Background: Factor-assisted co-transcriptional proofreading and precise selection of NTP substrates provide high transcription fidelity.
Results: Both processes can be achieved through active center tuning (ACT) from the inactive to catalytic state in response to establishing recognition contacts of the reaction substrates.
Conclusion: High transcription fidelity can be explained by ACT.
Significance: Suggested ACT mechanism represents an exceptional example of substrate recognition coupling to catalysis.
Abstract
Precise transcription by cellular RNA polymerase requires the efficient removal of noncognate nucleotide residues that are occasionally incorporated. Mis-incorporation causes the transcription elongation complex to backtrack, releasing a single strand 3′-RNA segment bearing a noncognate residue, which is hydrolyzed by the active center that carries two Mg2+ ions. However, in most x-ray structures only one Mg2+ is present. This Mg2+ is tightly bound to the active center aspartates, creating an inactive stable state. The first residue of the single strand RNA segment in the backtracked transcription elongation complex strongly promotes transcript hydrolytic cleavage by establishing a network of interactions that force a shift of stably bound Mg2+ to release some of its aspartate coordination valences for binding to the second Mg2+ thus enabling catalysis. Such a rearrangement that we call active center tuning (ACT) occurs when all recognition contacts of the active center-bound RNA segment are established and verified by tolerance to stress. Transcription factor Gre builds on the ACT mechanism in the same reaction by increasing the retention of the second Mg2+ and by activating the attacking water, causing 3000–4000-fold reaction acceleration and strongly reinforcing proofreading. The unified mechanism for RNA synthesis and degradation by RNA polymerase predicts that ACT also executes NTP selection thereby contributing to high transcription fidelity.
Transcription of DNA by RNA polymerase (RNAP)3 is a highly accurate templated synthesis. The fidelity of transcription relies on both efficient initial substrate selection and on proofreading that removes mis-incorporated residues.
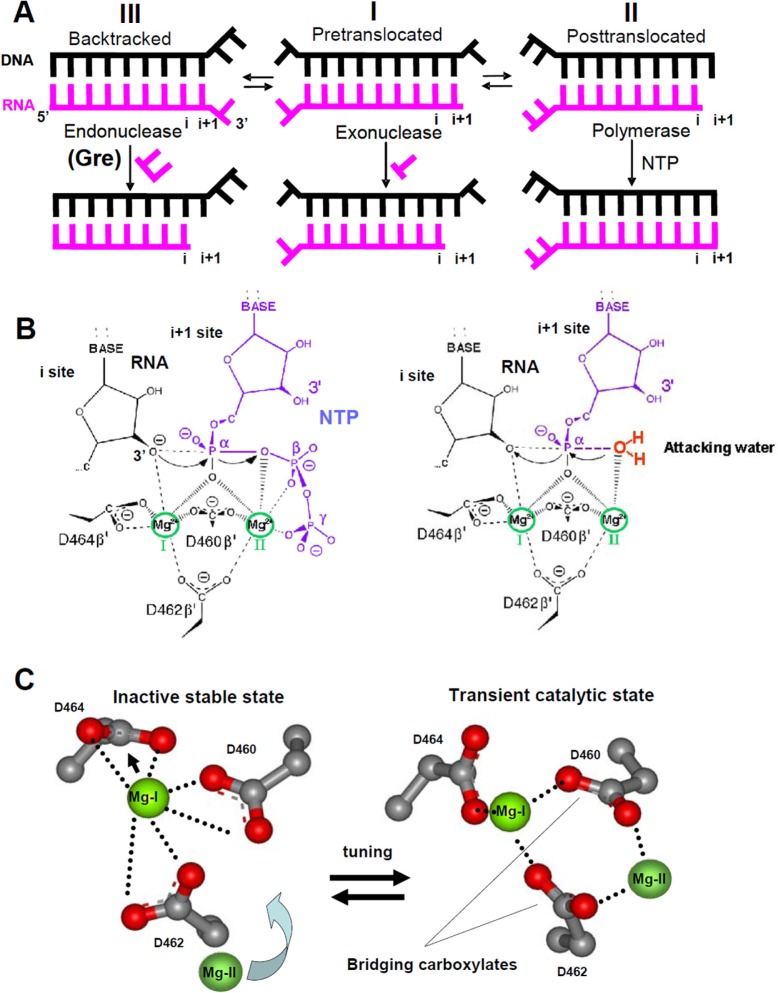
Addition of each nucleotide by RNAP yields two principal states (Fig. 1A) such that the enzyme moves forward along the DNA template from the pretranslocated state (I) to the post-translocated state (II), which vacates the active center for the next NTP substrate. The enzyme can also backtrack (state III), which releases the 3′-RNA segment from the RNA/DNA hybrid. Backtracking is strongly promoted when noncognate nucleotide residues are incorporated. The disengaged single strand RNA segment in a backtracked complex can be removed by RNAP in its endonucleolytic mode (1, 2), whereas the pre-translocated complex can release the 3′-terminal RNA residue (3) following exonuclease reaction (Fig. 1A). Intrinsic endonuclease activity of RNAP, strongly augmented by prokaryotic GreA and GreB proteins (4–6) and by eukaryotic SII factor (7, 8), is believed to provide proofreading that removes RNA segments containing a noncognate residue (9, 10), thereby enabling the enzyme to continue transcription from the newly formed RNA 3′ end.
FIGURE 1.
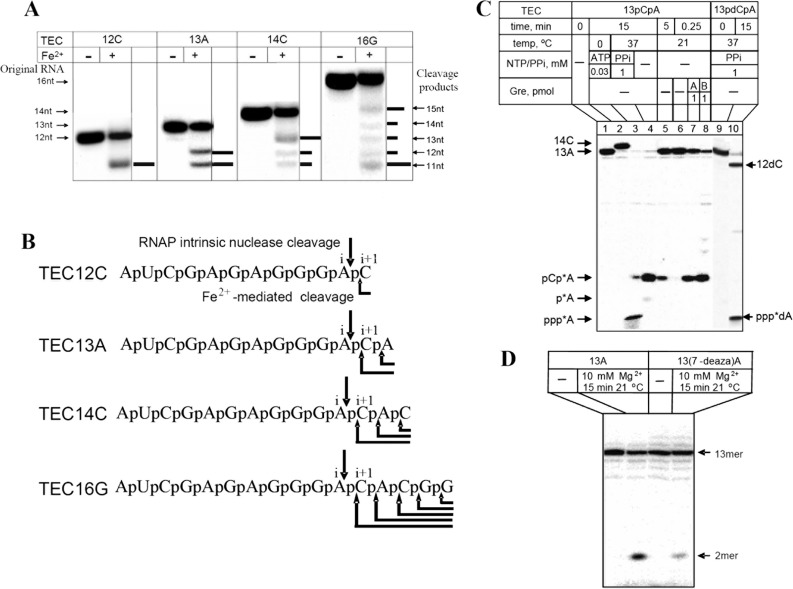
RNAP translocational states and catalytic reactions. A, states of TEC in which various RNAP catalytic activities are observed. B, reactions of NTP polymerization (left panel) and hydrolytic RNA cleavage (right panel). C, change of coordination geometry for active center Mg2+ and aspartate triads upon ACT. The configuration of proposed inactive stable (left panel) and transient catalytic (right panel) states of the active center are presented. The structure on the right is from 3GTG (24). Mg-II was modeled into this structure using Sn2 mechanism constraints. Mg2+ coordination bonds are shown as dotted lines.
All these reactions operate through a two-metal mechanism (3, 6) at the same active center (11) located at the intersection of the main channel that encloses the nucleic acid scaffold of transcription complex and the secondary channel, which supplies NTP substrates and accommodates a 3′-disengaged RNA segment in backtracked TEC. Also, a long α-helix of the β′ subunit (F-bridge or bridge helix) separating the main and secondary channels near the downstream edge of the RNA/DNA hybrid together with a β′G-loop (trigger loop) have been implicated in catalysis and RNAP translocation (12–16). Exactly how the two-metal mechanism operates in the case of endonuclease reaction and the role of various active center elements in catalysis remain obscure. Studies on Thermus aquaticus RNAP proposed direct involvement of the functional groups of the disengaged +2 RNA residue in endonuclease reaction through activation of the attacking water and coordination of catalytic Mg-II (17). However, molecular modeling shows that this is inconsistent with Sn2 reaction mechanism that strictly defines the position of two catalytic Me2+ ions, scissile phosphodiester group, and the attacking water.
Sn2 mechanism (3, 6, 11, 18–22) relies on two Mg2+ ions coordinated by the aspartate triad (β′ Asp-460, β′ Asp-462, and β′ Asp-464) of the active center (Fig. 1B). One of the Mg2+ ions (Mg-I) is coordinated by all three Asp residues, whereas the second Mg2+ ion (Mg-II) is coordinated by only two residues (Asp-460 and Asp-462). The latter are called “bridging” because they connect both Mg2+ ions. During RNA synthesis (Fig. 1B, left panel), Mg-I activates the 3′-attacking hydroxyl of the terminal RNA residue and the α-phosphate of an NTP substrate for nucleophilic attack; Mg-II facilitates release of pyrophosphate. In RNA hydrolytic cleavage (Fig. 1B, right panel), Mg2+ ions switch their role. In this reaction Mg-II activates the attacking water, whereas Mg-I promotes release of the leaving group. One of the ions (Mg-I) is bound with high affinity and the other (Mg-II) with low affinity and has to be stabilized by additional coordination with the following: (i) two terminal phosphates of incoming NTP in polymerization reaction; (ii) the same phosphates of NTP bound to an E site (which is believed to be involved in substrate loading (3, 20)) in exonuclease transcript cleavage, and (iii) carboxylate side chains of Gre or TFIIS proteins in factor-assisted RNA endonuclease cleavage (6, 18–21).
In the majority of x-ray structures of multisubunit RNAP, only one Mg2+ ion (Mg-I) is seen bound to all three aspartates of the active center. According to quantum mechanics calculations (23), this Mg2+ ion forms strong coordination bonds with the aspartates, which makes this state stable. However, all reactions of RNAP require two Mg2+ ions for catalysis, suggesting that this inactive stable state has to be tuned to a catalytically proficient state by inclusion of the second Mg2+ ion. To achieve this, Mg-I must be pushed aside to free the coordination valences of the bridging Asp residues so they can bind Mg-II for catalysis (Fig. 1C). The rearrangement involves repositioning of all three Asp side chains and a drastic change in Mg-I coordination geometry, causing rotation of the bridging aspartates to co-linear orientation (Fig. 1C, right panel). This assumption finds support in recently resolved x-ray structures (3GTG and 3PO3 (24, 25)) of TEC in which endonuclease reaction occurs. Analysis of these structures and our biochemical data show that the above rearrangement of the active center can be promoted by the recognition contacts of substrates in both RNA hydrolytic proofreading and RNA polymerization reactions. It is this rearrangement that we refer to herein as active center tuning (ACT).
