Abstract
Hormonal crosstalk is central for tailoring plant responses to the nature of challenges encountered. The role of antagonism between the two major defense hormones, salicylic acid (SA) and jasmonic acid (JA), and modulation of this interplay by ethylene (ET) in favor of JA signaling pathway in plant stress responses is well recognized, but the underlying mechanism is not fully understood. Here, we show the opposing function of two transcription factors, ethylene insensitive3 (EIN3) and EIN3-Like1 (EIL1), in SA-mediated suppression and JA-mediated activation of PLANT DEFENSIN1.2 (PDF1.2). This functional duality is mediated via their effect on protein, not transcript levels of the PDF1.2 transcriptional activator octadecanoid-responsive arabidopsis59 (ORA59). Specifically, JA induces ORA59 protein levels independently of EIN3/EIL1, whereas SA reduces the protein levels dependently of EIN3/EIL1. Co-infiltration assays revealed nuclear co-localization of ORA59 and EIN3, and split-luciferase together with yeast-two-hybrid assays established their physical interaction. The functional ramification of the physical interaction is EIN3-dependent degradation of ORA59 by the 26S proteasome.
These findings allude to SA-responsive reduction of ORA59 levels mediated by EIN3 binding to and targeting of ORA59 for degradation, thus nominating ORA59 pool as a coordination node for the antagonistic function of ET/JA and SA.
INTRODUCTION
To survive the broad range of frequently encountered environmental perturbations, plants have developed sophisticated stress responses tailored to the nature of the stress. Specifically, plants produce a blend of hormones, known as signal signature with varying quantities, compositions, and timing depending on the lifestyle and the nature of the stress (De Vos et al. 2005). In addition to the signal signature, plants also rearrange various hormone-signaling pathways to effectively counter these challenges while maintaining a fine balance for resources simultaneously required for growth and development.
The regulation of antagonistic and synergistic interplay between salicylic acid (SA) and jasmonic acid (JA) is one of the key strategies plants deploy to organize effective stress response signaling networks (Glazebrook 2005; Thaler et al. 2012; Van der Does et al. 2013). Superimposed on this, there are interactions with other hormones such as ethylene (ET), that further verify the complexity and significance of hormonal interaction in maximizing plants’ ability to effectively mount a well-designed adaptive response against a specific stress.
The plant hormone SA is a phenolic compound that plays a key role in plant defense responses primarily against biotrophic pathogens (Glazebrook 2005). Activation of the SA pathway at the site of infection triggers long distance resistance known as systemic acquired resistance (SAR) to prevent new infections and to limit the spread of the already existing infection (Vlot et al. 2009).
JA and its structurally related metabolites wild typelectively known as jasmonates are oxygenated lipid-derived metabolites produced rapidly by the oxylipin biosynthesis pathway in response to environmental and developmental signals (Wasternack and Hause 2013). The JA signaling pathway consists of two branches; the MYC branch coupled to wounding and defense against insect herbivores, and the ERF branch that is associated with enhanced resistance to necrotrophic pathogens (Lorenzo 2002; Lorenzo et al. 2003; Lorenzo et al. 2004; Kazan and Manners 2012). Similar to SA, activation of the JA pathway also leads to distal JA-dependent responses to further enhance plant protection against invasion (Howe and Jander 2008).
It is well established that the JA and SA response pathways interact mainly antagonistically although neutral and synergistic crosstalk are also described (Pieterse et al. 2009).
ET is a simple gaseous hormone regulating plant responses to developmental and environmental cues (Kieber and Ecker 1993). Ethylene binds to endoplasmic reticulum-localized ethylene receptors and further inhibits their interacting Raf-like kinase CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1) (Kieber et al. 1993; Chen et al. 2002). In the absence of ethylene, CTR1 directly phosphorylates another endoplasmic reticulum-localized transmembrane protein and a positive regulator in ethylene signaling, ETHYLENE INSENSITIVE 2 (EIN2) (Ju et al. 2012). The presence of ethylene, however, results in dephosphorylation of the EIN2 C-terminal end followed by its cleavage and transportation to the nucleus (Qiao et al. 2012; Wen et al. 2012). Downstream of EIN2 are two transcription factors, ETHYLENE INSENSITIVE 3 (EIN3) and its close homolog EIN3 LIKE 1 (EIL1) that control the majority of ethylene-mediated responses (Solano et al. 1998).
Both JA and ET are key hormones in enhancing plant resistance against necrotrophic fungus by eliciting expression of defense responsive transcription factors such as ETHYLENE RESPONSE FACTOR 1 (ERF1) and OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59). These two transcription factors are functional in the ERF branch of JA, necessary for induction of the downstream pathogen-responsive gene expression, including PLANT DEFENSIN1.2 (PDF1.2) (Dong 1998; Lorenzo et al. 2003; Pre et al. 2008; Zarei et al. 2011). Accordingly, both ERF1 and ORA59 are recognized as essential integrators of the JA and ethylene signal transduction pathways (Solano et al. 1998; Pre et al. 2008).
Recent findings have further unraveled the molecular mechanism of JA/ET interaction by providing evidence that JA augments EIN3/EIL1 transcriptional activity by reducing their interaction with HISTONE DEACETYLASE6 (HDAC6), while ET stimulates accumulation of EIN3/EIL1 protein levels. Hence, the integration of these two distinct mechanisms triggers induction of EIN3/EIL1 dependent gene expression (Zhu et al. 2011). As such, EIN3/EIL1 are implied as the direct molecular links for JA/ET interactions. However, the wounding-associated JA response pathway interacts antagonistically with ET, also through EIN3. In this case, MYC2 interacts with and inhibits EIN3 function, and reciprocally EIN3 represses MYC2 function in the regulation of wounding responses. Moreover, MYC2 directly induces the expression of F-box protein EIN3-BINDING F-BOX PROTEIN 1 (EBF1), which further targets EIN3 for degradation to provide an additional layer of repression of EIN3 (Song et al. 2014; Zhang et al. 2014). Collectively, these data identify EIN3 as a node for JA and ET crosstalk, and its central role in design of plant responses to the nature of stress encountered.
