectures and pattern diffusive gradients. Lab Chip. 2013;13:3246–3252. doi: 10.1039/c3lc50493j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso J. Integrated Approaches to Testing and Assessment (IATA) – ex. Skin Sensitisation 10th EPAA Annual Conference; Brussels. 2014; 2014. https://circabc.europa.eu/sd/a/e19c814a-39ac-43fa-8f75-5715185f16d9/03-barroso.pdf. [Google Scholar]
- Basketter D, Ashikaga T, Casati S, et al. Alternatives for skin sensitization: Hazard identification and potency categorization. Regul Toxicol Pharmacol; Report from an EPAA/CEFIC LRI/Cosmetics Europe cross-sector workshop; ECHA Helsinki. April 23/24, 2015; 2015. pp. 660–666. [DOI] [PubMed] [Google Scholar]
- Basketter DA, Clewell H, Kimber I, et al. A roadmap for the development of alternative (non-animal) methods for systemic toxicity testing. ALTEX. 2012;29:3–89. doi: 10.14573/altex.2012.1.003. [DOI] [PubMed] [Google Scholar]
- Basketter DA, Scholes EW. Comparison of the local lymph node assay with the guinea-pig maximization test for the detection of a range of contact allergens. Food Chem Toxicol. 1992;30:65–69. doi: 10.1016/0278-6915(92)90138-b. [DOI] [PubMed] [Google Scholar]
- Bauch C, Kolle SN, Ramirez T, et al. Putting the parts together: combining in vitro methods to test for skin sensitizing potentials. Regul Toxicol Pharmacol. 2012;63:489–504. doi: 10.1016/j.yrtph.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Baudoin R, Alberto G, Legendre A, et al. Investigation of expression and activity levels of primary rat hepatocyte detoxication genes under various flow rates and cell densities in microfluidic biochips. Biotechnol Prog. 2014;30:401–410. doi: 10.1002/btpr.1857. [DOI] [PubMed] [Google Scholar]
- Baudoin R, Legendre A, Jacques S, et al. Evaluation of a liver microfluidic biochip to predict in vivo clearances of seven drugs in rats. J Pharm Sci. 2014;103:706–718. doi: 10.1002/jps.23796. [DOI] [PubMed] [Google Scholar]
- Bauer KS, Dixon SC, Figg WD. Inhibition of angiogenesis by thalidomide requires metabolic activation, which is species-dependent. Biochem Pharmacol. 1998;55:1827–1834. doi: 10.1016/s0006-2952(98)00046-x. [DOI] [PubMed] [Google Scholar]
- Becker H, Gärtner C. Polymer microfabrication technologies for microfluidic systems. Anal Bioanal Chem. 2008;390:89–111. doi: 10.1007/s00216-007-1692-2. [DOI] [PubMed] [Google Scholar]
- Becker H, Gärtner C. Microfluidics and the life sciences. Sci Prog. 2012;95:175–198. doi: 10.3184/003685012X13361524970266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Benam KH, Dauth S, Hassell B, et al. Engineered in vitro disease models. Annu Rev Pathol. 2015a;10:195–262. doi: 10.1146/annurev-pathol-012414-040418. [DOI] [PubMed] [Google Scholar]
- Benam KH, Villenave R, Lucchesi C, et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2015b;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- Bersini S, Jeon JS, Dubini G, et al. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials. 2014;35:2454–2461. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier E, Surfus J, Verbsky J, et al. An arrayed high-content chemotaxis assay for patient diagnosis. Integr Biol. 2010;2:630–638. doi: 10.1039/c0ib00030b. [DOI] [PubMed] [Google Scholar]
- Berthier E, Young EW, Beebe D. Engineers are from PDMS-land, Biologists are from Polystyrenia. Lab Chip. 2012;12:1224–1237. doi: 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- BéruBé K, Aufderheide M, Breheny D, et al. In vitro models of inhalation toxicity and disease. Altern Lab Anim. 2009;37:89–141. [PubMed] [Google Scholar]
- Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32:760–762. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Bischel L, Young E, Mader B, Beebe D. Tubeless microfluidic angiogenesis assay with three-dimensional endothelial-lined microvessels. Biomaterials. 2013;34:1471–1477. doi: 10.1016/j.biomaterials.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol L, Galas JC, Hillaireau H, et al. A microdevice for parallelized pulmonary permeability studies. Biomed Microdevices. 2014;16:277–285. doi: 10.1007/s10544-013-9831-3. [DOI] [PubMed] [Google Scholar]
- Booth R, Kim H. Characterization of a microfluidic in vitro model of the blood-brain barrier. Lab Chip. 2012;12:1784–1792. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- Brandenberger C, Rothen-Rutishauser B, Mühlfeld C, et al. Effects and uptake of gold nanoparticles deposited at the air-liquid interface of a human epithelial airway model. Toxicol Appl Pharmacol. 2010;242:56–65. doi: 10.1016/j.taap.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Briasoulis E, Pavlidis N. Noncardiogenic pulmonary edema: an unusual and serious complication of anticancer therapy. Oncologist. 2001;6:153–161. doi: 10.1634/theoncologist.6-2-153. [DOI] [PubMed] [Google Scholar]
- Brischwein M, Grothe H, Wiest J, et al. Planar ruthenium oxide sensors for cell-on-a-chip metabolic studies. Chem Anal (Warsaw) 2009;54:1193–1201. [Google Scholar]
- Bristow JS. A treatise on the theory and practice of medicine. Philadelphia: H. C. Lea; 1876. [Google Scholar]
- Brown HS, Griffin M, Houston JB. Evaluation of cryopreserved human hepatocytes as an alternative in vitro system to microsomes for the prediction of metabolic clearance. Drug Metab Dispos. 2007;35:293–301. doi: 10.1124/dmd.106.011569. [DOI] [PubMed] [Google Scholar]
- Brown JA, Sherrod SD, Goodwin CR. Metabolic consequences of interleukin-6 challenge in developing neurons and astroglia. J Neuroinflamm. 2014;11:1–12. doi: 10.1186/s12974-014-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden N, Chapman K, Sewell F, Robinson V. Pioneering better science through the 3Rs: an introduction to the national centre for the replacement, refinement, and reduction of animals in research (NC3Rs) J Am Assoc Lab Anim Sci. 2015;54:198–208. [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busek M, Nötzel M, Polk C, Sonntag F. Characterization and simulation of peristaltic micropumps. J Sens Sens Syst. 2013;2:165–169. [Google Scholar]
- Bussek A, Wettwer E, Christ T, et al. Tissue slices from adult mammalian hearts as a model for pharmacological drug testing. Cell Physiol Biochem. 2009;24:527–536. doi: 10.1159/000257528. [DOI] [PubMed] [Google Scholar]
- Caldwell JC, Evans MV, Krishnan K. Cutting edge PBPK models and analyses: Providing the basis for future modeling efforts and bridges to emerging toxicology paradigms. J Toxicol. 2012;2012(852384):1–10. doi: 10.1155/2012/852384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro A, Hsu WM, Kulig KM, et al. In vitro analysis of a hepatic device with intrinsic microvascular-based channels. Biomed Microdevices. 2008;10:795–805. doi: 10.1007/s10544-008-9194-3. [DOI] [PubMed] [Google Scholar]
- Catapano G, Gerlach JC. Bioreactors for liver tissue engineering. In: Ashammakhi N, Reis RR, Chiellini E, editors. Topics in Tissue Engineering, (E-book) Vol. 3. 2007. pp. 1–42. [Google Scholar]
- Cheng W, Klauke N, Sedgwick H, et al. Metabolic monitoring of the electrically stimulated single heart cell within a microfluidic platform. Lab Chip. 2006;6:1424–1431. doi: 10.1039/b608202e. [DOI] [PubMed] [Google Scholar]
- Chung BG, Flanagan LA, Rhee SW, et al. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- Clair EWS. The calm after the cytokine storm: lessons from the TGN1412 trial. J Clin Invest. 2008;118:2006–2009. doi: 10.1172/JCI35382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Bender E. Q&A: Hans Clevers. Banking on organoids. Nature. 2015;521:S15. doi: 10.1038/521S15a. [DOI] [PubMed] [Google Scholar]
- Coecke S, Balls M, Bowe G, et al. Guidance on good cell culture practice. A report of the second ECVAM task force on good cell culture practice. Altern Lab Anim. 2005;33:261–287. doi: 10.1177/026119290503300313. [DOI] [PubMed] [Google Scholar]
- Cook D, Brown D, Alexander R, et al. Lessons learned from the fate of AstraZeneneca’s drug pipeline: a five-dimensional framework. Nat Rev Drug Discov. 2014;13:419–431. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- CORDIS: Community Research and Development Information Service. The Body-on-a-Chip (BoC) European Commission; 2011. http://cordis.europa.eu/project/rcn/104027_en.html. [Google Scholar]
- Cottrez F, Boitel E, Auriault C. Genes specifically modulated in sensitized skins allow the detection of sensitizers in a reconstructed human skin model. Development of the SENS-IS assay. Toxicol In Vitro. 2015;29:787–802. doi: 10.1016/j.tiv.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Cui ZF, Xu X, Trainor N, et al. Application of multiple parallel perfused microbioreactors and three-dimensional stem cell culture for toxicity testing. Toxicol in Vitro. 2007;21:1318–1324. doi: 10.1016/j.tiv.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Dance A. Correction for Dance, news feature: building benchtop human models. Proc Natl Acad Sci U S A. 2015;112:6773–6775. doi: 10.1073/pnas.1508841112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshian M, Busquet F, Hartung T, Leist M. Survey of European statistics on the use of animals in research & development, and on novel policy initiatives in the area of modern animal-free research. ALTEX. 2015;32:261–274. [Google Scholar]
- Dash A, Inman W, Hoffmaster K, et al. Liver tissue engineering in the evaluation of drug safety. Expert Opin Drug Metab Toxicol. 2009;5:1159–1174. doi: 10.1517/17425250903160664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmel F, Brischwein M, Wolf P, et al. Nutrient depletion and metabolic profiles in breast carcinoma cell lines measured with a label-free platform. Physiol Meas. 2015;36:1367–1381. doi: 10.1088/0967-3334/36/7/1367. [DOI] [PubMed] [Google Scholar]
- DHHS: U.S. Department of Health and Human Services. A Report of the Surgeon General. Atlanta: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2014. The Health Consequences of Smoking—50 Years of Progress. http://ash.org/wp-content/uploads/2014/01/full-report.pdf. [Google Scholar]
- Dieppe P, Ebrahim S, Martin RM, Jüni P. Lessons from the withdrawal of rofecoxib. Br Med J. 2004;329:867–868. doi: 10.1136/bmj.329.7471.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domansky K, Inman W, Serdy J, et al. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2010;10:51–58. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir T, Timko BP, Brigham MD, et al. Nanowired three dimensional cardiac patches. Nat Nanotechnol. 2012;6:720–725. doi: 10.1038/nnano.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BR, Hill DR, Ferguson MA, et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4 doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EASHW: European Agency for Safety and Health at Work. Regulation (EC) No. 1907/2006 – REACH of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restrictions of Chemicals (REACH) and establishing a European Chemicals Agency. 2006 https://osha.europa.eu/en/legislation/directives/regulation-ec-no-1907-2006-of-the-european-parliament-and-of-the-council.