Biochemical analysis of intrinsic endonuclease transcript cleavage in RNAP from yeast, mammals, eubacteria, and thermophilic bacteria revealed species-specific reaction features suggesting considerable differences in the reaction mechanism that have to be explained structurally. The examples are unusually high reaction efficacy in thermophiles (26) and the existence of specific endopyrophosphorolytic reaction pathway in pol II of yeast and mammals (27) not seen in bacteria.
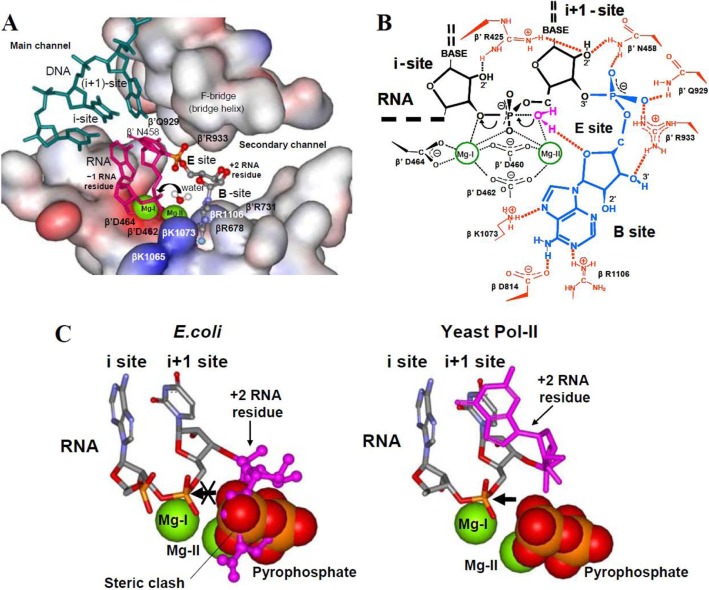
Exploring the mechanism of endonuclease reaction in Escherichia coli, we find that although consistent with biochemical data obtained on yeast RNAP the x-ray model had to be adjusted for E. coli RNAP to fit the experimental data. This was achieved by placing the nucleoside part of the 3′-disengaged +2 RNA residues into a cavity of the active center on the interface of β and β′ subunits (which we named B site, for base binding), while preserving the orientation of the phosphodiester group seen in x-ray structure 3GTG (Fig. 2). The resulting structural model combined with the ACT mechanism explains the pyrophosphorolytic endonuclease reaction pathway in the yeast enzyme and the absence of endonuclease stimulation by non-cognate NTP seen in exonuclease reaction (Fig. 2C).
FIGURE 2.
Molecular model of E. coli RNAP endonuclease reaction in backtracked TEC. The model (constructed using Protein Data Bank 3GTG structure) combines prior knowledge and present results as explained in the text. A, active center in surface representation performing 3′–5′-endonuclease reaction. Active center residues as well as RNA and DNA are indicated. 3′-Terminal disengaged +2 RNA residue (red-blue-metallic) is in the B site. Green spheres represent catalytic Mg-I and Mg-II. Attacking water is coordinated by Mg-II. The black arrow shows the line of attack. B, schematic representation of the endonuclease reaction mechanism shown in A. The black arrows indicate the direction for the electron density shift upon hydrolytic attack. C, relative arrangement of pyrophosphate and +2 RNA residue in the active center of yeast pol II and E. coli TEC. Orientation of +2 RNA residue in yeast enzyme is as in the structure 3GTG. Orientation of the same residue in E. coli TEC is modeled. Black arrow shows the direction of pyrophosphorolytic attack.
In the living cell, endonuclease transcription cleavage is assisted by Gre factors that strongly increase the reaction rate. We find that these factors build on the ACT mechanism by further increasing Mg-II retention and by activating the attacking water for hydrolytic reaction.
The above findings in combination with crystallographic data extend the two-metal active center model to a new level of understanding of unified catalytic mechanism of RNA synthesis and degradation by RNAP. The suggested mechanism for ACT is different from the active center adjustment caused by rearrangement of the trigger loop (15) or from NTP substrate re-location depicted in Thermus thermophilus RNAP (16), because ACT involves the loading (in hydrolytic RNA cleavage) or proper positioning (in RNA synthesis reaction) of catalytic Mg-II.
EXPERIMENTAL PROCEDURES
Reagents and Equipment
Unless otherwise stated, all chemistry reagents were purchased from Sigma, Acros Organics, TCI America, or Fluka. Nucleic acid enzymes were from New England Biolabs and United States Biochemical Corp. NTP substrates were from Amersham Biosciences; radioactive NTPs were from MP Biomedicals; oligonucleotide RNA primers were from Oligos, Etc. Preparative ion-exchange column chromatography was conducted on DE-53 cellulose and Dowex-1 × 2 (400 mesh). The radioactive RNA products resolved by electrophoresis were quantified by phosphorimagery using a Molecular Dynamics device (GE Healthcare) and Storm 60 software. Molecular modeling was performed using WebLab ViewerLight 4.0 (Molecular Simulations Inc.).
Modified Substrate Analogs
α-Methyl-ATP
Because the published synthetic procedure (28) did not yield sufficient amounts of the material, we developed new and more efficient and fast protocol. Twenty six milligrams (100 μmol) of dry adenosine were suspended in 90 μl of trimethylphosphate dried over barium oxide. The mixture was supplemented with 13 μl (20 mg, 150 μmol) of methylphosphonic dichloride and agitated for 1–1.5 h until the suspension became transparent. Three hundred microliters of 1 m solution of tributylammonium pyrophosphate, obtained as described previously (29), were mixed with 50 μl of tributylamine (dried over KOH) and added to the reaction mixture. The mixture was kept for 20 min at room temperature and supplemented with 9 ml of water. pH was adjusted to 3.0 with hydrochloric acid, and the product from the resulting solution was purified on the column (1.2 × 10 cm) with Dowex-1 × 2 using linear gradient (0–0.5 m, 200 ml) LiCl in 0.3 mm HCl. The elution rate was 50 ml/h. The fractions, corresponding to the product (eluted between 0.22 and 0.28 m LiCl) were collected, neutralized by LiOH, and concentrated to 0.3–0.4 ml by agitation with n-butanol. After addition of the equal volume of methanol, the product was precipitated with 15 ml of acetone. After centrifugation, the pellet was dissolved in water and additionally purified by re-chromatography in the same system. The fractions containing pure α-methyl-ATP (the main product) were treated as above. After precipitation with the acetone, the pellet was washed with 10 ml of acetone, dried in vacuo, and stored at −80 °C. Yield was 50%.
Cytidine 5′-Triphospho-3′-methylphosphate
Cytidine 3′-Methylphosphate—Solution of 35 mg (110 μmol) of 3′-cytidylic acid in 800 μl of 50% aqueous N,N-dimethylformamide was supplemented with 20 μl of 10 m LiOH and 100 μl of iodomethane. The mixture was kept at 40 °C in a tightly sealed tube. The pH was checked and adjusted to 7–8 by LiOH every 3 h, and iodomethane was added so that it was always in excess and formed a layer at the bottom of the tube. In about 30 h, the mixture was poured dropwise into 30 ml of acetone/ether (1:1); the residue was centrifuged, re-dissolved in water, and subjected to preparative TCL in an acetonitrile/water (3:1) developing system. The higher migrating product (Rf 0.32) was eluted with water and evaporated to dryness under reduced pressure. Yield was 35–40%. As expected, the product had UV absorption spectrum identical to CMP and displayed resistance to alkaline phosphatase.
Cytidine 5′-Phospho-3′-methylphosphate—Cytidine 3′-methylphosphate (40 μmol) was dissolved in 1 ml of polynucleotide kinase buffer and supplemented with 0.3 ml of 0.3 m ATP, pH 8.0. Fifty microliters of T4 phage polynucleotide kinase (Biolabs) was added, and the mixture was kept at 37 °C. After 30 h of incubation, TLC analysis revealed nearly complete conversion of the starting material to reaction product, which was purified by column chromatography on Dowex-1 × 2 and isolated as described above for α-methyl-ATP. Yield was 90%.
Cytidine-5′-Triphospho-3′-methyl Phosphate—It was obtained from cytidine-5′-phospho-3′-methyl phosphate using the procedure described previously (29) and purified on Dowex-1 × 2 as described above.
Transcriptional Complexes
Defined TECs were obtained by RNAP “walking” on the 5′-biotinylated T7A1 promoter fragment immobilized on neutravidin-coated agarose (Pierce). The 370-bp DNA promoter fragment with 5′-biotin tag was generated by PCR amplification from promoter-containing pTZ19 plasmid. The open promoter complex was obtained by mixing 1 μl of neutravidin-agarose suspension with immobilized T7A1 promoter with 1 pmol of RNAP in 20 μl of transcription buffer (TB) containing 50 mm Tris-HCl, pH 7.9, 100 mm KCl, 10 mm MgCl2, 1mm 2-mercaptoethanol. The mixture was incubated for 10 min at 37 °C and washed with TB (four times by 1 ml). To prepare TEC-13A with labeled penultimate residue, 10 μl of “start” mixture (10 μm ApUpC RNA primer and ATP, GTP 13 μm each) was added to obtain first TEC-11A stalled for the absence of the next substrate. After 2.5 min of incubation at 37 °C, the suspension was washed with cold TB (six times by 1 ml) and 1 μl of [α-32P]CTP (3000 Ci/mmol) was added to the reaction mixture. After 15 min of incubation at 0 °C, the beads were washed with cold TB without MgCl2, followed by addition of ATP and MgCl2 (to final concentrations of 30 μm and 1 mm, respectively) and incubation for 15 min at 0 °C. To remove the excess of ATP the mixture was washed by TB (four times by 1 ml). In the case of the internal labeling of RNA, the washed beads containing open complex were supplemented with 10 μl of the start mixture (see above) containing 1 μl of [α-32P]ATP (3000 Ci/mmol) and 13 μm GTP followed by incubation (2.5 min at 37 °C) to obtain TEC-11A. For characterization of the exonuclease reaction, the [α-32P]NTP was introduced into 3′-terminal RNA position of TEC-13A and TEC-21U.
