Abstract
RNAi has become an essential tool in C. elegans research. This unit describes procedures for RNAi in C. elegans by microinjecting with dsRNA, feeding with bacteria expressing dsRNA and soaking in dsRNA solution, as well as high-throughput methods for RNAi-based screens.
keywords for indexing: RNAi, microinjection, feeding, soaking, high-throughput screening assays, C. elegans
Introduction
The nematode C. elegans is a useful genetic model system to study a variety of biological processes (see Wood, 1988; Hope, 1999). Embryonic development, from fertilization to hatching, occurs in just 12 hours at room temperature, resulting in a first-stage larva with just over 550 cells. Newly hatched larvae progress through four larval molts to reach adulthood in ∼3 days, and adult hermaphrodites produce ∼300 self-progeny over the next three days. Both larval and adult worms have remarkably simple anatomies, consisting of an inner tube comprised of the pharynx and intestine, surrounded by a hydrostatic, fluid-filled body cavity and by an outer tube that consists of the musculature, hypodermis, and a collagenous cuticle. Remarkably, the diverse tissues that comprise the adult are specified by a total of only ∼1000 somatic cells. Due to the highly stereotyped cell division patterns, the origins and fates of each cell in the embryo and adult are known (Sulston and Horvitz, 1979; Sulston et al., 1983). Consequently, perturbations of the developmental program can be traced to changes in the fates of individual cells.
The ability to generate mutants in C. elegans has made this species a powerful model genetic organism. Since its discovery, RNA interference (RNAi) has become an important tool for C. elegans research and the scientific community in general. RNAi is a potent, specific, rapid, and simple method for determining loss-of-function phenotypes of genes in C. elegans, and has been the basis of several genome-wide screens (Fraser et al., 2000; Gonczy et al., 2000; Piano et al., 2000; Maeda et al., 2001; Ashrafi et al., 2003; Kamath et al., 2003, Watson et al., 2013). RNAi is also an excellent way to examine genetic interactions. For example, a dsRNA of interest can be delivered to a battery of mutants that might represent sensitized genetic backgrounds (Lehner et al., 2006b; Byrne et al., 2007). Thus, if RNAi in wild-type animals fails to give a phenotype, one may detect suppression or enhancement of phenotypes in mutant strains.
In C. elegans, RNAi can be induced by delivering dsRNA by microinjection (see Basic Protocol 1 and Alternate Protocol 1), feeding (see Basic Protocols 2, 3, and 4 and Alternate Protocol 2), and soaking (see Basic Protocol 5). Though not covered in this unit, RNAi can also be induced by in vivo transcription of dsRNA from hairpin or co-injected sense and antisense transgenes (promoter-driven RNAi; Tabara et al., 1999; Tavernarakis et al., 2000). In the microinjection and soaking protocols, in vitro preparations of dsRNA are delivered mechanically using a needle to inject dsRNA directly into the body or passively by soaking worms in dsRNA solution. In the feeding protocol, dsRNA is transcribed in E. coli and ingested by animals. The protocols provided in this unit are meant to help the complete novice at worm culture select and apply the appropriate RNAi delivery method. Towards that end, protocols are also provided for transferring and propagating (see Support Protocol 1) and for sexing and mating (see Support Protocol 2) C. elegans. References for more in depth discussion of these topics as well as C. elegans in general are provided at the end of the unit (see Key References and Internet Resources).
The RNAi method of choice depends on the experimental goal. Though it can be technically challenging at first, microinjection of dsRNA offers a quick turnaround, since dsRNA can be prepared in vitro using a PCR product as a template and stored at −80°C, and special bacterial strains or plates are not required. It is also possible to simultaneously inhibit two or more genes by coinjecting dsRNAs within a mixture (Dudley et al., 2002). Feeding dsRNA is simple and requires no special microinjection system. Furthermore, a sufficient volume of mutant lysate can be obtained for biochemical extracts or molecular analyses, such as immunoprecipitation, by feeding dsRNA to a large population of worms. RNAi-based genetic screens can be performed in microtiter format by feeding or soaking worms with dsRNA (Fraser et al., 2000; Maeda et al., 2001). In principal, promoter-driven RNAi can be used to silence a gene in a constitutive or regulated fashion, depending on the promoter used to drive dsRNA synthesis and if the transgene is integrated into the genome.
NOTE: All solutions and equipment coming into contact with organisms must be sterile, and proper sterile technique should be used accordingly.
Basic Protocol 1
Microinjecting Worms with dsRNA
Using a capillary needle, dsRNA can be microinjected anywhere into the body of a worm, and RNAi will spread systemically throughout the injected animal, including the germline, and into the next generation (Fire et al., 1998). To perform RNAi by microinjection, one will need to have access to a microscope equipped with a micromanipulator for holding and maneuvering the microinjection needle. Because it is not necessary to delivery dsRNA directly to the germline, injections need not be performed at high magnification or using differential interference contrast microscopy (Nomarski optics). Therefore, even if expertise in C. elegans microinjection is unavailable, it should be possible to master dsRNA microinjection with only a few hours of practice.
In this procedure, healthy L4 or adult hermaphrodites are immobilized under halocarbon oil on a dry agarose pad and microinjected with dsRNA, causing the body of the animal to noticeably inflate with the dsRNA solution. Injected animals are removed from the pad using a mouth pipet and a small amount of sterile M9 buffer, and then placed onto a C. elegans culture dish. After a period of recovery, animals that have moved (i.e., are alive) are transferred to individual plates and allowed to lay eggs for several days. The injected animals and their progeny are examined/scored for phenotypes resulting from gene silencing. In a successful RNAi by microinjection experiment, one should aim to recover ten viable injected hermaphrodites to obtain a high degree of confidence in the results.
In addition to the standard equipment for manipulating worms (e.g., stereomicroscope, platinum wire pick, media; Hope, 1999), this procedure requires microinjection equipment, including an inverted microscope equipped with a 10× objective, micromanipulator, N2 gas, and microinjection assembly to regulate the pressure and flow of the microinjection solution. The procedure described here utilizes a Nikon inverted microscope equipped with 10× objectives, gliding stage, and Narishige MN151 micromanipulator with fine z-axis adjustments, as well as a Tritech microinjector system for pressurizing the microinjection needle. Injections are normally carried out using the 10× objective for a total magnification of 100×. As noted above, it should be possible, with some practice, to perform microinjections with a range of different microinjection systems.
Important Note: The experimenter should become familiar with the microinjection setup before proceeding with the experiment.
Materials
0.1 to 1 μg/μl dsRNA
Halocarbon oil Series 700 (Halocarbon Products)
L4 to young-adult staged C. elegans (e.g., strain N2; CGC)
M9 buffer (see recipe)
NGM plates seeded with E. coli (see recipe)
Loading capillary (see recipe)
Microinjection needle (see recipe)
Inverted microscope with micromanipulator and microinjection assembly with N2 gas supply (e.g., TRITECH Research Microinjector System)
Agarose microinjection pads (see recipe)
30-mm Petri dish lid
Dissecting microscope
Platinum wire pick flattened at the end
Additional reagents and materials for transferring and propagating C. elegans (see Support Protocol 1)
Load and mount the microinjection needle
Pellet insoluble material that may clog the microinjection needle by centrifuging the 0.1 to 1 μg/μl dsRNA solution 5 min at 10,000 × g, 20°C.
-
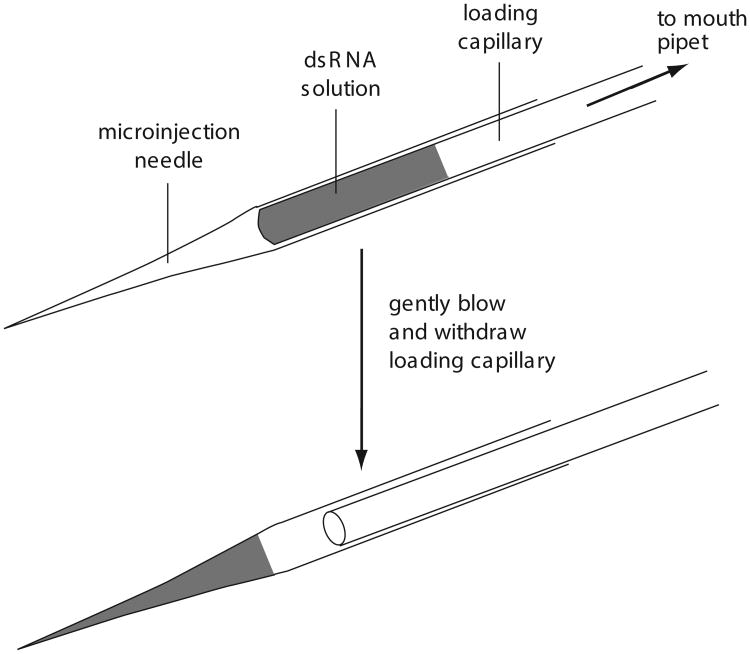
Touch the drawn-out end of the Pasteur pipet portion of the loading capillary to the dsRNA solution, allowing a small volume to be drawn into the pipet. Deposit the RNA solution just behind the drawn-out tip of the needle by threading the loading pipet all the way into the needle and gently blowing through the mouth pipet (Fig. 26.3.1).
Needles can be loaded and stored up to several hours inside a humidifying chamber constructed from a Petri dish containing a wet piece of filter paper. It is often prudent to load two needles for each microinjection solution and store one in the humidifying chamber as a backup in case of problems.
-
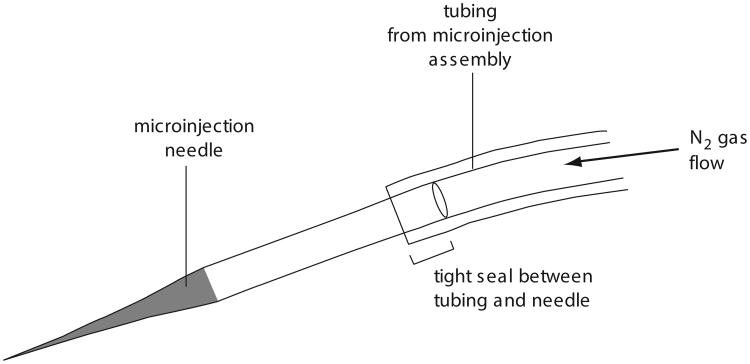
Attach the loaded microinjection needle to the microinjection assembly, sliding the tubing over the back end of the needle (Fig. 26.3.2).