- Ebrahimkhani MR, Young CL, Lauffenburger DA, et al. Approaches to in vitro tissue regeneration with application for human disease modeling and drug development. Drug Discov Today. 2014;19:754–762. doi: 10.1016/j.drudis.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimkhani MR, Young CL, Lauffenburger DA, et al. Approaches to in vitro tissue regeneration with application for human disease modeling and drug development. Drug Discov Today. 2014;19:754–62. doi: 10.1016/j.drudis.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EC: European Commission. European Parliament and Council Directive 2001/83/EC of 6 November 2001 on the Community code relating to medical products for human use. 2001 http://ec.europa.eu/health/files/eudralex/vol-1/dir_2001_83_consol_2012/dir_2001_83_cons_2012_en.pdf.
- EC: European Commission. European Parliament and Council Directive 2003/15/EC amending Council Directive 76/768/EEC on the approximation of the laws of the Member States relating to cosmetic products. OJ L66. 2003:26–35. [Google Scholar]
- EC: European Commission. European Parliament and Council regulation 1935/2004 of 27 October 2004 on materials and articles intended to come into contact with food and repealing Directives 80/590/EEC and 89/109/EEC. 2004 http://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32004R1935.
- EC: European Commission. European Parliament and Council regulation 1331/2008 of 16 December 2008 establishing a common authorisation procedure for food additives, food enzymes and food flavourings. 2008 http://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32008R1331.
- EC: European Commission. European Parliament and Council Regulation (EC) No. 1223/2009 on cosmetic products. OJ L342. 2009:59–209. [Google Scholar]
- Tralau T, Riebeling C, Pirow R, et al. Wind of change challenges toxicological regulators. Environ Health Perspect. 2012;120:1489–1494. doi: 10.1289/ehp.1104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EC: European Commission. European Parliament and Council regulation 528/2012 of 22 May 2012 concerning the making available on the market and use of biocidal products. 2012 http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=uriserv:OJ.L_.2012.167.01.0001.01.ENG.
- EC: European Commission. Environment Directorate: Joint Meeting of the Chemical Committee and the Working Party on Chemicals, Pesticides and Biotechnology. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins. 2012b http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono%282012%2910/part1&doclanguage=en.
- EC: European Commission. Seventh Report on the Statistics on the Number of Animals used for Experimental and other Scientific Purposes in the Member States of the European Union. Report from the Commission to the Council and the European Parliament. 2013 http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A52013DC0859.
- EC: European Commission. European Parliament and Council regulation 283/2013 setting out the data requirements for active substances, in accordance with Regulation (EC) No. 1107/2009 of the European Parliament and of the Council concerning the placing of plant protection products on the market. 2013a http://www.ecolex.org/ecolex/ledge/view/RecordDetails?index=documents&id=LEX-FAOC122301.
- EC: European Commission. European Parliament and Council regulation 284/2013 setting out the data requirements for plant protection products, in accordance with Regulation (EC) No. 1107/2009 of the European Parliament and of the Council concerning the placing of plant protection products on the market. 2013b http://faolex.fao.org/cgi-bin/faolex.exe?rec_id=122398&database=faolex&search_type=link&table=result&lang=eng&format_name=@ERALL.
- EC: European Commission. Commission Regulation (EU) 655/2013 of 10 July 2013 laying down common criteria for the justification of claims used in relation to cosmetic products. Official Journal of the European Union; 2013c. http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2013:190:0031:0034:EN:PDF. [Google Scholar]
- ECHA: European Chemicals Agency. The use of alternatives to testing on animals for the REACH regulation. Helsinki: European Chemicals Agency; 2011. ECHA-11-1R-004.2-EN. [Google Scholar]
- ECVAM: European Centre for the Validation of Alternative Methods. ECVAM Test Method Submissions 2008–2015. 2015 https://eurl-ecvam.jrc.ec.europa.eu/test-submission/ecvam-test-method-submissions-2008-2015.
- Edwards SJL. Research ethics committees and paternalism. J Med Ethics. 2004;30:88–91. doi: 10.1136/jme.2002.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- Emter R, Ellis G, Natsch A. Performance of a novel keratinocyte-based reporter cell line to screen skin sensitizers in vitro. Toxicol Appl Pharmacol. 2010;15:281–290. doi: 10.1016/j.taap.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Esch MB, Mahler GJ, Stokol T, Shuler ML. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 2014;14:3081–3092. doi: 10.1039/c4lc00371c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch MB, Sung JH, Yang J, et al. On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic “body-on-a-chip” devices. Biomed Microdevices. 2012;14:895–906. doi: 10.1007/s10544-012-9669-0. [DOI] [PubMed] [Google Scholar]
- Fabro S, Schumacher H, Smith RL, et al. The metabolism of thalidomide: some biological effects of thalidomide and it metabolites. Br J Pharmacol Chemother. 1965;25:352–362. doi: 10.1111/j.1476-5381.1965.tb02055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, editor. Label-free biosensors methods in drug discovery. New York: Humana Press; 2015. [Google Scholar]
- FDA: US Food and Drugs Administration. FDA Modernizing Regulatory Science. 2011a http://www.fda.gov/forconsumers/consumerupdates/ucm268201.htm.
- FDA: US Food and Drugs Administration. Advancing regulatory science at the FDA. 2011b http://www.fda.gov/downloads/ScienceResearch/SpecialTopics/RegulatoryScience/UCM268225.pdf.
- FDA: US Food and Drug Administration. Summary of NDA approvals & receipts, 1938 to the present. 2013 http://www.fda.gov/%20AboutFDA/WhatWeDo/History/ProductRegulation/SummaryofNDAApprovalsReceipts1938tothepresent/default.htm.
- FDA: US Food and Drug Administration. CDER new drug review: 2014 update. 2014a http://www.fda.gov/downloads/AboutFDA/CentersOffices/OfficeofMedicalProductsandTobacco/CDER/UCM426549.pdf.
- FDA: US Food and Drug Administration. Guidance for industry and FDA staff. Qualification process for drug development tools. 2014b http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM230597.pdf.
- Ferrell N, Ricci KB, Groszek J, et al. Albumin handling by renal tubular epithelial cells in a microfluidic bioreactor. Biotechnol Bioeng. 2012;109:797–803. doi: 10.1002/bit.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick S. Advancing Regulatory Science at the US Food and Drugs Administration: Modernizing Toxicology. 2012 http://www.fda.gov/downloads/advisorycommittees/committeesmeetingmaterials/scienceboardtothefoodanddrugadministration/ucm286069.pdf.