The studies here fill in the existing gap in our knowledge of the underlying molecular mechanism of ET/JA induction and SA suppression of PDF1.2 expression as a proxy for activation of the ERF/ORA59 branch of JA pathway. Specially, we reveal that the suppression of PDF1.2 expression by SA as well as its activation by JA both depends on EIN3/EIL1. In addition, in agreement with the previous report we identify ORA59 as mediator of this process (Leon-Reyes et al. 2010), and in corroboration with recent studies (Van der Does et al. 2013) we show that SA reduces ORA59 protein levels, and further demonstrate that this reduction is dependent on EIN3/EIL1. Moreover, we establish nuclear co-localization and physical interaction of EIN3/ORA59 as a mechanism for targeting ORA59 for degradation by 26S proteasome machinery. These data wild collectively uncover a new facet of ET-JA-SA interaction and identify fine-tuning of ORA59 pool as a platform poised delicately to balance the ET/JA synergism and ET/JA versus SA antagonism in tailoring stress-signaling pathways in Arabidopsis.
RESULTS
EIN3/EIL1 is required for SA-suppression and JA-induction of PDF1.2 expression
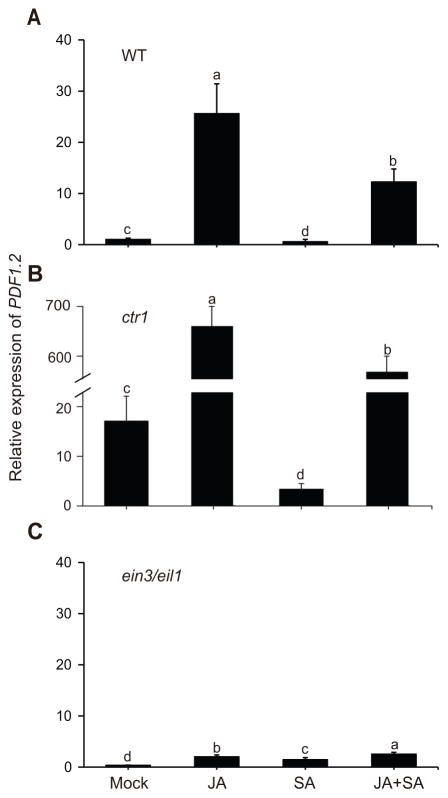
Regulation of the defense gene PDF1.2 is a clear example of interaction between hormones, as it is suppressed by SA and activated by coaction of JA/ET (Ndamukong et al. 2007; Pre et al. 2008). To examine the molecular links between PDF1.2 expression and hormonal interplay, we first confirmed the reported antagonistic impacts of SA and JA on PDF1.2 expression levels under our experimental conditions (Leon-Reyes et al. 2010; Van der Does et al. 2013). For these experiments, we specifically compared the data obtained both from soil and plate grown Wild type plants, and show JA-mediated induction and SA-mediated suppression of PDF1.2 expression levels are more pronounced in soil (Figure S1A, D), a condition closer to the natural conditions. In addition, using ctr1 and ein3 eil1 mutant lines furthered confirmed the already established dependency of JA-induction of PDF1.2 on functional EIN3/EIL1 (Zhu et al. 2011) (Figure S1B–F). Collectively, this data led us to test the involvement of these two transcription factors in SA-mediated suppression of PDF1.2 transcript levels using qRT-PCR in Wild type, ctr1, and ein3 eil1 genotypes grown on soil, and 24 h post mock, JA, SA, and JA+SA treatments. As expected, the results display clear JA-induction of PDF1.2 in Wild type, and an even stronger induction in ctr1 (Figure 1A, B). The ctr1 mutant line is dysfunctional in negative regulation of ET signaling events (Kieber et al. 1993), and accordingly displays higher basal as well as JA-induced PDF1.2 transcript levels as compared to the Wild type background. However, in agreement with the previous report (Zhu et al. 2011), the JA-induced PDF1.2 levels are notably compromised in ein3 eil1 mutant as compared to the other two backgrounds (Figures 1C, S1). Conversely, SA treated Wild type and ctr1 display reduced PDF1.2 transcript levels in Wild type and in the ctr1 backgrounds respectively, as compared to mock treated plants (Figures 1A, B, S1). Notably however, SA treatment no longer suppresses expression levels PDF1.2 in ein3 eil1 mutant background (Figure 1C). In fact, the observed further increase in the PDF1.2 transcript levels in response to combined SA/JA treatment is suggestive of the synergistic/additive function of the two hormones in the absence of a functional EIN3/EIL1 (Figures 1C, S1C, S1F). In line with this finding, overexpression of EIN3 in ein3 eil1 mutant background reestablishes strong JA-mediated induction and SA-mediated suppression of PFD1.2 (Figure 2A), thereby establishing EIN3 as the predominant regulator of the PDF1.2 expression.
Figure 1. EIN3/EIL1 is required for SA-suppression and JA-induction of PDF1.2 expression.
(A–C) Total RNA was extracted from three independently soil grown samples of Wild type (A), ctr1(B), and ein3 eil1 (C) genotypes 24 h after mock, JA, SA, and combined JA+SA treatments. Transcript levels of PDF1.2 were normalized to At4g34270 (T1P41-like family protein) and At4g26410 (M3E9) measured in the same samples. Data are mean ± SD of three independent biological replicates each with three technical repeats. Letters above bars indicate significant differences as determined by Tukey’s HSD (p <0.05).
Figure 2. Overexpression of EIN3 reestablishes SA-mediated suppression of PDF1.2 in ein3 eil1.