Incorporation of Modified Nucleotides in TECs
ATPαS and CTPαS were introduced in TEC-13A and TEC-14C at 300 μm substrate concentration upon incubation of the relevant TEC at 0 °C for 30 min. TEC-VI of Fig. 7A was obtained by incubation of TEC-11A with [α-32P]dCTP (3000 Ci/mmol, 3 μm) followed by elongation with 50 μm ATP for 20 min at 0 °C, whereas elongation of TEC-12dC with 150 μm ATP αCH3 (20 min at 37 °C) was used to generate TEC-VII of Fig. 7A. TEC-III was synthesized by elongation of natural TEC-11A with 150 μm ATP αCH3 for 20 min at 0 °C. Incubation of TEC-11A internally labeled at adenylate residues (see above) with 300 μm CTP-3′-methyl phosphate for 20 min at 37 °C yielded TEC-V of Fig. 7. The last four TECs were obtained in the presence of 1.5 mm MnCl2 and 10 mm MgCl2 to facilitate modified substrate incorporation. The modified TECs were washed with cold TB (without MnCl2 or MgCl2) and used in further experiments.
FIGURE 7.
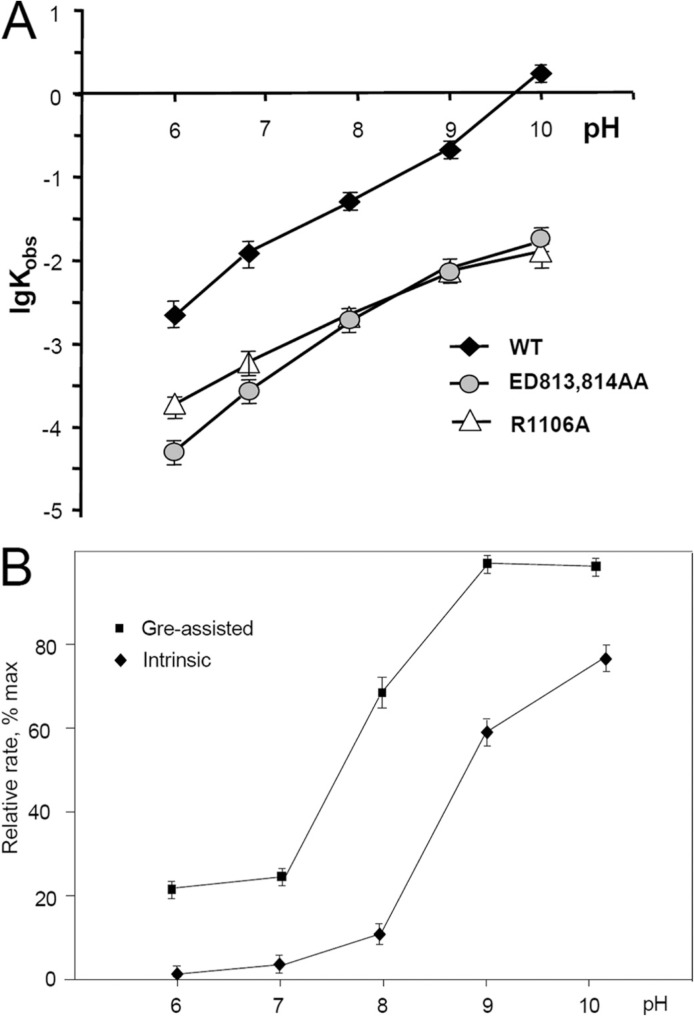
Dependence of the endonucleolytic transcript cleavage in TEC-13A on pH for WT and mutant enzymes in the presence of 10 m;1m Mg2+. A, intrinsic cleavage reaction. B, relative cleavage rate for intrinsic (with R1106A enzyme) and Gre-assisted reaction.
Intrinsic and Gre-assisted RNA Cleavage in TEC
For endonuclease cleavage, the sample of stalled 32P-labeled TEC in 10 μl of TB without MgCl2 was diluted by 10 μl of TB, containing various MgCl2 or MnCl2 concentrations, and the mixture was incubated at 21 °C. GreA- and GreB-induced cleavage was performed using an equimolar ratio of a factor to TEC. The cleavage was stopped by adding 1 volume of the loading buffer (7 m urea, 20 mm EDTA, bromphenol blue and xylene cyanol, 0.25% each), and reaction products were resolved on a 20% PAGE (20:3 acrylamide/N,N′-methylenebisacrylamide ratio). In the case of TECs with ATP αCH3 and CTP-3′-methylphosphonate, the reaction products were resolved on 30% PAGE (20:3 acrylamide/N,N′-methylenebisacrylamide ratio, 6 m urea, 1× TBE). The Arrhenius coefficient of 5.6 for endonuclease cleavage reaction was determined by measuring the rate of the reaction at 21 and 37 °C. Details of reaction conditions with PPi and rNTP/dNTP are presented in the figures. For testing the pH dependence of endonuclease cleavage, the stalled 32P-labeled TEC was washed (four times by 1 ml) with cold buffer (1 mm Tris-HCl, pH 7.9, 100 mm KCl2, and 1 mm 2-mercaptoethanol), and then 10 μl of the samples were mixed with the equal volumes of various pH buffers. The final concentrations of the buffers were as follows: 50 mm Pipes-HCl, pH 6.0, 50 mm Tris-HCl, pH 7.0–9.0, 50 mm CAPS-NaOH, pH 10.0. Other components were as in TB.
Hydroxyl Radical RNA Cleavage in TECs Mediated by Active Center-bound Fe2+
A stalled TEC in 10 μl of TB without MgCl2 was supplemented with 20 μm Fe(NH4)2(SO4)2 and 10 mm DTT. The reaction mixture was left at 21 °C. In 10 min, the reaction was stopped by adding of 1 volume of the loading buffer (7 m urea, 20 mm EDTA, bromphenol blue and xylene cyanol 0.25% each) and analyzed by PAGE.
Site-directed Mutagenesis of RNA Polymerase
Mutational substitutions in RNAP were introduced using the QuikChange mutagenesis kit following the supplier’s protocol.
RESULTS
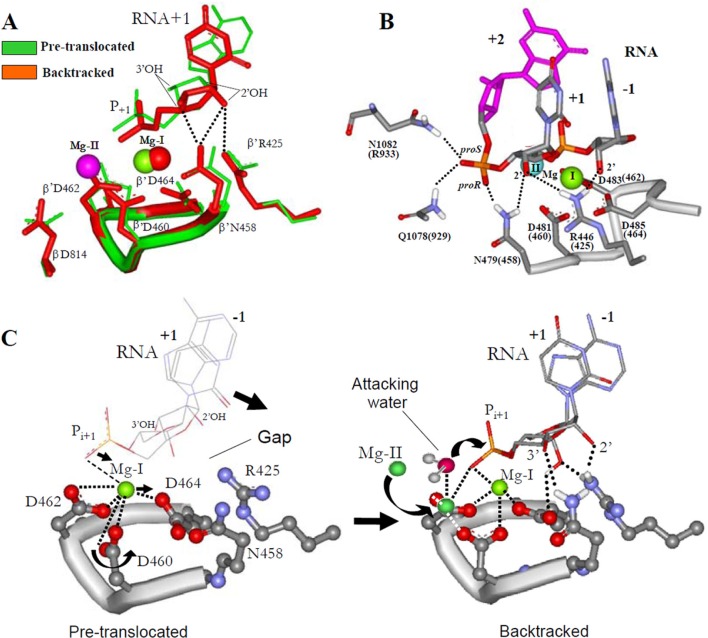
Structural Analysis of RNAP Suggests the Mechanism for Active Center Tuning in the Reaction of Hydrolytic RNA Cleavage—We analyzed the 14 available RNAP structures and found that in most cases the coordination geometry of Mg-I corresponds to the inactive state to which Mg-II cannot bind (data not shown); but in two of the structures the carboxylate residues are in the bridging orientation allowing coordination of Mg-II (which was modeled onto the structures as shown in Fig. 1C, right panel). Of particular note is that both of these exceptions represent backtracked RNAP complexes (24, 25). In Fig. 3A, we compare one of these, the backtracked complex of yeast pol II (Protein Data Bank 3GTG), with the closely related pretranslocated complex of that enzyme (PBD 1I6H (30)). In the backtracked complex, the contacts of +1 ribose residue with the active center side chains are established, and carboxylate residues Asp-460 and Asp-462 are in the active bridging orientation (Fig. 3, A and C, right panel). These ribose contacts are not established in pretranslocated complex of yeast pol II, in which the active center carboxylates are in inactive, nonbridging orientations (Fig. 3, A and C, left panel). This comparison points to a possible causal relationship between establishment of the ribose recognition contacts and catalytic orientation of bridging carboxylate residues. As seen from Fig. 3B, the disengaged i + 2 residue of 3′-RNA forms additional contacts with the active center that facilitate +1 ribose recognition interactions. In particular, the phosphate group of the +2 disengaged RNA residue forms hydrogen bonds with Rpb1 N479 (β′ Asn-458 in E. coli) and Rpb1 Q1078 (E. coli β′ Gln-929) side chains (24). Another interaction in yeast pol II includes hydrogen bonding with Rpb1 Asn-1082 (E. coli β′ Arg-933), which suggests a salt bridge with the phosphate in the bacterial enzyme (as determined from homology modeling). These additional interactions apparently bring the 2′-hydroxyl group of +1 RNA residue to hydrogen bonding distance with the guanidinium group of Rpb1 Arg-446 (E. coli β′ Arg-425), which has been implicated in catalysis of RNA synthesis (15). This movement of the +1 RNA residue associated with establishment of the recognition contacts (Fig. 3C) forces the shift of Mg-I, because it is tethered to the moving 5′-phosphodiester group of the +1 ribose residue through coordination. Movement of Mg-I is accompanied by re-orientation of two bridging carboxylates that remain bound to Mg-I through coordination but acquire free valences for binding of Mg-II, which is required for catalysis of subsequent hydrolytic reaction (Fig. 3C, right panel). We named the above rearrangement “active center tuning” because it sets the active center for catalysis. Next, we set up to find experimental support to ACT mechanism.
FIGURE 3.
ACT mechanism. A, structural alignment of pretranslocated (1I6H) and backtracked (3GTG) yeast pol II TEC. +2 RNA residue in backtracked TEC is not shown for the purpose of clarity. Mg-II (purple) was modeled as suggested by Sn2 reaction mechanism. Numbering of active center residues is as in E. coli. Dotted lines indicate hydrogen bonds. B, RNA contacts in the active center of backtracked yeast pol II TEC. Residues are numbered as both in yeast Rpb1 pol II and E. coli RNAP β′ subunits. C, dynamic model for ACT in RNA hydrolytic cleavage. Establishing of recognition contacts of RNA residue in the active center is accompanied by displacement of Mg-I and re-orientation of bridging carboxylates enabling Mg-II docking followed by coordination of the water and hydrolytic attack. Principal RNA contacts inducing ACT are indicated.