- Frey O, Misun PM, Fluri DA, et al. Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat Commun. 2014;5:4250. doi: 10.1038/ncomms5250. [DOI] [PubMed] [Google Scholar]
- Gao D, Liu H, Lin JM, et al. Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab Chip. 2013;13:978–985. doi: 10.1039/c2lc41215b. [DOI] [PubMed] [Google Scholar]
- Garcia-Canton C, Minet E, Anadon A, Meredith C. Metabolic characterization of cell systems used in in vitro toxicology testing: lung cell system BEAS-2B as a working example. Toxicol In Vitro. 2013;27:1719–1727. doi: 10.1016/j.tiv.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Ryan CA, Dearman RJ, Kimber I. Local lymph node assay (LLNA) for detection of sensitization capacity of chemicals. Methods. 2007;41:54–60. doi: 10.1016/j.ymeth.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Troutman JA, Foertsch LM, et al. Investigation of peptide reactivity of pro-hapten skin sensitizers using a peroxidase-peroxide oxidation system. Toxicol Sci. 2009;112:164–74. doi: 10.1093/toxsci/kfp192. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Vassallo JD, Bailey RE, et al. Development of a peptide reactivity assay for screening contact allergens. Toxicol Sci. 2004;81:332–343. doi: 10.1093/toxsci/kfh213. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Vassallo JD, Foertsch LM, et al. Quantification of chemical peptide reactivity for screening contact allergens: a classification tree model approach. Toxicol Sci. 2007;97:417–427. doi: 10.1093/toxsci/kfm064. [DOI] [PubMed] [Google Scholar]
- Gerlach JC. Development of a hybrid liver support system: a review. Int J Artif Organs. 1996;19:645–654. [PubMed] [Google Scholar]
- Ghaedi M, Calle EA, Mendez JJ, et al. Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. J Clin Invest. 2013;123:4950–4962. doi: 10.1172/JCI68793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese C, Demmler CD, Smmer R, et al. A human artificial lymph node in vitro – challenges and progress. Artif Organs. 2006;10:803–808. doi: 10.1111/j.1525-1594.2006.00303.x. [DOI] [PubMed] [Google Scholar]
- Giese C, Marx U. Human immunity in vitro – Solving immunogenicity and more. Adv Drug Deliv Rev. 2014;69–70:103–122. doi: 10.1016/j.addr.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Goral VN, Hsieh YC, Petzold ON, et al. Perfusion-based microfluidic device for three-dimensional dynamic primary human hepatocyte cell culture in the absence of biological or synthetic matrices or coagulants. Lab Chip. 2010;10:3380–3386. doi: 10.1039/c0lc00135j. [DOI] [PubMed] [Google Scholar]
- Gordon S, Daneshian M, Bouwstra J, et al. Non-animal models of epithelial barriers (skin, intestine and lung) in research, industrial applications and regulatory toxicology. ALTEX. 2015;32:327–378. doi: 10.14573/altex.1510051. [DOI] [PubMed] [Google Scholar]
- Gosselin B. Recent advances in neural recording microsystems. Sensors. 2011;11:4572. doi: 10.3390/s110504572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafton MMG, Wang L, Vidi PA, et al. Breast on-a-chip: Mimicry of the channeling system of the breast for development of theranostics. Integr Biol. 2011;3:451–459. doi: 10.1039/c0ib00132e. [DOI] [PubMed] [Google Scholar]
- Gramowski A, Jügelt K, Schröder OHU, et al. Acute functional neurotoxicity of lanthanum(III) in primary cortical networks. Toxicol Sci. 2011;120:173–183. doi: 10.1093/toxsci/kfq385. [DOI] [PubMed] [Google Scholar]
- Griep LM, Wolbers F, de Wagenaar B, et al. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed Microdevices. 2013;15:145–150. doi: 10.1007/s10544-012-9699-7. [DOI] [PubMed] [Google Scholar]
- Griffith LG, Swartz M. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- Grosberg A, Nesmith AP, Goss JA, et al. Muscle on a chip: in vitro contractility assays for smooth and striated muscle. J Pharmacol Toxicol Methods. 2012;65:126–135. doi: 10.1016/j.vascn.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhu X, Futai N, et al. Computerized microfluidic cell culture using elastomeric channels and Braille displays. Proc Natl Acad Sci U S A. 2004;101(15):861–15. 866. doi: 10.1073/pnas.0404353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther A, Yasotharan S, Vagaon A, et al. A microfluidic platform for probing small artery structure and function. Lab Chip. 2010;10:2341–2349. doi: 10.1039/c004675b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzardi MA, Vozzi F, Ahluwalia AD. Study of the crosstalk between hepatocytes and endothelial cells using a novel multicompartmental bioreactor: A comparison between connected cultures and cocultures. Tissue Eng Part A. 2009;15:3635–3644. doi: 10.1089/ten.TEA.2008.0695. [DOI] [PubMed] [Google Scholar]
- Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. JAMA. 2006;296:1731–1732. doi: 10.1001/jama.296.14.1731. [DOI] [PubMed] [Google Scholar]
- Haessler U, Teo JCM, Foretay D, et al. Migration dynamics of breast cancer cells in a tunable 3D interstitial flow chamber. Integr Biol (Camb) 2012;4:401–409. doi: 10.1039/c1ib00128k. [DOI] [PubMed] [Google Scholar]
- Hartung T. Food for thought … on alternative methods for cosmetics safety testing. ALTEX. 2008;25:147–162. doi: 10.14573/altex.2008.3.147. [DOI] [PubMed] [Google Scholar]
- Hartung T. Per aspirin ad astra …. Altern Lab Anim. 2009;37:45–47. doi: 10.1177/026119290903702S10. [DOI] [PubMed] [Google Scholar]
- Hartung T. Food for thought … on alternative methods for chemical safety testing. ALTEX. 2010a;27:3–14. doi: 10.14573/altex.2010.1.3. [DOI] [PubMed] [Google Scholar]
- Hartung T. Evidence based-toxicology – the toolbox of validation for the 21st century? ALTEX. 2010b;27:241–251. doi: 10.14573/altex.2010.4.253. [DOI] [PubMed] [Google Scholar]
- Hartung T. Look Back in anger – what clinical studies tell us about preclinical work. ALTEX. 2013;30:275–291. doi: 10.14573/altex.2013.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T. 3D - A new dimension of in vitro research. Adv Drug Deliv Rev. 2014;69–70:vi. doi: 10.1016/j.iddr.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T, Leist M. Food for thought … on the evolution of toxicology and the phasing out of animal testing. ALTEX. 2008;25:91–102. doi: 10.14573/altex.2008.2.91. [DOI] [PubMed] [Google Scholar]
- Hartung T, Luechtefeld T, Maertens A, Kleensang A. Integrated testing strategies for safety assessments. ALTEX. 2013;30:3–18. doi: 10.14573/altex.2013.1.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T, Rovida C. Chemical regulators have overreached. Nature. 2009;460:1080–1081. doi: 10.1038/4601080a. [DOI] [PubMed] [Google Scholar]
- Hartung T, Stephens M, Hoffmann S. Mechanistic validation. ALTEX. 2013a;30:119–130. doi: 10.14573/altex.2013.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T, Zurlo J. Alternative approaches for medical countermeasures to biological and chemical terrorism and warfare. ALTEX. 2012;29:251–260. doi: 10.14573/altex.2012.3.251. [DOI] [PubMed] [Google Scholar]
- Hasenberg T, Mühleder S, Dotzler A, et al. Emulating human microcapillaries in a multi-organ-chip platform. J Biotechnol. 2015;216:1–10. doi: 10.1016/j.jbiotec.2015.09.038. [DOI] [PubMed] [Google Scholar]
- Hay M, Thomas DW, Graighead JL, et al. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- Hennen J, Aeby P, Goebel C, et al. Cross talk between keratinocytes and dendritic cells: impact on the prediction of sensitization. Toxicol Sci. 2011;123:501–510. doi: 10.1093/toxsci/kfr174. [DOI] [PubMed] [Google Scholar]
- Hierlemann A, Frey U, Hafizovic S, Heer F. Growing cells atop microelectronic chips: interfacing electrogenic cells in vitro with CMOS-based microelectrode arrays. Proc IEEE. 2011;99:252–284. [Google Scholar]
- Himmel HM, Bussek A, Hoffmann M, et al. Field and action potential recordings in heart slices: Correlation with established in vitro and in vivo models. Br J Pharmacol. 2012;166:276–296. doi: 10.1111/j.1476-5381.2011.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota M, Kouzuki H, Ashikaga T, et al. Artificial neural network analysis of data from multiple in vitro assays for prediction of skin sensitization potency of chemicals. Toxicol In Vitro. 2013;27:1233–46. doi: 10.1016/j.tiv.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Ho CT, Lin RZ, Chang WY, et al. Rapid heterogeneous liver-cell on-chip patterning via the enhanced field-induced dielectrophoresis trap. Lab Chip. 2006;6:724–734. doi: 10.1039/b602036d. [DOI] [PubMed] [Google Scholar]
- Hockemeyer K, Janetopoulos C, Terekhov A, et al. Engineered threedimensional microfluidic device for interrogating cell cell interactions in the tumor microenvironment. Biomicrofluidics. 2014;8:044105. doi: 10.1063/1.4890330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelting L, Scheinhardt B, Bondarenko O, et al. A 3-dimensional human embryonic stem cell (hESC)-derived model to detect developmental neurotoxicity of nanoparticles. Arch Toxicol. 2013;87:721–733. doi: 10.1007/s00204-012-0984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Chang Y-J, Chen WC, et al. Enhancement of renal epithelial cell functions through microfluidic-based coculture with adipose-derived stem cells. Tissue Eng, Part A. 2013;19:2024–2034. doi: 10.1089/ten.tea.2012.0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Koo BK. Modeling mouse and human development using organoid cultures. Development. 2015;142:3113–3125. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- Huh DD. A human breathing lung-on-a-chip. Ann Am Thorac Soc. 2015;12(Suppl 1):S42–44. doi: 10.1513/AnnalsATS.201410-442MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Fujioka H, Tung Y-C, et al. Acoustically detectable cellular-Weselevel lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc Natl Acad Sci U S A. 104:18886–18891. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Hamilton G, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Kim HJ, Fraser JP, et al. Microfabrication of human organs-on-chips. Nat Protoc. 2013;8:2135–2157. doi: 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- Huh D, Leslie DC, Matthews BD, et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung PJ, Lee PJ, Sabounchi P, et al. A novel high aspect ratio microfluidic design to provide a stable and uniform microenvironment for cell growth in a high throughput mammalian cell culture array. Lab Chip. 2005a;5:44–48. doi: 10.1039/b410743h. [DOI] [PubMed] [Google Scholar]
- Hung PJ, Lee PJ, Sabounchi P, et al. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol Bioeng. 2005b;89:1e8. doi: 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- Hwa AJ, Fry RC, Sivaraman A, et al. Rat liver sinusoidal endothelial cells survive without exogenous VEGF in 3D perfused co-cultures with hepatocytes. FASEB J. 2007;21:2564–2579. doi: 10.1096/fj.06-7473com. [DOI] [PubMed] [Google Scholar]
- Hynes K, Menicanin D, Mrozik K, et al. Generation of functional mesenchymal stem cells from different induced pluripotent stem cell lines. Stem Cells Dev. 2014;15:1084–1096. doi: 10.1089/scd.2013.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illa X, Vila S, Yeste J, et al. A novel modular bioreactor to in vitro study the hepatic sinusoid. PLoS ONE. 2014;9:e111864. doi: 10.1371/journal.pone.0111864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMNA: Institute of Medicine of the National Academies. Scientific standards for studies on modified risk tobacco products. Washington, DC: National Academies Press; 2012. [Google Scholar]
- Imura Y, Sato K, Yoshimura E. Micro total bioassay system for ingested substances: Assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal Chem. 2010;82:9983–9988. doi: 10.1021/ac100806x. [DOI] [PubMed] [Google Scholar]
- Ingber DE, Whitesides GM. Lab on a chip: United States of America. Lab Chip. 2012;12:2089–2090. doi: 10.1039/c2lc90052a. [DOI] [PubMed] [Google Scholar]
- InSphero. [Accessed 18 January 2016];EU project “The Body-on-a-Chip”. A next generation drug testing device. 2012 http://www.insphero.com/company/eu-project-the-body-on-a-chip.