The tubing should fit snugly over the needle making a tight seal. If the seal is not tight enough, the N2 gas will escape around the needle when pressure is applied and the dsRNA solution will not flow from the needle.
-
Turn on the light source of an inverted microscope. Mount the loaded needle onto the micromanipulator and roughly align the tip of the needle into the center of the light beam.
The needle should be positioned such that it has a downward angle of ∼15° to 20°.
Figure 26.3.1.
Loading the microinjection needle.
Figure 26.3.2.
Attaching the microinjection needle to the microinjection assembly via tubing.
Prepare worms for microinjection
-
5. Working at a dissecting microscope, prepare a microinjection pad by placing a small (∼10-μl) drop of halocarbon oil Series 700 on the glass surface of the coverslip adjacent to the region coated with dry agarose. Place the microinjection pad onto an inverted 30-mm Petri dish lid resting on the stage of a dissecting microscope.
This will hold the microinjection pad at approximately the same height as the worms on a small plate, so that minimal focusing will be required while transferring worms to the microinjection pad.
6. Briefly flame a platinum wire pick that has been flattened at the end for picking up worms.
-
7. Touch the pick to the surface of the halocarbon oil on the pad. Using this viscous oil drop on the end of the wire, pick up several L4- to young adult-staged C. elegans for microinjection. Transfer the worms to a dry portion of the pad by gently touching the underside of the pick to the agarose surface of the pad and sliding the pick along to gently push the worm down to the surface of the pad, making a mental note of the configuration of the animals.
Pick up the worms by gently touching the oil-coated undersurface of the pick to the top of the worm on the Petri dish. When the pick is raised, the worm should be transferred from the plate to the underside of the pick by surface tension. Move to another worm and repeat the procedure.
Beginners should start with two or three worms, but experts will pick up as many as twenty worms on the same pick. If possible, choose worms that are either off the E. coli lawn or are on a thin area.
It is not necessary to place the worms in a row, or even to separate the worms from one another, but it is helpful if the majority of the worms are angled with the long axis of their bodies oriented in the same general direction.
8. Go back to the oil drop with the pick and obtain additional oil to spread over the worms as necessary, making sure that all of the worms are completely covered.
-
9. If the worms are not sticking to the microinjection pad, gently pat the worms along the length of the body to encourage them to stick to the agarose pad.
Important Note: Once the animals have been transferred to the microinjection pad, work as quickly as possible to prevent the animals from drying out.
Use pads that are thoroughly dry to immobilize the worms. If necessary, rebake pads 1 hr in a 50°C oven. Check to make sure that the agarose side of the pad is facing up. This can be accomplished by breathing lightly on the pad; if the correct side is up, the agar will block condensation on the coverslip. The worms will not stick to bare glass.
Inject the dsRNA into worms
10. Place the microinjection pad with immobilized worms onto the stage of the inverted microscope and focus on the worms.
11. Move the tip of the microinjection needle into the center of the field of view, being careful to keep the needle above the worms (Fig. 26.3.3A). Maintain this needle position and only move the needle into or out of the focal plane (i.e., toward or away from the microinjection pad) using the z-axis control knob on the micromanipulator.
-
12. Orient the pad so that the general body axis of the animals parallels the orientation of the needle (Fig. 26.3.3).
If the needle is perpendicular to the body axis of the animal, it may completely penetrate the animal or cause it to roll over during the microinjection procedure.
-
13. Using the 10× objective, choose a worm at one end of the cluster of worms and position the worm directly under the needle tip by moving the stage.
Keep in mind the configuration of worms placed on the pad (step 7). Move from one end of the cluster to the other systematically.
Some investigators prefer to move the stage by hand, while others prefer to use the stage dials. Try both and use whichever method is more comfortable.
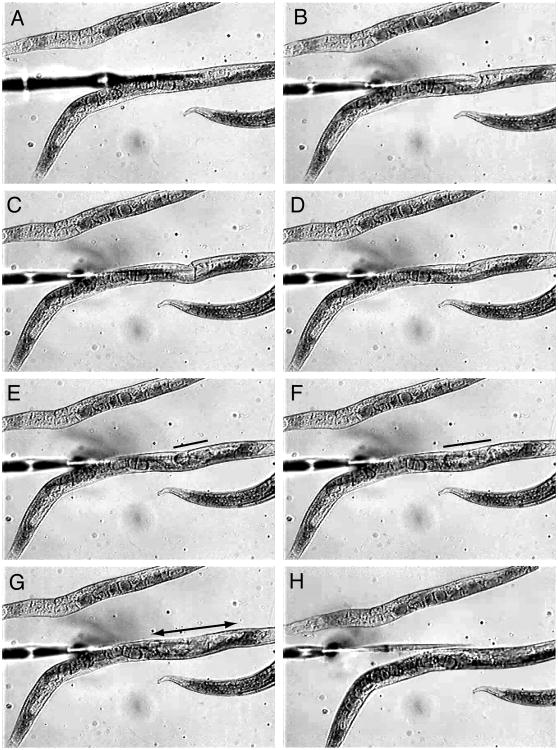
14. Focus carefully and lower the needle directly down on top of the first animal's body. With the needle pressing against the body of the animal (creating an indentation), push on the stage to gently move the worm into the needle tip (Fig. 26.3.3B to D).
-
15. Once the needle penetrates the cuticle, apply pressure to the needle and allow the dsRNA solution to flow into the animal.
With an ideal microinjection, the flow of the solution from the needle tip will be easily observable in both directions (i.e. towards the mouth and anus; Fig. 26.3.3E to G) and a slight increase in size near the point of injection may even be observed. Injecting too much solution will rapidly increase the size of the worm in both diameter and length, and the animal may not survive. Decrease the duration of the pulse and, if necessary, give several quick pulses to deliver the dsRNA into the animal.
16. Remove the needle from the worm by pulling back on the sliding stage (Fig. 26.3.3H). Raise the needle slightly using the z-axis knob on the micromanipulator and move to the next worm.
17. Repeat steps 13 to 16 until all of the worms are injected. Carefully remove the pad from the microinjection microscope and transfer to a dissecting microscope.
Figure 26.3.3.
Overview of the microinjection technique. (A) The injection needle enters the field of view from the left of the image and is oriented above and roughly parallel to the body axis of the worm. (B to D) The needle is lowered until the tip touches the worm forming a slight indentation. The worm is pushed against the needle until the tip of the needle penetrates the cuticle. (E to G) The dsRNA solution is injected into the body cavity of the worm and flows in both directions away from the tip (indicated by the bars and arrows). (H) The needle is then backed out of the worm by moving the worm away from the needle.
Recover injected animals for phenotype analysis
-
18. Recover the animals by placing a small drop of M9 buffer directly under the halocarbon oil so that it contacts the worms and gently floats them off of the agarose.
The loading capillary can be used for this purpose. Simply break off the very thin drawn-out end of the loading capillary, creating an opening just slightly larger in diameter than a worm. Using the capillary, blow M9 against the worms to loosen them from the pad and suck each worm up into the capillary.
Generally, it is wise to transfer worms in small groups, approximately five animals at a time, as occasionally worms become stuck to the glass inside the capillary.
-
19. Deposit the worms on a fresh NGM plate seeded with E. coli, and allow the worms to recover (i.e., incubate at 20°C) overnight (see Support Protocol 1).
Healthy worms that survive the microinjection should begin to move vigorously in the M9 buffer or shortly after transfer to the NGM plate.
20. Transfer each injected animal to a fresh NGM plate (day 1) and incubate an additional 24 hr at 20°C (see Support Protocol 1). Transfer the injected animals to a fresh NGM plate once more (day 2).
-
21. Monitor the injected animals and their progeny for phenotypes (see Hope, 1999; Wood, 1988).
The presence of embryos on day 1 plates 24 hr after removing the adult indicates that the microinjected dsRNA produces an embryonic lethal phenotype. However, always be sure to compare the injected animals and their progeny to uninjected or control-injected animals.
In some cases, postembryonic phenotypes can easily be determined by comparing the progeny of injected animals to the progeny of uninjected or control injected animals. These may include phenotypes such as uncoordinated (Unc), slow growth (Gro), sterility (Ste), larval lethal (Lvl), larval arrest (Lva), lethality (Let; any stage), egg-laying defective (Egl), high incidence of males (Him), dumpy (Dpy), small (Sma), long (Lon), blister (Bli), protruding vulva (Pvl), multivulva (Muv), and body morphology defects (Bmd).
Alternate Protocol 1
Microinjection into unc-42 rde-1 Animals Crossed to N2 Males to Examine Zygotic RNAi Phenotypes
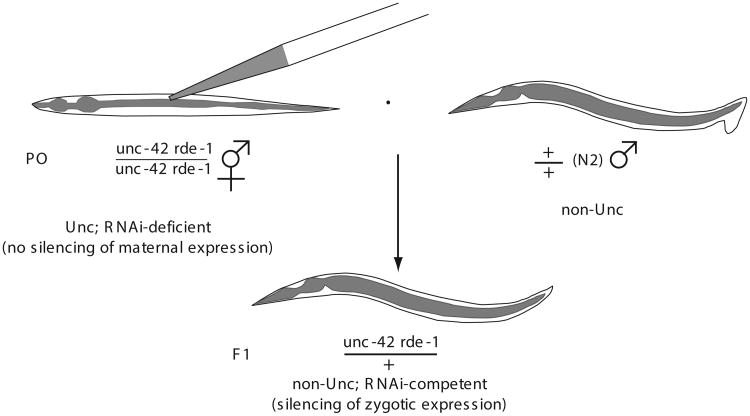
If RNAi by microinjection (see Basic Protocol 1) results in embryonic arrest, this could reflect a maternal function for the targeted gene. Zygotic activity of a targeted gene can be examined by injecting dsRNA into an RNAi-deficient (rde mutant) hermaphrodite mated to a wild-type N2 male (Fig. 26.3.4). In this scenario, maternal activity of the targeted gene will be unaffected since RNAi is not functional in the rde mutant hermaphrodite. However, wild-type males that successfully mate with the rde mutant hermaphrodite will provide RNAi activity in the cross progeny, thereby silencing zygotic expression of the targeted gene. To facilitate the identification of rde/+ cross progeny, an rde mutant that is marked with a recessive visible mutation—e.g., uncoordinated (Unc)—is mated to N2 males, injected with dsRNA, and the non-Unc cross progeny examined for phenotypes resulting from loss of zygotic activity of the targeted gene.