ACT Explains High Efficiency of Endonuclease Transcript Cleavage Compared with Exonuclease Reaction—The active center catalytic state apparently seen in backtracked TEC predicts that the endonuclease reaction (that proceeds in the backtracked complex) could be more efficient compared with exonuclease reaction occurring in a pretranslocated complex in which the active center is not tuned. For quantitative characterization of exo- and endonuclease hydrolytic activities, we examined the status of RNAP translocational equilibrium in various TECs assembled on the T7 A1 promoter using RNA cleavage by hydroxyl radicals generated by active center coordinated Fe2+, which substitutes Mg-I (6, 13, 21, 31, 32). The cleavage occurs between the RNA residues occupying the i and i + 1 site as shown in Fig. 4, A and B. Notably, the presence of 3′-terminal phosphate group increases electrophoretic mobility of the cleavage products (compare the size markers on the right and left of the Fig. 4A) because of two extra negative charges. As seen from the cleavage pattern in TEC-13A, RNA truncation to 12 and 11 nucleotides occurs with equal efficiency (Fig. 4, A and B) indicating that pretranslocated and backtracked by 1-nucleotide states are equally represented, thereby creating equal opportunities for exo- and endonuclease reactions. However, the efficiency of endonuclease reaction in this TEC is 50 times higher than that of exonuclease cleavage (which releases 3′-terminal pA residue) as determined from quantitation of corresponding reaction products (Fig. 4C, lane 4). In accordance with the prediction of the ACT mechanism, the bulk of this effect was due to enhanced Mg-II retention in the backtracked TEC (Table 1) as a result of freeing of the active center carboxylate coordination valences promoted by +2 RNA residue (as shown in Figs. 1C and 3C). High Me-II association with backtracked TECs was also seen for endonuclease cleavage of TEC-14C, TEC-16G, and TEC-22A assembled at the same promoter (data not shown) as in TEC formed of other DNA templates (33).
FIGURE 4.
Endo-, exonuclease, and pyrophosphorolytic RNA cleavage reactions of RNAP in TEC-13A. A, determination of backtracking equilibrium in TEC-12–16 by Fe2+-mediated hydroxyl radical RNA cleavage. Horizontal bars indicate the relative intensity of cleavage products. B, Fe2+-mediated and hydrolytic RNA cleavage sites in TEC-12–16. The arrows mark the cleavage sites. i and i +1 indicate the position of the active center sub-sites in TECs. C, PAGE analysis of RNA in natural and modified TEC-13 after incubation in different conditions. Products of endo- and exonuclease cleavage are indicated. Lane 1, original TEC-13A; lane 2, after elongation with CTP to TEC-14C; lane 3, after pyrophosphorolysis; lane 4, after incubation at 37 °C for 15 min; lanes 5 and 6, after incubation at 21 °C for 5 and 0.25 min, respectively; lanes 7 and 8, after incubation for 0.25 min with GreA and GreB, respectively; lanes 9 and 10 after incubation of deoxy-substituted TEC-13dCpA (structure VI of Fig. 5) with pyrophosphate. D, effect of 7-deazaA substitution in TEC-13pApTu (structure IV of Fig. 5) on endonuclease cleavage efficiency.
TABLE 1.
Reaction rates and Mg-II dissociation constants for hydrolytic RNA cleavage in TECs with nonmodified RNA
| TEC | Intrinsic cleavage | Factor-assisted cleavage | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Exonuclease | Endonuclease | GreA,B | GreA | GreB | ||||||
|
|
|
|
|
|
||||||
| Kd Mg2+ | kapp, min−1 at 10 mm Mg2+ | Kd Mg2+ | kapp, min−1 at 10 mm Mg2+ | Relative cleavage rate | Kd Mg2+ | kapp | Relative cleavage rate | kapp | Relative cleavage rate | |
| mm | mm | mm | min−1 | min−1 | ||||||
| TEC-13A (pCpAa) | NAb | NA | >100 (9–11c) | 0.042 | 1 | 0.3−0.5 | 1.5 | 1 | 3.1 | 1 |
| TEC-12C (pGpApCa) | NA | NA | >100 (8–12c) | 2.3 × 10−3 | 0.055 | 0.3–0.5 | 0.04 | 0.027 | 0.09 | 0.03 |
| TEC-13A (pAa) | NDd | 0.8 × 10−3 | NA | NA | ND | <10−3 | <6.7 × 10−4 | <10−3 | <3.2 × 10−4 | |
3′-terminal cleavage product.
NA means not applicable.
Done at pH 10.0.
ND means not detectable.
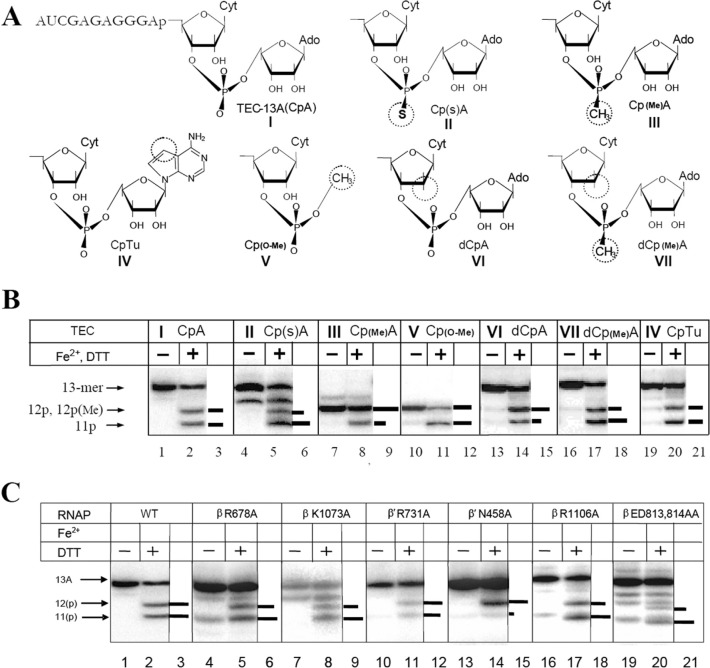
Effect of RNA Modifications on Transcript Cleavage and Pyrophosphorolysis Is Consistent with ACT—To explore further the ACT model predictions, we determined how various RNA substitutions (shown in Fig. 5A) affect the endonuclease reaction as summarized in Tables 1 and 2. The substitution of the R oxygen of phosphate group, which participates in tuning through hydrogen bonding with β′ Asn-458 of the active center (Figs. 2B and 3B) by bulkier and less electronegative sulfur caused a 6-fold reduction in endonuclease cleavage rate (Table 2). Methyl substitution of the same oxygen resulted in a 40-fold cleavage rate reduction evidently due to complete loss of the group’s hydrogen bonding (Table 2). Deletion of the entire nucleoside part of the disengaged +2 RNA residue (while preserving +2 phosphate) decreased the reaction rate by 20-fold. However, the most severe effect, a 300-fold rate reduction, was observed by 2′-deoxy substitution in the +1 RNA residue, which according to the model eliminates two hydrogen bonds with β′ Asn-458 and β′ Arg-425, involved in the tuning. This points to a crucial contribution of the 2′-hydroxyl group interactions to ACT, whereas the other RNA contacts involved in this mechanism play an auxiliary role. The implications of this finding in RNAP catalytic mechanism are presented under “Discussion.”
FIGURE 5.
Effect of structural modifications in RNA of TEC-13A and active center mutations of TEC translocational equilibrium (as revealed by hydroxyl radical transcript cleavage induced by active center bound Fe2+). A, structure of 3′-terminal dinucleotide RNA segment in tested TECs. Modifications are circled. B, Fe2+-induced cleavage of RNA in TEC. The TECs are numbered as in A. Horizontal bars represent the relative intensity of the cleavage products. C, effect of active center mutations on TEC translocational equilibrium.
TABLE 2.
Reaction rates and Mg-II dissociation constants for hydrolytic RNA cleavage in TECs with modified RNA
| TEC | Intrinsic cleavage | Factor-assisted cleavage | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Exonuclease | Endonuclease | GreA,B | GreA | GreB | ||||||
|
|
|
|
|
|
||||||
| Kd Mg2+ | kapp min−1 at mm Mg2+ | Kd Mg2+ | kapp, min−1 at mm Mg2+ | Relative cleavage rate | Kd Mg2+ | kapp | Relative cleavage ratea | kapp | Relative cleavage ratea | |
| mm | mm | mm | min−1 | min−1 | ||||||
| IIb TEC-13p(S)A (pCp(S)A)c | NDd | ND | >100 | 7 × 10−3 | 0.17 | 0.3–0.5 | 1.3 | 0.87 | 1.3 | 0.42 |
| III TEC-13p(Me)A (pCp(Me)A) | ND | ND | >100 | 10−3 | 0.024 | 0.3–0.5 | 0.09 | 0.07 | 0.09 | 0.03 |
| V TEC-12pCp(OMe) (pCp(OMe)) | ND | ND | >100 | 2.2 × 10−3 | 0.053 | 0.3–0.5 | 1.4 | 0.93 | 0.4 | 0.13 |
| VI TEC-13pdCpA) (pdCpA) | ND | ND | >100 | 1.3 × 10−4 | 3.0 × 10−3 | ND | 0.01 | 6.7 × 10−3 | 0.01 | 3.2 × 10−3 |
Data are as compared with nonmodified RNA in TEC-13.
Numbering is as in Fig. 5A.
3′-Terminal cleavage product.
ND mean not detectable.
As expected, the above deoxy-substitution also greatly affected pyrophosphorolysis (Fig. 4, lanes 9 and 10). As estimated, the substitution caused a 100–400-fold drop in pyrophosphorolysis rate, supporting the involvement of ACT in this reaction.
Importantly, the effect of the RNA substitutions was not due to the shift in backtracking equilibrium (see Fig. 5B). The above observations are in accord with our expectations from the ACT model. As suggested by x-ray structure of yeast pol II backtracked TEC, the other residues of disengaged 3′-RNA segment do not make interactions with RNAP. In agreement with this observation, introduction of phosphorothioate substitution in the +3 position of RNA did not affect RNA cleavage in backtracked TECs (data not shown).