- Itoh M, Umegaki-Arao N, Guo Z, et al. Generation of 3D skin equivalents fully reconstructed from human induced pluripotent stem cells (iPSCs) PLoS One. 2013;8:e77673. doi: 10.1371/journal.pone.0077673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K-J, Mehr AP, Hamilton GA, et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol. 2013;5:1119–1129. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- Jang M, Neuzil P, Volk T, et al. On-chip three-dimensional cell culture in phase guides improves hepatocyte functions in vitro. Biomicrofluidics. 2015;9:034113. doi: 10.1063/1.4922863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska JS, Natsch A, Ryan C, et al. Bayesian integrated testing strategy (ITS) for skin sensitization potency assessment: a decision support system for quantitative weight of evidence and adaptive testing strategy. Arch Toxicol. 2015;89:2355–2383. doi: 10.1007/s00204-015-1634-2. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Zervantonakis IK, Chung S, et al. In vitro model of tumor cell extravasation. PLoS One. 2013;8:e56910. doi: 10.1371/journal.pone.0056910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H, Albrekt AS, Borrebaeck CA, Lindstedt M. The GARD assay for assessment of chemical skin sensitizers. Toxicol In Vitro. 2013;27:1163–9. doi: 10.1016/j.tiv.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Johnstone AFM, Gross GW, Weiss DG, et al. Microelectrode arrays: A physiologically based neurotoxicity testing platform for the 21st century. Neurotoxicity. 2010;31:331–350. doi: 10.1016/j.neuro.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Jouan A. Drame de Rennes: le protocol de l’essai Clinique en accusation [Drama in Rennes : the protocol of the trial clinic blamed] Le Figaro; 2016. http://sante.lefigaro.fr/actualite/2016/01/21/24518-drame-rennes-protocole-lessai-clinique-accusation. [Google Scholar]
- Jun Y, Kim MJ, Hwang YH, et al. Microfluidics-generated pancreatic islet microfibers for enhanced immunoprotection. Biomaterials. 2013;34:8122–8130. doi: 10.1016/j.biomaterials.2013.07.079. [DOI] [PubMed] [Google Scholar]
- Kane BJ, Zinner MJ, Yarmush ML, Toner M. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal Chem. 2006;78:4291–4298. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- Kauffman AL, Gyurdieva AV, Mabus JR, et al. Alternative functional in vitro models of human intestinal epithelia. Front Pharmacol. 2013;4:79. doi: 10.3389/fphar.2013.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman BE, Kim HJ, Padmanabhan K, et al. In vitro myelin formation using embryonic stem cells. Development. 2015;142:2213–2225. doi: 10.1242/dev.116517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel M, Frank F. A better prescription for drug-development financing. Nat Biotechnol. 2007;25:859–866. doi: 10.1038/nbt0807-859. [DOI] [PubMed] [Google Scholar]
- Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ingber DE. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol. 2013;5:1130–1140. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-Y, Fluri DA, Kelm JM, et al. 96-well format-based microfluidic platform for parallel interconnection of multiple multicellular spheroids. J Lab Autom. 2015a;20:274–82. doi: 10.1177/2211068214564056. [DOI] [PubMed] [Google Scholar]
- Kim J-Y, Fluri DA, Marchan R, et al. 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J Biotechnol. 2015b;205:24–35. doi: 10.1016/j.jbiotec.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Kim L, Toh YC, Voldman J, Yu H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip. 2007;7:681–694. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- Kim S, Lee H, Chung M, Jeon NL. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip. 2013;13:1489–1500. doi: 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- Kim S, Takayama S. Organ-on-a-chip and the kidney. Kidney Res Clin Pract. 2015;34:165–169. doi: 10.1016/j.krcp.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lee JW, Choi I, et al. A microfluidic device with 3D hydrogel villi scaffold to simulate intestinal absorption. J Nanosci Nanotechnol. 2013;13:7220–7228. doi: 10.1166/jnn.2013.8088. [DOI] [PubMed] [Google Scholar]
- Kimura H, Yamamoto T, Sakai H, et al. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip. 2008;8:741–746. doi: 10.1039/b717091b. [DOI] [PubMed] [Google Scholar]
- Klauke N, Smith GL, Cooper JM. Stimulation of single isolated adult ventricular myocytes within a low volume using a planar microelectrode array. Biophys J. 2003;85:1766–1774. doi: 10.1016/S0006-3495(03)74606-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke N, Smith GL, Cooper JM. Extracellular recordings of field potentials from single cardiomyocytes. Biophys J. 2006;91:2543–2551. doi: 10.1529/biophysj.106.085183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke N, Smith GL, Cooper JM. Microfluidic systems to examine intercellular coupling of pairs of cardiac myocytes. Lab Chip. 2007;7:731–739. doi: 10.1039/b706175g. [DOI] [PubMed] [Google Scholar]
- Klein SG, Serchi T, Hoffmann L, et al. An improved 3D tetraculture system mimicking the cellular organisation at the alveolar barrier to study the potential toxic effects of particles on the lung. Part Fibre Toxicol. 2013;10:31. doi: 10.1186/1743-8977-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinhans R, Brischwein M, Wang P, et al. Sensor-based cell and tissue screening for personalized cancer chemotherapy. Med Biol Eng Comput. 2012;50:117–26. doi: 10.1007/s11517-011-0855-7. [DOI] [PubMed] [Google Scholar]
- Knazek RA. Cell culture on artificial capillaries: An approach to tissue growth in vitro. Science. 1972;178:3–5. doi: 10.1126/science.178.4056.65. [DOI] [PubMed] [Google Scholar]
- Khoshfetrat Pakazad S, Savov A, van de Stolpe A, Dekker R. A novel Stretchable Micro-Electrode Array (SMEA) design for directional stretching of cells. J Micromech Microeng. 2014;24:034003. [Google Scholar]
- Kosten IJ, Spiekstra SW, de Gruijl TD, Gibbs S. MUTZ-3 derived Langerhans cells in human skin equivalents show differential migration and phenotypic plasticity after allergen or irritant exposure. Toxicol Appl Pharmacol. 2015;15:35–42. doi: 10.1016/j.taap.2015.05.017. [DOI] [PubMed] [Google Scholar]
- Krewski D, Acosta D, Jr, Andersen M, et al. Toxicity testing in the 21st century: A vision and a strategy. J Toxicol Environ Health B Crit Rev. 2010;13:51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahar N, Lei NY, Wang J, et al. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PloS One. 2011;6:e26898. doi: 10.1371/journal.pone.0026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;10:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc E, Baudoin R, Corlu A, et al. Selective control of liver and kidney cells migration during organotypic cocultures inside fibronectin-coated rectangular silicone microchannels. Biomaterials. 2007;28:1820–1829. doi: 10.1016/j.biomaterials.2006.11.044. [DOI] [PubMed] [Google Scholar]
- Leclerc E, Hamon J, Claude I, et al. Investigation of acetaminophen toxicity in HepG2/C3a microscale cultures using a system biology model of glutathione depletion. Cell Biol Toxicol. 2015;31:173–185. doi: 10.1007/s10565-015-9302-0. [DOI] [PubMed] [Google Scholar]
- Leclerc E, Hamon J, Legendre A, Bois FY. Integration of pharmacokinetic and NRF2 system biology models to describe reactive oxygen species production and subsequent glutathione depletion in liver microfluidic biochips after flutamide exposure. Toxicol In Vitro. 2014;28:1230–1241. doi: 10.1016/j.tiv.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Leclerc E, Sakai Y, Fujii T. Microfluidic PDMS (polydimethylsiloxane) bioreactor for large-scale culture of hepatocytes. Biotechnol Prog. 2004;20:750–755. doi: 10.1021/bp0300568. [DOI] [PubMed] [Google Scholar]
- Ledford H. Translational research: 4 ways to fix the clinical trial. Nature. 2011;477:526–528. doi: 10.1038/477526a. [DOI] [PubMed] [Google Scholar]
- Lee D, Wang Y, Mendoza-Elias JE, et al. Dual microfluidic perifusion networks for concurrent islet perifusion and optical imaging. Biomed Microdevices. 2012;14:7–16. doi: 10.1007/s10544-011-9580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kim S, Chung M, et al. A bioengineered array of 3D microvessels for vascular permeability assay. Microvasc Res. 2014;91:90–98. doi: 10.1016/j.mvr.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Lee PJ, Ghorashian N, Gaige TA, Hung PJ. Microfluidic system for automated cell-based assays. JALA Charlottesv VA. 2007a;12:363–367. doi: 10.1016/j.jala.2007.07.001. j.jala.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PJ, Hung PJ, Lee LP. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol Bioeng. 2007b;97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- Lees JG, Lim SA, Croll T, et al. Transplantation of 3D scaffolds seeded with human embryonic stem cells: biological features of surrogate tissue and teratoma-forming potential. Regen Med. 2007;2:289–300. doi: 10.2217/17460751.2.3.289. [DOI] [PubMed] [Google Scholar]
- Legendre A, Baudoin R, Alberto G, et al. Metabolic characterization of primary rat hepatocytes cultivated in parallel microfluidic biochips. J Pharm Sci. 2013;102:3264–3276. doi: 10.1002/jps.23466. [DOI] [PubMed] [Google Scholar]
- Legendre A, Jacques S, Dumont F, et al. Investigation of the hepatotoxicity of flutamide: Pro-survival/apoptotic and necrotic switch in primary rat hepatocytes characterized by metabolic and transcriptomic profiles in microfluidic liver biochips. Toxicol In Vitro. 2014;28:1075–1087. doi: 10.1016/j.tiv.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Lehmann AD, Daum N, Bur M, et al. An in vitro triple cell co-culture model with primary cells mimicking the human alveolar epithelial barrier. Eur J Pharm Biopharm. 2011;77:398–406. doi: 10.1016/j.ejpb.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Leist M, Bremer S, Brundin P, et al. The biological and ethical basis of the use of human embryonic stem cells for in vitro test systems or cell therapy. ALTEX. 2008;25:163–190. [PubMed] [Google Scholar]