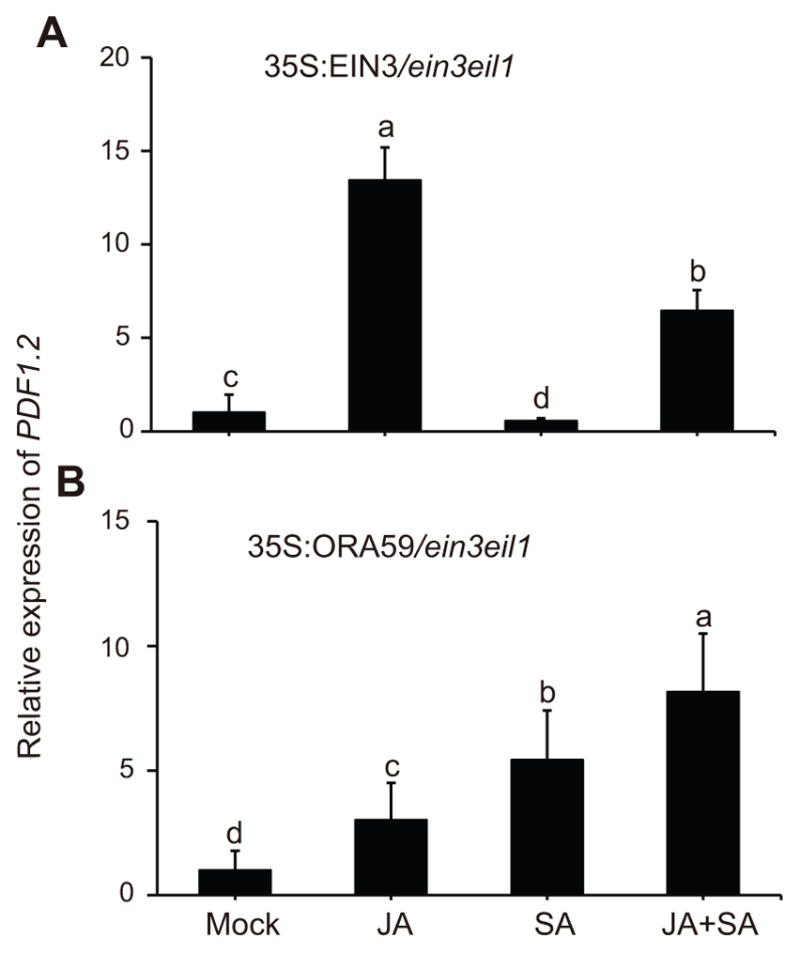
Relative expression levels of PDF1.2 24 h post mock, JA, SA, and JA+SA treatments in ein3 eil1 loss of function mutant expressing 35S:EIN3 (A) and 35S:ORA59 (B). The transcript levels of PDF1.2 were normalized as described in the Figure 1. Data are mean ±SD of three independently soil grown biological replicates each with three technical repeats. Letters above bars indicate significant differences as determined by Tukey’s HSD method (p <0.05).
In light of the ORA59 function, as the key regulator in ET/JA activation of PDF1.2 (Pre et al. 2008; Leon-Reyes et al. 2010; Zarei et al. 2011), we questioned whether SA application might alter PDF1.2 expression in ein3 eil1 mutant plants overexpressing ORA59. To address this question, we generated 35S:ORA59:LUC fusion construct and introduced it into the ein3 eil1 mutant background. Subsequently, we examined PDF1.2 expression levels in these soil grown plants after treatment with JA, SA or the combination (Figure 2B). These data clearly show differentially and strongly enhanced PDF1.2 expression levels in response to individual or combined JA and SA application as compared to the mock treatment, with the highest induction in plants treated with both hormones. The highly induced PDF1.2 in response to SA/JA combined application imply synergistic/additive roles of these hormones, and additionally display a molecular link between their crosstalk via ORA59 and the EIN3/EIL1 transcription factors.
Taken together these results extend the previous finding (Zhu et al. 2011) and further establishes opposing functions of EIN3/EIL1 in SA-suppression and JA-induction of PDF1.2.
SA-mediated reduced stability of ORA59 is EIN3/EIL1 dependent
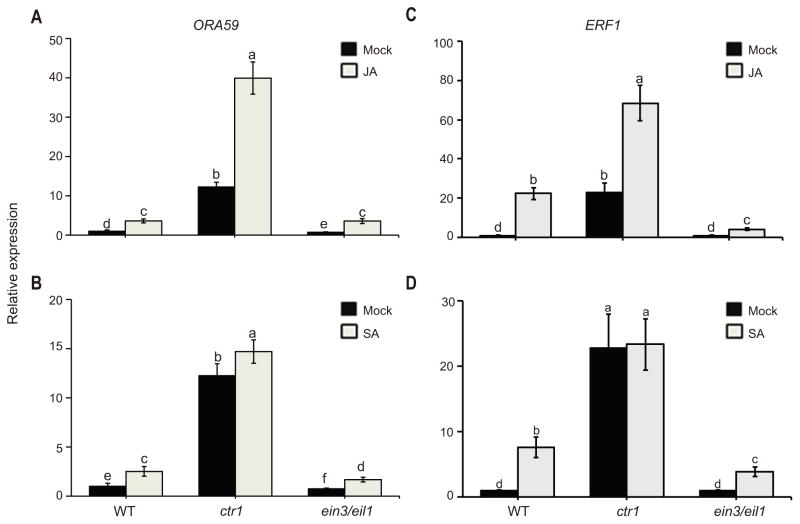
Next, we examined the possible link between altered PDF1.2 transcript levels and EIN3/EIL1 mediated modulation of ORA59 expression. We also extended our query by examining the EIN3/EIL1-dependent alteration of transcript levels of the transcription factor ERF1, known involves in regulation of PDF1.2 expression in the ERF branch of JA (Dong 1998; Lorenzo et al. 2003; Pre et al. 2008; Zarei et al. 2011). For these studies, we examined the transcript levels of ORA59 and ERF1 in soil-grown Wild type, ctr1 and ein3 eil1 backgrounds in response to mock, JA and treatments. This data confirmed the earlier report on EIN3/EIL1 dependent JA-induced transcriptional activation of ORA59 and ERF1 (Zhu et al. 2011), and further established the presence of high basal and higher JA-induced ORA59 and ERF1 expression levels in ctr1 as compared to Wild type and ein3 eil1 lines (Figures 3, 2A, 2C). Furthermore, we show that SA treatment results in significant induction of ORA59 transcript levels in all three genotypes at different degrees, but independently of EIN3/EIL1 (Figures 3B, S2B, S2D). In contrast, SA treatment as compared to mock resulted to markedly increased levels of the ERF1 transcript levels both in the Wild type and ein3 eil1 mutant lines, albeit partially EIN3/EIL1 dependent and most notably in Wild type (Figure 3D). Collectively, this finding is suggestive of requirement for EIN3/EIL1 in SA-mediated regulation of expression levels of EFR1, but not ORA59.
Figure 3. ERF1 transcript level is partially EIN3/EIL1-dependent.
(A–D) Total RNA was extracted from three independently soil grown Wild type, ctr1, and ein3 eil1 genotypes 24 h after mock (black bars), JA (A and C grey bars) or SA (B and D grey bars) treatments. Transcript levels of ORA59 (A–B) and ERF1 (C–D) were normalized to At4g34270 (T1P41-like family protein) and At4g26410 (M3E9) measured in the same samples. Data are mean ± SD of three independent biological samples each with three technical repeats. Letters above bars indicate significant differences as determined by Tukey’s HSD method (p <0.05).