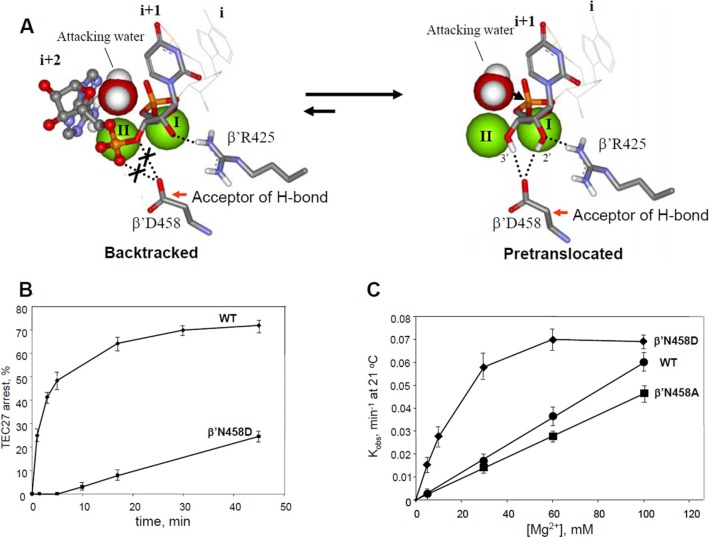
Effect of β′N458D Mutation Supports ACT Mechanism—Important evidence for ACT is provided by the properties of a β′ N458D substitution (Fig. 6). According to the model, it was expected that this mutation would suppress backtracking by reinforcing the interactions of the 3′-terminal RNA residue with recognition protein side chains that trigger ACT. In detail, the β′ N458D substitution introduces an ionized oxy-group in place of a neutral amide that is expected to affect crucially the hydrogen bonding network in TEC. An Asp residue cannot stabilize the backtracked state because it cannot hydrogen bond to a phosphate group (Fig. 6A) of the +2 RNA nucleotide the way the Asn residue in WT backtracked TEC does (Fig. 3B). Instead, an Asp residue can form strong hydrogen bonds simultaneously with the 2′- and 3′-hydroxyl groups of the +1 3′-terminal RNA residue in the TEC pretranslocated state (Fig. 6B), because the oxo-group of Asp is a better hydrogen bond acceptor than the amide of Asn. Both of the above factors should stabilize the pretranslocated state. Indeed, the substitution strongly suppressed backtracking in TEC-27 (Fig. 6C) as seen from the reduced rate for accumulation of arrested fraction of the TEC. As a result, this mutant displayed only the exonuclease type of activity typical of pretranslocated TEC. The most striking effect of the substitution was observed in the exonuclease reaction, which compared with WT had a higher reaction rate and a lower dissociation constant for Mg-II (10 mm versus >100 mm) (Fig. 6C) as expected from the ACT model. Remarkably, control β′ N458A substitution displayed an even slightly opposite effect on exonuclease cleavage compared with the WT enzyme. Alternatively, the N458D substitution could increase overall Mg2+ binding by introduction of a residue capable of additional coordination of Mg-II. However, considering the large distance (9.2 Å) from Mg-II catalytic position to N458D carboxylate group, this hypothesis looks unlikely. Therefore, the β′ N458D substitution properties also support the ACT mechanism predictions.
FIGURE 6.
Effect of β[prime]N458D substitution on hydrolytic RNA cleavage. A, inability of β′ Asp-458 residue of the active center to hydrogen bond to the phosphate of 3β′-disengaged +2 RNA residue destabilizes backtracked state and shifts the equilibrium to pretranslocated state, which is stabilized by strong hydrogen bonding of terminal RNA residue to β′ Asp-458. This enhanced ribose contacts promote ACT that increases the Mg-II retention and the reaction rate. B, time course for arrested state accumulation for WT and the mutant enzyme in TEC-27U. The complex was incubated at 37 °C in the absence of Mg2+ for the indicated periods of time followed by elongation in the presence of Mg2+ and four NTP substrates. C, dependence of the hydrolytic exonuclease cleavage rate on Mg2+ concentration for WT and mutant enzymes in TEC-21.
Adenine Residue in +2 Position of Disengaged RNA Segment in Backtracked TEC Is Strongly Preferred in Endonuclease Cleavage—To further investigate the mechanism of endonuclease reaction in E. coli TEC, we explored the sensitivity of endonuclease hydrolysis to the size and sequence of 3′-disengaged RNA segment in backtracked TEC. For these experiment, we used the same set of TECs assembled on the T7 A1 promoter containing RNAs 12–16 nucleotides long (Fig. 4). As seen from Fig. 4A, hydroxyl radicals generated by active center bound Fe2+ cut the 3′-terminal nucleoside residue in TEC-12C, indicating that this complex is pretranslocated. TEC-13A was truncated by one and by dinucleotide segments with the hydroxyl radicals suggesting that this TEC exists in pretranslocated and backtracked by one nucleotide states. RNA cleavage patterns in TEC-14C and TEC-16G revealed the states with different degrees of backtracking (Fig. 4, A and B) along with the pretranslocated state. Surprisingly, despite the variety of backtracked states randomly populated in the tested TECs, in all these TECs the cleavage occurred between A11 and C12 of the RNA transcript (data not shown), pointing to sequence preference of endonuclease reaction. The cleavage preference is not due to the identity of RNA residues occupying i and i + 1 sites of the active center, because the cleavage between A13 and C14 in TEC-16G (allowed by backtracking equilibrium) is also expected should the above be the case. Another possible specificity determinant could be a residue in the register +2 (first 3′-unpaired single strand RNA residue of backtracked TEC). To test this proposal, we introduced tubercidin residue (containing a –CH 3 segment instead of –N = at position 7 of the purine base as shown in Fig. 5A, structure IV) instead of adenosine in position 13 of TEC-13A. Importantly, this substitution does not change the base pairing interactions in DNA scaffold of the TEC, thereby excluding the influence of RNA/DNA sequence on RNAP translocational equilibrium and the cleavage. In agreement with this expectation, the efficiency of endonuclease cleavage in the modified TEC was significantly (∼6–7-fold) reduced (Fig. 4D). As follows from Fe2+ affinity cleavage (Fig. 5B, lanes 19–21), this was not due to the change in backtracking status of the TEC. The above sequence preference of endonuclease cleavage is also observed in the other TECs in which adenosine residue is the first in disengaged RNA segment. The examples are TEC-12C (Table 1), arrested TEC-21U and TEC-27U (cleavage between A11 and C12 (3)), as well as TEC-22A, in which the cleavage occurs between A20 and U21 (data not shown).
Modeling of Endonuclease Reaction for E. coli RNAP—Strong adenine preference (>30-fold) of the cleavage reaction described above as well as our previous cross-linking data (34) could not be explained by x-ray structure of yeast pol II backtracked TEC, suggesting different orientation of +2 RNA residue in E. coli TEC.
We modified the x-ray model (3GTG) by placing a base of +2 RNA residue in the active center cleft in the vicinity of the residues βArg-678, βArg-1107, and β′ Asp-460–Asp-464 segment (as suggested by cross-linking (34)), while preserving the orientation of +2 phosphate seen in x-ray structure as shown in Fig. 2, A and B. This was achieved through the rotation of +2 RNA residue around P–5′O and 5′O–5′C bonds using the above cross-linking constrains for E. coli RNAP as well as those imposed by the aperture of the active center (Fig. 2A). This allowed unambiguous placing of the nucleoside residue in the narrow pocket at the interface of β and β′ subunits formed by evolutionary invariant residues βLys-1073, βArg-678, βArg-1106, βAsp-814, and the catalytic β′ DDD-loop of the active center (Fig. 2, A and B), which we named B site (for base binding). In this arrangement, the 6-amino group of adenine is in a position to form a hydrogen bond with βAsp-814, whereas N-7 and N-1 of the base can H-bond to side chains of βLys-1073 and βArg-1106, respectively (Fig. 2B). Remarkably, the endocyclic oxygen of the +2 RNA residue ribose bound in the B site is within hydrogen bonding distance to the attacking water, which can position the water for the attack and therefore contribute to catalysis. Notably (Fig. 2B), only adenine can establish a full set of interactions with active center side chains, which explains the preference of the base in endonuclease reaction. The placing of the residue in the B site also explains the inability of E. coli enzyme to perform endopyrophosphorolytic RNA cleavage and the absence of the stimulating effect of noncognate NTP (3) on the reaction through steric blockage of the PPi-binding site and E site by the base of the +2 RNA residue(Fig. 2C).
Effect of the Substitutions in B Site on Transcript Cleavage Activity as Explained by the Model—Further support to the model is provided by the effect of the substitutions of the evolutionarily conserved side chains surrounding the +2 RNA residue on endonuclease and exonuclease reactions (Fig. 2,A and B). Surprisingly, we observed dramatic differences in response of these reactions to the same mutations (Table 3). Indeed, double substitution βE813A/D814A had no effect on exonuclease cleavage, but it reduced the rate of endo-reaction by a factor 20. Mutations βK1073A, βR1106A, and βR678A stimulated exonuclease cleavage by factors 4.2, 55, and 95, respectively (Table 3), but strong inhibition (10–20 times) was seen in endonuclease cleavage for the first two mutations, although the last substitution had no effect on the reaction. β′ N458A and β′ R731A substitutions marginally (1.2–4-fold) affected the rate of both reactions (Table 3).
TABLE 3.
Effect of active center mutations on exo- and endonuclease activity
| Enzyme | Exonuclease (TEC-21) | Endonuclease (TEC-13) | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| kapp, min−1 at 10 mm Mg2+ | Relative cleavage rate | Mg-II Kd, mm at pH 8.0 | kapp, min−1 at 10 mm Mg2+ | Relative cleavage rate | Mg-II Kd, mm at pH 8.0 | |
| WT | 5.5 × 10−3 | 1.0 | >100 | 40 × 10−3 | 1.0 | >60 |
| E813A/D814A | 5.5 × 10−3 | 1.0 | >100 | 1.8 × 10−3 | 0.047 | >60 |
| R1106A | 300 × 10−3 | 55 | 10 | 2.2 × 10−3 | 0.054 | 1 |
| N458A | 4.5 × 10−3 | 0.82 | >100 | 10 × 10−3 | 0.25 | >60 |
| K1073A | 14 × 10−3 | 4.2 | >100 | 3.5 × 10−3 | 0.09 | >60 |
| R678A | 520 × 10−3 | 95 | >100 | 42 × 10−3 | 1.05 | >60 |
| R731A | 4.5 × 10−3 | 0.82 | >100 | 19.5 × 10−3 | 0.5 | 15 |
| N458D | 30 × 103 | 5.5 | 15 | Absent | ||
These specific effects of the substitutions on endonuclease RNA cleavage can be rationally explained by the suggested model. In particular, mutation of βAsp-814, βLys-1073, and βArg-1106 resulting in a significant drop of endonuclease activity should weaken hydrogen bonding with adenine of +2 RNA residues in the B site (Fig. 2B), affecting the residue orientation crucial for positioning of the attacking water. In our previous studies (3), stimulatory effect of βArg-1106 substitution on exonuclease cleavage was attributed to increased retention of Mg-II in the mutant due to additional coordination by βAsp-814 carboxylate (which is not available for coordination in WT enzyme due to engagement in salt bridge with βArg-1106 as shown in Fig. 2B). The same effect of the mutation on Mg-II retention was seen in endonuclease reaction (Table 3). However, this stimulating effect was apparently negated by elimination of water positioning through disruption of adenosine interactions in the B site caused by mutation. As a result, the substitution increased the competing exonuclease pathway in TEC-13 so that the rates of both hydrolytic reactions became comparable (data not shown). The lower Mg-II dissociation constant for the mutant in endonuclease reaction (1 mm) compared with exo-cleavage (10 mm) can be explained by the additional synergistic effect of ACT.