- Leist M, Hartung T. Inflammatory findings on species extrapolations: humans are definitely no 70-kg mice. Arch Toxicol. 2013;87:563–567. doi: 10.1007/s00204-013-1038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Hasiwa N, Rovida C, et al. Consensus report on the future of animal-free systemic toxicity testing. ALTEX. 2014;31:341–356. doi: 10.14573/altex.1406091. [DOI] [PubMed] [Google Scholar]
- Leist M, Hasiwa M, Daneshian M, Hartung T. Validation and quality control of replacement alternatives – current status and future challenges. Toxicol Res. 2012;1:8. doi: 10.1039/C2TX20011B. [DOI] [Google Scholar]
- Leng L, Tan Y, Gong F, et al. Differentiation of primordial germ cells from induced pluripotent stem cells of primary ovarian insufficiency. Hum Reprod. 2015;30:737–738. doi: 10.1093/humrep/deu358. [DOI] [PubMed] [Google Scholar]
- Li E, Xu Z, Liu F, et al. Continual exposure to cigarette smoke extracts induces tumor-like transformation of human non-tumor bronchial epithelial cells in a microfluidic chip. J Thorac Oncol. 2014;9:1091–1100. doi: 10.1097/JTO.0000000000000219. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu C, Ma T. In vitro organogenesis from pluripotent stem cells. Organogenesis. 2014;10:159–163. doi: 10.4161/org.28918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch G, Klimant I, Frank B, et al. Luminescence lifetime imaging of oxygen, pH, and carbon dioxide distribution using optical sensors. Appl Spectrosc. 2000;54:548–559. [Google Scholar]
- Lippmann ES, Al-Ahmad A, Azarin SM, et al. A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci Rep. 2014;4:4160. doi: 10.1038/srep04160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wu C, Cai H, et al. Cell-based biosensors and their application in biomedicine. Chem Rev. 2014;114:6423–6461. doi: 10.1021/cr2003129. [DOI] [PubMed] [Google Scholar]
- Loskill P, Marcus SG, Mathur A, et al. μOrgano: a Lego®-like plug & play system for modular multi-organ-chips. PLoS One. 2015;10:e0139587. doi: 10.1371/journal.pone.0139587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechtefeld T, Maertens A, McKim J, Hartung T, Kleensang A, Sá-Rocha V. Probabilistic hazard assessment for skin sensitization potency by dose response modeling using feature elimination instead of QSAR. J Appl Toxicol. 2015;35:1361–1371. doi: 10.1002/jat.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler GJ, Esch MB, Glahn RP, Shuler ML. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol Bioeng. 2009a;104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- Mahler GJ, Esch MB, Tako E, et al. Oral exposure to polystyrene nanoparticles affects iron absorption. Nat Nanotechnol. 2012;7:264–271. doi: 10.1038/nnano.2012.3. [DOI] [PubMed] [Google Scholar]
- Mahler GJ, Shuler ML, Glahn RP. Characterization of Caco-2 and HT29-MTX cocultures in an in vitro digestion/cell culture model used to predict iron bioavailability. J Nutr Biochem. 2009b;20:494–502. doi: 10.1016/j.jnutbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Manuppello JR, Sullivan KM. Toxicity assessment of tobacco productsin vitro. ATLA. 2015;43:39–67. doi: 10.1177/026119291504300108. http://www.piscltd.org.uk/wp-content/uploads/2015/04/ATLA-Manupello-2015.pdf. [DOI] [PubMed] [Google Scholar]
- Manz A, Graber N, Widmer HM. Miniaturized total chemical analysis systems: a novel concept for chemical sensing. Sensors Actuat B-Chem. 1990;1:244–248. [Google Scholar]
- Marks JD, Luce JM, Lazar NM, et al. Effect of increases in lung volume on clearance of aerosolized solute from human lungs. J Appl Physiol. 1985;59:1242–1248. doi: 10.1152/jappl.1985.59.4.1242. [DOI] [PubMed] [Google Scholar]
- Marx U. Trends in cell culture technology. In: Balls M, Combes RD, Bhogal N, editors. New technologies for toxicity testing. Austin, New York: Landes Bioscience & Springer Science+Business Media; 2012. pp. 26–46. [Google Scholar]
- Marx U, Walles H, Hoffmann S, et al. “Human-on-a-chip” developments: a translational cutting-edge alternative to systemic safety assessment and efficiency evaluation of substances in laboratory animals and man? Altern Lab Anim. 2012;40:235–257. doi: 10.1177/026119291204000504. [DOI] [PubMed] [Google Scholar]
- Maschmeyer I, Hasenberg T, Jaenicke A, et al. Chip-based human live-intestine and liver-skin co-cultures – A first step toward systemic repeated dose substabce testing in vitro. Eur J Pharm Biopharm. 2015a;95:77–87. doi: 10.1016/j.ejpb.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschmeyer I, Lorenz AK, Schimek K, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015b;15:2688–2699. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- Mason GR, Peters AM, Bagdades E, et al. Evaluation of pulmonary alveolar epithelial integrity by the detection of restriction to diffusion of hydrophilic solutes of different molecular sizes. Clin Sci (Lond) 2001;100:231–236. [PubMed] [Google Scholar]
- Materne EM, Ramme AP, Terrasso AP, et al. A multi-organ chip co-culture of neurospheres and liver equivalentsfor long-term substance testing. J Biotechnol. 2015;205:36–46. doi: 10.1016/j.jbiotec.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Matthews RA. Medical progress depends on animal models – doesn’t it? J R Soc Med. 2008;101:95–98. doi: 10.1258/jrsm.2007.070164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe GJ, Chang JY, Glahn RP, Shuler ML. Development of a gastrointestinal tract microscale cell culture analog to predict drug transport. Mol Celullar Biomech. 2008;5:119–132. [PubMed] [Google Scholar]
- McCracken KW, Howell JC, Wells JM, Spence JR. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLimans WF, Blumenson LE, Tunnah KV. Kinetics of gas diffusion in mammalian cell culture systems. II. Theory. Biotechnol Bioeng. 1968;10:741–763. [Google Scholar]
- Meyvantsson I, Warrick JW, Hayes S, et al. Automated cell culture in high density tubeless microfluidic device arrays. Lab Chip. 2008;8:717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- Mileni M, Johnson DS, Wang Z, et al. Structure-guided inhibitor design for human FAAH by interspecies active site conversion. Proc Natl Acad Sci U S A. 2008;105:12820–12824. doi: 10.1073/pnas.0806121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno EL, Hachi S, Hemmer K, et al. Differentiation of neuroepithelial stem cells into functional dopaminergic neurons in 3D microfluidic cell culture. Lab Chip. 2015;15:2419–2428. doi: 10.1039/c5lc00180c. [DOI] [PubMed] [Google Scholar]
- Mu X, Zheng W, Xiao L, et al. Engineering a 3D vascular network in hydrogel for mimicking a nephron. Lab Chip. 2013;13:1612–1618. doi: 10.1039/c3lc41342j. [DOI] [PubMed] [Google Scholar]
- Mummery C, Wilmut I, Stolpe A, van de and Roelen BAJ. Stem cells Scientific facts and fiction. London: Elsevier; 2011. [Google Scholar]
- Nakayama H, Kimura H, Komori K, et al. Development of a disposable three-compartment micro-cell culture device for toxicokinetic study in humans and its preliminary evaluation. AATEX. 2008;14:619–622. [Google Scholar]
- Nalayanda DD, Puleo CM, Fulton WB, et al. Characterization of pulmonary cell growth parameters in a continuous perfusion microfluidic environment. Exp Lung Res. 2007;33:321–335. doi: 10.1080/01902140701557754. [DOI] [PubMed] [Google Scholar]
- Nalayanda DD, Wang Q, Fulton WB, et al. Engineering an artificial alveolar-capillary membrane: A novel continuously perfused model within microchannels. J Pediatr Surg. 2010;45:45–51. doi: 10.1016/j.jpedsurg.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsch A, Ryan CA, Foertsch L, et al. A dataset on 145 chemicals tested in alternative assays for skin sensitization undergoing prevalidation. J Appl Toxicol. 2013;33:1337–1352. doi: 10.1002/jat.2868. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Gleghorn JP. Sculpting organs: Mechanical regulations of tissue development. Annu Rev Biomed Eng. 2011;14:129–154. doi: 10.1146/annurev-bioeng-071811-150043. [DOI] [PubMed] [Google Scholar]
- Nery FC, da Hora CC, Yaqub U, et al. New methods for investigation of neuronal migration in embryonic brain explants. J Neurosci Methods. 2015;239:80–84. doi: 10.1016/j.jneumeth.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JE, Niles JA, Vega SP, et al. Modeling the lung: Design and development of tissue engineered macro- and micro-physiologic lung models for research use. Exp Biol Med. 2014;239:1135–1169. doi: 10.1177/1535370214536679. [DOI] [PubMed] [Google Scholar]
- Nukada Y, Miyazawa M, Kazutushi S, et al. Data integration of non-animal tests for the development of a test battery to predict the skin sensitizing potential of potency of chemicals. Toxicol In Vitro. 2013;25:609–618. doi: 10.1016/j.tiv.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Obach SR. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: An examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos. 1999;27:1350–1359. [PubMed] [Google Scholar]
- OECD: Organisation for Economic Cooperation and Development. Guidelines for the Testing of Chemicals, Section 4. 1992 http://www.oecd-ilibrary.org/environment/test-no-406-skin-sensitisation_9789264070660-en.
- OECD: Organisation for Economic Cooperation and Development. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins. Part 2: Use of the AoP to Develop Chemical Categories and Integrated Assessment and Testing Approaches. 2012 Series on Testing and Assessment No. 168. http://www.oecd.org/chemicalsafety/testing/seriesontestingandassessmentpublicationsbynumber.htm.