Figure 26.3.4.
Schematic of the zygotic RNAi procedure.
Additional Materials (also see Basic Protocol 1)
Male N2 C. elegans (CGC)
unc-42 rde-1 C. elegans (WM36; CGC)
Mating plates (see recipe)
Additional reagents and equipment for transferring and propagating (see Support Protocol 1), and mating C. elegans
Mate wild-type (N2) C. elegans males with an unc-42 rde-1 mutant strain overnight (see Support Protocol 2; Hope, 1999).
Inject dsRNA into the mated Unc hermaphrodites (see Basic Protocol 1).
Transfer (see Support Protocol 1) each injected hermaphrodite to individual mating plates with males to recover (i.e., incubate at 20°C) overnight and to continue mating (see Support Protocol 2).
Transfer the hermaphrodite alone to a fresh NGM plate and score non-Unc cross progeny for RNAi phenotypes as described above.
Basic Protocol 2
Feeding Worms dsRNA
RNAi can be induced by feeding worms bacteria expressing dsRNA (Timmons et al., 2001). First, a cDNA corresponding to the gene of interest is cloned into a bacterial expression vector between opposing phage T7 polymerase promoter sites. The feeding vector is then transformed into the E. coli HT115 strain carrying the DE3 lysogen (unit 1.8), providing IPTG inducible expression of the phage T7 RNA polymerase. The HT115(DE3) strain also lacks the Rnc gene, which encodes RNase III and is therefore deficient in degrading dsRNA. The RNAi food is prepared and seeded onto NGM plates containing IPTG. Animals are placed onto the RNAi food and both the hermaphrodite and progeny are examined and scored for phenotypes.
To simultaneously target two genes by feeding, we and others (Min et al., 2010) have found it best to fuse two cDNA fragments, one from each gene, into a single feeding vector. Mixing of two E. coli strains also works, but is more prone to error caused by unequal mixing or RNA expression in the two different feeding strains. Do not attempt to feed animals that were previously injected with dsRNA; for unknown reasons the microinjection of dsRNA (even nonspecific dsRNA) will suppress RNAi by feeding.
This protocol can be easily scaled-up to examine the molecular and biochemical consequences of depleting a particular gene product.
RNAi by feeding in microtiter format–the topic of Basic Protocols 3 and 4 and Alternate Protocol 2–allows systematic screens to be performed (Fraser et al., 2000, Sonnichsen et al., 2005, Lehner et al., 2006c, Watson et al., 2013).
Materials
cDNA
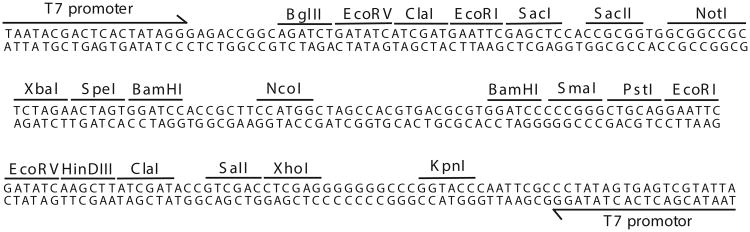
L4440 double T7 RNAi feeding vector (Fig. 26.3.5; Addgene)
Figure 26.3.5.
Multiple cloning site of the double T7 RNAi feeding vector L4440. Opposing phage T7 promoters flank the multiple cloning site of L4440. The restriction sites specific to the MCS are shown, and unique sites are indicated in bold.
E. coli RNAi feeding strain HT115(DE3) (CGC)
Terrific broth (TB; unit 1.1) containing 50 μg/ml each ampicillin and tetracycline (unit 1.4)
TB containing 50 μg/ml ampicillin
M9 buffer (see recipe)
M9/15% glycerol (see recipe)
NGM/amp/IPTG plates (see recipe)
Gravid/young adult, L1 larvae, or L4 larvae C. elegans of appropriate strain(s) (e.g., N2, CGC)
NGM/amp/IPTG plates (see recipe) seeded with HT115 containing empty L4440 RNAi feeding vector
Additional reagents and equipment for subcloning DNA fragments (unit 3.16), transforming E. coli (unit 1.8), growing E. coli (unit 1.2), and transferring and propagating C. elegans (see Support Protocol 1)
NOTE: Perform each RNAi experiment in triplicate.
Prepare the RNAi feeding strain
-
Clone a cDNA for the gene of interest into L4440 double T7 RNAi feeding vector (unit 3.16).
Avoid introns—the presence of introns in the feeding construct may reduce the efficiency of interference. Alternatives to L4440 include Litmus 28i and 38i (NEB) and Gateway-compatible versions of L4440, L4440-dest-RNAi (Rual et al., 2004) and L4440-gtwy (Caldwell et al., 2006). L4440 and L4440-gtwy are available through Addgene.
Transform the RNAi feeding construct into the E. coli feeding strain HT115(DE3) (unit 1.8).
Pick a colony and inoculate 2 ml terrific broth (TB) containing 50 μg/ml each ampicillin and tetracycline, and grow overnight at 37°C (unit 1.2).
-
Seed the starter culture into 1 liter TB containing 50 μg/ml ampicillin and incubate with shaking at 37°C for 8 to 16 hr (unit 1.2).
Tetracycline in this culture is unnecessary and, in fact, may reduce the efficiency of RNAi (Kamath et al., 2001).
Cultures can be scaled down; however, excess food from large cultures can be stored at –80°C, for several months.
Harvest the bacteria
-
5a. For centrifugation: Centrifuge bacteria 10 min at 800 × g, 4°C. Resuspend the bacterial pellets in M9 buffer (use 25 ml for each liter of culture) and transfer to a 50-ml conical tube.
Harvest food by centrifugation if food is needed urgently.
If time permits, allow the bacteria to precipitate passively to reduce the likelihood of contaminating the dsRNA food.
- 5b. For passive precipitation: Replace TB medium with M9 buffer as follows:
- Allow bacteria to settle to the bottom of the culture flask overnight at 4°C. Carefully aspirate as much of the medium from the flask as possible without disturbing the bacteria, which forms a soft layer at the bottom of the flask. Swirl the flask to suspend the bacteria in the remaining volume of TB and pour into a 50-ml conical tube.
-
Pellet the bacteria in a clinical centrifuge 10 min at 800 × g (4500 rpm), 20°C. Remove broth by aspiration.These pellets tend to be soft and pouring off the medium could result in loss of the bacterial pellet.
- Wash the bacteria in 5 pellet volumes M9 buffer by vortexing until the clumps are dissolved.
6. Pellet the bacteria in a clinical centrifuge 10 min at 800 × g (4500 rpm), 20°C.
-
7a. For immediate/short term use: Resuspend washed bacteria in 5 pellet volumes of M9 buffer. Store up to 1 month at 4°C.
RNAi food is ready to use after resuspension in either buffer.
7b. For storage: Resuspend washed bacteria in M9/15% glycerol. Store in 10-ml aliquots for several months at –70°C.
8. Seed an appropriate number of NGM/amp/IPTG plates with one drop (∼100 μl) each RNAi food.
-
9. Allow plates to dry overnight at room temperature.
This also allows induction of dsRNA production and consistently produces the most penetrant RNAi phenotypes by feeding.
Seeded plates can be stored in a sealed container up to several weeks at 4°C. Warm the RNAi plates to room temperature prior to use.
Cultivate worms on RNAi food and analyze phenotypes
-
10a. Test for embryonic and post-embryonic phenotypes: Place single gravid adult C. elegans onto NGM/amp/IPTG plates. As a control, place single gravid adults onto HT115(DE3) containing the empty L4440 RNAi feeding vector. After 24 hr, transfer each RNAi-fed or control-fed adult to a fresh plate of the same type and count the number of progeny (eggs and larvae) on each plate.
By placing gravid adults onto RNAi plates, one can assess the ability of the RNAi to produce embryonic as well as postembryonic phenotypes on the same plate.
10b. Test for embryonic lethal phenotypes: Place single L4 animals onto NGM/amp/IPTG (RNAi) and control—i.e., seeded with HT115(DE3) containing the empty L4440 RNAi feeding vector—plates. After 24 hr, transfer each adult to a fresh plate of the same type onto which it was seeded (i.e. RNAi or control) and count the number of progeny (eggs and larvae) on each plate.
10c. Test for postembryonic phenotypes: Place 20 newly hatched L1 larvae onto RNAi and control plates.
-
11. Examine the plates each day to determine whether the RNAi food produces any embryonic and/or postembryonic phenotypes.
The presence of embryos on RNAi plates 24 hr after removing the adult indicates that the RNAi produces an embryonic lethal phenotype. However, always be sure to compare RNAi plates to control plates.
In some cases, postembryonic phenotypes can easily be determined by comparing the RNAi-fed progeny with the control-fed progeny.
Basic Protocol 3
RNAi by Feeding i 96-Well Agar Plates
The ability to perform RNAi knockdown by feeding makes genome-scale RNAi screens feasible. The principles are the same as described in Basic Protocol 2, but the protocol must be adapted for use in high-throughput 96-well format. Because a single hermaphrodite can produce ∼300eggs, however, the wells would become overcrowded and the animals would quickly consume all of the food, making it unfeasible to examine second-generation adults in 96-well format. High-throughput screens therefore are typically performed within a single generation or focus on phenotypes that can be scored in early larval stages, before the population runs out of food.
Genome-scale RNAi screens can easily be performed in 96-well liquid or agar or 24-well agar plates. The choice of platform depends on many factors including the ability to visualize the phenotype of interest in liquid vs agar and the variability and penetrance of phenotypes to be scored. For screens in 96-well format, phenotypes to be screened should be robust, because the number of animals per well (10 to 20) is limited. Worms grown on solid substrate appear healthier and are easier to visualize than those grown in liquid, where the turbidity and thrashing can obstruct cellular detail and fluorescence.