Another substitution that increased Mg-II retention in endonuclease reaction was β′ R731A, whose influence can be similar to that of βR1106A (see above). Smaller magnitude of the effect can be explained by the larger distance of β′ Arg-731 residue from β′ Asp-814 (4.5–5.6 Å in different structures). The other example of striking difference is represented by the effect of βR678A substitution. The enhanced activity of the mutation in the exo-cleavage (Table 3) does not allow simple explanation, because the substitution did not change Mg-II retention. One of the possible effects could be Mg-II re-positioning to a more optimal location due to elimination of electrostatic repulsion between the metal and positively charged Arg side chain. In the model for endonuclease reaction (Fig. 2, A and B), the βArg-678 residue is completely shielded from the active center by the base of +2 RNA residue, which could reduce the influence of the substitution on Mg-II re-positioning.
Transitions in the active center caused by pH have been previously demonstrated for exo- and endonuclease reactions (2, 3). It is envisioned that deprotonation of the attacking water at elevated pH should generally stimulate nuclease activity. Such stimulation should plateau out upon the pH approaching system’s pKa value. Instead, like in the case of exonuclease reaction(3), the pH dependence curve for wild-type enzyme in endonuclease cleavage displayed the abrupt upturn in the range 9–10 (Fig. 7A), suggesting that the additional pH-dependent factor contributes to catalysis. Similarly to exonuclease hydrolysis (3), the upturn can be explained by deprotonation of βArg-1106, which destroys its salt bridge with βAsp-814, enabling additional Mg-II coordination by the latter (see above). This is supported by the upturn absence for βE813A/D814A and βR1106A mutants in endonuclease reaction. Indeed, the first mutation eliminates additional Mg-II coordination by βAsp-814, thus making Mg-II binding pH-independent as does the second substitution by eliminating the salt bridge.
As seen from Fe2+-mediated hydroxyl radical RNA cleavage in TEC-13A, most of the tested mutations did not change significantly the pretranslocated-to-backtracked state ratio (Fig. 5C). Notably, β′ N458A substitution increased the ratio to 4:1 from 1:1 in WT TEC, suggesting an important contribution of the residue to stabilization of the backtracked state. The change in the equilibrium can be explained by loss of the hydrogen bonding of the side chain with the +2 RNA residue phosphate in backtracked state, whereas in pretranslocated state the β′ Asn-458 residue does not contact RNA (30).Substitution β′ R933S suppressed backtracking, re-directing the main RNA cleavage site in TEC-26A from between A11 and C12 to between A20 and U21 (data not shown). This change can be explained by stabilization of the backtracked state in wild-type RNAP through hydrogen bonding and the salt bridge of β′ Arg-933 side chain with the phosphate of the +2 RNA residue as suggested by homology modeling (Figs. 2B and 3B).
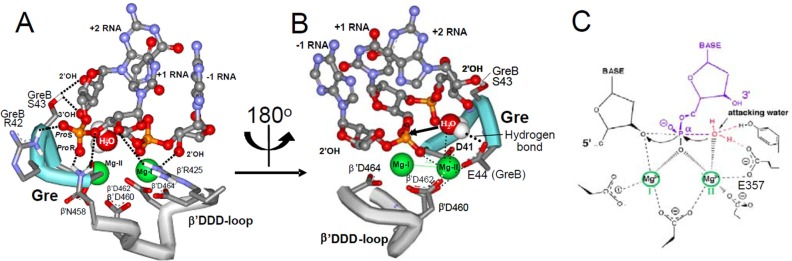
Molecular Modeling of Gre-assisted Transcript Cleavage Reaction—Gre-assisted endonuclease RNA hydrolysis is believed to provide co-transcriptional proofreading in the bacterial cell, because this reaction is much more efficient compared with intrinsic cleavage. For the absence of x-ray structure of TEC with Gre factors and to explain our results, we built a working model, which compiles high resolution structures of T. aquaticus RNAP and GreB factor modeled into the EM structure for the GreB-T. aquaticus RNAP complex (19), as well as the structure of nucleic acid scaffold and active center DDD loop of yeast pol II elongation complex (24), in which factor-assisted reaction proceeds. The modeling was accomplished by: i) pasting of the DNA/RNA scaffold of backtracked transcription complex of yeast Pol-II onto a high resolution model of Gre/Taq RNAP transcription complex derived from low-resolution EM image (19); ii) placing of β′ DDD-loop segment of yeast Pol-II backtracked TEC (PDB 3GTG) into Taq RNAP structure of Gre complex (19); iii) fitting of catalytic Mg-II (not seen in yeast Pol-II x-ray image) and the attacking water into the active center using constraints of Sn2 hydrolytic mechanism, and iv) adjusting of the resulting model through GreB moving towards the active center by ∼0.5 nm to enable coordination of Mg-II by two carboxylate residues of Gre tip as suggested in previous studies (6, 19, 21).
For the model shown in Fig. 8, docking of the Gre factor into the RNAP secondary channel caused minor clashes with the RNAP protein, suggesting structural adjustment in the complex (19), also seen in the co-crystal of Gfh1, Gre-like inhibitor with T. thermophilus RNAP (35). Remarkably, no clashes were observed at the active center. In the model Gre tip contacts the β′ DDD-loop of the active center (residues 458–464), β′ F-bridge residues 781–785, β′ 729–736 segment (GARG motif), β regions 677–681 (NRAP motif), and 1104–1107 (PSRM motif). Such placement of the factor is supported by biochemical evidence. In particular, cross-linking data reflect ∼1 nm distance of Gre factor tip from the RNA base in i + 1 site of the active center (6) consistent with the model. Yet binding of the factor to TEC causes protection of all of the above listed active center sites against hydroxyl radicals generated by active center coordinated Fe2+ (21). The important conclusion from the Gre docking model was displacement of the nucleoside part of disengaged +2 RNA residue from the B site due to steric clashes of the residue with the Glu-44 side chain of the Gre tip that has to coordinate Mg-II. This prediction is in accord with the loss of adenine specificity of Gre-stimulated reaction observed in nonstimulated cleavage (6, 36). The same conclusion follows from the absence of the effect of +2 RNA base elimination on GreA-mediated cleavage (Table 2, TEC-V). Remarkably, the spatial arrangement of the +2 RNA phosphate residue catalytically competent in intrinsic cleavage reaction is allowed in the Gre complex, which is consistent with the observed effects of ACT (mediated through the phosphate residue) on Gre-stimulated cleavage (see below). However, docking of Gre factor changes the environment of this phosphate residue (Fig. 8). Specifically, contacts of the phosphate with β′ Asn-458 are preserved, although suggested interactions with β′ Gln-929 and β′ Arg-933 are lost due to displacement by Gre. Instead, hydroxyl groups of β′ Thr-928 of the G-helix and β′ Thr-786 of the F-bridge move to hydrogen bonding distance to S oxygen of the phosphate residue. GreB Arg-42 residue comes to close proximity to the phosphate allowing the salt bridge, thereby mimicking β′ Arg-933. These extensive contacts suggest functional importance of the interactions and are consistent with the absolute requirement of the phosphate group of +2 RNA residue for Gre-assisted cleavage.
FIGURE 8.
Model for Gre/RNAP TEC. A and B, different views of the complex. The Gre tip is shown in blue. Principal contacts of Gre side chains with RNA, Mg-II, and attacking water are indicated. C, scheme for exonuclease active center of DNA polymerase I.s
Possible Involvement of ACT in Gre-assisted Transcript Cleavage—To determine whether the same ACT mechanism operates in the Gre-mediated reaction, we examined the effect of the RNA modifications described above on factor-assisted cleavage, which was generally the same as for nonassisted cleavage (Tables 1 and 2). Importantly, as in the case of nonassisted reaction, removal of the entire disengaged +2 RNA residue reducing TEC-13A to TEC-12C (which does not backtrack), abolished the cleavage, underscoring the crucial role of disengaged +2 RNA residues in this process. Also, as in the intrinsic reaction, the 2′ deoxy substitution in +1 RNA residue of TEC-13A decreased the rate of the reaction 100- and 300-fold for GreA and GreB, respectively (Tables 1 and 2), strongly suggesting that the same ACT mechanism also operates in Gre-assisted cleavage.
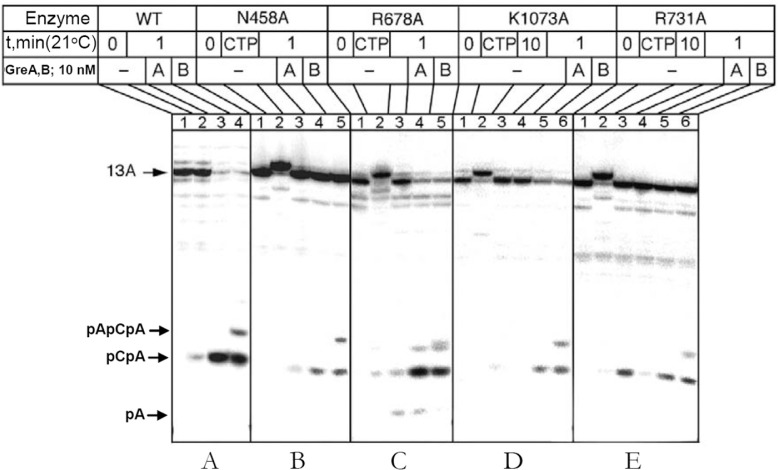
Effects of RNAP Active Center Mutations on Gre-stimulated Cleavage as Explained by the Model—Sensitivity of the intrinsic endonuclease reaction in TEC to the substitutions of the side chains adjacent to catalytic DDD-loop of the active center (Table 3) was also observed in the Gre-assisted reaction (Fig. 9). In contrast to intrinsic reaction, substitution of β′ N458A had a dramatic effect (∼20–30-fold) on both GreA- and GreB-assisted cleavage (Fig. 9B), reflecting more significant involvement of the residue in factor-mediated catalysis. Our model explains this effect by tight contact of the Gre tip with β′ Asn-458, which may act as a platform for positioning catalytic Gre Asp-41 and Glu-44 carboxylates that hold Mg-II. The substitution of β′ Asn-458 by a smaller alanine side chain can shift Gre tip and mis-align the carboxylates as can it affect the ACT due to the loss of the side chain interactions with +1 and +2 RNA residues (Fig. 3B). Another mutation that strongly affected Gre reaction was β′ R731A (Fig. 9E). Specific effects of this substitution on factor-depended cleavage can be similarly explained by mis-alignment of the factor’s tip in the active center. Indeed, in our model β′ Arg-731 is within the salt bridging distance to Asp-47 of GreB (Glu-47 in GreA), which can be involved in tip positioning. In support of this suggestion, E47K substitution (expected to affect Gre-TEC interaction through change of electrostatic attraction to repulsion with β′ Arg-731) caused dramatic drop in GreA activity (21). Mutations βK1073A (Fig. 9D) and βR1106A (data not shown) had a marginal effect on the Gre-assisted reaction, reflecting the loss of the residues contacts with the displaced +2 RNA adenine base, which are crucial for intrinsic cleavage.