- OECD: Organisation for Economic Cooperation and Development. In the Series on Testing and Assessment. GD 184. Paris: Organisation for Economic Cooperation and Development; 2013. Guidance Document on Developing and Assessing Adverse Outcome Pathways. http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono%282013%296&doclanguage=en. [Google Scholar]
- Ogawa M, Ogawa S, Bear CE, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol. 2015;33:853–861. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- Oleago C, Bernabini C, Smith AS, et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. 2016;6:20030. doi: 10.1038/srep20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson H, Betton G, Robinson D, et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- Ootani A, Li X, Sangiorgi E, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2010;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwehand K, Spiekstra SW, Waaijman T, et al. Technical advance: Langerhans cells derived from a human cell line in a full-thickness skin equivalent undergo allergen-induced maturation and migration. J Leukoc Biol. 2011;90:1027–1033. doi: 10.1189/jlb.0610374. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gürtler M, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Li Y, Toner M. Radial flow hepatocyte bioreactor using stacked microfabricated grooved substrates. Biotechnol Bioeng. 2008;99:455–467. doi: 10.1002/bit.21572. [DOI] [PubMed] [Google Scholar]
- Park JY, Hwang CM, Lee SH. Ice-lithographic fabrication of concave microwells and a microfluidic network. Biomed Microdevices. 2009;11:129–133. doi: 10.1007/s10544-008-9216-1. [DOI] [PubMed] [Google Scholar]
- Park TH, Shuler ML. Integration of cell culture and microfabrication technology. Biotechnol Prog. 2003;19:243–253. doi: 10.1021/bp020143k. [DOI] [PubMed] [Google Scholar]
- Park YK, Tu T-Y, Lim SH. In vitro microvessel growth and remodeling within a threedimensional microfluidic environment. Cell Mol Bioeng. 2014;7:15–25. doi: 10.1007/s12195-013-0315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasirayi G, Scott SM, Islam M, et al. Low cost microfluidic cell culture array using normally closed valves for cytotoxicity assay. Talanta. 2014;129:491–498. doi: 10.1016/j.talanta.2014.06.020. [DOI] [PubMed] [Google Scholar]
- Patsch C, Challet-Meylan L, Thoma EC, et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SM, Mytelka DS, Dunwiddie CT, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- Pearce TM, Williams JJ, Kruzel SP, et al. Dynamic control of extracellular environment in in vitro neural recording systems. IEEE Trans Neural Syst Rehabil Eng. 2005a;13:207–212. doi: 10.1109/TNSRE.2005.848685. [DOI] [PubMed] [Google Scholar]
- Pearce TM, Wilson JA, Oakes SG, et al. Integrated microelectrode array and microfluidics for temperature clamp of sensory neurons in culture. Lab Chip. 2005b;5:97–101. doi: 10.1039/b407871c. [DOI] [PubMed] [Google Scholar]
- Perel P, Roberts I, Sena E, et al. Comparison of treatment effects between animal experiments and clinical trials: systematic review. Br Med J. 2007;334:197. doi: 10.1136/bmj.39048.407928.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perestrelo AR, Águas ACP, Rainer A, Forte G. Microfluidic organ/body-on-a-chip devices at the convergence of Biology and microengineering. Sensors (Basel) 2015;15:31142–31170. doi: 10.3390/s151229848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pexton SR, Kalcioglu ZI, Cohen JC, et al. Marrow-derived stem cell motility in 3D synthetic scaffold is governed by geometry along with adhesivity and stiffness. Biotechnol Bioeng. 2011;108:1181–1193. doi: 10.1002/bit.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PhARMA: Pharmaceutical Research and Manufacturers of America. 2015 profile Biopharmaceutical Research Industry. 2015 http://www.phrma.org/sites/default/files/pdf/2015_phrma_profile.pdf.
- Piroird C, Ovigne JM, Rousset F, et al. The Myeloid U937 Skin Sensitization Test (U-SENS) addresses the activation of dendritic cell event in the adverse outcome pathway for skin sensitization. Toxicol In Vitro. 2015;29:901–916. doi: 10.1016/j.tiv.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Polacheck WJ, Li R, Uzel SGM, Kamm RD. Microfluidic platforms for mechanobiology. Lab Chip. 2013;13:2252–2267. doi: 10.1039/c3lc41393d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MJ, Domansky K, Kaazempur-Mofrad MR, et al. A microfabricated array bioreactor for perfused 3D liver culture. Biotechnol Bioeng. 2002;78:257–269. doi: 10.1002/bit.10143. [DOI] [PubMed] [Google Scholar]
- Prieto P, Kinsner-Ovaskainen A, Stanzel S, et al. The value of selected in vitro and in silico methods to predict acute oral toxicity in a regulatory context: Results from the European Project A Cute Tox. Toxicol In Vitro. 2013;27:357–376. doi: 10.1016/j.tiv.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Prot JM, Maciel L, Bricks T, et al. First pass intestinal and liver metabolism of paracetamol in a microfluidic platform coupled with a mathematical modeling as a means of evaluating ADME processes in humans. Biotechnol Bioeng. 2014;111:2027–2040. doi: 10.1002/bit.25232. [DOI] [PubMed] [Google Scholar]
- Prytherch Z, Job C, Marshall H, et al. Tissue-specific stem cell differentiation in an in vitro airway model. Macromol Biosci. 2011;11:1467–1477. doi: 10.1002/mabi.201100181. [DOI] [PubMed] [Google Scholar]
- Ramadan Q, Jafarpoorchekab H, Huang C, et al. NutriChip: nutrition analysis meets microfluidics. Lab Chip. 2013;13:196–203. doi: 10.1039/c2lc40845g. [DOI] [PubMed] [Google Scholar]
- Ramirez T, Mehling A, Kolle SN, et al. LuSens: a keratinocyte based ARE reporter gene assay for use in integrated testing strategies for skin sensitization hazard identification. Toxicol In Vitro. 2014;28:1482–1497. doi: 10.1016/j.tiv.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Reuter H, Spieker J, Gerlach S. In vitro detection of contact allergens: development of an optimized protocol using human peripheral blood monocyte-derived dendritic cells. Toxicol In Vitro. 2011;25:315–323. doi: 10.1016/j.tiv.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Rhee SW, Taylor AM, Tu CH, et al. Patterned cell culture inside microfluidic devices. Lab Chip. 2005;5:102–107. doi: 10.1039/b403091e. [DOI] [PubMed] [Google Scholar]
- Richter A, Schmucker SS, Esser PR, et al. Human T cell priming assay (hTCPA) for the identification of contact allergens based on naive T cells and DC--IFN-γ and TNF-α readout. Toxicol In Vitro. 2013;27:1180–1185. doi: 10.1016/j.tiv.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Rismani Yazdi S, Shadmani A, Bürgel SC, et al. Adding the ‘heart’ to hanging drop networks for microphysiological multi-tissue experiments. Lab Chip. 2015;7:4138–4147. doi: 10.1039/c5lc01000d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rookmaaker MB, Schutgens F, Verhaar MC, Clevers H. Development and application of human adult stem or progenitor cell organoids. Nat Rev Nephrol. 2015;11:546–554. doi: 10.1038/nrneph.2015.118. [DOI] [PubMed] [Google Scholar]
- Rothen-Rutishauser B, Blank F, Mühlfeld C, Gehr P. In vitro models of the human epithelial airway barrier to study the toxic potential of particulate matter. Expert Opin Drug Metab Toxicol. 2008;4:1075–1089. doi: 10.1517/17425255.4.8.1075. [DOI] [PubMed] [Google Scholar]
- Rovida C, Asakura C, Daneshian M, et al. Toxicity testing in the 21st century beyond environmental chemicals. ALTEX. 2015a;32:25–40. doi: 10.14573/altex.1506201. http://dx.doi.org/10.14573/altex.1411011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida C, Alépée N, Api AM, et al. Integrated testing strategies (ITS) for safety assessment. ALTEX. 2015b;32:25–40. doi: 10.14573/altex.1411011. http://dx.doi.org/10.14573/altex.1411011. [DOI] [PubMed] [Google Scholar]
- Rovida C, Hartung T. Re-evaluation of animal numbers and costs for in vivo tests to accomplish REACH legislation requirements for chemicals – a report by the transatlantic think tank for toxicology (t(4)) ALTEX. 2009;26:187–208. [PubMed] [Google Scholar]
- Russian Government. Innovation-based expansion of the pharmaceutical and medical industry; Meeting of the Presidium of the Council for Edonomic Modernisation and Innovative Development of Russia; 6 May; 2014. http://government.ru/en/news/12433/ [Google Scholar]
- Russian Ministry of Science. SRC Bioclinicum. 2011. Grant No. 16.522.12.2015 (10.10.2011) [Google Scholar]
- Samatov TR, Shkurnikov MU, Tonevitskaya SA, Tonevitsky AG. Modelling the metastatic cascade by in vitro microfluidic platforms. Prog Histochem Cytochem. 2015;49:21–29. doi: 10.1016/j.proghi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Sandig V, Jordan I. Designer cells derived from primary tissue and designed cell lines as a suitable cell source for drug discovery and safety assessment. In: Marx U, Sandig V, editors. Drug testing in vitro. Breakthroughs and trends in cell culture technology. Berlin: Wiley-VCH; 2007. pp. 177–203. [Google Scholar]
- Sarkar U, Rivera-Burgos D, Large EM, et al. Metabolite profiling and pharmacokinetic evaluation of hydrocrotisone in a perfused three-dimensional human liver bioreactor. Drug Metab Dispos. 2015;43:1091–1099. doi: 10.1124/dmd.115.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Clevers H. SnapShot: Growing Organoids from Stem Cells. Cell. 2015;161:1700–1700. e1. doi: 10.1016/j.cell.2015.06.028. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- Schimek K, Busek M, Brincker S, et al. Integrating biological vasculature into a multi-organ-chip microsystem. Lab Chip. 2013;13:3588–3598. doi: 10.1039/c3lc50217a. [DOI] [PubMed] [Google Scholar]
- Schimek K, Busek M, Brincker S, et al. Integrating biological vasculature into a multi-organchip microsystem. Lab Chip. 2013;13:3588–3598. doi: 10.1039/c3lc50217a. [DOI] [PubMed] [Google Scholar]
- Schleh C, Rothen-Rutishauser BM, Blank F, et al. Surfactant Protein D modulates allergen particle uptake and inflammatory response in a human epithelial airway model. Respir Res. 2012;13:8. doi: 10.1186/1465-9921-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmälzlin E, van Dongen JT, Klimant I, et al. An optical multifrequency phase modulation method using microbeads for measuring intracellular oxygen concentrations in plants. Biophys J. 2005;89:1339–1345. doi: 10.1529/biophysj.105.063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnerch A, Cerdan C, Bhatia M. Distinguishing between mouse and human pluripotent stem cell regulation: the best laid plans of mice and men. Stem Cells. 2010;28:419–430. doi: 10.1002/stem.298. [DOI] [PubMed] [Google Scholar]