A genome-scale RNAi screen requires an RNAi clone library, equipment and space to incubate 96-well deep-well plates, and access to a suitable microscope for scoring phenotypes of interest. Genome-scale libraries have been created (Kamath et al., 2003, Rual et al., 2004) and can purchased as a collection of clones carried in E. coli HT115(DE3) and frozen in 96- or 384-well formats (System Bioscience and GE Dharmacon). These libraries should be maintained at –80°C. Mini-libraries are also available for specific gene sets, including transcription factors, phosphatases, and chromatin factors (Ackerman and Gems, 2012, and System Bioscience), and custom libraries can be created by hand picking the desired clones from available libraries and re-arraying them into new 96-well plates. A genome-scale screen requires the growth and examination of over two hundred 96-well plates, and thus the screen is usually broken up into a series of smaller screens, staggered over multiple days. The number of plates to set up and screen in a single round will depend both on the available space for growth of bacteria and the number of plates that can be screened in a single day.
The protocol below outlines large-scale screening on solid media within a single generation, focusing on phenotypes that can be observed on a dissection microscope. We use this protocol extensively to identify regulators of gene expression using fluorescent reporter strains and to examine post-embryonic phenotypes. In this procedure, eggs or early larval animals (L1s) are placed on the RNAi feeding strains and screened for phenotypes after 2-4 days, once control animals have reached the adult stage.
This protocol can be made more efficient through the use of high-throughput liquid handling or pipetting systems. The Viaflow 96 (Integra Biosciences), or similar 96 well pipetting tools can be used to dispense media and bacteria. In addition, a COPAS biosort (Union Biometrica) can be used to add animals to plates. However, these high-tech approaches are not essential. Although more laborious, an 8-channel repeat pipettor can be used to dispense media, and multichannel pipettes can be used to inoculate and dispense bacteria and worms. Thus, genome-scale screens need not be limited to labs equipped with expensive liquid handling systems.
Materials
LB broth
Tetracycline
Ampicilin or carbenicillin
NGM broth and agar (see recipe)
M9 buffer (see recipe)
Buffered bleach (see recipe)
IPTG
RNAi library frozen at –80°C
Flat-bottom 96-well plates
96-well deep-well plates (2 mL)
Breathable lids, such as Airpore tape sheets (Qiagen)
Large-volume reservoirs for media (purchased or home-made using autoclaved micropipette tip boxes)
50-ml reagent reservoirs for eggs and L1s
Aluminum seal for frozen stocks
Grow worms to collect eggs
Calculate the number of eggs required for the experiment. For each 96-well plate to be screened, you will need approximately 2500 eggs. A 10-cm plate containing 1000 adult wild-type animals fed E. coli OP50 will provide at least 50,000 eggs. Prepare an appropriate number of 10-cm NGM plates for the experiment, and seed each 10-cm NGM plate with 1000 hatched L1s. Incubate the worms at 20°C for about 3 days. The cultures will be ready for hypochlorite bleaching when you see a lot of embryos on the plate or in the mothers.
Prepare 96-well NGM-agar/amp/IPTG screening plates
-
Prepare NGM agar plus 50 μg/ml ampicillin and 5 mM IPTG as described. Keep the NGM/amp/IPTG media hot while plates are being poured by placing the media in a 60°C waterbath and pouring into reservoirs for dispensing as needed.
96-well NGM/amp/IPTG-agar plates should be prepared just prior to use, one day before seeding them with bacteria. The small volume of these wells makes them prone to dehydration, which can cause the agar to crack or pull away from the sides of wells, allowing worms to leave the agar surface.
25μg/ml carbenicillin can be used in lieu of ampicillin. Although more expensive than ampicillin, carbenicilin is more stable.
-
Dispense 200 μl NGM/amp/IPTG-agar media into each well of a 96-well plate. Move quickly to ensure that media remains hot during dispensing. Media can be dispensed using liquid handling systems, 96-channel pipetors, such as the Viaflow 96, or with an 8-channel repeat pipettor using large-bore (P1000) tips.
Tips need to be changed regularly as they will start to clog resulting in uneven dispensing of agar. We recommend using barrier tips to ensure that agar is not sucked into the dispensing machinery.
Prepare bacteria
-
3. Prepare the required number of 96-well deep-well plates by filling each well with 1 ml LB broth containing 50 μg/ml ampicillin and 15 μg/ml tetracycline using a liquid handling system or 96-channel pipetting system.
If controls are not present within the library plates, prepare a separate plate for controls. The L4440 vector without insert in HT115(DE3) is used as a negative control and positive controls can be phenotypic, such as unc-22 (twitching) or lin-26 (larval arrest, lethality), or target fluorescence markers such as GFP.
4. Remove library stock plates from the −80°C freezer and place on dry ice.
5. Remove aluminum seal carefully to avoid the generation of aerosols that could cross contaminate stock wells.
-
6. Inoculate starter cultures by gently scraping frozen bacteria from the wells of the stock plate using a 96-well pipetting system or multichannel pippette equipped with sterile pipette tips, and then dip the scraped bacterial cells into the deep-well plates containing LB plus ampicillin and tetracycline prepared in step 3.
Depending on the number of screens to be done, it may be more economical to purchase a pin tool, which can be washed, bleached and flamed with ethanol to sterilize tool between plates. If purchasing a pinner, be certain to purchase one with pins long enough to be used with deep-well plates.
Repeated freezing and thawing of stocks is not recommended. However, we find that it is often easier and more reliable to inoculate from thawed stocks than frozen and thus, we recommend making a working copy if the library will undergo repeated cycles of freezing and thawing. Working copies can then be regenerated regularly from original stocks.
7. Cover library stock plates with a new aluminum seal, and return the plates to the −80°C freezer immediately.
-
8. Cover the starter cultures with breathable sealing film, incubate at 37°C overnight on a vibrating platform or shaking incubator.
Vibrating platforms are available that can be placed in an incubator or warm room, allowing multiple 96-well deep-well plates to be processed simultaneously. Stacking adapters are available for shaking platforms that allow four to five 96-well deep-well plates to be stacked together in the incubator.
9. The next day, prepare an appropriate number of 96-well deep-well plates (one for each starter plate) containing 1 ml LB plus 50 μg/ml ampicillin.
10. Remove 50 μl from each well of starter culture and inoculate into the corresponding wells of deep-well plates containing LB/amp. Incubate with shaking at 37°C for 6 hours; A600 should be approximately 0.8.
11. Pellet bacteria in a table-top centrifuge 15 min at 2700 rpm, and remove supernatant by rapidly inverting the plate over a container to catch the supernatant, which should be disposed of appropriately.
12. Suspend each bacterial pellet in 100 μl (1/10th volume) M9 by pipetting up and down 5-6 times. This can be done using the mix function on many electronic pipetting systems or by hand using a multichannel pipette.
Seed NGM-agar/amp/IPTG screening plates with bacteria
-
13. Using a 96-well liquid dispenser or a multichannel pipette, transfer 10-15 μl of the bacterial suspension from each well of the deep-well plates to the corresponding wells of a 96-well NGM/amp/IPTG-agar plate, being careful not to touch the top of the agar with the pipette tips.
Touching the agar surface can create holes or depressions in the agar that worms will crawl into.
-
14. Dry plates in a laminar flow hood checking central wells for liquid at 15 min intervals. Remove plates as soon as central wells are dry.
Overdrying will cause agar to crack. If cracking of peripheral wells is a problem, an additional 5 to 10 μl of sterile water or M9 can be added to these wells to prevent overdrying.
15. Once plates are sufficiently dry, wrap plates with plastic film or seal in zip-lock bags to prevent further drying.
16. Leave plates overnight at room temperature to induce expression of dsRNA.
Add worms to screening plates
-
17. Collect eggs from worms growing on 10-cm plates by hypochlorite bleaching (see Support Protocol 3). Animals can be added to screening plates as eggs or L1s. If L1s will be used, allow the embryos to hatch overnight in M9.
A benefit of using L1s is that the viability of embryos can be monitored after bleaching and prior to plating, ensuring that an intended number of viable worms are plated. In addition, the cultures will be more synchronous.
18. Determine the number of embryos or L1s per 7 μl of suspension, and dilute the suspension of worms so that 7 μl contains between 20 and 25 embryos or L1s.
19. Put the worm suspension into a sterile 50 ml reagent reservoir and using a multichannel pipette dispense 7 μl (20 to 25 embryos or L1s) to each well of each screening plate. Agitate the reservoir frequently to prevent embryos or larvae from settling to the bottom.
20. Allow the plates seeded with worms to dry in a laminar flow hood as described in step 14
21. Once the plates with worms are dry, invert plates, seal them in zip lock bags to maintain humidity, and incubate 3 to 4 days.
22. Use the remaining worm suspension to inoculate 10-cm NGM plates for the next round of screening.
23. After 3 to 4 days, screen plates for worms with desired phenotypes using a dissecting microscope.
-
24. For wells that give a positive phenotype, streak the bacterial clones from frozen stocks onto LB plates containing ampicillin and isolate single colonies for retesting and sequencing.
Due to the risk of cross-contamination between wells of RNAi libraries stored in 96- or 384-well plates, we suggest that positive clones be streaked for single colonies and that individual colonies be picked, grown, and used for retesting.
25. Individual clones (including the Arhinger and ORFeome library clones) that retest as positive can be amplified from bacteria by PCR using the primers: RNAiRv:TGGATAACCGTATTACCGCC and RNAiFw: GTTTTCCCAGTCACGACGTT.
Alternate Protocol 2
Screening in 24-Well Agar Plates
For some screens, the expected phenotypes (e.g., those with variable penetrance) require examination of a larger number of animals than can be grown in 96-well format. These screens can be adapted to high-throughput using 24-well plates, as described below.
Additional Materials (see also Basic Protocol 4)
24-well plates
Wheaton unispence
3-mm tubing
12-channel pipette
Prepare NGM agar plus 50 μg/ml ampicillin and 5mM IPTG.
Using a Wheaton unispence equipped with 3-mm tubing, dispense 2 ml NGM/amp/IPTG agar into each well of a 24-well plate. Prepare four 24-well plates for each 96-well plate from the library to be screened. 2 L NGM agar will produce 40 plates (enough to screen ten 96-well plates).
Prepare bacterial RNAi feeding strains in 96-well deep well plates as described in steps 3 to 6 of Basic Protocol 4.
-
After resuspending bacteria in M9, add 30 μl of each bacterial suspension to the 24-well plates.