FIGURE 9.
Effect of active center mutations on Gre-assisted hydrolytic RNA cleavage. Principal cleavage products are indicated.
Effect of pH on Gre-assisted RNA Cleavage Reveals General Base Catalysis Predicted by the Model—An additional factor involved in catalysis of hydrolytic RNA cleavage could be activation of the attacking water. Indeed, in the exonuclease active center of DNA polymerase (DNAP) the activation is achieved through H-bonding of the water with the carboxylate group of Glu-357 (37), which also coordinates Mg-II (Fig. 8C). As seen from our model (Fig. 8B), Glu-44 of Gre could assume the same orientation as Glu-357 in DNAP allowing simultaneous coordination of Mg-II and activation of the attacking water. To test this prediction, we examined the efficiency of intrinsic and Gre-stimulated RNA cleavage at various pH values. Previously (3), we observed that pH had a dual effect on nonassisted RNA cleavage affecting both Mg-II binding and ionization of the attacking water. To exclude the effect of pH on Mg-II retention as a reference, we used βR1106A mutant enzyme in which the binding of magnesium is pH-independent (3). As seen from Fig. 7B, the response of the reaction rates to pH was strikingly different. Specifically, pH elevation from 6 to 10 caused an ∼100-fold increase in the rate of the intrinsic reaction, although the rate of Gre reaction increased only five times, suggesting that attacking water ionization is a much less crucial factor in Gre-stimulated reaction. Indeed, the apparent pKa value for the water deduced from the pH dependence was 8.9 and 7.8 (for intrinsic and Gre-assisted cleavage respectively) reflecting 10–15-fold water activation by the Gre factor. In accordance with this mechanism, the mutational substitution of the Glu-44 residue in Gre simultaneously decreased retention of Mg-II (6, 21) and restored intrinsic cleavage activity of RNAP (in Gre-TEC complex), manifested by high pH dependence (21), thereby reflecting elimination of the attacking water activation by the substitution. These data are in agreement with our model, because the loss of coordination contact of the mutated residue with Mg-II allowed the base binding in the B site in the mode competent for intrinsic reaction. The above observations support the constructed structural model for Gre-assisted catalysis.
DISCUSSION
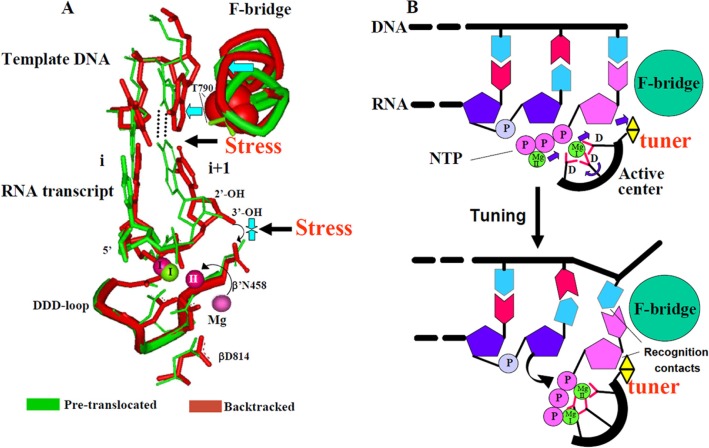
Role of ACT Mechanism in Substrate Selection by RNA Polymerase—The common Sn2 mechanism views RNA hydrolysis and pyrophosphorolysis as nucleotidyl transfer reaction operating in reverse, suggesting that the same substrate recognition principles (through ACT) could operate in both RNA synthesis and RNA degradation. This idea is supported by the strong negative effect of +1 RNA residue deoxy-substitution on both pyrophosphorolysis and endonuclease reaction rate (200–400-fold), which is comparable with the discrimination factor (120–1200-fold) between ribo- and deoxy-NTP incorporation by RNAP (15, 33). The common mechanism paradigm would posit that in the first step of substrate selection the nucleoside part of the NTP occupies the same position as the 3′-terminal +1 RNA residue in the pretranslocated complex (Fig. 10, A and B) in which the pairing with a DNA base is in place, but the ribose contacts are not established and the active center is not tuned. At the next selection step, the ribose recognition contacts of NTP substrate must be established so that the nucleoside part of NTP will assume its orientation as in the +1 RNA residue of the backtracked complex in which both base pairing and ribose recognition contacts are in place and the active center is tuned (Fig. 10, A and B). In the backtracked complex, simultaneous formation of a base pair and ribose recognition contacts causes significant angling of the base pair compared with the pretranslocated complex (Fig. 10A). This angling weakens hydrogen bonds and affects stacking interaction of the bases, thereby creating a stress that challenges both base pairing and ribose contacts so that noncomplementary or imperfectly base-paired NTPs are rejected (due to loss of the contacts with a template DNA base), as NTPs with inappropriate ribose (e.g. 2′-, or 3′-deoxy-NTPs). Therefore, tuning of the catalytic part of the active center is possible only when all recognition contacts of the NTP substrate are established and tested by tolerance to the stress. In support of this selection mechanism, introduction of a bulkier substituent to the part of the F-bridge (bridge helix) that crashes into the base pair (β′ T790V) increased the fidelity of transcription (33) by imposing more stress (Fig. 10, A and B). The suggested ACT mechanism for substrate selection is fundamentally different from that proposed for DNAP, whereby discrimination between deoxy- and ribo-substrate is achieved through strict fitting requirements for the sugar (38) rather than through active center rearrangement. Remarkably, in the DNAP active center carboxylates stem from rigid scaffolds, whereas in multi-subunit RNAP they reside in a flexible loop (DDD-loop). As follows from Ramachandran plots (data not shown), ACT is accompanied by significant re-shaping of the DDD-loop, which underscores the importance of the active center flexibility in multisubunit RNAP catalytic mechanism. Such re-shaping would not be possible in DNAP.
FIGURE 10.
Insights into substrate selection mechanism by RNAP derived from transcript cleavage reaction. A, superposition of the structures of pretranslocated and backtracked TECs. +2 RNA residue in backtracked TEC is omitted for clarity. Mg-I and Mg-II are marked. Curved arrow shows Mg-II capture by the active center upon ACT. Blue arrows indicate the pressure point exerted by F-bridge (bridge helix) on the 3′-terminal base pair of RNA/DNA hybrid and +1 RNA ribose recognition point. β′ Thr-790 side chain, which intrudes into terminal RNA/DNA base pair, is shown in space fill representation. Black arrows indicate stress points that challenge base pairing and ribose recognition in tuned TEC. B, model for NTP substrate recognition coupled with ACT. Simultaneous recognition of the base and ribose of correct NTP substrate creates stressed intermediate complex and triggers ACT enabling catalysis. Tuner is represented by β′ Arg-425 and β′ Asn-458 residues that recognize the substrate’s ribose. Active center catalytic carboxylates are indicated as D. Arrows show repositioning of catalytic Mg2+ ions accompanying ACT.
Species-specific Mechanisms for RNA Cleavage in Various RNAP
Possible Effect of RNAP Trigger Loop Transitions on Endonuclease Cleavage—Structural and biochemical studies have implicated the transitions of the β′-trigger loop in the catalytic mechanism of RNA polymerization (13, 14, 16, 17, 39, 40). The absence of an effect of α -amanitin (which blocks trigger loop transition), on endonuclease cleavage in yeast pol II (41), as well as insensitivity of the same reaction to trigger loop deletion in E. coli (40) argues against the involvement of this transition in endonuclease reaction at least in the E. coli and yeast enzymes. In accordance with this conclusion, deletion of the entire trigger loop did not affect substrate discrimination by RNAP (40). However, one cannot exclude the indirect effect of trigger loop transitions on endonuclease cleavage. Indeed, the status of the loop can affect backtracking, which is the first step leading to endonuclease transcript cleavage. In accord with this suggestion, we demonstrate that mutation of β′ Arg-933 residue of the loop detectably reduced backtracking. Notably, testing with T. thermophilus RNAP revealed a significant effect of trigger loop deletion on endonuclease cleavage, suggesting involvement of the loop in the reaction in thermophiles (42).
Catalytic Mechanism of Endonuclease Transcript Cleavage in E. coli—This study demonstrates a significant difference in the features of exo- and endonucleolytic RNA cleavage in TEC despite apparent mechanistic similarity of the reactions. This difference is evidenced by the following: (i) absolute reaction rate, which is 20–50 times higher for endonuclease hydrolysis; (ii) response to active center mutations, which generally have the opposite effect on the reaction rates, and (iii) response to noncognate NTP, which greatly stimulates exonuclease cleavage, while having no effect on endonuclease.
Our data attribute the observed differences between the reactions to the influence of the +2 RNA residue in backtracked TEC. This conclusion is strongly supported by the crystal structure of yeast pol II backtracked TEC in which only this residue of disengaged transcript segment is seen in defined orientation (24), suggesting specific interactions with active center side chains.
Although the x-ray structure is consistent with the results of biochemical studies on yeast pol II, fitting the data obtained for E. coli RNAP required adjustment of the structural model. Thus, re-positioning of the nucleoside part of +2 RNA residue from the orientation seen in yeast pol II into the active center cleft (B site) between DDD-loop and the “basic rim” was required.
Besides ACT, other factors that can increase the efficiency of endonuclease reaction are positioning and/or activation of the attacking water. Both modes involve hydrogen bonding of the water’s hydrogen atom with an acceptor group. In the positioning mode, establishing the hydrogen bond results in stabilization of the attacking water, whereas in the activation process, an acceptor group (general base) also lowers the apparent pKa value of the attacking water facilitating ionization, which can be detected by measuring the reaction rate at various pH values. Such testing performed in this study did not reveal activation of the attacking water in intrinsic endonuclease reaction. However, water “positioning” in this reaction is consistent with the deleterious effect of active center substitutions that are expected to disrupt the positioning by loosening the contacts with the adenine nitric base of +2 RNA residue, which in turn hydrogen bonds with the attacking water through the endocyclic oxygen (Fig. 2B). Surprisingly, we observed activation of the attacking water in a Gre-stimulated reaction.