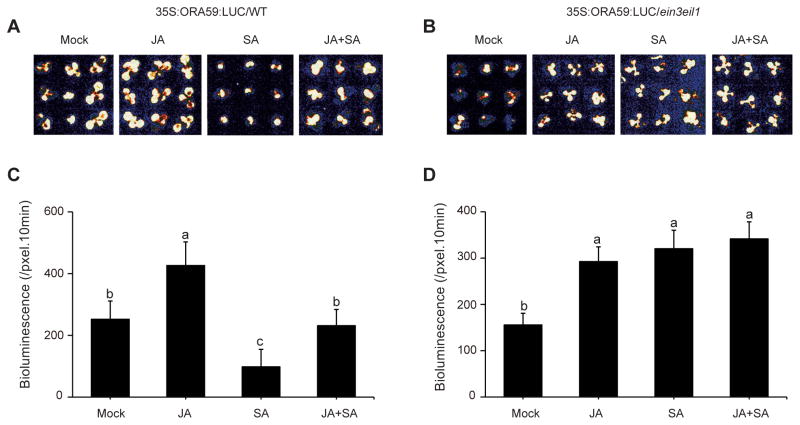
The disparity between the SA-mediated alteration of expression levels of the two above transcription factors, namely marginally enhanced ORA59 expression and increased ERF1 expression levels, as opposed to the suppression of PDF1.2 led us to question whether under our experimental conditions modulation of ORA59 protein levels could contribute to PDF1.2 suppression, as previously reported (Van der Does et al. 2013). To address this question, in an expanded study we employed Wild type and ein3 eil1 mutant backgrounds expressing 35S:ORA59:LUC fusion construct. The 35S:ORA59:LUC activity as a measure of ORA59 protein level was determined in the homozygous transgenic mock, JA, SA or JA+SA treated seedlings (Figure 4A–D). This data show that SA treatment reduced the LUC activity below the levels of the mock treated seedlings in Wild type background, while JA treatment enhanced the LUC activity, and that combined JA/SA application notably reduced the JA-mediated enhancement (Figure 4A, C). In ein3eil1 background, the LUC activity levels in response to singular or combinatorial application of SA or JA are equally enhanced as compared to the corresponding mock treatment (Figure 4B, D).
Figure 4. SA-mediated reduction of ORA59 protein abundance is dependent on EIN3/EIL1.
Representative images of Wild type (A) and ein3 eil1 (B) seedlings expressing 35S:ORA59:LUC after 24 h of mock (control), JA, SA, and JA+SA treatments. ORA59 protein levels as a measure of LUC intensity in the WT (C) and ein3 eil1 backgrounds (D) using Andor Solis image analysis software. Data are mean ± SEM (n = 36). Letters above bars indicate significant differences as determined by Tukey’s HSD (p <0.05).
Together the data illustrate that SA-mediated reduced stability of ORA59 protein is dependent on EIN3/EIL1, but the JA-induced enhanced stability of ORA59 is not.
EIN3 and ORA59 are nuclear co-localized and physically interact
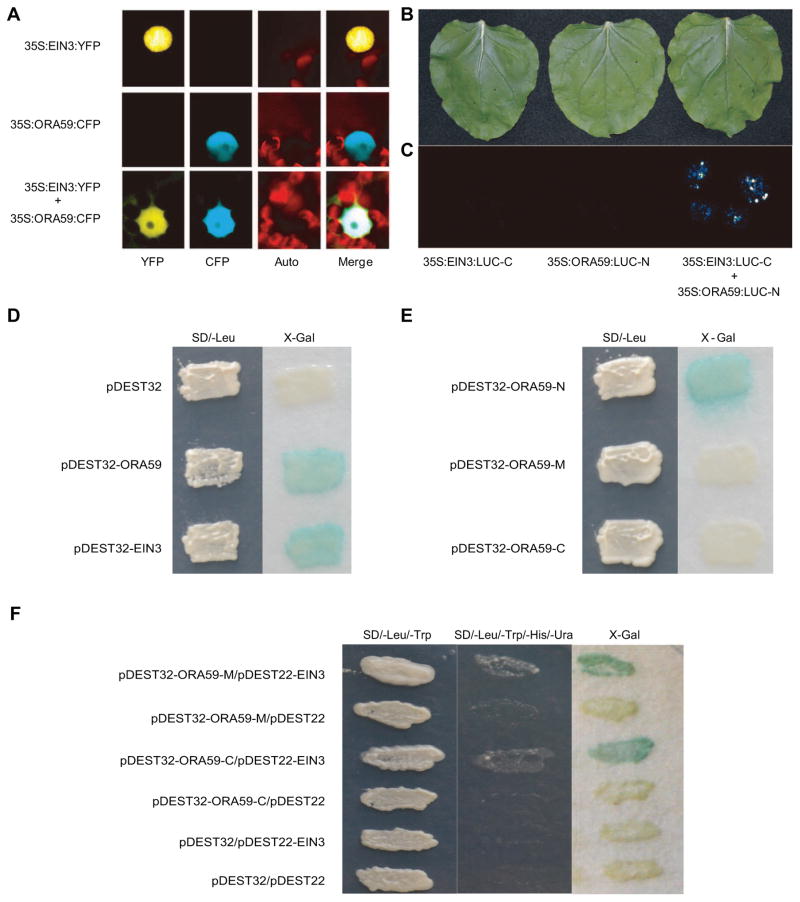
To gain insight into the mechanism(s) underlying the requirement for EIN3 in SA-reduced ORA59 levels, we first questioned the potential co-localization of these two transcription factors. Thus, we generated 35S:EIN3:YFP and 35S:ORA59:CFP constructs and subsequently used them individually or in combination for agro-infiltration-based transient assays in Nicotiana benthamiana (tobacco) leaves (Figure 5A). These data show the expected nuclear localization of the two transcription factors, and further established their nuclear co-localization.
Figure 5. EIN3 and ORA59 are nuclear co-localized and physically interact.