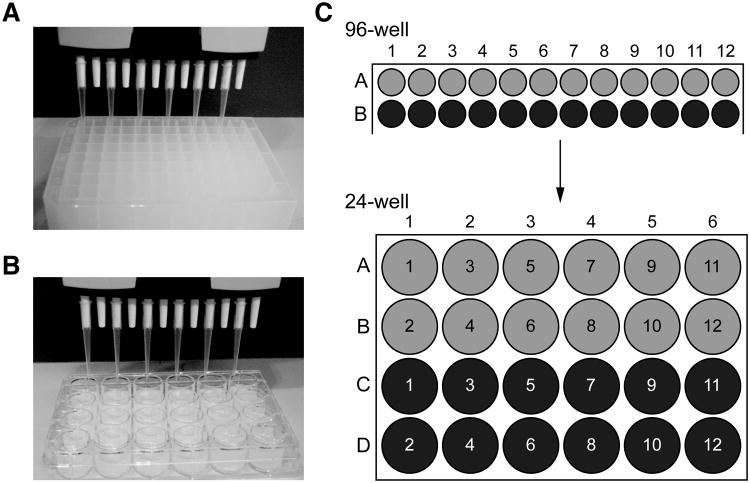
Although there are a number of strategies to go from 96-well to 24-well plates, using a 12-channel pipette with tips added to every other channel is the simplest. In this configuration, each row of a 96-well plate is transferred to two rows of a 24-well plate: for example, samples from the odd-numbered wells in row A of the 96-well plate are transferred to row A of a 24-well plate, and samples from the even-numbered wells in row A from the 96-well plate are transferred to row B of the 24-well plate, and so on (Fig. 26.3.6)
Seed plates with 50 eggs or L1s per well.
Figure 26.3.6.
Transferring a library from 96-well to 24-well plates. A 12-channel pipette loaded with tips on alternating channels is used to withdraw bacterial culture from the odd number wells in the top row of a 96-well deep-well plate (A) and transfer the cultures to the top row of a 24-well plate (B). Bacteria from the even number wells in the top row of the 96-well plate are then transferred to the second row of the 24-well plate. (C) Following this pattern, it takes two rows of a 96-well plate to fill a 24-well plate.
Basic Protocol 4
RNAi in 96-Well Liquid Culture
Performing large-scale RNAi knockdown in liquid has many advantages over solid-media based assays, including the ease of set up, the ability to transfer animals more rapidly for further processing and the ease of compound addition. High-throughput image aquision pipelines have been developed using liquid cultures and for some applications, RNAi feeding and image analysis can be done in a single plate without having to transfer animals (Cipriani and Piano, 2011, Rohde and Yanik, 2011). Some strains do not grow well in liquid, so it is prudent to test mutants for their ability to grow in liquid before using them to set up a large-scale screen. The level of detail that can be observed in liquid is also decreased relative to solid media. Thrashing of animals in liquid can make it difficult to see cellular details.
To perform these assays we use an adaptation of the protocol described by Lehner et al., 2006c. This protocol requires the use of a 15-25°C shaking incubator, room for incubation of 96-well deep-well plates, and access to a suitable microscope for scoring phenotypes of interest.
Materials
LB broth (see recipe)
Tetracycline
Ampicilin or carbenicillin
NGM broth and Agar (see recipe)
M9 buffer (see recipe)
Buffered bleach (see recipe)
IPTG
RNAi library frozen at -80°C
Flat-bottom 96-well plates
96-well deep-well plates (2 mL)
Breathable lids, such as Airpore tape sheets (Qiagen)
Reservoirs (purchased or home-made using autoclaved micropipette tip boxes)
Aluminum seal for frozen stocks
Inoculate an overnight culture from frozen library stocks as described in steps 3 to 6 of Basic Protocol 4, but do not use tetracycline in the overnight cultures.
Following overnight incubation of bacteria, induce expression of dsRNA by addition of IPTG to a final concentration of 5 mM. Incubate for 4 hrs at 37°C with shaking.
Pellet bacteria by centrifugation for 15 min at 2700 rpm and suspend in 200 μl of NGM liquid media containing 50 μg/ml ampicillin plus 1 mM IPTG.
-
Transfer 90 μl of bacteria from each well to 96-well flat-bottom plates.
Do not use round-bottom plates.
Anti-fungals can be added (fungizone) if desired.
Bacteria can be suspended in a smaller volume to generate a more concentrated bacterial suspension if food becomes limiting for the assay.
Dilute eggs or L1s to approximately 1 animal per μl. Using a multi-channel pipette to add 10μl of egg suspension per well.
-
Seal 96-well plates with breathable sealing film and then cover with plate lids. The breathable film will limit condensation and evaporation.
Although we find that covering the plate with both a breathable sealing film and a plate lid limits evaporation, how much will likely depend on the air flow and the relative humidity in the incubator. Others have suggested placing 96-well plates into plastic containers or humid chambers to combat evaporation (Lehner et al., 2006c).
Incubate with shaking at 100 rpm, 20°C. After four to five days, phenotypes can be scored.
Basic Protocol 5
Soaking Worms in dsRNA
Soaking worms in dsRNA solution can also induce RNAi (Tabara et al., 1998). In this simple procedure, L4-stage animals are placed in soaking solution containing dsRNA corresponding to the gene to be silenced. Animals are soaked for up to 24 hr and recovered on Petri dishes. The exposed animals and their progeny are then examined for phenotypes.
The soaking procedure is capable of being scaled up or performed in microtiter format (Maeda et al., 2001). Another variation of the procedure is to soak L1-stage animals to examine postembryonic phenotypes.
Materials
dsRNA
Nonspecific dsRNA—i.e., dsRNA that is nonhomologous to any C. elegans sequence and which is comparable in length to the experimental dsRNA (e.g., gfp; optional)
Soaking solution (see recipe)
C. elegans (e.g., N2, CGC)
M9 buffer (see recipe)
NGM plates (see recipe) with and without seeded E. coli OP50 (see recipe)
65°C water bath
0.2- and 2.0-ml microcentrifuge tubes
5-ml glass pipet
Additional reagents and equipment for ethanol precipitation (unit 2.1a)
-
1. Ethanol precipitate (unit 2.1a) an appropriate amount of dsRNA sample and, if available, nonspecific dsRNA. Dissolve the pellet in soaking solution to a final concentration of 0.5 to 5.0 mg/ml.
It may be necessary to try several different concentrations of dsRNA to determine the concentration required to generate a phenotype.
2. Denature the RNA by heating 15 min at 65°C. Anneal the dsRNA by cooling the sample to room temperature for ∼15 to 30 min.
-
3. Pipet 5 μl sample dsRNA into an appropriately labeled 0.2-ml microcentrifuge tube. Do the same for the nonspecific dsRNA control, or if this is not available, use soaking solution alone.
It may also be useful to set up a positive control (i.e., a dsRNA proven to demonstrate a phenotype) to determine that the procedure is working as expected.
4. Using a 5-ml glass pipet, rinse a healthy mixed-stage population of C. elegans with 2 ml M9 buffer and transfer to a 2.0-ml microcentrifuge tube.
5. Pellet the animals by microcentrifuging 30 sec at 800 × g, 20°C. Remove M9 buffer with a 1-ml pipet tip and wash again.
6. Transfer the worms to an unseeded NGM plate and let them crawl around for 5 to 10 min at 20°C to allow complete digestion of bacteria in the gut.
-
7a. To test for embryonic lethal phenotypes: Pick and transfer five to ten L4-stage animals to each sample or control tube (step 3). Incubate 24 hr at 20°C. Recover the soaked worms by plating them onto an NGM plate seeded with E. coli OP50, transfer each worm to an individual plate, and incubate 24 hr at 20°C. Transfer each worm to a fresh plate and count the number of progeny.
7b. To test for postembryonic phenotypes: Place 20 newly hatched L1 larvae into each sample or control tube and incubate 24 hr at 20°C. Recover soaked worms by plating them onto an NGM plate seeded with E. coli OP50. When the L1-soaked animals reach L4 stage, transfer each animal to an individual plate.
8. Monitor the treated animals and their progeny for phenotypes resulting from dsRNA treatment.
Support Protocol 1
Picking and Propagating C. Elegans
C. elegans can be transferred from one plate to another using a worm pick—a short piece of platinum wire with one end flattened and the other end mounted to a Pasteur pipet. Commercially manufactured worm picks are also available (Genesee Scientific). Platinum wire heats and cools rapidly, so the wire can and should be flamed before and after every pick to avoid contamination.
Label the bottom half of an NGM plate (see recipe) with the appropriate strain name and the date. Working under a dissecting microscope, pick up a dab of E. coli OP50 food (see recipe) on the underside of the flattened end of the worm pick. Gently touch the food on the bottom of the pick to the top of a worm. The worm should stick to the food on the worm pick by surface tension. Quickly transfer the worm to a new NGM plate, gently touching the worm to the plate and allowing the worm to crawl off of the worm pick.
For those unfamiliar with C. elegans, propagating worms may be difficult at first, but with practice picking and maintaining worms will be routine. See Support Protocol 2, Hope (1999), and Wood (1988) for additional details on propagating, staging, cleaning, synchronizing, and mating C. elegans.
Support Protocol 2
Sexing and Mating C. Elegans
C. elegans comes in two sexes, hermaphrodite and male, which are determined by the number of X chromosomes. Males can be distinguished from hermaphrodites under the dissecting microscope as they lack hermaphrodite sex organs (e.g., vulva and uterus containing developing embryos) and the male tail has a barbed appearance. Hermaphrodites (XX) produce both sperm and oocytes and can reproduce by self-fertilization. Hermaphrodites can also be cross-fertilized by C. elegans males (XO), which produce sperm only. If a C. elegans male successfully mates with a hermaphrodite, the majority of the progeny will be cross-progeny because male sperm out competes hermaphrodite sperm. As such, one half of the cross-progeny will be males and the other half will be hermaphrodites. For a detailed description of hermaphrodites and males and their identification, refer to Wood (1988) and Riddle et al. (1997).
Males arise by spontaneous X chromosome nondisjunction at a frequency of ∼0.1% (Riddle et al., 1997). This frequency can be increased by heat-shock treatment of L4 hermaphrodites at 30°C for 4 hr. Once a male stock is obtained, the population should be maintained by periodically setting up crosses. In addition, we find that we can also freeze animals from these plates once starved using standard protocols (Steirnagle, 2006), and recover males once thawed. Thus, once the population of males has been expanded, frozed stocks containing males can be generated for later use.