Implications for Transcript Cleavage Mechanisms in Yeast pol II and RNAP from Thermophiles—Despite the common origin, the multisubunit RNAP evolved to develop specific mechanisms for regulation of RNA synthesis and transcription-coupled RNA processing, associated with RNAP endonuclease activity. Endonuclease reaction of RNA degradation in TEC displayed by all cellular RNAP is an intrinsic property of the enzymes. However, there is a striking difference in the relative rates of cleavage, which are ∼1, 102, and 104 for yeast pol II, E. coli, and T. aquaticus RNAP, respectively (17, 41, and this study). Contrary to E. coli, the endonuclease reaction in yeast pol II enzyme can yet proceed through the pyrophosphorolytic pathway (27). The biological significance of this pathway is unclear (due to low intracellular pyrophosphate concentration); however, conceptually, the absence of this reaction in E. coli points to the important difference in the structural mechanism of hydrolytic RNA cleavage. The model for the reaction suggested in this study attributes this phenomenon to steric block of the pyrophosphate-binding site by the base of +2 RNA residue in E. coli, although the x-ray structure of yeast pol II TEC allows pyrophosphate binding (Fig. 2C). Obstruction of B site of the active center by +2 RNA residue also explains the absence of the stimulatory effect of noncognate NTP on endonuclease in E. coli (3) and T. aquaticus (17). A higher reaction rate by E. coli RNAP compared with yeast enzyme could be explained by the influence of the base of +2 RNA residue, which according to our model facilitates the reaction by positioning the attacking water (Fig. 2, A and B). The difference in the reaction mechanism in yeast and E. coli is surprising given high evolutional conservation of the active center. The possible explanation involves stabilization of the orientation of the +2 RNA residue seen in x-ray structure by hydrogen bonding with the side chain of Rpb2 Tyr-679 highly conserved in eukaryotes. The counterpart of Tyr-679 in E. coli RNAP is methionine (βMet-681) that cannot hydrogen bond. Another explanation for this phenomenon is impossibility of the interactions of G or U residues in the +2 RNA position with the adenine-specific B site in the crystal of the yeast pol II complex, resulting in alternative orientations of the residue. The issue can be addressed by crystallization of the appropriate complexes.
Specific effects of +2 RNA residue based on Mg-II retention in T. aquaticus RNAP (17) and not observed in the E. coli enzyme underscore the difference in catalytic mechanisms. Unlike in the E. coli RNAP N-7 substitution in the +2 adenine base did not change the reaction rate in T. aquaticus TEC, but it significantly reduced the binding constant for Mg-II. Other variations of the structure of the +2 nucleoside also effected Mg-II retention in T. aquaticus TEC, although in E. coli TEC the change of the whole nucleoside residue to the methyl group did not alter the Kd values for Mg-II. Yet, pH values increasing from 9 to 10 only marginally stimulated endonuclease reaction in the T. aquaticus case, whereas in E. coli TEC the abrupt upturn of pH dependence curve is seen in this range (Fig. 7A). The upturn in E. coli TEC is explained by elimination of the salt bridge of βAsp-814 with βArg-1106 (which enables Mg-II coordination) due to deprotonation of the latter. These facts suggest higher occupancy of the coordination site by Mg-II in T. aquaticus than in E. coli TEC, which is consistent with the higher endonuclease activity of parent T. thermophilus TEC (26) and confirmed by direct measurement of Mg-II Kd values in T. aquaticus TEC (17).
Stronger retention of Mg-II in T. aquaticus RNAP can be explained by the higher degree of ACT in this enzyme. Another explanation involves the contribution of the structure seen in T. aquaticus core RNAP (43), in which the βArg-1106–βAsp-814 salt bridge is broken, thereby facilitating Mg-II binding.
Along with the above differences, there are also remarkable similarities in the catalytic mechanism of E. coli and T. aquaticus enzymes, as evidenced by the same effect of thio-substitution at +2 RNA residue on the reaction rate and by the fact that +2 adenylate is the most catalytically proficient residue in endonuclease reaction for both T. aquaticus (17) and E. coli TEC.
Catalytic Mechanism for Transcript Cleavage Factors—Gre factors, found in all known bacteria, are required for normal growth. They execute proofreading and facilitate propagation of RNAP along DNA during RNA synthesis by both preventing transcriptional arrest and rescuing arrested TEC. This is achieved through hydrolytic cleavage of the disengaged RNA segment in reversibly backtracked and/or arrested transcription complexes. Gre factors strongly (∼3000–4000-fold) stimulate intrinsic RNase H type activity of RNAP by increasing retention of Mg-II required for catalysis (6, 19, 21). However, the magnitude of the enhancement of Mg2+ retention is only ∼300-fold (6), pointing to another catalytic factor, which according to this work is 10–15-fold activation of the attacking water that when combined with Mg2+ retention enhancement factor accounts for the whole magnitude of the catalytic Gre effect. These findings clearly demonstrate how Gre factors build up on the RNAP active center to enhance the enzyme’s rudimentary RNA cleavage activity to the level of typical nuclease.
Variations in the mechanism of RNA cleavage by cellular RNAP can account for different arsenals of auxiliary protein factors assisting this reaction in the living cell. Indeed, in yeast and E. coli intrinsic transcript cleavage activity of RNAP is low and has to be stimulated by protein factors TFIIS and GreA and GreB. The importance of Gre factors for E. coli is underscored by poor fitness of a bacterial strain lacking both factors (2). Notably, in T. thermophilus enzyme basal endonuclease activity is nearly 2 orders of magnitude higher than in E. coli, which could be sufficient on its own to maintain the vital function of transcript cleavage. This concept is supported by the absence of GreB-like factor in T. aquaticus and T. thermophilus. Interestingly, the stimulation effect of the GreA-like factor from T. thermophilus on T. thermophilus RNAP is only ∼30-fold (26), which is about 2 orders of magnitude less than that for E. coli RNAP with GreB, which makes the rate of factor-stimulated RNA cleavage nearly equal for both enzymes.
Supplementary Material
Acknowledgments
We are grateful to Drs. Albert Weixlbaumer, Andrey Feklistov, Seth Darst, Karl Drlica, Loren Day, JBC Editorial Board, as well as to anonymous reviewers for support, numerous suggestions, and valuable comments. Additional data and experimental details are available from the authors upon request.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM-30717-21.
The abbreviations used are: RNAP, RNA polymerase; TEC, transcription elongation complex; ACT, active center tuning; CAPS, 3-(cyclohexylamino)propanesulfonic acid; TB, transcription buffer; pol, polymerase; DNAP, DNA polymerase; ATPαS, adenosine 5′-(α-thio)-triphosphate; CTPαS, cytidine 5′-(α-thio)-triphosphate.
This article contains supplemental ACT files.
Supplemental material: http://www.jbc.org/content/suppl/2013/01/15/M112.424002.DC1.html
REFERENCES
- 1.Surratt CK, Milan SC, Chamberlin M. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci USA. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 5.Stebbins CE, Borukhov S, Orlova M, Polyakov A, Goldfarb A, Darst SA. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- 6.Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reines D, Ghanouni P, Li QQ, Mote J., Jr The RNA polymerase II elongation complex. Factor-dependent transcription elongation nascent RNA cleavage. J Biol Chem. 1992;267:15516–15522. [PMC free article] [PubMed] [Google Scholar]
- 8.Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 33–53 direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 9.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA. Control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 10.Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P. Structural basis of transcription. Mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol. Cell. 2009;34:710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Sosunov V, Zorov S, Sosunova E, Nikolaev A, Zakeyeva I, Bass I, Goldfarb A, Nikiforov V, Severinov K, Mustaev A. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic Acids Res. 2005;33:4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription. RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 13.Epshtein V, Mustaev A, Markovtsov V, Bereshchenko O, Nikiforov V, Goldfarb A. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol. Cell. 2002;10:623–634. doi: 10.1016/s1097-2765(02)00640-8. [DOI] [PubMed] [Google Scholar]
- 14.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription. Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 17.Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 18.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 19.Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 20.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription. Nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steitz T. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 23.Zhu R, Janetzko F, Zhang Y, van Duin ACT, Goddard WA, 3rd, Salahub DR. Characterization of the active center of yeast RNA polymerase II by DFT and ReaxFF calculations. Theor Chem Account. 2008;120:479–489. [Google Scholar]
- 24.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 26.Hogan BP, Hartsch T, Erie DA. Transcript cleavage by Thermus thermophilus RNA polymerase. Effects of GreA and anti-GreA factors. J Biol Chem. 2002;277:967–975. doi: 10.1074/jbc.M108737200. [DOI] [PubMed] [Google Scholar]
- 27.Rudd MD, Izban MG, Luse DS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci USA. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi H, Endo T, Kaji A. Enzymatic synthesis of oligonucleotides containing methylphosphonate internucleotide linkages. Biochemistry. 1990;29:8747–8753. doi: 10.1021/bi00489a035. [DOI] [PubMed] [Google Scholar]
- 29.Hoard DE, Ott DG. Conversion of mono-and oligodeoxyribonucleotides to 5-thiphosphates. J Am Chem Soc. 1965;87:1785–1788. doi: 10.1021/ja01086a031. [DOI] [PubMed] [Google Scholar]
- 30.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription. An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 31.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 32.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 33.Svetlov V, Vassylyev DG, Artsimovitch I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J Biol Chem. 2004;279:38087–38090. doi: 10.1074/jbc.C400316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markovtsov V, Mustaev A, Goldfarb A. Protein-RNA interactions in the active center of transcription elongation complex. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 36.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 37.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 38.Joyce CM. Choosing the right sugar. How polymerases select a nucleotide substrate. Proc Natl Acad Sci USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chlenov M, Masuda S, Murakami KS, Nikiforov V, Darst SA, Mustaev A. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase β′ subunit. J Mol Biol. 2005;353:138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weilbaecher RG, Awrey DE, Edwards AM, Kane CM. Intrinsic transcript cleavage in yeast RNA polymerase II elongation complexes. J Biol Chem. 2003;278:24189–24199. doi: 10.1074/jbc.M211197200. [DOI] [PubMed] [Google Scholar]
- 42.Yuzenkova Y, Zenkin N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc Natl Acad Sci USA. 2010;107:10878–10883. doi: 10.1073/pnas.0914424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.