(A) Representatives of transient assays in Nicotiana benthamiana expressing 35S:EIN3:YFP, or 35S:ORA59:CFP individually or in a combination, displaying nuclear co-localization of these two transcription factors. Representative images of split luciferase complementation assays in Nicotiana benthamiana displayed by (B) bright-field, and (C) dark-field of leaves expressing ORA59 and EIN3fused to amino (N)- and carboxyl (C)-terminal fragments of luciferase (LUC) alone and in combination. (D) Display of the auto activation of ORA59 and EIN3 in Y2H assays. (E) Diploid yeast cells containing truncated ORA59 polypeptide fragments identifies amino acids 1–80 (ORA59-N) as the auto activation domain which is absent in mid segment containing amino acids 81–200 (ORA59-M), and C-terminus containing amino acids 201–244 (ORA59-C). (F) Colonies were gown on Leu, Trp, His and Ura (middle panel) and with X-Gal (right panel). Negative controls contained empty bait or pray vector.
In view of this co-localization, we next tested if EIN3 and ORA59 physically interact, using a split luciferase complementation assay in tobacco (Figure 5B, C). To perform the split luciferase assay we generated fusion constructs of ORA59 and EIN3 to amino- and carboxyl-terminal fragments of LUC (LUC-N or LUC-C), respectively. Subsequently, the constructs, individually or together, were used in agro-infiltration-based transient assays in tobacco leaves, as described previously (Wang et al. 2014a; Wang et al. 2015). The outcome of these studies clearly display the reconstitution of LUC activity only in leaves co-infiltrated with both constructs, thereby illustrating the physical interaction of the two transcription factors.
To further verify the protein-protein interaction data, we utilized Y2H assays, and established that the strong auto activation of both ORA59 and EIN3 hamper the use of this approach (Figure 5D). To potentially resolve the auto activation issue, we generated three constructs representing segments of the ORA59 protein displaying amino acids 1–80 (ORA59-N)-, and the middle amino acids 81–200 (ORA59-M)-, and carboxyl terminus amino acids, 201–244, (ORA59-C). This approach identified ORA59-N as the region responsible for auto activation (Figure 5E). Next, we introduced ORA59-C and -M as the bait for EIN3 along with various controls including the empty vector pDEST32 (Figure 5F). Collectively this data verified protein-protein interactions between ORA59-M and -C and EIN3 (Figure 5F). As such these analyses identified the auto activation domain of ORA59, and further confirmed protein-protein interaction between EIN3 and ORA59.
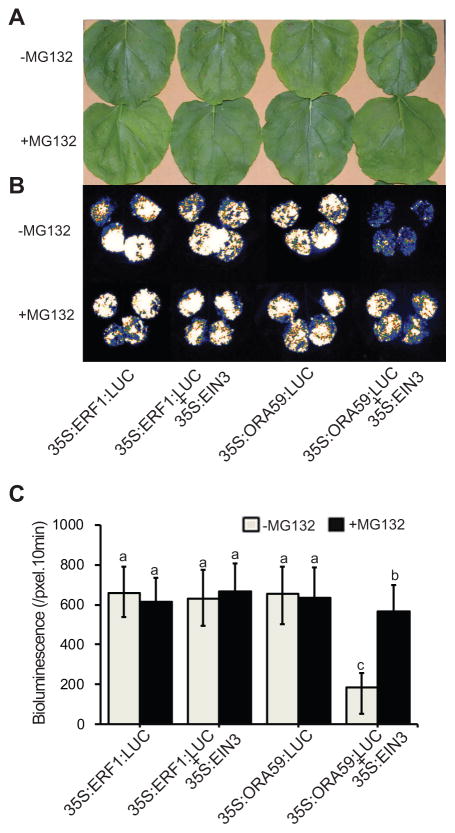
EIN3-dependent degradation of ORA59
The physical interaction between ORA59 and EIN3 led us to question whether the underlying mechanism of SA-mediated suppression of PDF1.2 is via regulation of the ORA59 protein levels necessary for binding to and activation of the PDF1.2 promoter. We reasoned that interaction with EIN3 might modify the protein stability of ORA59, or sequester most of ORA59 available and necessary for binding to the PDF1.2 promoter, or alter the DNA binding capacity of ORA59. Thus, we examined the stability of ORA59 using co-infiltration assays in tobacco leaves. Leaves were infiltrated with various constructs including 35S:ORA59:LUC alone or in combination with 35S:EIN3 in the presence and absence of the 26S proteasome inhibitor, MG132. As the control, 35S:ERF1:LUC construct assayed alone or in combination with 35S:EIN3 were included. Selection of ERF1 is based on the combination of the established function of this transcription factor in the ERF branch of JA for induction of PDF1.2 (Dong 1998; Lorenzo et al. 2003; Pre et al. 2008; Zarei et al. 2011), in conjunction with its EIN3/EIL1-dependent expression. Additionally, the choice of this control is further justified by the absence of protein-protein interaction between EIN3 and ERF1 as evidenced by the outcome of split luciferase assays using fusion constructs of 35S:EIN3:LUC-C and 35S:ERF1:LUC-N in agro-infiltration-based transient assays in tobacco leaves (Figure S3).
In the protein stability assays, the visual and quantitative LUC activity analyses, as a proxy for ERF protein levels, confirms similar levels in all the assays regardless of whether it is infiltrated alone or together with EIN3, and irrespective of the presence or the absence of MG132 (Figure 6A–C). This data further confirms that the ERF1 transcript and not the protein levels are mainly altered by EIN3 (Figures 3C, D and 6). In contrast, the LUC activity of ORA59 is strongly reduced in the presence of 35S:EIN3 and in the absence of MG132, however addition of MG132 rescues the ORA59:LUC activity (Figure 6A–C). This data support the notion that EIN3-medaited regulation of ORA59 is predominantly at the protein level, whereby EIN3 specifically bind to and destabilizes ORA59 protein and that MG132 abolishes the destabilizing effect of EIN3 on ORA59 (Figure 6).
Figure 6. EIN3-dependent degradation of ORA59.
Representatives of transient expression assays in Nicotiana benthamiana displayed by bright- (A) and dark-field (B) of leaves expressing 35S:ERF1:LUC alone or together with 35S:EIN3, 35S:ORA59:LUC alone, or together with 35S:EIN3 in the presence or absence of 26S proteasome inhibitor MG132. (C) Intensity of LUC bioluminescence of the aforementioned leaves quantified using Andor Solis image analysis software. Data are mean ± SD (n = 20). Letters above bars indicate significant differences as determined by Tukey’s HSD method (p <0.05).