Genetic crosses in C. elegans are typically set up between a phenotypically wild-type male and a hermaphrodite that is homozygous for a visible mutation—e.g., uncoordinated (Unc). This allows cross-progeny (non-Unc) to be easily distinguished from self-progeny (Unc). Multiple (e.g., ten) young healthy males and multiple (e.g., five) L4 or young adult hermaphrodites should be chosen when crossing C. elegans strains.
Support Protocol 3
Collection of C. Elegans Eggs by Hypochlorite Treatment
C. elegans embryos, but not larvae or adults, are resistant to hypochlorite, and thus eggs can be collected by hypochlorite treatment to generate a semi-synchronous population.
Materials
10-cm plates with gravid adult worms (Basic Protocol 3)
15-ml falcon tubes
Buffered bleach, prepared fresh (see recipe)
M9 buffer (see recipe)
Wash worms and eggs off 10-cm agar plates using 5 ml of water, and transfer to a 15-ml conical tube.
Check the plates under a microscope to ensure that you have washed all the eggs off.
Pellet worms and eggs in a tabletop centrifuge for 2 min at 1200 rpm, and discard supernatant.
Add buffered bleach and mix gently (do not vortex). Check worms regularly by placing tube under a dissecting microscope. When most adult carcasses have broken open and dissolved, add M9 up to 10 ml, mix by inversion, and pellet by centrifugation as above. Hypochlorite treatment typically takes 4 to 5 min and should not exceed 7 min. Overbleaching will kill embryos.
-
Wash eggs 3 times with 10 ml M9, and suspend the embryos in 5 ml M9.
Animals can be further synchronized to L1 arrest by incubating them in M9 with gentle rocking for 12 to 18 hours. To prevent hypoxia, the buffer should not exceed 7 ml.
Reagents and Solutions
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2; for suppliers, see APPENDIX 4.
Agarose microinjection pads
Prepare injection pads by sandwiching a couple of drops of melted 2% (w/v) agarose between two 22 × 50–mm glass coverslips. Once the agarose hardens (<1 min), carefully remove one of the coverslips. Dry the agarose pad onto the remaining coverslip by baking at 50°C for 1 hr. A large number of microinjection pads can be made at one time and stored indefinitely at room temperature in a closed container (e.g., a Petri dish or empty coverslip box) to prevent dust from accumulating on the surface.
Baking dries the agarose to the coverslip. One of the coverslips must be removed before baking. The popped off coverslip can then be reused for another microinjection pad.
Buffered Bleach (prepared fresh)
7 ml H2O
2 ml Bleach (5-6% sodium hypochlorite)
1 ml 5M NaOH
Commercial bleach varies in strength: be sure to check the percent sodium hypochlorite.
E. coli OP50 food
Inoculate (unit 1.2) 100 ml of L broth (unit 6.12) with a single colony of OP50 (CGC) using sterile technique. Incubate the culture overnight at 37°C with shaking. Store the liquid culture at 4°C for several weeks
The uracil auxotrophic E. coli OP50 strain is typically used as a food source for C. elegans, as the limited growth of E. coli OP50 on NGM plates provides easier observation of the worms.
Loading capillary
The loading capillary consists of a drawn-out Pasteur pipet connected via tubing to a mouth pipet. The diameter of the drawn-out end of the Pasteur pipet must be less than that of the inner diameter of the nondrawn-out portion of the microinjection needle (see recipe) to facilitate loading.
M9 buffer with and without 15% glycerol
3.0 g monobasic potassium phosphate (KH2PO4)
6.0 g dibasic sodium phosphate (Na2HPO4)
5.0 g NaCl
1 ml 1 M magnesium sulfate (MgSO4)
Add 150 ml glycerol (15%) if appropriate
Adjust volume to 1 liter with H2O
If using glycerol, prepare 100-ml aliquots
Sterilize by autoclaving
Store up to 2 months at 20°C
Mating plates
Flame the end of a Pasteur pipet until the edges of the tip curl in and nearly close the opening. This rounded end will be less likely to scratch the surface of the NGM plate during seeding. Using the Pasteur pipet, withdraw some E. coli OP50 food (see recipe) from an overnight culture. Touch the rounded tip of the Pasteur pipet to the center of a 30-mm NGM plate (see recipe), dispensing a very small droplet of the E. coli OP50 food. Incubate the plates at room temperature 2 to 3 hr and store at 4°C in a sealed container for several weeks.
Mating plates are NGM plates seeded with a very thin and small (∼5 mm) spot of E. coli OP50 in the center.
Alternatively, a small dab of E. coli OP50 food can be transferred from a dry seeded plate to an unseeded 30-mm NGM plate just prior to setting up a cross.
See unit 1.2 for more information regarding culture of E. coli.
Microinjection needles
The ideal microinjection needle (i.e., borosilicate glass capillaries, World Precision Instruments) should taper to a sharp and stiff point and can be pulled using 1.2- or 1-mm capillary blanks and a wide variety of needle-pulling devices (e.g., Narishige PN-30). After pulling the first two needles, examine them under a dissecting microscope. The needles should taper gradually and steadily to a very sharp point. Using one of the first two needles, gently touch the other needle just behind the tip while watching through the dissecting scope. If the needle tips bend easily without breaking then it is probably not stiff enough for penetrating the cuticle of the worm. A common problem is a needle that is hair-like at the tip. To correct this problem, reduce the heat setting on the needle puller. It may also be necessary to adjust the pull strength. Get advice from someone experienced in pulling capillaries if problems persist. Most needles will be closed at the tip after pulling and thus, it will be necessary to gently break the needle tip prior to microinjection. Alternatively, the needle tips can be etched with acid (Mello and Fire, 1995).
NGM plates or NGM/amp/IPTG plates
Prepare the following in a 2-liter Erlenmeyer flask:
3.0 g NaCl
17 g Agar
2.5 g Bacto-peptone
975 ml H2O
Add a stir bar and sterilize by autoclaving. Cool to ∼55°C and add 25 ml of 1 M potassium phosphate buffer, pH 6 (see recipe), 1 ml of 1 M CaCl2, 1 ml of 5 mg/ml cholesterol in 100% ethanol, and 1 ml of 1 M MgSO4. For NGM/amp/IPTG plates, also add 50 μg/ml ampicillin and 1 mM (final) IPTG (unit 1.4). Dispense into Petri dishes (∼3 ml per 30-mm plate). Dry the plates at room temperature for 1 to 2 days before seeding with OP50. Store up to 1 month at 4°C in a sealed container.
NGM plates seeded with OP50
Seed each 30-mm NGM plate (see recipe) with one drop (∼100 μl) of E. coli OP50 food (see recipe). Allow the E. coli OP50 drop to dry into the plate and to grow overnight at room temperature or for 8 hr at 37°C (unit 1.2). Store seeded plates in a sealed container for several weeks at 4°C.
Potassium phosphate buffer, 1 M, pH 6.0
108.3 g monobasic potassium phosphate (KH2PO4)
35.6 g dibasic potassium phosphate (K2HPO4)
Adjust volume to 1 liter with H2O
Adjust pH to 6.0 if necessary
Store up to 2 months at 20°C
Soaking buffer, 10×
1.55 g dibasic sodium phosphate (Na2HPO4)
0.75 g monobasic potassium phosphate (KH2PO4)
0.12 g NaCl
0.25 g ammonium chloride (NH4Cl)
0.5 g gelatin
Adjust volume to 100 ml with H2O
Sterilize by autoclaving
Store up to 2 months at 20°C
Soaking solution
0.5 ml 10× soaking buffer (see recipe)
15 μl 1 M spermidine (Sigma)
4.5 ml H2O
Use immediately
Commentary
Background Information
RNA interference (RNAi) is a form of sequence-specific, post-transcriptional gene silencing (PTGS) that is triggered by the introduction of dsRNA (unit 26.1; Fire et al., 1998). RNAi is related to a number of gene-silencing phenomena in a variety of organisms. Genetic and biochemical studies have revealed that the underlying mechanisms of these silencing phenomena are remarkably conserved (for reviews, see unit 26.1 and Hannon, 2002). The specificity and potency of RNAi provided researchers with an efficient means of inactivating gene expression in model organisms where conventional gene-replacement techniques were unavailable, such as C. elegans, Drosophila, fungi, plants, and trypanosomes (Bosher and Labouesse, 2000). The ability to perform RNAi in human cells (Elbashir et al., 2001) has rapidly transformed basic research, and a number of clinical trials are underway using RNAi as a therapy for human disease.
RNAi can inhibit gene function in worms injected with dsRNA (see Basic Protocol 1 and Alternate Protocol; Fire et al., 1998), soaked in solution containing dsRNA (see Basic Protocol 3; Tabara et al., 1998), fed bacteria expressing dsRNA (see Basic Protocol 2; Timmons et al., 2001), or engineered to express dsRNA in vivo (Tabara et al., 1999). Interference by dsRNA is sequence-specific and potent (Fire et al., 1998). Worms that are injected in the gut with ∼60,000 dsRNA molecules targeting the muscle-specific unc-22 gene exhibit an unc-22(–) twitching phenotype (Fire et al., 1998). Moreover, F1 progeny of injected worms also exhibit a severe twitching phenotype. These findings indicate that dsRNA, or a secondary silencing factor, is transported from one tissue to another and that interference can be inherited. The specific, potent, systemic, and heritable features of RNAi make this an ideal method for assaying the function of genes in C. elegans.
The ability to induce RNAi by a variety of methods allows researchers of different skill levels to apply the technique and to characterize gene function under a variety of conditions. Typically, investigators determine whether a particular gene has an RNAi phenotype first by microinjection, simply because the preparation of dsRNA is straightforward and takes less time to prepare than RNAi food and plates (see Time Considerations). However, this does not necessarily mean that RNAi by microinjection is superior to other methods. While some genes are indeed silenced more effectively by microinjection than by feeding, the converse is also true (Kamath et al., 2001), and other genes are equally sensitive to both methods (Kamath et al., 2003; unpub. observ.). Therefore, if microinjection does not work, feeding and/or soaking should be attempted. For RNAi by feeding, Kamath et al. (2003) generated a library of 16,757 RNAi bacterial feeding clones, which has since been expanded to 19,763 clones (available from Source Bioscience). An independent library of 11,560 ORFeome RNAi clones (available from GE Dharmacon), targeting 10,499 individual genes, has also been generated (Rual et al., 2004). Together, the coverage of libraries is approaching 90% of the 20,466 predicted protein-coding genes, based on the recent genome release (WS244).