- Schuster D, Laggner C, Langer T. Why drugs fail – a study on side effects in new chemical entities. Curr Pharm Des. 2005;11:3545–3559. doi: 10.2174/138161205774414510. [DOI] [PubMed] [Google Scholar]
- Sellgren KL, Butala EJ, Gilmour BP, et al. A biomimetic multicellular model of the airways using primary human cells. Lab Chip. 2014;14:3349–3358. doi: 10.1039/c4lc00552j. [DOI] [PubMed] [Google Scholar]
- Sena ES, van der Worp HB, Bath PM, et al. Publication bias in reports of animal stroke studies leads to major overstatement of efficacy. PLoS Biol. 2010;8:e1000344. doi: 10.1371/journal.pbio.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyavina NV, Trushkin EV, Rusanov AL, et al. Current technologies for in vitro testing of medicines: use of microbioreactors. Biotechnol Russia. 2013;1:51–58. [Google Scholar]
- Seok J, Warren HS, Cuenca AG, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;26:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergachev I, Rusanov A, Trushkin E, et al. Fluorescent optical fiber sensors for cell viability monitoring. Analyst. 2013;138:4066–4069. doi: 10.1039/c3an00248a. [DOI] [PubMed] [Google Scholar]
- Sheldon LS, Cohen Hubal EA. Exposure as part of a systems approach for assessing risk. Environ Health Perspect. 2009;117:119–1194. doi: 10.1289/ehp.0800407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi TF, Kimmel DW, Cliffel DE. Multianalyte microphysiometry reveals changes in cellular bioenergetics upon exposure to fluorescent dyes. Anal Chem. 2013;85:11677–11680. doi: 10.1021/ac402764x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuler ML. Modeling life. Ann Biomed Eng. 2012;40:1399–1407. doi: 10.1007/s10439-012-0567-7. [DOI] [PubMed] [Google Scholar]
- Silva PN, Green BJ, Altamentova SM, Rocheleau JV. A microfluidic device designed to induce media flow throughout pancreatic islets while limiting shear-induced damage. Lab Chip. 2013;13:4374–4384. doi: 10.1039/c3lc50680k. [DOI] [PubMed] [Google Scholar]
- Simão D, Pinto C, Fernandes P, et al. Evaluation of helper-dependent canine adenovirus vectors in a 3D human CNS model. Gene Ther. 2016;23:86–94. doi: 10.1038/gt.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CS, Petzold BC, Pruitt BL. Microsystems for biomimetic stimulation of cardiac cells. Lab Chip. 2012;12:3235–3248. doi: 10.1039/c2lc40308k. [DOI] [PubMed] [Google Scholar]
- Sin A, Chin KC, Jamil MF, et al. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog. 2004;20:338–345. doi: 10.1021/bp034077d. [DOI] [PubMed] [Google Scholar]
- SLAS: Society for Laboratory Automation and Screening. [Accessed 17 January 2016];Microplate Standards, ANSI/SLAS 1-2004 through ANSI/SLAS 4-2004. 2014 http://www.slas.org/resources/information/industry-standards/
- Smith GD, Takayama S, Swain JE. Rethinking in vitro embryo culture: New developments in culture platforms and potential to improve assisted reproductive technologies. Biol Reprod. 2012;86:62. doi: 10.1095/biolreprod.111.095778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snouber LC, Aninat C, Grsicom L, et al. Investigation of ifosfamide nephrotoxicity induced in a liver-kidney co-culture biochip. Biotechnol Bioeng. 2013;110:597–608. doi: 10.1002/bit.24707. [DOI] [PubMed] [Google Scholar]
- Snouber LC, Bunescu A, Naudot M, et al. Metabolomics-on-a-chip of hepatotoxicity induced by anticancer drug flutamide and its active metabolite hydroxyflutamide using HepG2/C3a microfluidic biochips. Toxicol Sci. 2013;132:8–20. doi: 10.1093/toxsci/kfs230. [DOI] [PubMed] [Google Scholar]
- Snouber LC, Jacques S, Monge M, et al. Transcriptomic analysis of the effect of ifosfamide on MDCK cells cultivated in microfluidic biochips. Genomics. 2012;100:27–34. doi: 10.1016/j.ygeno.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Snouber LC, Letourneur F, Chafey P, et al. Analysis of transcriptomic and proteomic profiles demonstrates improved Madin-Darby canine kidney cell function in a renal microfluidic biochip. Biotechnol. 2011;28:474–484. doi: 10.1002/btpr.743. [DOI] [PubMed] [Google Scholar]
- Somia N, Verma IM. Gene therapy: trials and tribulations. Nat Rev Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- Song B, Smink AM, Jones CV, et al. The directed differentiation of human iPS cells into kidney podocytes. PLoS One. 2012;7:e46453. doi: 10.1371/journal.pone.0046453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F, Gruchow M, Wagner I, et al. Miniatirisierte humane organtypische Zellund Gewebekulturen. BIOspektrum. 2011;17:418–421. [Google Scholar]
- Sonntag F, Schilling N, Mader K, et al. Design and prototyping of a chip-based multi-micro-organoid culture system for substance testing, predictive to human (substance) exposure. J Biotechnol. 2010;148:70–75. doi: 10.1016/j.jbiotec.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Spira ME, Hai A. Multi-electrode array technologies for neuroscience and cardiology. Nat Nanotechnol. 2013;8:83–94. doi: 10.1038/nnano.2012.265. [DOI] [PubMed] [Google Scholar]
- Stebbings R, Findlay L, Edwards C, et al. ‘Cytokine storm’ in the phase I trial of monoclonal antibody TGN1412: Better understanding the causes to improve preclinical testing of immunotherapeutics. J Immunol. 2012;179:3325–3331. doi: 10.4049/jimmunol.179.5.3325. [DOI] [PubMed] [Google Scholar]
- Stephens TD, Fillmore BJ. Hypothesis: Thalidomide embryopathy-proposed mechanism of action. Teratology. 2000;61:189–195. doi: 10.1002/(SICI)1096-9926(200003)61:3<189::AID-TERA6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Stewart CE, Torr EE, Mohd Jamili NH, et al. Evaluation of differentiated human bronchial epithelial cell culture systems for asthma research. J Allergy (Cairo) 2012;2012:943982. doi: 10.1155/2012/943982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes C, Cirit M, Lauffenburger D. Physiome-on-a-chip: The challenge of “scaling” in design, operation, and translation of microphysiological systems. CPT Pharmacometrics Syst Pharmacol. 2015;4:559–562. doi: 10.1002/psp4.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Stolpe A, den Toonder J. Workshop meeting report Organs-on-Chips: human disease models. Lab Chip. 2013;13:3449–3470. doi: 10.1039/c3lc50248a. [DOI] [PubMed] [Google Scholar]
- Stucki AO, Stucki JD, Hall SR, et al. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip. 2015;15:1302–1310. doi: 10.1039/c4lc01252f. [DOI] [PubMed] [Google Scholar]
- Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- Sung JH, Shuler ML. A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip. 2009;9:1385–1394. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- Sung JH, Shuler ML. In vitro microscale systems for systematic drug toxicity study. Bioprocess Biosyst Eng. 2010;33:5–19. doi: 10.1007/s00449-009-0369-y. [DOI] [PubMed] [Google Scholar]
- Sung JH, Yu J, Luo D, et al. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip. 2011;11:389–392. doi: 10.1039/c0lc00273a. [DOI] [PubMed] [Google Scholar]
- Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Szkolnicka D, Farnworth SL, Lucendo-Villarin B, Hay DC. Deriving functional hepatocytes from pluripotent stem cells. Curr Protoc Stem Cell Biol. 2014;30:1G.5.1–1G.5.12. doi: 10.1002/9780470151808.sc01g05s30. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tatosian D, Shuler ML. A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnol Bioeng. 2009;103:187–198. doi: 10.1002/bit.22219. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, et al. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrasso AP, Pinto C, Serra M, et al. Novel scalable 3D cell based model for in vitro neurotoxicity testing: Combining human differentiated neurospheres with gene expression and functional endpoints. J Biotechnol. 2015;10:82–92. doi: 10.1016/j.jbiotec.2014.12.011. [DOI] [PubMed] [Google Scholar]
- TissUse: TissUse GmbH. Human-on-a-chip technology. 2014 http://www.tissuse.com/technology.html.
- Toh YC, Lim TC, Tai D, et al. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip. 2009;9:2026–2035. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- Toh YC, Zhang C, Zhang J, et al. A novel 3D mammalian cell perfusion-culture system in microfluidic channels. Lab Chip. 2007;7:302–309. doi: 10.1039/b614872g. [DOI] [PubMed] [Google Scholar]
- Tollefsen KE, Scholz S, Cronin MT, et al. Applying Adverse Outcome Pathways (AOPs) to support Integrated Approaches to Testing and Assessment (IATA) Reg Toxicol Pharmacol. 2014;70:629–640. doi: 10.1016/j.yrtph.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Tourovskaia A, Fauver M, Kramer G, et al. Tissue-engineered microenvironment systems for modeling human vasculature. Exp Biol Med (Maywood) 2014;239:1264–1271. doi: 10.1177/1535370214539228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tralau T, Riebeling C, Pirow R, et al. Wind of change challenges toxicological regulators. Environ Health Perspect. 2012;120:1489–1494. doi: 10.1289/ehp.1104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tralau T, Luch A. Moving from rats to cellular omics in regulatory toxicology: great challenge toward sustainability or “up-shit-creek without a paddle”? Arch Toxicol. 2015;89:819–821. doi: 10.1007/s00204-015-1511-z. [DOI] [PubMed] [Google Scholar]
- Tralau T, Oelgeschläger M, Gürtler R, et al. Regulatory toxicology in the twenty-first century: challenges, perspectives and possible solutions. Arch Toxicol. 2015;89:823–850. doi: 10.1007/s00204-015-1510-0. [DOI] [PubMed] [Google Scholar]
- Trietsch SJ, Israëls GD, Joore J, et al. Microfluidic titer plate for stratified 3D cell culture. Lab Chip. 2013;13:3548–3554. doi: 10.1039/c3lc50210d. [DOI] [PubMed] [Google Scholar]
- Tsujita-Inoue K, Atobe T, Hirota M, et al. In silico risk assessment for skin sensitization using artificial neural network analysis. J Toxicol Sci. 2015;40:193–209. doi: 10.2131/jts.40.193. [DOI] [PubMed] [Google Scholar]
- Tsujita-Inoue K, Hirota M, Ashikaga T, et al. Skin sensitization risk assessment model using artificial neural network analysis of data from multiple in vitro assays. Toxicol In Vitro. 2014;28:626–639. doi: 10.1016/j.tiv.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Tufts CSDD: Center for the Study of Drug Development. Cost to developing and win marketing approval for a new drug in $2.6 billion. 2014 Published 18 November 2014. http://csdd.tufts.edu/news/complete_story/pr_tufts_csdd_2014_cost_study.