Collectively, these results illustrate the EIN3 mediated regulation of the protein and not the transcript levels of ORA59 (Figures 3A, B and 6). Specifically, our data support the notion that EIN3 binds to and mediates targeting of ORA59 for degradation by the 26S proteasome machinery.
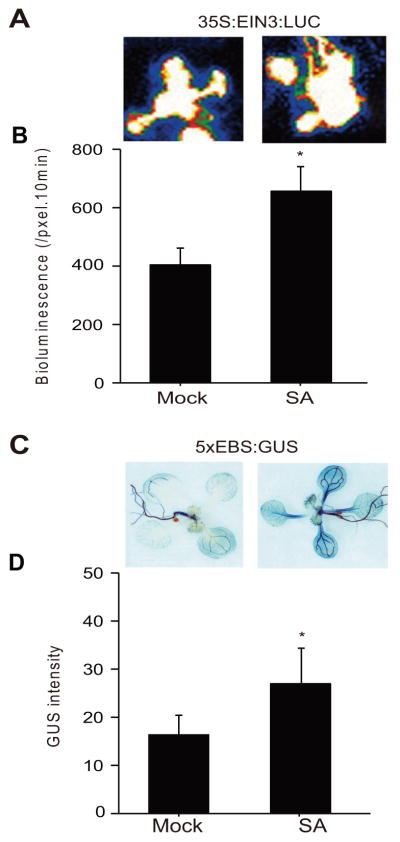
SA-mediated regulation of EIN3 protein abundance
The aforementioned results raises the question of whether SA mediates regulation of EIN3 protein level. To address this question, 35S:EIN3:LUC expression lines were employed as a proxy for the EIN3 protein levels in response to SA treatment. The result shows that SA application enhances EIN3 protein abundance (Figure 7A, B).
Figure 7. SA enhances EIN3 protein abundance and induces ethylene-responsive reporter.
(A) Representative images of seedlings expressing 35S:EIN3:LUC after 24 h of mock (control), and SA treatments. (B) EIN3 protein levels as a measure of LUC intensity using Andor Solis image analysis software. Data are mean ± SEM (n = 40). Asterisk above bars indicate significant differences as determined by Tukey’s HSD (p<0.05). (C) Representative images of seedlings expressing 5XEBS:GUS seedlings after 24 h of mock (control), and SA treatments. (D) Histograms display GUS intensity measurements. Data are mean ± SEM (n = 34). Asterisk above bars indicate significant differences as determined by Tukey’s HSD (p <0.05).
The central role of EIN3 in ethylene signaling pathway led us to question of whether SA-regulation of EIN3 may extend to SA affecting the ethylene-signaling pathway. To answer this, plants expressing ethylene-responsive reporter 5XEBS:GUS were employed and exogenously mock or SA treated (Figure 7C, D). The enhanced GUS signal in SA as compared to mock treated seedlings (Figure 7C) followed by quantification of the GUS intensity (Figure 7D) clearly display SA positive regulation of the reporter gene and by extension a proxy for activation of ethylene-signaling pathway.
DISCUSSION
Optimal and effective responses to ever-changing environment is central to plant fitness, and dependent on tightly controlled signaling cascades integrating external and internal signals to craft an optimal response. Amongst the various response pathways, the interconnection between SA, JA and ET constitute a regulatory signaling hub that integrates inputs and tailors plant responses to the natures of stresses encountered (Koornneef et al. 2008; Clarke et al. 2009; Leon-Reyes et al. 2010; Ritsema et al. 2010; Cerrudo et al. 2012).
In Arabidopsis, the SA-mediated suppression and ET/JA-induction of PDF1.2 establish this marker of the ERF branch of the JA pathway as an ideal target for examining the underlying mechanism of the hormonal crosstalk regulating effective and tailored stress responses. Here we demonstrate opposing functions of EIN3/EIL1 in SA suppression and JA activation of PDF1.2. The sharp contrast between EIN3/EIL1 dependent JA activation of PDF1.2 versus mutual inhibition of JA-activated transcription factors MYC2 and EIN3/EIL1 (Song et al. 2014; Zhang et al. 2014), further signifies the delicate balance of these interactions in tailoring plant stress responses. Furthermore, this finding confirms the major role of EIN3/EIL1 transcription factors in integration of signaling cascades most appropriate for the stress encountered.
Here we specifically establish that the SA-mediated induction of ORA59 transcript level is independent of EIN3/EIL. Furthermore, our results clearly show that EIN3/EIL dependent SA repression of PDF1.2 is due to the reduced abundance of ORA59 protein rather than the altered ORA59 transcript levels. In accordance with this finding, the expression level of PDF1.2 is enhanced in SA-treated 35S:ORA59/ein3 eil1 lines, while it is suppressed in 35S:EIN3/ein3 eil1 lines. The co-infiltration assays demonstrate nuclear co-localization and the combination of split-luciferase and Y2H assays established physical interaction of ORA59 and EIN3 proteins. This physical interaction tags ORA59 for degradation that is abolished in the presence 26S proteasome inhibitor, MG132.
We further demonstrate that exogenous application of SA results in increased EIN3 protein abundance, and affects the ethylene-signaling pathway. Additionally, SA promotes EIN3 binding to and targeting of ORA59 for 26S proteasome degradation, thereby depleting the ORA59 pool required for transactivation of PDF1.2. On the other hand, JA does not promote this physical interaction, thus allowing the transactivation of PDF1.2 by ORA59. Accordingly we present the model (Figure 8) depicting the available pool of ORA59 as a coordination node for synergistic action of JA/ET and the antagonistic function of SA. The EIN3 physical interaction with ORA59 presents an additional level of complexity considering the capability of EIN3 to interact with at the very least three other proteins JAZ, MYC2 and DELLA (Hou et al. 2010; An et al. 2012; Hong et al. 2012; Yang et al. 2012). All together these findings position protein-protein interaction at the heart of integration of signaling pathways, and as such examining their dynamics a priority for future studies.
Figure 8. Simplified schematic model of EIN3/EIL1 opposing functions in JA-induced and SA-reduced expression of PDF1.2.