Due to the requirement of a balancer chromosome for viability, a null mutation in an essential gene limits the ability to perform biochemistry or applications that require large populations. The ability to silence a gene in large populations of C. elegans using RNAi by feeding or soaking should make C. elegans more amenable to such applications.
RNAi is also a simple way to examine genetic interactions between genes. Sequential microinjection, microinjection or soaking with multiple dsRNAs simultaneously, exposure of mutant strains to dsRNA, as well as feeding constructs containing more than one insert have all been successfully used to determine whether two genes interact (Grishok et al., 2001; Bei et al., 2002; Dudley et al., 2002; Byrne et al., 2007; Lehner et al., 2006b).
Despite its success, RNAi in C. elegans does have its limitations. RNAi does not work for every gene. In particular, pharyngeal, vulval, sperm, and neuronal genes are often difficult to silence (Fire et al., 1998). The reasons for this are not entirely known, but could include inefficient transport or maintenance of trigger dsRNA or secondary signals to these tissues. In some cases, efficient silencing can be triggered for refractory genes by driving the expression of hairpin dsRNA from a transgene in the worm (Tavernarakis et al., 2000). The enhancement of RNAi in the nervous system has been of particular interest. For example, expression of sid-1 in the nervous system is reported to enhance the efficiency of neuronal RNAi (Calixto et al., 2010), and mutations that enhance RNAi, including rrf-3, eri-1 and lin-15 mutants, as well as combinations of these mutations, have also proved useful (Simmer et al., 2002; Lehner et al., 2006a; Schmitz et al., 2006). In the latter case, the enhancement of RNAi is not restricted to the neuronal tissue, and these RNAi-sensitized strains have been used to uncover RNAi-induced phenotypes not observed in wild-type backgrounds.
A large amount of RNAi data is being generated in C. elegans. The data can be accessed and searched from Wormbase (http://www.wormbase.org) to determine whether a particular gene exhibits an RNAi phenotype or to identify genes that show a particular phenotype. In general, the quality of the data is good, but keep in mind that a negative result does not necessarily mean that a particular gene does not exhibit an RNAi phenotype. It simply means that under the conditions tested, no phenotype was observed. Investigators should try the experiment by additional methods before accepting a negative RNAi result.
A common problem with all RNAi methods is the possibility of inadvertently silencing related genes. Generally, so-called off-target silencing only occurs when a few segments of at least 25 nucleotides identity exist within the gene (Parrish et al., 2000). As a standard control for this problem in RNAi studies, it is always important to identify related genes in the genome and assay each of these for similar or overlapping phenotypes (Rual et al., 2007).
Although RNAi can be inherited (Fire et al., 1998; Grishok et al., 2000), it is not permanent. While this may be considered as a benefit to some, to the classical geneticist this is a drawback: genetic mutants remain the gold standard in C. elegans research. Variability is typically not a problem with RNAi, but a range of phenotypes is possible, so animals should be observed carefully. Because the feeding strategy (see Basic Protocol 2) requires less actual worm manipulation, it may be the more consistent protocol, provided care is taken when preparing the dsRNA food and media. When possible, it is a good idea to confirm RNAi phenotypes by obtaining a genetic mutant, and the advent of CRISPR-mediated genome engineering in C. elegans (Friedland et al., 2013) leaves little excuse. In fact, RNAi and CRISPR-mediated genome engineering provide worm researchers with an outstanding one-two punch. For example, RNAi can be used as a rapid approach to assign gene function or to screen for candidate genes in a pathway (Basic Protocols 3 and 4 and Alternate Protocol 2), and CRISPR-mediated genome engineering can be used to quickly obtain genetic mutants (Friedland et al., 2013; Dickinson et al., 2013; Kim et al., 2014).
Critical Parameters
Target Selection
There are few hard and fast rules for determining the region of a gene to target. Since introns and untranslated regions are not efficient targets of the RNAi machinery (Montgomery et al., 1998), the dsRNA should be designed to target the coding region of the mature mRNA. Although 500 to 1000 bp is sufficient, if feasible, target the entire coding region of the gene of interest or several different regions. The sensitivity of worms to RNAi targeting different regions of a particular gene can be determined in a relatively short period of time by microinjection.
As mentioned above, care should be taken when performing RNAi on members of a gene family. Experiments should be designed such that the targeted region of a gene of interest does not overlap with the other member(s) of the gene family. This may be difficult if extensive homology exists, but in most cases this should not be a problem. Use BLAST (unit 19.3) to identify related sequences in C. elegans and to determine whether there is significant identity to complicate the analysis. If there is significant identity between two sequences, attempt to find and target regions that are divergent.
Obtaining cDNA Clones
Gateway-compatible open-reading-frame (ORF) clones have been generated for a many C. elegans genes (Reboul et al., 2003; Lamesch et al., 2004). Individual clones can be purchased from Source BioScience or GE Dharmacon, and the entire library is available from GE Dharmacon (see Internet Resources). Information related to the amplification and cloning of ORFeome clones can be found on the WorfDB website (see Internet Resources). The laboratory of Yuji Kohara (ykohara@lab.nig.ac.jp) has also generated a library of C. elegans cDNAs. In addition, they have generated sequence tags for a large number of these clones and assigned them to the corresponding genes. If cDNAs for a particular gene have been identified, a clone or yk (for Yuji Kohara) number will be listed on the Gene report page in Wormbase (http://www.wormbase.org; see Internet Resources) under the subheading Reagents: Matching cDNAs. The Kohara cDNA clones are made available to the C. elegans community and are provided as a small aliquot of Lambda ZAP II bacteriophage. A kit provided by Stratagene can be used to amplify the phage and to excise the pBluescript SK(–) phagemid. Each insert is flanked by the phage T3 and T7 RNA polymerase promoters, which can be used to synthesize sense and antisense RNA strands in vitro (unit 3.8). If a cDNA clone is not available, a cDNA can be generated by RT-PCR from total worm RNA or polyA-selected mRNA using sequence-specific oligonucleotides and cloned into an appropriate expression vector (e.g., pBluescript).
Transcription templates can also be generated by PCR from genomic DNA using oligonucleotides with 5′ tails containing phage T7 RNA polymerase promoter sequences. In this case, try to identify and amplify the longest contiguous stretch of exon sequence possible, minimizing the inclusion of introns. The resultant PCR product will contain a portion of the gene of interest flanked by opposing T7 promoter sites. Using such a template, both sense and antisense strands of RNA can be produced simultaneously in a single in vitro transcription reaction.
When generating clones for RNAi by feeding, inserts should be exon rich. The authors have had success with a variety of feeding constructs, targeting sequences from 300 to 2400 nt long. The optimal length of the targeted region for RNAi by feeding is likely to be gene specific and should be determined empirically; however, the parameters discussed above represent a good starting point.
dsRNA Preparation
Sense and antisense RNA strands are prepared using standard protocols and kits, and then annealed to make dsRNA. Several companies now provide high yield in vitro transcription kits. The Megascript Kit from Ambion is outstanding, and the instruction manual provides a lot of useful information, from designing oligonucleotides containing phage promoters to troubleshooting transcription reactions.
Efficient RNAi will be achieved for many genes over a very broad range of dsRNA concentrations (0.01 to 5 mg/ml). Initially, higher concentrations (i.e., 1 mg/ml) are preferred because some genes are less sensitive to RNAi than others for unknown reasons. A small percentage (<5%) of nonspecific embryonic lethality among progeny of the injected animals is sometimes observed; however, there are no other noticeable side effects from injecting high dsRNA concentrations.
At very low concentrations of dsRNA, one may be able to induce hypomorphic phenotypes. Mixing two different RNAi feeding strains has been shown to dramatically reduce the penetrance of mutant phenotypes observed with one feeding strain alone (Kamath et al., 2001). These authors also found that a 1:1 mixture of unc-37 RNAi food and normal OP50 (which does not express the dsRNA) completely abolished the embryonic lethality induced by unc-37 RNAi; however, 100% of the progeny displayed hypomorphic postembryonic phenotypes, which was also observed when the RNAi food was induced with very low levels of IPTG. Therefore, adjusting the levels of dsRNA delivered to the animals can alter the penetrance and spectra of RNAi phenotypes.
Silencing Multiple Genes Simultaneously
As discussed above, care should be taken when designing RNAi experiments so that simultaneous silencing of related genes does not occur inadvertently. Nevertheless, there may be instances when it is desirable to silence multiple genes simultaneously. For instance, two related genes that function redundantly in a particular process might show no phenotype or produce an incompletely penetrant phenotype when silenced individually by RNAi. On the other hand, simultaneous RNAi targeting of both genes may dramatically produce a completely penetrant phenotype (Grishok et al., 2001). As mentioned above, RNAi is also a simple and rapid method for examining genetic interactions. In principle, each of the dsRNA delivery methods is amenable to simultaneous RNAi; however, microinjection is the most widely used technique for this purpose. Simultaneous RNAi by feeding also works, although it is somewhat limited relative to microinjection. As mentioned above, mixing two different RNAi feeding strains together dramatically reduces the penetrance of phenotypes resulting from RNAi, but by engineering the feeding vector to contain two inserts, the authors and others (Min et al., 2010) have been able to silence two genes simultaneously by double RNAi feeding without an apparent reduction in the penetrance of RNAi relative to either single RNAi food. The combined inserts so far have not exceeded 1.5 kb in length, so presumably additional genes could be targeted, for example in a triple RNAi feeding construct.
Cell- or Tissue-specific RNAi
Silencing a gene in a specific cell or tissue may be desirable when the gene of interest is essential or expressed in all tissues and the function in a particular tissue is being investigated. Tissue-specific RNAi strains have been developed by tissue-specific transgene rescue of an RNAi-deficient mutant. For example, rde-1 mutants are completely resistant to RNAi in all tissues (Tabara et al., 1999), but tissue-specific expression of rde-1 has been used to rescue RNAi in intestinal, muscle, neuronal, and hypodermal tissues (Espelt et al., 2005, Qadota et al., 2007, Firnhaber and Hammerlund, 2013).
Troubleshooting
Table 26.3.1 presents a guide to some of the common problems of achieving RNAi in C. elegans, as well as their causes and solutions.