- van der Jagt K, Munn S, Tørsløv J, de Bruijn J. Alternative approaches can reduce the use of test animals under REACH. Institute for Health and Consumer Protection: European Commission Joint Research Centre; 2004. Report EUR 21405 EN. http://publications.jrc.ec.europa.eu/repository/bitstream/JRC29111/EUR%2021405%20EN.pdf. [Google Scholar]
- van der Veen JW, Rorije E, Emter R, et al. Evaluating the performance of integrated approaches for hazard identification of skin sensitizing chemicals. Regul Toxicol Pharmacol. 2014;69:371–379. doi: 10.1016/j.yrtph.2014.04.018. [DOI] [PubMed] [Google Scholar]
- Van der Worp HB, Howells DW, Sena ES, et al. Can animal models of disease reliably inform human studies? PLoS Med. 2010;7:e1000245. doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duinen V, Trietsch SJ, Joore J, et al. Microfluidic 3D cell culture: from tools to tissue models. Curr Opin Biotechnol. 2015;35:118–126. doi: 10.1016/j.copbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
- van Midwoud PM, Groothuis GM, Me3rema MT, Verpoorte E. Microfluidic biochip for the perfusion of precision-cut rat liver slices for metabolism and toxicology studies. Biotechnol Bioeng. 2010;105:184–194. doi: 10.1002/bit.22516. [DOI] [PubMed] [Google Scholar]
- van Midwoud PM, Merema MT, Verpoorte E, Groothuis GMM. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip. 2010;10:2778–2786. doi: 10.1039/c0lc00043d. [DOI] [PubMed] [Google Scholar]
- van Midwoud PM, Merema MT, Verweij N, et al. Hydrogel embedding of precision-cut liver slices in a microfluidic device improves drug metabolic activity. Biotechnol Bioeng. 2011;108:1404–1412. doi: 10.1002/bit.23053. [DOI] [PubMed] [Google Scholar]
- van Midwoud PME, Verpoorte E, Groothuis GMM. Microfluidic devices for in vitro studies on liver drug metabolism and toxicity. Integr Biol (Camb) 2011;3:509–521. doi: 10.1039/c0ib00119h. [DOI] [PubMed] [Google Scholar]
- Villa-Diaz LG, Torisawa YS, Uchida T, et al. Microfluidic culture of single human embryonic stem cell colonies. Lab Chip. 2009;9:1749–1755. doi: 10.1039/b820380f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viravaidya K, Sin A, Shuler ML. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog. 2004;20:316–323. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- Vivares A, Salle-Lefort S, Arabeyre-Fabre C, et al. Morphological behaviour and metabolic capacity of cryopreserved human primary hepatocytes cultivated in a perfused multiwell device. Xenobiotica. 2015;45:29–44. doi: 10.3109/00498254.2014.944612. [DOI] [PubMed] [Google Scholar]
- Vozzi F, Heinrich J, Bader A, Ahluwalia AD. Connected culture of murine hepatocytes and human umbilical vein endothelial cells in a multicompartmental bioreactor. Tissue Eng Part A. 2009;15:1291–1299. doi: 10.1089/ten.tea.2008.0066. [DOI] [PubMed] [Google Scholar]
- Vunjak-Novakovic G, Bhatia S, Chen C, Hirschi K. HeLiVa platform: Integrated heart-liver-vascular systems for drug testing in human health and disease. Stem Cell Res Ther. 2013;4:S8. doi: 10.1186/scrt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I, Materne EM, Brinckner S, et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip. 2013;13:3538–3547. doi: 10.1039/c3lc50234a. [DOI] [PubMed] [Google Scholar]
- Wai SN, Large LM, Hughes DJ, et al. Long-term persistence of patient-derived hepatitis B virus infection in 3D microfluidic primary hepatocyte cultures. Proceedings of the 65th meeting of the American Association for the Study of Liver Disease; 2014 7–11 November; Boston, USA. 2014. [Google Scholar]
- Wang G, McCain ML, Yang L, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Liu Q. Cell-based biosensors principles and applications. Boston: Artech House; 2010. [Google Scholar]
- Wang XY, Jin ZH, Gan BW, et al. Engineering interconnected 3D vascular networks in hydrogels using molded sodium alginate lattice as the sacrificial template. Lab Chip. 2014;14:2709–2716. doi: 10.1039/c4lc00069b. [DOI] [PubMed] [Google Scholar]
- Wang Y, Susando T, Lei X, et al. Current development of bioreactors for extracorporeal bioartificial liver (Review) Biointerphases. 2010;5:FA116–131. doi: 10.1116/1.3521520. [DOI] [PubMed] [Google Scholar]
- Waring MJ, Arrowsmith J, Leach AR, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discov. 2015;14:475–486. doi: 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- Weiss D, Brischwein M, Grothe H, et al. Label-free monitoring of whole cell vitality. 35th Annual International Conference of the IEEE EMBS; Osaka, Japan. 3–7 July 2013; 2013. pp. 1607–1610. [DOI] [PubMed] [Google Scholar]
- Werdich AA, Lima EA, Ivanov B, et al. A microfluidic device to confine a single cardiac myocyte in a sub-nanoliter volume on planar microelectrodes for extracellular potential recordings. Lab Chip. 2004;4:357–362. doi: 10.1039/b315648f. [DOI] [PubMed] [Google Scholar]
- Westein E, van der Meer AD, Kuijpers MJE, et al. Atherosclerotic geometries exacerbate pathological thrombus formation poststenosis in a von Willebrand factor-dependent manner. Proc Natl Acad Sci U S A. 2013;110:1357–1362. doi: 10.1073/pnas.1209905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SE, Clark AM, Taylor DP, et al. Spontaneous dormancy of metastatic breast cancer cells in an all human liver microphysiologic system. Br J Cancer. 2014;111:2342–2350. doi: 10.1038/bjc.2014.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Wiest J. 28-day microphysiological monitoring of human hepatocellular cells. ALTEX Proc. 2015;4:2, 12. [Google Scholar]
- Wikswo JP, Block FE, Cliffel DE, et al. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans Biomed Eng. 2013;60:682–690. doi: 10.1109/TBME.2013.2244891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikswo JP, Curtis EL, Eagleton ZE, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip. 2013;13:3496–3511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett C. Testimony of The Human Society of the United States and the Humane Society Legislative Fund before the House Committee on Energy and Commerce Subcommittee on Environment and the Economy on Issues Pertaining to Reauthorizing the Toxic Substances Control Act 2014 [Google Scholar]
- Wolf P, Hartl F, Brischwein M, Wolf B. Determination of dynamic doxorubicin-EC50 value in an automated high-content workstation for cellular assays. Toxicol In Vitro. 2011;25:1889–1894. doi: 10.1016/j.tiv.2011.05.031. [DOI] [PubMed] [Google Scholar]
- Woollam DH. Principles of teratogenesis: mode of action of thalidomide. Proc R Soc Med. 1965;58:497–501. [PMC free article] [PubMed] [Google Scholar]
- Wu MH, Huang SB, Cui Z, et al. A high throughput perfusion-based microbioreactor platform integrated with pneumatic micropumps for three-dimensional cell culture. Biomed Microdevices. 2008;10:309–319. doi: 10.1007/s10544-007-9138-3. [DOI] [PubMed] [Google Scholar]
- Wu MH, Huang SB, Lee GB. Microfuidic cell culture systems for drug research. Lab Chip. 2010;10:939–956. doi: 10.1039/b921695b. [DOI] [PubMed] [Google Scholar]
- Xia Y, Sancho-Martinez I, Nivet E, et al. The generation of kidney organoids by differentiation of human pluripotent cells to ureteric bud progenitor-like cells. Nat Protoc. 2014;9:2693–2704. doi: 10.1038/nprot.2014.182. [DOI] [PubMed] [Google Scholar]
- Ye M, Sanchez HM, Hultz M, et al. Brain microvascular endothelial cells resist elongation due to curvature and shear stress. Sci Rep. 2014;4:4681. doi: 10.1038/srep04681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young EWK, Simmons C. Macro- and microscale fluid flow systems for endothelial cell biology. Lab Chip. 2010;10:143–160. doi: 10.1039/b913390a. [DOI] [PubMed] [Google Scholar]
- Yu J, Cilfone N, Large E, et al. Quantitative systems pharmacology approaches applied to microphysiological systems (MPS): Data interpretation and multi-MPS integration. CPT Pharmacometrics Syst Pharmacol. 2015;4:585–594. doi: 10.1002/psp4.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Peng S, Luo D, March JC. In vitro 3D human small intestinal villous model for drug permeability determination. Biotechnol Bioeng. 2012;109:2173–2178. doi: 10.1002/bit.24518. [DOI] [PubMed] [Google Scholar]
- Zervantonakis IK, Hughes-Alford SK, Charest JL, et al. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc Natl Acad Sci U S A. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhao Z, Abdul Rahim NA, et al. Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip. 2009;9:3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- Zhang MY, Lee PJ, Hung PJ, et al. Microfluidic environment for high density hepatocyte culture. Biomed Microdevices. 2008;10:117–121. doi: 10.1007/s10544-007-9116-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shen W, Sun B, et al. Plasticity of marrow mesenchymal stem cells from human first-trimester fetus: from single-cell clone to neuronal differentiation. Cell Reprogram. 2011;13:57–64. doi: 10.1089/cell.2010.0044. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Teoh SH, Teo EY, et al. A comparison of bioreactors for culture of fetal mesenchymal stem cells for bone tissue engineering. Biomaterials. 2010;31:8684–8695. doi: 10.1016/j.biomaterials.2010.07.097. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chen J, Craven M, et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci U S A. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziaie B, Baldi A, Lei M, et al. Hard and soft micromachining for BioMEMS: review of techniques and examples of applications in microfluidics and drug delivery. Adv Drug Deliv Rev. 2004;56:145–172. doi: 10.1016/j.addr.2003.09.001. [DOI] [PubMed] [Google Scholar]