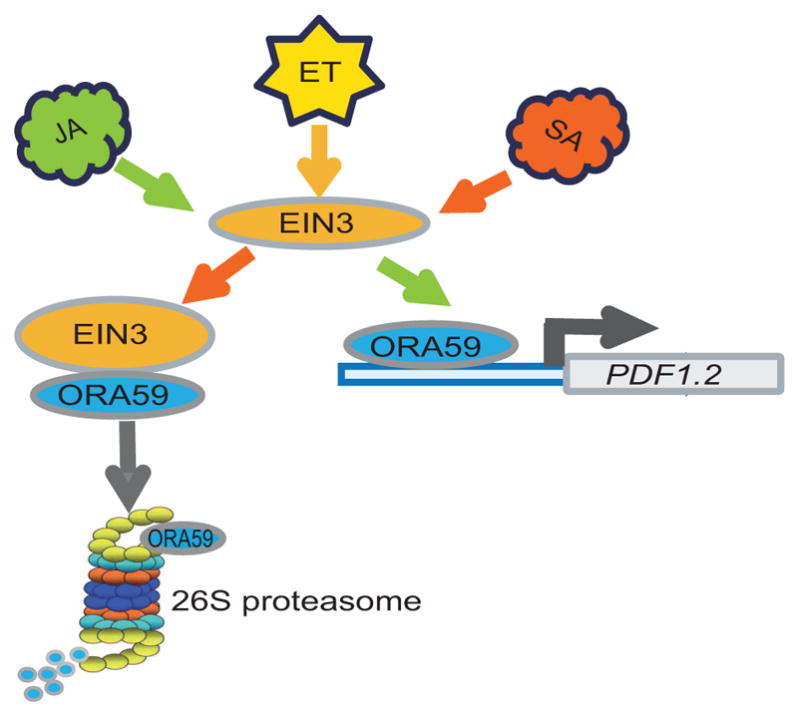
SA treatment promotes EIN3 binding to, and targeting of ORA59 for degradation, while JA treatment boosts EIN3 transactivation of ORA59 and consequently induction of PDF1.2.
MATERIALS AND METHODS
Plant growth condition and hormone treatment
All the mutants and transgenic lines used in this study are from Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Wild type) background that for simplicity here is designated Wild type. The ctr1 (CS8057) was obtained from the ABRC stocks, ein3/eil1 was kindly provided by Hongwei Guo (Peking University, China). Homozygous plants generated from various crosses identified by PCR-based genotyping using primers listed in Supplemental Table 1.
The various genotypes (Wild type, ctr1 and ein3/eil1) were grown in LD (16/8 h photoperiod) condition at 22°C. Exogenous application of jasmonic acid (JA) 100 μM, salicylic acid (SA) 1 mM, or JA (100 μM) + SA (1 mM) combination in 0.001% Triton X-100 were conducted by spraying 2-week-old plants, and 0.001% Triton X-100 spraying was used as mock. Analyses were performed on rosette leaves 24 hours post treatment. All experiments were conducted on soil grown plants unless otherwise noted.
To grow plants on plates, surface-sterilized seeds were planted on 1/2 Murashige and Skoog (MS) medium (2.16 g/L MS salts, pH 5.75, and 8 g/L agar) and imbibed for 3 d at 4°C to improve germination uniformity.
Plasmid construction and generation of transgenic plants
Overexpression constructs were prepared by amplifying EIN3 (AT3G20770) and ORA59 (AT1G06160) from the cDNA of Wild type using listed primers (Supplementary Table 1), followed by cloning the genes into pENTR™-D-TOPO vector (Invitrogen), and subsequent recombination with pEN-L4-p35S-R1 and pEN-R2-Luc-L3 using multiple LR reactions together with pB7m34GW as a destination vector according to MultiSite Gateway® Pro (Invitrogen). The resulting binary vectors contained 35S:ORA59:LUC and 35S:EIN3:LUC sequences were used to transform Wild type and ein3 eil1 by the floral dip method (Clough and Bent 1998).
Nuclear localization, luciferase split assay and yeast two-hybrid analyses
The coding regions of EIN3 and ORA59 were cloned from Arabidopsis (Wild type) cDNA, followed by split luciferase assay and yeast-two-hybrid (Y2H) assays for protein-protein interaction, and co-filtration assays in Nicotiana benthamiana for nuclear localization performed as previously described (Wang et al. 2014a).
The initial experiments using Y2H were performed with constructs containing pDEST32-ORA59, pDEST32-EIN3 and pDEST32 (empty vector as control) transformed into the yeast strain MaV203. The outcome displayed auto activation by ORA59 and EIN3. To identify the auto activation region of ORA59, we divided ORA59 into three segments, that is amino acids 1–80 (ORA59-N)-, and the middle amino acids 81–200 (ORA59-M)-, and carboxyl terminus amino acids, 201–244, (ORA59-C), and cloned them individually into pDEST32, followed by their introduction into yeast strain MaV203. These experiments identified ORA59-N as the auto activation domain. To examine the protein-protein interaction between EIN3 and ORA59, we constructed and transformed pDEST32-ORA59-M and pDEST32-ORA59-C into MAV203, and introduced pDEST22-EIN3 and pDEST22 empty vector into MAV103. The subsequent yeast mating and selection were performed as previously described (Wang et al. 2014a; Wang et al. 2014b).
Expression analyses
Expression analyses using qRT-PCR were conducted as previously described (Walley et al. 2008). Briefly, total RNA from rosette leaves was isolated by TRIzol extraction (Life Technologies) and further purified using the Qiagen RNeasy kit with on column DNase treatment (Qiagen) to eliminate DNA contamination. 2 μg RNA was reverse transcribed using Superscript III (Invitrogen). qRT-PCR was performed using At4g34270 and At4g26410 as reference genes for the internal controls as previously described for transcript normalization (Walley et al. 2008). Primers used are listed in Table S1.
Luciferase imaging
The luciferase imaging was performed as previously described (Walley et al. 2007) using a CCD camera (Andor Technology) 72 h post infiltration. Images were acquired every 10 min for 120 min, and luciferase activity was quantified as mean counts pixel−1 exposure time−1 using Andor Solis image analysis software (Andor Technology).
Histochemical GUS staining
GUS histochemical staining of 5XEBS:GUS transgenic Arabidopsis plants was performed according to the previously described method (Jefferson 1987).
Statistical Analyses
To determine statistical significance we employed Tukey’s HSD (Honestly Significant Difference) test. The difference was considered significant if p < 0.05.