Table 26.3.1. Troubleshooting Guide for C. elegans RNAi Studies.
| Problem | Possible cause | Solution |
|---|---|---|
| RNAi by microinjection | ||
| dsRNA solution does not flow from the needle | Needle is closed | Etch or break the needles after pulling to open |
| Needle is clogged | Break the tip of the needle to restore flow | |
| Use a new needle | ||
| Centrifuge dsRNA solution prior to use | ||
| Worms do not stick to agarose injection pad | Agarose is not dry enough | Bake agarose pads at least 1 hr at 50°C |
| No RNAi phenotype observed | dsRNA is degraded | Prepare fresh dsRNA and analyze on 1% agarose gel to check quality (unit 2.5a) |
| Perform positive control for RNAi by injection | ||
| Target gene is refractory to RNAi | Try another region of the gene | |
| Attempt to achieve RNAi by feeding or soaking | ||
| RNAi by feeding | ||
| No RNAi phenotype observed | dsRNA production is not induced | Test the plates by performing a positive control for RNAi |
| Prepare fresh NGM/Amp/IPTG plates | ||
| RNAi food is contaminated | Prepare fresh RNAi food. Be sure to include both ampicillin and tetracycline in the starter culture to prevent contamination with another bacterial species. Perform all food preparation carefully. | |
| RNAi by soaking | ||
| No RNAi phenotype observed | dsRNA is degraded | Prepare fresh dsRNA and soaking solution |
Anticipated Results
Regardless of the delivery method, if RNAi produces a phenotype, the animals will typically be affected for one generation—i.e., the F1 progeny of animals exposed to dsRNA. However, for RNAi by feeding, as long as the animals are cultivated on RNAi food, they are exposed to dsRNA and the phenotype will persist. Investigators have observed persistent RNAi by microinjection into the F2 generation, although the penetrance is reduced.
In a typical RNAi microinjection experiment, early embryos developing within the uterus may be unaffected, but may carry the silencing signal into the subsequent generation. These unaffected and carrier progeny will be laid on the initial recovery plate after microinjection. After transferring an injected animal from the recovery plate to a fresh plate (∼16 hr), the remaining portion of the brood should be completely affected by the RNAi.
For uncharacterized genes, it is difficult to know what to expect from an RNAi experiment; however, if the expression pattern or localization is known (e.g., from in situ hybridization), then an educated guess can be made about the type of phenotype for which to look. For example, if in situ analysis indicates that the gene product is expressed in the germline, one might expect to see an embryonic-lethal phenotype or postembryonic-sterile phenotype. For a gene product expressed in muscle cells, one might expect defects in motility. In situ expression data can be searched from Wormbase or from the Nematode Expression Pattern Database (NEXTDB; see Internet Resources).
It is recommended that the novice enlist the aid of individuals with expertise in C. elegans biology when phenotyping animals.
Time Considerations
RNAi by Microinjection
A complete microinjection experiment from injecting animals to scoring phenotypes will take 3 to 6 days. On day 0, the novice should expect to spend about 1 to 2 hr at the injection microscope performing microinjection. With practice, this time can be shortened to ∼15 min for each dsRNA and strain. The amount of time to transfer animals to individual NGM plates on days 1 and 2 will vary depending on the number of worms to transfer (5 to 30 min). Expect to spend several hours counting progeny and analyzing phenotypes.
RNAi by Feeding
Growth, preparation, and induction of the RNAi food will take 2 to 4 days. The remainder of the feeding experiment will take 3 to 6 days to complete. As for microinjection, plan to spend several hours counting progeny and analyzing phenotypes.
RNAi-library Screening
A typical experiment will take 2 to 3 days to set up, followed by 3 to 4 days of growth. The amount of time it will take to screen for a desired phenotype will depend on the number of plates to be screened. Conversely, the number of plates that can be screened in a reasonable amount of time will vary, depending on the ability to screen for the desired phenotype. Typically, we stagger the start of large-scale screens over several days, and screen accordingly.
RNAi by Soaking
The amount of time it will take to perform the entire soaking procedure will be similar to that of microinjection.
Acknowledgments
The authors wish to thank Amy Holdorf, Gina Caldas, and Krishna Ghanta for helful comments on the manuscript. This work was supported in part by Ruth L. Kirschstein fellowship GM63348 (D.C.), a Canadian Institutes of Health Research postdoctoral fellowship (L.T.M.), and by National Institutes of Health grants DK068429 (A.J.M.W) from the N.I.D.D.K. and GM58800 (C.C.M.) from the N.I.G.M.S. C.C.M is a Howard Hughes Medical Institute Investigator.
Footnotes
Internet Resources: http://www.wormbase.org
Wormbase is an essential tool for C. elegans biologists, and provides an abundance of useful information regarding the genome and biology of C. elegans. The curators at Wormbase have provided annotations of each of the predicted genes including RNAi phenotypes and expression data as well as links to useful sites. The curators are constantly updating and upgrading the site.
Worm atlas is an online database featuring behavioural and structural anatomy of the worm, C. elegans.
The RNAi database is a searchable database of ovary or germline-enriched genes by gene or by phenotype.
http://c.elegans.tripod.com/RNAi.htm
The RNAi@elegans.net page has many informative links for RNAi in C. elegans and other organisms.
http://nematode.lab.nig.ac.jp/
NEXTDB is an expression pattern database based on in situ hybridizations using expressed sequence tags.
http://worfdb.dfci.harvard.edu
Worm ORFeome DataBase (WorfDB) is a searchable database for C. elegans ORF clones.
http://www.cbs.umn.edu/research/resources/cgc
The Caenorhabditis Genetics Center (CGC), funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440), is the central repository for C. elegans strains and distributes strains at a minimal cost to researchers.
http://www.lifesciences.sourcebioscience.com/clone-products/non-mammalian/c-elegans/
Source Bioscience sells ORFeome and RNAi reagents, including the genome-wide RNAi feeding library and sub-libraries.
GE Dharmacon provides ORFeome reagents, including the ORFeome RNAi feeding collection.
Literature Cited
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, Collins J, Mello CC. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell. 2002;3:113–125. doi: 10.1016/s1534-5807(02)00185-5. [DOI] [PubMed] [Google Scholar]
- Bosher JM, Labouesse M. RNA interference: Genetic wand and genetic watchdog. Nat Cell Biol. 2000;2:E31–36. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- Byrne AB, Weirauch MT, Wong V, Koeva M, Dixon SJ, Stuart JM, Roy PJ. A global analysis of genetic interactions in Caenorhabditis elegans. J Biol. 2007;6:8. doi: 10.1186/jbiol58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell GA, Williams SN, Caldwell KA. Integrated Genomics:A discovery-based laboratory Course. Wiley; 2006. [Google Scholar]
- Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cipriani PG, Piano F. RNAi methods and screening: RNAi based high-throughput genetic interaction screening. Methods Cell Biol. 2011;106:89–111. doi: 10.1016/B978-0-12-544172-8.00004-9. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley NR, Labbe JC, Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc Natl Acad Sci USA. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Espelt MV, Extevez AY, Yin X, Strange K. Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-triphosphate receptor and phospholipases C beta and gamma. J Gen Physiol. 2005;126:379–392. doi: 10.1085/jgp.200509355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Firnhaber C, Hammarlund M. Neuron-specific feeding RNAi in in C. elegans and its use in a screen for essential genes required for GABA neuron function. PLoS Genet. 2013;9:e1003921. doi: 10.1371/journal.pgen.1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Exvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume AM, Martin C, Ozlu N, Bork P, Hyman AA. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hope IA, editor. C elegans: A Practical Approach The Practical Approach Series. Oxford University Press; New York: 1999. A very good source for general information on culturing and maintaining worms as well as a comprehensive microinjection protocol for germline transformation. [Google Scholar]
- Jansen G, Hazendonk E, Thijssen KL, Plasterk RH. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. This manuscript provides an in depth analysis of the RNAi by feeding technique as well as a comparison to RNAi by microinjection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. comment 220-221. [DOI] [PubMed] [Google Scholar]
- Lehner B, Calixto A, Crombie C, Tischler J, Fortunato A, Chalfie M, Fraser AG. Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol. 2006a;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006b;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Lehner B, Tischler J, Fraser AG. RNAi screens in Caenorhabditis elegans in 96-well liquid format and their application to the systematic identification of genetic interactions. Nat Protoc. 2006c;1:1617–1620. doi: 10.1038/nprot.2006.245. [DOI] [PubMed] [Google Scholar]
- Liu LX, Spoerke JM, Mulligan EL, Chen J, Reardon B, Westlund B, Sun L, Abel K, Armstrong B, Hardiman G, King J, McCague L, Basson M, Clover R, Johnson CD. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 1999;9:859–867. doi: 10.1101/gr.9.9.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Min K, Kang J, Lee J. A modified feeding RNAi method for simultaneous knock-down of more than one gene in Caenorhabditis elegans. Biotechniques. 2010;48:229–232. doi: 10.2144/000113365. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:15502–1557. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol. 2000;10:1619–1622. doi: 10.1016/s0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- Qadota H, Inoue M, Hikita T, Koppen M, Hardin JD, Amano M, Moerman DG, Kaibushi K. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–173. doi: 10.1016/j.gene.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR. C ELEGANS II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1997. C ELEGANS II provides an extensive overview of worm biology. [PubMed] [Google Scholar]
- Rohde CB, Yanik MF. Subcellular in vivo time-lapse imaging and optical manipulation of Caenorhabditis elegans in standard multiwell plates. Nat Commun. 2011;2:271. doi: 10.1038/ncomms1266. [DOI] [PubMed] [Google Scholar]
- Rual JF, Klitgord N, Achaz G. Novel insights into RNAi off-target effects using C. elegans paralogs. BMC Genomics. 2007;8:106. doi: 10.1186/1471-2164-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, vanderLinden AM, Kuijk E, van der Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Röder M, Finell J, Häntsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gönczy P, Coulson A, Hyman AA, Echeverri CJ. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–9. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Steirnagle T. Maintenance of C. elegans. Wormbook. 2006 Feb;11:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tabara H, Grishok A, Mello CC. RNAi in C. elegans: Soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Arda HE, Zhu LJ, Walhout AJM. Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response. Cell. 2013;153:253–66. doi: 10.1016/j.cell.2013.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1988. The predecessor of C ELEGANS II (Riddle et al., 1997) The Nematode is full of important information about the biology of C elegans. [Google Scholar]