Supplementary Material
Relative expression levels of PDF1.2 24 h after JA, SA, and JA+SA to mock treatments of the three genotypes of WT, ctr1, and ein3 eil1 grown either on plates (A–C), or on soil (D–F). The transcript levels of PDF1.2 were normalized to At4g34270 (T1P41-like family protein) and At4g26410 (M3E9) measured in the same samples. Data are mean ± SD of three independent biological replicates each with three technical repeats. Letters above bars indicate significant differences as determined by Tukey’s HSD (p <0.05).
This Figure is extracted from the data in Figure 3 to show the relative expression levels of ORA59 in response to JA and SA versus mock treatment in WT, ctr1 and ein3/eil1 genotypes. Asterisks above bars indicate significant differences as determined by Tukey’s HSD (p <0.05).
Representative of transient expression assays in Nicotiana benthamiana displayed by bright- (A) and dark-field (B) of a leaf expressing 35S: LUC (control), and 35S:EIN3:LUC-C together with 35S:ERF1:LUC-N.
Table S1. List of primers
Acknowledgments
The authors wish to thank DRs Marta Bjornson, Amancio de Souza and Jin-Zheng Wang for their comments on the manuscript. This work was supported by National Institutes of Health (R01GM107311), and National Science Foundation (IOS-1036491and IOS-1352478) grants awarded to KD. We have no conflict of interest.
Footnotes
AUTHORS CONTRIBUTIONS
X. H. has performed all the luciferase split assay and yeast two-hybrid analyses, J. J. carried out the expression analyses and assisted in manuscript preparation, C.W. performed cloning and generation of transgenic plants, and K.D. wrote the manuscript.
References
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012;22:915–927. doi: 10.1038/cr.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo I, Keller MM, Cargnel MD, Demkura PV, de Wit M, Patitucci MS, Pierik R, Pieterse CM, Ballaré CL. Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 2012;158:2042–2052. doi: 10.1104/pp.112.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem. 2002;277:19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- Clarke SM, Cristescu SM, Miersch O, Harren FJM, Wasternack C, Mur LAJ. Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 2009;182:175–187. doi: 10.1111/j.1469-8137.2008.02735.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux JP, Van Loon LC, Dicke M, Pieterse CM. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- Dong XN. SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 2012;24:2635–2648. doi: 10.1105/tpc.112.098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XL, Lee LYC, Xia KF, Yen YY, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Ju CL, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang JH, Garrett WM, Kessenbrock M, Groth G, Tucker ML, Cooper B, Kieber JJ, Chang C. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:19486–19491. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012;17:22–31. doi: 10.1016/j.tplants.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Ecker JR. Ethylene gas - Its not just for ripening any more. Trends Genet. 1993;9:356–362. doi: 10.1016/0168-9525(93)90041-f. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, Van Loon LC, Pieterse CMJ. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant physiol. 2008;147:1358–1368. doi: 10.1104/pp.108.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, Du Y, Koornneef A, Proietti S, Korbes AP, Memelink J, Pieterse CM, Ritsema T. Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol Plant Microbe Interact. 2010;23:187–197. doi: 10.1094/MPMI-23-2-0187. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. Jasmonate-insensitive1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50:128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- Pre M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen ZX, Huang SSC, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338:390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsema T, van Zanten M, Leon-Reyes A, Voesenek LACJ, Millenaar FF, Pieterse CMJ, Peeters AJM. Kinome profiling reveals an interaction between jasmonate, salicylate and light control of hyponastic petiole growth in Arabidopsis thaliana. PLoS ONE. 2010;5:e14255. doi: 10.1371/journal.pone.0014255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao QM, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Gene Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, Xie D. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell. 2014;26:263–279. doi: 10.1105/tpc.113.120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Humphrey PT, Whiteman NK. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012;17:260–270. doi: 10.1016/j.tplants.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Korbes AP, Memelink J, Ritsema T, Van Wees SC, Pieterse CM. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell. 2013;25:744–761. doi: 10.1105/tpc.112.108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 2007;3:1800–1812. doi: 10.1371/journal.pgen.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ, Wagner D, Dehesh K. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 2008;4:e1000237. doi: 10.1371/journal.ppat.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Guthrie C, Sarmast MK, Dehesh K. BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell. 2014a;26:3589–3602. doi: 10.1105/tpc.114.130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Sarmast MK, Jiang J, Dehesh K. The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell. 2015;27:1128–1139. doi: 10.1105/tpc.15.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang J, Jiang J, Chen S, Guan Z, Liao Y, Chen F. Reference genes for normalizing transcription in diploid and tetraploid Arabidopsis. Sci Rep. 2014b;4:6781. doi: 10.1038/srep06781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Zhang CL, Ji YS, Zhao Q, He WR, An FY, Jiang LW, Guo HW. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22:1613–1616. doi: 10.1038/cr.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li JG, Deng XW, Lee CM, Thomashow MF, Yang YN, He ZH, He SY. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A, Korbes AP, Younessi P, Montiel G, Champion A, Memelink J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol. 2011;75:321–331. doi: 10.1007/s11103-010-9728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu Z, An F, Hao D, Li P, Song J, Yi C, Guo H. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell. 2014;26:1105–1117. doi: 10.1105/tpc.113.122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZQ, An FY, Feng Y, Li PP, Xue L, Mu A, Jiang ZQ, Kim JM, To TK, Li W, Zhang XY, Yu Q, Dong Z, Chen WQ, Seki M, Zhou JM, Guo HW. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative expression levels of PDF1.2 24 h after JA, SA, and JA+SA to mock treatments of the three genotypes of WT, ctr1, and ein3 eil1 grown either on plates (A–C), or on soil (D–F). The transcript levels of PDF1.2 were normalized to At4g34270 (T1P41-like family protein) and At4g26410 (M3E9) measured in the same samples. Data are mean ± SD of three independent biological replicates each with three technical repeats. Letters above bars indicate significant differences as determined by Tukey’s HSD (p <0.05).
This Figure is extracted from the data in Figure 3 to show the relative expression levels of ORA59 in response to JA and SA versus mock treatment in WT, ctr1 and ein3/eil1 genotypes. Asterisks above bars indicate significant differences as determined by Tukey’s HSD (p <0.05).
Representative of transient expression assays in Nicotiana benthamiana displayed by bright- (A) and dark-field (B) of a leaf expressing 35S: LUC (control), and 35S:EIN3:LUC-C together with 35S:ERF1:LUC-N.
Table S1. List of primers